Abstract
The transforming growth factor-beta (TGFβ) superfamily encompasses widespread and evolutionarily conserved polypeptide growth factors that regulate and orchestrate growth and differentiation in all cell types and tissues. While they regulate asymmetric cell division and cell fate determination during early development and embryogenesis, TGFβ family members play a major regulatory role in hormonal and immune responses, cell growth, cell death and cell immortalization, bone formation, tissue remodeling and repair, and erythropoiesis throughout adult life. The biological and physiological functions of TGFβ, the founding member of this family, and its receptors are of central importance to human diseases, particularly cancer. By regulating cell growth, death, and immortalization, TGFβ signaling pathways exert tumor suppressor effects in normal cells and early carcinomas. Thus, it is not surprising that a high number of human tumors arise due to mutations or deletions in the genes coding for the various TGFβ signaling components. As tumors develop and progress, these protective and cytostatic effects of TGFβ are often lost. TGFβ signaling then switches to promote cancer progression, invasion, and tumor metastasis. The molecular mechanisms underlying this dual role of TGFβ in human cancer will be discussed in depth in this paper, and it will highlight the challenge and importance of developing novel therapeutic strategies specifically aimed at blocking the prometastatic arm of the TGFβ signaling pathway without affecting its tumor suppressive effects.
1. Introduction
The transforming growth factor-beta (TGFβ) was discovered more than two decades ago and was isolated as a secreted factor from sarcoma virus-infected cells [1–3]. TGFβ was shown to transiently confer on normal fibroblasts phenotypic properties of transformed cells, as demonstrated by their acquired ability to grow in soft agar in an anchorage-independent manner [2].
Since then, more than 40 different family members have been identified, including the activin/inhibin subfamily, the bone morphogenetic proteins (BMPs), nodal, myostatin, and the mullerian inhibitory substance (MIS) [4–7]. As for the TGFβ subfamily, three distinct isoforms have been identified (TGFβ-1, -2, -3), each encoded by a different gene [4, 8–10]. Of the three different types of TGFβ, which share around 70% homology within their sequence, TGFβ-1 has been the most studied isoform and will hereinafter be referred to as TGFβ.
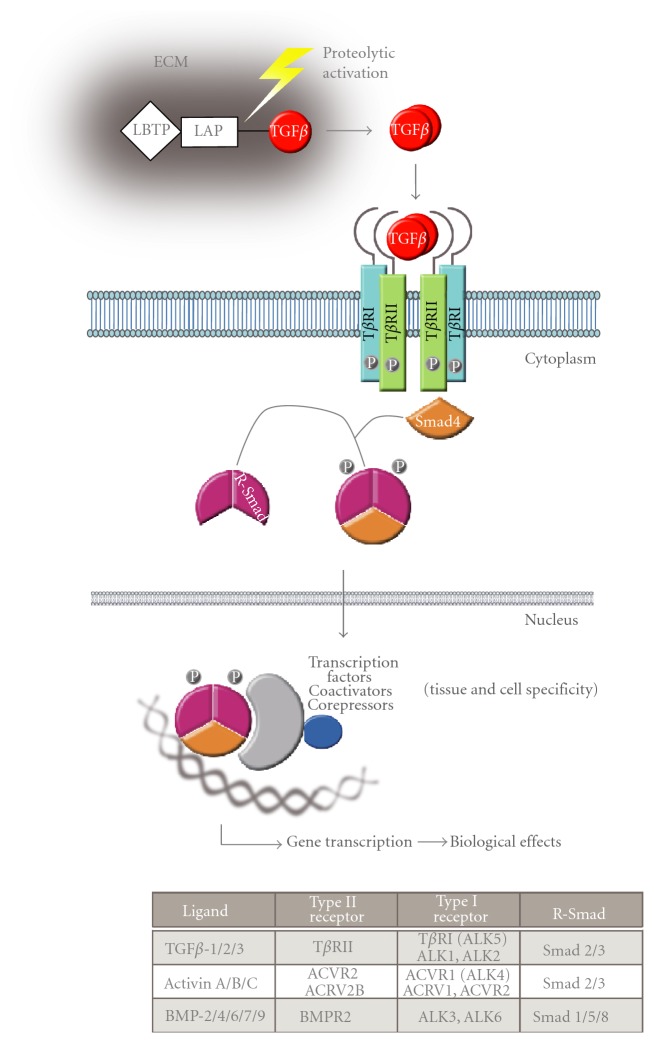
The active TGFβ molecule is a homodimer stabilized by hydrophobic interactions strengthened by a disulfide bond. Each monomer contains β strands interlocked by disulfide bonds that form the cysteine knot [11]. This active form of TGFβ is synthesized from a large inactive precursor molecule, called latent TGFβ. As shown in Figure 1, latent TGFβ is composed of a TGFβ dimer in a noncovalent complex with the TGFβ propeptide or latency-associated peptide (LAP) that remains bound to TGFβ after secretion, retaining TGFβ in an inactive form and the latent TGFβ-binding protein (LTBP) which is linked to LAP by a disulfide bond [12]. This precursor molecule is stored in the extracellular matrix that acts as a reservoir for TGFβ. The activation of the TGFβ precursor is controlled by multiple processes, such as proteolytic enzymatic activity (furins, plasmin, calpain, etc.) but also acid, alkali, and heat-induced proteolysis [12]. Moreover, TGFβ can be activated by glycosidases, thrombospondin, and by some therapeutic molecules, such as antiestrogens and retinoic acid [13, 14]. The mature TGFβ is a homodimeric protein composed of two monomeric subunits linked by a single disulfide bond strengthened by some hydrophobic interactions [11].
Figure 1.
The TGFβ/Smad canonical signaling pathway. TGFβ belongs to a superfamily of growth factors that also includes the activins and BMPs. The active TGFβ ligand is a dimeric molecule composed of two monomers linked by a disulfide bridge and hydrophobic interactions. Each TGFβ subunit is synthesized as a large inactive precursor molecule bound to accessory proteins (LAP and LTBP). This precursor is stored in the extracellular matrix (ECM) and can be rapidly cleaved and activated by several proteolytic mechanisms to become bioavailable. Signal transduction starts with ligand binding to a complex of specific serine/threonine kinase receptors (type I, type II). The type II receptor is constitutively autophosphorylated and, upon ligand binding, transphosphorylates the juxtamembrane region of the type I receptor. This is followed by phosphorylation and recruitment of the R-Smads to the type I receptor and phospho-R-Smad complex formation with common partner Smad4 in the cytoplasm. The Smad complex is then translocated to the nucleus where it interacts with various transcription factors, coactivators, or corepressors to regulate target gene expression. The table lists the different ligands from the superfamily and their interactions with specific receptors and R-Smad proteins.
In 1982, Massagué et al. identified a 60 kDa high-affinity cell surface receptor (type I receptor) for TGFβ [15]. Subsequently, using affinity cross-linking approaches, other TGFβ receptors were discovered and identified (type II and type III receptors) [16]. Following identification of its specific receptors, TGFβ was shown to control and modulate a plethora of biological effects, ranging from cell growth and differentiation, embryogenesis, hormonal synthesis and secretion, immunity, reproduction, bone formation, tissue remodeling and repair, and erythropoiesis, among others [5, 6, 8, 17, 18].
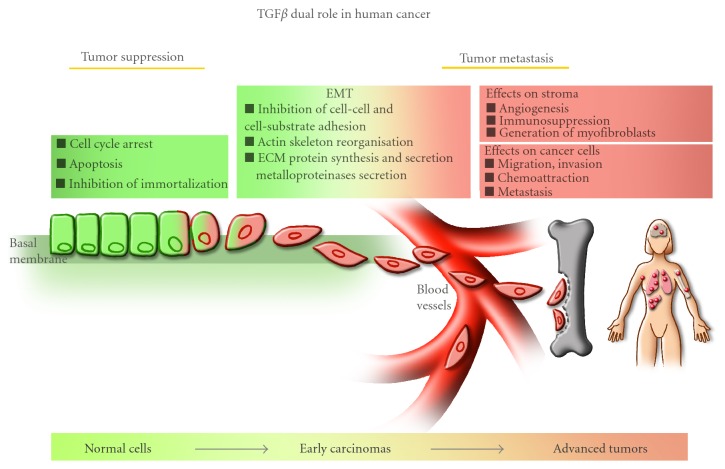
TGFβ and its receptors are widely expressed in all tissues and TGFβ signal transduction pathways play a major role in human diseases. Indeed, while loss of function has been implicated in hyperproliferative disorders, tumor formation, inflammation, and autoimmune diseases, gain of function leads to immunosuppression and tumor metastasis [6, 9, 19, 20]. Thus, TGFβ plays a dual role in human cancers, acting both as a tumor suppressor and as a promoter of tumor metastasis. The tumor suppressive effects of TGFβ, which include inhibition of cell proliferation, induction of apoptosis, and inhibition of cell immortalization, are observed in normal cells and early carcinomas. Conversely, the tumor promoting effects of this growth factor, which include induction of Epithelial-Mesenchymal Transition (EMT), cell adhesion, migration, invasion, chemoattraction, and tumor metastasis, are more specifically observed in aggressive and invasive tumors [6, 21–25]. In addition, as tumors grow and progress, they generally produce and secrete a large amount of autocrine TGFβ that is then released in the tumor vicinity [26]. These increased TGFβ levels not only affect the tumor cells themselves but also the surrounding stroma by inhibiting cell adhesion, inducing immunosuppression and angiogenesis, and by promoting the degradation of the extracellular matrix, further contributing to the metastatic process. Thus, the dual role played by TGFβ and particularly its prometastatic effects make it an attractive target for the development of novel therapies aimed at specifically blocking the pro-metastatic arm of its signaling pathway.
2. TGFβ Signal Transduction
TGFβ ligands interact with a complex of two transmembrane serine/threonine kinase receptors [5, 8, 27]. Signaling starts with ligand binding to the extracellular domain of the type II TGFβ receptor (TβRII), a constitutively autophosphorylated serine/threonine kinase receptor (Figure 1). Of the three TGFβ isoforms, TGFβ2 has the lowest affinity for the type II receptor. As such, TGFβ2 requires binding to an accessory receptor (type III receptor/betaglycan) first to efficiently bind TβRII [28]. TGFβ binding to TβRII is followed by the recruitment of the type I into the complex receptor and its transphosphorylation by the TβRII kinase domain. TGFβ interacts with three distinct type I receptors, including the Activin-Like-Kinase 1 (ALK1), ALK2 or ALK5 [29]. Of note, ALK5 is the predominant form expressed in epithelial cells and is commonly referred as TβRI, being the preferred partner for TGFβ. Given the dimeric nature of mature TGFβ, the resulting receptor complex is in fact a tetramer, composed of two molecules of TβRII associated with two molecules of type I receptor [18]. Phosphorylation of the type I receptors occurs mainly in the juxtamembrane region of the intracellular domain of the receptor, called the GS domain as it is rich in glycine and serine residues [5, 7]. The penultimate residue in the GS domain of the type I receptor, adjacent to the kinase domain, is always a threonine or a glutamine residue. Mutation of this residue to aspartate or glutamate confers elevated kinase activity on the receptor in vitro and constitutive signaling activity in the cell, allowing the type I receptor to fully transmit signals in the absence of ligand or type II receptor. The activated type I receptor is the main component of the TGFβ receptor complex and controls various downstream signaling pathways, including the canonical Smad-dependent pathway [18] as well as non-Smad signaling mechanisms [30].
2.1. Smad-Dependent Pathway
The activated type I receptor recruits and phosphorylates the Smad proteins, the main known effector molecules for these serine kinase receptors. The two receptor-regulated Smads (R-Smads), Smad2 and Smad3, are phosphorylated by the TGFβ and activin type I receptors (ALK5 and ALK4, resp.) on their C-terminal serine residues (SxS motif). While activin and TGFβ share the same R-Smad signaling molecules and mostly signal through Smad2 and Smad3, other members of the TGFβ superfamily, such as the BMPs, signal through distinct R-Smad proteins (Smad1, 5 and 8) following activation of their specific receptors by ligand binding (Figure 1). Once phosphorylated, Smad2 and Smad3 detach from the receptor complex and associate with the common partner Smad4 within the cytoplasm [8, 31–34]. Thus, the activated Smad complex is heterotrimeric, composed of two phospho-Smad2 or phospho-Smad3 moieties with a Smad4 molecule. The Smad complex is then translocated to the nucleus where it acts as a DNA site-specific transcriptional regulator [35]. Nuclear translocation of the Smad complex is regulated by both importin-dependent and -independent mechanisms [36, 37]. Smad proteins recognize the DNA sequence CAGAC, termed the Smad binding element (SBE), as well as some GC rich sequences, but their affinity for DNA is low [38]. In order to achieve a high-affinity DNA binding, the Smads associate with various DNA binding partners [39]. The Smads and associated cofactors bind in concert with their respective cognate recognition sites on DNA, thus ensuring specific selection of the targeted gene promoters and of the TGFβ-mediated transcription response. These co-factors may be functionally expressed in different cell types thus providing another basis for tissue and cell type-specific functions for TGFβ ligands [40, 41]. Furthermore, the Smad complex associates with transcriptional coactivators or corepressors, resulting in the induction or repression, respectively, of a given TGFβ-Smad target gene [42, 43].
2.2. Non-Smad Pathways
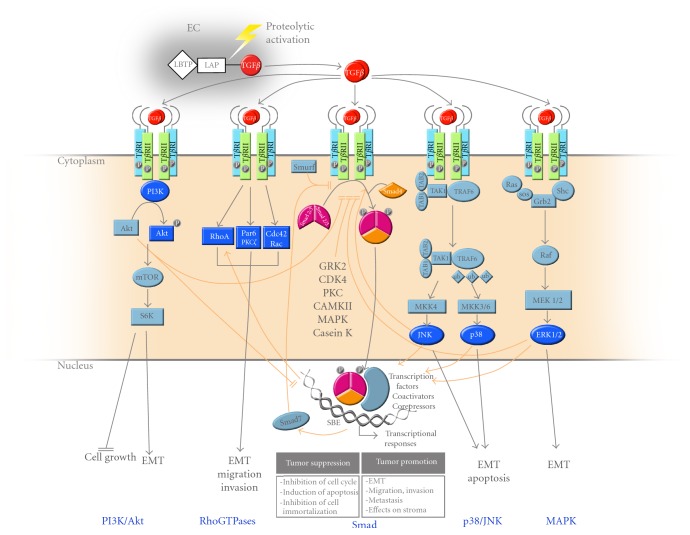
While the Smad pathway represents the canonical signaling pathway for TGFβ ligands, other intracellular signaling cascades have been shown to be activated in response to these ligands (Figure 2). In particular, the stress-activated kinases p38 and JNK (Jun N-terminal Kinase) have been shown to be induced by TGFβ ligands and synergize with Smad signaling to lead to apoptosis and epithelial-mesenchymal transition (EMT) [44–49]. The p38 kinase pathway also plays an important role downstream of activin signaling and was shown to be required for activin-mediated cell growth arrest in breast cancer [47] and activin-mediated inhibition of human Pit-1 gene expression in pituitary tumors [49]. TGFβ can also signal through the mitogen activated protein kinase (MAPK) pathway by activating the extracellular-signal-regulated kinases 1 and 2 (ERK1 and ERK2), further leading to the induction of EMT [45, 50, 51]. Rho GTPases have been shown to relay the TGFβ signals leading to cytoskeleton reorganization, cell motility, and invasion, through activation of RhoA, Cdc42, and Rac [52, 53]. Finally, TGFβ was also shown to signal through the mTOR and the phosphoinositide 3-kinase (PI3 K)/Akt pathway to regulate cell growth inhibition [54] and induction of EMT [55, 56].
Figure 2.
The multiple TGFβ signaling pathways. The canonical Smad pathway is responsible for most of the TGFβ biological responses leading to tumor suppression (growth arrest, apoptosis, and prevention of immortalization) and tumor promotion (EMT, migration, invasion, and metastasis). Even though Smads are central to TGFβ signaling, ligands from this family also signal through other non-Smad pathways. As indicated, TGFβ can activate the PI3 K/Akt, RhoGTPase, MAPK, and stress-activated kinase (p38/JNK) pathways, leading to various biological effects. Depicted by the orange arrows, these pathways also cross-talk or synergise with the Smad pathway to antagonize or potentiate TGFβ signaling, respectively. Several Smad inhibitory pathways are also indicated, including TGFβ-induced gene expression of the inhibitory Smad7, and R-Smad linker phosphorylation by intracellular protein kinases (GRK2, CDK4, PKC, CamKII, MAPK, and Casein kinase).
2.3. Shutting off TGFβ Signaling
Although mechanistically simple, the TGF/Smad signaling cascade is regulated by multiple autoregulatory mechanisms that exist to maintain tightly regulated TGFβ-induced responses. The first TGFβ/Smad inhibitory pathway that has been described involves a Smad family member, Smad7. The inhibitory Smad7 functions through a negative feedback loop mechanism to terminate signaling by sterically preventing access of Smad2/3 to the kinase domain of the type I receptor [57, 58]. In addition, Smad7 also recruits protein phosphatases and ubiquitin ligases (Smurf1/2) to the activated TGFβ receptor, further contributing to the termination of TGFβ signaling [59–62]. Receptor internalization and receptor downregulation are also important means of regulating TGFβ signaling [63]. The TGFβ receptors can be constitutively internalized by clathrin-independent or -dependent mechanisms, through the recruitment of endocytic adaptors like AP-2 and βarrestins to the TGFβ receptor [64, 65]. At the Smad level, nuclear translocated Smad2 and Smad3 can be subject to ubiquitin-proteasome-mediated degradation or dephosphorylation, leading to termination of signaling in both cases [66–69]. Cross-talk from other signaling pathways, such as epidermal growth factor (EGF) and oncogenic Ras signaling, which interfere with the nuclear translocation of the Smads, can further act to negatively regulate Smad-dependent signal transduction [70–72].
While phosphorylation of the C-terminal MH2 domain of the Smad by the type I receptor leads to activation of the R-Smads, phosphorylation of the linker domain by various nonreceptor intracellular kinases inhibits Smad signaling. As shown in Figure 2, such kinases include the MAPK kinases [70, 73], calcium-calmodulin-dependent protein kinase II [74], cyclin-dependent kinase CDK2/4 [75], casein kinase [76], protein kinase C [77], and G protein-coupled receptor kinase 2 (GRK2) [78]. These kinases specifically target the linker region of the R-Smads on multiple distinct serine and threonine residues, leading to termination of Smad signaling. Thus, the linker domain appears as a primary site for negative regulation of Smad signaling. In the case of GRK2, the kinase itself is regulated by TGFβ signaling and acts in a negative feedback loop [78]. Because GRK2 plays a central role in modulating G-protein coupled receptor signaling, we also found TGFβ-induced GRK2 expression antagonizes angiotensin II-regulated vascular smooth muscle cell proliferation and migration [79]. GRK2 physically interacts with the MH1 and MH2 domains of the receptor-regulated Smads and phosphorylates their linker region on a specific single serine/threonine residue [78]. GRK2-induced Smad phosphorylation then leads to complete inhibition of TGFβ-induced Smad activation, nuclear translocation, and target gene expression and inhibits the TGFβ antiproliferative and pro-apoptotic responses. Thus, GRK2 appears as a novel TGFβ antagonist that strongly inhibits cell growth arrest and apoptosis in both normal and cancer cells [78]. Interestingly, mutating the GRK2 phosphorylation site within the Smad linker domain to an aspartate residue to mimic a constitutively phosphorylated Smad generates dominant negative Smads that efficiently inhibit TGFβ responses [80].
