Abstract
Replication of HIV-1 and N-tropic murine leukemia virus (N-MLV) is restricted in a number of different primate cells. In some cell lines, cross-saturation experiments suggest that the two viruses are interacting with the same restriction factor. Recently, Trim5α protein from rhesus monkey was found to restrict HIV-1. We have confirmed this result and have shown that Trim5α from two African green monkey cell lines, Vero and CV-1, also restricts HIV-1. In addition, we show that human, rhesus, and African green monkey Trim5α can restrict N-MLV. By using a panel of MLV capsid mutants, subtle differences in the anti-MLV activity were identified among the different primate Trim5α cDNAs. Trim1 isolated from humans and green monkeys was also found to restrict N-MLV. We hypothesize that the Trim family of proteins plays a widespread role in innate immunity to viral infection.
The fossil record left by endogenous retroviruses (1) provides convincing evidence that retroviruses and their vertebrate hosts have coexisted for a period of tens of millions of years (2–4). During this time, evolution of host defense mechanisms might be expected. Such mechanisms would include those acting over the whole organism such as genes regulating immune responses (5), as well as those like Fv4 (6) and APOBEC3G (7), that act in a cell-autonomous fashion directed against specific viruses or viral gene products.
One class of the latter group is known as restriction factors (8). Members of this class interact with the retroviral capsid (CA) protein in a specific and saturable manner and block one or more after entry, preintegration steps in the viral life cycle. The mouse Fv1 gene provides the prototype for this class. First described >30 years ago (9, 10), it has two alleles, Fv1n and Fv1b, targeting B- and N-tropic murine leukemia viruses (MLVs), respectively. Viral tropism is determined mainly by CA amino acid 110 (11). The Fv1 gene is found only in mice (12), but a MLV restriction activity called Ref1, also CA amino acid 110-specific, is present in a variety of different species, including human (13). A similar restriction activity directed against CA of lentiviruses is described in refs. 14–17. This activity, called Lv1, is present in cells from a variety of primate species. Cross-saturation experiments suggest that Ref1 and Lv1 share the same target (18). However, there are significant differences in restriction patterns shown by Lv1 in cells from different primate species (14–16), raising the question as to whether one gene is responsible for all of the observed effects (19). Very recently, Sodroski and coworkers (20) have shown that the α isoform of the Trim5 gene from rhesus monkeys can restrict HIV-1 replication. We report below experiments designed to investigate whether this gene also encodes anti-MLV activity. Furthermore, we have examined whether the properties of Trim5α cloned from different species account for the characteristic restriction patterns seen in corresponding cell lines.
Materials and Methods
Restriction Assays. Assays for Ref1/Lv1 function were carried out essentially as described for Fv1 by two-color fluorescence-activated cell sorter (FACS) analysis (21, 22). Cells were first transduced with restriction factor and enhanced yellow fluorescent protein by means of a “delivery” virus, in most cases adding sufficient virus to infect 35–40% of the cells. After 48 h in culture, cells were transduced with a “tester” virus carrying the CA target for restriction and enhanced GFP (EGFP). After another 48 h, cells were analyzed by two-color FACS in a Becton Dickinson LRS1 machine. Restriction was assessed by comparing the percentage of infected cells (green) in the populations of cells with and without restriction factor (yellow). In some experiments, cyclosporine A (3 μg/ml) was added 2 h before tester virus transduction.
