Abstract
Trauma- and stress-related disorders are clinically heterogeneous and associated with substantial genetic risk. Understanding the biological origins of heterogeneity of key intermediate phenotypes such as cognition and emotion can provide novel mechanistic insights into disorder pathogenesis. Performing quantitative genetics in animal models is a tractable strategy for examining both the genetic basis of intermediate phenotypes and functional testing of candidate quantitative traits genes (QTGs). Here, existing and newly collected data were used for collaborative genome-wide mapping of cued fear acquisition and expression in 65 mouse strains from the BXD genetic reference panel. For fear acquisition, we identified a significant locus on chromosome (Chr) 10 and eight suggestive loci on Chr 2, 4, 5, 11, 13, and 15. For fear expression, we identified one significant and another highly suggestive locus on Chr 13, as well as four suggestive loci on Chr 10, 11, and X. Across these loci, 60 putative QTGs were identified. The quantitative trait locus on distal Chr 13 contained a single, highly promising gene at the location of the peak likelihood ratio statistic score. The gene, hyperpolarization-activated cyclic nucleotide-gated channel 1 (Hcn1), regulates neuronal excitability. Validation experiments using behavioral pharmacology revealed that functional Hcn channels in the basolateral amygdala are necessary for conditioned fear acquisition and expression. Hcn1, together with the other candidate QTGs, thus provide new targets for neurobiological and treatment studies of fear learning and trauma- and stress-related disorders.
SIGNIFICANCE STATEMENT There is a knowledge gap in understanding the genetic contributions to behavioral heterogeneity in typical and atypical populations. Mouse genetic reference panels (GRPs) provide one approach for identifying genetic sources of variation. Here, we identified three loci for conditioned fear acquisition and expression in a mouse GRP. Each locus contained candidate quantitative trait genes (QTGs). One locus had a single QTG, Hcn1 (hyperpolarization-activated cyclic nucleotide-gated channel 1), which has been implicated in neuronal excitability and learning. This discovery was validated using behavioral pharmacology, revealing that Hcn channels in the basolateral amygdala are required for fear acquisition and expression. The study thus identifies novel candidate QTGs that may contribute to variation in emotional learning and highlight the utility of mouse GRPs for the identification of genes underlying complex traits.
Keywords: anxiety, associative learning, HCN1, individual differences, quantitative trait gene, threat detection
Introduction
Identifying genetic risk factors for trauma- and stress-related disorders is a high priority for psychiatric genetics (Sullivan et al., 2012; Logue et al., 2015; Smoller, 2016). Twin and family studies have established substantial genetic risk (30–60%) for posttraumatic stress disorder, anxiety disorders, and major depression (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, 2013; Smoller, 2016). Moreover, candidate gene and genome-wide association studies have identified a number of genetic risk factors (Smoller, 2016), with large collaborative mapping studies increasing statistical power to detect common, rare or shared variants (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; Gatt et al., 2015; Logue et al., 2015). However, across psychiatric conditions, there have been challenges with replication, “missing heritability,” deciphering molecular mechanisms that affect brain function, and determining the role of risk variants across related and comorbid disorders (Sullivan et al., 2012; Zuk et al., 2014; Logue et al., 2015; Smoller, 2016).
As proposed by the National Institute of Mental Health (National Institutes of Health) Research Domain Criteria initiative and others, understanding the biological origins of heterogeneity in key intermediate phenotypes such as cognition and emotion that are affected in trauma- and stress-related disorders has the potential to bring mechanistic precision to psychiatric medicine (Kas et al., 2007; Casey et al., 2013; Norrholm et al., 2014; Hariri and Holmes, 2015). In this context, individual differences in emotional memory and conditioned fear are well established endophenotypes (Norrholm et al., 2014; Wilker et al., 2014) that are conserved across species (Holmes and Singewald, 2013; Bukalo et al., 2014; Bowers and Ressler, 2015). Therefore, a quantitative genetics approach in animal genetic reference panels (GRPs) provides a tractable approach for examining the genetic basis of intermediate phenotypes and enables efficient functional testing of candidate quantitative traits genes (QTGs) (Kas et al., 2007; Plomin et al., 2009; Holmes and Singewald, 2013).
Mouse GRPs are composed of hundreds of recombinant inbred (RI) strains and offer a number of advantages for examining the genetic architecture of intermediate phenotypes (Philip et al., 2010; Overall et al., 2015)—particularly collaborative approaches that increase power to detect QTGs. Here, we designed experiments to both expand and exploit preliminary fear acquisition and expression datasets previously collected in the BXD mouse GRP (Yang et al., 2008; Brigman et al., 2009). Using genome-wide mapping, we identified significant and highly suggestive loci for fear acquisition [chromosome (Chr) 10] and expression (Chr 13) and several other suggestive loci for both traits. Collectively, these loci contain 60 putative QTGs, with 26 candidates that are particularly promising based on their reported involvement in relevant functions and disorders.
We found that a highly suggestive quantitative trait locus (QTL) for fear expression on Chr 13 contained a single candidate QTG, hyperpolarization-activated cyclic nucleotide-gated channel 1 (Hcn1)—previously shown to regulate neuronal excitability, synaptic plasticity, and several forms of learning and memory (Nolan et al., 2003, 2004; Wang et al., 2007; Shah, 2014; Maroso et al., 2016). We next confirmed that the Hcn1 protein was richly expressed in multiple brain regions implicated in fear, including the cortex, hippocampus, and basolateral amygdala (BLA). To establish a causal connection between Hcn1 and conditioned fear, we blocked Hcn channels in the BLA pharmacologically, which robustly attenuated fear. Together, our findings confirm and identify genetic loci for learned fear and reveal a novel functional role for Hcn1 in this behavior.
Materials and Methods
Mice.
The BXD GRP comprises >100 RI strains generated from progressive matings of C57BL/6J (B6) and DBA/2J (D2) offspring (B x D) (Peirce et al., 2004). The B6 and D2 parental lines show broad differences in many neurobiological and behavioral traits and high levels of sequence variation (∼5 m SNPs, 500K INDELS, 55K CNVs). These are catalogued at >7500 informative SNPs in the BXD offspring strains (Mozhui et al., 2007). Studies were performed at the University of Southern California (USC) and in the Laboratory of Behavioral and Genomic Neuroscience at the National Institute on Alcohol Abuse and Alcoholism (NIH) using a study design similar to that described below and in Yang et al. (2008) and Brigman et al. (2009). The B6 and D2 parental strains, F1 cross (B6D2F1), and 62 BXD strains were tested in an overlapping manner across sites, with 15 BXD strains (11, 12, 13, 14, 22, 23, 34, 38, 48a, 51, 61, 65b, 70, 81, and 95) tested exclusively at NIH, 24 BXD strains (6, 15, 27, 29, 42, 43, 44, 48, 49, 50, 53, 62, 63, 64, 67, 68, 73, 74, 77, 79, 83, 85, 89, and 91) tested exclusively at USC, and the parental strains, F1 cross and 23 BXD strains (8, 9, 16, 19, 21, 28, 31, 32, 39, 45, 55, 60, 65, 65a, 66, 69, 75, 86, 87, 90, 98, 99, and 100) tested at both sites. For studies conducted at USC and in Yang et al. (2008), adult male mice were obtained from The Jackson Laboratory at 6–10 weeks of age. For studies conducted in Brigman et al. (2009), adult male and female mice were obtained from the University of Tennessee Health Science Center at ∼8 weeks of age. Mice were housed in pairs (USC) or 1–4 per cage (NIH) by strain and were allowed to acclimate to the facilities for 2–4 weeks before testing. Mice were maintained on a 12 h light/dark cycle (lights on 6:00 A.M.) with ad libitum access to food and water except during testing. Testing at both sites occurred during the light phase (between 8:00 A.M. and 5:00 P.M.). Adult mice (3–5 months of age) were tested at USC (9–12 mice/BXD strain; 70–75 mice/parental strain) and NIH (2–12 mice/strain). All procedures were approved by the Institutional Animal Care and Use Committee at USC and NIH and conformed to NIH guidelines.
Fear conditioning.
Similar auditory-cued fear conditioning tasks were used at both sites and the strains that were tested at both sites showed similar levels of conditioned freezing during training and testing (see Results).
Fear-conditioning paradigm at USC.