3. TGFβ as a Tumor Suppressor
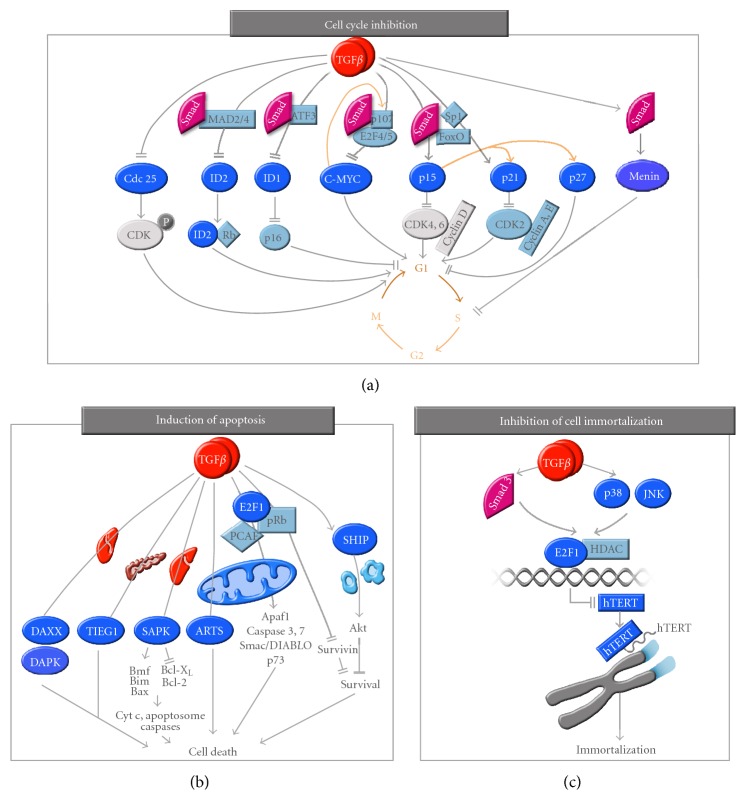
The most well-characterized effects of TGFβ are its tumor suppressor effects in epithelial, endothelial, myeloid, and lymphoid cell types. Expression of the TGFβ type II receptor (TβRII) in breast cancer cells prevents tumor formation [81], while inactivating mutations or overexpression of a dominant negative form of the receptor abolish TGFβ tumor suppressive effects and increase tumorigenicity [82–84]. Moreover, low expression levels of TβRII are correlated with more advanced and aggressive tumor stages, suggesting that the TGFβ signaling pathway acts as a tumor suppressor in the early stages of tumor development [85]. As shown in Figure 3, TGFβ exerts strong cytostatic effects on most of its target tissues by inhibiting cell cycle in the G1 phase. In addition, TGFβ induces apoptosis and prevents cell immortalization in numerous target tissues [4, 24, 48, 86].
Figure 3.
TGFβ and tumor suppression. (a) Cell cycle inhibition. TGFβ exerts strong cytostatic effects and induces cell cycle arrest in the G1 phase by increasing the expression of the small cyclin-dependent kinase inhibitors p15, p21, and p27. These effects are Smad-dependent but also require the transcription factors Sp1 and FoxO. p15 directly inhibits CDK4/6 and displaces p21 and p27 from their preexisting CDK4/6 complexes allowing them to bind and inhibit CDK2-cyclin A/E complexes (orange arrows). TGFβ-induced cell cycle arrest also relies on the downregulation of the oncogene c-myc through Smads and repressor E2F4/5. The transcription factors from the ID family are also repressed by TGFβ through Smads, MAD2/4, and ATF3, further contributing to TGFβ-mediated cell cycle arrest. Finally, other pathways, potentially more tissue specific, have been described, including upregulation of the tumor suppressor menin in pituitary adenomas, leading to G1 arrest, and downregulation of the tyrosine phosphatase CDC25A in mammary epithelial cells, also leading to G1 arrest. (b) Induction of apoptosis. A central pathway in the mediation of the TGFβ proapoptotic effects involves the E2F1-pRb-P/CAF pathway that leads to gene transcription of multiple TGFβ proapoptotic target genes in various types of normal and cancer cells. In hematopoietic cells, TGFβ specifically induces expression of the lipid phosphatase SHIP, which in turn decreases second messenger PIP3 level and blocks Akt-mediated survival pathways, leading to cell death in both B and T lymphocytes. Other tissue specific proapoptotic pathways have been described downstream of TGFβ, including the TGFβ-mediated induction of the two proapoptotic proteins DAXX and DAPK in liver cells, the transcription factor TIEG1 in pancreatic cells, and the mitochondrial protein ARTS. TGFβ also promotes apoptosis in an SAPK-dependent manner by inducing pro-apoptotic target gene expression (Bmf, Bim and Bax) and by repressing antiapoptotic gene expression (Bcl-Xl and Bcl-2), further inducing mitochondrial release of cytochrome C and activation of the apoptosome, leading to caspase-dependent apoptosis in hepatocytes and B-lymphocytes. In colon cancer, TGFβ was also shown to inhibit expression of the prosurvival protein survivin. (c) Inhibition of cell immortalization. TGFβ also exerts its tumor suppressive effects through inhibition of cell immortalization in normal and cancer cells. This effect is mediated through the Smad, p38, and JNK pathways and requires recruitment of histone deacetylases (HDAC) to the telomerase (hTERT) gene promoter, further leading to inhibition of telomerase expression, and thereby preventing cell immortalization.
3.1. Cell Cycle Inhibition
Cell cycle progression is controlled by intracellular protein kinases, called cyclin dependent kinases (CDKs). Once activated and associated to their regulatory cyclin subunits, the CDKs induce gene transcription of a number of cell cycle regulators (DNA polymerases, oncogenes, etc.), allowing for cell cycle progression from G1 to S phase. As shown in Figure 3, TGFβ induces cell cycle arrest in G1 by inducing the expression of small inhibitory molecules, the cyclin-dependent kinase inhibitors (CDKIs) p15INK4B [87] and/or p21KIP1 [88], which in turn inhibit specific CDK activity. TGFβ-induced gene transcription of p15 and p21 is mediated by Smad association with specific transcription factors, such as FoxO forkhead [89] and Sp1 [90, 91]. p15INK4B interacts with either CDK4 or CDK6 or with CDK4-cyclin D or CDK6-cyclin D complexes, while p21CIP1 interacts with CDK2-cyclin A or CDK2-cyclin E complexes [92]. TGFβ-induced p15INK4B expression leads to p15INK4B binding to CDK4 and CDK6, blocking their association with their regulatory cyclins, thereby inhibiting their function and inducing G1 arrest. Moreover, p15INK4B binding to the cyclin D-CDK4/6 complexes also displaces p21CIP1 or the related p27KIP1 from these complexes, thus allowing these proteins to bind and inactivate CDK2-cyclin A and CDK2-cyclin E complexes [92, 93].
In addition, TGFβ represses the expression of growth promoting factors such as the oncogene c-MYC [78, 94], and the ID family of helix-loop-helix transcription factors (ID1, ID2, and ID3) [95–97]. These proteins regulate angiogenesis, cell growth, and differentiation and are often upregulated in human cancer [95, 96, 98]. Thus, inhibition of their expression by TGFβ largely contributes to this growth factor's antiproliferative effect. TGFβ-mediated c-MYC downregulation is mediated by a transcriptional regulatory complex including Smad3, Smad4, the repressor E2F4/5, and p107 [99] and directly leads to cell growth arrest. Interestingly, expression of the two CDKIs p15INK4B and p21CIP1 is normally restrained by the binding of c-MYC and the zinc-finger protein MIZ1, in the proximal region of their promoters [100, 101]. Inhibition of c-MYC expression by TGFβ thus further contributes to increased expression of p15INK4B and p21CIP1 and induction of G1 arrest. The ID proteins interact with the retinoblastoma tumor suppressive protein (pRB) to promote cell proliferation and have been implicated in promoting tumorigenesis [96, 98]. ID1 also delays cellular senescence in primary mammalian cells through inhibition of the cell cycle regulatory protein p16INK4a. TGFβ inhibits ID1 expression in a Smad3-dependent manner through induction of the activating transcription factor-3 (ATF-3), a well-known ID1 repressor [95]. As c-MYC binds the ID2 gene promoter and activates ID2 gene expression, its downregulation by TGFβ also leads to inhibition of ID2 gene transcription [96, 97]. In ovarian cancer cells, we also found the engulfment protein GULP to act as a key regulator of TGFβ-mediated growth inhibition [102]. Finally, TGFβ was also shown to specifically inhibit expression of the tyrosine phosphatase CDC25A in normal mammary epithelial cells, by means of a Smad3/E2F4/5/p130 inhibitory complex [103]. CDC25A normally dephosphorylates an inhibitory site on CDK4 and CDK6. Thus, TGFβ-mediated inhibition of CDC25A expression allows for sustained CDK4/6 phosphorylation on their inhibitory sites and further induces cell cycle arrest (Figure 3(a)) [103].
Recent work from our laboratory has also revealed a role for the tumor suppressor menin downstream of TGFβ/Smad signaling in pituitary adenoma cells [5, 104, 105]. Pituitary adenomas are common monoclonal neoplasms accounting for approximately twenty percent of primary intracranial tumors with prolactin-secreting pituitary adenomas (prolactinomas) and are the most common form of pituitary tumors in humans [106, 107]. They are associated with very high levels of the hormone prolactin, exhibit increased tumor growth, and they give rise to severe endocrine disorders, including amenorrhea, infertility issues associated with galactorrhea in females, and impotence in males [106–108]. Multiple endocrine neoplasia type 1 (MEN1) is an autosomal dominant disorder characterized by endocrine tumours of the parathyroid, pancreatic islets, and anterior pituitary, particularly prolactinomas [5, 109]. We found menin to physically interact with Smad3 in somatolactotrope cells and showed that inactivating menin expression antagonizes TGFβ signaling [104]. We found that menin suppresses TGFβ-induced transcriptional activity by inhibiting the binding of the Smads to DNA [104]. The role of menin is not restricted to TGFβ as we also found menin to be required for activin signaling in pituitary cells [110]. Results from our laboratory indicate that activin negatively regulates prolactin gene expression through reduction of Pit-1 expression in a Smad- and menin-dependent manner and that menin is required for activin-induced cell growth inhibition in somatolactotrope cells, highlighting a critical role for activin in mediating pituitary cell growth and Pit-1/prolactin gene expression through the Smads and menin [5, 110].
3.2. Induction of Apoptosis
TGFβ stimulates cell death in various target tissues and these effects have been particularly well documented in the various epithelium, liver, and immune system [111–116]. However, the molecular mechanisms and signaling pathways underlying these pro-apoptotic effects of TGFβ remain largely uncharacterized. Several apoptotic regulators have been implicated downstream of the TGFβ signaling pathway, often in a cell- or tissue-specific manner. For instance, in hepatocarcinomas, the Daxx adaptor protein couples the TGFβ signaling pathway to the cell death machinery through its interaction with the type II TGFβ receptor (TβRII) [117]. Interaction of the Daxx protein to TβRII leads to its stabilization and further activation of the JNK and Fas-mediated apoptotic pathways. In liver cancer, TGFβ also induces gene expression of the death-associated protein kinase DAPK, which promotes cell death, in a Smad-dependent manner, thereby linking the Smad proteins to the mitochondrial proapoptotic processes [118]. The TGFβ-inducible early-response gene (TIEG1) is a Krüppel-like zinc finger transcription factor that mediates apoptosis in pancreatic epithelial cells [119]. Another mitochondrial protein that has been shown to mediate some of the TGFβ responses is the septin-like protein ARTS (apoptosis-related protein in the TGFβ signaling pathway), which can potentiate apoptosis induced by TGFβ, even in cells resistant to TGFβ-mediated cell death [120]. The stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) signaling pathway also plays a critical role in mediating TGFβ, through Smad interaction with the activator protein AP1 [121, 122]. TGFβ causes Smad- and the SAPK/p38-dependent transcriptional induction of the pro-apoptotic Bcl-2 family members Bmf and Bim, which in turn activate the pro-apoptotic factor Bax that induces mitochondrial release of cytochrome c and activation of the apoptosome, leading to caspase-dependent apoptosis in hepatocytes and B-lymphocytes [123, 124]. Conversely, the anti-apoptotic proteins Bcl-XL and Bcl-2 have been demonstrated to be down-regulated by TGFβ in various cell types [125–128]. Each of these signaling events eventually couples TGFβ to the cell death machinery, leading to changes of expression, localization, and activation of members of the Bcl-2 family and caspases [116]. Interestingly, the pro-apoptotic effects of TGFβ are particularly strong in immune cells. Work from our laboratory showed that in lymphocytes, both TGFβ and activin induce the expression of the Src homology 2 domain-containing 5′ inositol phosphatase (SHIP), leading to immune cell death [113]. TGFβ-induced SHIP expression is Smad-dependent and results in intracellular changes in the pool of phospholipids. Upon TGFβ stimulation, the increased SHIP expression leads to decreased levels of second messenger PIP3 (Phosphatidyl Inositol triphosphate), further contributing to inhibition of the Akt survival pathway and resulting in cell death in both B and T lymphocytes [113]. TGFβ also antagonizes survival signaling by inhibiting expression of the prosurvival protein survivin through the physical interaction of Smad3 with Akt, leading to apoptosis in colon cancer [129–131]. In prostate epithelial cells, inhibition of survivin by TGFβ is Smad-dependent and involves recruitment of a pRb/E2F4 repressive complex to the survivin promoter [132]. Finally, we recently uncovered a central mechanism by which TGFβ induces apoptosis in both normal and cancer cells of various origins [133]. Indeed, we found TGFβ to increase expression of the transcription factor E2F1, further leading to the formation and binding of a transcriptionally active E2F1-pRb-P/CAF complex on multiple TGFβ pro-apoptotic target gene promoters, thereby activating their transcription and highlighting E2F1 as a central mediator of the TGFβ apoptotic program (Figure 3(b)) [133].
3.3. Prevention of Cellular Immortalization
Normal cells are only able to replicate a defined number of times, called the Hayflick limit [134], after which cells enter senescence and die. This limited number of replication cycles is due to the progressive shortening of the ends of the chromosomes, called telomeres, as DNA polymerases fail to completely replicate genetic material at each cell division. As a result, after a number of cell divisions, the length of the telomeres shortens to a critical point, eventually leading to chromosome instability, senescence, and cell death. Interestingly, cancer cells are not subjected to this limitation and thus achieve immortalization, due to the reactivation of an enzymatic program, the telomerase activity. In fact, elevated telomerase activity is so commonly observed in cancer cells that it is being used as a prognostic marker for cancer. Telomerase is a reverse transcriptase that adds telomeric DNA repeats at the end of chromosomes, thereby preventing their shortening. Interestingly, TGFβ regulates the levels of human telomerase reverse transcriptase (hTERT), the protein component of the telomerase enzyme, by repressing its expression in normal and cancer cells [48, 135, 136]. This TGFβ-mediated repression of telomerase is Smad3-specific and requires the transcription factor E2F1 as well as the stress-activated kinase and histone deacetylase activities (Figure 3(c)) [48].
In summary, TGFβ acts as a tumor suppressor, acting through three different signaling arms (cell cycle inhibition, induction of apoptosis, and prevention of cell immortalization) and it is by the combined effects of these three separate signaling axes that TGFβ exerts its potent tumor suppressive effects in most cell types and tissues.
4. Genetic Defects in the TGFβ Signaling Components and Human Cancer
The role of TGFβ as a potent tumor suppressor is further highlighted by the fact that many inactivating mutations in TGFβ receptors and Smad genes have been found to be an underlying cause for human cancer [4, 9, 24, 137]. Multiple genetic and epigenetic alterations of the TGFβ signaling pathway components have been reported to inhibit TGFβ tumor suppressive effects, thereby favoring tumor development [137]. These are often found in human cancers of various origin (Table 1) [4, 6, 9, 24, 137] and clearly illustrate the critical role played by the TGFβ signaling pathway in preventing tumor formation.
Table 1.
Mutations and deletions in the TGFβ signaling pathway. While expression of TGFβ itself is often increased in human tumors, expression of the genes encoding various components of the TGFβ signaling cascade (receptor type I and type II, Smad2, Smad3, and Smad4) are often mutated or deleted in human cancer. Occurrence of mutation and deletion and incidence rates in different human cancers are indicated in percentages. Loss of heterozygosity (LH) is also indicated.
| Molecules | Cancers |
|---|---|

|
Increased expression:
breast (68%), lung (48%), pancreas (47%), esophagus (37%), stomach (23%), colon, prostate. |
|
| |

|
Mutations/deletions:
colon (28%), ovary (25%), head and neck carcinoma (21%), stomach (15%), breast (12%), lung, endometrium, liver, uterus, biliary track, glyomas. |
|
| |

|
Mutations/deletions:
ovary (30%), head and neck carcinoma (17%, LH 53%), bladder (LH 31%), prostate (25%), breast (6%), biliary track. |
|
| |

|
Mutations/deletions:
colon (8%), uterus (8%), liver, lung. |
|
| |

|
Mutations/deletions:
lymphoblastic leukemia, stomach. |
|
| |

|
Mutations/deletions:
pancreas (50%, LH 90%, deletion 30%), colon (LH 60%), stomach (LH 60%), lung (LH 56%), breast (12%, LH 30%), head and neck carcinoma (LH 40%), prostate (LH 30%), biliary track (16%), uterus (4%), bladder, oesophagus, kidney, liver, ovary |
4.1. Mutations in the TGFβ Receptor Genes
Mutations in either alleles of the TGFβ type II receptor (T βRII), leading to the formation of truncated or kinase inactive mutant forms of the receptor, are frequently found in colorectal, gastric, biliary, pulmonary, ovarian, esophageal, head and neck cancers, and gliomas [137, 138]. They also occur in other types of tumors, such as those of the endometrium, pancreas, liver, and breast cancers, though with a lower frequency [137]. These inactivating mutations of TβRII are more frequently observed in tumors with microsatellite instability, due to mutations in mismatch repair genes. The type I TGFβ receptor (TβRI) also often harbors frameshift and missense mutations in ovarian, breast, esophageal, pancreatic, and head and neck cancers. Epigenetic alterations of the TGFβ receptor genes, such as promoter hypermethylation or altered/defective expression of the transcription factors that regulate their expression, also lead to decreased receptor expression and inefficient receptor activity [85].
4.2. Smad Mutations
Like the TGFβ receptors, the genes coding for the Smad proteins are often mutated or deleted in human cancers [137]. Smad mutations are due to loss of chromosome regions, deletions, frameshift mutations, nonsense, and missense mutations [139]. Of the three domains that compose the Smad molecules, it is within the carboxy-terminal MH2 domain of the Smads that these mutations preferentially occur [137, 140]. These mutations are mostly found in Smad2 and Smad4 and either prevent complex formation with the Smad partners or block activation of Smad-mediated gene transcription [139, 140]. As shown in Table 1, the tumor suppressor Smad4, also known as dpc4 (deleted in pancreatic cancer), is particularly affected by these genetic alterations, being mutated or deleted in no less than half of human pancreatic cancer, where it was originally characterized [141]. Since then, mutations in the Smad4 gene have been characterized in other type of tumors (Table 1) [137]. Mutations in the Smad2 gene also have a relatively high occurrence in lung, liver, and colorectal cancers [137, 142], while the rate of mutation in the Smad3 gene is much lower. In fact, to date there are only few examples of such defects in Smad3 expression, found in some gastric cancers and certain types of leukemia [143].
