Abstract
The C-terminal region of cardiac troponin I (cTnI) is known to be important in cardiac function, as removal of the last 17 C-terminal residues of human cTnI has been associated with myocardial stunning. To investigate the C-terminal region of cTnI, three C-terminal deletion mutations in human cTnI were generated: Δ1 (deletion of residue 210), Δ3 (deletion of residues 208-210), and Δ5 (deletion of residues 206-210). Mammalian two-hybrid studies showed that the interactions between cTnI mutants and cardiac troponin C (cTnC) or cardiac troponin T (cTnT) were impaired in Δ3 and Δ5 mutants when compared to wild-type cTnI. Troponin complexes containing 2-[4’-(iodoacetamido) anilino] naphthalene-6-sulfonic acid (IAANS) labeled cTnC showed that the troponin complex containing cTnI Δ5 had a small increase in Ca2+ affinity (P < 0.05); while the cTnI Δ1- and Δ3 troponin complexes showed no difference in Ca2+ affinity when compared to wild-type troponin. In vitro motility assays showed that all truncation mutants had increased Ca2+ dependent motility relative to wild-type cTnI. These results suggest that the last 5 C-terminal residues of cTnI influence the binding of cTnI with cTnC and cTnT, affects the Ca2+-dependence of filament sliding, and demonstrate the importance of this region of cTnI.
Keywords: Heart, troponin I, mammalian two-hybrid, in vitro motility assay, unloaded filament sliding, calcium
INTRODUCTION
Cardiac troponin (cTn) is located at regular intervals along the thin filament and consists of three subunits: cardiac troponin C (cTnC), troponin I (cTnI), and troponin T (cTnT). The cardiac troponin complex plays an important role in the regulation of striated muscle contraction. Interaction between these proteins regulates the contraction (systole) and relaxation (diastole) of cardiac muscle [1]. During diastole, cTnI contributes to the inhibition of actin-myosin cross-bridge formation by interacting with actin. When Ca2+ binds to cTnC during systole, the affinity of cTnI for Ca2+-cTnC is increased, resulting in weaker interactions between cTnI and actin. These changes together with changes in cTnT structure lead to changes in the position of tropomyosin (Tm) on the thin filament, causing an activation of contraction by promoting the conversion of cross-bridges from a blocked or weak binding state to a strong binding (force-generating) state. The cTnI inhibitory region (residues 139–150 in mouse cTnI) binds to either TnC or actin, but not to both simultaneously [2]. This region has been shown to inhibit actomyosin ATPase activity and is a vital part of the molecular switch responsible for initiating muscle contraction. The cTnI C-terminal region binds to the N-terminal region of cTnC [3].
Both the N- and C-terminal ends of TnI are not as well investigated as its inhibitory region. The N-terminal region (cardiac specific region) has unique alternatively spliced regions which are important for modulating Ca2+ sensitivity of force development [4, 5]. The N-terminal region of cTnI has been shown to modulate myofilament activation [6]. The role of the last 5 residues of the C-terminus of cTnI (residues 205 to 210) is not well understood. Mutations within this region of cTnI are associated with hypertrophic cardiomyopathy (HCM), restrictive cardiomyopathy (RCM), and dilated cardiomyopathy (DCM). At least five mutations associated with HCM (K206Q [7], K206I [8], K207T [9], E209A [10] and E209K [11]) occur within the last 5 C-terminal residues of human cTnI [12]. The K207T mutation was discovered in an American patient and was classified as likely pathogenic but little data is available about patients with this mutation [9]. The E209A mutation was found in five index patients in the Netherlands and is currently considered an unclassified variant of unknown pathological significance [10]. The E209K was found during genetic testing of 2,912 probands with HCM [11].
A 17 residue C-terminal deletion in human cTnI was found to be associated with myocardial stunning and increased Ca2+ sensitivity in an in vitro motility assay [13]. In another investigation, removal of 17 amino acids from the C-terminus of human cTnI (cTnI 1–192) showed that these residues perform an important stabilizing role ensuring proper conformational switching of Tn–Tm [14]. Previous reports using affinity chromatography and actin sedimentation experiments suggest that cTnI mutants (cTnI 1-209 and cTnI 1-192) did not affect Tn assembly and were not sufficiently strong to affect interactions between the Tn subunits or between cTnI and actintropomyosin (Tm) [13]. However, Ferrieres et al. using 20-mer peptides of cTnI found that the C-terminal peptide, residues 191-210, interacted weakly with TnC (KD 1.69 ± 0.89 × 10−6), suggesting that under some conditions this region of cTnI may potentially interact with TnC [15]. Other studies also suggest that the last 17 C-terminal residues in cTnI are important for Ca2+ regulation of the thin filament through its interaction with Tm and/or actin [14, 16]. cTnI 1-192 increased Ca2+ sensitivity of both the actin-Tm-activated S1 ATPase activity and the Ca2+-dependent sliding velocity of reconstituted thin filaments (in an in vitro motility assay) compared with cTnI 1-209 [13].
During ischemia, myofibrillar α-actinin and TnI are both degraded, with TnI being degraded at its C-terminus. After ischemia/reperfusion, myosin light chain-1 (MLC1) is also degraded at its N-terminus [17]. Other studies have shown that cTnI C-terminal degradation is associated with reduced maximal force and altered sensitivity to Ca2+. Wild-type mouse cTnI (211 residues), and the C-terminal mutants cTnI-(1–199) (missing 12 residues), cTnI-(1–188) (missing 23 residues), and cTnI-(1–151) (missing 60 residues) were investigated by Rarick et al. [6]. Ureapolyacrylamide gel-shift analysis and size exclusion chromatography showed that each cTnI C-terminal deletion mutant was able to bind to cTnC, and co-sedimentation assays showed that these mutants co-sediment with actin. The cTnI-(1–188) and cTnI-(1–151) mutants inhibited maximum ATPase activity to approximately 75% and 50% of that of the WT cTnI, respectively. The cTnI-(1–188)·cTnC complex only partially restored Ca2+ sensitivity, while the cTnI-(1–151)·cTnC complex did not restore any Ca2+ sensitivity. These results all suggest that residues 152–199 (C-terminal to the inhibitory region) of cTnI are essential for full inhibitory activity and Ca2+ sensitivity of cardiac myofibrillar ATPase.
The only cTnI residue in the last 5 C-terminal residues that was previously investigated was K206 [8, 18]. The HCM associated mutation K206Q was found to enhance filament sliding [18, 19], while another HCM mutation, K206I, was found to increase myofilament Ca2+ sensitivity and possibly impair cTnI interactions with actin and cTnC [8]. Since no direct results currently suggest that the last 4 C-terminal residues of cTnI are important for the physiological function of cTnI, but several HCM mutations occur within this region, the functional role of the last 5 residues of cTnI was investigated. Using a mammalian two hybrid system deletion of just three residues from the C-terminal of cTnI was found to affect its interaction with both cTnC and cTnT. Fluorescent binding studies suggest that deletion of the last 5 residues of TnI caused a small increase in the Ca2+ affinity of the Tn complex. Removal of the last 3 or last 5 residues from cTnI significantly increased the Ca2+sensitivity in in vitro motility assays. Taken together, these results show that the last 5 residues of cTnI are functionally important for muscle contraction.
MATERIALS AND METHODS
Mutation, expression and purification of cardiac troponin I, troponin T and troponin C
Detailed methods for expression and purification of the wild-type and mutated Tn subunits are similar to what was previously described (11, 19, 20). Briefly, for cTnT, bacterially expressed and extracted cTnT proteins were purified on a fast flow Q-sepharose column, followed by a NaCl gradient elution. The semi-purified cTnT were then further purified on an S-sepharose column. Purified cTnT was eluted from the column with a NaCl gradient. All steps were performed at 4°C unless otherwise indicated.