Behavioral sessions were conducted in an automated four-chamber near-infrared video fear conditioning system (Med Associates). Each chamber (30 × 25 × 21 cm, L × W × H) had stainless steel walls and floor bars and a transparent acrylic door and was housed within a ventilated sound-attenuating chamber (64 × 76 × 36 cm, L × W × H). Mice were acclimated for 30 minutes to the training context “A” (stainless-steel walls and floor bars, 70% ethanol cleaning solution) and trained the following day using 5 paired presentations of a tone-conditioned stimulus (CS; 30 s, 5 kHz, 85 dB) and a foot-shock unconditioned stimulus (US; 2 s, 0.5 mA; CS and US coterminated). There was a 180 s stimulus-free period before the first CS–US pairing (baseline period) and after each pairing (interstimulus interval, ISI). Mice were tested for cued-fear 24 h later in a novel context “B” (textured plastic walls, smooth plastic floor, 70% ethanol cleaning solution) using 10 CS presentations (30 s CS, 180 s baseline period, 60 s ISI). Behavioral sessions were videotaped (30 frames/s) under near-infrared light and freezing times were scored automatically using VideoFreeze software (Med Associates).
Fear conditioning paradigm at NIH.
Behavioral sessions were conducted using Med Associates or San Diego Instruments fear-conditioning systems, as described in Yang et al. (2008). During training, mice received 3–5 pairings of the tone CS and foot-shock US and were tested for cued-freezing 24 h later in a novel context. Mice were trained in context “A” (transparent walls, stainless-steel floor bars, 19.5% ethanol and 1% vanilla extract cleaning solution) using 3 paired presentations of a tone CS (30 s, 3 kHz, 80 dB) and a foot-shock US (2 s, 0.6 mA; CS and US coterminated). There was a 120 s baseline period before the first CS–US pairing and a 60–120 s ISI after each pairing. Mice were tested for cued-fear 24 h later in a novel context “B” (black and white checkered walls, smooth plastic floor, 50% ethanol cleaning solution) using one CS presentation (180 s continuous CS, 180 s baseline period). Behavioral sessions were videotaped and scored manually in 5 s intervals for freezing, which was defined as the absence of all movement except that required for breathing, by an observer who was blinded to strain identity.
Freezing during the final CS presentation was unavailable for the NIH training datasets, so we assessed learning during training (fear acquisition) as the percentage of time mice spent freezing immediately after the final CS–US pairing in the NIH and USC datasets. This measure of fear acquisition was positively correlated with freezing during the final CS in the USC dataset (n = 50 strains, R2 = 0.55, p < 0.0001). Although correlated, these are not identical measures, thus loci mapping with additional datasets, when available, would be warranted. Fear learning during testing (fear expression) was computed as the average percentage of freezing during the first three (USC) or one (NIH) CS presentation(s), which reflects learning magnitudes before behavioral evidence of extinction. Of the 26 strains that were tested at both sites, 70% (18 of 26), including the parental strains, showed high concordance in the percentage of freezing measured during training and testing (fear acquisition: R2 = 0.89, slope = 0.88 ± 0.08; fear expression: R2 = 0.79, slope = 1.00 ± 0.13), whereas 30% (8 of 26) showed discordant freezing levels across sites (fear acquisition: R2 = 0.64, slope = −0.41 ± 0.12; fear expression: R2 = 0.15, slope = 0.22 ± 0.21). The results of this comparison provide evidence that, even in different experimental environments at different times and with distinct experimenters handling the test mice, the training and testing protocols generated similar levels of conditioned fear. For all overlapping strains, we averaged data collected across sites. In separate analyses, we conducted mapping in three ways by either omitting or using NIH or USC data for the eight discordant strains and, under all conditions, mapping results were highly similar (data not shown).
QTL mapping.
Genome-wide simple interval mapping (SIM) was performed using the genome mapping tools within GeneNetwork (GN; http://www.genenetwork.org) to identify suggestive and significant QTLs. We used composite interval mapping (CIM) to identify additional suggestive QTLs masked by linkage and pair scanning to identify epistatic interactions. The B6 and D2 parental strains show substantial sequence variation (Shifman et al., 2006). The BXD RI strains have been genotyped at ∼14,000 markers, including 7636 informative SNPs and microsatellite markers (Shifman et al., 2006). A subset of 3811 informative markers is used for QTL mapping within GN (intermarker interval, 0.66 Mb; Shifman et al., 2006). All mapping was performed without parental strain data and with strain weighting to account for variations in SEM across strains and studies. We performed permutation tests (5000) in GN to determine the likelihood ratio statistic (LRS) thresholds for suggestive and significant QTLs, corresponding to genome-wide probabilities of 0.63 and 0.05, respectively (Lander and Kruglyak, 1995). The proportion of phenotypic variance accounted for by each significant locus was estimated using the square of the Pearson correlation coefficient (r) between the trait and the peak marker. Within the 1.5 logarithm of the odds (LOD) drop support intervals, we selected candidate QTGs based on their involvement in human neurological disorders (NCBI databases: OMIM, ClinVar, and MedGen) or phenotypes related to anxiety, stress, conditioned fear, and learning and memory.
Behavioral pharmacology.
Adult male B6 mice were implanted stereotaxically with indwelling cannulas directed bilaterally at the BLA (n = 9–10/group). Mice were anesthetized with 2% isoflurane and immobilized in a stereotaxic instrument (David Kopf Instruments). Bilateral guide cannulas (26-gauge; Plastics One) were targeted at the BLA (relative to bregma in millimeters: anteroposterior −1.5, mediolateral ±3.2, dorsoventral −4.45). After surgery, the guide cannulas were kept patent using stainless steel obturators (33-gauge) that extended 1 mm beyond the guide cannula. Drug microinjections were performed with stainless steel infusion stylets (33-gauge) that also extended 1 mm beyond the guide cannula. Cannulas were implanted and affixed to the skull with dental cement and mice were allowed to recover for ∼1 week. Mice were conditioned in Context A using 3 CS–US pairings and 24 h later received a cued-fear test using 5 30 s CS presentations in Context B. Forty-eight hours later, mice received a contextual fear test (8 min) in Context A. To examine the effect of Hcn channel antagonism on fear acquisition and expression, the Hcn channel antagonist ZD7288 (4-ethylphenylamino-1,2-dimethyl-6-methylaminopyrimidinium chloride; Tocris Bioscience) was infused (0.3 μg/0.5 μl/side) 30 min before conditioning (day 1) or before cued fear testing (day 2) using a syringe pump and 10 μl Hamilton syringes connected to polyethylene tubing at a rate of 0.1 μl/min. Infusion stylets were left in place for 2 min to allow for drug diffusion and obturators were then replaced and mice were returned to their home cage before training or testing. The dose and time point of ZD7288 administration was chosen based on prior microinjection studies (Chevaleyre and Castillo, 2002; Kocsis and Li, 2004) and drug dose was based on the salt form and dissolved in 0.9% saline with 1% dimethyl sulfoxide (DMSO; Sigma-Aldrich).
Immunohistochemistry (IHC) and confocal microscopy.
IHC was performed on forebrain tissue and dendritic arbors in the hippocampus, amygdala, and cortex were imaged using fluorescent microscopy (Zeiss lsm700 scope). Adult mice were briefly sedated with isofluorane, overdosed with ketamine-xylazine (80 mg/kg ketamine, 20 mg/kg xylazine, IP), and intracardially perfused with 0.1 m PBS (15 ml) followed by 4% paraformaldehyde (60–75 ml). Brains were removed and postfixed in 4% paraformaldehyde for 18–24 h, cryoprotected in 30% sucrose, and then stored at −80°C until sectioning. A cryostat was used to collect 14 μm coronal brain sections and slide-mounted sections were allowed to dry for 60–120 min at room temperature and then stored at −80°C. The day of IHC processing, slide-mounted sections were allowed to thaw and dry for several hours. During the IHC procedure, slides were rinsed in 0.1 m TBS-0.05% Tween 20 (TBS-T), washed in 0.3% hydrogen peroxide in methanol, washed in TBS-T, incubated for 30 min in fab fragment donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, 715-007-003, 1:50), and blocked for 30 min in TNB blocking buffer [0.1 m Tris-HCl, pH 7.5, 0.15 m NaCl, and 0.5% (w/v) blocking reagent (PerkinElmer, FP1020)] at room temperature. Slides were then incubated in primary antibody in blocking buffer (1:500, UC Davis/NIH NeuroMab Facility, Clone No. N70/28) overnight at 4°C, washed in 0.1 m TBS-T, incubated in HRP-labeled secondary antibody (1:500, Jackson ImmunoResearch Laboratories, 715-035-150), washed in TBS-T, incubated in TSA Plus Cyanine-3 amplification buffer (PerkinElmer, NEL744001KT), washed in TBS, and coverslipped with Vectashield.
Statistical analyses.