Finally, inhibition of Smad expression in human tumors can also result from gene amplification of Smad transcriptional repressors. In particular, the two Smad repressors SnoN and Ski are often found activated in in human cancers, highlighting their potent oncogenic properties [144]. Similarly, overexpression of the inhibitory Smad family member, Smad7 has been reported in several types of human cancers, including pancreatic [145], endometrial [146], and thyroid follicular [147] tumors, resulting in inhibition of TGFβ/Smad signaling.
4.3. Mutations/Alterations in the Non-Smad TGFβ Signaling Pathways
Besides the known mutations in the TGFβ receptors and canonical Smad pathway, other types of genetic alterations have also been reported to affect TGFβ signaling and tumor formation. For instance, oncogenic activation of the Ras-Raf-MAPK pathway and c-Jun NH2-terminal kinase in hepatocellular carcinoma has been reported to induce phosphorylation of the Smad3 linker domain by MAPK, further preventing C-terminal phosphorylation of the Smad by the TβRI kinase domain and inhibiting TGFβ cytostatic effects [72]. Moreover, epigenetic alterations of other signaling components can also favor the TGFβ pro-metastatic effects. Indeed, hypomethylation of the Platelet-Derived Growth Factor β (PDGFβ) gene promotes glioblastoma cell proliferation in response to TGFβ [148]. Epigenetic downregulation of human disabled homolog 2 (DAB2) also switches TGFβ from a tumor suppressor to a tumor promoter in head and neck carcinomas [149]. Finally, the Sine oculis homeobox homolog 1 (Six1) homeoprotein was also shown to induce human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGFβ signaling [150].
5. TGFβ as a Prometastatic Factor
As described in the previous sections, the tumor suppressive role of TGFβ has been well described in multiple target tissues. Interestingly, while TGFβ acts as a tumor suppressor in normal cells and early carcinoma, its cytostatic effects are often lost during the progression of the disease. Indeed, as indicated above, many different human tumors are either resistant to the TGFβ cytostatic effects, due to genetic and epigenetic modifications in the TGFβ signaling components, or become resistant, due to the activation of prooncogenic signaling pathways (MAPK, PI3 K, Ras, c-MYC), which then simply override any growth inhibitory signaling pathways, including TGFβ/Smad [9, 20, 72, 151]. Meanwhile, other TGFβ responses prevail, unrelated to the TGFβ cytostatic effects, which favor tumor progression and metastasis [6, 9, 20, 151].
These pro-metastatic effects of TGFβ were best characterized in breast cancer, the most common form of cancer in women in North America. In 2012, 1.8 million new cases of cancer will be diagnosed in the US and Canada alone, including 250,000 new cases of breast cancer. For 2012 alone, the breast cancer-related death toll is estimated at 45,000 individuals in the US/Canada (http://www.cancer.org/, http://www.cancer.ca/). There are several classifications of breast tumors, depending on the tumor size, the presence of tumor cells outside the primary site either in the lymph nodes or distant metastatic sites, and the growing speed rate of the cancer cells (established from biopsies) [152]. While the rate of remission and overall survival are high in the case of localized primary tumors, they are dramatically lower for metastatic tumors that have propagated to distant sites. The growth of these tumors is independent of hormonal and human epidermal growth factor receptor 2 (HER2) levels. As a result, they usually are insensitive to hormone-based therapies (Tamoxifen, aromatase inhibitors) and therapies targeting the HER2 receptor (Trastuzumab, Herceptin). These aggressive metastatic tumors are responsible for the large majority of breast cancer-related deaths [153].
Although typically associated with the TGFβ cytostatic responses, the Smad proteins are also critical for TGFβ-mediated tumor metastasis. Indeed, expression of dominant negative Smad3 or expression of a mutant form of the TGFβ type I receptor that fails to recruit the Smad proteins, in human mammary epithelial cells, significantly diminished their ability to colonize the lungs [154, 155]. Moreover, overexpression of the inhibitory Smad7 impaired mammary carcinoma cell invasion [156]. Finally, gene silencing of the common partner Smad4 in MDA-MB231 invasive breast cancer cells impaired their ability to form osteolytic lesions and the formation of bone metastasis [157, 158].
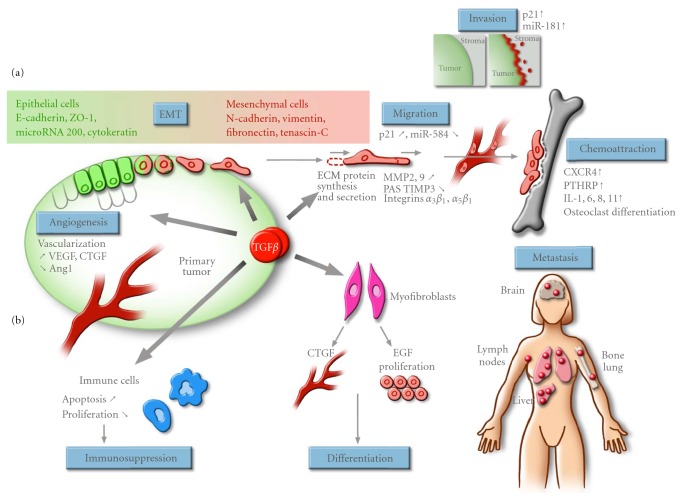
As shown in Figure 4, TGFβ plays a major role in promoting breast cancer migration, invasion, and metastasis by acting at various levels: (a) on the stroma and neighboring cells surrounding the tumor and (b) directly on the cancer cells themselves. These pro-metastatic responses of TGFβ include the ability to remodel the surrounding extracellular matrix (ECM), through stimulation of matrix metalloproteinase (MMP) expression and modulation of the plasminogen activation system, resulting in TGFβ-mediated matrix degradation and, consequently, an increasing release of stored TGFβ from the ECM that acts as a TGFβ reservoir [159]. Indeed, in many types of cancer, increased production of TGFβ correlates with higher tumor grade [159, 160]. Increased TGFβ expression is observed in breast tumors and correlates with the aggressiveness and advanced stage of the tumor [26]. Interestingly, in mammary carcinoma patients, immunocytochemical analysis revealed that secreted TGFβ strongly localizes to the advancing edges of the primary tumor and to lymph node metastases [161, 162]. This tumor-derived TGFβ can exert autocrine effects, that is, effects on the tumor cells themselves, as well as paracrine effects on components of the tumor milieu such as stromal fibroblasts, endothelial cells, and immune cells. Increased secretion of TGFβ affects and stimulates angiogenesis, contributes to myofibroblast differentiation and causes local and systemic immunosuppression, further contributing to tumor progression and metastasis [9, 20, 137, 151]. As outlined above, cancer cells themselves respond to and are affected by the increased TGFβ levels, which leads to phenotypic change of the cancer cells from epithelial to mesenchymal (EMT transition), loss of polarity and adhesion, associated with increased migration and invasion cell properties, as well as increased chemoattraction to distant tissues (e.g., bone), thereby favoring and inducing metastatic development [9, 20].
Figure 4.
TGFβ prometastatic effects. Tumor cells synthesize and secrete a significant amount of TGFβ, which affects both the cancer cell and the stroma. As a result, TGFβ promotes tumor progression and metastasis by acting directly on the cancer cells themselves and by affecting the stroma and surrounding environment. (a) In cancer cells, TGFβ promotes the epithelial to mesenchymal transition (EMT) by decreasing cell adhesion and by blocking expression of epithelial proteins (E-Cadherin, ZO-1, etc.) while increasing the expression of mesenchymal proteins (N-Cadherin, vimentin, fibronectin, tenascin-C). TGFβ also promotes cell migration and invasion through multiple signalling pathways (increased p21 expression, microRNA regulation, increased synthesis and secretion of metalloproteinase expression, activation of RhoGTPases, decreased TIMP3 expression, and regulation of the plasminogen activator system (PAS)). TGFβ also promotes tumor metastasis by potentiating chemoattraction of the cancer cells to distant organs (bone, lymph node, lung, liver, and brain) and by increasing expression of cytokines (CXCR4, IL-11 and PTHrP) that will promote osteoclast differentiation and the development of osteolytic lesions. (b) TGFβ affects the stroma and the surrounding environment to varying degrees. TGFβ induces angiogenesis and stimulates the vascularisation surrounding the tumor by increasing VEGF and CTGF expression in epithelial cells and fibroblasts. Furthermore, TGFβ also inhibits expression of angiopoetin-1 in fibroblasts, thus increasing permeability of blood vessels associated to the tumor. By inducing hematopoietic cell death, TGFβ induces local and systemic immunosuppression, preventing the immune cells from infiltrating the tumor and allowing the tumor to escape host immunosurveillance. TGFβ also promotes myofibroblast differentiation, further promoting tumor growth.
Thus, the tumor-permissive effects of TGFβ provide for a unique therapeutic opportunity in that specifically blocking this signaling network may interrupt mechanisms that are essential for tumor metastasis. However, it is important to note that because of the dual role played by TGFβ, acting as both tumor suppressor and tumor promoter, elucidation of the molecular events and components leading to both arms of TGFβ signaling will be critical to further design therapeutic strategies aimed at specifically blocking TGFβ-mediated tumor metastasis without affecting the tumor suppressive effects of this growth factor.
5.1. Paracrine Activity of Tumor-Derived TGFβ: Effects on the Tumor Microenvironment
5.1.1. Immunosuppression
Elevated TGFβ levels released from the ECM reservoir exert a profound effect on the immune system. Indeed, as mentioned above, TGFβ acts as a potent inducer of apoptosis in immune cells. Through up-regulation of the lipid phosphatase SHIP and subsequent inhibition of the PI3kinase/Akt survival pathway, TGFβ can induce cell death in both B and T lymphocytes [113]. TGFβ directly targets cytotoxic T cell functions during tumor evasion of immune surveillance by suppressing production of cytolytic factors (pore-forming protein perforin, caspase-activating factors granzymes A and B, and pro-apoptotic cytokines Fas-ligand and interferon γ) [163]. TGFβ also inhibits expression and activity of interleukin-2 and its receptors [164] and blocks T lymphocyte stimulation by dendritic cells during an immune response [165]. TGFβ inhibits proliferation and differentiation of T lymphocytes, lymphokine-activated killer cells, natural killer cells (NK), neutrophils, macrophages, and B cells [166]. Finally, TGFβ also decreases tumor cell surface immunogenicity by inhibiting expression of major histocompatibility complex class II antigens, through a Smad3-dependent mechanism [167–169]. These immunosuppressive effects of TGFβ are best illustrated by the blockade of the TGFβ signaling cascade through overexpression of a dominant-negative receptor (DN-TβRII), which restored both CD8+ and CD4+ mediated immune response [170]. Thus, the increased concentration of released TGFβ in the tumor vicinity dramatically contributes to the tumor progression process, as local and systemic immunosuppression induced by TGFβ allows the tumor to escape host immunosurveillance [4, 6, 137].
5.1.2. Angiogenesis
The process of angiogenesis is essential to tumor growth as it allows blood vessels to deliver nutrients and oxygen to the tumor cells, and allows cancer cells that have detached from the primary tumor to reach and intravasate into the blood system. The TGFβ signaling pathway is a potent inducer of fibrosis and angiogenesis in vivo [171] and promotes chemoattraction of angiogenic cytokine-secreting monocytes [172]. TGFβ signaling in endothelial cells is rather complex as these cells express two TGFβ type I receptors (ALK1 and ALK5) [173]. The classic ALK5-mediated pathway leads to Smad2/3 activation, resulting in vessel maturation and angiogenic resolution, while ALK1-mediated signaling antagonizes TGFβ/ALK5 responses by inducing Smad1/5 and generates transcriptional responses that are linked to angiogenesis [174–177]. The TGFβ effects in angiogenesis are best illustrated by the multiple germline mutation mice models (ligand, receptors (I, II and III) and Smad1/5) that all lead to vascular and endothelial cell defects [178–185]. Increased expression of TGFβ correlates with increased microvessel density and with poor prognosis in various tumor types, such as breast cancer and nonsmall cell lung carcinoma [186, 187]. There are many ways in which TGFβ contributes to the angiogenic process. TGFβ stimulates the expression of angiogenic factors such as vascular endothelial growth factor (VEGF) and connective-tissue growth factors (CTGF) in epithelial cells and fibroblasts [188, 189]. TGFβ-induced expression, secretion, and activity of MMPs also contribute to the dissolution of mature vessels around the tumor and the release of endothelial cells from the basement membrane, allowing them to further migrate and invade [190]. Moreover, TGFβ represses expression of angiopoietin-1, a critical factor in maintaining vessel integrity, in fibroblasts thereby contributing to the permeable properties of tumor-associated blood vessels [191].
5.1.3. Myofibroblast Generation
Many recent studies have focused on the emerging role of tumor-stroma interactions, which are essential for supporting tumor progression. Myofibroblasts, also known as cancer-associated fibroblasts (CAFs), are mesenchymal cells harboring characteristics from both fibroblasts and smooth muscle cells [192]. These cells can secrete numerous cytokines, growth factors, and ECM components and have the ability to substantially promote tumorigenesis and their appearance precedes the invasive stage of cancer. In a coimplantation breast tumor xenograft model, resident human mammary fibroblasts progressively converted into CAF myofibroblasts during the course of tumor progression [193]. During this process, they displayed increased autocrine signaling loops, mediated by TGFβ and SDF-1 cytokines, which acted in both auto-stimulatory and cross-communicating fashions. These autocrine-signaling loops initiated and maintained the differentiation of fibroblasts into myofibroblasts and the concurrent tumor-promoting phenotype [193]. TGFβ also significantly increased the percent of myofibroblasts and invasion rate in CAF cultures [194] and increased the production and secretion of urokinase-type plasminogen activator (uPA) by human breast myofibroblasts [195]. Thus, TGFβ induces the generation and maturation of myofibroblasts from precursor fibroblasts which, in turn, stimulate invasion of the tumor cells through secretion of proliferative, proinvasive and proangiogenic factors.
As summarized in Figure 4, TGFβ plays an important role in promoting tumor growth and development as well as tumor metastasis by allowing tumor cells to survive, detach and migrate away from the primary tumor to invade the surrounding tumor environment and metastasize to distant organs.
5.2. Autocrine Activity of Tumor-Derived TGFβ: Effects on the Tumor Cells
The increased TGFβ levels produced by the tumor cells also contribute to the formation of a favorable microenvironment for tumor growth and spread by acting directly on the tumor cells themselves. Tumor-produced TGFβ stimulates EMT, cell migration, and invasion and promotes chemoattraction of the tumor cell towards distant organs (Figure 4).
5.2.1. Epithelial to Mesenchymal Transition
The EMT process characterizes the differentiation of highly organized and tightly connected networks of epithelial cells into disorganized and mobile mesenchymal cells with stem cell-like properties. The EMT process involves a loss of cell-to-cell contact and the acquisition of fibroblastic characteristics by the epithelial cells as well as the acquisition of migratory and invasive properties by the cancer cells [196, 197]. EMT is a naturally occurring process that takes place during embryogenesis and development. EMT drives and governs morphogenesis by inducing the differentiation of the epithelium into mesenchymal cell types to generate the different embryonic territories. The EMT process is characterized by the dissolution of epithelial tight junctions and basolateral adherens junctions, resulting in the loss of epithelial cell polarity. This is best exemplified by the loss of epithelial gene expression (E-cadherin, ZO-1, occludin, claudin, cytokeratins 8, 18, and 19, desmoplakin) and the induction of more mesenchymal markers (N-Cadherin, vimentin, fibronectin, tenascin-C, and vitronectin). During EMT, the actin cytoskeleton is also reorganized from a cortical adherens-associated location into actin stress fibers anchored to focal adhesion complexes that contribute to the formation of filopodia and promote cell migration. EMT also contributes to cancer cell invasion and dissemination. Indeed, down-regulation of E-cadherin allows for the release of β-catenin leading to increased expression of c-MYC, cyclin D1, and MMP7, thereby promoting the invasive behavior of the cells. Moreover, during EMT, increased secretion of extracellular proteases and reduced expression of ECM proteins further contribute to cancer cell invasion. EMT is under the control of several key transcription factors that regulate expression of mesenchymal markers and repression of epithelial genes. These include the zinc-finger proteins Snail and Slug, the basic helix-loop-helix factor Twist, the zinc-finger/homeodomain proteins ZEB-1 and -2, as well as the forkhead factor FoxC3 [4], which are all regulated and under the control of TGFβ [198].
The role of TGFβ in EMT has been relatively well characterized. TGFβ induces reversible EMT in both normal and cancer contexts [199, 200]. Reciprocally, blocking TGFβ signaling by overexpression of a dominant-negative TβRII efficiently prevents skin squamous cancer cells from undergoing EMT in vivo [201]. Interestingly, the tumor cells located at the invasion front, which contain high levels of TGFβ, show enhanced EMT features. The canonical Smad pathway plays a central role in mediating the TGFβ-induced EMT effects. Smad-dependent activation of transcription leads to expression of the EMT regulatory factors, such as Snail, Slug, ZEB-2, and Twist through induction of the expression of high-mobility group A2 (HMGA2) protein [198], resulting in repression of E-cadherin expression [202] and dissociation of desmosomes [203]. Phosphorylation of the cell polarity protein Par6 by TβRII also leads to the dissolution of cell junction complexes [204]. While Smad-dependent TGFβ-induced EMT is enhanced by Ras signaling [205], other Smad-independent TGFβ downstream signaling pathways, including the Ras/PI3 K [206–208], RhoA [53], mTOR [56, 209], Erk MAPK [50, 210–212], and p38 stress-activated kinase [44] pathways also contribute to TGFβ-induced EMT.
MicroRNAs (small noncoding RNAs) also play an important role in the regulation and maintenance of EMT downstream of TGFβ [6, 213]. MicroRNAs (miRNAs) have eluded researchers for decades, stealthily regulating many of the major biological processes in eukaryotic cells. miRNAs regulate gene expression posttranscriptionally by guiding the RNA-induced silencing complex (RISC) to their cognate site of the 3′untranslated region (3′UTR) of the target mRNA. While miRNAs represent only 1% of all human genes, over a third of the transcriptome is regulated by these miRNAs [214]. Individual miRNAs can regulate hundreds of genes directly and thousands indirectly [215, 216]. By controlling and regulating the expression of so many genes, it clearly became apparent that miRNAs play a central and critical role in the pathogenesis of human diseases, including cancer [217–222]. Moreover, about half of the miRNA encoding genes are located in chromosomal regions that are being altered during tumorigenesis [223]. Numerous miRNA signatures have been characterized in human cancers and implicated in the tumorigenic process [6, 224–228]. While some miRNAs exert their effects as classical oncogenes or tumor suppressors [229], others act in the advanced stages of the disease by promoting cancer progression and tumor metastasis [230–233]. Many of these miRNAs regulate TGFβ-mediated tumor metastasis [6]. As an example, TGFβ represses expression of miR-200 leading to increased levels of the miR-200 target, ZEB2/SIP1 [213]. As ZEB2/SIP1 acts as the main repressor of E-Cadherin expression, TGFβ-mediated down-regulation of miR-200 leads to decreased E-cadherin levels and EMT in breast [234], pancreatic [235], and colorectal cancer [236]. In turn, ZEB2/SIP1 targets TGFβ and miR-200 transcription in a feedforward loop which stabilizes EMT [236].
By inducing EMT and modifying the cell phenotype, TGFβ alters cell-to-cell contact and communication as well as the adhesive, migratory and invasive properties of the tumor cells (Figure 4).