Cells and Viruses. Mus dunni (mouse), TE671 and HT1080 (human), Vero and CV-1 [African green monkey (AGM)], and owl monkey kidney (OMK) cells, were cultivated in DMEM containing 10% FCS and antibiotics, whereas LLCMK2 (rhesus monkey) cells were passaged in Eagle's MEM containing 5% FCS, 1% nonessential amino acids, 1% glutamine, and antibiotics. Viruses were generated by simultaneous CaPO4-mediated transient transfections of 293T cells with three plasmids providing vector, gag-pol, and env functions (21, 22). Virus-containing supernatants were filtered, frozen in aliquots at -70°C and titrated on Fv1-null (23) M. dunni or human TE671 cells. The delivery retroviral vector (pLGatewayIY) was a derivative of pLFv1nIEG modified to contain a multiple cloning site as well as a Gateway reading frame cassette A in place of the Fv1 gene (M.W.Y., unpublished data), thereby allowing introduction of restriction genes cloned in a Gateway entry vector (Invitrogen). Delivery vectors were packaged by using Mo-MLV gag-pol (24) and vesicular stomatitis virus G protein (21). Plasmids specifying N-, B- and NB (Moloney virus-derived)-tropic MLVs CA are described in ref. 21. Derivatives carrying the CA N82D and L117H changes on N-tropic MLV background and E110A on a B-tropic background are described in ref. 25. Gag-pol of HIV-1 was provided by p8.91 (26). The HIV-1 CA G89V mutation was introduced by site-directed mutagenesis using the QuikChange system (Stratagene). The MLV tester viruses contained pLNCG (M.W.Y., unpublished data), a derivative of pLNCX vector with EGFP inserted in the multiple cloning site; the HIV-1 vector was SIN CSGW (27).
Trim5α Cloning. The human, green monkey, and rhesus Trim5α cDNAs were cloned by RT-PCR using primers directed to the 5′ end (CACCATGGCTTCTGGAATCCTGGTTAATG; includes added sequence of CACC, which provides Kozak sequence and allows directional cloning into a Gateway entry vector) and 3′ end (TCAAGAGCTTGGTGAGCACAG) of the published human sequence (20). mRNA was isolated from TE671, Vero and CV-1, and LLCMK2 cells respectively, by using the Oligotex-direct mRNA mini kit (Qiagen, Valencia, CA), reverse transcribed with the First-Strand cDNA Synthesis Kit (Roche Diagnostics) using an oligo(dT) primer, and PCR amplified using PfuUltra (Stratagene) with the 3′ and 5′ primers. PCR products were cloned into the pENTR TOPO vector (Invitrogen), and were sequenced and transferred to pLGatewayIY. cDNA to Trim1 mRNA was prepared similarly by using forward and reverse primers to the 5′ end (CACCATGGAAACACTGGAGTC) and 3′ end (TTAATGACAGGTTTTCATCCC) of the published human coding sequences (28).
Short Interfering (si)RNA Experiments. siRNA was designed against the region of Trim5α targeted by siRNA1 in the study by Stremlau et al. (20): Two oligonucleotides (GATCCCCGCTCAGGGAGGTCA AGT TGT TCA AGAGACA ACT TGACCTCCCTGAGCTTTTTGGAAA and AGCTTTTCCAAAAAGCTCAGGGAGGTCAAGTTGTCTCTTGAACAACTTGACCTCCCTGAGCGGG) were annealed in a buffer containing 100 mM potassium acetate, 30 mM Hepes-KOH (pH 7.4), and 2 mM magnesium acetate and was cloned into BglII/HindIII-digested retroviral vector pLHiSN (M. Dodding, unpublished work). pLHiSN was constructed by inserting the EcoRI/XhoI fragment from pSUPER, which contained the histone promoter followed by BglII and HindIII sites, into pLXSN from which the original BglII and HindIII sites were removed by site-directed mutagenesis. The vectors were packaged into MLV-based virions and were used to transduce target cells, at a multiplicity of infection of >5, that were originally seeded in 12-well plates. Three days after transduction, the cells were replated into 24-well plates and challenged with tester viruses carrying the EGFP marker. The percentage of cells transduced with tester virus was measured 3 days later by FACS. To assess effects of siRNA on stable mRNA levels, mRNA from ≈5 × 106 transduced or untransduced cells were prepared by using the Oligotex Direct mRA mini kit (Qiagen). One hundred nanograms of each mRNA was reverse-transcribed by using AMV RT (Roche) and 5-fold serial dilutions of RT products were submitted to PCR. Trim5α was amplified by using the primers CACCATGGCTTCTGGAATCCTGGTTAATG and AGGAACGTGTGGTGACCA. The GAPDH gene was also amplified as a control by using primers CGAGATCCCTCCAAAATCAA and TGTGGTCATGAGTCCTTCCA.