Data are presented as the mean ± SEM. Student's t tests were used to examine differences in means between two groups. Differences in the means of three or more groups were tested using one-way ANOVA followed by Bonferroni post hoc tests. Broad-sense heritability was estimated for each phenotype using one-way ANOVA to determine the proportion of phenotypic variance accounted for by strain membership compared with the total phenotypic variance (SSStrain/SSTotal, determined from R2 values and reported as h2).
Results
Heterogeneity in fear acquisition and expression
Across 65 strains of mice from the BXD panel, we found broad heterogeneity in fear behavior (Figs. 1; 2). The magnitude of freezing after the final training trial (“fear acquisition”) varied 30-fold across the panel (3.0 ± 1.0, BXD92 to 87.5 ± 4.2, BXD13; strain-effect: F(64,839) = 12.69, p < 0.001; Fig. 1A). There was continuous variation in conditioned freezing during testing (“fear expression”), with values varying 10-fold across the panel (9.75 ± 6.95, BXD92 to 97.22 ± 2.78, BXD13; strain-effect: F(64,837) = 11.29, p < 0.001; Fig. 2A). The fear acquisition and expression traits were positively correlated (R2 = 0.53, p < 0.001) and moderately heritable (acquisition, h2 = 0.49; expression, h2 = 0.46), consistent with previous reports (Brigman et al., 2009; Carhuatanta et al., 2014).
Figure 1.
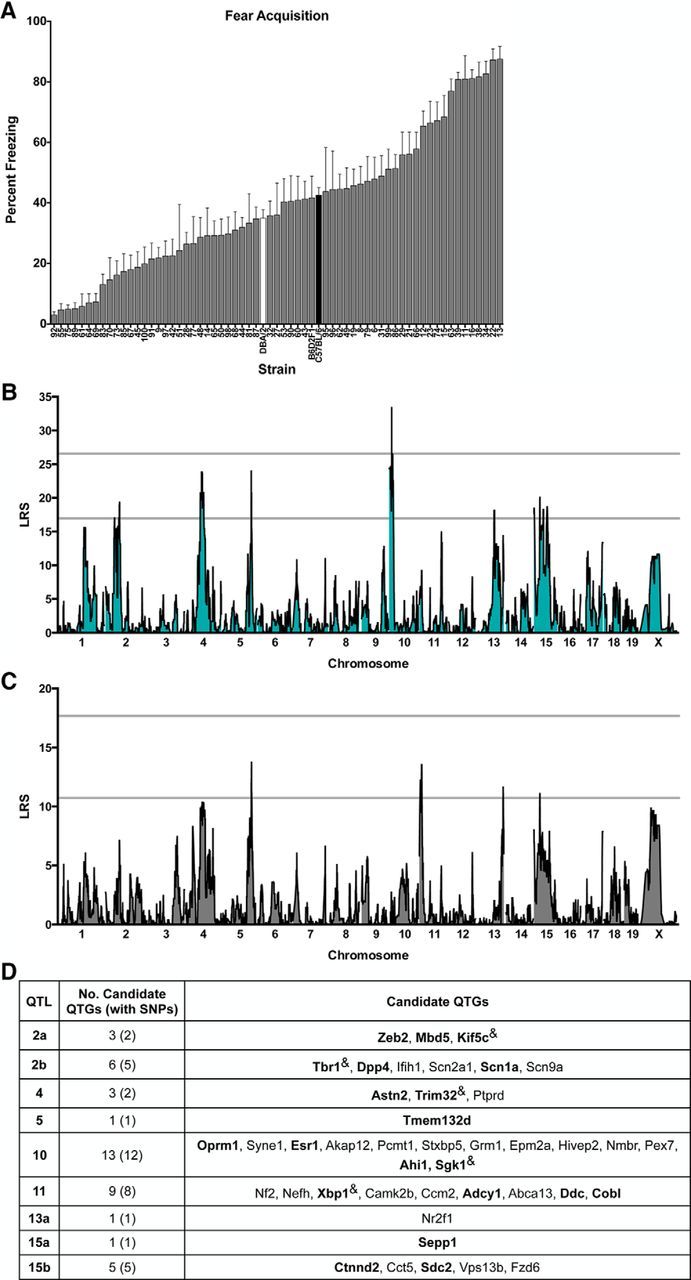
SIM and CIM to identify QTLs for conditioned fear acquisition in the BXD mouse GRP. A, BXD panel shows continuous variation in fear acquisition as assessed by the percentage of time strains spent freezing in the training context after the final pairing of the CS and US. Data are shown as means ± SEM. Parental strains are shown in black (B6) and white (D2). The F1 cross (B6D2F1) and 61 BXD strains are shown in gray. B, SIM reveals a significant QTL (LRS > 26.56; top gray line) on proximal Chr 10 (peak LRS at 14.12 Mb). There are also nine suggestive loci (LRS > 16.95; bottom gray line) located on Chr 2, 4, 5, 13, and 15. C, CIM controlling for QTL 10 using marker rs3686911 reveals an additional suggestive QTL on Chr 11 (peak LRS at 14.12 Mb). D, Candidate protein-coding genes listed in order of location on each on each chromosome and selected based on evidence of involvement in human neurological disorders or key phenotypes related to fear and anxiety, learning and memory, or anxiety and mood disorders. Bolded genes are of special interest based on meeting both criteria or evidence of involvement in conditioned fear or anxiety disorders. “&” indicates genes with no known single-nucleotide polymorphisms (SNPs) between the parental strains.
Figure 2.
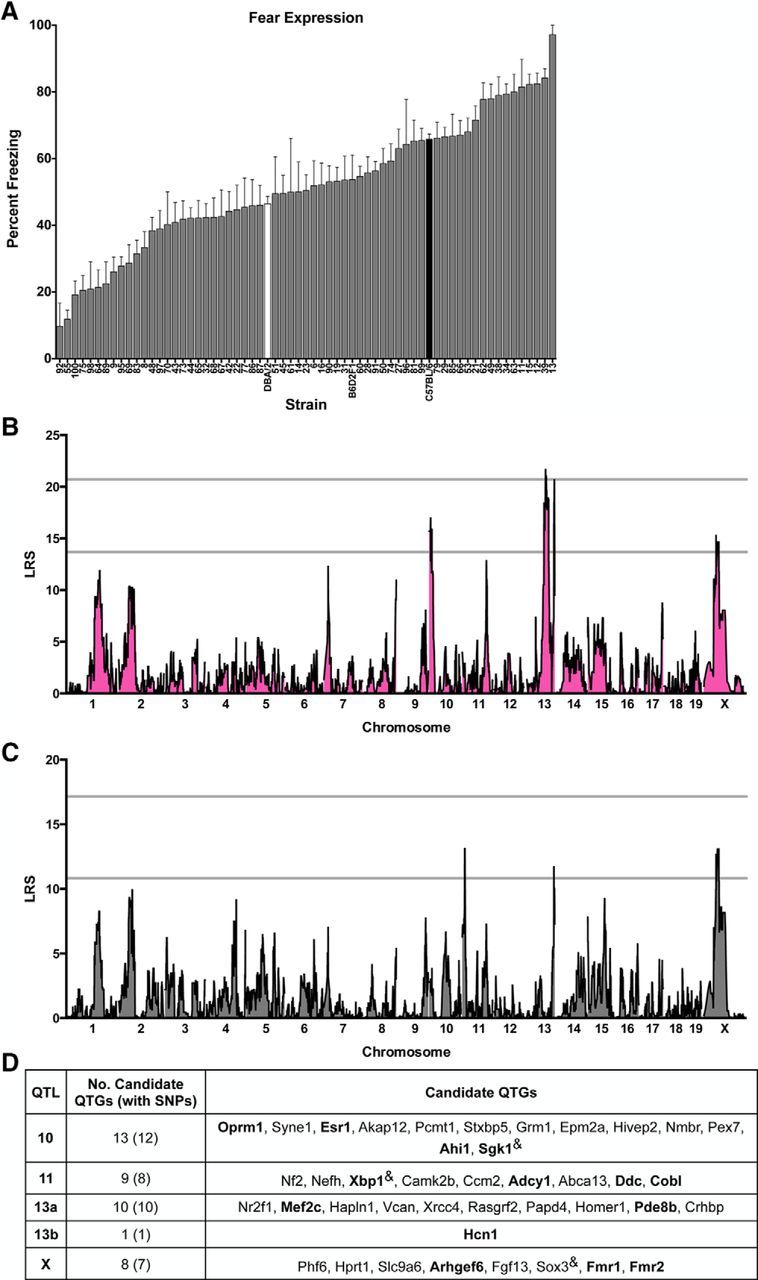
SIM and CIM to identify QTLs for conditioned fear expression in the BXD mouse GRP. A, BXD panel shows continuous variation in fear expression as assessed by the percentage of time strains spent freezing in response to the CS in the testing context. Data are shown as means ± SEM. Parental strains are shown in black (B6) and white (D2). The F1 cross (B6D2F1) and 62 BXD strains are shown in gray. B, SIM reveals a significant QTL (LRS > 21.70) on Chr 13 (peak LRS at 84.65 Mb), a highly suggestive QTL (LRS = 20.67) on distal Chr 13 (peak LRS at 118.39 Mb), and two suggestive loci (LRS > 13.68) on Chr 10 and Chr X. C, CIM controlling for QTL 13a using marker mCV24625340 reveals an additional suggestive QTL on Chr 11 (peak LRS at 13.57 Mb). D, Candidate protein-coding genes listed in order of location on each on each chromosome and selected based on evidence of involvement in human neurological disorders or key phenotypes related to fear and anxiety, learning and memory, or anxiety and mood disorders. Bolded genes are of special interest based on meeting both criteria or evidence of involvement in conditioned fear or anxiety disorders. “&” indicates genes with no known single-nucleotide polymorphisms (SNPs) between the parental strains.