Formation of Troponin Complexes
Formation of the human cardiac Tn complexes containing human recombinant cTnT, cTnI, cTnC was as previously described (11, 19, 20). Proper stoichiometry was verified by SDS-PAGE (Figure 1) and aliquots of complexes stored at – 80°C. Proper stoichiometry of the Tn complex was based upon the relative amounts of TnT, TnI, and TnC in the final Tn complex compared against the relative amounts of TnT, TnI, and TnC in HPLC purified porcine Tn complex used as a standard.
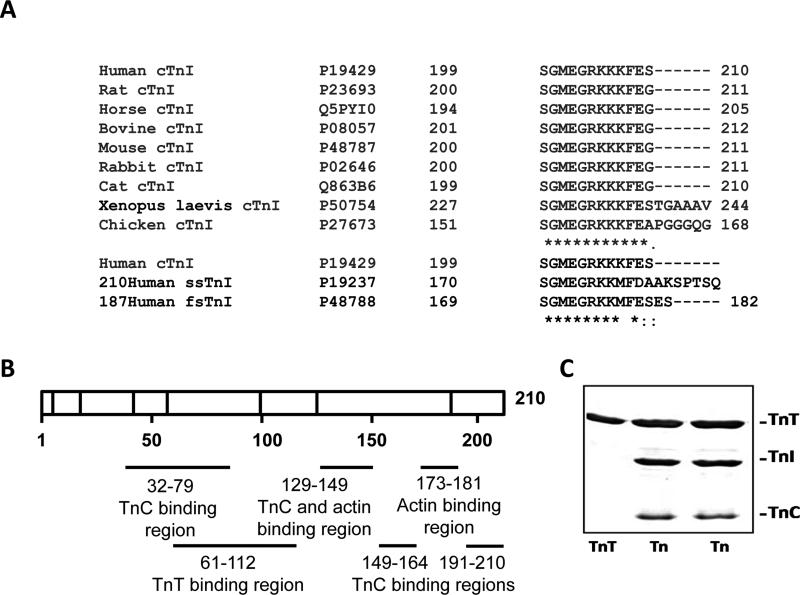
Figure 1.
C-terminal homology of cTnI in different animals and the regions of cTnI known to interact with other thin filament proteins. A, C-terminal homology between cTnI from different animals. B, Regions of cTnI that interact with other thin filament proteins. C, SDS-PAGE of cTnT and Tn complexes. Lanes 2 and 3 contain different preparations of Tn complex. Lane 1 contains 2 μg of protein while lanes 2 and 3 contain 5 μg of protein.
Mammalian Two Hybrid
The Checkmate Mammalian Two-Hybrid System (Promega) was utilized to measure protein-protein interaction. WT cTnI and cTnI deletion mutants as well as WT cTnT, and cTnC were subcloned into pACT and pBIND vectors. CV1 cells were transfected with pACT and pBind DNA constructs as well as the pG5luc Vector. TransIT-LT1 transfection reagent (Mirus) was used for all experiments and cells were incubated for 36-48 h following transfection with no media change. Cells were lysed and analyzed using the dual luciferase reporter assay (Promega). Control experiments using different combinations of cTnI, cTnC, and cTnT subcloned vectors and empty vectors were also carried out to validate the reliability of the experiments.
Fluorescence labeling of cTnC
WT cTnC was fluorescently labeled at both cysteines 35 and 84 using 2-(4′-(iodoacetomido)aniline)naphthalene-6-sulfonic acid (IAANS). Fluorescent incorporation and subsequent purification of labeled cTnC was carried out as previously described [20]. Briefly, cTnC protein solutions were first made in 2 mM EDTA and 7.5 mM DDT and then exhaustively dialyzed against 50 mM Tris, pH 7.5, 150 mM KCl, 0.2 mM DTT, 0.2 mM EDTA. The dialyzed proteins were adjusted to a concentration of 1–2 mg/mL, and labeling was initiated by addition of IAANS at a concentration which saturated the DTT and was in 4-fold molar excess over Cys residues. Excess label was removed by gel filtration chromatography. The IAANS labeled cTnC was used to make the Tn complex with cTnI and cTnT as previously described [20]. All protein concentrations were determined using the Bradford assay (Bio-Rad) with purified cTnC as the standard. The amount of IAANS associated with the protein was determined by UV absorbance at 325 nm using an extinction coefficient of 24,900 M−1.
Measurement of the effect of calcium binding on fluorescence from IAANS-labeled cTnC in troponin complex
Steady state fluorescence was measured at 25 °C using a Lumina fluorescence spectrometer (Thermo Scientific). IAANS fluorescence was excited at 330nm, and emission monitored at 450nm after different Ca2+ concentrations were applied (pCa 9.0 to pCa 4.0). The concentrations of free and titrated Ca2+ were calculated using a previously described program (pCa calculator [21]). The program made corrections for the dilution effects that occurred during titration of Ca2+. The data were fitted to the Hill equation and plotted using SigmaPlot 11.0. The apparent Ca2+ affinities are reported as pCa50 values ± SEM.
Actin-Tm-activated Myosin-ATPase Assay
Myofilament components such as porcine cardiac myosin and porcine cardiac Tm were prepared as previously described [22]. The ATPase inhibitory assay was performed in a 100 μl reaction mixture of 100 mM KCl, 4 mM MgCl2, 1.0 mM EGTA, 2.5 mM ATP (Mg2+-ATP), 0.1 mM dithiothreitol, 10 mM MOPS, pH 7 as previously described [23].
In Vitro Motility Assays
In vitro motility assays were performed to measure the unloaded sliding speed of fluorescently labeled F-actin or reconstituted thin filaments propelled by rabbit skeletal heavy meromyosin (HMM). Motility assays were carried out essentially as described [24-26].
Rabbit skeletal muscle myosin, HMM, and actin were prepared for motility assays as described [27-30]; all animal procedures associated with obtaining muscle tissue for myosin and actin for motility assay were approved by Florida State University's Institutional Animal Care and Use Committee. Purified F-actin was fluorescently labeled with rhodamine phalloidin (RhPh). Recombinant human cardiac tropomyosin (homodimeric αTm) was bacterially expressed and purified as published [31, 32].
Flow cells for motility assays were constructed as described [25, 33-36]. Solutions for assays with regulated thin filaments was calculated to have a composition of 2 mM MgATP, 1 mM Mg2+, 10 mM ethylene glycol tetraacetic acid (EGTA) total, sufficient Ca(CH3COO)2 to achieve the desired pCa (pCa 9 – 4), 50 mM K+, 15 mM Na+, 20 mM 3-(N-morpholino) propanesulfonic acid (MOPS) total, pH 7.00 at 30°C, 0.3% methyl cellulose, and ionic strength was adjusted to 0.085 mM with TrisOH/CH3COOH) [18, 32, 37]; stock motility buffers were made at pCa 9, 8, 7, 6, 5 and 4, and intermediate pCa solutions were obtained by mixing appropriate, calculated volumes of the two stock solutions at the nearest, whole pCa values. To obtain regulated thin filaments, equimolar concentrations (25 nM each) of cTn and cTm were applied to RhPh-labeled F-actin in the flow cell immediately preceding addition of the motility buffer, as described [18, 19, 24, 31, 32, 36]; cTn and cTm (25 nM each) were also added to motility buffers for assays with regulated thin filaments. To minimize photo-oxidative damage, motility buffers additionally contained 3 mg ml−1 glucose, 100 mg ml−1 glucose oxidase, 18 mg ml−1 catalase and 40 mM DTT. All motility assays were conducted at 30°C.