5.2.2. Cell Adhesion
TGFβ-induced EMT leads to changes in the expression profiles of adhesion molecules that profoundly diminish cell-to-cell and cell-to-substrate adhesion. These inhibitory effects on cell adhesion promote the detachment of the cancer cells from the primary tumor and their dissemination throughout the stroma. For instance, in the skin, melanocytes are tightly connected to keratinocytes through surface expression of E-cadherin. In melanoma, TGFβ-induced EMT leads to downregulation of E-cadherin and alters the communication between keratinocytes and melanocytes and further allows melanoma cells to attach and communicate with fibroblasts from the stroma and endothelial cells, thereby favoring their propagation throughout the derma. In osteosarcomas, TGFβ inhibits cell adhesion to the substrate laminin, by down-regulating expression of the laminin receptor, α 3 β 1 integrin [237]. Interestingly, TGFβ specifically inhibits cell interaction with laminin, as receptors for other substrates, such as collagen (α 2 β 1 integrin) and fibronectin (α 5 β 1 integrin) are not affected by TGFβ [237].
5.2.3. Cell Migration
A direct consequence of EMT is the acquisition of migratory and invasive properties by the cancer cells [238]. As mentioned above, TGFβ regulates the expression of several transcription factors such as HMGA2, Snail, Slug, and Twist during the EMT process. Expression of HMGA2, Snail or Twist alone can induce EMT and increase cell migration [238]. Expression of a dominant negative TβRII prevents TGFβ-induced EMT and blocks migration [239]. Overexpression of constitutively active TβRI restores cellular motility through the activation of PI3 K/Akt and MAPK pathways. In order to migrate, cells generate lamellipodia protrusions at the front end while retracting the trailing end. These events are coordinated by Rho-family GTPases which are themselves activated by TGFβ [240]. Although the induction of Rho signaling by TGFβ is not fully understood, it has recently been shown that RhoA activator is up-regulated by TGFβ in a Smad4-dependent manner [240]. While TGFβ exerts an important role in breast cancer progression as a pro-metastatic factor, notably through enhancement of cell migration, it is becoming clear that microRNAs also play a crucial role in the mediation of these effects [6]. For instance, miR-155 (which is regulated by TGFβ targets RhoA, thus directly contributing to epithelial plasticity [241]. Interestingly, we recently found TGFβ-mediated regulation of several microRNAs to be critical for TGFβ-induced cell migration [242, 243]. In particular, our results highlight a novel signaling route whereby TGFβ silences expression of the microRNA miR-584, further leading to actin re-arrangement and breast cancer cell migration [242]. TGFβ down-regulation of miR-584 in breast cancer cells leads to increased expression of its downstream target, the actin-binding protein PHACTR1, resulting in enhanced cellular migration (Figure 4). Accordingly, over-expressing miR-584 or knocking down expression of the target PHACTR1 resulted in a drastic reorganization of the actin cytoskeleton and impaired TGFβ-induced cell migration [242].
5.2.4. Cell Invasion
In addition to its promigratory role, TGFβ also contributes to the ECM remodeling and invasiveness of the cells by increasing the expression of metalloproteinases and the generation of plasmin which, in turn, contributes to the release of stored TGFβ from the ECM, further increasing cell invasion [244]. The increased TGFβ levels allow cancer cells to cross through the ECM to reach distant metastatic sites. In invasive hepatocellular carcinomas, TGFβ induces transcriptional expression of α3β1-integrin, a key player in basement membrane invasion [245, 246]. Moreover, TGFβ has been shown to inhibit expression of tissue inhibitor of metalloproteinase 3, TIMP3, further contributing to hepatocellular carcinoma cell invasion [247]. Though often associated together, TGFβ-induced EMT can be dissociated from TGFβ-induced invasion and metastasis. Indeed, using a skin carcinogenesis mouse model, it was found that TGFβ-mediated EMT requires a functional TGFβ type II receptor (TβRII), whereas TGFβ-mediated tumor invasion is associated with reduced TβRII signaling in tumor epithelia [248]. As outlined above, microRNAs are important regulators of the metastatic process. We and others have demonstrated the mir-181 family of microRNAs to be up-regulated by TGFβ and activin, a closely related TGFβ family member [243, 247, 249]. In hepatocellular carcinoma, TGFβ-induced miR-181 targets TIMP3 for degradation, thereby increasing invasiveness of the cells [247]. We also found miR-181 to be a downstream regulator of activin/TGFβ-induced cellular migration and invasion in breast cancer (Figure 4). As a critical regulator of tumor cell migration and invasion and breast cancer progression in vitro, miR-181 could potentially be an important therapeutic target [243].
Finally, a recent study from our laboratory identified p21Cip1 (p21), a member of the core cell cycle machinery, as a key regulator of TGFβ-mediated breast cancer cell migration and invasion [250] (Figure 4). We found p21 expression to correlate with poor overall and distant metastasis-free survival in breast cancer patients. Furthermore, using in vivo xenograft animal models, we found p21 to be essential for local tumor invasion [250]. p21 interacts with Smad3 and the acetyltransferase P/CAF to regulate Smad acetylation and transcriptional activity, as well as gene transcription of downstream TGFβ-induced pro-metastatic genes [250, 251]. Our data also showed a significant association between TGFβ/Smad3 signaling, p21, and P/CAF expression with lymph node positivity, making them potential useful prognosis markers for lymph node metastasis. Together these findings highlight an important role for the p21-P/CAF-Smad3 signaling axis in promoting breast cancer cell migration and invasion at the transcriptional level, and support the notion of a direct oncogenic role for p21 in the progression of breast cancer to a metastatic disease [250].
5.2.5. Contribution to Distant Metastasis and Chemoattraction
While TGFβ directly contributes to local invasion, this is only the first event in a multistep process which will eventually lead to the formation and establishment of secondary tumors [252]. As shown in Figure 4, TGFβ also contributes to the establishment of metastasis by contributing to the growth of these secondary lesions [4]. Tumor cells that have migrated through and invaded the matrix can penetrate blood vessels, through a mechanism called intravasation. Once in the circulation, tumor cells will then disseminate to distant sites and organs by exiting blood vessels (extravasation) and form new colonies in a new favorable distant microenvironment. TGFβ stimulates the secretion of osteolytic cytokines, which further contribute to the metastatic process by digesting the bone matrix [253]. These events are Smad-dependent as the formation of osteolytic lesions in mice by breast cancer, melanoma, and renal carcinoma cells can be blocked by overexpressing the inhibitory Smad7 or a dominant negative TGFβ receptor [254–256]. TGFβ stimulates the secretion of the parathyroid hormone related protein (PTHrP) which promotes the differentiation of osteoclast precursors and bone resorption [253] and induces expression of the bone homing receptor C-X-C chemokine receptor type 4 (CXCR4) [257]. Association of the stroma-derived factor-1 (SDF-1) ligand to its receptor CXCR4 promotes chemoattraction of the breast cancer cells to the bone secondary sites [4, 258]. TGFβ also induces the expression of the interleukin proteins IL-1, IL-6, IL-11, and connective tissue growth factor (CTGF), leading to osteoclastic differentiation and angiogenesis, and further contributing to bone resorption and the formation of osteolytic lesions [95, 158, 259–261]. TGFβ-mediated IL-11 and PTHrP by the cancer cells leads to increased expression of receptor activator of nuclear factor kappa-B ligand (RANKL) at the osteoclast cell surface, further enhancing progenitor cell differentiation into osteoclasts and bone demineralization [159, 160]. In addition to the development of bone osteolytic lesions, TGFβ also contributes to the development of metastases by directing cells to specific tissues and by enhancing the extravasation of breast cancer cells into the lung parenchyma [262], by inducing expression of cyclooxygenase-2 (COX2), epidermal growth factor receptor (EGFR), and angiopoietin-like 4 (ANGPTL4), and promoting development of lung metastasis [263]. Some of these genes (COX2 and EGFR) have also been associated with brain metastasis [264].
5.2.6. Antagonizing Suppressor of Metastasis Pathways in Breast Cancer
Mammary gland growth and differentiation is a complex process regulated by steroids, polypeptide hormones, and growth factors, among which TGFβ and the hormone prolactin play major roles. While prolactin is required for lobuloalveolar formation and functional differentiation of mammary epithelial cells, TGFβ exerts an opposite effect, inducing apoptosis during mammary gland involution and inhibiting milk protein expression [265, 266]. Prolactin signaling is mediated by the interaction of its specific receptor with the intracellular tyrosine kinase Jak2 [267–273]. Once phosphorylated, tyrosine residues on both Jak2 and the prolactin receptor create docking sites for the recruitment and activation of the transcription factor Stat5 which will then activate gene transcription of target genes such as those encoding milk proteins and cell growth regulators [274–276]. The importance of Stat5 in mammary gland development is further highlighted by the Stat5a knockout mouse which show no lobuloalveolar development during pregnancy and a complete absence of lactation [277]. Interestingly, an elegant study from the Ali laboratory, revealed prolactin to also act as a suppressor of metastasis in breast cancer [278]. Indeed, prolactin and Jak2 were shown to play a critical role in regulating epithelial-mesenchymal transition. Activation of the prolactin/Jak2 signaling pathway in mesenchymal-like breast cancer cells suppressed their mesenchymal properties and reduced their invasive behavior while blocking prolactin autocrine function in epithelial-like breast cancer cells induced mesenchymal-like phenotypic changes and enhanced their invasive capacity [278]. Interestingly, blocking prolactin signaling led to activation of the two major prometastatic pathways, the mitogen-activated protein kinase and the TGFβ/Smad signaling pathways, highlighting prolactin as a critical regulator of epithelial plasticity and defining a new role for prolactin as an invasion suppressor hormone in breast cancer [278]. TGFβ is expressed in each phase of postnatal mammary gland development [279] and all three isoforms of TGFβ are up-regulated during mammary gland involution [265, 280, 281]. TGFβ inhibits alveolar formation, milk protein synthesis and induces apoptosis during involution of the mammary gland [282–284], suggesting that TGFβ signaling may also antagonize prolactin-induced signals in mammary cells [285, 286]. In a recent study, we identified a novel antagonistic crosstalk mechanism by which TGFβ/Smad signaling inhibits prolactin signaling and Stat5-mediated gene transcription and mammary epithelial cell differentiation, by preventing Stat5 binding to its coactivator CBP [287]. These studies indicate that the prolactin and TGFβ signaling cascades oppose their effects not only to regulate differentiation of mammary epithelial cells and lactation but also to modulate tumor formation and breast cancer metastasis [278, 287].
6. Targeting TGFβ in Cancer Therapy
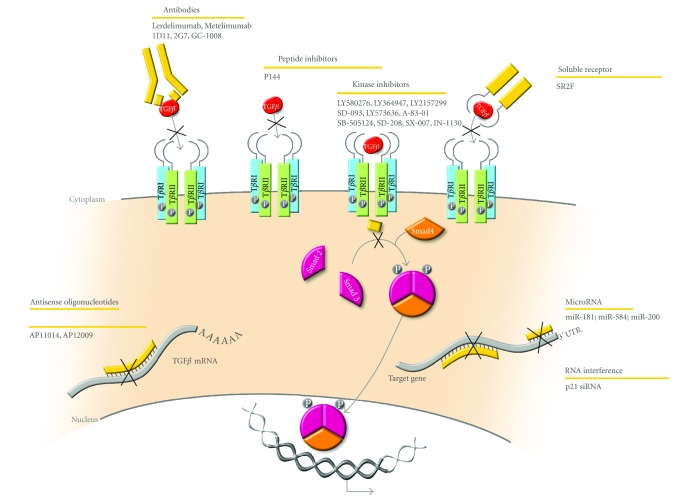
As outlined in this paper, TGFβ tumor suppressive effects are often lost in aggressive tumors, while tumor promoting and pro-invasive responses remain and prevail, leading to the development of distant metastases. Moreover, since TGFβ expression is increased in many cancers and correlates with the stage of the tumor [159, 160], blocking the TGFβ signaling pathway may provide for a unique therapeutic opportunity against tumor metastasis. As such, several approaches to develop new therapeutic tools that would interfere with the TGFβ pathway have been undertaken in recent years (Figure 5) [6, 159, 160, 288–291].
Figure 5.
TGFβ antagonists and inhibitors. Blocking the TGFβ signaling pathway provides for a unique therapeutic opportunity against tumor metastasis. As such, several approaches to develop new therapeutic tools that would interfere with TGFβ signaling have been undertaken in recent years. Blocking antibodies, peptide inhibitors, kinase inhibitors, soluble receptors, and antisense oligonucleotides are all being tested, some of which are at different phases of clinical trials. In future, more specific approaches may involve targeting specific microRNAs or making use of RNA interference approaches to block expression of specific downstream TGFβ pro-metastatic targets (e.g., p21).
6.1. Preventing Ligand-Receptor Interaction Using Blocking Monoclonal Antibodies, Soluble Receptors, and Peptide Inhibitors
Blocking antibodies against specific TGFβ isoforms, such as TGFβ-1 (Metelimumab or CAT-192; Cambridge Antibody Technology and Genzyme) [289] and TGFβ-2 (Lerdelimumab or CAT-152 or Trabio; Cambridge Antibody Technology) [292–294] have been developed and tested. However, both proved unsuccessful in clinical trials and were subsequently discontinued [295–298] (reviewed in [6]). As all TGFβ isoforms influence tumors, pan-TGFβ antibodies are also being developed, as they may prove more efficient than isoform-specific antibodies [6, 288, 291, 299–302]. Targeting TGFβ signaling with soluble receptors has also been investigated, using exogenous expression of a soluble TβRII [303, 304], soluble recombinant TβRIII (betaglycan) [305], decorin, a proteoglycan induced by TGFβ [306–308] or even a chimeric soluble receptor was constructed by fusing the extracellular domain of TβRII to the Fc regions of human immunoglobulin IgG1 (Fc:TβRII or SR2F) [307, 308]. Synthetic short peptides derived from TGFβ receptors that block TGFβ binding to its receptors are also an interesting avenue. One such promising candidate, P144 (DigNA Biotech), inhibits TGFβ signaling and collagen type I synthesis in cardiac fibroblasts and potentially prevents myocardial fibrosis in hypertensive rats [309].
6.2. Blocking TGFβ Production at the Translational Level, Using Antisense Oligonucleotides
Antisense oligonucleotides (ASO) are 13–25-nt single-stranded nucleic acids, chemically modified or not, that are complementary to the target mRNA [6, 310, 311]. The compound AP 11014, currently in advanced preclinical studies, specifically targets the TGFβ-1 mRNA and has been shown to significantly reduce TGFβ-1 secretion in multiple cancer cell lines, impeding TGF-β1-induced immunosuppression [6, 312, 313]. Another compound, known as AP 12009 (or Trabedersen; Antisense Pharma), was designed to target the TGFβ-2 mRNA and showed some efficacy in pancreatic cancer and glioblastoma [6, 302, 314–317].
6.3. Blocking the Downstream Receptor-Mediated Signaling Cascade, by Interfering with the Receptor Kinase Activity, Using Small-Molecule Inhibitors
The development of TGFβ receptor kinase inhibitors has primarily focused on TβRI, due to extensive knowledge of the effect of this receptor on Smad phosphorylation and since targeting TβRI would not disrupt potential TβRI-independent pathways initiated by TβRII [23, 296]. Multiple TβRI kinase inhibitor compounds have been developed and tested and showed various levels of efficacy (reviewed in [6]). Some were tested in xenograft models in breast and non-small cell lung cancers and showed tumor growth delay in vivo [318]. Oral administration of LY2157299 with advanced malignancies was determined to be safe and well tolerated in a phase I clinical trial [319]. Moreover, SD-093 strongly inhibits the TGFβ-induced motility and invasiveness of pancreatic carcinoma cells [320] and TGFβ-induced EMT in mammary epithelial cells [321, 322]. SD-208 and SX-007 efficiently inhibit TGFβ-induced migration and invasion and prolonged survival in murine glioma tumors [323, 324]. SB-431542, a selective inhibitor of Smad3 phosphorylation by TβRI, inhibits TGFβ-induced fibronectin and type I collagen synthesis in renal epithelial carcinoma cells [325]. LY2109761 inhibits both Smad-dependent and -independent TGFβ responses and attenuates TGFβ-induced cell migration, invasion, and tumorigenicity in colon adenocarcinoma [326] and decreases liver metastases and prolonged survival in a murine pancreatic cancer model [327].
6.4. Importance of the Specificity of Blocking TGFβ Signaling
One of the main concerns in targeted therapy is the off-target effects. Because TGFβ exerts a dual role in cancer, targeted therapy to block TGFβ signaling raises a major concern. Indeed, blocking the TGFβ pro-metastatic effects will only be beneficial if the therapy does not affect the tumor suppressive arm of TGFβ signaling. In a Neu-driven breast cancer model, constitutively active TβRI increased the latency of the primary mammary tumor but also increased pulmonary metastasis whereas a dominant negative TβRII decreased the latency of the primary tumor but also significantly decreased the number of lung metastases [262]. This finding indicates that although TGFβ acts as a tumor suppressor on the primary tumor, it may act on the ability of the breast cancer cells to extravasate from lung vessels to the parenchyma. Although loss of TβRII correlates with poor prognosis in esophageal cancer [328] and renal carcinoma [329], it also correlates with better survival rate in colon cancer [330] and gastric cancer [331], clearly indicating that the role of TβRII in carcinogenesis may be stage and tissue specific.
Overall it seems that the beneficial effects of TGFβ could be context dependent. These potential risks give rise to the necessity to identify patients in which the pro-metastatic arm of the TGFβ signaling pathway is predominant. Assessing the levels of TGFβ in the serum or the tumor has been studied as a tool for screening patients, showing a correlation between high serum levels and tumor progression and metastasis [302]. Many efforts have been made to target TGFβ in cancer due to its crucial role in cancer progression. Several strategies have been developed and some molecules have shown encouraging and promising results. However, these strategies still have very important challenges to overcome. Alternatively, new strategies aimed at specifically targeting the pro-metastatic arm of the TGFβ signaling pathway may prove more useful and safer. For this, identifying the downstream signaling components and elucidating the molecular mechanisms by which this growth factor promotes cell migration, invasion, EMT, and metastasis will be critical for establishing successful specific therapies. For instance, RNA interference approaches specifically targeting intracellular downstream molecules relaying these tumor promoting effects of TGFβ could prove useful. We recently found the cell cycle regulator, p21 to play a central role in the mediation of TGFβ-mediated local tumor cell invasion [250]. Using in vitro approaches and in vivo xenograft animal models, we found that blocking p21 expression with specific shRNAs and siRNAs could significantly alter the TGFβ tumor promoting effects, without affecting cell growth or tumor formation [250]. Thus, designing therapeutic strategies aiming at knocking down p21 expression in breast cancer patients may prove useful to prevent or circumvent the metastatic disease in this tissue (Figure 5).
Another important parameter to consider relates to the tissue specificity of the therapy. Indeed, while the TGFβ pro-metastatic effects have been relatively well characterized in breast cancer, the role of TGFβ in tumor cell invasion and metastasis in other tissues remain elusive. For instance, in melanoma, which is the leading cause of death due to cancer in young adults from 25 to 30 years old [332], the TGFβ effects on tumor progression remain unclear. While some reports suggested that TGFβ signaling could promote the metastatic potential of the cells [150, 217], other studies, including recent work from our laboratory, indicate that TGFβ potently inhibits cell migration and invasion of melanoma cells issued from various patients with different clinical backgrounds [333, 334]. Thus, in this particular context, clinical strategies aiming at mimicking the TGFβ antiproliferative, antimigratory and anti-invasive effects may prove beneficial for the treatment of melanomas at different stages of their progression, including primary and metastatic tumors.
