Abstract
DNA assemblies containing a pendant dipyridophenazine complex of Ru(II) along with two oxidative traps, a site containing the nucleoside analog methylindole (5′-GMG-3′) and a 5′-GGG-3′ site, have been constructed to explore long-range charge transport through the base pair stack. With these chemically well defined assemblies, in combination with the flash/quench technique, formation of the methylindole cation radical and the neutral guanine radical is monitored directly by using transient absorption spectroscopy, and yields of oxidative damage are quantitated biochemically by gel electrophoresis. In these assemblies the base radicals form with a rate of ≥107 s–1. The rate of base radical formation does not change upon the addition of a second radical trap, the 5′-GGG-3′ site; however, the yield of methylindole oxidation is significantly lower. This observation indicates that the 5′-GGG-3′ site is effective in competing for the migrating charge and provides a second trapping site. Switching the orientation of the two trapping sites does not affect the yield of oxidized products at either site. Therefore, in DNA both forward and reverse charge transport occur so as to provide equilibration across the duplex on a timescale that is fast compared with trapping at a particular site. Further evidence of charge equilibration results from incorporating an intervening base-stacking perturbation and monitoring the fate of the injected charge. These experiments underscore the dynamic nature of DNA charge transport and reveal the importance of considering radical propagation in both directions along the DNA duplex.
Numerous spectroscopic and biochemical experiments have shown that the base stack of DNA can mediate charge transport (CT) reactions (1–4). Chemically well defined assemblies, consisting of DNA duplexes with covalently bound oxidants, have been particularly useful in exploring the effects of base-stacking perturbations (5–8), intervening DNA sequence (9, 10), and donor–acceptor distance (11–13) on CT. Long-range oxidative DNA damage has been demonstrated over a distance of 200 Å (14, 15). Indeed, DNA either packaged in nucleosome core particles (16) or inside the cell nucleus (17) has been found to be susceptible to long-range oxidative damage.
Based on spectroscopic and biochemical experiments using Ru and Rh intercalating oxidants along with temperature-dependent base–base CT chemistry, we have proposed a model for CT involving conformationally gated charge hopping among DNA domains (18–20). Domains over which charge may be delocalized are defined by sequence and dynamics; a domain size of ≈4 bp has been characterized in assemblies containing repetitive tracts of adenines. Earlier, using measurements only of oxidative DNA damage yield as a function of intervening sequence, Giese, Jortner, and coworkers proposed a model involving a mixture of base hopping and tunneling (21–23). Also based on oxidative yield determinations, Schuster and coworkers (24) proposed phonon-assisted polaron hopping between guanine bases. However, studies measuring only damage yield do not provide a complete picture as to the dynamic nature of the CT process.
DNA assemblies containing dipyridophenazine (dppz) complexes of Ru(II) are particularly useful for probing the mechanism of CT, because they allow both for spectroscopic studies to monitor formation of DNA radicals on a short timescale and for biochemical analysis to determine the yield of oxidative damage occurring on a longer timescale. With these Ru complexes, a flash/quench technique is typically used (25, 26). The cycle is initiated by visible light, which excites the intercalated Ru(II) complex. This excited Ru(II) complex, *Ru(II), is then quenched by a nonintercalating electron acceptor, Q, such as [Ru(NH3)6]3+ or [Co(NH3)5Cl]2+, so as to form Ru(III) in situ. It is this Ru(III) species that can oxidize guanines from a distance. The oxidized guanine radical can then undergo further reaction to form a family of oxidative products (27).
Assemblies containing 4-methylindole (M) as the electron donor and a tethered Ru intercalator were constructed to explore long-range DNA CT spectroscopically and biochemically (28, 29). The methylindole moiety is particularly amenable as an artificial base in these studies because of its relatively low oxidation potential [1 V vs. normal hydrogen electrode (NHE)] and the strong absorptivity at 600 nm of its cation radical. Formation of the M cation radical 17–37 Å away from the tethered intercalating oxidant occurs with a rate ≥107 s–1 and is found to be coincident with quenching of the ruthenium excited state to form the Ru(III) oxidant (28); CT is not rate-limiting. The extent of radical localization at the injection site was found to affect the yield of long-range oxidative damage by modulating the extent of a nonproductive backreaction with reduced quencher (29).
Recent photophysical experiments have also demonstrated that back electron transfer (BET) can play a significant role in diminishing the yield of charge propagation out to a distant site (30, 31). If BET with the reduced DNA-bound photooxidant is faster than trapping of the guanine radical by H2O and/or O2, fast BET can prevent formation of a permanent guanine lesion. The consequences of rapid BET demonstrate that in DNA reverse CT must be as carefully considered as in studies of forward CT. Given radical migration in both directions through DNA, yields of oxidative damage will necessarily reflect some extent of charge equilibration before radical localization and trapping.
To explore this issue, we have prepared duplexes containing a tethered dppz complex of Ru(II), the nucleoside analog methylindole, and a second radical trap, a GGG site; the two trapping sites have been varied in position with and without an intervening stacking perturbation to limit migration between these low-potential sites. Using these assemblies, here we report results demonstrating charge equilibration across the DNA duplex.
