Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor that plays a pivotal role in obesity and diabetes. PPARγ has two isoforms, PPARγ1 and PPARγ2. We investigated the functional differences between PPARγ1 and PPARγ2 by selectively disrupting PPARγ2 in mice. In contrast to the embryonic lethality of PPARγ-deficient mice, PPARγ2-/- mice survived. Although normal development was identified in other tissues we examined, PPARγ2-/- mice exhibited an overall reduction in white adipose tissue, less lipid accumulation, and decreased expression of adipogenic genes in adipose tissue. In addition, insulin sensitivity was impaired in male PPARγ2-/- mice, with dramatically decreased expression of insulin receptor substrate 1 and glucose transporter 4 in the skeletal muscle, but thiazolidinediones were able to normalize this insulin resistance. Consistent with in vivo data, PPARγ2-/- mouse embryonic fibroblasts showed a dramatically reduced capacity for adipogenesis in vitro compared with wild-type mouse embryonic fibroblasts. Taken together, our data demonstrate that PPARγ2 deficiency impairs the development of adipose tissue and insulin sensitivity. PPARγ2-/- mice may provide a tool to study the role of PPARγ2 in obesity and diabetes.
Keywords: adipogenesis, obesity, diabetes
Obesity poses a major public health problem by predisposing individuals to coronary heart disease, congestive heart failure, and stroke. Recent results have indicated that an estimated 61% of U.S. adults are either overweight or obese (www.cdc.gov/nccdphp/dnpa/obesity/defining.htm). A large proportion of obese individuals exhibit concomitant insulin resistance, hypertension, and dyslipidemia as part of a complex dysmetabolic syndrome (also called syndrome X) (1). Among the genes that have been associated with syndrome X, the Pro12Ala variant of the peroxisome proliferator-activated receptor γ2 (PPARγ2) gene has emerged as a leading candidate on the basis of its significant association with diabetes (2).
Mounting data have documented that PPARγ is the molecular target of the thiazolidinedione (TZD) class of antidiabetic drugs, such as troglitazone (Rezulin), rosiglitazone (Avandia), and pioglitazone (Actos) (1). PPARγ is highly expressed in adipose tissues but is expressed at much lower levels in other tissues, including major insulin target tissues, skeletal muscle, and liver (3, 4). This expression pattern suggests that adipose tissue may be the primary target for the insulin-sensitizing effect of TZDs. Indeed, recent reports have documented that the selective reduction of PPARγ in adipose tissue demonstrates its essential role in adipogenesis (5, 6). In addition, liver- and muscle-specific PPARγ deficiency did not affect the antidiabetic action of TZDs (7, 8), which suggests that TZDs target adipose tissue and markedly change the gene expression of adipocytokines that may improve insulin resistance in other tissues. Therefore, studying the molecular basis of the effect of PPARγ on adipocyte differentiation and insulin sensitivity is an important goal for understanding the causes, prevention, and treatment of obesity.
PPARγ is a key regulator of adipose cell differentiation, fatty acid uptake, and lipogenesis through its influence on the production of the enzymes required for lipid storage and metabolism (1, 9). The PPARγ gene is composed of nine exons that span >100 kb on chromosome 3p25–24 (10). It generates two isoforms, PPARγ1 and PPARγ2, by using alternative promoters and differential splicing (11). PPARγ2 has an additional 30 amino acids at the N terminus that render its ligand-independent activation domain, which is somewhat more effective in activating the transcription of the PPARγ reporter gene than PPARγ1 (3). Other properties of PPARγ1 and PPARγ2, including DNA binding, ligand binding, and interaction with coactivators, are mediated by identical domains and are quite similar.
Although PPARγ1 and PPARγ2 are expressed at comparable levels in adipocytes, the relative importance of the two PPARγ isoforms for adipogenesis has remained an open question. In vitro studies of the ability of each isoform to stimulate the differentiation of adipose tissue have yielded controversial results (12, 13). In the present study, we have generated PPARγ2-specific knockout (KO) mice by selectively targeting the PPARγ2-specific exon B of the PPARγ gene. Here we show that the development of adipose tissue and insulin sensitivity in PPARγ2-/- mice are impaired.
Materials and Methods
Generation of PPARγ2-/- Mice. A genomic clone that included the 5′ end of the murine PPARγ2 gene was isolated from a SV129 mouse library. The targeting vector was generated in the pKO plasmid, which contains the PGK-neo-poly(A) and pol2-DTA-poly(A) cassettes for positive and negative selection, respectively (see Supporting Text, which is published as supporting information on the PNAS web site).
Metabolic Parameters. Serum glucose levels were measured by using Glucometer Elite (Bayer, Mishawaka, IN). Plasma insulin, leptin, and adiponectin levels were measured by radio immunoassay kits from Linco Research (St. Charles, MO). Plasma triglyceride and total cholesterol levels were determined by enzymatic assays (Boehringer–Mannheim). i.p. glucose tolerance tests (2 g/kg) were performed as described (14). Insulin sensitivity was measured by injecting insulin into the abdominal cavity at a dose of 1.5 units/kg (15).