Results
Cloning of Trim5α from Different Primate Species. Cells from a panel of primate species show specific differences in their restriction properties for HIV-1 and MLV (13–16). Recently, restriction of HIV-1 in monkey cells has been attributed to the action of the Trim5α gene product. To test whether the differences in restriction patterns could be explained on the basis of genetic differences between Trim5α genes in primates, we first set out to compare the sequences of the human, AGM, rhesus monkey, and OMK genes. By using primers to the human sequence, we cloned the human, rhesus monkey, and AGM genes by RT-PCR from RNA isolated from TE671, LLCMK2, and both CV-1 and Vero cells, respectively. However, for reasons that remain to be elucidated, Trim5α could not be cloned in this way from OMK cells.
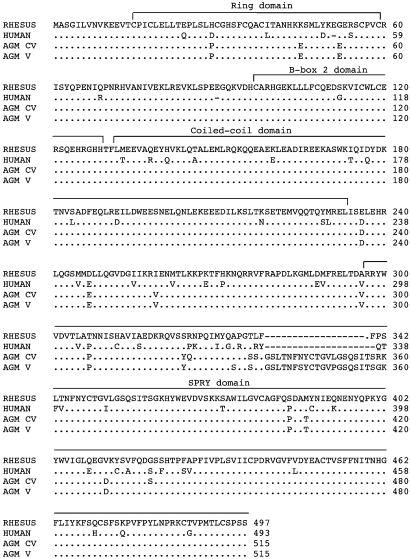
A comparison of the predicted proteins is shown in Fig. 1. The human and rhesus monkey sequences were essentially identical to those published in ref. 20; they share 87% sequence identity. As predicted from evolutionary trees, the two AGM-derived sequences are less highly related to the human sequence. In particular, they contain a 20-aa insertion within the SPRY domain compared with human (18 aa compared with rhesus). The two AGM sequences differ by 7 aa from one another, with six of the differences lying in or immediately adjacent to the insertion.
Fig. 1.
Comparison of amino acid sequence of Trim5α from different primates. Predicted amino acid sequences of the Trim5α protein from human (TE671), rhesus monkey (LLCMK2), AGM-V (Vero), and AGM-CV (CV-1). The positions of the motifs characteristic of Trim proteins are indicated. The sequences of the first eight and last six amino acids were determined by the (human) primers used for amplification.
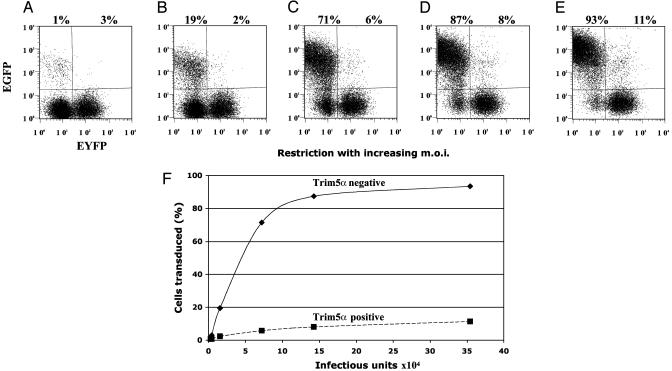
Trim5α Restriction of HIV-1. To examine the restriction properties of Trim5α we used a rapid two-color FACS assay originally developed for the study of Fv1 (21, 22). In this assay, cells are first transduced with a retroviral vector expressing the restriction gene, as well as, from an internal ribosome initiation site, enhanced yellow fluorescent protein, followed by infection with a second retroviral vector, including EGFP. Restriction can be quantified by comparing the percentage of infected cells (green) in cells with and without restriction factor (yellow). Fig. 2 shows data from one typical experiment in which the effect of the Trim5α gene from rhesus monkey on HIV-1 infection of human HT1080 cells was measured. Restriction of HIV-1 was clearly evident over a wide range of initial multiplicities of infection (Fig. 2 A–E). Even with the highest dose of virus, an ≈10-fold difference in the number of infected cells was seen after Trim5α introduction (data summarized in Fig. 2F).