Methods
DNA Synthesis. Oligonucleotides were synthesized on an ABI DNA synthesizer by using phosphoramidite chemistry (32, 33), purified by reverse-phase HPLC and characterized by matrix-assisted laser desorption ionization–time-of-flight MS. The ligands 4′-methylbipyridine-4-butyric acid (bpy') and dppz along with the complex [Ru(bpy')(dppz)(phen)]Cl2 were synthesized as described (34–38). The synthesis of ruthenium-modified oligonucleotides was carried out with a racemic Ru mixture as described (39). Purification of the Ru-modified DNA by HPLC yields four isomers, which were characterized by UV-visible spectroscopy and matrix-assisted laser desorption ionization–time-of-flight MS. A mixture of all four diastereomers was used for the laser spectroscopy and biochemical experiments.
Assay of Oxidized Products. Unmetalated oligonucleotide strands were labeled at the 5′ end with 32P by using standard procedures (40). DNA duplexes were formed by mixing equal concentrations of complementary strands and heating to 90°C followed by slow cooling to 20°C. DNA duplexes (5 μM) and bulge-containing assemblies (1 μM) were irradiated at room temperature in the presence of [Co(NH3)5Cl]2+ by using a HeCd laser (≈13 mW at 442 nm). For the intermolecular control experiments, equimolar amounts of non-32P-labeled Ru-DNA were mixed with their corresponding 32P-labeled assembly, which did not contain tethered ruthenium. For the intermolecular control experiments the inosine at the ruthenium-binding site was replaced by guanine to render this site susceptible to singlet oxygen. Additionally, in the intermolecular control experiments, M was replaced with guanine, yielding two GGG sites. After irradiation, all samples were treated with 10% (vol/vol) piperidine at 90°C for 30 min, dried, and subjected to electrophoresis through a 20% denaturing polyacrylamide gel. The levels of damage were quantitated by using phosphorimagery (imagequant).
Laser Spectroscopy. DNA duplexes (20 μM) were formed by mixing equal concentrations of complementary strands and heating to 90°C followed by slow cooling to room temperature. The [Ru(NH3)6]3+ quencher concentration was 300 μM. Time-resolved emission and transient absorption experiments were carried out on a Nd:YAG laser with an excitation wavelength of 470 nm. The emission of the intercalated Ru complexes was monitored at 610 nm.
Results
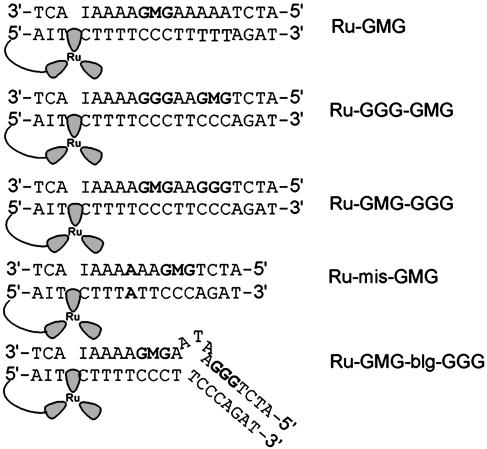
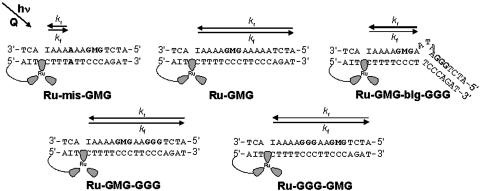
Ruthenium-DNA Assemblies. The ruthenium-DNA assemblies used in this study are shown in Fig. 1. Each DNA assembly contains a 5′ tethered [Ru(bpy')(dppz)(phen)]2+ and a methylindole flanked on either side by guanines to afford stability. Ru-GMG is the control assembly containing only one site of low oxidation potential, the GMG site. Ru-GGG-GMG contains a GGG positioned between the ruthenium-binding site and the GMG site, and Ru-GMG-GGG has the GGG positioned distal to the ruthenium-binding site. Ru-mis-GMG contains an AA mismatch intervening between the ruthenium and the methylindole. Ru-GMG-blg-GGG contains a 5′-ATA-3′ bulge between the proximal GMG and distal GGG sites. In all cases, inosine is incorporated near the ruthenium-binding site to allow for maximal charge injection and propagation (29).
Fig. 1.
Schematic illustrations of the DNA assemblies used in this study. Ru, [Ru(bpy')(dppz)(phen)]2+; I, inosine nucleotide; M, methylindole nucleotide.
CT Chemistry Is an Intraduplex Reaction. To initiate CT chemistry with dppz complexes of ruthenium the flash/quench technique is used. However, when these same ruthenium complexes are excited in the absence of quencher, *Ru(II) sensitizes the formation of singlet oxygen. Because singlet oxygen reacts preferentially with guanines (41) within diffusional reach, this chemistry can be used to mark the binding site of the oxidant.
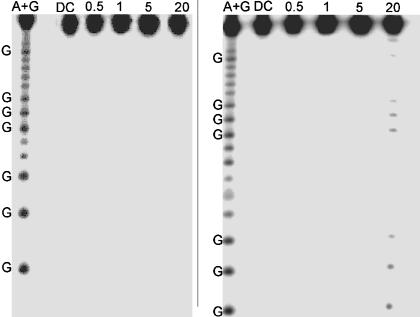
To confirm the intraduplex nature of the CT chemistry, we irradiated an unlabeled ruthenium-modified duplex in the presence of a 32P-end-labeled duplex containing no ruthenium. If interduplex associations occur, damage will be observed on the 32P-end-labeled duplex even though this assembly does not contain ruthenium. Over a concentration range of 0.5–20 μM, the 32P-end-labeled duplexes show no damage at all (Fig. 2). This lack of reaction indicates that over this concentration range all CT chemistry is intraduplex and the assemblies do not form higher-order aggregates.