QTL mapping
Using genome-wide mapping for fear acquisition, we identified a significant locus on proximal Chr 10 (14.1232 Mb, LRS = 33.49), which explains 30% of trait variation across strains, and nine suggestive loci located on Chr 2, 4, 5, 13, and 15 (Fig. 1B). For fear expression, we identified one significant locus on proximal Chr 13 (13a: 84.6480 Mb, LRS = 21.72) and another highly suggestive locus on distal Chr 13 (13b: 118.3938 Mb, LRS = 20.67), which independently explain 27% and 19% of trait variation across strains, respectively (Fig. 2B). There are also four suggestive loci located on Chr 10 and X for this trait (Fig. 2B). Interestingly, QTLs 10 and 13a are detected at the suggestive or significant level for both acquisition and expression, consistent with the hypothesis that shared or neighboring genes within these loci affect both traits. To further understand possible linkage between Chr 10 and Chr 13a, we conducted CIM of the acquisition and expression datasets using the markers with the highest LRS scores. For fear acquisition, controlling for QTL 10 using marker rs3686911 completely eliminated QTL 13a, revealed a new suggestive QTL on Chr 11 (peak LRS at 14.1232 Mb), and unmasked QTL 13b (Fig. 1C). In a similar manner, in the fear expression dataset, controlling for QTL 13a using marker mCV24625340 completely eliminated QTL 10 and revealed QTL 11 (same location as in fear acquisition; Fig. 2C). Therefore, CIM analysis for both datasets provides evidence of linkage between QTLs 10 and 13a and reveals a suggestive QTL on Chr 11. Pair-scan mapping of the fear acquisition and expression datasets did not provide evidence of epistatic interactions (data not shown).
Table 1 summarizes the location of the suggestive and significant QTLs for fear acquisition and fear expression. Analysis of linkage disequilibrium provides evidence of linkage of peaks on Chr 10, 15, and X. For clarity, in Table 1, we show the mapping results and gene information for each individual locus; linked loci are given the same QTL name (superscript indicates position) and unlinked loci on the same Chr are listed as QTL “a” and “b.” For fear acquisition, B6 alleles at QTL 10 have an additive effect size of 17.9% increased freezing. For fear expression, D2 alleles at QTL 13a have an additive effect size of 11.7% increased freezing and B6 alleles at QTL 13b have an additive effect size of 11.3%. All other loci, with the exception of QTL 5, had additive effects associated with B6 alleles. To centralize a resource of specific and related datasets, we include citations of previously published studies, as well as unpublished quantitative genetics findings with significant or suggestive loci within 10 Mb of the loci reported here. With the exception of QTLs 11 and X, all loci overlap with those previously reported as significant or suggestive in quantitative genetics studies of fear learning, anxiety, or depression-like behavior in rodents (Table 1). In particular, QTL 10 was identified for contextual fear (Parker et al., 2012) and spatial learning (Shea et al., 2015) and in unpublished datasets available on GN for contextual fear, anxiety, and passive avoidance. QTL 13a has been identified for cued fear acquisition and expression and contextual fear (Philip et al., 2010; Carhuatanta et al., 2014), anxiety (Philip et al., 2010) and amygdala and hippocampal morphology (Mozhui et al., 2007; Krebs et al., 2011). Some of these studies were performed in small numbers of RI strains. Our identification of significant QTLs 10 and 13a supports the findings of the previous work and highlights the broad consistency of mapping results, especially in similarly powered studies (Philip et al., 2010; Carhuatanta et al., 2014).
Table 1.
Quantitative trait locus locations and features for fear acquisition and expression
| Trait | QTL | Chr | QTL location (Mb) | 1.5 LOD interval (Mb) | Peak LRS score | Peak LRS location (Mb) | Peak markerc | No. of genesb | Genes with SNPs | Genes with human ortholog | Genotype, additive effect size | Previous reports of QTLd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fear acquisition | 2a | 2 | 43.54–44.06 | 41.30–50.50 | 16.99 | 43.5383 | rs13476464 | 11 | 9 | 11 | B6, 13.1 | Anxiety (Yang et al., 2008; Philip et al., 2010) |
| 2b | 2 | 62.00–65.50 | 57.64–68.79 | 19.47 | 65.1401 | D2Mit205 | 42 | 39 | 42 | B6, 14.0 | Contextual fear (Yang et al., 2008; Parker et al., 2014) | |
| 4 | 4 | 66.84–81.93 | 65.61–81.93 | 23.84 | 71.8013 | rs6222684 | 15 | 14 | 14 | B6, 15.3 | Contextual fear (Brigman et al., 2009) | |
| 5 | 5 | 127.04–128.05 | 126.62–128.27 | 24.08 | 127.0399 | gnf05.120.578 | 4 | 4 | 4 | D2, 15.3 | Contextual fear (Parker et al., 2012), Cued fear acquisition (Carhuatanta et al., 2014); Anxiety (Yang et al., 2008; Cook, Unpub, 2009) | |
| 10 | 10 | 3.34–21.83 | 3.13–21.83 | 33.49** | 14.1232 | rs3686911 | 71 | 66 | 71 | B6, 17.9 | Anxiety (Cook, Unpub, 2009), passive avoidance (Paylor, Unpub, 2011), contextual fear (Parker et al., 2012; Liao, Unpub, 2013), spatial learning (Shea et al., 2015) | |
| 11 | 11 | 8.06–14.90 | 4.41–16.85 | 13.55a | 13.5740 | rs3673722 | 62 | 40 | 62 | B6, 13.5 | Neocortex volume (Beatty and Laughlin, 2006) | |
| 13a | 13 | 78.42–80.88 | 78.26–81.06 | 18.16 | 78.4146 | rs13481896 | 2 | 1 | 2 | D2, 13.5 | Anxiety (Cook, Unpub, 2009; Philip et al., 2010), contextual fear and cued fear acquisition and expression (Philip et al., 2010; Carhuatanta et al., 2014), hippocampus morphology (Krebs et al., 2011), amygdala morphology (Mozhui et al., 2007) | |
| 15a | 15 | 3.23–4.35 | 3.22–7.27 | 18.44 | 3.9279 | rs13459177 | 18 | 12 | 18 | B6, 13.6 | Depression assay (Lad et al., 2007) | |
| 15b1 | 15 | 27.87–28.88 | 27.29–44.21 | 20.07 | 28.2515 | rs13482497 | 24 | 19 | 24 | B6, 14.5 | Anxiety (Philip et al., 2010) | |
| 15b2 | 15 | 42.12–44.18 | 27.29–44.21 | 18.26 | 42.1234 | rs6339552 | 40 | 28 | 39 | B6, 13.6 | See linked loci 15b1 and 15b3 | |
| 15b3 | 15 | 57.99–60.99 | 55.00–63.15 | 18.77 | 58.9910 | rs3702158c | 28 | 14 | 27 | B6, 15.2 | Anxiety (Melloni, Unpub, 2009; Philip et al., 2010) | |
| Fear expression | 101 | 10 | 3.34–9.13 | 3.13–19.10 | 17.03 | 9.1332 | D10Mit28 | 33 | 30 | 33 | B6, 10.5 | See QTL 10 (above) |
| 102 | 10 | 14.122–14.123 | 3.13–19.10 | 15.95 | 14.1225 | rs3686911 | 24 | 24 | 24 | B6, 10.3 | See QTL 10 (above) | |
| 11 | 11 | 13.57–14.90 | 4.41–16.48 | 13.11a | 13.5740 | rs3673722 | 61 | 39 | 61 | B6, 12.0 | See QTL 11 (above) | |
| 13a | 13 | 78.42–96.28 | 78.26–96.47 | 21.72* | 84.6480 | mCV24625340c | 54 | 41 | 54 | D2, 11.7 | See QTL 13a (above) | |
| 13b | 13 | 116.72–119.53 | 116.58–119.97 | 20.67∧ | 118.3938 | rs13482033 | 6 | 5 | 6 | B6, 11.3 | Depression assay (Chen, Unpub, 2013) | |
| X1 | X | 56.49 | 47.53–68.69 | 15.37 | 56.4887 | rs13483770 | 37 | 28 | 33 | B6, 10.2 | See QTL X2 below | |
| X2 | X | 61.60–66.92 | 47.53–68.69 | 14.65 | 61.5953 | rs13483785 | 15 | 9 | 14 | B6, 9.8 | Hippocampus morphology (Lassalle et al., 1994) |
**Highly significant LRS (p < 0.01);
*significant LRS (p < 0.05);
∧highly suggestive LRS (p < 0.068); all other loci are suggestive (p < 0.63). Fear acquisition: suggestive LRS > 16.95, significant LRS > 26.56; fear expression: suggestive LRS > 13.68, significant LRS > 21.70.