Fig. 2.
Titration of HIV-1 in HT1080 cells transduced with Trim5α from rhesus monkey. (A–E) The FACS profiles of cell populations challenged with increasing amounts of HIV-1 vector carrying the EGFP marker. (A) A total of 4 × 103 infectious units. (B) A total of 16 × 103 infectious units. (C) A total of 70 × 103 infectious units. (D) A total of 140 × 103 infectious units. (E) A total of 350 × 103 infectious units. Enhanced yellow fluorescent protein (EYFP) identifies Trim5α transduced cells, whereas EGFP follows HIV-1 infection. The percentages of HIV-1 infected cells without and with Trim5α are shown above the profiles. m.o.i., multiplicity of infection. (F) The percentage of infected cells from A–E as a function of the amount of added HIV-1. Data shown are from one typical experiment.
By using this assay, we tested the restriction properties of Trim5α cloned from human, AGM, and rhesus monkey after transfer to human HT1080 cells (Table 1). The AGM and rhesus monkey forms clearly inhibited HIV-1, as shown by the 5- to 10-fold reduction in infection of Trim5α-transduced cells. Little difference was seen in HIV-1 restriction between Trim5α from Vero and CV-1 cells. Trim5α isolated from LLCMK2 cells restricted HIV-1, despite the fact that these cells show little or no restriction (M.W.Y., unpublished observations). The human gene had a small but reproducible effect. Under the conditions of our assay, restriction appeared largely independent of cyclophillin A binding, because essentially identical results were obtained with HIV-1 carrying the G89V change in CA or after cyclosporine A treatment, both conditions that prevent cyclophillin A from binding CA (29). Because cyclophillin A exclusion has been associated with low levels of HIV-1 restriction in human cells (29), it is possible that factors other than Trim5α are acting on cyclophillin A-depleted virions. It thus appears that Trim5α can account for some but perhaps not all of the Lv1 activity shown by human, AGM, and rhesus monkey cells.
Table 1. Restriction of HIV-1 by introduced Trim5α.
| Source of Trim5α | HIV-1 | G89V | CsA-treated* |
|---|---|---|---|
| Human | 0.77 ± 0.03 | 0.77 ± 0.05 | 0.83 ± 0.03 |
| AGM (CV-1) | 0.10 ± 0.02 | 0.20 ± 0.04 | 0.28 ± 0.08 |
| AGM (Vero) | 0.06 ± 0.01 | 0.08 ± 0.02 | 0.11 ± 0.01 |
| Rhesus | 0.11 ± 0.03 | 0.14 ± 0.02 | 0.12 ± 0.01 |
Results shown are the mean of three experiments.
CsA, cyclosporine A. Ratio of Trim5α+ cells infected/Trim5α— cells infected
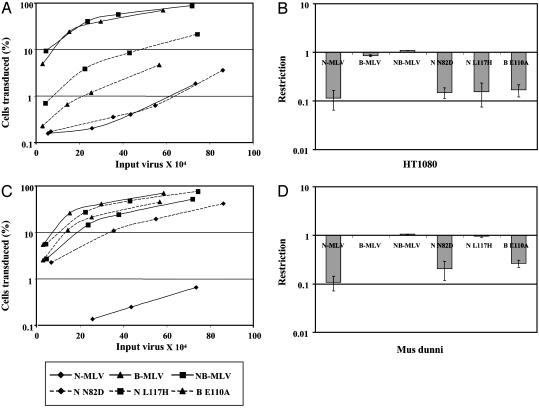
Trim5α Restriction of MLV. To investigate whether Trim5α also encodes Ref1 activity, we performed analogous experiments with a panel of MLVs. Ref1 has previously been shown to restrict N-tropic MLV but not B- or NB-tropic MLV (13). To further define Ref1 specificity, we have now included three additional viruses, carrying mutations at the three positions in CA (82, 110, and 117) that differentiate N-tropic and NB-tropic MLVs. All three variants show altered susceptibility to different alleles of Fv1 (25) and might be expected to allow us to distinguish closely related forms of Trim5α. The six MLVs were titrated on M. dunni as well as human, AGM, and rhesus monkey cells. On human TE671 cells, NB- and B-tropic viruses yielded the highest titers, whereas all of the others were significantly reduced but to varying degrees with N-MLV and N82D showing the greatest extent of restriction (Fig. 3A). Only N-MLV was restricted to the same extent in Vero cells, but N82D was also reduced to an appreciable degree (Fig. 3C). CV-1 gave results indistinguishable from Vero cells but no restriction was seen in LLCMK2 cells (data not shown).