Fig. 2.
Control experiments for interduplex reaction. Shown is PAGE after irradiation of Ru-GGG-GGG (Left) and bulge-containing duplex Ru-GGG-blg-GGG (Right) in the absence of quencher. Samples contained equimolar amounts of nonlabeled duplex containing the Ru oxidant and 32P-labeled duplexes without Ru. Lane 1 shows the sequencing reaction A+G. Lane 2 shows damage of 5 μM duplex in the absence of light. Lanes 3, 4, 5, and 6 display damage after 60 min of irradiation of 0.5, 1, 5, and 20 μM Ru-DNA, respectively. All samples contained 15 mM sodium phosphate (pH 7) with 50 mM NaCl.
Similar results are obtained for the bulge-containing assembly at low concentrations (0.5–5 μM). No damage is observed at concentrations of ≤5 μM in the assemblies that do not contain ruthenium; under these conditions the assemblies do not aggregate and the CT is intraduplex. However, at 20 μM, the bulge-containing assembly does yield singlet oxygen damage even though it does not contain tethered ruthenium (Fig. 2). Thus, at this high concentration interduplex interactions do occur. The singlet oxygen damage is observed predominately at the guanines in both GGG sites on either side of the bulge. The metallointercalator binds most likely at the bulge itself. However, compared with guanine, adenine and thymine are far less susceptible to damage by 1O2, and, therefore, damage is not observed immediately at the bulge.
Because of the interduplex associations at 20 μM, experiments using the bulge-containing assembly were conducted only at low concentration (1 μM) where no damage is detectable. This interduplex reaction observed at 20 μM clearly results from a species present in equilibrium with the free ruthenium-modified duplex and the 32P-end-labeled duplex. The high sensitivity of phosphorimagery, in combination with guanine damage by 1O2 sensitization by the DNA-bound intercalator, provides critical information regarding the binding site of the oxidant. It is therefore quite straightforward to identify and confirm conditions under which solely an intraduplex CT reaction occurs. Some percentage of aggregation of DNA duplexes would instead yield detectable damage. Under the conditions used for transient absorption and biochemical experiments, no aggregation is observed and all CT reactions are intraduplex.
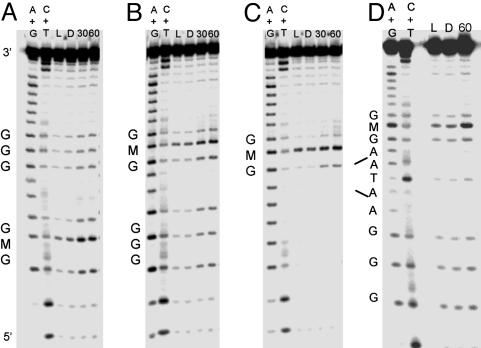
Oxidative Damage Products Observed by Gel Electrophoresis. Oxidative damage assays were carried out with denaturing PAGE, which allows for quantitation of DNA damage products resulting from flash/quench experiments. The PAGE results reveal the damage pattern for Ru-GMG, Ru-GGG-GMG, Ru-GMG-GGG, and Ru-GMG-blg-GGG (Fig. 3). These assemblies have two oxidatively sensitive sites, GMG and GGG, which on one-electron oxidation yield different damage products. Damage at guanine is expected to include 8-oxo-guanine, formamidopyrimidine, oxazalone, and imidazalone (27); damage at M also produces a piperidine-sensitive lesion, but the final products have not been characterized.
Fig. 3.
Comparison of oxidative damage in different assemblies. Shown are PAGE results after irradiation of Ru-GGG-GMG (A), Ru-GMG-GGG (B), Ru-GMG (C), and Ru-GMG-blg-GGG (D) in the presence of quencher. For each assembly, lanes 1 and 2 represent the sequencing reactions A + G and C + T, respectively. Lane 3 is a light control irradiated for 30 min in the absence of quencher. Lanes 4, 5, and 6 show damage after 0, 30, and 60 min of irradiation, respectively. Samples consisted of 5 μM duplex (1 μM for bulge assembly), 15 mM sodium phosphate (pH 7), 50 mM NaCl, and 125 μM [Co(NH3)5Cl]2+.
In all assemblies examined, damage at the methylindole-containing site is predominant. For Ru-GGG-GMG, Ru-GMG-GGG, and Ru-GMG-blg-GGG damage is also observed at the low-energy GGG site. The ratio of damage at the distal GGG compared with the proximal GMG is 0.4 ± 0.1 for Ru-GMG-blg-GGG. In contrast, a ratio of 0.8 ± 0.1 is observed for the assembly lacking a bulge. This decrease in damage out to the distal GGG site for Ru-GMG-blg-GGG is consistent with earlier studies where intervening base-stacking perturbations, such as the 5′-ATA-3′ base bulge, were found to inhibit CT (5, 6).