7. Conclusion
TGFβ plays a major role in regulating cancer formation and progression. While acting as a tumor suppressor in normal cells and early carcinomas, TGFβ switches roles to in fact promote tumor progression in more advanced invasive cancers (Figure 6). Understanding and elucidating the intracellular and molecular mechanisms that trigger the TGFβ tumorigenic effects will thus be critical for the development of novel anticancer therapies based on the use of TGFβ antagonists. Combined research from academia and industry has led to the development of such new therapeutic tools, some of which have demonstrated promising results. Although the available TGFβ antagonists tested so far have shown some relative efficacy in different types of cancer, their use may also be limited. Indeed, even though several antagonists are currently being tested in clinical trials, their long-term efficiency and potential adverse side effects remain to be determined. In particular, it is difficult to predict whether blocking all TGFβ effects, as with the current strategies, will allow for sufficient blockage of the pro-metastatic arm of the TGFβ pathway without affecting the tumor suppressive arm, thereby giving rise to spontaneous tumors elsewhere in the organism. In that respect, it will be vitally important to focus efforts on the development of novel strategies aimed at specifically manipulating the downstream signaling components of the TGFβ tumor promoting effects, as they may prove more effective and safer in the long run.
Figure 6.
The dual role of TGFβ in human cancer. In summary, TGFβ acts as a tumor suppressor in normal cells and early carcinomas, while it promotes tumor metastasis in more advance stages of cancer. The tumor suppressive effects of TGFβ include cell cycle arrest, apoptosis and prevention of cell immortalization. TGFβ also induces EMT, marked by a decrease in cell-cell and cell-substrate contact, a reorganization of the actin cytoskeleton, as well as an increase in metalloproteinase synthesis and secretion. In human cancers, inactivating mutations in the TGFβ signaling components or activating mutations in oncogenic signaling pathways are often observed and provide an underlying basis for tumor development. These mutations attenuate the TGFβ tumor suppressive effects but do not affect its tumor promoting effects on cancer cells and on the surrounding environment, including EMT, cell migration and invasion, angiogenesis, immunosuppression, myofibroblast generation, chemoattraction, and tumor metastasis, further promoting TGFβ-induced tumor progression to secondary distant sites.
Acknowledgments
The author thanks Ms. Juliana Korah for critical reading of this paper. This work was supported by grants from the Canadian Institutes for Health Research. J.-J. Lebrun is the recipient of a McGill Sir William Dawson Research Chair.
References
- 1.Fabricant R. N., Todaro C. J. Nerve growth factor and malignant melanomas. New England Journal of Medicine. 1978;298(7):p. 402. doi: 10.1056/NEJM197802162980713. [DOI] [PubMed] [Google Scholar]
- 2.De Larco J. E., Todaro G. J. Growth factors from murine sarcoma virus-transformed cells. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(8):4001–4005. doi: 10.1073/pnas.75.8.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts A. B., Anzano M. A., Meyers C. A., et al. Purification and properties of a type β transforming growth factor from bovine kidney. Biochemistry. 1983;22(25):5692–5698. doi: 10.1021/bi00294a002. [DOI] [PubMed] [Google Scholar]
- 4.Massagué J. TGFβ in cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebrun J.-J. Activin, TGF-β and menin in pituitary tumorigenesis. Advances in Experimental Medicine and Biology. 2009;668:69–78. doi: 10.1007/978-1-4419-1664-8_7. [DOI] [PubMed] [Google Scholar]
- 6.Humbert L., Neel J. C., Lebrun J.-J. Targeting TGF-beta signaling in human cancer therapy. Trends in Cell & Molecular Biology. 2010;5:69–107. [Google Scholar]
- 7.Lebrun J.-J., Chen Y., Vale W. W. Activin and Follistatin, Regulatory Functions in System and Cell Biology. New York, NY, USA: Serono Symposia Publication, Springer; 1997. Receptor serine kinases and signaling by activins and inhibins; pp. 1–20. [Google Scholar]
- 8.Massagué J. TGF-β signal transduction. Annual Review of Biochemistry. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 9.Derynck R., Akhurst R. J., Balmain A. TGF-β signaling in tumor suppression and cancer progression. Nature Genetics. 2001;29(2):117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 10.Chin D., Boyle G. M., Parsons P. G., Coman W. B. What is transforming growth factor-beta (TGF-β)? British Journal of Plastic Surgery. 2004;57(3):215–221. doi: 10.1016/j.bjps.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Sun P. D., Davies D. R. The cystine-knot growth-factor superfamily. Annual Review of Biophysics and Biomolecular Structure. 1995;24:269–291. doi: 10.1146/annurev.bb.24.060195.001413. [DOI] [PubMed] [Google Scholar]
- 12.Miyazono K., Ichijo H., Heldin C. H. Transforming growth factor-β: latent forms, binding proteins and receptors. Growth Factors. 1993;8(1):11–22. doi: 10.3109/08977199309029130. [DOI] [PubMed] [Google Scholar]
- 13.Koli K., Saharinen J., Hyytiäinen M., Penttinen C., Keski-Oja J. Latency, activation, and binding proteins of TGF-β . Microscopy Research and Technique. 2001;52(4):354–362. doi: 10.1002/1097-0029(20010215)52:4<354::AID-JEMT1020>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 14.Khalil N. TGF-β: from latent to active. Microbes and Infection. 1999;1(15):1255–1263. doi: 10.1016/S1286-4579(99)00259-2. [DOI] [PubMed] [Google Scholar]
- 15.Massague J., Czech M. P., Iwata K., DeLarco J. E., Todaro G. J. Affinity labeling of a transforming growth factor receptor that does not interact with epidermal growth factor. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(22 I):6822–6826. doi: 10.1073/pnas.79.22.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massague J., Like B. Cellular receptors for type β transforming growth factor. Ligand binding and affinity labeling in human and rodent cell lines. Journal of Biological Chemistry. 1985;260(5):2636–2645. [PubMed] [Google Scholar]
- 17.Massague J. The transforming growth factor-β family. Annual Review of Cell Biology. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y., Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 19.Massagué J., Blain S. W., Lo R. S. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 20.Wakefield L. M., Roberts A. B. TGF-β signaling: positive and negative effects on tumorigenesis. Current Opinion in Genetics and Development. 2002;12(1):22–29. doi: 10.1016/S0959-437X(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 21.Xu J., Lamouille S., Derynck R. TGF-β-induced epithelial to mesenchymal transition. Cell Research. 2009;19(2):156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Their J. P. Epithelial-mesenchymal transitions in tumor progression. Nature Reviews Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 23.Yingling J. M., Blanchard K. L., Sawyer J. S. Development of TGF-β signalling inhibitors for cancer therapy. Nature Reviews Drug Discovery. 2004;3(12):1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- 24.Siegel P. M., Massagué J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nature Reviews Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 25.Dumont N., Arteaga C. L. Targeting the TGFβ signaling network in human neoplasia. Cancer Cell. 2003;3(6):531–536. doi: 10.1016/S1535-6108(03)00135-1. [DOI] [PubMed] [Google Scholar]
- 26.Gorsch S. M., Memoli V. A., Stukel T. A., Gold L. I., Arrick B. A. Immunohistochemical staining for transforming growth factor β1 associates with disease progression in human breast cancer. Cancer Research. 1992;52(24):6949–6952. [PubMed] [Google Scholar]
- 27.Lebrun J.-J., Vale W. W. Activin and inhibin have antagonistic effects on ligand-dependent heteromerization of the type I and type II activin receptors and human erythroid differentiation. Molecular and Cellular Biology. 1997;17(3):1682–1691. doi: 10.1128/mcb.17.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massagué J., Chen Y. G. Controlling TGF-β signaling. Genes and Development. 2000;14(6):627–644. [PubMed] [Google Scholar]
- 29.Mehra A., Wrana J. L. TGF-β and the Smad signal transduction pathway. Biochemistry and Cell Biology. 2002;80(5):605–622. doi: 10.1139/o02-161. [DOI] [PubMed] [Google Scholar]
- 30.Moustakas A., Heldin C. H. Non-Smad TGF-β signals. Journal of Cell Science. 2005;118(16):3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 31.Jayaraman L., Massagué J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-β pathway. Journal of Biological Chemistry. 2000;275(52):40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- 32.Chacko B. M., Qin B., Correia J. J., Lam S. S., De Caestecker M. P., Lin K. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nature Structural Biology. 2001;8(3):248–253. doi: 10.1038/84995. [DOI] [PubMed] [Google Scholar]
- 33.Chacko B. M., Qin B. Y., Tiwari A., et al. Structural basis of heteromeric Smad protein assembly in TGF-β signaling. Molecular Cell. 2004;15(5):813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Lebrun J.-J., Takabe K., Chen Y., Vale W. Roles of pathway-specific and inhibitory Smads in activin receptor signaling. Molecular Endocrinology. 1999;13(1):15–23. doi: 10.1210/me.13.1.15. [DOI] [PubMed] [Google Scholar]
- 35.Xu L., Chen Y. G., Massagué J. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFβ-dependent phosphorylation. Nature Cell Biology. 2000;2(8):559–562. doi: 10.1038/35019649. [DOI] [PubMed] [Google Scholar]
- 36.Reguly T., Wrana J. L. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends in Cell Biology. 2003;13(5):216–220. doi: 10.1016/S0962-8924(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 37.Xu L., Massagué J. Nucleocytoplasmic shuttling of signal transducers. Nature Reviews Molecular Cell Biology. 2004;5(3):209–219. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y., Wang Y. F., Jayaraman L., Yang H., Massagué J., Pavletich N. P. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell. 1998;94(5):585–594. doi: 10.1016/S0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 39.Massagué J., Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO Journal. 2000;19(8):1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Weisberg E., Fridmacher V., Watanabe M., Naco G., Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389(6646):85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 41.Hata A., Seoane J., Lagna G., Montalvo E., Hemmati-Brivanlou A., Massagué J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP- Smad and Olf signaling pathways. Cell. 2000;100(2):229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 42.Feng X. H., Derynck R. Specificity and versatility in TGF-β signaling through smads. Annual Review of Cell and Developmental Biology. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 43.Massagué J., Seoane J., Wotton D. Smad transcription factors. Genes and Development. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 44.Bakin A. V., Rinehart C., Tomlinson A. K., Arteaga C. L. p38 mitogen-activated protein kinase is required for TGFβ-mediated fibroblastic transdifferentiation and cell migration. Journal of Cell Science. 2002;115(15):3193–3206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 45.Yan Z., Winawer S., Friedman E. Two different signal transduction pathways can be activated by transforming growth factor β1 in epithelial cells. Journal of Biological Chemistry. 1994;269(18):13231–13237. [PubMed] [Google Scholar]
- 46.Hanafusa H., Ninomiya-Tsuji J., Masuyama N., et al. Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-β-induced gene expression. Journal of Biological Chemistry. 1999;274(38):27161–27167. doi: 10.1074/jbc.274.38.27161. [DOI] [PubMed] [Google Scholar]
- 47.Cocolakis E., Lemay S., Ali S., Lebrun J.-J. The p38 MAPK pathway is required for cell growth inhibition of human breast cancer cells in response to activin. Journal of Biological Chemistry. 2001;276(21):18430–18436. doi: 10.1074/jbc.M010768200. [DOI] [PubMed] [Google Scholar]
- 48.Lacerte A., Korah J., Roy M., Yang X. J., Lemay S., Lebrun J.-J. Transforming growth factor-β inhibits telomerase through SMAD3 and E2F transcription factors. Cellular Signalling. 2008;20(1):50–59. doi: 10.1016/j.cellsig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 49.De Guise C., Lacerte A., Rafiei S., et al. Activin inhibits the human Pit-1 gene promoter through the p38 kinase pathway in a smad-independent manner. Endocrinology. 2006;147(9):4351–4362. doi: 10.1210/en.2006-0444. [DOI] [PubMed] [Google Scholar]
- 50.Davies M., Robinson M., Smith E., Huntley S., Prime S., Paterson I. Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-β1 involves MAPK, Smad and AP-1 signalling pathways. Journal of Cellular Biochemistry. 2005;95(5):918–931. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 51.Lee M. K., Pardoux C., Hall M. C., et al. TGF-β activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO Journal. 2007;26(17):3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edlund S., Landström M., Heldin C. H., Aspenström P. Transforming growth factor-β-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Molecular Biology of the Cell. 2002;13(3):902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhowmick N. A., Ghiassi M., Bakin A., et al. Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Molecular Biology of the Cell. 2001;12(1):27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen R. H., Su Y. H., Chuang R. L. C., Chang T. Y. Suppression of transforming growth factor-β-induced apoptosis through a phosphatidylinositol 3-kinase/Akt-dependent pathway. Oncogene. 1998;17(15):1959–1968. doi: 10.1038/sj.onc.1202111. [DOI] [PubMed] [Google Scholar]
- 55.Péron P., Rahmani M., Zagar Y., Durand-Schneider A. M., Lardeux B., Bernuau D. Potentiation of Smad transactivation by Jun proteins during a combined treatment with epidermal growth factor and transforming growth factor-β in rat hepatocytes: role of phosphatidylinositol 3-kinase-induced AP-1 activation. Journal of Biological Chemistry. 2001;276(13):10524–10531. doi: 10.1074/jbc.M005919200. [DOI] [PubMed] [Google Scholar]
- 56.Lamouille S., Derynck R. Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. Journal of Cell Biology. 2007;178(3):437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi H., Abdollah S., Qiu Y., et al. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89(7):1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 58.Nakao A., Afrakhte M., Morén A., et al. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature. 1997;389(6651):631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 59.Shi W., Sun C., He B., et al. GADD34-PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. Journal of Cell Biology. 2004;164(2):291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kavsak P., Rasmussen R. K., Causing C. G., et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGFβ receptor for degradation. Molecular Cell. 2000;6(6):1365–1375. doi: 10.1016/S1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki C., Murakami G., Fukuchi M., et al. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. Journal of Biological Chemistry. 2002;277(42):39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- 62.Tajima Y., Goto K., Yoshida M., et al. Chromosomal region maintenance 1 (CRM1)-dependent nuclear export of Smad ubiquitin regulatory factor 1 (Smurf1) is essential for negative regulation of transforming growth factor-β signaling by Smad7. Journal of Biological Chemistry. 2003;278(12):10716–10721. doi: 10.1074/jbc.M212663200. [DOI] [PubMed] [Google Scholar]
- 63.Di Guglielmo G. M., Le Roy C., Goodfellow A. F., Wrana J. L. Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nature Cell Biology. 2003;5(5):410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 64.Chen W., Kirkbride K. C., How T., et al. β-arrestin 2 mediates endocytosis of type III TGF-β receptor and down-regulation of its signaling. Science. 2003;301(5638):1394–1397. doi: 10.1126/science.1083195. [DOI] [PubMed] [Google Scholar]
- 65.Yao D., Ehrlich M., Henis Y. I., Leof E. B. Transforming growth factor-β receptors interact with AP2 by direct binding to β2 subunit. Molecular Biology of the Cell. 2002;13(11):4001–4012. doi: 10.1091/mbc.02-07-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lo R. S., Massague J. Ubiquitin-dependent degradation of TGF-beta-activated smad2. Nature Cell Biology. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- 67.Lin X., Duan X., Liang Y. Y., et al. PPM1A functions as a smad phosphatase to terminate TGFβ signaling. Cell. 2006;125(5):915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu L., Kang Y., Çöl S., Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Molecular Cell. 2002;10(2):271–282. doi: 10.1016/S1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- 69.Inman G. J., Nicolás F. J., Hill C. S. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Molecular Cell. 2002;10(2):283–294. doi: 10.1016/S1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 70.Kretzschmar M., Doody J., Massagué J. Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature. 1997;389(6651):618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 71.Kretzschmar M., Doody J., Timokhina I., Massagué J. A mechanism of repression of TGFfβ/Smad signaling by oncogenic Ras. Genes and Development. 1999;13(7):804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagata H., Hatano E., Tada M., et al. Inhibition of c-Jun NH2-terminal kinase switches Smad3 signaling from oncogenesis to tumor-suppression in rat hepatocellular carcinoma. Hepatology. 2009;49(6):1944–1953. doi: 10.1002/hep.22860. [DOI] [PubMed] [Google Scholar]
- 73.Kretzschmar M., Liu F., Hata A., Doody J., Massagué J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes and Development. 1997;11(8):984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 74.Wicks S. J., Stephen L., Abdel-Wahab N., Mason R. M., Chantry A. Inactivation of Smad-transforming growth factor β signaling by Ca2+-calmodulin-dependent protein kinase II. Molecular and Cellular Biology. 2000;20(21):8103–8111. doi: 10.1128/MCB.20.21.8103-8111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 76.Waddell D. S., Liberati N. T., Guo X., Frederick J. P., Wang Y. F. Casein kinase Iε plays a functional role in the transforming growth factor-β signaling pathway. Journal of Biological Chemistry. 2004;279(28):29236–29246. doi: 10.1074/jbc.M400880200. [DOI] [PubMed] [Google Scholar]
- 77.Yakymovych I., Engström U., Grimsby S., Heldin C. H., Souchelnytskyi S. Inhibition of transforming growth factor-β signaling by low molecular weight compounds interfering with ATP- or substrate-binding sites of the TGFβ type I receptor kinase. Biochemistry. 2002;41(36):11000–11007. doi: 10.1021/bi025936u. [DOI] [PubMed] [Google Scholar]
- 78.Ho J., Cocolakis E., Dumas V. M., Posner B. I., Laporte S. A., Lebrun J.-J. The G protein-coupled receptor kinase-2 is a TGFβ-inducible antagonist of TGFβ signal transduction. EMBO Journal. 2005;24(18):3247–3258. doi: 10.1038/sj.emboj.7600794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo J., Chen H., Ho J., et al. TGFβ-induced GRK2 expression attenuates AngII-regulated vascular smooth muscle cell proliferation and migration. Cellular Signalling. 2009;21(6):899–905. doi: 10.1016/j.cellsig.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 80.Ho J., Chen H., Lebrun J.-J. Novel dominant negative Smad antagonists to TGFβ signaling. Cellular Signalling. 2007;19(7):1565–1574. doi: 10.1016/j.cellsig.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 81.Sun L., Wu G., Willson J. K. V., et al. Expression of transforming growth factor β type II receptor leads to reduced malignancy in human breast cancer MCF-7 cells. Journal of Biological Chemistry. 1994;269(42):26449–26455. [PubMed] [Google Scholar]