Histology. Tissues were fixed in 10% formalin, dehydrated, embedded in paraffin, and sectioned for hematoxylin/eosin staining. Images were captured by using an Olympus BX60 camera (Tokyo) at different magnifications and processed in adobe photoshop (Adobe Systems, San Jose, CA).
RNA and Protein Preparation and Analysis. RNA and protein expression levels were analyzed by Northern and Western blot analysis, as described (16).
Preparation of Mouse Embryonic Fibroblast (MEF) Cells and the Induction of Adipocyte Differentiation. Mouse embryos were carefully collected between embryonic day (E)12.5 and E13.5, and MEFs were prepared as described (17). Fat accumulation was scored by determination of mRNA and protein levels of adipocyte-specific genes and by staining of lipids with Oil red O (18) (see Supporting Text for detailed methods).
Statistical Analysis. All data were evaluated with a two-tailed unpaired Student's t test or compared by one-way ANOVA followed by Fisher's t test and are expressed as the mean ± SD. A value of P < 0.05 was considered statistically significant.
Results
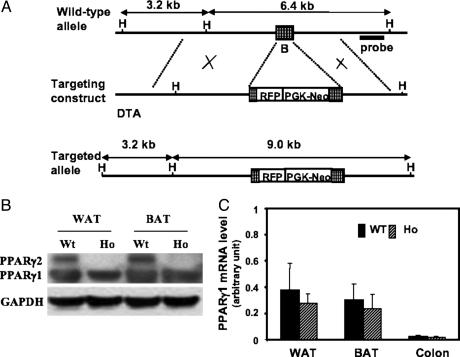
Generation of PPARγ2-/- Mice. To study the functional role of PPARγ in vivo, we produced a selective disruption of the PPARγ2 isofrom by using a targeting vector that was designed to replace the initiation codon and the partial exon B with the red fluorescence protein (RFP) coding sequence and the neomycin-resistance gene cassette (Fig. 1A). RFP was driven by the PPARγ2 promoter. Embryonic stem cell clones containing the correct replacement (Fig. 6A, which is published as supporting information on the PNAS web site) were injected into C57BL/6 blastocysts. Germ-line transmission was achieved, and heterozygous (PPARγ2+/-) mice were crossed to produce homozygous (PPARγ2-/-) offspring (Fig. 6B). The absence of PPARγ2 expression in adipose tissue of PPARγ2-/- mice was confirmed by RT-PCR for the mRNA (Fig. 6C) and by Western blotting for the protein (Fig. 1B). As expected, the expression of PPARγ1 in adipose tissues and in other organs, such as colon, was not significantly affected by PPARγ2 KO, as determined by quantitative real-time PCR analyses (Fig. 1C). Taken together, our data show that we have successfully generated PPARγ2-specific KO mice.
Fig. 1.
Specific disruption of the PPARγ2 gene. (A) The structures of the endogenous PPARγ gene, targeting vector, and mutated chromosomes. The position of the B exon of the gene is indicated as an open box. H, restriction sites of HindIII; RFP, red fluorescence protein gene; PGK-Neo, neomycin resistance gene cassette driven by the phosphoglycerate kinase (PGK) promoter; DTA, diphtheria toxin-A chain gene. (B) Western blot analysis of PPARγ1 and PPARγ2 in BAT and WAT from wild-type or homozygous mice. (C) Quantitative real-time PCR was performed by using specific primers for PPARγ1, as described in Materials and Methods. The RNA tested was from WAT and BAT, and colon samples were from wild-type and homozygous mice. The relative PPARγ1 mRNA levels normalized by GAPDH are expressed as mean ± SD (n = 6).
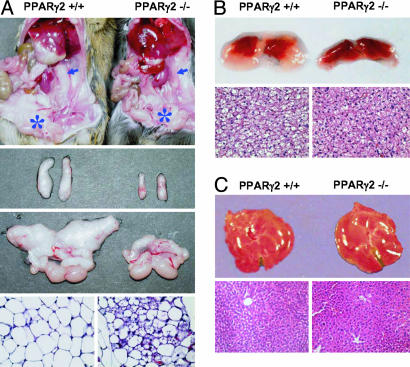
Reduced White Adipose Tissue (WAT) Mass and Lipid Accumulation in PPARγ2-/- Mice. In contrast to the embryonic lethality of PPARγ null mice (19–21), PPARγ2-/- mice survived. There were 61 PPARγ2-/- mice of 247 live-born progeny of the PPARγ2+/- mice. This number of PPARγ2-/- offspring was consistent with Mendelian inheritance. There was no evidence of gross abnormality or tumor development in PPARγ2-/- mice up to age 24 weeks. After weaning, PPARγ2+/+ and PPARγ2-/- mice gained weight similarly when they were fed regular chow. The body weights of 12- to 15-week-old wild-type and PPARγ2-/- littermates were indistinguishable (Fig. 7A, which is published as supporting information on the PNAS web site, n = 7–11). However, PPARγ2-/- animals were protected against obesity induced by a high-fat diet (Fig. 7B, n = 5–7). In addition, food intake was similar in wild-type (3.1 ± 0.3 g per mouse per day) and PPARγ2-/- mice (3.2 ± 0.3 g per mouse per day). Of interest, the PPARγ2-/- mice exhibited an overall reduction in the weight of WAT. Compared with tissues from PPARγ2+/+ mice, the weights of reproductive, inguinal, and retroperitoneal fat pads had significantly decreased by 74%, 25%, and 37% respectively (Fig. 7C). In the animals fed the high-fat diet, fat pads in PPARγ2-/- mice showed no significant changes, but these pads showed dramatic increases in the wild-type littermates, leading to an even more dramatic difference (data not shown). This overall reduction in adipose tissue in PPARγ2-/- mice was observed in males and females. In contrast to WAT, other tissues, including brown adipose tissue (BAT), liver, heart, and spleen, had similar weights (Fig. 7C) and exhibited no gross abnormalities when PPARγ2-/- mice were compared with their wild-type littermates.