Sliding of RhPh-labeled thin filaments and F-actin was observed by fluorescence microscopy on a Nikon Eclipse TE2000-U inverted microscope (Nikon USA, Melville, NY) at 100x or 150x magnification [24, 38]. Temperature was maintained by a heating coil (model OH30 heater and B600 temperature controller, 20/20 Technology, Inc, Wilmington, NC, USA) placed on the microscope objective. Field images were recorded onto DVDs using a VE1000SIT camera (Dage-MTI, Michigan City, IN). Movie clips were processed as described to obtain sliding speeds (in units of μm s−1) for all tracked filaments using ImageJ (National Institutes of Health, Bethesda, MD) and custom motility analysis software from the University of Washington; mean sliding speed (s) and standard error of the mean (SEM) were calculated for each flow cell [24]. Note that each flow cell corresponds to one of the four Tns tested at one Ca2+ level (one pCa). The mean, unloaded sliding speed data (s, in units of μm s−1) for each Tn (WT or mutant) were fit to the following, 4-parameter form of the Hill Equation:
Where smin is the speed at very low concentrations of Ca2+ (high pCa values), smax is the Ca2+-activatable increment in speed, (smax + smin) is the speed at high concentrations of Ca2+ (low pCa values), pCa50 is the pCa at which s = smax/2 + smin, and cooperativity parameter nH reflects the steepness of the speed-pCa relationship around pCa50. The data were normalized to (smax + smin) to allow comparisons between data collected with the two different batches of myosin used in this study. The normalized speed data for each Tn were then fit to one of two modified versions of the Hill Eq. For smin/(smax + smin) > 0.02, the normalized data were fit to a 3-parameter form of the Hill Eq in which smax was substituted with the normalized form 1-s’min:
Alternatively, for smin/(smax + smin) < 0.02, the normalized data were fit to a 2-parameter form of the Hill Equation in which smax and smin were constrained to be the normalized values 1.0 and 0.0, respectively:
Statistical Analysis
Significant differences between samples were calculated using the student's t-test (P < 0.05, Sigma Plot 11.0). Nonlinear least squares regressions for analyzing motility data were accomplished using SigmaPlot software (version 11.2.0.5; Systat Software, Inc., Chicago, IL, USA) and verified with the NLS function in the R programming environment (R Foundation for Statistical Computing). Regression parameter errors are standard error of the mean (SEM).
RESULTS
Comparison of TnI C-terminal region
The C-terminus of cTnI is highly conserved among different species and is also well conserved between the three different TnI isoforms (Figure 1A). Eleven of the last 12 residues (SGMEGRKKKFE) are identical in cTnI from 9 different species (Figure 1). The C-terminal residue of human cTnI (S) is different from the C-terminal residue in rat, mouse, horse, bovine, rabbit and cat (all have G as the C-terminal residue). The highly conserved nature of the C-terminal residues suggests that this region is likely to be important for the biological function of TnI. TnI contains several binding sites for TnC, TnT, Tm and actin (Figure 1B). Little is known about the last 5 C-terminal residues of cTnI. However, mutations at K206 enhance filament sliding (K206Q) [18, 19], increase myofilament Ca2+ sensitivity, and possibly affect cTnI interactions with cTnC and actin [8]. Tn complexes containing WT or C-terminal deletion mutants of TnI were as made described in the methods. SDS-PAGE of WT Tn complexes and TnT are shown in Figure 1C. cTnC is weakly stained with Coomassie blue relative to other Tn subunits because of its acidic nature.
Effect of C-terminal cTnI deletions on the interaction between cTnI and other thin filament proteins
Interactions between cTnI and other thin filament components were determined using the Checkmate mammalian two-hybrid system from Promega. Two-hybrid systems are one of the most powerful methods for detecting protein-protein interactions in vivo [39, 40]. In the Checkmate mammalian two-hybrid system, genes are cloned into pBIND and pACT vectors to form fusion proteins with the DNA-binding domain of GAL4 and the activation domain of VP16, respectively. The vector pBIND expresses Renilla reniformis luciferase which allows transfection efficiency to be normalized. Cells were transfected with pBIND and pACT vectors together with the pG5luc vector which contains GAL4 binding sites upstream of a firefly luciferase gene. Because of the high sensitivity of the Checkmate mammalian two-hybrid system, differences in relative affinity between test proteins could be readily detected. This system has already been shown to be able to detect changes in functional interactions between Tn subunits [40].
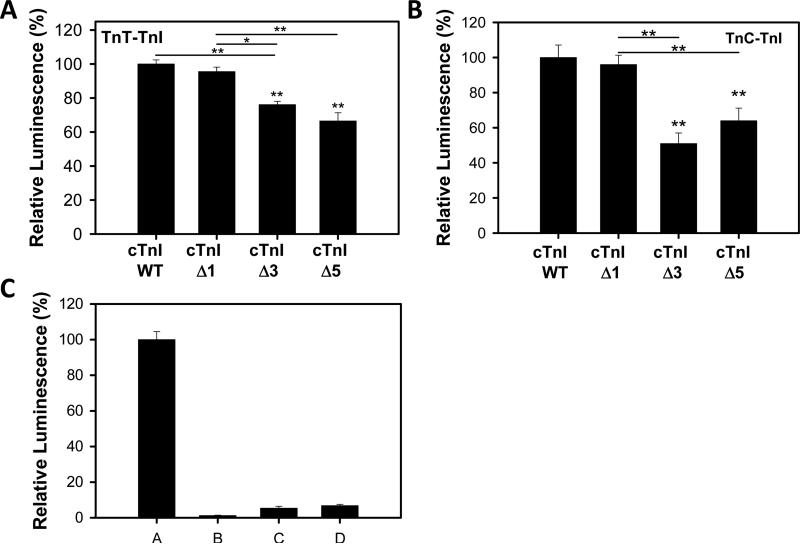
Interaction between cTnT (pACT) and different cTnIs (pBIND) measured using the two-hybrid system showed that the binding between cTnT-cTnI was decreased when cTnI was lacking either 3 or 5 C-terminal residues (cTnI Δ3 or Δ5) (Fig. 2A). Removal of the cTnI C-terminal residue serine (cTnI Δ1) did not affect its interaction with cTnT relative to wild-type cTnI. The binding of cTnC (pACT) and cTnI Δ3 and Δ5 truncations were also impaired relative to wild-type cTnI and cTnI Δ1 (Fig. 2B). Controls showing the low levels of luminescence for empty pACT and pBIND vectors are shown in Fig. 2C. This is the first report of the last five residues of cTnI affecting the interaction with other myofilament protein. Attempts to determine the effects of these cTnI truncations on TnI's interaction with actin were unsuccessful using the mammalian two hybrid system.
Figure 2.
Effects of C-terminal deletions on cTnI-cTnT and cTni-cTnC interactions. Mammalian two-hydrid was utilized to determine disruptions in the interactions between different cTnI's and wild-type cTnT. * P < 0.05 (n=4-6). A) Effect of C-terminus deletion mutations in cTnI on interactions with cTnC. B) Interactions between cTnI deletion mutations and wild-type cTnC. * P < 0.05, ** P < 0.001 (n=4). C) Control experiments to determine the interactions between empty vectors and different combinations of pACT and pBIND vectors. A, cTnC (pACT) and wild-type cTnI (pBIND), B, pACT and pBIND, C, cTnC (pACT) and pBIND, pACT and wild-type cTnI (pBIND). Data are expressed as mean ± SEM.