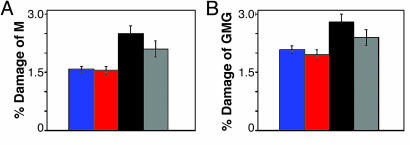
For all assemblies, the amount of damage at the M site after 60 min of irradiation was quantitated with respect to the parent band and corrected for damage observed in the absence of irradiation (Fig. 4A). Here, the greatest amount of M damage is observed for Ru-GMG. The assemblies containing an additional low-energy site, Ru-GMG-GGG and Ru-GGG-GMG, show ≈40% less oxidative damage at the M. However, on insertion of a base bulge, a partial restoration of damage at the M, ≈50% compared with Ru-GMG, is observed. As seen in Fig. 3, damage at the guanine bases flanking the methylindole site is also seen. These guanines were incorporated specifically to provide stability at the methylindole site by base stacking. However, this stacking may also render the guanines more susceptible to oxidative damage. If the damage at these flanking guanine sites is quantitated and included with the damage at the M, a pattern similar to that in Fig. 4A is obtained (Fig. 4B).
Fig. 4.
Quantitation of damage observed by PAGE. Shown are data for M alone (A) or M and flanking guanines (B) compared with parent band for Ru-GGG-GMG (blue), Ru-GMG-GGG (red), Ru-GMG (black), and Ru-GMG-blg-GGG (gray).
Emission and Transient Absorption Spectroscopy on Ruthenium-Modified Assemblies. Time-resolved luminescence measurements at 610 nm indicate that the excited-state ruthenium complex, *[Ru(bpy')(dppz)(phen)]2+, decays biexponentially with τ1 = 71 ns (76%) and τ2 = 279 ns (26%) for Ru-GMG (Table 1). Similar values are obtained for all assemblies examined. These values are consistent with those reported earlier for dppz complexes of Ru(II) bound to DNA (42, 43). On addition of 15 eq of [Ru(NH3)6]3+, the luminescence is quenched by 80% in all assemblies.
Table 1. Kinetic data for [Ru(bpy')(dppz)(phen)]2+ and the methylindole radical (Mrad).
| Measurement | Ru-GMG | Ru-GMG-GGG | Ru-GGG-GMG | Ru-mis-GMG |
|---|---|---|---|---|
| Luminescence lifetime of *[Ru]2+ -DNA* | τ1 = 71 ns (76%) | τ1 = 68 ns (76%) | τ1 = 72 ns (79%) | τ1 = 80 ns (74%) |
| τ2 = 279 ns (26%) | τ2 = 227 ns (26%) | τ2 = 269 ns (26%) | τ2 = 280 ns (29%) | |
| Mrad formation†‡ | 4 × 107 s-1 | 4 × 107 s-1 | 4 × 107 s-1 | N/A |
| Mrad decay†§ | 1 × 106 s-1 | ¶ | ¶ | N/A |
All samples contained 20 μM Ru-duplex, 10 mM NaCl, 10 mM potassium phosphate (pH 7). N/A, not applicable.
The luminescence traces were fit to a biexponential function by a nonlinear least-squares method with convolution of the instrument response function. Uncertainties in values are ±10%.
Samples contained 15 eq of [Ru(NH3)6]3+.
The rise of the signal at 600 nm was fit to a monoexponential function by a nonlinear least-squares method. Considering bandwidth used and region fit, these values represent lower limits.
The transient absorption decay at 600 nm corresponding to the Mrad was fit to a monoexponential function by a nonlinear least-squares method. Uncertainties in values are ±10%.
No significant decay was observed over 50 μs.
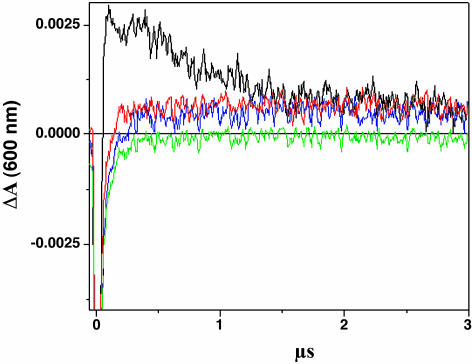
Fig. 5 shows the kinetic development of the transient absorption spectra monitored at 600 nm after laser excitation of Ru-GMG, Ru-GGG-GMG, Ru-GMG-GGG, or Ru-mis-GMG in the presence of [Ru(NH3)6]3+. Previous transient absorption and EPR studies provide evidence for formation of the methylindole cation radical that absorbs in this region (28). The transient signals for all assemblies are initially negative, attributed to emission from residual *Ru(II) generated in the flash/quench technique. For Ru-GMG, a large positive signal at 600 nm, consistent with formation of the methylindole radical, is observed. Positive signals of roughly equal height are also observed at 600 nm for Ru-GGG-GMG and Ru-GMG-GGG, but they are considerably smaller than that for Ru-GMG. Although these signals are small, they are significant and clearly represent a positive signal compared with that obtained with the mismatch-containing assembly. In Ru-mis-GMG, which contains an AA mismatch intervening between the ruthenium oxidant and methylindole, no significant positive signal is observed. This attenuated yield in radical formation is consistent with decreased yields of CT products seen previously in mismatch-containing DNA (6, 28).
Fig. 5.
Time-resolved transient absorption data. Shown are traces at 600 nm for Ru-GMG (black), Ru-GGG-GMG (blue), Ru-GMG-GGG (red), and Ru-mis-GMG (green). The samples contained 20 μM Ru-DNA, 300 μM [Ru(NH3)6]3+, 10 mM NaCl, and 10 mM potassium phosphate (pH 7).