To further characterize the phenotype of adipose tissue, we examined histological sections of WAT and BAT, as well as other organs. Only the adipose tissues showed striking morphological changes (Fig. 2). In control littermates, white-fat adipocytes were uniform in size, whereas adipocytes in PPARγ2-/- mice, especially for reproductive fat deposits, were smaller and heterogeneous in size, which indicates that the reduction in total fat mass may be caused by less triglyceride accumulation (Fig. 2 A). The detailed analysis of adipocyte size and the DNA content of WAT from wild-type and PPARγ2-/- mice documented that the cell size of adipocytes in reproductive fat pad decreased ≈2.6-fold and the DNA content per milligram of WAT increased ≈1.5-fold (data not shown). On the basis of the ≈4-fold weight loss of reproductive fat pad in PPARγ2 KO mice (Fig. 7C), we conclude that the decrease in both the size and number of adipocytes contributed to the lipodystropy seen in PPARγ2-/- mice.
Fig. 2.
Reduced lipid accumulation in WAT and BAT in PPARγ2-/- mice. (A Top) A comparison of WAT samples. A ventral view of a PPARγ2-/- mouse and its control littermate. (Middle) A comparison of retroperitoneal and reproductive fat pads. (Bottom) Hematoxylin/eosin (H&E)-stained sections of reproductive fat from a male PPARγ2-/- mouse and its control littermate. (B Upper) A comparison of BAT samples. (Lower) H&E-stained sections of brown fat from a male PPARγ2-/- mouse and its control littermate. (C Upper) A comparison of liver samples. (Lower) H&E-stained sections of liver from a PPARγ2-/- male mouse and its control littermate.
To address whether PPARγ2 deficiency in WAT represents the development of BAT, we examined the expression of a BAT-specific gene, UCP-1. Our results document that UCP-1 was not expressed in the WAT of PPARγ2-/- mice (data not shown). A reduction in lipid accumulation was also seen in BAT, both the number and size of lipid droplets were reduced in PPARγ2-/- mice, compared with their wild-type littermates, in which brown fat adipocytes normally contained multiple lipid droplets (Fig. 2B). Therefore, this may account, at least in part, for the darker appearance of PPARγ2-/- BAT. Unlike PPARγ-deficient mice (20) and the adipocyte-selective reduction in PPARγ (5, 6) that increased the deposition of fat in the liver, PPARγ2-/- mice had a normal liver appearance and no appreciable fatty acid accumulation in the liver (Fig. 2C).
Consistent with reduced adipose tissue mass, circulating leptin and adiponectin levels were decreased by 28∼43% and 45∼46% in male and female PPARγ2-/- mice, respectively (Fig. 8 A and B, which is published as supporting information on the PNAS web site; n = 7–11, P < 0.05). No statistically significant differences between wild-type and KO mice were observed for circulating triglyceride and cholesterol levels (Fig. 8 C and D, n = 6–7).
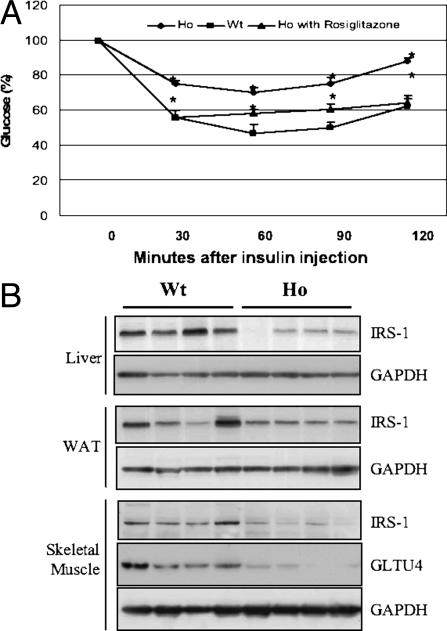
Increased Insulin Resistance in PPARγ2-/- Mice. To further characterize the phenotype of PPARγ2-/- mice, we examined their metabolic parameters. Both fasting plasma glucose and insulin levels of PPARγ2-/- mice were normal, compared with those of wild-type mice (Fig. 8 E and F). Similarly, there was no difference in the nonfasting state (data not shown). In addition, we found that glucose tolerance between PPARγ2-/- and wild-type littermates was similar according to the results of i.p. glucose tolerance tests (data not shown). Intriguingly, insulin tolerance tests showed that PPARγ2-/- male mice were severely resistant to the glucose-lowering effect of exogenous insulin (Fig. 3A) but not their female counterparts (data not shown). Of interest, the impaired insulin sensitivity in male PPARγ2-/- mice was normalized by treatment for 2 weeks with the PPARγ agonist rosiglitazone at 4 mg/kg per day (Fig. 3A). To further define the molecular mechanism, we examined the expression levels of the proteins responsible for glucose transportation and insulin signaling in the liver, skeletal muscle, and WAT. As shown in Fig. 3B, the expression of insulin receptor (IR) substrate 1 in PPARγ2-/- mice was significantly reduced in the skeletal muscle, liver, and WAT, and the expression level of GLUT4 was also dramatically decreased in the skeletal muscle of PPARγ2-/- mice. Taken together, these results suggest that PPARγ2-/- may impair insulin-stimulated glucose disposal in skeletal muscle and insulin signaling in skeletal muscle, liver, and WAT. Although the mechanisms causing the sex difference in insulin resistance are unknown, it will be interesting to study whether the estrogen-signaling pathway has any compensatory effect to PPARγ2 deficiency in female mice.