Fig. 3.
Restriction of MLV by Trim5α. The graphs on the left show titration curves for a panel of MLVs on TE671 (A) and Vero (C) cell lines. Cells were infected with increasing amounts of viruses (previously titrated on M. dunni cells). Viruses tested were N-MLV (♦, solid line), B-MLV (▴, solid line), NB-MLV (▪, solid line), N-N82D (♦, dashed line), N-L117H (▪, dashed line), and B-E110A (▴, dashed line). The titrations come from one typical experiment. The plots in the right column show the fold reduction (percentage of Trim5α+ cells infected/percentage of Trim5α- cells infected) in MLV titer as measured by the two-color FACS assay upon introduction of human (B) and AGM (D) Trim5α into M. dunni cells. Means and SD from three experiments are shown.
Upon introduction of human Trim5α into M. dunni cells, we observed restriction of N-MLV, N82D, L117H, and E110A, but not B-MLV or NB-MLV (Fig. 3B), which is consistent with the results from TE671 cells. However, the extent of restriction did not show as much variation as seen in TE671, suggesting that other factors, expression levels, for instance, can influence this parameter. AGM Trim5α showed highest restriction activity against N-MLV, but also showed significant inhibition of N82D and E110A (Fig. 3D). Identical results were obtained with CV-1 Trim5α. In contrast to the human form, AGM Trim5α did not restrict L117H. Upon introduction into M. dunni cells, rhesus monkey Trim5α inhibited only N-MLV with no effect on N82D or the other viruses (data not shown). Overall, these studies show that Trim5α can block MLV replication, with susceptibility to restriction conferred by the CA protein. In addition, the results also suggested that there might be subtle differences in the CA affinity of the different primate Trim5αs.
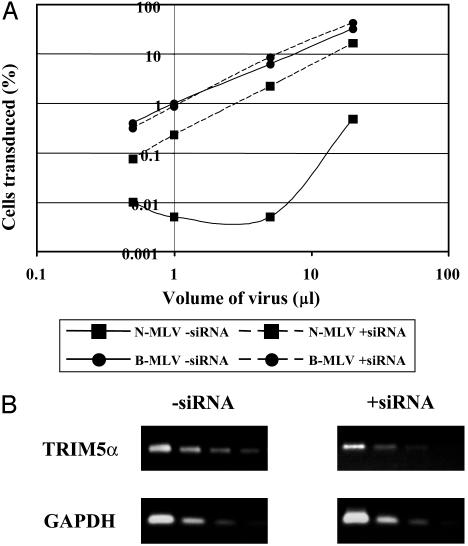
siRNA Experiments. To further address the role of Trim5α, we have examined the effect of mRNA levels on restriction. SiRNA to human Trim5α was introduced into TE671 cells through a retroviral vector. This treatment had no effect on infection with B-MLV but increased N-MLV titers >10-fold (Fig. 4A). RT-PCRs showed a 5- to 10-fold reduction in Trim5α mRNA expression with no effect on a GAPDH control (Fig. 4B). Similar results were obtained with siRNA treated CV-1 cells (data not shown). These observations suggest that Trim5α expression levels may control the patterns of restriction seen and may provide an explanation for the apparent lack of restriction in cells from which it is possible to clone active Trim5α cDNAs.
Fig. 4.