- 82.Wang J., Sun L., Myeroff L., et al. Demonstration that mutation of the type II transforming growth factor β receptor inactivates its tumor suppressor activity in replication error- positive colon carcinoma cells. Journal of Biological Chemistry. 1995;270(37):22044–22049. doi: 10.1074/jbc.270.37.22044. [DOI] [PubMed] [Google Scholar]
- 83.Go C., Li P., Wang X. J. Blocking transforming growth factor β signaling in transgenic epidermis accelerates chemical carcinogenesis: a mechanism associated with increased angiogenesis. Cancer Research. 1999;59(12):2861–2868. [PubMed] [Google Scholar]
- 84.Böttinger E. P., Jakubczak J. L., Haines D. C., Bagnall K., Wakefield L. M. Transgenic mice overexpressing a dominant-negative mutant type II transforming growth factor β receptor show enhanced tumorigenesis in the mammary gland and lung in response to the carcinogen 7,12-dimethylbenz-[a]- anthracene. Cancer Research. 1997;57(24):5564–5570. [PubMed] [Google Scholar]
- 85.Kim S. J., Im Y. H., Markowitz S. D., Bang Y. J. Molecular mechanisms of inactivation of TGF-β receptors during carcinogenesis. Cytokine and Growth Factor Reviews. 2000;11(1-2):159–168. doi: 10.1016/S1359-6101(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 86.Moses H. L., Yang E. Y., Pietenpol J. A. TGF-β stimulation and inhibition of cell proliferation: new mechanistic insights. Cell. 1990;63(2):245–247. doi: 10.1016/0092-8674(90)90155-8. [DOI] [PubMed] [Google Scholar]
- 87.Li J. M., Nichols M. A., Chandrasekharan S., Xiong Y., Wang X. F. Transforming growth factor β activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. Journal of Biological Chemistry. 1995;270(45):26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- 88.Datto M. B., Li Y., Panus J. F., Howe D. J., Xiong Y., Wang X. F. Transforming growth factor β induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(12):5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seoane J., Le H. V., Shen L., Anderson S. A., Massagué J. Integration of smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117(2):211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 90.Pardali K., Kurisaki A., Morén A., Ten Dijke P., Kardassis D., Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21Waf1/Cip1 regulation by transforming growth factor-β . Journal of Biological Chemistry. 2000;275(38):29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- 91.Ho J., De Guise C., Kim C., Lemay S., Wang X. F., Lebrun J.-J. Activin induces hepatocyte cell growth arrest through induction of the cyclin-dependent kinase inhibitor p15INK4B and Sp1. Cellular Signalling. 2004;16(6):693–701. doi: 10.1016/j.cellsig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 92.Reynisdottir I., Polyak K., Iavarone A., Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β . Genes and Development. 1995;9(15):1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 93.Sandhu C., Garbe J., Bhattacharya N., et al. Transforming growth factor β stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes, and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Molecular and Cellular Biology. 1997;17(5):2458–2467. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coffey R. J., Bascom C. C., Sipes N. J., Graves-Deal R., Weissman B. E., Moses H. L. Selective inhibition of growth-related gene expression in murine keratinocytes by transforming growth factor β . Molecular and Cellular Biology. 1988;8(8):3088–3093. doi: 10.1128/mcb.8.8.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang Y., Chen C. R., Massagué J. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Molecular Cell. 2003;11(4):915–926. doi: 10.1016/S1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 96.Lasorella A., Noseda M., Beyna M., Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407(6804):592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 97.Kowanetz M., Valcourt U., Bergström R., Heldin C. H., Moustakas A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor β and bone morphogenetic protein. Molecular and Cellular Biology. 2004;24(10):4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Norton J. D. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. Journal of Cell Science. 2000;113(part 22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 99.Chen C. R., Kang Y., Siegel P. M., Massagué J. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell. 2002;110(1):19–32. doi: 10.1016/S0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 100.Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., Massagué J. TGFβ influences myc, miz-1 and smad to control the CDK inhibitor p15INK4b . Nature Cell Biology. 2001;3(4):400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 101.Staller P., Peukert K., Kiermaier A., et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nature Cell Biology. 2001;3(4):392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 102.Ma C.-I. J., Martin C., Ma Z., et al. Engulfment protein GULP is regulator of transforming growth factor-β response in ovarian cells. Journal of Biological Chemistry. 2012;287(24):20636–20651. doi: 10.1074/jbc.M111.314997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iavarone A., Massagué J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature. 1997;387(6631):417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 104.Kaji H., Canaff L., Lebrun J.-J., Goltzman D., Hendy G. N. Inactivation of menin, a Smad3-interacting protein, blocks transforming growth factor type β signaling. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(7):3837–3842. doi: 10.1073/pnas.061358098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hendy C. N., Kaji H., Sowa H., Lebrun J.-J., Canaff L. Menin and TGF-β superfamily member signaling via the Smad pathway in pituitary, parathyroid and osteoblast. Hormone and Metabolic Research. 2005;37(6):375–379. doi: 10.1055/s-2005-870152. [DOI] [PubMed] [Google Scholar]
- 106.Ben-Jonathan N., Hnasko R. Dopamine as a prolactin (PRL) inhibitor. Endocrine Reviews. 2001;22(6):724–763. doi: 10.1210/er.22.6.724. [DOI] [PubMed] [Google Scholar]
- 107.Heaney A. P., Melmed S. Molecular targets in pituitary tumours. Nature Reviews Cancer. 2004;4(4):285–295. doi: 10.1038/nrc1320. [DOI] [PubMed] [Google Scholar]
- 108.Asa S. L., Ezzat S. The cytogenesis and pathogenesis of pituitary adenomas. Endocrine Reviews. 1998;19(6):798–827. doi: 10.1210/edrv.19.6.0350. [DOI] [PubMed] [Google Scholar]
- 109.Guo S. S., Sawicki M. P. Molecular and genetic mechanisms of tumorigenesis in multiple endocrine neoplasia type-1. Molecular Endocrinology. 2001;15(10):1653–1664. doi: 10.1210/me.15.10.1653. [DOI] [PubMed] [Google Scholar]
- 110.Lacerte A., Lee E. H., Reynaud R., et al. Activin inhibits pituitary prolactin expression and cell growth through Smads, Pit-1 and menin. Molecular Endocrinology. 2004;18(6):1558–1569. doi: 10.1210/me.2003-0470. [DOI] [PubMed] [Google Scholar]
- 111.Oberhammer F. A., Pavelka M., Sharma S., et al. Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor β1. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(12):5408–5412. doi: 10.1073/pnas.89.12.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chaouchi N., Arvanitakis L., Auffredou M. T., Blanchard D. A., Vazquez A., Sharma S. Characterization of transforming growth factor-β1 induced apoptosis in normal human B cells and lymphoma B cell lines. Oncogene. 1995;11(8):1615–1622. [PubMed] [Google Scholar]
- 113.Valderrama-Carvajal H., Cocolakis E., Lacerte A., et al. Activin/TGF-β induce apoptosis through Smad-dependent expression of the lipid phosphatase SHIP. Nature Cell Biology. 2002;4(12):963–969. doi: 10.1038/ncb885. [DOI] [PubMed] [Google Scholar]
- 114.Yanagisawa K., Osada H., Masuda A., et al. Induction of apoptosis by Smad3 and down-regulation of Smad3 expression in response to TGF-β in human normal lung epithelial cells. Oncogene. 1998;17(13):1743–1747. doi: 10.1038/sj.onc.1202052. [DOI] [PubMed] [Google Scholar]
- 115.Rotello R. J., Lieberman R. C., Purchio A. F., Gerschenson L. E. Coordinated regulation of apoptosis and cell proliferation by transforming growth factor β1 in cultured uterine epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(8):3412–3415. doi: 10.1073/pnas.88.8.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schuster N., Krieglstein K. Mechanisms of TGF-β-mediated apoptosis. Cell and Tissue Research. 2002;307(1):1–14. doi: 10.1007/s00441-001-0479-6. [DOI] [PubMed] [Google Scholar]
- 117.Perlman R., Schiemann W. P., Brooks M. W., Lodish H. F., Weinberg R. A. TGF-β-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nature Cell Biology. 2001;3(8):708–714. doi: 10.1038/35087019. [DOI] [PubMed] [Google Scholar]
- 118.Jang C. W., Chen C. H., Chen C. C., Chen J. Y., Su Y. H., Chen R. H. TGF-β induces apoptosis through Smad-mediated expression of DAP-kinase. Nature Cell Biology. 2002;4(1):51–58. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 119.Tachibana I., Imoto M., Adjei P. N., et al. Overexpression of the TGFβ-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. Journal of Clinical Investigation. 1997;99(10):2365–2374. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Larisch S., Yi Y., Lotan R., et al. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nature Cell Biology. 2000;2(12):915–921. doi: 10.1038/35046566. [DOI] [PubMed] [Google Scholar]
- 121.Atfi A., Buisine M., Mazars A., Gespach C. Induction of apoptosis by DPC4, a transcriptional factor regulated by transforming growth factor-β through stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) signaling pathway. Journal of Biological Chemistry. 1997;272(40):24731–24734. doi: 10.1074/jbc.272.40.24731. [DOI] [PubMed] [Google Scholar]
- 122.Yamamura Y., Hua X., Bergelson S., Lodish H. F. Critical role of Smads and AP-1 complex in transforming growth factor-β-dependent apoptosis. Journal of Biological Chemistry. 2000;275(46):36295–36302. doi: 10.1074/jbc.M006023200. [DOI] [PubMed] [Google Scholar]
- 123.Ohgushi M., Kuroki S., Fukamachi H., et al. Transforming growth factor β-dependent sequential activation of Smad, Bim, and caspase-9 mediates physiological apoptosis in gastric epithelial cells. Molecular and Cellular Biology. 2005;25(22):10017–10028. doi: 10.1128/MCB.25.22.10017-10028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wildey G. M., Patil S., Howe P. H. Smad3 potentiates transforming growth factor β (TGFβ)-induced apoptosis and expression of the BH3-only protein Bim in WEHI 231 B lymphocytes. Journal of Biological Chemistry. 2003;278(20):18069–18077. doi: 10.1074/jbc.M211958200. [DOI] [PubMed] [Google Scholar]
- 125.Saltzman A., Munro R., Searfoss G., Franks C., Jaye M., Ivashchenko Y. Transforming growth factor-β-mediated apoptosis in the Ramos B-lymphoma cell line is accompanied by caspase activation and Bcl-X(L) downregulation. Experimental Cell Research. 1998;242(1):244–254. doi: 10.1006/excr.1998.4096. [DOI] [PubMed] [Google Scholar]
- 126.Francis J. M., Heyworth C. M., Spooncer E., Pierce A., Dexter T. M., Whetton A. D. Transforming growth factor-β1 induces apoptosis independently of p53 and selectively reduces expression of Bcl-2 in multipotent hematopoietic cells. Journal of Biological Chemistry. 2000;275(50):39137–39145. doi: 10.1074/jbc.M007212200. [DOI] [PubMed] [Google Scholar]
- 127.Chipuk J. E., Bhat M., Hsing A. Y., Ma J., Danielpour D. Bcl-xL Blocks transforming growth factor-β1-induced apoptosis by inhibiting cytochrome c release and not by directly antagonizing Apaf-1-dependent caspase activation in prostate epithelial cells. Journal of Biological Chemistry. 2001;276(28):26614–26621. doi: 10.1074/jbc.M100913200. [DOI] [PubMed] [Google Scholar]
- 128.Kanamaru C., Yasuda H., Fujita T. Involvement of Smad proteins in TGF-β and activin A-induced apoptosis and growth inhibition of liver cells. Hepatology Research. 2003;23(3):211–219. doi: 10.1016/S1386-6346(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 129.Wang J., Yang L., Yang J., et al. Transforming growth factor β induces apoptosis through repressing the phosphoinositide 3-kinase/AKT/survivin pathway in colon cancer cells. Cancer Research. 2008;68(9):3152–3160. doi: 10.1158/0008-5472.CAN-07-5348. [DOI] [PubMed] [Google Scholar]
- 130.Conery A. R., Cao Y., Thompson E. A., Townsend C. M., Ko T. C., Luo K. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-β-induced apoptosis. Nature Cell Biology. 2004;6(4):366–372. doi: 10.1038/ncb1117. [DOI] [PubMed] [Google Scholar]
- 131.Remy I., Montmarquette A., Michnick S. W. PKB/Akt modulates TGF-β signalling through a direct interaction with Smad3. Nature Cell Biology. 2004;6(4):358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- 132.Yang J., Song K., Krebs T. L., Jackson M. W., Danielpour D. Rb/E2F4 and Smad2/3 link survivin to TGF-β-induced apoptosis and tumor progression. Oncogene. 2008;27(40):5326–5338. doi: 10.1038/onc.2008.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Korah J., Falah N., Lacerte A., Lebrun J.-J. A transcriptionally active pRb-E2F1-P/CAF signaling pathway is central to TGFβ-mediated apoptosis. Cell Death and Disease Cell Death and Disease. 2012;3(article e407) doi: 10.1038/cddis.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shay J. W., Wright W. E. Hayflick, his limit, and cellular ageing. Nature Reviews Molecular Cell Biology. 2000;1(1):72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- 135.Rama S., Suresh Y., Rao A. J. Regulation of telomerase during human placental differentiation: a role for TGFβ1. Molecular and Cellular Endocrinology. 2001;182(2):233–248. doi: 10.1016/s0303-7207(01)00550-0. [DOI] [PubMed] [Google Scholar]
- 136.Yang H., Kyo S., Takatura M., Sun L. Autocrine transforming growth factor β suppresses telomerase activity and transcription of human telomerase reverse transcriptase in human cancer cells. Cell Growth and Differentiation. 2001;12(2):119–127. [PubMed] [Google Scholar]
- 137.Blobe G. C., Schiemann W. P., Lodish H. F. Role of transforming growth factor β in human disease. New England Journal of Medicine. 2000;342(18):1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 138.Levy L., Hill C. S. Alterations in components of the TGF-β superfamily signaling pathways in human cancer. Cytokine and Growth Factor Reviews. 2006;17(1-2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 139.Hata Phd A., Massagué Phd J., Shi Phd Y. TGF-β signaling and cancer: structural and functional consequences of mutations in Smads. Molecular Medicine Today. 1998;4(6):257–262. doi: 10.1016/S1357-4310(98)01247-7. [DOI] [PubMed] [Google Scholar]
- 140.Maliekal T. T., Antony M. L., Nair A., Paulmurugan R., Karunagaran D. Loss of expression, and mutations of Smad 2 and Smad 4 in human cervical cancer. Oncogene. 2003;22(31):4889–4897. doi: 10.1038/sj.onc.1206806. [DOI] [PubMed] [Google Scholar]
- 141.Hahn S. A., Schutte M., Shamsul Hoque A. T. M., et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271(5247):350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 142.Sjöblom T., Jones S., Wood L. D., et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 143.Wolfraim L. A., Fernandez T. M., Mamura M., et al. Loss of Smad3 in acute T-cell lymphoblastic leukemia. New England Journal of Medicine. 2004;351(6):552–559. doi: 10.1056/NEJMoa031197. [DOI] [PubMed] [Google Scholar]
- 144.Zhu Q., Krakowski A. R., Dunham E. E., et al. Dual role of SnoN in mammalian tumorigenesis. Molecular and Cellular Biology. 2007;27(1):324–339. doi: 10.1128/MCB.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kleeff J., Ishiwata T., Maruyama H., et al. The TGF-β signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene. 1999;18(39):5363–5372. doi: 10.1038/sj.onc.1202909. [DOI] [PubMed] [Google Scholar]
- 146.Dowdy S. C., Mariani A., Reinholz M. M., et al. Overexpression of the TGF-β antagonist Smad7 in endometrial cancer. Gynecologic Oncology. 2005;96(2):368–373. doi: 10.1016/j.ygyno.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 147.Cerutti J. M., Ebina K. N., Matsuo S. E., Martins L., Maciel R. M. B., Kimura E. T. Expression of Smad4 and Smad7 in human thyroid follicular carcinoma cell lines. Journal of Endocrinological Investigation. 2003;26(6):516–521. doi: 10.1007/BF03345213. [DOI] [PubMed] [Google Scholar]
- 148.Bruna A., Darken R. S., Rojo F., et al. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 149.Hannigan A., Smith P., Kalna G., et al. Epigenetic downregulation of human disabled homolog 2 switches TGF-β from a tumor suppressor to a tumor promoter. Journal of Clinical Investigation. 2010;120(8):2842–2857. doi: 10.1172/JCI36125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Micalizzi D. S., Christensen K. L., Jedlicka P., et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-β signaling. Journal of Clinical Investigation. 2009;119(9):2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Akhurst R. J., Derynck R. TGF-β signaling in cancer—a double-edged sword. Trends in Cell Biology. 2001;11(11):S44–S51. doi: 10.1016/S0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 152.Compton C. C., Byrd D. R., Garcia-Aguilar J., Kurtzman S., Olawaiye A., Washington M. K., editors. AJCC Staging Manual. 7th. Springer; 2009. [Google Scholar]
- 153.Arslan C., Altundag K., Dizdar O. Emerging drugs in metastatic breast cancer: an update. Expert Opinion on Emerging Drugs. 2011;16(4):647–667. doi: 10.1517/14728214.2011.640672. [DOI] [PubMed] [Google Scholar]
- 154.Tian F., DaCosta Byfield S., Parks W. T., et al. Smad-binding defective mutant of transforming growth factor β type I receptor enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Research. 2004;64(13):4523–4530. doi: 10.1158/0008-5472.CAN-04-0030. [DOI] [PubMed] [Google Scholar]
- 155.Tian F., Byfield S. D., Parks W. T., et al. Reduction in Smad2/3 signaling enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Research. 2003;63(23):8284–8292. [PubMed] [Google Scholar]
- 156.Azuma H., Ehata S., Miyazaki H., et al. Effect of Smad7 expression on metastasis of mouse mammary carcinoma JygMC(A) cells. Journal of the National Cancer Institute. 2005;97(23):1734–1746. doi: 10.1093/jnci/dji399. [DOI] [PubMed] [Google Scholar]
- 157.Deckers M., Van Dinther M., Buijs J., et al. The tumor suppressor Smad4 is required for transforming growth factor β-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Research. 2006;66(4):2202–2209. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 158.Kang Y., He W., Tulley S., et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Derynck R., Goeddel D. V., Ullrich A. Synthesis of messenger RNAs for transforming growth factors α and β and the epidermal growth factor receptor by human tumors. Cancer Research. 1987;47(3):707–712. [PubMed] [Google Scholar]
- 160.Dickson R. B., Kasid A., Huff K. K. Activation of growth factor secretion in tumorigenic states of breast cancer induced by 17β-estradiol or v-Ha-ras oncogene. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(3):837–841. doi: 10.1073/pnas.84.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dalal B. I., Keown P. A., Greenberg A. H. Immunocytochemical localization of secreted transforming growth factor- β1 to the advancing edges of primary tumors and to lymph node metastases of human mammary carcinoma. American Journal of Pathology. 1993;143(2):381–389. [PMC free article] [PubMed] [Google Scholar]
- 162.Walker R. A., Dearing S. J., Gallacher B. Relationship of transforming growth factor β1 to extracellular matrix and stromal infiltrates in invasive breast carcinoma. British Journal of Cancer. 1994;69(6):1160–1165. doi: 10.1038/bjc.1994.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Thomas D. A., Massagué J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 164.Kehrl J. H., Wakefield L. M., Roberts A. B. Production of transforming growth factor β by human T lymphocytes and its potential role in the regulation of T cell growth. Journal of Experimental Medicine. 1986;163(5):1037–1050. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Geissmann F., Revy P., Regnault A., et al. TGF-β1 prevents the noncognate maturation of human dendritic Langerhans cells. Journal of Immunology. 1999;162(8):4567–4575. [PubMed] [Google Scholar]