Fig. 3.
Impaired insulin sensitivity in PPARγ2-/- male mice. (A) Plasma glucose levels after an acute injection of insulin (1.5 units/kg) in chow-fed male mice. Treatment of PPARγ2-/- mice with rosiglitazone normalized the action of insulin action **, P < 0.05. (B) Reduced IR substrate 1 (IRS1) in the skeletal muscle, liver, and WAT of PPARγ2-/- mice. The level of GLUT4 expression was dramatically decreased in skeletal muscle of PPARγ2-/- mice.
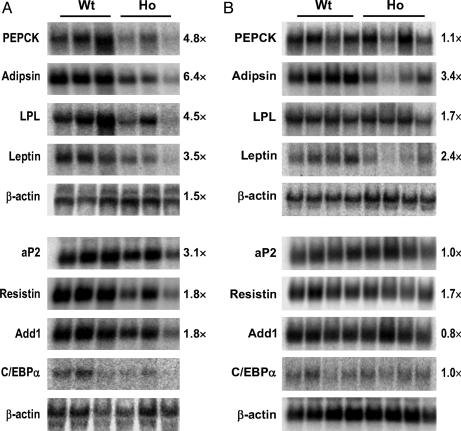
Decreased Expression of Adipogenic Genes in PPARγ2-/- Mice. The development of adipose tissue involves a differentiation switch that activates a new program of gene expression, followed by the accumulation of lipids in a hormone-sensitive manner (9). To further explore the molecular basis through which PPARγ2 deficiency impairs the development of fat tissue and insulin sensitivity, we examined the expression levels of a set of genes involved in adipogenesis by Northern blot analyses. We examined the expression levels of a set of adipocyte marker genes, including phosphoenolpyruvate carboxykinase, adipsin, lipoprotein lipase (LPL), leptin, adipocyte fatty acid-binding protein (aP2), resistin, adipocyte determination and differentiation factor 1 (Add1), and CCAAT/enhancer-binding protein α (C/EBPα). Consistent with the morphological changes of PPARγ2-/- WAT, the expression levels of these genes were significantly decreased by 2- to 6-fold in PPARγ2-/- WAT compared with PPARγ2+/+ WAT (Fig. 4A), which suggests that adipogenesis and lipogenesis are impaired in PPARγ2-/- mice. In contrast, the changes in expression levels of these genes were less significant in BAT than in WAT (Fig. 4B).
Fig. 4.
Decreased adipogenic gene expression in PPARγ2-/- mice. Northern blot analyses of gene expression in WAT (A) and BAT (B)of PPARγ2-/- male mice and their control littermates. Total RNA (20 μg/lane) was hybridized with the indicated probes. The intensity of the signals was quantitated by PhosphorImager analysis. The number beside each image represents the average value of fold reduction in the PPARγ2-/- mice, which was calculated after normalization by β-actin signal. PEPCK, phosphoenolpyruvate carboxykinase; LPL, lipoprotein lipase; aP2, adipocyte fatty acid-binding protein; Add1, adipocyte determination and differentiation factor 1; C/EBPα, CCAAT/enhancer-binding protein α.
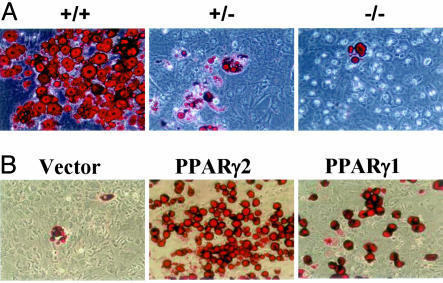
Impaired in Vitro Adipogenesis of PPARγ2-Deficient MEFs. The adipogenesis of MEFs by hormonal induction is a well established model system for the study of adipocyte differentiation in vitro. To further examine the contribution of the two PPARγ isoforms to adipogenesis, we isolated MEFs from days 12.5 to 13.5 of PPARγ2+/+ and PPARγ2-/- embryos. The adipogenesis of MEFs was induced by hormonal stimulation (10 μg/ml insulin/1 μM dexamethasone/0.25 mM isobuthylmethylxanthine) for 2 days, plus incubation with the PPARγ ligand troglitazone (10 μmol/liter). At day 9 after hormonal induction, there was extensive lipid accumulation in PPARγ2+/+ MEFs (15–20%), reduced lipid accumulation in PPARγ2+/- MEFs (2–5%), and barely any lipid accumulation in PPARγ2-/- MEFs (0.1–0.5%) (Fig. 5A). In agreement with these morphological changes, the markers of adipogenesis, including aP2 and CCAAT/EBPα, were also significantly reduced in the hormone-induced PPARγ2-/- MEFs, compared with those in PPARγ2+/+ MEFs (Fig. 9, which is published as supporting information on the PNAS web site). These results indicate that PPARγ2 plays the dominant role in adipogenesis in vitro.