The rise of the traces at 600 nm (<0.2 μs) was fit to a monoexponential function, indicating that the formation of the transient occurs with a rate of ≈4 × 107 s–1 for all three assemblies (Table 1). This rate is consistent with those obtained previously for formation of the methylindole cation radical by CT from a bound Ru oxidant (28, 29). This value is comparable with the timescale of the quenched emission of the Ru complex bound to DNA and approaches the instrument response. For all assemblies, the return to the Ru(II) ground state is also observed by monitoring the bleach of the metal-to-ligand charge transfer band at 440 nm. The disappearance of the negative signal at 440 nm, reflecting the conversion from *Ru(II) to Ru(III), is concomitant with the rise of the positive signal at 600 nm. Hence, the rate of formation of Ru(III) and the base radical are coincident so that the rate of CT is greater than or equal to these values.
Also evident in Fig. 5 is the difference in the decay rate of the positive signal at 600 nm for Ru-GMG, Ru-GGG-GMG, and Ru-GMG-GGG. The radical formed in Ru-GMG appears to contain two components, one of which decays significantly faster than those formed in Ru-GGG-GMG and Ru-GMG-GGG. Indeed, fitting the first part of the decay of the 600-nm signal to a monoexponential function reveals a rate ≈106 s–1 for Ru-GMG, whereas no significant decay is observed for the long-lived component nor for Ru-GGG-GMG and Ru-GMG-GGG over 50 μs. This long-lived species may reflect generation of the guanine radical; the transient assigned to the neutral guanine radical was found to decay on the order of 104 s–1 in analogous Ru-tethered duplexes (26). The finding of oxidative damage at the guanine bases flanking M in the biochemical experiments supports this formation of guanine radical in Ru-GMG and in Ru-GGG-GMG and Ru-GMG-GGG. The neutral guanine radical does absorb at 600 nm but with an extinction coefficient 3.5-fold lower than that for the methylindole cation radical (44–46).
Discussion
Singlet Oxygen Chemistry to Confirm an Intraduplex CT Reaction. The ruthenium photochemistry (26) is valuable not only in initiating charge injection into the DNA base pair stack but also in providing a reaction to mark the intercalator position along the DNA duplex. It has been suggested that duplexes containing a tethered Ru or Rh oxidant may aggregate (8, 47). In appropriately designed control experiments with a mixture of unlabeled ruthenium-tethered duplexes and 32P-end-labeled duplexes containing no ruthenium, any interduplex association can be detected. No damage is observed on the 32P-end-labeled duplex lacking the pendant intercalator. Hence, the data presented here clearly show that, at the concentrations used, the duplex assemblies do not aggregate. Thus, all CT chemistry is intraduplex. For these assays of intermolecularity, the bulge-containing assembly serves as a useful positive control. Here, a small amount of interduplex reaction is detectable at 20 μM, consistent with higher binding of the dppz complex at the bulged site. Transient absorption experiments, requiring higher concentrations, thus were not carried out with the bulge-containing assembly. Instead all CT experiments with this assembly were conducted at 1 μM.
Formation of DNA CT Intermediates. In Ru-GGG-GMG, the presence of the GGG site clearly diminishes the yield of methylindole cation radical. After charge injection, the hole has two low-energy sites for localization in Ru-GGG-GMG but only one in Ru-GMG. The oxidation potential of M is estimated to be 1.0 V vs. normal hydrogen electrode (NHE) (28), whereas the oxidation potential of a 5′-GGG-3′ site is 1.0 (48, 49) to 1.2 (50) V vs. NHE. However, the presence of the GGG site does not affect the rate of radical formation. The signal at 600 nm of Ru-GMG has two components, with contributions likely from the methylindole cation radical and the neutral guanine radical; this signal is considerably smaller for Ru-GGG-GMG and Ru-GMG-GGG, reflecting a greater contribution from the more weakly absorbing guanine radical. However, the rise of the signal at 600 nm occurs with a rate of 107 s–1 for all assemblies. This lack of sensitivity of the rate of radical formation is consistent with earlier studies demonstrating that CT is not rate-limiting over this distance (28).
Perhaps most interesting is that the relative orientations of the GGG and GMG sites do not affect methylindole cation radical formation. Lewis et al. (51) have suggested a hopping rate of 106 s–1 among guanine sites. If such a rate were operative, one would expect to see differences in the transient profiles for these two assemblies on the microsecond timescale. Instead, the insensitivity to orientation we find when the product radical is monitored directly supports a faster diffusive hopping mechanism.
The transient absorption data also provide insight into the different reactivity of the two sites. In Ru-GMG, which contains only one low potential site, the rate of methylindole cation radical decay is ≈106 s–1. The rate of decay of the neutral guanine radical in similar DNA assemblies is much slower, ≈104 s–1 (26). The transient signals obtained for assemblies containing both the GMG and GGG sites, Ru-GGG-GMG and Ru-GMG-GGG, are consistent with decay profiles for assemblies containing either GMG or GGG. For these assemblies the signal heights are smaller, as expected from the smaller extinction coefficient for the neutral guanine radical, than for the methylindole cation radical (44–46). Moreover, the signals do not decay appreciably over the 50-μs time window, consistent with our earlier measurements of guanine radical decay in flash/quench experiments.