Effect of C-terminal cTnI deletions on Ca2+ binding to Troponin
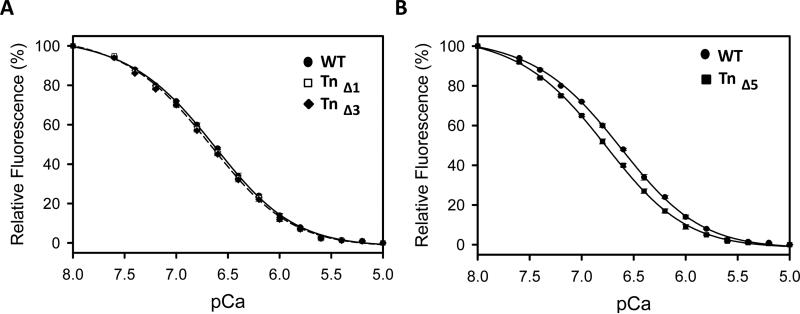
cTnC was purified and labeled with IAANS fluorescent probe. The fluorescently labeled cTnC was then incorporated into Tn complexes containing either wild-type or truncated cTnI. Measurement of the fluorescence changes of these complexes with different Ca2+ concentrations showed that Tn complexes containing cTnI Δ1 or Δ3 had pCa50s which were similar to wild-type Tn (Fig. 4, Table 1). However, the Tn containing cTnI Δ5 showed significantly increased Ca2+ sensitivity relative to Tn containing wild-type cTnI. Since the 5 C-terminal residues of cTnI do not directly interact with cTnC or cTnT, these results suggest that the 5 residues at the C-terminus of cTnI indirectly affect binding of Ca2+ to cTnC.
Figure 4.
Effect of C-terminus deletion mutations in cTnI on the calcium binding to TnC IAANS-Tn complexes. Apparent Ca2+-affinities of Tn complexes were determined by steady-state fluorescence measurements as described in the materials and methods. Data are expressed as mean ± SEM, n=4.
Table 1.
Effect of Cardiac Troponin I C-terminal deletions on calcium binding to Troponin.
| Troponin Complex | pCa50 | Number of samples (n) |
|---|---|---|
| WT Tn | 6.66 ± 0.01 | 4 |
| Tn containing cTnI Δ1 | 6.67 ± 0.01 | 4 |
| Tn containing cTnI Δ3 | 6.71 ± 0.02 | 4 |
| Tn containing cTnI Δ5 | 6.81 ± 0.02* | 4 |
p< 0.05 for deletion mutant relative to wild-type troponin.
Actin-Tm-activated Myosin-ATPase Assay
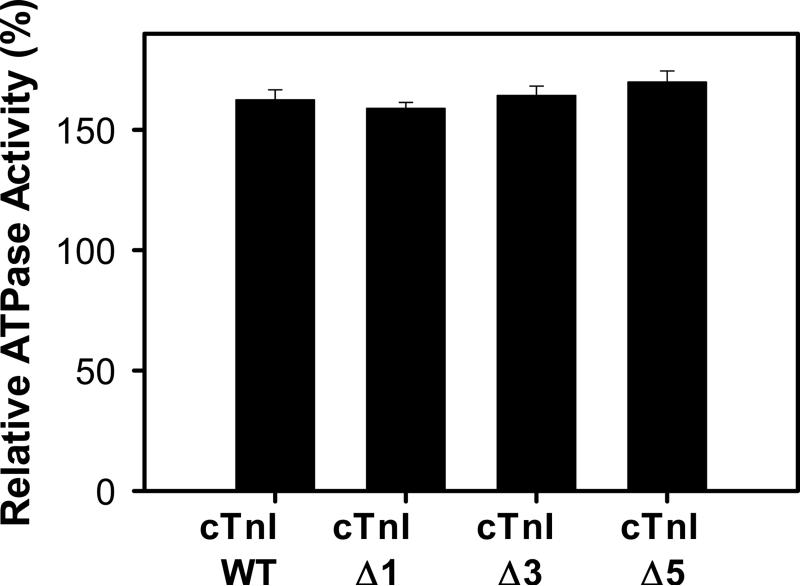
In the presence of 0.5 mM Ca2+, the actin-Tm-activated myosin-ATPase activity was similar for all the cTnI mutants investigated (Fig. 4). The data shown in Fig. 4 utilizes 1 μM Tn. At higher concentrations (1.5 μM) the maximum ATPase activity was not further increased (data not shown), which is consistent with the amount of Tn needed for maximal ATPase activity being 1 μM which would result in a 1:1 ratio of Tn:Tm.
Effect of C-terminal cTnI deletions on in vitro Motility
Maximum speed
For all four Tns examined in this study, the maximum Ca2+-activated sliding speed for regulated thin filaments was ~ 20 – 40% faster than that for unregulated F-actin measured on the same day; this enhancement (super-activation) of filament sliding by the Ca2+ regulatory proteins is consistent with previous results using cardiac Tn and Tm [19, 24, 31, 32, 41]. There was no systematic effect of the deletion mutants on maximum Ca2+-activated sliding speed. However, the maximum speed for the three mutants was ~ 10 – 20% faster than WT. This is consistent with the enhancement obtained at the same concentration (25 nM) of human cardiac Tn containing cTnI1-163 truncation mutant that is missing the entire C-terminal mobile domain [24]. It is also consistent with the enhancement of maximum speed observed with FHC-related mutants that affect single amino acids at the C-terminus of cTnI (R206K) [18].
Filament sliding in the absence of Ca2+
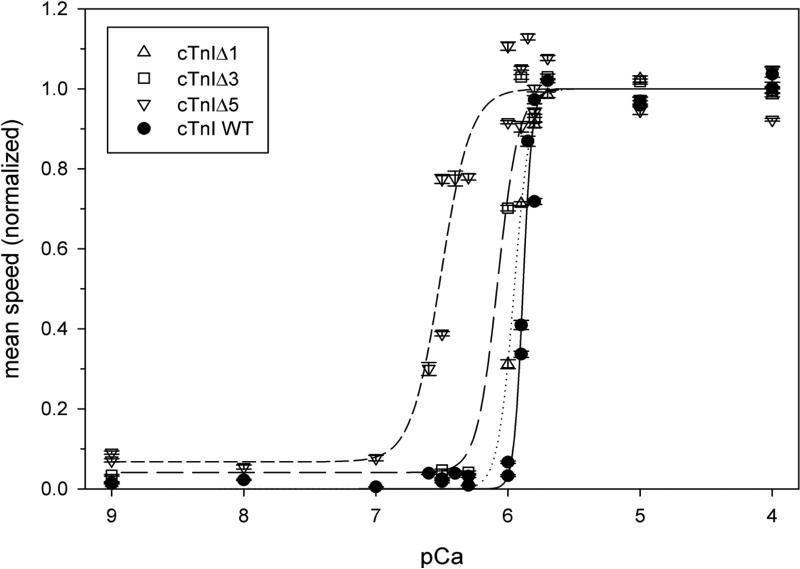
In high pCa (low Ca2+) conditions, little or no significant, directed sliding was measured with WT or the Δ1 mutant protein. Average sliding speed (smin) for the Δ3 and Δ5 mutants at high pCa was ~ 4% or 7%, respectively, of the maximum speed for the same protein (Fig. 5).
Figure 5.
In vitro Motility assay for Troponin containing C-terminus deletion mutations in cTnI.
Enhancement of Ca2+-sensitivity of regulated thin filament sliding speed relative to WT control (filled circles and solid line) after removing 1 (open triangles and dotted line), 3 (open squares and long-dashed line) or 5 (open inverted triangles and short-dashed line) C-terminal residues from cTnI. In vitro motility assays were conducted and filament sliding speeds were quantified as described in Materials and Methods. Each point represents the mean ± SEM sliding speed for all filaments quantified in one flow cell, and each flow cell represents one of the four troponin constructs tested at one pCa. All speeds were normalized to nonlinear least squares regression smax + smin (in μm s−1) for the same troponin. Lines represent nonlinear least squares regressions of the Hill Eq for normalized speed data, as described in Materials and Methods; see Table 2 for regression parameter estimates.