- 166.Letterio J. J., Roberts A. B. Regulation of immune responses by TGF-β . Annual Review of Immunology. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 167.Czarniecki C. W., Chiu H. H., Wong G. H. W., McCabe S. M., Palladino M. A. Transforming growth factor-β1 modulates the expression of class II histocompatibility antigens on human cells. Journal of Immunology. 1988;140(12):4217–4223. [PubMed] [Google Scholar]
- 168.Geiser A. G., Letterio J. J., Kulkarni A. B., Karlsson S., Roberts A. B., Sporn M. B. Transforming growth factor β1 (TGF-β1) controls expression of major histocompatibility genes in the postnatal mouse: aberrant histocompatibility antigen expression in the pathogenesis of the TGF-β1 null mouse phenotype. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(21):9944–9948. doi: 10.1073/pnas.90.21.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dong Y., Tang L., Letterio J. J., Benveniste E. N. The Smad3 protein is involved in TGF-β inhibition of class II transactivator and class II MHC expression. Journal of Immunology. 2001;167(1):311–319. doi: 10.4049/jimmunol.167.1.311. [DOI] [PubMed] [Google Scholar]
- 170.Gorelink L., Flavell R. A. Immune-mediated eradication of tumors through the blockade of transforming growth factor-β signaling in T cells. Nature Medicine. 2001;7(10):1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 171.Roberts A. B., Sporn M. B., Assoian R. K. Transforming growth factor type β: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(12):4167–4171. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sunderkotter C., Goebeler M., Schulze-Osthoff K., Bhardwaj R., Sorg C. Macrophage-derived angiogenesis factors. Pharmacology and Therapeutics. 1991;51(2):195–216. doi: 10.1016/0163-7258(91)90077-Y. [DOI] [PubMed] [Google Scholar]
- 173.Pepper M. S., Vassalli J. D., Orci L., Montesano R. Biphasic effect of transforming growth factor-β1 on in vitro angiogenesis. Experimental Cell Research. 1993;204(2):356–363. doi: 10.1006/excr.1993.1043. [DOI] [PubMed] [Google Scholar]
- 174.Goumans M. J., Valdimarsdottir G., Itoh S., et al. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFβ/ALK5 signaling. Molecular Cell. 2003;12(4):817–828. doi: 10.1016/S1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- 175.Goumans M. J., Valdimarsdottir G., Itoh S., Rosendahl A., Sideras P., Ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO Journal. 2002;21(7):1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wu X., Ma J., Han J. D., Wang N., Chen Y. G. Distinct regulation of gene expression in human endothelial cells by TGF-β and its receptors. Microvascular Research. 2006;71(1):12–19. doi: 10.1016/j.mvr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 177.Ota T., Fujii M., Sugizaki T., et al. Targets of transcriptional regulation by two distinct type I receptors for transforming growth factor-β in human umbilical vein endothelial cells. Journal of Cellular Physiology. 2002;193(3):299–318. doi: 10.1002/jcp.10170. [DOI] [PubMed] [Google Scholar]
- 178.Dickson M. C., Martin J. S., Cousins F. M., Kulkarni A. B., Karlsson S., Akhurst R. J. Defective haematopoiesis and vasculogenesis in transforming growth factor-β1 knock out mice. Development. 1995;121(6):1845–1854. doi: 10.1242/dev.121.6.1845. [DOI] [PubMed] [Google Scholar]
- 179.Larsson J., Goumans M. J., Sjöstrand L. J., et al. Abnormal angiogenesis but intact hematopoietic potential in TGF-β type I receptor-deficient mice. EMBO Journal. 2001;20(7):1663–1673. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Goumans M. J., Zwijsen A., Van Rooijen M. A., Huylebroeck D., Roelen B. A. J., Mummery C. L. Transforming growth factor-β signalling in extraembryonic mesoderm is required for yolk sac vasculogenesis in mice. Development. 1999;126(16):3473–3483. doi: 10.1242/dev.126.16.3473. [DOI] [PubMed] [Google Scholar]
- 181.Oshima M., Oshima H., Taketo M. M. TGF-β receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Developmental Biology. 1996;179(1):297–302. doi: 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- 182.Brown C. B., Boyer A. S., Runyan R. B., Barnett J. V. Requirement of type III TGF-β receptor for endocardial cell transformation in the heart. Science. 1999;283(5410):2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 183.Compton L. A., Potash D. A., Brown C. B., Barnett J. V. Coronary vessel development is dependent on the type III transforming growth factor β receptor. Circulation Research. 2007;101(8):784–791. doi: 10.1161/CIRCRESAHA.107.152082. [DOI] [PubMed] [Google Scholar]
- 184.Lechleider R. J., Ryan J. L., Garrett L., et al. Targeted mutagenesis of Smad1 reveals an essential role in chorioallantoic fusion. Developmental Biology. 2001;240(1):157–167. doi: 10.1006/dbio.2001.0469. [DOI] [PubMed] [Google Scholar]
- 185.Chang H., Huylebroeck D., Verschueren K., Guo Q., Matzuk M. M., Zwijsen A. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development. 1999;126(8):1631–1642. doi: 10.1242/dev.126.8.1631. [DOI] [PubMed] [Google Scholar]
- 186.De Jong J. S., Van Diest P. J., Van Der Valk P., Baak J. P. A. Expression of growth factors, growth-inhibiting factors, and their receptors in invasive breast cancer. II: correlations with proliferation and angiogenesis. Journal of Pathology. 1998;184(1):53–57. doi: 10.1002/(SICI)1096-9896(199801)184:1<53::AID-PATH6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 187.Hasegawa Y., Takanashi S., Kanehira Y., Tsushima T., Imai T., Okumura K. Transforming growth factor-β1 level correlates with angiogenesis, tumor progression, and prognosis in patients with nonsmall cell lung carcinoma. Cancer. 2001;91(5):964–971. [PubMed] [Google Scholar]
- 188.Pertovaara L., Kaipainen A., Mustonen T., et al. Vascular endothelial growth factor is induced in response to transforming growth factor-β in fibroblastic and epithelial cells. Journal of Biological Chemistry. 1994;269(9):6271–6274. [PubMed] [Google Scholar]
- 189.Shimo T., Nakanishi T., Nishida T., et al. Involvement of CTGF, a hypertrophic chondrocyte-specific gene product, in tumor angiogenesis. Oncology. 2001;61(4):315–322. doi: 10.1159/000055339. [DOI] [PubMed] [Google Scholar]
- 190.Hagedorn H. G., Bachmeier B. E., Nerlich A. G. Synthesis and degradation of basement membranes and extracellular matrix and their regulation by TGF-beta in invasive carcinomas (Review) International Journal of Oncology. 2001;18(4):669–681. doi: 10.3892/ijo.18.4.669. [DOI] [PubMed] [Google Scholar]
- 191.Enholm B., Paavonen K., Ristimäki A., et al. Comparison of VEGF, VEGF-B, VEGF-C and Ang-1 mRNA regulation by serum, growth factors, oncoproteins and hypoxia. Oncogene. 1997;14(20):2475–2483. doi: 10.1038/sj.onc.1201090. [DOI] [PubMed] [Google Scholar]
- 192.De Wever O., Mareel M. Role of tissue stroma in cancer cell invasion. Journal of Pathology. 2003;200(4):429–447. doi: 10.1002/path.1398. [DOI] [PubMed] [Google Scholar]
- 193.Kojima Y., Acar A., Eaton E. N., et al. Autocrine TGF-β and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(46):20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Casey T. M., Eneman J., Crocker A., et al. Cancer associated fibroblasts stimulated by transforming growth factor beta1 (TGF-β1) increase invasion rate of tumor cells: a population study. Breast Cancer Research and Treatment. 2008;110(1):39–49. doi: 10.1007/s10549-007-9684-7. [DOI] [PubMed] [Google Scholar]
- 195.Sieuwerts A. M., Klijn J. G. M., Henzen-Logmans S. C., et al. Urokinase-type-plasminogen-activator(UPA) production by human breast (MYO)fibroblasts in vitro: influence of transforming growth factor-β 1 (TGFβ 1) compared with factor(S) released by human epithelial-carcinoma cells. International Journal of Cancer. 1998;76(6):829–835. doi: 10.1002/(sici)1097-0215(19980610)76:6<829::aid-ijc11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 196.Thiery J. P. Epithelial-mesenchymal transitions in development and pathologies. Current Opinion in Cell Biology. 2003;15(6):740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 197.Derynck R., Akhurst R. J. Differentiation plasticity regulated by TGF-β family proteins in development and disease. Nature Cell Biology. 2007;9(9):1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 198.Thuault S., Valcourt U., Petersen M., Manfioletti G., Heldin C. H., Moustakas A. Transforming growth factor-β employs HMGA2 to elicit epithelial-mesenchymal transition. Journal of Cell Biology. 2006;174(2):175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Miettinen P. J., Ebner R., Lopez A. R., Derynck R. TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. Journal of Cell Biology. 1994;127(6):2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Oft M., Peli J., Rudaz C., Schwarz H., Beug H., Reichmann E. TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes and Development. 1996;10(19):2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 201.Portella G., Cumming S. A., Liddell J., et al. Transforming growth factor β is essential for spindle cell conversion of mouse skin carcinoma in vivo: implications for tumor invasion. Cell Growth and Differentiation. 1998;9(5):393–404. [PubMed] [Google Scholar]
- 202.Hajra K. M., David Y-S. Chen, Fearon E. R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Research. 2002;62(6):1613–1618. [PubMed] [Google Scholar]
- 203.Savagner P., Yamada K. M., Thiery J. P. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. Journal of Cell Biology. 1997;137(6):1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ozdamar B., Bose R., Barrios-Rodiles M., Wang H. R., Zhang Y., Wrana J. L. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science. 2005;307(5715):1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 205.Oft M., Akhurst R. J., Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nature Cell Biology. 2002;4(7):487–494. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- 206.Janda E., Lehmann K., Killisch I., et al. Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. Journal of Cell Biology. 2002;156(2):299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gotzmann J., Huber H., Thallinger C., et al. Hepatocytes convert to a fibroblastoid phenotype through the cooperation of TGF-β1 and Ha-Ras: steps towards invasiveness. Journal of Cell Science. 2002;115(6):1189–1202. doi: 10.1242/jcs.115.6.1189. [DOI] [PubMed] [Google Scholar]
- 208.Larue L., Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24(50):7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 209.Zavadil J., Bitzer M., Liang D., et al. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β . Proceedings of the National Academy of Sciences of the United States of America. 2001;98(12):6686–6691. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xie L., Law B. K., Chytil A. M., Brown K. A., Aakre M. E., Moses H. L. Activation of the Erk pathway is required for TGF-β1-induced EMT in vitro. Neoplasia. 2004;6(5):603–610. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ellenrieder V., Hendler S. F., Boeck W., et al. Transforming growth factor β1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Research. 2001;61(10):4222–4228. [PubMed] [Google Scholar]
- 212.Grände M., Franzen Å., Karlsson J.-O., Ericson L. E., Heldin N.-E., Nilsson M. Transforming growth factor-β and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes. Journal of Cell Science. 2002;115(22):4227–4236. doi: 10.1242/jcs.00091. [DOI] [PubMed] [Google Scholar]
- 213.Christoffersen N. R., Silahtaroglu A., Ørom U. A., Kauppinen S., Lund A. H. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13(8):1172–1178. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ahmed F. E. Role of miRNA in carcinogenesis and biomarker selection: a methodological view. Expert Review of Molecular Diagnostics. 2007;7(5):569–603. doi: 10.1586/14737159.7.5.569. [DOI] [PubMed] [Google Scholar]
- 215.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 216.Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Burrow A. A., Williams L. E., Pierce L. C. T., Wang Y. H. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10, article no. 59 doi: 10.1186/1471-2164-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PloS One. 2010;5(5) doi: 10.1371/journal.pone.0010615.e10615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Krützfeldt J., Rajewsky N., Braich R., et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 220.Calin G. A., Dumitru C. D., Shimizu M., et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Iorio M. V., Ferracin M., Liu C. G., et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Research. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 222.Calin G. A., Ferracin M., Cimmino A., et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. New England Journal of Medicine. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 223.Calin G. A., Sevignani C., Dumitru C. D., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Qian B., Katsaros D., Lu L., et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-β1. Breast Cancer Research and Treatment. 2009;117(1):131–140. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 225.Schaefer A., Jung M., Mollenkopf H. J., et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. International Journal of Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 226.Jiang L., Mao P., Song L., et al. miR-182 as a prognostic marker for glioma progression and patient survival. American Journal of Pathology. 2010;177(1):29–38. doi: 10.2353/ajpath.2010.090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhang Y., Li M., Wang H., et al. Profiling of 95 MicroRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World Journal of Surgery. 2009;33(4):698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Calin G. A., Croce C. M. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 229.Ventura A., Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gregory P. A., Bracken C. P., Bert A. G., Goodall G. J. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7(20):3112–3117. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 231.Bracken C. P., Gregory P. A., Kolesnikoff N., et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Research. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 232.Park S. M., Gaur A. B., Lengyel E., Peter M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes and Development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Valastyan S., Reinhardt F., Benaich N., et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 234.Korpal M., Lee E. S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. Journal of Biological Chemistry. 2008;283(22):14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Javor J., Ferencik S., Bucova M., et al. Polymorphisms in the genes encoding TGF-β1, TNF-α, and IL-6 show association with type 1 diabetes mellitus in the Slovak population. Archivum Immunologiae et Therapiae Experimentalis. 2010;58(5):385–393. doi: 10.1007/s00005-010-0092-z. [DOI] [PubMed] [Google Scholar]
- 236.Burk U., Schubert J., Wellner U., et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Reports. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Heino J., Ignotz R. A., Hemler M. E., Crouse C., Massague J. Regulation of cell adhesion receptors by transforming growth factor-β. Concomitant regulation of integrins that share a common β1 subunit. Journal of Biological Chemistry. 1989;264(1):380–388. [PubMed] [Google Scholar]
- 238.Moustakas A., Heldin C. H. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Science. 2007;98(10):1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dumont N., Bakin A. V., Arteaga C. L. Autocrine transforming growth factor-β signaling mediates Smad-independent motility in human cancer cells. Journal of Biological Chemistry. 2003;278(5):3275–3285. doi: 10.1074/jbc.M204623200. [DOI] [PubMed] [Google Scholar]
- 240.Tsapara A., Luthert P., Greenwood J., Hill C. S., Matter K., Balda M. S. The RhoA activator GEF-H1/Lfc is a transforming growth factor-β target gene and effector that regulates α-smooth muscle actin expression and cell migration. Molecular Biology of the Cell. 2010;21(6):860–870. doi: 10.1091/mbc.E09-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kong W., Yang H., He L., et al. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Molecular and Cellular Biology. 2008;28(22):6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Fils-Aimé N., Dai M., Guo J., Neel J. C., Lebrun J.-J. Micro-RNA-584 and PHACTR1: new signaling route through which transforming growth factor-beta mediates the migration and actin dynamics of breast cancer cells. doi: 10.1074/jbc.M112.430934. The Journal of Biological Chemistry. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Neel J. C., Lebrun J.-J. TGFβ/Activin A-induced miR-181 induces cellular migration and invasion in breast cancer cells. doi: 10.1016/j.cellsig.2013.03.013. Cellular Signalling. In press. [DOI] [PubMed] [Google Scholar]
- 244.Wiercinska E., Naber H. P. H., Pardali E., Van Der Pluijm G., Van Dam H., Ten Dijke P. The TGF-β/Smad pathway induces breast cancer cell invasion through the up-regulation of matrix metalloproteinase 2 and 9 in a spheroid invasion model system. Breast Cancer Research and Treatment. 2011;128(3):657–666. doi: 10.1007/s10549-010-1147-x. [DOI] [PubMed] [Google Scholar]
- 245.Giannelli G., Fransvea E., Marinosci F., et al. Transforming growth factor-β1 triggers hepatocellular carcinoma invasiveness via α3β1 integrin. American Journal of Pathology. 2002;161(1):183–193. doi: 10.1016/s0002-9440(10)64170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Giannelli G., Astigiano S., Antonaci S., et al. Role of the α3β1 and α6β4 integrins in tumor invasion. Clinical and Experimental Metastasis. 2002;19(3):217–223. doi: 10.1023/A:1015579204607. [DOI] [PubMed] [Google Scholar]
- 247.Wang B., Hsu S. H., Majumder S., et al. TGFβ-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29(12):1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Han G., Lu S.-L., Li A. G., et al. Distinct mechanisms of TGF-β 1-mediated epithelial-to- mesenchymal transition and metastasis during skin carcinogenesis. Journal of Clinical Investigation. 2005;115(7):1714–1723. doi: 10.1172/JCI24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wang Y., Yu Y., Tsuyada A., et al. Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30(12):1470–1480. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Dai M., Al-Odaini A. A., Arakelian A., Rabbani S. A., Ali S., Lebrun J.-J. A novel function for p21Cip1 and acetyltransferase p/CAF as critical transcriptional regulators of TGFβ-mediated breast cancer cell migration and invasion. Breast Cancer Research. 2012;14(5, article no. R127) doi: 10.1186/bcr3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.AlOdaini A., Dai M., Wang Y., Ali S., Lebrun J.-J. KiSS1 gene as a novel mediator of TGFβ pro-invasive effects in triple negative breast cancer. In press. [Google Scholar]
- 252.Gupta G. P., Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 253.Kingsley L. A., Fournier P. G. J., Chirgwin J. M., Guise T. A. Molecular biology of bone metastasis. Molecular Cancer Therapeutics. 2007;6(10):2609–2617. doi: 10.1158/1535-7163.MCT-07-0234. [DOI] [PubMed] [Google Scholar]
- 254.Yin J. J., Selander K., Chirgwin J. M., et al. TGF-β signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. Journal of Clinical Investigation. 1999;103(2):197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Javelaud D., Mohammad K. S., McKenna C. R., et al. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Research. 2007;67(5):2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- 256.Kominsky S. L., Doucet M., Brady K., Weber K. L. TGF-β promotes the establishment of renal cell carcinoma bone metastasis. Journal of Bone and Mineral Research. 2007;22(1):37–44. doi: 10.1359/jbmr.061005. [DOI] [PubMed] [Google Scholar]
- 257.Bertran E., Caja L., Navarro E., et al. Role of CXCR4/SDF-1α in the migratory phenotype of hepatoma cells that have undergone epithelial-mesenchymal transition in response to the transforming growth factor-β . Cellular Signalling. 2009;21(11):1595–1606. doi: 10.1016/j.cellsig.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 258.Wang J., Loberg R., Taichman R. S. The pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis. Cancer and Metastasis Reviews. 2006;25(4):573–587. doi: 10.1007/s10555-006-9019-x. [DOI] [PubMed] [Google Scholar]