Competition Between Two Oxidatively Sensitive Sites in DNA. The biochemical experiments confirm the competition between the GMG and GGG sites, irrespective of orientation, and provide quantitative information regarding the yield of oxidized products at both sites. Again these data support the idea that the GGG site is effective in competing for the migrating hole and provides an additional trapping site. Thus, by both biochemical and spectroscopic analysis, switching the orientation of the GMG and GGG does not affect the yield of CT products. Both forward and back CT occurs through the DNA base pair stack so as to provide equilibration across the duplex on a timescale that is fast compared with localization at a particular site(s).
Further evidence of charge equilibration results from incorporating an intervening base-stacking perturbation and monitoring the fate of the injected hole. Upon insertion of a 5′-ATA-3′ base bulge, a 50% decrease in CT to the distal GGG site is observed compared with the non-bulge-containing assembly. A decrease in CT to a distal site has been observed previously with base mismatches (6), nonaromatic protein side chains (7), and a variety of base bulges (5).
Most interestingly, with the decrease in CT to the distal GGG site as a result of the base bulge, an increase in oxidative damage at M relative to Ru-GMG-GGG is observed. Thus, we can consider this increase as a recovery in oxidative damage compared with the control assembly containing no GGG site. The recovery, however, is not complete. The intervening 5′-ATA-3′ base bulge does not eliminate CT to the distal GGG site; it decreases damage at the distal site by 50%. Thus we find also a 50% recovery of M damage upon insertion of the base bulge. These data suggest that the 50% of radicals that do not initially traverse the bulge to reach the GGG site instead return to the proximal M site to yield oxidative damage. Equilibration across the duplex is therefore achieved before trapping at an individual site(s).
Fig. 6 illustrates our model to account for the transient absorption and biochemical data. After flash/quench and hole injection, charge propagation occurs in a forward direction with a rate kf. In Ru-GMG, Ru-GGG-GMG, and Ru-GMG-GGG, the hole can sample the entire length of the duplex before returning with a rate kr.In Ru-GMG-blg-GGG, the bulge presents a barrier to CT and most of the injected charge can only sample the region of the helix proximal to the ruthenium oxidant. Charge that might have been trapped at the distal GGG site in the absence of a bulge instead returns to M and is trapped as oxidative damage. Some charge may be trapped during forward propagation; however, a portion of charges certainly is trapped after initially sampling the entire duplex. For Ru-mis-GMG, a base mismatch prevents charge propagation to yield the methylindole cation radical and the charge equilibrates only over the region of the duplex proximal to the ruthenium.
Fig. 6.
Proposed model of charge equilibration in DNA.
Kinetic and Themodynamic Traps of CT Damage. To observe competition between two different sites in DNA, neither site can behave as an absolute kinetic or thermodynamic trap. If one site presents a very deep thermodynamic well, damage may be funneled exclusively to this site. If on the other hand, one site is a fast kinetic trap of oxidative damage, the second site may not be sampled.
In the data presented here both biochemical and spectroscopic analyses reveal comparable amounts of oxidative damage at the methylindole site in Ru-GMG-GGG and Ru-GGG-GMG, regardless of orientation of the two oxidative traps. This lack of dependence on orientation of the trapping sites is in contrast to previous biochemical studies using an internally tethered anthraquinone oxidant (15). An intervening 8-oxo-G was found to reduce significantly the yield of oxidative damage at a distal GG site. However, when 8-oxo-G was not intervening and instead was positioned distal to the GG site, the 8-oxo-G was not nearly as efficient in competing for and trapping oxidative damage. This contrasting behavior may result from differences in oxidation potential for M and 8-oxo-G; the oxidation potential of M is considerably higher than that of 8-oxo-G (E0 = 0.6–0.8 V) (52–54). Additionally, based on calculations, the ionization potential of a GGG site is 0.2 eV lower than that of a GG site (47).
Precedence also exists for a kinetic factor in the trapping of oxidative damage at two distinct sites in DNA. Selective and exclusive oxidation at a 7-deaza-guanine (ZG) base in DNA was observed in the presence of a GGG site by using a cyanobenzophenone-2′-deoxyuridine oxidant (10), despite the fact that theoretical calculations predict that the ionization potential of ZG is 0.38 eV higher than that of GGG. A kinetic factor was suggested to explain the observed oxidative damage at only ZG. Damage was not examined in an assembly where the ZG and GGG sites were switched in orientation relative to the oxidant.
Another example of a kinetic trap for oxidizing equivalents in duplex DNA is a thymine dimer. The repair of this DNA lesion, which forms as a result of a [2 + 2] photocycloaddition between adjacent thymine bases, can be triggered by one-electron oxidation with a standard oxidation potential of approximately +2.0 V (55). Thermodynamically, the thymine dimer is considerably more difficult to oxidize than a 5′-GG-3′ site. Furthermore, the radicals formed at the two sites have distinctively different lifetimes; the neutral guanine radical has a lifetime of 104 s–1, whereas the thymine dimer cation radical is estimated to have a lifetime of 109 s–1 (56). Nonetheless, when both a thymine dimer and a 5′-GG-3′ site are present in a rhodium-tethered duplex, both repair of the dimer and oxidation of the 5′-GG-3′ site are observed (57). Whereas the 5′-GG-3′ site provides a thermodynamic trap, the dimer provides a kinetic trap, resulting in competition between the two sites.