Effects of Trim5α siRNA on restriction in primate cells. (A) Titration curves of MLV and HIV-1. ▪, TE671: N-MLV; ▪, B-MLV. Continuous lines show titers in the absence of siRNA and broken lines show titers in the presence of siRNA. (B) PCR of 5-fold serial dilutions of Trim5α cDNA isolated from the cells in A. GAPDH was used as a negative control. Data from one representative experiment are shown.
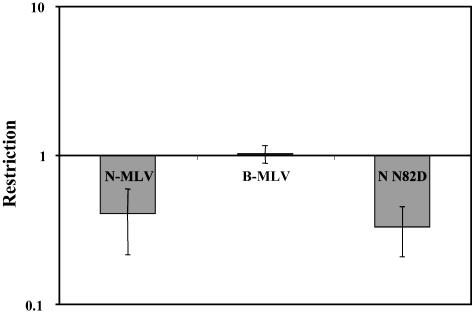
Trim1 Restriction of MLV. The Trim motif is shared by >30 genes in the human genome, but little is known about their physiological roles (30). This fact raises the possibility that other Trim genes might have antiretroviral activity. To begin to address this question, we have introduced cDNA to Trim1 into our delivery vector and tested for restriction activity. Trim1 cDNAs from human, AGM, or OMK cells did not show any activity against HIV-1 (data not shown). However, AGM Trim1 showed significant restriction of N-MLV and N82D (Fig. 5). Trim1 from human and OMK cells also restricted these two viruses. Although the extent of restriction by Trim1 was lower than Trim5α, the CA dependence of restriction argues strongly for specific inhibition.
Fig. 5.
Restriction of MLV by Trim1. The graph shows the fold reduction (percentage of Trim1+ cells infected/percentage of Trim1- cells infected) in MLV titer as measured by the two-color FACS assay upon introduction of Vero Trim1 into M. dunni cells. Means and SD from three experiments are shown.
Discussion
The most important conclusion from the studies presented above is that Trim5α shows restriction activity against both HIV-1 and MLV. The target for restriction is the CA molecule of HIV-1 and MLV that share <20% identity with one another. At the same time, as inferred from previous infectivity studies (14–16), Trim5α does not seem to interact with the relatively closely related simian immunodeficiency virus CA, but is sensitive to single amino acid changes in the MLV CA. The mechanism of Trim5α restriction is as yet poorly explored, but it will clearly be important to probe the interaction between Trim5α and CA. The differences between the different forms of Trim5α will provide a useful starting point for genetic studies to map specificity determinants on Trim5α, but demonstration of a direct interaction will be more informative. However, this undertaking may not prove to be a trivial one, as judged by our experiences in attempting to show such an interaction between Fv1 and MLV CA. Ultimately, detailed structural studies will be required to understand specificity patterns. The consequences of this interaction also remain to be explored; possible hypotheses include a direct effect on uncoating of viral capsid (20) and the sequestering of subviral structures in inappropriate cellular compartments by means of the Trim5α localization domain(s) (30).
Because Trim5α can restrict both HIV-1 and MLV, it seems reasonable to ask whether it is responsible for the entire variety of restriction activities previously described in primate cells. A definitive answer to this question is complicated by the observation that, in several cases, we have demonstrated antiviral activity with Trim5α cloned from a cell line apparently lacking the corresponding restriction activity. We attribute this finding to effects on transcription levels in a manner analogous to that seen previously with Fv1 (21). Alternatively, the relative expression levels of the various isoforms of the gene might regulate the degree of restriction, because the γ-isoform of Trim5 can act in a dominant-negative manner (20). This result raises the formal possibility that a gene cloned from a restricting cell is not actually responsible for all of the restriction seen in that cell. In cases where two restricting genes can be cloned from one cell, e.g., Trim1 and Trim5α, it may be difficult to assess the relative contribution of the two genes in that cell type. We note that the two viruses susceptible to both Trim5α and Trim1 restriction, N-MLV and N82D, show the most pronounced restriction in TE671 and Vero cells. Therefore, although we can confidently conclude that Trim5α has both Ref1 and Lv1 activities, there may be cases where Trim5α obscures the contribution of other restriction genes. Such putative additional factors may also help explain the cyclosporine A effect reported in human cells (29).