Fig. 5.
Reduced lipid accumulation in PPARγ2-/- MEFs. (A) At day 9 after induction of adipocyte differentiation, cells were fixed and stained for neutral lipids with Oil red O. (B) Either PPARγ1 or PPARγ2 promotes the in vitro adipogenesis of PPARγ2-/- MEFs. We generated the PPARγ2-KO MEF-based cell lines by stable transfection with the puromycin resistance vector (pQCXIP), pQCXIP containing mouse wild-type PPARγ1, or PPARγ2 cDNA. At day 9 after induction, cells were fixed and stained for neutral lipids with Oil red O (×200).
To define whether the overexpression of PPARγ1 and PPARγ2 in PPARγ2-/- MEFs can rescue adipogenesis, we generated PPARγ2-/- MEF-based stable cell lines by stable transfection with puromycin resistance vectors that contained mouse wild-type PPARγ1 or PPARγ2 cDNAs. Of interest, both PPARγ1 and PPARγ2 can promote the in vitro adipogenesis of PPARγ2-/- MEFs (Fig. 5B). Consistent with a previous report (13), PPARγ2 was more powerful than PPARγ1 in adipogenesis in vitro.
Discussion
PPARγ has emerged in recent years as a key regulator of adipogenesis. The existence of PPARγ1 and PPARγ2 isoforms that differ in their N terminus has raised the question whether functional differences existed between them. To date, only in vitro studies have been conducted to address this question, but the data reported were controversial (10, 11). The results presented here indicate that PPARγ1 and PPARγ2 in vivo can drive adipose tissue development, but that PPARγ2 plays the dominant role in adipogenesis.
We report here that PPARγ2-deficient mice survived according to Mendelian inheritance. There was no evidence of gross abnormality in PPARγ2-/- mice up to age 24 weeks. This observation is different from that of a recent study (>40% of PPARγ hyp/hyp mice died by the time of weaning) that used a different strategy to target PPARγ2 (6). Koutnikova et al. (6) replaced the proline residue at position 12 of PPARγ2 gene with alanine by use of homologous recombination. Theoretically, the integrity of PPARγ2 gene expression should be retained in their approach. However, the PPARγ hyp/hyp mice generated by Koutnikova et al. (6) had both PPARγ1 and PPARγ2 knocked out in WAT but elevated PPARγ1 expression in BAT. We realized that one of three loxP sites was inserted into the -45 position of PPARγ2 gene, which might have impaired the initiation site of the PPARγ2 gene transcription. Indeed, our KO configuration did not affect PPARγ2 gene promoter activity, because we did not modify the promoter (data not shown).
In vivo, WAT can be found in a variety of locations, including reproductive, inguinal, retroperitoneal, and s.c. deposits. Although it has been known for many years that the metabolic behavior of mature fat cells differs from deposit to deposit (22–24), the molecular mechanisms that underlie those differences are poorly understood. In addition, it has been documented that increased visceral adiposity in humans is associated with a higher risk of insulin resistance, dyslipidemia, and cardiovascular disease compared with the increase of s.c. adiposity (25). In the present study, we found that the degree of reduced fat tissue pads in different locations was significantly different in PPARγ2-/- WAT, which suggests that PPARγ2 has different adipogenic potential in different locations of adipose tissue. The isolation of preadipocytes from different areas of PPARγ2-/- WAT may provide a useful tool to explore the mechanisms of regional differences in the behavior of fat cells.
It has been well known that the development of adipose tissue consists of two distinct processes, the formation of new adipocytes from precursor cells and the increase in adipocyte size due to fat storage. According to the histological examination of PPARγ2-/- WAT (Fig. 2 A), there are many clusters of small-sized adipocytes scattered among fully developed adipocytes, which indicates that the reduction of adipose tissue in PPARγ2-/- WAT resulted from less fat storage and/or new adipocyte formation. One explanation for the heterogeneity in the size of fat cells in PPARγ2-/- mice might be that lipogenesis is impaired. Indeed, we found reduced expression levels of lipogenic genes, including phosphoenolpyruvate carboxykinase, adipsin, lipoprotein lipase, and lipoprotein lipase, in PPARγ2-/- WAT (Fig. 4). Further studies are required to approach the underlying mechanisms of this heterogeneity. Another explanation for the differences in the size of fat cells in PPARγ2-/- WAT might be different stages of adipocyte differentiation. During adipocyte differentiation, there is a positive-feedback loop between PPARγ and C/EBPα. These two transcriptional factors cooperate to activate the full program of adipogenesis and insulin sensitivity (26). This hypothesis is supported by our results that show the reduced expression of C/EBPα in PPARγ2-/- WAT. In addition, the expression of Add1, the mouse homologue of human sterol regulatory element-binding protein 1 (SREBP1), was also significantly decreased in PPARγ2-/- WAT. Add1/SREBP1 has been shown not only to stimulate many genes involved in fatty acid and cholesterol metabolism (27) but also to potentiate the transcriptional activity of PPARγ, probably through the production of endogenous ligands for PPARγ (28).