Significantly, by selecting appropriate sites, namely GMG and GGG, that are comparable energetically and kinetically, we can observe charge equilibration across the DNA duplex. Charge migration leads to a sampling of the entire duplex on a timescale that is fast compared with localization and trapping. CT through DNA cannot be considered statically with conclusions based on measurements of yield. Instead, the reaction must be viewed dynamically with rates of CT across the duplex in both forward and reverse directions being considered.
Acknowledgments
This study was supported by the National Institutes of Health.
Abbreviations: CT, charge transport; dppz, dipyridophenazine; M, 4-methylindole; BET, back electron transfer; bpy', 4′-methylbypyridine-4-butyric acid.
References
- 1.Delaney, S. & Barton, J. K. (2003) J. Org. Chem. 68, 6475–6483. [DOI] [PubMed] [Google Scholar]
- 2.Schuster, G. B. (2000) Acc. Chem. Res. 33, 253–260. [DOI] [PubMed] [Google Scholar]
- 3.Giese, B. (2002) Annu. Rev. Biochem. 71, 51–70. [DOI] [PubMed] [Google Scholar]
- 4.Lewis, F. D., Letsinger, R. L. & Wasielewski, M. R. (2001) Acc. Chem. Res. 34, 159–170. [DOI] [PubMed] [Google Scholar]
- 5.Hall, D. B. & Barton, J. K. (1997) J. Am. Chem. Soc. 119, 5045–5046. [Google Scholar]
- 6.Bhattacharya, P. K. & Barton, J. K. (2001) J. Am. Chem. Soc. 123, 8649–8656. [DOI] [PubMed] [Google Scholar]
- 7.Wagenknecht, H.-A., Rajski, S. R., Pascaly, M., Stemp, E. D. A. & Barton, J. K. (2001) J. Am. Chem. Soc. 123, 4400–4407. [DOI] [PubMed] [Google Scholar]
- 8.Schlientz, N. W. & Schuster, G. B. (2003) J. Am. Chem. Soc. 125, 15732–15733. [DOI] [PubMed] [Google Scholar]
- 9.Williams, T. T., Odom, D. T. & Barton, J. K. (2000) J. Am. Chem. Soc. 122, 9048–9049. [Google Scholar]
- 10.Nakatani, K., Dohno, C. & Saito, I. (2000) J. Am. Chem. Soc. 122, 5893–5894. [Google Scholar]
- 11.Furrer, E. & Giese, B. (2003) Helv. Chim. Acta 86, 3623–3632. [Google Scholar]
- 12.Kawai, K., Takada, T., Tojo, S. & Majima, T. (2004) J. Am. Chem. Soc. 126, 1125–1129. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, F. D., Liu, J. Q., Weigel, W., Rettig, W., Kurnikov, I. V. & Beratan, D. N. (2002) Proc. Natl. Acad. Sci. USA 99, 12536–12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nunez, M. E., Hall, D. B. & Barton, J. K. (1999) Chem. Biol. 6, 85–97. [DOI] [PubMed] [Google Scholar]
- 15.Ly, D., Sanii, L. & Schuster, G. B. (1999) J. Am. Chem. Soc. 121, 9400–9410. [Google Scholar]
- 16.Nunez, M. E., Noyes, K. T. & Barton, J. K. (2002) Chem. Biol. 9, 403–415. [DOI] [PubMed] [Google Scholar]
- 17.Nunez, M. E., Holmquist, G. P. & Barton, J. K. (2001) Biochemistry 40, 12465–12471. [DOI] [PubMed] [Google Scholar]
- 18.Kelley, S. O. & Barton, J. K. (1999) Science 283, 375–381. [DOI] [PubMed] [Google Scholar]
- 19.O'Neill, M. A., Becker, H.-C., Wan, C., Barton, J. K. & Zewail, A. H. (2003) Angew. Chem. Int. Ed. Engl. 42, 5896–5900. [DOI] [PubMed] [Google Scholar]
- 20.Williams, T. T., Dohno, C., Stemp, E. D. A. & Barton, J. K. (2004) J. Am. Chem. Soc., in press. [DOI] [PubMed]
- 21.Bixon, M., Giese, B., Wessely, S., Langenbacher, T., Michel-Beyerle, M. E. & Jortner, J. (1999) Proc. Natl. Acad. Sci. USA 96, 11713–11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giese, B., Wessely, S., Spormann, M., Lindemann, U., Meggers, E. & Michel-Beyerle, M. E. (1999) Angew. Chem. Int. Ed. Engl. 38, 996–998. [DOI] [PubMed] [Google Scholar]
- 23.Kendrick, T. & Giese, B. (2002) Chem. Commun. 18, 2016–2017. [DOI] [PubMed] [Google Scholar]
- 24.Henderson, P. T., Jones, D., Hampikian, G., Kan, Y. & Schuster, G. B. (1999) Proc. Natl. Acad. Sci. USA 96, 8353–8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang, I.-J., Gray, H. B. & Winkler, J. R. (1991) J. Am. Chem. Soc. 113, 7056–7057. [Google Scholar]