More than 30 Trim genes have been described in the human genome but there is little information about their functional roles (30). A few appear important in regulating cell growth and differentiation (31, 32) but most are poorly characterized. The findings that Trim5α inhibits both HIV-1 and N-MLV and that Trim1 inhibits N-MLV raise the possibility that still more Trim genes might play a role in determining the susceptibility of a cell to retroviral infection. So far, we have not carried out an exhaustive study of different Trim genes. We have tested Trim18, which forms heterodimers with Trim1 (33), but this combination lacks anti-MLV activity (data not shown). However, there is good evidence that Trim19 (promyelocytic leukemia or PML protein) responds to HIV-1 infection (34); hence, it does not seem unreasonable to speculate that additional members of the family of genes containing Trim motifs might have antiretroviral activity, perhaps directed against other, as-yet-untested viruses. Furthermore, it has been reported that mRNA levels of several Trim genes rise after influenza virus infection (35). Indeed, Trim19 (PML) has been shown to confer resistance to influenza A and vesicular stomatitis virus by interfering with viral mRNA and protein synthesis (36). Together, these observations raise the possibility that Trim proteins play a hitherto unsuspected, but widespread, role in innate immunity to viral infection. Thus, the susceptibility of any given individual to infection may show a significant genetic component determined by a combination of factors controlling the activity and expression levels of a variety of normal intracellular proteins with antiviral activity such as members of the APOBEC (7) or Trim families.
Acknowledgments
We thank Ekaterini Kotsopoulou and Mark Dodding for providing pSUPER and pLHiSN and Graham Preece and Chris Atkins for assistance with FACS. This work was supported by the U.K. Medical Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AGM, African green monkey; OMK, owl monkey kidney; CA, capsid; EGFP, enhanced GFP; MLV, murine leukemia virus; FACS, fluorescence-activated cell sorter/sorting; siRNA, short interfering RNA.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY625000–AY625007).
See Commentary on page 10496.
References
- 1.Boeke, J. D. & Stoye, J. P. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 343-435. [PubMed]
- 2.Goodchild, N. L., Wilkinson, D. A. & Mager, D. L. (1993) Virology 196, 778-788. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, W. E. & Coffin, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshaw, R., Pereira, V., Katzourakis, A., Paces, J., Burt, A. & Tristem, M. (2004) Proc. Natl. Acad. Sci. USA 101, 4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasenkrug, K. J. & Chesebro, B. (1997) Proc. Natl. Acad. Sci. USA 94, 7811-7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner, M. B., Kozak, C. A. & O'Brien, S. J. (1991) Trends Genet. 7, 22-27. [DOI] [PubMed] [Google Scholar]
- 7.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. V., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 116, 803-809. [DOI] [PubMed] [Google Scholar]
- 8.Bieniasz, P. D. (2003) Trends Microbiol. 11, 286-291. [DOI] [PubMed] [Google Scholar]
- 9.Lilly, F. (1970) J. Natl. Cancer Inst. 45, 163-169. [PubMed] [Google Scholar]
- 10.Hartley, J. W., Rowe, W. P. & Huebner, R. J. (1970) J. Virol. 5, 221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak, C. A. & Chakraborti, A. (1996) Virology 225, 300-306. [DOI] [PubMed] [Google Scholar]
- 12.Best, S., Le Tissier, P., Towers, G. & Stoye, J. P. (1996) Nature 382, 826-829. [DOI] [PubMed] [Google Scholar]