The disruption of PPARγ2 in mice leads to reduced adipose tissue. This phenotype is similar to that of fat-specific IR KO mice (FIRKO) (29), which also have low fat mass but normal glucose, insulin, triglyceride, cholesterol, and i.p. glucose tolerance test results. In FIRKO mice, leptin and adiponectin in plasma are significantly increased, which suggests that the IR KO in WAT has a protective effect over the glucose metabolism and aging (29, 30). Because the PPARγ2-/- KO mice show insulin resistance and reduced levels of leptin and adiponectin in plasma, it is unlikely that PPARγ2-/- and FIRKO mice developed lipodystrophy through the same mechanism. However, investigating possible crosstalk between the PPARγ and the IR signaling pathways in WAT may provide insights into the understanding of adipocyte differentiation. The specific reduction of PPARγ in adipose tissue showed the essential role of PPARγ in adipogenesis and also highlighted its role in maintaining the integrity and function of mature adipocytes (5, 6).
A recent study documented that the fat mass reduction in both BAT and WAT was observed in fat-selective PPARγ-KO (FKO) mice that was accompanied by hyperlipidemia and liver steatosis (5). In contrast, our PPARγ2-/- mice, which did not have liver steatosis or dyslipidemia, showed fat reduction only in WAT. It is of interest that the impairment of insulin sensitivity was dramatically improved by TZD in FKO and PPARγ2-/- mice. This finding raises the question of where the sites of TZD action are in these mice. The sensitization effects in FKO and PPARγ2-/- mice may occur as a direct activation of PPARγ within liver and skeletal muscle. Indeed, our data in the present study document that the expression of IR substrate 1 in PPARγ2-/- mice was significantly reduced in skeletal muscle, liver, and WAT, and the expression level of glucose transporter 4 was also dramatically decreased in skeletal muscle of PPARγ2-/- mice. Intriguingly, TZD also improves insulin sensitivity in liver- or muscle-selective PPARγ-KO mice (7, 8, 31, 32), which suggests that the fat is essential for this response. Taken together, these results indicate that WAT, liver, and muscle are crosstalking with each other, and all contribute to insulin sensitivity.
To explore the molecular mechanisms of insulin resistance in PPARγ2-/- KO mice, we have documented reduced levels of leptin and adiponectin in plasma. Leptin can decrease the desire for food intake and stimulate energy metabolism, and leptin treatment of ob/ob mice improves insulin sensitivity even before significantly reducing body weight (33). Adiponectin is a potent insulin enhancer that links adipose tissue and whole-body glucose metabolism (34). Indeed, a recent provocative report documented that decreased expression levels of adiponectin and leptin were associated with insulin resistance in murine models of lipodystrophy (35). Of interest, insulin resistance was completely reversed by a combination of physiological doses of adiponectin and leptin but only partially by either adiponectin or leptin alone (35), which suggests that reduced levels of adiponectin and leptin play critical roles in the development of insulin resistance in a lack of adipose tissue.
PPARγ2-/- mice provide a model for studying the role of PPARγ2 in the regulation of resistin secretion from adipose tissue in vivo. Initial studies have documented that resistin induces insulin resistance, and that the suppressive effect of TZDs on resistin secretion may contribute to the insulin-sensitizing effect of this class of drugs (36). However, our data demonstrate that expression levels of resistin were reduced in the WAT of PPARγ2-/- mice. This finding agrees that resistin (like lipoprotein lipase, leptin, and adipsin) tracks the differentiation of adipose tissue. Indeed, the activation of PPARγ induces adipogenesis, which leads to the induction of resistin (37). Of interest, reduced levels of resistin mRNA in the WAT of obese and diabetic mice, including ob/ob, db/db, and KKAy mice, have been reported (38, 39). Taken together, these results indicate that the role of PPARγ in the regulation of resistin expression is more complex than was originally believed. The underlying molecular mechanism remains to be further explored.
Although changes in the phenotype, including reduction of fat mass, less lipid accumulation, and decreased adipogenic gene expression, in female mice were similar to those of male PPARγ2-/- mice, the insulin sensitivity in PPARγ2-/- female mice was not affected. Similar phenotypes have been previously documented in other mouse models of type 2 diabetes (40, 41). In addition, only male mice exhibited a reduction in atherosclerosis in response to PPAR-specific ligands (42). Intriguingly, metabolic responses to TZDs in female mice that had their ovaries removed were more similar to those of male mice (42). Although the basis for these sex differences is not fully understood, they are likely to relate to the influence of estrogens and progestins. Indeed, it has been reported that estrogens are able to induce the production of PPARγ endogenous ligands (43).