Ca2+ dependence of filament sliding—Hill parameters pCa50 and nH
There was a general trend for the apparent cooperativity (nH) of filament sliding to decrease as more residues were removed from the C-terminus of cTnI (Fig. 5 and Table 2). For example, nH decreased from 12.7 ± 2.1 for WT to 3.9 ± 1.2 for the Δ5 mutant. In addition to the reduction in apparent cooperativity of filament sliding, speed also became progressively more sensitive to Ca2+ (i.e., pCa50 shifted leftward) as more residues were removed from the C-terminus of cTnI (Fig. 5 and Table 2). For example, pCa50 for the Δ5 mutant was 6.51 ± 0.03, compared with 5.89 ± 0.01 for WT; this corresponds to a large, 0.62 pCa unit shift in the direction of enhanced Ca2+ sensitivity for the mutant. This shift is comparable to the greatest shift in motility speed-pCa relations with an FHC-related, single amino acid mutation (K206Q) in the C-terminus of cTnI [18]. These results extend our insights into the role(s) of cTnI's C-terminus in control of unloaded sliding of regulated thin filaments [18, 24, 42].
Table 2.
Effect of Cardiac Troponin I C-terminal deletions on speed-pCa relations in motility assays.
| Troponin Complex | Regression parameter estimates ± SE | |||
|---|---|---|---|---|
| pCa50 | nH | Smin/(Smin + Smax) | R2 | |
| WT Tn | 5.89 ± 0.01 | 12.7 ± 2.1 | 0 | 0.981 |
| Tn containing cTnI Δ1 | 5.95 ± 0.01 | 7.1 ± 0.4 | 0 | 0.999 |
| Tn containing cTnI Δ3 | 6.08 ± 0.02 | (5.5) | 0.04 ± 0.02 | 0.987 |
| Tn containing cTnI Δ5 | 6.51 ± 0.03 | 3.9 ± 1.2 | 0.07 ± 0.05 | 0.947 |
Regression parameter estimates for motility data shown in Fig. 5 were obtained as described in Materials and Methods. Note that nH for Tn containing cTnI Δ3 was constrained to be 5.5 because the data did not adequately define this slope parameter.
DISCUSSION
TnI function is critical for muscle physiology as this protein is important in regulating muscle contraction [43, 44]. At regions that interact with other thin filament proteins, cTnI contains: the switch peptide region (residues 148–163) which interacts with the N-terminal domain of TnC in the presence of Ca2+, the inhibitory peptide region (residues 128–147) which binds actin and inhibits actomyosin ATPase activity, and a second actin binding region (residues 168–188). However, relatively little is known about the last 5 C-terminal residues of cTnI. The N- and C-terminals of TnI were unresolved in crystal structures of the Tn complex [45, 46]. Homology modeling based upon the crystal structures of Tn using threading predicts that the C-terminus of cTnI has the potential to be alpha helical (data not shown). In 1997, cTnI was identified as a disease gene associated with HCM [7]. cTnI has also more recently been shown to be associated with RCM [47]. A female patient with a one nucleotide deletion that resulted in a premature stop codon at amino acid 176 (cTnI lacking the last 35 C-terminal residues) showed approximately 50% total TnI content in myocardial tissue [47]. It is possible that the truncated form of cTnI was rapidly degraded. This patient died at 28 years and showed right ventricular hypertrophy and a RCM phenotype. The patient's father died from sudden cardiac death at 29 years of age [47].
Since the discovery of cTnI as a HCM associated gene, five HCM mutations (K206Q [7], K206I [18], K207T [9], E209A [10] and E209K [11]) have been found to occur within the last 5 C-terminal residues of human cTnI [12]. Although the effect of these HCM mutations on Tn function are unknown, the presence of these mutations in this region of cTnI and the high sequence conservation in this region of cTnI both suggest that the last 5 C-terminal residues of cTnI are important in cTnI function. Using the mammalian two hybrid system, which is a very sensitive technique, we found that removal of the last 3 or last 5 residues of cTnI decreased the ability of cTnI to interact with both cTnT and cTnC. Removal of the last C-terminal cardiac residue of cTnI did not affect cTnI's ability to interact with cTnC or cTnT. These results strongly suggest that the last 3 residues affect cTnI's ability to interact with other Tn subunits. The removal of 5 residues from the C-terminus (cTnI Δ5) did not significantly affect the ability of cTnI to interact with cTnT or cTnC relative to cTnI Δ3, suggesting that residues 208-210 (FES) may be more important than residues 206 and 207 (S and K) for the interaction between TnI and TnC or TnT. The last residue of human cTnI (S) is not as well conserved as residues 208-209 (FE), and may explain why the C-terminal residue (S, residue 210) does not affect the interaction between cTnI and cTnT or cTnC.
Tn complexes containing cTnI 1-192 (lacking the last 17 residues of cTnI) exchanged into human cardiac myofibrils and permeabilized cardiomyocytes from healthy donor hearts showed increased Ca2+ sensitivity and slower rates of force activation and redevelopment compared to myofibrils and cardiomyocytes containing control wild-type [48]. Several investigations suggest that the last 17 residues of cTnI are important for cTnI interactions with other Tn subunits and actin-Tm [14, 49-51]. Galinska et al. [14], using electron microscopy and 3D image reconstruction, showed that the last 17 amino acids of cTnI (residues 193-210) are important in stabilizing Tm in the Ca2+-activated state (C-state). Galinska et al. data supports the “fly-casting mechanism” originally proposed for unstructured proteins [52], and then proposed for the C-terminal region of TnI [53]. The “fly-casting mechanism” has also been supported by other techniques such as fluorescence anisotropy [54]. The “fly-casting” model suggests that the C-terminus of TnI is mobile and remains unfolded when not interacting with actin, but as sufficient interaction between TnI and actin occurs, the C-terminal region starts folding and moving (or “reeling in”) the Tn complex to the position occupied by TnI fully bound to actin, ensuring proper conformational switching of Tn-Tm. Earlier studies using reconstituted Tn complex containing mini-TnT (the two Tm-binding sites were deleted) demonstrated a saturable binding of TnI to Tm at pCa 9, which was abolished when TnI lacking the C-terminal 19 residues was utilized instead of wild-type TnI, suggesting that the C-terminal end of TnI is involved in the Ca2+ regulation of the thin filament through interaction with Tm [16].
The mobility of the C-terminus of TnI is supported by several studies [55, 56]. At low Ca2+ concentrations cTnI residues 167-210 (the mobile domain) have been shown to stabilize the interaction between actin and cTnI [57]. Using single-molecule Förster resonance energy transfer (FRET) combined with computational approaches, Metskas and Rhoades found that the C-terminal domain of TnI (residues 147-210) in the Tn complex is likely extended, with moderate helical propensity around residues 169–175 and weaker helical propensity closer to the C-terminus [56]. Using steady-state FRET, cTnI residues 197 and 210 were found to move away from Cys-190 of Tm, Cys-374 of actin, and the phalloidin binding site on the F-actin filament on binding of Ca2+ to the thin filaments [55]. Myosin S1 binding to relaxed thin filaments also induced movement of residues 197 and 210 similar to that observed when Ca2+ binds to the thin filaments suggesting that Ca2+ and myosin S1 binding to thin filament causes movement of the C-terminal domain of cTnI from the F-actin outer domain towards the inner domain [55].