- 13.Towers, G., Bock, M., Martin, S., Takeuchi, Y., Stoye, J. P. & Danos, O. (2000) Proc. Natl. Acad. Sci. USA 97, 12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, W., Schubert, D., LaBonte, J., Munson, L., Gibson, S., Scammell, J., Ferrigno, P. & Sodroski, J. (1999) J. Virol. 73, 10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besnier, C., Takeuchi, Y. & Towers, G. (2002) Proc. Natl. Acad. Sci. USA 99, 11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan, S., Hatziioannou, T., Cunningham, T., Musing, M. A., Gottlinger, H. G. & Bieniasz, P. D. (2002) Proc. Natl. Acad. Sci. USA 99, 11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Münk, C., Brandt, S. M., Lucero, G. & Landau, N. R. (2002) Proc. Natl. Acad. Sci. USA 99, 13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatziioannou, T., Cowan, S., Goff, S. P., Bieniasz, P. D. & Towers, G. J. (2003) EMBO J. 22, 385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoye, J. P. (2002) Proc. Natl. Acad. Sci. USA 99, 11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissler, P. & Sodroski, J. (2004) Nature 427, 848-853. [DOI] [PubMed] [Google Scholar]
- 21.Bock, M., Bishop, K. N., Towers, G. & Stoye, J. P. (2000) J. Virol. 74, 7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop, K. N., Bock, M., Towers, G. & Stoye, J. P. (2001) J. Virol. 75, 5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi, C. F., Bonhomme, F., Buckler-White, A., Buckler, C., Orth, A., Lander, M. R., Chattopadhyay, S. K. & Morse, H. C., III (1998) Mamm. Genome 9, 1049-1055. [DOI] [PubMed] [Google Scholar]
- 24.Soneoka, Y., Cannon, P. M., Ramsdale, E. E., Griffiths, J. C., Romano, G., Kingsman, S. M. & Kingsman, A. J. (1995) Nucleic Acids Res. 23, 628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens, A., Bock, M., Ellis, S., LeTissier, P., Bishop, K. N., Yap, M. W., Taylor, W. & Stoye, J. P. (2004) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 26.Naldini, L., Blömer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M. & Trono, D. (1996) Science 272, 263-267. [DOI] [PubMed] [Google Scholar]
- 27.Bainbridge, J. W., Stephens, C., Parsley, K., Demaison, C., Halfyard, A., Thrasher, A. J. & Ali, R. R. (2001) Gene Ther. 8, 1665-1668. [DOI] [PubMed] [Google Scholar]
- 28.Buchner, G., Montini, E., Andolfi, G., Quaderi, N., Cainarca, S., Messali, S., Bassi, M. T., Ballabio, A., Meroni, G. & Franco, B. (1999) Hum. Mol. Genet. 8, 1397-1407. [DOI] [PubMed] [Google Scholar]
- 29.Towers, G. J., Hatziioannou, T., Cowan, S., Goff, S. P., Luban, J. & Bieniasz, P. D. (2003) Nat. Med. 9, 1138-1143. [DOI] [PubMed] [Google Scholar]
- 30.Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., et al. (2001) EMBO J. 20, 2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry, J., Mather, I. H., McDermott, M. F. & Pontarotti, P. (1998) Mol. Biol. Evol. 15, 1696-1705. [DOI] [PubMed] [Google Scholar]
- 32.Seto, M. H., Liu, H.-L., Zajchowski, D. A. & Whitlow, M. (1999) Proteins Struct. Funct. Genet. 35, 235-249. [PubMed] [Google Scholar]
- 33.Short, K. M., Hopwood, B., Yi, Z. & Cox, T. C. (January 4, 2002) BMC Cell Biol., 10.1186/1471-2121-3-1. [DOI] [PMC free article] [PubMed]
- 34.Turelli, P., Doucas, V., Craig, E., Mangeat, B., Klages, N., Evans, R., Kalpana, G. & Trono, D. (2001) Mol. Cell 7, 1245-1254. [DOI] [PubMed] [Google Scholar]
- 35.Geiss, G. K., Salvatore, M., Tumpey, T. M., Carter, V. S., Wang, X., Basler, C. F., Taubenberger, J. K., Bumgarner, R. E., Palese, P., Katze, M. G. & Garcia-Sastre, A. (2002) Proc. Natl. Acad. Sci. USA 99, 10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chelbi-Alix, M. K., Quignon, F., Pelicano, L., Koken, M. H. M. & de Thé, H. (1998) J. Virol. 72, 1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]