Conclusion
We have shown that PPARγ2-specific KO leads to a reduction in fat mass, lower lipid accumulation, a decrease in adipogenic gene expression, and impaired insulin sensitivity. The results of our study clearly have demonstrated that, without PPARγ2, PPARγ1 alone is able to drive the development of adipose tissue. The PPARγ2-deficient mouse model may provide a tool to study the functional differences between γ2 and γ1 in many aspects and to evaluate the effect of new antiobesity and antidiabetes drugs.
Supplementary Material
Acknowledgments
This work was partially supported by National Institutes of Health Grants HL068878, HL03676, and S06GM08248. M.F. is supported by postdoctoral fellowships from the American Heart Association Southeast Affiliate.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PPAR, peroxisome proliferator-activated receptor; TZD, thiazolidinedione; MEF, mouse embryonic fibroblast; KO, knockout; WAT, white adipose tissue; BAT, brown adipose tissue; IR, insulin receptor.
References
- 1.Kersten, S., Desvergne, B. & Wahli, W. (2000) Nature 405, 421-424. [DOI] [PubMed] [Google Scholar]
- 2.Altshuler, D., Hirschhorn, J. N., Klannemark, M., Lindgren, C. M., Vohl, M. C., Nemesh, J., Lane, C. R., Schaffner, S. F., Bolk, S., Brewer, C., et al. (2000) Nat. Genet. 26, 76-80. [DOI] [PubMed] [Google Scholar]
- 3.Vidal-Puig, A. J., Considine, R. V., Jimenez-Linan, M., Werman, A., Pories, W. J., Caro, J. F. & Flier, J. S. (1997) J. Clin. Invest. 99, 2416-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escher, P., Braissant, O., Basu-Modak, S., Michalik, L., Wahli, W. & Desvergne, B. (2001) Endocrinology 142, 4195-4202. [DOI] [PubMed] [Google Scholar]
- 5.He, W., Barak, Y., Hevener, A., Olson, P., Liao, D., Le, J., Nelson, M., Ong, E., Olefsky, J. M. & Evans, R. M. (2003) Proc. Natl. Acad. Sci. USA 100, 15712-15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koutnikova, H., Cock, T. A., Watanabe, M., Houten, S. M., Champy, M. F., Dierich, A. & Auwerx, J. (2003) Proc. Natl. Acad. Sci. USA 100, 14457-14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norris, A. W., Chen, L., Fisher, S. J., Szanto, I., Ristow, M., Jozsi, A. C., Hirshman, M. F., Rosen, E. D., Goodyear, L. J., Gonzalez, F. J., et al. (2003) J. Clin. Invest. 112, 608-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsusue, K., Haluzik, M., Lambert, G., Yim, S. H., Gavrilova, O., Ward, J. M., Brewer, B., Reitman, M. L. & Gonzalez, F. J. (2003) J. Clin. Invest. 111, 737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen, E. D., Walkey, C. J., Puigserver, P. & Spiegelman, B. M. (2000) Genes Dev. 14, 1293-1307. [PubMed] [Google Scholar]
- 10.Zhu, Y., Qi, C., Korenberg, J. R., Chen, X. N., Noya, D., Rao, M. S. & Reddy, J. K. (1995) Proc. Natl. Acad. Sci. 92, 7921-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fajas, L., Auboeuf, D., Raspe, E., Schoonjans, K., Lefebvre, A. M., Saladin, R., Najib, J., Laville, M., Fruchart, J. C., Deeb, S., et al. (1997) J. Biol. Chem. 272, 18779-18789. [DOI] [PubMed] [Google Scholar]
- 12.Ren, D., Collingwood, T. N., Rebar, E. J., Wolffe, A. P. & Camp, H. S. (2002) Genes Dev. 16, 27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller, E., Drori, S., Aiyer, A., Yie, J., Sarraf, P., Chen, H., Hauser, S., Rosen, E. D., Ge, K., Roeder, R. G. & Spiegelman, B. M. (2002) J. Biol. Chem. 277, 41925-41930. [DOI] [PubMed] [Google Scholar]
- 14.Rocchi, S., Picard, F., Vamecq, J., Gelman, L., Potier, N., Zeyer, D., Dubuquoy, L., Bac, P., Champy, M. F., Plunket, K. D., et al. (2001) Mol. Cell 8, 737-747. [DOI] [PubMed] [Google Scholar]
- 15.Lee, K., Villena, J. A., Moon, Y. S., Kim, K. H., Lee, S., Kang, C. & Sul, H. S. (2003) J. Clin. Invest. 111, 453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu, M., Zhu, X., Wang, Q., Zhang, J., Song, Q., Zheng, H., Ogawa, W., Du, J. & Chen, Y. E. (2001) Circ. Res. 89, 1058-1064. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J. B., Petersen, R. K., Larsen, B. M., Bartkova, J., Alsner, J. & Kristiansen, K. (1999) J. Biol. Chem. 274, 2386-2393. [DOI] [PubMed] [Google Scholar]