aCIM analysis LRS. Fear acquisition: suggestive LRS >10.82, significant LRS > 17.33; Fear expression: suggestive LRS > 10.74, significant LRS > 16.85.
bLoci within Chr 10, 15 and X are linked and show overlapping drop support intervals. Candidate genes were selected across the entire drop support interval and separated by approximate peak locations: 101–3.13-12.0 Mb, 102–12.1-19.10 Mb, 15b1–27.29-35.0 Mb, 15b2–35.1-44.18 Mb, X1–47.53-58.0 Mb and X2–58.1-68.69 Mb.
cNo marker available for peak LRS location; closest marker is listed. rs3702158 is located at 56.9920 Mb, mCV24625340 is located at 84.8411 Mb.
dPrior QTLs reported in Pubmed or GeneNetwork for traits related to fear and anxiety, learning and memory, stress, or emotional regulation. Reports were required to have a suggestive or significant LRS score within 10 Mb of the present QTLs and to be from datasets with at least 15 strains. “Unpub” refers to specific investigators depositing unpublished datasets in GeneNetwork (www.genenetwork.org).
QTG identification
We generated candidate gene lists from the 1.5 LOD support intervals and narrowed our analysis to protein encoding genes. There are 317 protein-coding genes in the QTLs identified for fear acquisition and 230 in the QTLs identified for fear expression, with 120 genes present in shared QTLs 10, 11, and 13a, for a total of 427 protein-encoding genes. Of these 427 genes, 76.8% (328) have SNPs between the parental strains and 98.1% (419) have a human ortholog. We selected a prioritized list of 60 candidate genes based on their involvement in a human neurological disorder (38 genes) or prioritized brain-based phenotypes (41 genes)—with 19 genes meeting both criteria (bolded in Figs. 1D, 2D). As described above, the most significant and recurring loci for fear acquisition and expression are QTLs 10, 13a, and 13b. QTL 10 contained 13 genes that met one or both criteria, several of which have been implicated previously in conditioned fear (Oprm1, Good and Westbrook, 1995; Sanders et al., 2005; Sgk1, Lee et al., 2007; Kim and Richardson, 2009), mood and anxiety disorders (Oprm1, Liberzon et al., 2007; Esr1, Sundermann et al., 2010; Ryan et al., 2011; Sgk1, Licznerski et al., 2015), intellectual disability (Hivep2, Takagi et al., 2015), and learning and memory (Nmbr, Yamada et al., 2003; Sgk1, Tyan et al., 2008; Roesler et al., 2012; Hivep2, Takao et al., 2013). QTL 13a contained 11 genes that met one or both criteria, several of which have been implicated previously in conditioned fear (Mef2c, Cole et al., 2012; Pde8b, Tsai et al., 2012), learning and memory (Mef2c, Barbosa et al., 2008; Zweier et al., 2010; Homer1, Wagner et al., 2014), and stress response and anxiety disorder risk (Crhbp, Westphal and Seasholtz, 2006; Enoch et al., 2008). These various QTGs represent interesting candidates for further study.
Nomination and characterization of Hcn1 in fear
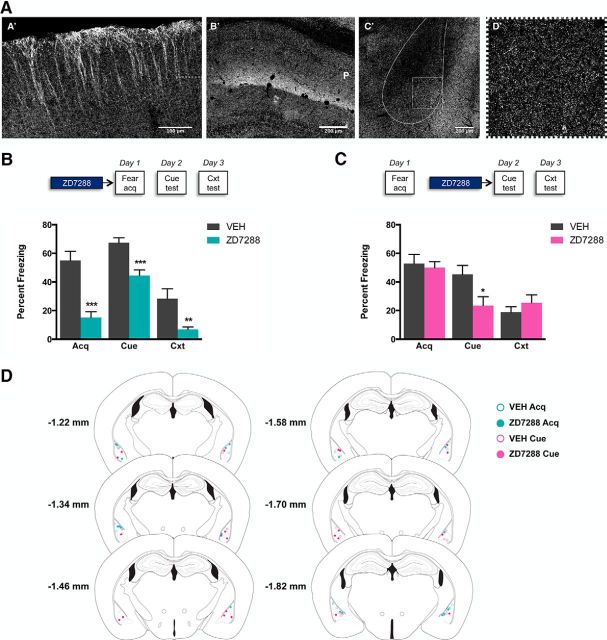
Notably, we found that one of the two major fear expression QTLs (13b) is a gene-sparse region that harbors a single candidate QTG, hyperpolarization-activated cyclic nucleotide-gated nonselective cation channel subunit 1 (Hcn1), located at the peak LRS score. Hcn1 is one of four subunits that make up Hcn channels. These channels are present at high levels in the brain and have a well established role in regulating resting membrane potential, neuronal excitability, and synaptic plasticity (He et al., 2014; Shah, 2014) and are implicated in various forms of learning and memory (Nolan et al., 2003, 2004; Wang et al., 2007). Interestingly, recent studies have shown that Hcn channels increase the excitability of pyramidal neurons in the principal fear-mediating region of the brain, the BLA (Giesbrecht et al., 2010; but see Park et al., 2011). However, whereas one prior study found that forebrain-wide deletion of Hcn1 in a hybrid B6x129SvEv background impaired spatial memory but not conditioned fear (Nolan et al., 2004), the specific role of Hcn1 in the BLA is currently unknown. Therefore, using immunocytochemistry, we first confirmed earlier reports that Hcn1 has moderate to high expression in putative pyramidal neurons in the mouse BLA, as well as the cortex and hippocampus (Fig. 3A; Moosmang et al., 1999; Notomi and Shigemoto, 2004). We then infused the Hcn channel antagonist ZD7288 into the BLA (Fig. 3C) of B6 mice before conditioning or cued-fear testing. ZD7288 infusion before fear conditioning significantly reduced freezing relative to vehicle-infused controls during the final CS presentation during training (t(16) = 5.22, p = 0.000084; Fig. 3B) and during the tests for cued (t(16) = 4.36, p = 0.00049) and contextual (t(16) = 2.98, p = 0.0087) fear expression the following 2 d (Fig. 3B). ZD7288 infusion before cued-fear testing significantly reduced freezing during the cued-fear test (t(17) = 2.49, p = 0.023), but did not affect freezing during the contextual fear test (t(17) = 0.94, p = 0.36) (Fig. 3C). These results demonstrate a novel and important functional role for Hcn channels expressed in the BLA in both fear acquisition and cued-fear expression.
Figure 3.