The impaired interactions between TnI deletion (Δ3 and Δ5) mutants and TnC suggest that either direct interactions between the TnC and the C-terminal of TnI are affected or the Δ5 mutants indirectly affects other TnI regions that interact with TnC. The possibility of a direct interaction is based upon data which suggests that the TnI C-terminal peptide, residues 191-210, may interact weakly with TnC [15]. Since no report has shown that the C-terminal region of TnI directly interacts with TnT, the impaired interactions between TnI Δ3 and Δ5 mutants and TnT are possibly due to conformational changes in other regions of TnI that interact with TnT. It is also possible that in the Tn complex, the impaired interactions between TnI Δ3 and Δ5 mutants and TnC may amplify or reduce the effect of the truncations on TnT's interaction with TnI. However, the mammalian two hybrid system utilized in these studies, like other two hybrid systems, has some caveats including the limitation of only detecting the interaction between two subunits and not more complicated protein-protein interactions, false positives due to expression of high levels of proteins which are not normally present in the same cell at the same time, and potential interactions with the GAL4 or VP16 domains that are used to quantify the protein-protein interactions, which necessitates the use of proper controls. The benefits of using the mammalian two-hybrid system are that it is an extremely powerful method for detecting protein-protein interactions in vivo [39] and has an advantage over the yeast two hybrid system by allowing mammalian protein interactions to be investigated in an environment that is more comparable to that in vivo [58].
Cardiomyopathy mutations in the C-terminal region of cTnI show increased myofilament Ca2+-sensitivity and impaired binding of cTnI with actin and/or Tm [50, 59]. Mathur et al. showed that some cardiomyopathy causing cTnI mutations such as R191H may function by stabilizing a functional intermediate state of actin [59]. Mathur et al. also showed that the R193H mutation may stabilize the active state of thin filaments [59]. A transgenic mouse model containing the R193H mutation showed increased myofibrillar Ca2+ sensitivity and impaired relaxation in the mutant cardiac myocytes [60]. Recently, the HCM cTnI mutation K206I was investigated and found to significantly increase myofilament Ca2+ sensitivity with possible impaired interactions between cTnI K206I and actin and cTnI K206I and cTnC [8]. Using site specific monoclonal antibodies as a probe and mouse cTnI containing the RCM associated mutations R193H or R205H showed conformational changes in the TnT-TnI interface of TnI (residues 114-134) and increased binding affinity for TnT [49]. These mutants also showed increased binding affinity for TnC at pCa 4 [49]. Tn complexes containing TnI mutants, TnC, and mini- TnT lacking Tm binding sites showed increase binding affinity for Tm at pCa 9 and pCa 5.5 when compared to Tn complexes containing wild-type TnI suggesting that conformational changes in the C-terminal of cTnI alter its interaction with Tm and other Tn subunits [49].
Post-translational modifications may also affect the C-terminal region of cTnI [50]. In human cardiomyocytes, low levels of cTnI-Ser199 pseudo-phosphorylation (~6%) were found to increase myofilament Ca2+-sensitivity [50]. The increase in myofilament Ca2+-sensitivity was suggested to be due to a decrease in the binding affinity of cTnI for actin-Tm as determined by co-sedimentation assays [50]. These results all suggest that the C-terminal region is important for proper TnI function and that the C-terminal of TnI is capable of altering the interaction between cTnI and actin and/or Tm.
NMR spectroscopy of skeletal TnI suggests that residues 135-182 of TnI are flexible in solution with no stable secondary structure [53]. Interestingly, the C-terminal region of skeletal TnI is not affected by Ca2+ binding to TnC [53]. Investigating the Ca2+ sensitivity of IAANS-labeled Tn complexes containing WT and C-terminus deletion mutants of cTnI showed that only Tn containing cTnI Δ5 showed significant changes in Ca2+ sensitivity relative to WT Tn. Tn containing the cTnI Δ5 mutation had a small increase in Ca2+ affinity (P<0.05) compared to wild-type Tn; while the cTnI Δ1- and Δ3- containing Tn complexes didn't show a significant difference in Ca2+ affinity when compared to wild-type Tn. These results suggest that cTnI residues 206 and 207 (S and K) may be needed to affect Ca2+ binding at the Tn complex level.
To evaluate the functional significance of the last 5 C-terminal residues of cTnI, in vitro motility assays were carried out. Although there was no systematic effect of the deletion mutants on maximum Ca2+-activated sliding speed, the maximum speed for the three mutants was ~ 10 – 20% faster than WT. The Ca2+-sensitivity of the filaments containing cTnI Δ1, cTnI Δ3 and cTnI Δ5 mutants was significantly increased relative to wild-type cTnI in a trend that was consistent with the changes observed in Ca2+-binding affinity (Table 1), although the effects on pCa50 for motility were quantitatively larger (Table 2). The increased Ca2+ sensitivity was also associated with a reduction in apparent cooperativity of filament sliding for the cTnI mutants. These results, together with the previous results which show that the K206Q mutation enhanced filament sliding with in vitro motility assays [18], suggest that residues 206-209 are functionally important for cTnI. The large increase in Ca2+-sensitivity observed for the cTnI Δ5 mutant was similar to the largest shift in Ca2+-sensitivity of isometric force-pCa relations [61] and motility speed-pCa relations observed with an FHC-related point mutation (E180G) in α-Tm [31]. Interestingly, the in vitro motility results for the cTnI Δ5 is similar to the enhancement obtained at the same concentration (25 nM) of human cTn containing the cTnI1-163 truncation mutant that was missing the entire C-terminal mobile domain [24]. It may be that a significant proportion of the enhanced sliding filament observed in the cTnI1-163 truncation mutant was due to the last of the last 5 C-terminal residues. The enhanced sliding speed (“superactivation”) at saturating Ca2+ in the in vitro motility assays is likely due to some part of troponin other than the C-terminus of TnI [24].
Our results suggest that deletion of the last 5 residues from the C-terminal of cTnI resulted in a small change in the Ca2+ binding properties of the Tn complex, while deletion of the last 3 C-terminal residues had no effect on the Ca2+ binding properties of the Tn complex. Since the effect of cTnI Δ5 was relatively small in relation to the large increases in Ca2+ sensitivity observed in the motility assays, the results are consistent with previous results which suggest that most of the changes in the Ca2+ binding properties of the Tn complex occur only when Tn is reconstituted with actin and Tm in a thin filament [62]. Since no changes were observed for 3 in the fluorescence studies but significant changes in calcium sensitivity were observed for the motility studies, the small conformational changes in troponin containing the 5 mutant may not be critical for the sensitivity observed in the motility studies. In the motility assays, the reconstitution of the troponins into thin filaments, plus the presence of cross-bridges, are likely to expose greater effects of the deletions, as the structural changes in TnC in the troponin complex are likely to be distinctly different from myosin-propelled sliding of thin filaments. It is possible that the altered interaction between TnI 3 and 5 mutants and TnT could lead to altered interaction of the Tn complex with Tm. Another report showed that HCM mutations in the C-terminal region of cTnI (L198P, and R204H) impair TnI's interactions with TnC and TnT using a mammalian two hybrid system similar to the one utilized for our studies [40]. The enhanced Ca2+ sensitivity observed in motility studies may be due to altered TnI interactions with TnC and actin, resulting in TnI interacting more predominantly with TnC than with actin-Tm.
The C-terminal deletion mutants investigated in this study are likely altering the conformation and interaction between the cTnI and/or Tn complex with actin-Tm. As previously suggested by another group, changes in TnI interaction with actin-Tm may impair the ability of the TnI switch region to interact with the hydrophobic pocket of TnC, resulting in reduced Ca2+ sensitivity of TnC [62].