- 18.Chawla, A. & Lazar, M. A. (1994) Proc. Natl. Acad. Sci. USA 91, 1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosen, E. D., Sarraf, P., Troy, A. E., Bradwin, G., Moore, K., Milstone, D. S., Spiegelman, B. M. & Mortensen, R. M. (1999) Mol. Cell 4, 611-617. [DOI] [PubMed] [Google Scholar]
- 20.Barak, Y., Nelson, M. C., Ong, E. S., Jones, Y. Z., Ruiz-Lozano, P., Chien, K. R., Koder, A. & Evans, R. M. (1999) Mol. Cell 4, 585-595. [DOI] [PubMed] [Google Scholar]
- 21.Kubota, N., Terauchi, Y., Miki, H., Tamemoto, H., Yamauchi, T., Komeda, K., Satoh, S., Nakano, R., Ishii, C., Sugiyama, T., et al. (1999) Mol. Cell 4, 597-609. [DOI] [PubMed] [Google Scholar]
- 22.Ostman, J., Arner, P., Engfeldt, P. & Kager, L. (1979) Metabolism 28, 1198-1205. [DOI] [PubMed] [Google Scholar]
- 23.Djian, P., Phillips, M. & Green, H. (1985) J. Cell Physiol. 124, 554-556. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto, C., Tsujita, T. & Okuda, H. (1997) J. Lipid Res. 38, 132-138. [PubMed] [Google Scholar]
- 25.Reaven, G. M. (1988) Diabetes 37, 1595-1607. [DOI] [PubMed] [Google Scholar]
- 26.Rosen, E. D. & Spiegelman, B. M. (2001) J. Biol. Chem. 276, 37731-37734. [DOI] [PubMed] [Google Scholar]
- 27.Shimano, H., Shimomura, I., Hammer, R. E., Herz, J., Goldstein, J. L., Brown, M. S. & Horton, J. D. (1997) J. Clin. Invest. 100, 2115-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. B., Wright, H. M., Wright, M. & Spiegelman, B. M. (1998) Proc. Natl. Acad. Sci. USA 95, 4333-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bluher, M., Michael, M. D., Peroni, O. D., Ueki, K., Carter, N., Kahn, B. B. & Kahn, C. R. (2002) Dev. Cell 3, 25-38. [DOI] [PubMed] [Google Scholar]
- 30.Bluher, M., Kahn, B. B. & Kahn, C. R. (2003) Science 299, 572-574. [DOI] [PubMed] [Google Scholar]
- 31.Gavrilova, O., Haluzik, M., Matsusue, K., Cutson, J. J., Johnson, L., Dietz, K. R., Nicol, C. J., Vinson, C., Gonzalez, F. J. & Reitman, M. L. (2003) J. Biol. Chem. 278, 34268-34276. [DOI] [PubMed] [Google Scholar]
- 32.Hevener, A. L., He, W., Barak, Y., Le, J., Bandyopadhyay, G., Olson, P., Wilkes, J., Evans, R. M. & Olefsky, J. (2003) Nat. Med. 9, 1491-1497. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y., Proenca, R., Maffei, M., Barone, M., Leopold, L. & Friedman, J. M. (1994) Nature 372, 425-432. [DOI] [PubMed] [Google Scholar]
- 34.Berg, A. H., Combs, T. P., Du, X., Brownlee, M. & Scherer, P. E. (2001) Nat. Med. 7, 947-953. [DOI] [PubMed] [Google Scholar]
- 35.Yamauchi, T., Kamon, J., Waki, H., Terauchi, Y., Kubota, N., Hara, K., Mori, Y., Ide, T., Murakami, K., Tsuboyama-Kasaoka, N., et al. (2001) Nat. Med. 7, 941-946. [DOI] [PubMed] [Google Scholar]
- 36.Steppan, C. M., Bailey, S. T., Bhat, S., Brown, E. J., Banerjee, R. R., Wright, C. M., Patel, H. R., Ahima, R. S. & Lazar, M. A. (2001) Nature 409, 307-312. [DOI] [PubMed] [Google Scholar]
- 37.Li, Y. & Lazar, M. A. (2002) Mol. Endocrinol. 16, 1040-1048. [DOI] [PubMed] [Google Scholar]
- 38.Way, J. M., Gorgun, C. Z., Tong, Q., Uysal, K. T., Brown, K. K., Harrington, W. W., Oliver, W. R., Willson, T. M., Kliewer, S. A. & Hotamisligil, G. S. (2001) J. Biol. Chem. 276, 25651-25653. [DOI] [PubMed] [Google Scholar]
- 39.Moore, G. B., Chapman, H., Holder, J. C., Lister, C. A., Piercy, V., Smith, S. A. & Clapham, J. C. (2001) Biochem. Biophys. Res. Commun. 286, 735-741. [DOI] [PubMed] [Google Scholar]
- 40.Kim, J. H., Sen, S., Avery, C. S., Simpson, E., Chandler, P., Nishina, P. M., Churchill, G. A. & Naggert, J. K. (2001) Genomics 74, 273-286. [DOI] [PubMed] [Google Scholar]
- 41.Lan, H., Rabaglia, M. E., Stoehr, J. P., Nadler, S. T., Schueler, K. L., Zou, F., Yandell, B. S. & Attie, A. D. (2003) Diabetes 52, 688-700. [DOI] [PubMed] [Google Scholar]
- 42.Li, A. C., Brown, K. K., Silvestre, M. J., Willson, T. M., Palinski, W. & Glass, C. K. (2000) J. Clin. Invest. 106, 523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma, H., Sprecher, H. W. & Kolattukudy, P. E. (1998) J. Biol. Chem. 273, 30131-30138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.