A, Immunohistochemical staining for Hcn1 reveals dense localization in distal cortical dendrites (A′; dashed line marks separation between layer I/II and layer III) and hippocampal CA1 stratum lacunosum moleculare (B′; P marks the location of the pyramidal cell layer). Hcn1 immunolocalization is evident in the BLA (C′; outlined), with higher magnification of the expected punctate labeling in the BLA (D′; boxed). B, Behavioral pharmacology to examine the role of Hcn channels in the BLA on cued and contextual fear acquisition. Top, Drug administration time course. Bottom, Microinjection of the Hcn channel antagonist ZD7288 (0.3 μg/0.5 μl/side) into the BLA 30 min before cued-fear training impairs fear acquisition in Context A (Acq), cued fear expression 24 h later in Context B (Cue), and contextual fear expression 48 h later in Context A (Cxt). C, Behavioral pharmacology to examine the role of Hcn channels in the BLA on cued and contextual fear expression. Top, Drug administration time course. Bottom, Microinjection of the Hcn channel antagonist ZD7288 (0.3 μg/0.5 μl/side) into the BLA 30 min before cued-fear testing impairs cued fear expression in Context B (Cue), but does not affect contextual fear expression 48 h later in Context A (Cxt). Data are shown as mean ± SEM. n = 9–10/group. D, Summary of cannula tip placements in the BLA in mice treated with vehicle (VEH) or Hcn channel antagonist (ZD7288) (illustration of coronal brain sections from Paxinos and Watson, 1996).
Discussion
Using a collaborative quantitative genetics approach, we identified multiple QTLs for conditioned fear behavior in mice. Within these loci, we identified 60 candidate QTGs of high biological rationale based on publications indicating their involvement in human neurological disorders or key phenotypes, including conditioned fear, learning and memory, and anxiety disorders. QTL 13b was particularly noteworthy because it contained a single candidate QTG, Hcn1, previously implicated in learning (Nolan et al., 2003, 2004; Wang et al., 2007) and anxiety- and depression-related behaviors in rodents (Giesbrecht et al., 2010; Park et al., 2011; Kim et al., 2012; He et al., 2014). Validating the QTL-mapping data, we found that Hcn1 was richly expressed in the BLA and that pharmacologic blockade of Hcn channels in this brain region impaired fear acquisition and cued-fear expression. Together, these data establish Hcn1 as a novel QTG for learned fear and, more generally, support the utility of quantitative genetics for identifying QTLs and QTGs for disorder-relevant intermediate phenotypes.
Conditioned fear is a polygenic trait and simple and composite interval analyses revealed complex linkage across loci and genotypes. In particular, QTLs 10 (B6 additive effect) and 13a (D2 additive effect) show substantial linkage and were detected at the suggestive or significant level for both fear acquisition and expression using SIM. Similarly, the Hcn1 locus (QTL 13b; B6 additive effect) was detected at the suggestive level after CIM for fear acquisition and SIM for fear expression. Overlapping yet distinct molecular mechanisms support fear acquisition, consolidation, and expression (Johansen et al., 2011). Therefore, the identification of overlapping loci suggests either shared genetic regulation of these processes or the influence of unique genes that are in close physical proximity or a combination of the two. RI mice and parental strains provide a genetically and behaviorally tractable system for future studies aimed at parsing these possibilities.
We mapped a number of promising candidate QTGs implicated previously in conditioned fear, learning and memory, stress, or human neurological disorders. To our knowledge, QTL 13b containing Hcn1 has not been reported previously as being related to fear. How might genetic variation in Hcn1 affect fear learning? The Hcn1 gene is large (378 kb, 8 exons) and contains >3500 intronic SNPs and INDELs of unknown biological influence and few SNPs in coding regions. Examination of microarray datasets in GN did not reveal compelling correlations between fear phenotypes and regional Hcn1 gene expression, albeit in a smaller (and potentially underpowered) set of strains than used in our current QTL analysis. However, because Hcn1 modulates neuronal excitability as a function of its differentially localized distribution on dendritic or somal regions (Magee, 1998; Lörincz et al., 2002; Lewis et al., 2011; Kim et al., 2012), measures of gene expression may reflect poorly what could be robust differences in Hcn1 function. More precisely delineated protein expression/distribution analyses would be valuable in determining whether intronic or coding variants affect the stability and/or trafficking of Hcn1 channels.
Nevertheless, there is a biologically compelling rationale for Hcn1 involvement in this behavior. Hcn1 channels are expressed in brain regions that are critically involved in fear learning, including the amygdala, PFC, and hippocampus, where they have an important role is establishing the resting membrane potential (Moosmang et al., 1999; Notomi and Shigemoto, 2004; Shah, 2014). Moreover, Hcn channels have profound effects on neuronal excitability and synaptic plasticity, which depend upon presynaptic and somatodendritic trafficking (Lörincz et al., 2002; Huang et al., 2011), the complement of other ion channels (George et al., 2009), and interactions with cAMP and the auxiliary subunit TRIP8b (Huang et al., 2011; Hu et al., 2013; Shah, 2014). Last, previous work shows that Hcn1 channels modulate spatial learning and working memory (Nolan et al., 2003, 2004; Wang et al., 2007; Maroso et al., 2016), as well as anxiety-, depression-, and stress-related behaviors in rodents (Giesbrecht et al., 2010; Park et al., 2011; Kim et al., 2012). One prior study found a subtle abnormality in eye-blink conditioning but normal cued and contextual fear (freezing) in mutant mice on a mixed 129xB6 background with forebrain-wide Hcn1 gene deletion (Nolan et al., 2003, 2004). The reason for the apparent discrepancy with our QTL and pharmacological data is not clear, but there are a number of plausible explanations. First, given the importance of Hcn channels in setting resting membrane potential, genetic deletion of Hcn1 could induce strong compensatory effects that mask the channel's normal function. Second, the Cre driver line used to delete Hcn1 may have incompletely deleted the gene in the BLA or Hcn1 deletion at multiple regions within the wider fear circuitry may have produced different effects to loss of function in the BLA alone. In lieu of studies to test for these various possibilities, based on prior and current findings, we posit a role for Hcn1 in modulating neuronal and synaptic processes in the BLA necessary for the formation and expression of fear memories. This could have implications for the treatment of anxiety-, stress-, and learning-related disorders (McIntosh et al., 2012; Bukalo et al., 2014; Omrani et al., 2015). For example, selective pharmacologic agents targeting Hcn channels are currently in use to improve cardiac function and brain-specific compounds are under development (Han et al., 2015; Sartiani et al., 2015).
The collaborative and open access nature of GRPs thus provides exceptional opportunities for quantitative genetic analysis of intermediate phenotypes in animal models. Conditioned fear is a complex polygenic trait and the functional testing in a parental strain performed here provides a tractable simplifying model in which the genotype of all relevant genes is held constant. With both replication of previously reported QTLs and the discovery of new ones, a goal for future studies in the field is to identify the QTGs within the other loci and to understand epistatic mechanisms by which the function of individual QTGs is revealed or masked.
Footnotes
This work was supported by the Simms/Mann Chair in Developmental Neurogenetics and National Institute of Mental Health, National Institutes of Health (Grant R01 MH080759 to P.L.), the National Institute on Alcohol Abuse and Alcoholism Intramural Program (A.J.H.), and Autism Speaks (Translational Postdoctoral Fellowship 7595 to A.T.K.). We thank Dr. Megan Mulligan and Dr. Robert Williams for helpful comments on this manuscript.
The authors declare no competing financial interests.