CONCLUSION
Taken together, these results suggest that the last 3 C-terminal residues of cTnI are important contributors to the proper functioning of cTnI. Similar to what was observed with cTnI truncation mutants lacking the last 17 C-terminal residues, cTnI 3 mutants also increase the Ca2+ sensitivity of thin filament sliding speed. Removal of the last 3 residues from the C-terminus of cTnI is enough to considerably increase the apparent sensitivity to Ca2+ in motility assays, suggesting that conformational changes in cTnI affect its interactions with other myofilament components, resulting in enhanced Ca2+ sensitivity when the Tn complex is part of the thin filament. It is likely that long-range interactions between TnI and other regulatory regions present in TnC, TnT, Tm and/or actin in the thin filament are affected by the last three residues of cTnI. The results also suggest that the last 3 C-terminal residues of cTnI affect protein-protein interactions between cTnI and cTnT or cTnC. The interactions between Tn subunits are complex since while both cTnI Δ3 and cTnI Δ5 affect the interaction between cTnI and cTnT or cTnC, only cTnI Δ5 affect the Ca2+ sensitivity of Tn complexes. The results from these studies predict that cTnI mutations resulting in stop codons that result in the loss of three or more C-terminal residues would have significantly altered functional properties. Mutations in this region of cTnI are all likely to have functional consequences. Overall, these results indicate that the last 5 residues of cTnI are more important that previously known.
Figure 3.
Effect of the cTnI truncations on the activation (+ 0.5 mM Ca2+) of the actin-Tm-activated myosin-ATPase activity. The protein concentrations used in this assay were: 3.5 μm actin, 1 μM Tm, 1 μM Tn, and 0.6 μM myosin. Data is presented as mean ± SEM, n=4.
ACKNOWLEDGMENTS
We thank Dr. Michael Regnier, University of Washington, for permission to use the HAMM Lab's custom-designed, automated analysis program for motility data. We thank Dr. Myriam A. Badr, Nancy L. Meyer, Dr. Campion K. P. Loong, and especially Dr. Aya Kataoka Takeda for assistance with motility assays and/or protein preparations for motility assays.
FUNDING
This work was supported by a Hellman Fellowship (AVG) and NIH/NHLBI grant HL63974 (PBC). Funding sources did not have any role in: study design; collection, analysis and interpretation of data; writing or decision to submit this article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Xu Q, Dewey S, Nguyen S, Gomes AV. Journal of molecular and cellular cardiology. 2010;48:899–909. doi: 10.1016/j.yjmcc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Talbot JA, Hodges RS. The Journal of biological chemistry. 1981;256:2798–2802. [PubMed] [Google Scholar]
- 3.Kleerekoper Q, Howarth JW, Guo X, Solaro RJ, Rosevear PR. Biochemistry. 1995;34:13343–13352. doi: 10.1021/bi00041a010. [DOI] [PubMed] [Google Scholar]
- 4.Gomes AV, Guzman G, Zhao J, Potter JD. The Journal of biological chemistry. 2002;277:35341–35349. doi: 10.1074/jbc.M204118200. [DOI] [PubMed] [Google Scholar]
- 5.Gomes AV, Venkatraman G, Davis JP, Tikunova SB, Engel P, Solaro RJ, Potter JD. The Journal of biological chemistry. 2004;279:49579–49587. doi: 10.1074/jbc.M407340200. [DOI] [PubMed] [Google Scholar]
- 6.Rarick HM, Tang HP, Guo XD, Martin AF, Solaro RJ. Journal of molecular and cellular cardiology. 1999;31:363–375. doi: 10.1006/jmcc.1998.0870. [DOI] [PubMed] [Google Scholar]
- 7.Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, Hiroi S, Sasaoka T, Ohbuchi N, Nakamura T, Koyanagi T, Hwang TH, Choo JA, Chung KS, Hasegawa A, Nagai R, Okazaki O, Nakamura H, Matsuzaki M, Sakamoto T, Toshima H, Koga Y, Imaizumi T, Sasazuki T. Nature genetics. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 8.Warren CM, Karam CN, Wolska BM, Kobayashi T, de Tombe PP, Arteaga GM, Bos JM, Ackerman MJ, Solaro RJ. Circulation. Cardiovascular genetics. 2015;8:765–773. doi: 10.1161/CIRCGENETICS.115.001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppini R, Ho CY, Ashley E, Day S, Ferrantini C, Girolami F, Tomberli B, Bardi S, Torricelli F, Cecchi F, Mugelli A, Poggesi C, Tardiff J, Olivotto I. Journal of the American College of Cardiology. 2014;64:2589–2600. doi: 10.1016/j.jacc.2014.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Wijngaard A, Volders P, Van Tintelen JP, Jongbloed JD, van den Berg MP, Lekanne Deprez RH, Mannens MM, Hofmann N, Slegtenhorst M, Dooijes D, Michels M, Arens Y, Jongbloed R, Smeets BJ. Netherlands heart journal : monthly journal of the Netherlands Society of Cardiology and the Netherlands Heart Foundation. 2011;19:344–351. doi: 10.1007/s12471-011-0135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL. Genetics in medicine : official journal of the American College of Medical Genetics. 2015 [Google Scholar]
- 12.Mogensen J, Hey T, Lambrecht S. The Canadian journal of cardiology. 2015 doi: 10.1016/j.cjca.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Foster DB, Noguchi T, VanBuren P, Murphy AM, Van Eyk JE. Circulation research. 2003;93:917–924. doi: 10.1161/01.RES.0000099889.35340.6F. [DOI] [PubMed] [Google Scholar]
- 14.Galinska A, Hatch V, Craig R, Murphy AM, Van Eyk JE, Wang CL, Lehman W, Foster DB. Circulation research. 2010;106:705–711. doi: 10.1161/CIRCRESAHA.109.210047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrieres G, Pugniere M, Mani JC, Villard S, Laprade M, Doutre P, Pau B, Granier C. FEBS letters. 2000;479:99–105. doi: 10.1016/s0014-5793(00)01881-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Akhter S, Mottl S, Jin JP. The FEBS journal. 2011;278:3348–3359. doi: 10.1111/j.1742-4658.2011.08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Eyk JE, Powers F, Law W, Larue C, Hodges RS, Solaro RJ. Circulation research. 1998;82:261–271. doi: 10.1161/01.res.82.2.261. [DOI] [PubMed] [Google Scholar]
- 18.Köhler J, Chen Y, Brenner B, Gordon AM, Kraft T, Martyn DA, Regnier M, Rivera AJ, Wang C-K, Chase PB. Physiological Genomics. 2003;14:117–128. doi: 10.1152/physiolgenomics.00101.2002. [DOI] [PubMed] [Google Scholar]
- 19.Brunet NM, Mihajlović G, Aledealat K, Wang F, Xiong P, von Molnár S, Chase PB. Journal of Biomedicine and Biotechnology. 2012;2012:657523. doi: 10.1155/2012/657523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putkey JA, Liu W, Lin X, Ahmed S, Zhang M, Potter JD, Kerrick WG. Biochemistry. 1997;36:970–978. doi: 10.1021/bi9617466. [DOI] [PubMed] [Google Scholar]
- 21.Dweck D, Reyes-Alfonso A, Jr., Potter JD. Analytical biochemistry. 2005;347:303–315. doi: 10.1016/j.ab.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Lang R, Gomes AV, Zhao J, Housmans PR, Miller T, Potter JD. The Journal of biological chemistry. 2002;277:11670–11678. doi: 10.1074/jbc.M108912200. [DOI] [PubMed] [Google Scholar]
- 23.Gomes AV, Liang J, Potter JD. The Journal of biological chemistry. 2005;280:30909–30915. doi: 10.1074/jbc.M500287200. [DOI] [PubMed] [Google Scholar]