- 259.Franchimont N., Rydziel S., Canalis E. Transforming growth factor-β increases interleukin-6 transcripts in osteoblasts. Bone. 2000;26(3):249–253. doi: 10.1016/S8756-3282(99)00275-6. [DOI] [PubMed] [Google Scholar]
- 260.Wahl S. M., Costa G. L., Corcoran M., Wahl L. M., Berger A. E. Transforming growth factor-β mediates IL-1-dependent induction of IL-1 receptor antagonist. Journal of Immunology. 1993;150(8 I):3553–3560. [PubMed] [Google Scholar]
- 261.Steeve K. T., Marc P., Sandrine T., Dominique H., Yannick F. IL-6, RANKL, TNF-alpha/IL-1: interrelations in bone resorption pathophysiology. Cytokine and Growth Factor Reviews. 2004;15(1):49–60. doi: 10.1016/j.cytogfr.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 262.Siegel P. M., Shu W., Cardiff R. D., Muller W. J., Massagué J. Transforming growth factor β signaling impairs neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Padua D., Zhang X. H. F., Wang Q., et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Bos P. D., Zhang X. H.-F., Nadal C., et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Nguyen A. V., Pollard J. W. Transforming growth factor β3 induces cell death during the first stage of mammary gland involution. Development. 2000;127(14):3107–3118. doi: 10.1242/dev.127.14.3107. [DOI] [PubMed] [Google Scholar]
- 266.Mieth M., Boehmer F. D., Ball R., Groner B., Grosse R. Transforming growth factor-β inhibits lactogenic hormone induction of β-casein expression in HC11 mouse mammary epithelial cells. Growth Factors. 1990;4(1):9–15. doi: 10.3109/08977199009011005. [DOI] [PubMed] [Google Scholar]
- 267.Rui H., Lebrun J.-J., Kirken R. A., Kelly P. A., Farrar W. L. JAK2 activation and cell proliferation induced by antibody-mediated prolactin receptor dimerization. Endocrinology. 1994;135(4):1299–1306. doi: 10.1210/en.135.4.1299. [DOI] [PubMed] [Google Scholar]
- 268.Rui H., Kirken R. A., Farrar W. L. Activation of receptor-associated tyrosine kinase JAK2 by prolactin. Journal of Biological Chemistry. 1994;269(7):5364–5368. [PubMed] [Google Scholar]
- 269.Dusanter-Fourt I., Muller O., Ziemiecki A., et al. Identification of JAK protein tyrosine kinases as signaling molecules for prolactin. Functional analysis of prolactin receptor and prolactin—erythropoietin receptor chimera expressed in lymphoid cells. EMBO Journal. 1994;13(11):2583–2591. doi: 10.1002/j.1460-2075.1994.tb06548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lebrun J.-J., Ali S., Ullrich A., Kelly P. A. Proline-rich sequence-mediated Jak2 association to the prolactin receptor is required but not sufficient for signal transduction. Journal of Biological Chemistry. 1995;270(18):10664–10670. doi: 10.1074/jbc.270.18.10664. [DOI] [PubMed] [Google Scholar]
- 271.Lebrun J.-J., Ali S., Goffin V., Ullrich A., Kelly P. A. A single phosphotyrosine residue of the prolactin receptor is responsible for activation of gene transcription. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(9):4031–4035. doi: 10.1073/pnas.92.9.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lebrun J.-J., Ali S., Sofer L., Ullrich A., Kelly P. A. Prolactin-induced proliferation of nb2 cells involves tyrosine phosphorylation of the prolactin receptor and its associated tyrosine kinase JAK2. Journal of Biological Chemistry. 1994;269(19):14021–14026. [PubMed] [Google Scholar]
- 273.Ali S., Chen Z., Lebrun J.-J., et al. PTP1D is a positive regulator of the prolactin signal leading to β-casein promoter activation. EMBO Journal. 1996;15(1):135–142. [PMC free article] [PubMed] [Google Scholar]
- 274.Schmitt-Ney M., Doppler W., Ball R. K., Groner B. β-Casein gene promoter activity is regulated by the hormone-mediated relief of transcriptional repression and a mammary-gland-specific nuclear factor. Molecular and Cellular Biology. 1991;11(7):3745–3755. doi: 10.1128/mcb.11.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kirch H. C., Putzer B., Brockmann D., Esche H., Kloke O. Formation of the early-region-2 transcription-factor-1—retinoblastoma-protein (E2F-1-RB) transrepressor and release of the retinoblastoma protein from nuclear complexes containing cyclin A is induced by interferon α in U937V cells but not in interferon-α-resistant U937VR cells. European Journal of Biochemistry. 1997;246(3):736–744. doi: 10.1111/j.1432-1033.1997.00736.x. [DOI] [PubMed] [Google Scholar]
- 276.Chughtai N., Schimchowitsch S., Lebrun J.-J., Ali S. Prolactin induces SHP-2 association with Stat5, nuclear translocation, and binding to the β-casein gene promoter in mammary cells. Journal of Biological Chemistry. 2002;277(34):31107–31114. doi: 10.1074/jbc.M200156200. [DOI] [PubMed] [Google Scholar]
- 277.Liu X., Robinson G. W., Wagner K. U., Garrett L., Wynshaw-Boris A., Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes and Development. 1997;11(2):179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 278.Nouhi Z., Chughtai N., Hartley S., Cocolakis E., Lebrun J.-J., Ali S. Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Research. 2006;66(3):1824–1832. doi: 10.1158/0008-5472.CAN-05-2292. [DOI] [PubMed] [Google Scholar]
- 279.Daniel C. W., Robinson S., Silberstein G. B. The transforming growth factors beta in development and functional differentiation of the mouse mammary gland. Advances in Experimental Medicine and Biology. 2001;501:61–70. doi: 10.1007/978-1-4615-1371-1_7. [DOI] [PubMed] [Google Scholar]
- 280.Faure E., Heisterkamp N., Groffen J., Kaartinen V. Differential expression of TGF-β isoforms during postlactational mammary gland involution. Cell and Tissue Research. 2000;300(1):89–95. doi: 10.1007/s004410050050. [DOI] [PubMed] [Google Scholar]
- 281.Strange R., Li F., Saurer S., Burkhardt A., Friis R. R. Apoptotic cell death and tissue remodelling during mouse mammary gland involution. Development. 1992;115(1):49–58. doi: 10.1242/dev.115.1.49. [DOI] [PubMed] [Google Scholar]
- 282.Robinson S. D., Silberstein G. B., Roberts A. B., Flanders K. C., Daniel C. W. Regulated expression and growth inhibitory effects of transforming growth factor-β isoforms in mouse mammary gland development. Development. 1991;113(3):867–878. doi: 10.1242/dev.113.3.867. [DOI] [PubMed] [Google Scholar]
- 283.Robinson S. D., Roberts A. B., Daniel C. W. TGFβ suppresses casein synthesis in mouse mammary explants and may play a role in controlling milk levels during pregnancy. Journal of Cell Biology. 1993;120(1):245–252. doi: 10.1083/jcb.120.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Sudlow A. W., Wilde C. J., Burgoyne R. D. Transforming growth factor-β1 inhibits casein secretion from differentiating mammary-gland explants but not from lactating mammary cells. Biochemical Journal. 1994;304(part 2):333–336. doi: 10.1042/bj3040333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Jhappan C., Geiser A. G., Kordon E. C., et al. Targeting expression of a transforming growth factor β1 transgene to the pregnant mammary gland inhibits alveolar development and lactation. EMBO Journal. 1993;12(5):1835–1845. doi: 10.1002/j.1460-2075.1993.tb05832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Pierce D. F., Johnson M. D., Matsui Y., et al. Inhibition of mammary duct development but not alveolar outgrowth during pregnancy in transgenic mice expressing active TGF-β1. Genes and Development. 1993;7(12 A):2308–2317. doi: 10.1101/gad.7.12a.2308. [DOI] [PubMed] [Google Scholar]
- 287.Cocolakis E., Dai M., Drevet L., et al. Smad signaling antagonizes STAT5-mediated gene transcription and mammary epithelial cell differentiation. Journal of Biological Chemistry. 2008;283(3):1293–1307. doi: 10.1074/jbc.M707492200. [DOI] [PubMed] [Google Scholar]
- 288.Grütter C., Wilkinson T., Turner R., et al. A cytokine-neutralizing antibody as a structural mimetic of 2 receptor interactions. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(51):20251–20256. doi: 10.1073/pnas.0807200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Benigni A., Zoja C., Corna D., et al. Add-on anti-TGF-β antibody to ACE inhibitor arrests progressive diabetic nephropathy in the rat. Journal of the American Society of Nephrology. 2003;14(7):1816–1824. doi: 10.1097/01.ASN.0000074238.61967.B7. [DOI] [PubMed] [Google Scholar]
- 290.Ananth S., Knebelmann B., Grüning W., et al. Transforming growth factor β1 is a target for the von Hippel-Lindau tumor suppressor and a critical growth factor for clear cell renal carcinoma. Cancer Research. 1999;59(9):2210–2216. [PubMed] [Google Scholar]
- 291.Morris J. C., Shapiro G. I., Tan A. R. Phase I/II study of GC1008: a human anti-transforming growth factor-beta (TGFβ) monoclonal antibody (MAb) in patients with advanced malignant melanoma (MM) or renal cell carcinoma (RCC) Journal of Clinical Oncology. 2008;26(supplement, abstract no. 9028) [Google Scholar]
- 292.Thompson J. E., Vaughan T. J., Williams A. J., et al. A fully human antibody neutralising biologically active human TGFβ2 for use in therapy. Journal of Immunological Methods. 1999;227(1-2):17–29. doi: 10.1016/S0022-1759(99)00060-5. [DOI] [PubMed] [Google Scholar]
- 293.Mead A. L., Wong T. T. L., Cordeiro M. F., Anderson I. K., Khaw P. T. Evaluation of anti-TGF-β2 antibody as a new postoperative anti-scarring agent in glaucoma surgery. Investigative Ophthalmology and Visual Science. 2003;44(8):3394–3401. doi: 10.1167/iovs.02-0978. [DOI] [PubMed] [Google Scholar]
- 294.Hill C., Flyvbjerg A., Rasch R., Bak M., Logan A. Transforming growth factor-β2 antibody attenuates fibrosis in the experimental diabetic rat kidney. Journal of Endocrinology. 2001;170(3):647–651. doi: 10.1677/joe.0.1700647. [DOI] [PubMed] [Google Scholar]
- 295.Denton C. P., Merkel P. A., Furst D. E., et al. Recombinant human anti-transforming growth factor β1 antibody therapy in systemic sclerosis: a multicenter, randomized, placebo-controlled phase I/II trial of CAT-192. Arthritis and Rheumatism. 2007;56(1):323–333. doi: 10.1002/art.22289. [DOI] [PubMed] [Google Scholar]
- 296.Bonafoux D., Lee W. C. Strategies for TGF-β modulation: a review of recent patents. Expert Opinion on Therapeutic Patents. 2009;19(12):1759–1769. doi: 10.1517/13543770903397400. [DOI] [PubMed] [Google Scholar]
- 297.Khaw P., Grehn F., Holló G., et al. A phase III study of subconjunctival human anti-transforming growth factor β 2 monoclonal antibody (CAT-152) to prevent scarring after first-time trabeculectomy. Ophthalmology. 2007;114(10):1822–1830. doi: 10.1016/j.ophtha.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 298. Cambridge Antibody Technology, website, 2010.
- 299.Dasch J. R., Pace D. R., Waegell W., Inenaga D., Ellingsworth L. Monoclonal antibodies recognizing transforming growth factor-β: bioactivity neutralization and transforming growth factor β2 affinity purification. Journal of Immunology. 1989;142(5):1536–1541. [PubMed] [Google Scholar]
- 300.Arteaga C. L., Hurd S. D., Winnier A. R., Johnson M. D., Fendly B. M., Forbes J. T. Anti-transforming growth factor (TGF)-β antibodies inhibit breast cancer cell tumorigenicity and increase mouse spleen natural killer cell activity. Implications for a possible role of tumor cell/host TGF-β interactions in human breast cancer progression. Journal of Clinical Investigation. 1993;92(6):2569–2576. doi: 10.1172/JCI116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Genzyme Corporation. Genzyme Corporate Pipeline. 2010, http://www.genzyme.com/research/pipeline/pipe_home.asp.
- 302.Lahn M., Kloeker S., Berry B. S. TGF-β inhibitors for the treatment of cancer. Expert Opinion on Investigational Drugs. 2005;14(6):629–641. doi: 10.1517/13543784.14.6.629. [DOI] [PubMed] [Google Scholar]
- 303.Rowland-Goldsmith M. A., Maruyama H., Kusama T., Ralli S., Korc M. Soluble type II transforming growth factor-β (TGF-β) receptor inhibits TGF-β signaling in COLO-357 pancreatic cancer cells in vitro and attenuates tumor formation. Clinical Cancer Research. 2001;7(9):2931–2940. [PubMed] [Google Scholar]
- 304.Rowland-Goldsmith M. A., Maruyama H., Matsuda K., et al. Soluble type II transforming growth factor-beta receptor attenuates expression of metastasis-associated genes and suppresses pancreatic cancer cell metastasis. Molecular Cancer Therapeutics. 2002;1(3):161–167. [PubMed] [Google Scholar]
- 305.Lopez-Casillas F., Payne H. M., Andres J. L., Massague J. Betaglycan can act as a dual modulator of TGF-β access to signaling receptors: mapping of ligand binding and GAG attachment sites. Journal of Cell Biology. 1994;124(4):557–568. doi: 10.1083/jcb.124.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Border W. A., Noble N. A., Yamamoto T., et al. Natural inhibitor of transforming growth factor-β protects against scarring in experimental kidney disease. Nature. 1992;360(6402):361–364. doi: 10.1038/360361a0. [DOI] [PubMed] [Google Scholar]
- 307.Komesli S., Vivien D., Dutartre P. Chimeric extracellular domain of type II transforming growth factor (TGF)-β receptor fused to the Fc region of human immunoglobulin as a TGF-β antagonist. European Journal of Biochemistry. 1998;254(3):505–513. doi: 10.1046/j.1432-1327.1998.2540505.x. [DOI] [PubMed] [Google Scholar]
- 308.Yang Y.-A., Dukhanina O., Tang B., et al. Lifetime exposure to a soluble TGF-β antagonist protects mice against metastasis without adverse side effects. Journal of Clinical Investigation. 2002;109(12):1607–1615. doi: 10.1172/JCI200215333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Hermida N., López B., González A., et al. A synthetic peptide from transforming growth factor-β 1 type III receptor prevents myocardial fibrosis in spontaneously hypertensive rats. Cardiovascular Research. 2009;81(3):601–609. doi: 10.1093/cvr/cvn315. [DOI] [PubMed] [Google Scholar]
- 310.Crooke S. T. Molecular mechanisms of action of antisense drugs. Biochimica et Biophysica Acta. 1999;1489(1):31–43. doi: 10.1016/S0167-4781(99)00148-7. [DOI] [PubMed] [Google Scholar]
- 311.Tamm I., Dörken B., Hartmann G. Antisense therapy in oncology: new hope for an old idea? Lancet. 2001;358(9280):489–497. doi: 10.1016/S0140-6736(01)05629-X. [DOI] [PubMed] [Google Scholar]
- 312.Schlingensiepen K. H. The TGF-beta1 antisense oligonucleotide AP, 11014 for the treatment of non-small cell lung, colorectal and prostate cancer: preclinical studies. Journal of Clinical Oncology. 2004;22(abstract no. 3132) [Google Scholar]
- 313.Fischer D., Bischof A., Egger T., et al. Simultaneous reversal of immunosuppression, inhibition of tumor growth and migration by the TGF-beta1 antisense oligonucleotide AP, 11014 in prostatic, colorectal and non-small cell lung cancer. Proceedings of the American Association for Cancer Research Meeting; 2005; [Google Scholar]
- 314.Stauder G., Bischof A., Egger T., et al. TGF-β2 suppression by the antisense oligonucleotide AP, 12009 as treatment for pancreatic cancer: preclinical efficacy data. Journal of Clinical Oncology. 2004;22(14s) supplement, abstract no. 4106 [Google Scholar]
- 315.Schlingensiepen K. H., Schlingensiepen R., Steinbrecher A., et al. Targeted tumor therapy with the TGF-β2 antisense compound AP 12009. Cytokine and Growth Factor Reviews. 2006;17(1-2):129–139. doi: 10.1016/j.cytogfr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 316.Hau P., Jachimczak P., Schlingensiepen R., et al. Inhibition of TGF-β2 with ap 12009 in recurrent malignant gliomas: from preclinical to phase I/II studies. Oligonucleotides. 2007;17(2):201–212. doi: 10.1089/oli.2006.0053. [DOI] [PubMed] [Google Scholar]
- 317.Antisense Pharma. 2010, http://www.antisense-pharma.com/products/f_products.htm.
- 318.Yingling C. D. Dihydropyrrolopyrazoles as TGFβ receptor kinase inhibitors for cancer therapy. Proceedings of the 16th EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics; 2004; [Google Scholar]
- 319.Calvo-Aller E., Baselga J., Glatt S., et al. First human dose escalation study in patients with metastatic malignancies to determine safety and pharmacokinetics of LY2157299, a small molecule inhibitor of the transforming growth factor-beta receptor I kinase. Journal of Clinical Oncology. 2008;26(supplement, abstract no. 14554) [Google Scholar]
- 320.Subramanian G., Schwarz R. E., Higgins L., et al. Targeting endogenous transforming growth factor β receptor signaling in Smad4-deficient human pancreatic carcinoma cells inhibits their invasive phenotype. Cancer Research. 2004;64(15):5200–5211. doi: 10.1158/0008-5472.CAN-04-0018. [DOI] [PubMed] [Google Scholar]
- 321.Peng S. B., Yan L., Xia X., et al. Kinetic characterization of novel pyrazole TGF-β receptor I kinase inhibitors and their blockade of the epithelial-mesenchymal transition. Biochemistry. 2005;44(7):2293–2304. doi: 10.1021/bi048851x. [DOI] [PubMed] [Google Scholar]
- 322.Ge R., Rajeev V., Subramanian G., et al. Selective inhibitors of type I receptor kinase block cellular transforming growth factor-β signaling. Biochemical Pharmacology. 2004;68(1):41–50. doi: 10.1016/j.bcp.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 323.Uhl M., Aulwurm S., Wischhusen J., et al. SD-208, a novel transforming growth factor β receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Research. 2004;64(21):7954–7961. doi: 10.1158/0008-5472.CAN-04-1013. [DOI] [PubMed] [Google Scholar]
- 324.Tran T.-T., Uhl M., Jing Y. M., et al. Inhibiting TGF-β signaling restores immune surveillance in the SMA-560 glioma model. Neuro-Oncology. 2007;9(3):259–270. doi: 10.1215/15228517-2007-010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Laping N. J., Grygielko E., Mathur A., et al. Inhibition of transforming growth factor (TGF)-β1-induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity: SB-431542. Molecular Pharmacology. 2002;62(1):58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 326.Zhang B., Halder S. K., Zhang S., Datta P. K. Targeting transforming growth factor-β signaling in liver metastasis of colon cancer. Cancer Letters. 2009;277(1):114–120. doi: 10.1016/j.canlet.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Melisi D., Ishiyama S., Sclabas G. M., et al. LY2109761, a novel transforming growth factor β receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Molecular Cancer Therapeutics. 2008;7(4):829–840. doi: 10.1158/1535-7163.MCT-07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Fukai Y., Fukuchi M., Masuda N., et al. Reduced expression of transforming growth factor-β receptors is an unfavorable prognostic factor in human esophageal squamous cell carcinoma. International Journal of Cancer. 2003;104(2):161–166. doi: 10.1002/ijc.10929. [DOI] [PubMed] [Google Scholar]
- 329.Miyajima A., Asano T., Seta K., Asano T., Kakoi N., Hayakawa M. Loss of expression of transforming growth factor-beta receptor as a prognostic factor in patients with renal cell carcinoma. Urology. 2003;61(5):1072–1077. doi: 10.1016/S0090-4295(02)02553-0. [DOI] [PubMed] [Google Scholar]
- 330.Watanabe T., Wu T. T., Catalano P. J., et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. New England Journal of Medicine. 2001;344(16):1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Tateishi M., Kusaba I., Masuda H., Tanaka T., Matsumata T., Sugimachi K. The progression of invasiveness regarding the role of transforming growth factor β receptor type II in gastric cancer. European Journal of Surgical Oncology. 2000;26(4):377–380. doi: 10.1053/ejso.1999.0902. [DOI] [PubMed] [Google Scholar]
- 332.Houghton A. N., Polsky D. Focus on melanoma. Cancer Cell. 2002;2(4):275–278. doi: 10.1016/S1535-6108(02)00161-7. [DOI] [PubMed] [Google Scholar]
- 333.Ramont L., Pasco S., Hornebeck W., Maquart F. X., Monboisse J. C. Transforming growth factor-β1 inhibits tumor growth in a mouse melanoma model by down-regulating the plasminogen activation system. Experimental Cell Research. 2003;291(1):1–10. doi: 10.1016/S0014-4827(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 334.Humbert L., Lebrun J.-J. TGF-beta inhibits human cutaneous melanoma cell migration and invasion through regulation of the plasminogen activator system. Cellular Signalling. 2012;25(2):490–500. doi: 10.1016/j.cellsig.2012.10.011. [DOI] [PubMed] [Google Scholar]