References
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty J, Laughlin RE. Genomic regulation of natural variation in cortical and noncortical brain volume. BMC Neurosci. 2006;7:16. doi: 10.1186/1471-2202-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers ME, Ressler KJ. An overview of translationally informed treatments for posttraumatic stress disorder: animal models of pavlovian fear conditioning to human clinical trials. Biol Psychiatry. 2015;78:E15–E27. doi: 10.1016/j.biopsych.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigman JL, Mathur P, Lu L, Williams RW, Holmes A. Genetic relationship between anxiety-related and fear-related behaviors in BXD recombinant inbred mice. Behav Pharmacol. 2009;20:204–209. doi: 10.1097/FBP.0b013e32830c368c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukalo O, Pinard CR, Holmes A. Mechanisms to medicines: elucidating neural and molecular substrates of fear extinction to identify novel treatments for anxiety disorders. Br J Pharmacol. 2014;171:4690–4718. doi: 10.1111/bph.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carhuatanta KA, Shea CJ, Herman JP, Jankord R. Unique genetic loci identified for emotional behavior in control and chronic stress conditions. Front Behav Neurosci. 2014;8:341. doi: 10.3389/fnbeh.2014.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Craddock N, Cuthbert BN, Hyman SE, Lee FS, Ressler KJ. DSM-5 and RDoC: progress in psychiatry research? Nat Rev Neurosci. 2013;14:810–814. doi: 10.1038/nrn3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevaleyre V, Castillo PE. Assessing the role of Ih channels in synaptic transmission and mossy fiber LTP. Proc Natl Acad Sci U S A. 2002;99:9538–9543. doi: 10.1073/pnas.142213199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, Vetere G, Pekar T, Ross PJ, Neve RL, Frankland PW, Josselyn SA. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat Neurosci. 2012;15:1255–1264. doi: 10.1038/nn.3189. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, Neale BM, et al. International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Ducci F, Yuan Q, Liu J, White KV, Albaugh B, Hodgkinson CA, Goldman D. Common genetic origins for EEG, alcoholism and anxiety: the role of CRH-BP. PLoS One. 2008;3:e3620. doi: 10.1371/journal.pone.0003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt JM, Burton KL, Williams LM, Schofield PR. Specific and common genes implicated across major mental disorders: a review of meta-analysis studies. J Psychiatr Res. 2015;60:1–13. doi: 10.1016/j.jpsychires.2014.09.014. [DOI] [PubMed] [Google Scholar]
- George MS, Abbott LF, Siegelbaum SA. HCN hyperpolarization-activated cation channels inhibit EPSPs by interactions with M-type K(+) channels. Nat Neurosci. 2009;12:577–584. doi: 10.1038/nn.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbrecht CJ, Mackay JP, Silveira HB, Urban JH, Colmers WF. Countervailing modulation of Ih by neuropeptide Y and corticotrophin-releasing factor in basolateral amygdala as a possible mechanism for their effects on stress-related behaviors. J Neurosci. 2010;30:16970–16982. doi: 10.1523/JNEUROSCI.2306-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AJ, Westbrook RF. Effects of a microinjection of morphine into the amygdala on the acquisition and expression of conditioned fear and hypoalgesia in rats. Behav Neurosci. 1995;109:631–641. doi: 10.1037/0735-7044.109.4.631. [DOI] [PubMed] [Google Scholar]
- Han Y, Lyman K, Clutter M, Schiltz GE, Ismail QA, Prados DB, Luan CH, Chetkovich DM. Identification of small-molecule inhibitors of hyperpolarization-activated cyclic nucleotide-gated channels. J Biomol Screen. 2015;20:1124–1131. doi: 10.1177/1087057115589590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Holmes A. Finding translation in stress research. Nat Neurosci. 2015;18:1347–1352. doi: 10.1038/nn.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Chen F, Li B, Hu Z. Neurophysiology of HCN channels: from cellular functions to multiple regulations. Prog Neurobiol. 2014;112:1–23. doi: 10.1016/j.pneurobio.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Holmes A, Singewald N. Individual differences in recovery from traumatic fear. Trends Neurosci. 2013;36:23–31. doi: 10.1016/j.tins.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Santoro B, Saponaro A, Liu H, Moroni A, Siegelbaum S. Binding of the auxiliary subunit TRIP8b to HCN channels shifts the mode of action of cAMP. J Gen Physiol. 2013;142:599–612. doi: 10.1085/jgp.201311013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Lujan R, Kadurin I, Uebele VN, Renger JJ, Dolphin AC, Shah MM. Presynaptic HCN1 channels regulate Cav3.2 activity and neurotransmission at select cortical synapses. Nat Neurosci. 2011;14:478–486. doi: 10.1038/nn.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen JP, Cain CK, Ostroff LE, LeDoux JE. Molecular mechanisms of fear learning and memory. Cell. 2011;147:509–524. doi: 10.1016/j.cell.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas MJ, Fernandes C, Schalkwyk LC, Collier DA. Genetics of behavioural domains across the neuropsychiatric spectrum; of mice and men. Mol Psychiatry. 2007;12:324–330. doi: 10.1038/sj.mp.4001979. [DOI] [PubMed] [Google Scholar]
- Kim CS, Chang PY, Johnston D. Enhancement of dorsal hippocampal activity by knockdown of HCN1 channels leads to anxiolytic- and antidepressant-like behaviors. Neuron. 2012;75:503–516. doi: 10.1016/j.neuron.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richardson R. The effect of the mu-opioid receptor antagonist naloxone on extinction of conditioned fear in the developing rat. Learn Mem. 2009;16:161–166. doi: 10.1101/lm.1282309. [DOI] [PubMed] [Google Scholar]
- Kocsis B, Li S. In vivo contribution of h-channels in the septal pacemaker to theta rhythm generation. Eur J Neurosci. 2004;20:2149–2158. doi: 10.1111/j.1460-9568.2004.03678.x. [DOI] [PubMed] [Google Scholar]
- Krebs J, Römer B, Overall RW, Fabel K, Babu H, Brandt MD, Williams RW, Jessberger S, Kempermann G. Adult hippocampal neurogenesis and plasticity in the infrapyramidal bundle of the mossy fiber projection: II. Genetic covariation and identification of Nos1 as linking candidate gene. Front Neurosci. 2011;5:106. doi: 10.3389/fnins.2011.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad HV, Liu L, Payá-Cano JL, Fernandes C, Schalkwyk LC. Quantitative traits for the tail suspension test: automation, optimization, and BXD RI mapping. Mamm Genome. 2007;18:482–491. doi: 10.1007/s00335-007-9029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lassalle JM, Halley H, Roullet P. Analysis of behavioral and hippocampal variation in congenic albino and pigmented BALB mice. Behav Genet. 1994;24:161–169. doi: 10.1007/BF01067820. [DOI] [PubMed] [Google Scholar]
- Lee CT, Ma YL, Lee EH. Serum- and glucocorticoid-inducible kinase1 enhances contextual fear memory formation through down-regulation of the expression of Hes5. J Neurochem. 2007;100:1531–1542. doi: 10.1111/j.1471-4159.2006.04284.x. [DOI] [PubMed] [Google Scholar]
- Lewis AS, Vaidya SP, Blaiss CA, Liu Z, Stoub TR, Brager DH, Chen X, Bender RA, Estep CM, Popov AB, Kang CE, Van Veldhoven PP, Bayliss DA, Nicholson DA, Powell CM, Johnston D, Chetkovich DM. Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. J Neurosci. 2011;31:7424–7440. doi: 10.1523/JNEUROSCI.0936-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon I, Taylor SF, Phan KL, Britton JC, Fig LM, Bueller JA, Koeppe RA, Zubieta JK. Altered central micro-opioid receptor binding after psychological trauma. Biol Psychiatry. 2007;61:1030–1038. doi: 10.1016/j.biopsych.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Licznerski P, Duric V, Banasr M, Alavian KN, Ota KT, Kang HJ, Jonas EA, Ursano R, Krystal JH, Duman RS, Traumatic Stress Brain Study Group Decreased SGK1 expression and function contributes to behavioral deficits induced by traumatic stress. PLoS Biol. 2015;13:e1002282. doi: 10.1371/journal.pbio.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Amstadter AB, Baker DG, Duncan L, Koenen KC, Liberzon I, Miller MW, Morey RA, Nievergelt CM, Ressler KJ, Smith AK, Smoller JW, Stein MB, Sumner JA, Uddin M. The Psychiatric Genomics Consortium Posttraumatic Stress Disorder Workgroup: posttraumatic stress disorder enters the age of large-scale genomic collaboration. Neuropsychopharmacology. 2015;40:2287–2297. doi: 10.1038/npp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lörincz A, Notomi T, Tamás G, Shigemoto R, Nusser Z. Polarized and compartment-dependent distribution of HCN1 in pyramidal cell dendrites. Nat Neurosci. 2002;5:1185–1193. doi: 10.1038/nn962. [DOI] [PubMed] [Google Scholar]