- 26.Stemp, E. D. A., Arkin, M. R. & Barton, J. K. (1997) J. Am. Chem. Soc. 119, 2921–2925. [Google Scholar]
- 27.Burrows, C. J. & Muller, J. G. (1998) Chem. Rev. 98, 1109–1151. [DOI] [PubMed] [Google Scholar]
- 28.Pascaly, M., Yoo, J. & Barton, J. K. (2002) J. Am. Chem. Soc. 124, 9083–9092. [DOI] [PubMed] [Google Scholar]
- 29.Yoo, J., Delaney, S., Stemp, E. D. A. & Barton, J. K. (2003) J. Am. Chem. Soc. 125, 664–665. [DOI] [PubMed] [Google Scholar]
- 30.Dohno, C., Stemp, E. D. A. & Barton, J. K. (2003) J. Am. Chem. Soc. 125, 9586–9587. [DOI] [PubMed] [Google Scholar]
- 31.O'Neill, M. A., Dohno, C. & Barton, J. K. (2004) J. Am. Chem. Soc. 126, 1316–1317. [DOI] [PubMed] [Google Scholar]
- 32.Beaucage, S. L. & Caruthers, M. H. (1981) Tetrahedron Lett. 22, 1859–1862. [Google Scholar]
- 33.Goodchild, J. (1990) Bioconjug. Chem. 1, 165–187. [DOI] [PubMed] [Google Scholar]
- 34.Della Ciena, L., Hamachi, I. & Meyer, T. J. (1989) J. Org. Chem. 54, 1731–1735. [Google Scholar]
- 35.Dupueur, C. M. & Barton, J. K. (1997) Inorg. Chem. 36, 33–43. [Google Scholar]
- 36.Amouyal, E., Homsi, A., Chambron, J.-C. & Sauvage, J.-P. (1990) J. Chem. Soc. Dalton Trans. 6, 1841–1845. [Google Scholar]
- 37.Strouse, G. F., Anderson, P. A., Schoonover, J. R., Meyer, T. J. & Keene, F. R. (1992) Inorg. Chem. 31, 3004–3006. [Google Scholar]
- 38.Anderson, P. A., Deacon, G. B., Haarman, K. H., Keene, F. R., Meyer, T. J., Reitsma, D. A., Skelton, B. W., Strouse, G. F., Thomas, N. C. & Treadway, J. A. (1995) Inorg. Chem. 34, 6145–6157. [Google Scholar]
- 39.Holmlin, R. E., Dandliker, P. J. & Barton, J. K. (1999) Bioconjug. Chem. 10, 1122–1130. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) in Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 41.Rougee, M. & Bensasson, R. V. (1986) C. R. Acad. Sci. Ser. II 302, 1223–1226. [Google Scholar]
- 42.Delaney, S., Pascaly, M., Bhattacharya, P., Koun, H. & Barton, J. K. (2002) Inorg. Chem. 41, 1966–1974. [DOI] [PubMed] [Google Scholar]
- 43.Lincoln, P., Broo, A. & Norden, B. (1996) J. Am. Chem. Soc. 118, 2644–2653. [Google Scholar]
- 44.Candeias, L. P. & Steenken, S. (1989) J. Am. Chem. Soc. 111, 1094–1099. [Google Scholar]
- 45.Steenken, S. & Jovanovic, S. V. (1997) J. Am. Chem. Soc. 119, 617–618. [Google Scholar]
- 46.Solar, S., Getoff, N., Surdar, P. S., Armstrong, D. A. & Gingh, A. (1991) J. Phys. Chem. 95, 3639–3643. [Google Scholar]
- 47.Fahlman, R. P., Sharma, R. D. & Sen, D. (2002) J. Am. Chem. Soc. 124, 12477–12485. [DOI] [PubMed] [Google Scholar]
- 48.Saito, I., Takayama, M., Sugiyama, H., Nakatani, K., Tsuchida, A. & Yamamoto, M. (1995) J. Am. Chem. Soc. 117, 6406–6407. [Google Scholar]
- 49.Prat, F., Houk, K. N. & Foote, C. S. (1998) J. Am. Chem. Soc. 120, 845–846. [Google Scholar]
- 50.Lewis, F. D., Liu, X., Liu, J., Hayes, R. T. & Wasielewski, M. R. (2000) J. Am. Chem. Soc. 122, 12037–12038. [Google Scholar]
- 51.Lewis, F. D., Liu, X., Liu, J., Miller, S. E., Hayes, R. T. & Wasielewski, M. R. (2000) Nature 406, 51–53. [DOI] [PubMed] [Google Scholar]
- 52.Berger, M., Anselmino, C., Mouret, J.-F. & Cadet, J. (1990) J. Liq. Chromatogr. 13, 929–940. [Google Scholar]
- 53.Goyal, R. N. & Dryhurst, G. (1982) J. Electroanal. Chem. 135, 75–91. [Google Scholar]
- 54.Yanagawa, H., Ogawa, Y. & Ueno, M. (1992) J. Biol. Chem. 267, 13320–13326. [PubMed] [Google Scholar]
- 55.Pac, C., Kubo, J., Tetsuro, M. & Sakurai, H. (1982) Photochem. Photobiol. 36, 273–282. [Google Scholar]
- 56.Kemmink, J., Eker, A. P. M. & Kaptein, R. (1986) Photochem. Photobiol. 44, 137–142. [DOI] [PubMed] [Google Scholar]
- 57.Dandliker, P. J., Nunez, M. E. & Barton, J. K. (1998) Biochemistry 37, 6491–6502. [DOI] [PubMed] [Google Scholar]