- 24.Schoffstall B, LaBarbera VA, Brunet NM, Gavino BJ, Herring L, Heshmati S, Kraft BH, Inchausti V, Meyer NL, Moonoo D, Takeda AK, Chase PB. DNA and Cell Biology. 2011;30:653–659. doi: 10.1089/dna.2010.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chase PB, Chen Y, Kulin K, Daniel TL. American Journal of Physiology: Cell Physiology. 2000;278:C1088–C1098. doi: 10.1152/ajpcell.2000.278.6.C1088. [DOI] [PubMed] [Google Scholar]
- 26.Kron SJ, Toyoshima YY, Uyeda TQP, Spudich JA. Methods in Enzymology. 1991;196:399–416. doi: 10.1016/0076-6879(91)96035-p. [DOI] [PubMed] [Google Scholar]
- 27.Gordon AM, LaMadrid M, Chen Y, Luo Z, Chase PB. Biophysical Journal. 1997;72:1295–1307. doi: 10.1016/S0006-3495(97)78776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoffstall B, Kataoka A, Clark A, Chase PB. Journal of Pharmacology and Experimental Therapeutics. 2005;312:12–18. doi: 10.1124/jpet.104.073445. [DOI] [PubMed] [Google Scholar]
- 29.Margossian SS, Lowey S. Methods in Enzymology. 1982;85:55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 30.Pardee JD, Spudich JA. Methods in Enzymology. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang F, Brunet NM, Grubich JR, Bienkiewicz E, Asbury TM, Compton LA, Mihajlović G, Miller VF, Chase PB. Journal of Biomedicine and Biotechnology. 2011;2011:435271. doi: 10.1155/2011/435271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoffstall B, Brunet NM, Wang F, Williams S, Barnes AT, Miller VF, Compton LA, McFadden LA, Taylor DW, Dhanarajan R, Seavy M, Chase PB. Journal of Physiology. 2006;577:935–944. doi: 10.1113/jphysiol.2006.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butcher MT, Chase PB, Hermanson JW, Clark AN, Brunet NM, Bertram JEA. American Journal of Physiology - Regulatory, Ingtegrative and Comparative Physiology. 2010;299:R996–R1005. doi: 10.1152/ajpregu.00510.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grove TJ, McFadden LA, Chase PB, Moerland TS. Journal of Muscle Research and Cell Motility. 2005;26:191–197. doi: 10.1007/s10974-005-9010-0. [DOI] [PubMed] [Google Scholar]
- 35.Regnier M, Rivera AJ, Chen Y, Chase PB. Circulation research. 2000;86:1211–1217. doi: 10.1161/01.res.86.12.1211. [DOI] [PubMed] [Google Scholar]
- 36.Brunet NM, Chase PB, Mihajlovic G, Schoffstall B. Archives of biochemistry and biophysics. 2014;552-553:11–20. doi: 10.1016/j.abb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang B, Chen Y, Wang C-K, Luo Z, Regnier M, Gordon AM, Chase PB. Biophysical Journal. 2003;85:1775–1786. doi: 10.1016/S0006-3495(03)74607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoffstall B, Clark A, Chase PB. Biophysical Journal. 2006;91:2216–2226. doi: 10.1529/biophysj.105.079061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fearon ER, Finkel T, Gillison ML, Kennedy SP, Casella JF, Tomaselli GF, Morrow JS, Van Dang C. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:7958–7962. doi: 10.1073/pnas.89.17.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doolan A, Tebo M, Ingles J, Nguyen L, Tsoutsman T, Lam L, Chiu C, Chung J, Weintraub RG, Semsarian C. Journal of molecular and cellular cardiology. 2005;38:387–393. doi: 10.1016/j.yjmcc.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Homsher E, Kim B, Bobkova A, Tobacman LS. Biophysical Journal. 1996;70:1881–1892. doi: 10.1016/S0006-3495(96)79753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moonoo D, Meyer NL, Inchausti V, Brunet NM, LaBarbera V, Chase PB, Schoffstall B. Biophysical Journal. 2010;98:357a. [Google Scholar]
- 43.Tardiff JC. Circulation research. 2011;108:765–782. doi: 10.1161/CIRCRESAHA.110.224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomes AV, Potter JD. Molecular and cellular biochemistry. 2004;263:99–114. doi: 10.1023/B:MCBI.0000041852.42291.aa. [DOI] [PubMed] [Google Scholar]
- 45.Takeda S, Yamashita A, Maeda K, Maeda Y. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 46.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kostareva A, Gudkova A, Sjoberg G, Morner S, Semernin E, Krutikov A, Shlyakhto E, Sejersen T. International journal of cardiology. 2009;131:410–412. doi: 10.1016/j.ijcard.2007.07.108. [DOI] [PubMed] [Google Scholar]
- 48.Narolska NA, Piroddi N, Belus A, Boontje NM, Scellini B, Deppermann S, Zaremba R, Musters RJ, dos Remedios C, Jaquet K, Foster DB, Murphy AM, van Eyk JE, Tesi C, Poggesi C, van der Velden J, Stienen GJ. Circulation research. 2006;99:1012–1020. doi: 10.1161/01.RES.0000248753.30340.af. [DOI] [PubMed] [Google Scholar]
- 49.Akhter S, Bueltmann K, Jr., Huang X, Jin JP. Archives of biochemistry and biophysics. 2014;552-553:3–10. doi: 10.1016/j.abb.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Wijnker PJ, Li Y, Zhang P, Foster DB, dos Remedios C, Van Eyk JE, Stienen GJ, Murphy AM, van der Velden J. Journal of molecular and cellular cardiology. 2015;82:93–103. doi: 10.1016/j.yjmcc.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tachampa K, Kobayashi T, Wang H, Martin AF, Biesiadecki BJ, Solaro RJ, de Tombe PP. The Journal of biological chemistry. 2008;283:15114–15121. doi: 10.1074/jbc.M801636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoemaker BA, Portman JJ, Wolynes PG. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blumenschein TM, Stone DB, Fletterick RJ, Mendelson RA, Sykes BD. Biophys J. 2006;90:2436–2444. doi: 10.1529/biophysj.105.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z, Li KL, Rieck D, Ouyang Y, Chandra M, Dong WJ. The Journal of biological chemistry. 2012;287:7661–7674. doi: 10.1074/jbc.M111.281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Chalovich JM, Marriott G. PloS one. 2012;7:e50420. doi: 10.1371/journal.pone.0050420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metskas LA, Rhoades E. Journal of the American Chemical Society. 2015;137:11962–11969. doi: 10.1021/jacs.5b04471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami K, Yumoto F, Ohki SY, Yasunaga T, Tanokura M, Wakabayashi T. Journal of molecular biology. 2005;352:178–201. doi: 10.1016/j.jmb.2005.06.067. [DOI] [PubMed] [Google Scholar]
- 58.Dang CV, Barrett J, Villa-Garcia M, Resar LM, Kato GJ, Fearon ER. Molecular and cellular biology. 1991;11:954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathur MC, Kobayashi T, Chalovich JM. Biophys J. 2009;96:2237–2244. doi: 10.1016/j.bpj.2008.12.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Zhang L, Jean-Charles PY, Nan C, Chen G, Tian J, Jin JP, Gelb IJ, Huang X. Journal of molecular and cellular cardiology. 2013;62:227–236. doi: 10.1016/j.yjmcc.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai F, Weis A, Takeda AK, Chase PB, Kawai M. Biophysical Journal. 2011;100:1014–1023. doi: 10.1016/j.bpj.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu B, Tikunova SB, Kline KP, Siddiqui JK, Davis JP. PloS one. 2012;7:e38259. doi: 10.1371/journal.pone.0038259. [DOI] [PMC free article] [PubMed] [Google Scholar]