- Magee JC. Dendritic hyperpolarization-activated currents modify the integrative properties of hippocampal CA1 pyramidal neurons. J Neurosci. 1998;18:7613–7624. doi: 10.1523/JNEUROSCI.18-19-07613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S, Wray NR, Lewis CM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroso M, Szabo GG, Kim HK, Alexander A, Bui AD, Lee SH, Lutz B, Soltesz I. Cannabinoid control of learning and memory through HCN channels. Neuron. 2016;89:1059–1073. doi: 10.1016/j.neuron.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AM, Simen AA, Evans KL, Hall J, Macintyre DJ, Blackwood D, Morris AD, Smith BH, Dominiczak A, Porteous D, Deary HI, Thomson PA. Genetic variation in hyperpolarization-activated cyclic nucleotide-gated channels and its relationship with neuroticism, cognition and risk of depression. Front Genet. 2012;3:116. doi: 10.3389/fgene.2012.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmang S, Biel M, Hofmann F, Ludwig A. Differential distribution of four hyperpolarization-activated cation channels in mouse brain. Biol Chem. 1999;380:975–980. doi: 10.1515/BC.1999.121. [DOI] [PubMed] [Google Scholar]
- Mozhui K, Hamre KM, Holmes A, Lu L, Williams RW. Genetic and structural analysis of the basolateral amygdala complex in BXD recombinant inbred mice. Behav Genet. 2007;37:223–243. doi: 10.1007/s10519-006-9122-3. [DOI] [PubMed] [Google Scholar]
- Nolan MF, Malleret G, Lee KH, Gibbs E, Dudman JT, Santoro B, Yin D, Thompson RF, Siegelbaum SA, Kandel ER, Morozov A. The hyperpolarization-activated HCN1 channel is important for motor learning and neuronal integration by cerebellar Purkinje cells. Cell. 2003;115:551–564. doi: 10.1016/S0092-8674(03)00884-5. [DOI] [PubMed] [Google Scholar]
- Nolan MF, Malleret G, Dudman JT, Buhl DL, Santoro B, Gibbs E, Vronskaya S, Buzsáki G, Siegelbaum SA, Kandel ER, Morozov A. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119:719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Norrholm SD, Glover EM, Stevens JS, Fani N, Galatzer-Levy IR, Bradley B, Ressler KJ, Jovanovic T. Fear load: the psychophysiological over-expression of fear as an intermediate phenotype associated with trauma reactions. Int J Psychophysiol. 2014 doi: 10.1016/j.ijpsycho.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Shigemoto R. Immunohistochemical localization of Ih channel subunits, HCN1–4, in the rat brain. J Comp Neurol. 2004;471:241–276. doi: 10.1002/cne.11039. [DOI] [PubMed] [Google Scholar]
- Omrani A, van der Vaart T, Mientjes E, van Woerden GM, Hojjati MR, Li KW, Gutmann DH, Levelt CN, Smit AB, Silva AJ, Kushner SA, Elgersma Y. HCN channels are a novel therapeutic target for cognitive dysfunction in Neurofibromatosis type 1. Mol Psychiatry. 2015;20:1311–1321. doi: 10.1038/mp.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall RW, Williams RW, Heimel JA. Collaborative mining of public data resources in neuroinformatics. Front Neurosci. 2015;9:90. doi: 10.3389/fnins.2015.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Yi JH, Kim H, Choi K, Kang SJ, Shin KS. HCN channel activity-dependent modulation of inhibitory synaptic transmission in the rat basolateral amygdala. Biochem Biophys Res Commun. 2011;404:952–957. doi: 10.1016/j.bbrc.2010.12.087. [DOI] [PubMed] [Google Scholar]
- Parker CC, Sokoloff G, Cheng R, Palmer AA. Genome-wide association for fear conditioning in an advanced intercross mouse line. Behav Genet. 2012a;42:437–448. doi: 10.1007/s10519-011-9524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Carbonetto P, Sokoloff G, Park YJ, Abney M, Palmer AA. High-resolution genetic mapping of complex traits from a combined analysis of F2 and advanced intercross mice. Genetics. 2014;198:103–116. doi: 10.1534/genetics.114.167056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Ed 3. New York: Academic; 1996. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–878. doi: 10.1038/ni.1747. [DOI] [PubMed] [Google Scholar]
- Roesler R, Kent P, Schroder N, Schwartsmann G, Merali Z. Bombesin receptor regulation of emotional memory. Rev Neurosci. 2012;23:571–586. doi: 10.1515/revneuro-2012-0046. [DOI] [PubMed] [Google Scholar]
- Ryan J, Scali J, Carrière I, Scarabin PY, Ritchie K, Ancelin ML. Estrogen receptor gene variants are associated with anxiety disorders in older women. Psychoneuroendocrinology. 2011;36:1582–1586. doi: 10.1016/j.psyneuen.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Sanders MJ, Kieffer BL, Fanselow MS. Deletion of the mu opioid receptor results in impaired acquisition of Pavlovian context fear. Neurobiol Learn Mem. 2005;84:33–41. doi: 10.1016/j.nlm.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Sartiani L, Romanelli MN, Mugelli A, Cerbai E. Updates on HCN channels in the heart: function, dysfunction and pharmacology. Curr Drug Targets. 2015;16:868–876. doi: 10.2174/1389450116666150531152047. [DOI] [PubMed] [Google Scholar]
- Shah MM. Cortical HCN channels: function, trafficking and plasticity. J Physiol. 2014;592:2711–2719. doi: 10.1113/jphysiol.2013.270058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea CJ, Carhuatanta KA, Wagner J, Bechmann N, Moore R, Herman JP, Jankord R. Variable impact of chronic stress on spatial learning and memory in BXD mice. Physiol Behav. 2015;150:69–77. doi: 10.1016/j.physbeh.2015.06.022. [DOI] [PubMed] [Google Scholar]
- Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW. The genetics of stress-related disorders: PTSD, depression and anxiety disorders. Neuropsychopharmacology. 2016;41:297–319. doi: 10.1038/npp.2015.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermann EE, Maki PM, Bishop JR. A review of estrogen receptor alpha gene (ESR1) polymorphisms, mood, and cognition. Menopause. 2010;17:874–886. doi: 10.1097/gme.0b013e3181df4a19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Nishizaki Y, Matsui F, Wakamatsu N, Higashi Y. De novo inbred heterozygous Zeb2/Sip1 mutant mice uniquely generated by germ-line conditional knockout exhibit craniofacial, callosal and behavioral defects associated with Mowat-Wilson syndrome. Hum Mol Genet. 2015;24:6390–6402. doi: 10.1093/hmg/ddv350. [DOI] [PubMed] [Google Scholar]
- Takao K, Kobayashi K, Hagihara H, et al. Deficiency of schnurri-2, an MHC enhancer binding protein, induces mild chronic inflammation in the brain and confers molecular, neuronal, and behavioral phenotypes related to schizophrenia. Neuropsychopharmacology. 2013;38:1409–1425. doi: 10.1038/npp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LC, Chan GC, Nangle SN, Shimizu-Albergine M, Jones GL, Storm DR, Beavo JA, Zweifel LS. Inactivation of Pde8b enhances memory, motor performance, and protects against age-induced motor coordination decay. Genes Brain Behav. 2012;11:837–847. doi: 10.1111/j.1601-183X.2012.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyan SW, Tsai MC, Lin CL, Ma YL, Lee EH. Serum- and glucocorticoid-inducible kinase 1 enhances zif268 expression through the mediation of SRF and CREB1 associated with spatial memory formation. J Neurochem. 2008;105:820–832. doi: 10.1111/j.1471-4159.2007.05186.x. [DOI] [PubMed] [Google Scholar]
- Wagner KV, Häusl AS, Pöhlmann ML, Hartmann J, Labermaier C, Müller MB, Schmidt MV. Hippocampal Homer1 levels influence motivational behavior in an operant conditioning task. PLoS One. 2014;9:e85975. doi: 10.1371/journal.pone.0085975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Ramos BP, Paspalas CD, Shu Y, Simen A, Duque A, Vijayraghavan S, Brennan A, Dudley A, Nou E, Mazer JA, McCormick DA, Arnsten AF. Alpha2A-adrenoceptors strengthen working memory networks by inhibiting cAMP-HCN channel signaling in prefrontal cortex. Cell. 2007;129:397–410. doi: 10.1016/j.cell.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Westphal NJ, Seasholtz AF. CRH-BP: the regulation and function of a phylogenetically conserved binding protein. Front Biosci. 2006;11:1878–1891. doi: 10.2741/1931. [DOI] [PubMed] [Google Scholar]
- Wilker S, Elbert T, Kolassa IT. The downside of strong emotional memories: how human memory-related genes influence the risk for posttraumatic stress disorder–a selective review. Neurobiol Learn Mem. 2014;112:75–86. doi: 10.1016/j.nlm.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Yamada K, Santo-Yamada Y, Wada K. Stress-induced impairment of inhibitory avoidance learning in female neuromedin B receptor-deficient mice. Physiol Behav. 2003;78:303–309. doi: 10.1016/S0031-9384(02)00979-4. [DOI] [PubMed] [Google Scholar]
- Yang RJ, Mozhui K, Karlsson RM, Cameron HA, Williams RW, Holmes A. Variation in mouse basolateral amygdala volume is associated with differences in stress reactivity and fear learning. Neuropsychopharmacology. 2008;33:2595–2604. doi: 10.1038/sj.npp.1301665. [DOI] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier M, Gregor A, Zweier C, Engels H, Sticht H, Wohlleber E, Bijlsma EK, Holder SE, Zenker M, Rossier E, Grasshoff U, Johnson DS, Robertson L, Firth HV, Firth HV, Ekici AB, Reis A, Rauch A. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat. 2010;31:722–733. doi: 10.1002/humu.21253. [DOI] [PubMed] [Google Scholar]