Abstract
We report on a new strategy for identifying highly specific aptamers against a predetermined epitope of a target. Termed “ligand-guided selection” (LIGS), this method uniquely exploits the selection step, the core of SELEX (Systematic Evolution Exponential enrichment). LIGS uses a naturally occurring stronger and highly specific bivalent binder, an antibody (Ab) interacting with its cognate antigen to outcompete specific aptamers from a partially enriched SELEX pool, as a strategy. We demonstrate the hypothesis of LIGS by utilizing an Ab binding to membrane-bound Immunoglobulin M (mIgM) to selectively elute aptamers that are specific for mIgM from a SELEX pool that is partially enriched toward mIgM expressing Ramos cells. The selected aptamers show specificity toward Ramos cells. We identified three aptamer candidates utilizing LIGS that could be outcompeted by mIgM Ab, demonstrating that LIGS can be successfully applied to select aptamers from a partially evolved cell-SELEX library, against predetermined receptor proteins using a cognate ligand. This proof-of-concept study introduces a new biochemical-screening platform that exploits the binding of a secondary stronger molecular entity to its target as a partition step, to identify highly specific artificial nucleic acid ligands.
Introduction
Nucleic acid aptamers (nucleic acid-based antibody analogs) are being investigated to develop therapeutic molecules for the treatment of a variety of diseases [1]. The synthetic nature of aptamers makes them attractive for the introduction of elegant chemistries to engineer molecular tools, especially compared with the use of antibodies, their protein-based rival [2]. The process by which aptamers are selected is referred to as SELEX [3,4]. The SELEX process is a screening method that combines in vitro evolution and combinatorial chemistry [5].
Recently, considerable efforts have been aimed at improving SELEX to generate aptamers that are suitable for applications in translational research. For example, SELEX methods have been introduced to select aptamers against whole cells to identify cell-surface proteins; modified nucleic acids have been introduced to enhance the diversity of SELEX libraries to produce high-affinity aptamers; and methods have been introduced to increase the efficiency of the polymerase chain reaction (PCR) of SELEX against proteins [6–10]. However, no biochemical techniques have thus far been introduced to select specific aptamers against a predetermined epitope of a receptor protein in its endogenous state with no prior manipulation of the target.
Herein, we report a novel biochemical technique for identifying specific aptamers from a partially evolved library directed by binding of a pre-existing secondary ligand with its cognate receptor. This strategy, termed ligand-guided selection (LIGS), takes advantage of the evolutionary selection step of SELEX. The aptamers are evolved in the SELEX process based on the survival of high-affinity ligands by outcompeting the low-affinity ligands during the partition step followed by PCR amplification.
We exploited this feature of the partition step to isolate specific aptamers. This is accomplished by introducing a stronger secondary high-affinity ligand, in this example, an antibody against IgM expressed on Burkitt's lymphoma cells (Ab) to outcompete and replace the aptamer candidates binding to the same target of the Ab. Based on the specificity of Ab toward its target, the aptamers identified by LIGS are also expected to show specificity toward Ab's target. The selected aptamers show specificity toward Ramos cells. As expected, the identified specific aptamers for membrane-bound Immunoglobulin M (mIgM) compete with the cognate Ab binding to its target. This proof-of-concept study introduces a new biochemical-screening platform that exploits the binding of a secondary stronger molecular entity to its target as a partition step, to identify highly specific artificial nucleic acid ligands.
Materials and Methods
Cell culture
Cell lines, Ramos (Burkitt's lymphoma) and Jurkat.E6 (T lymphocyte), were a generous gift from David Scheinberg lab and Morgan Huse lab, Memorial Sloan Kettering Cancer Center. All cells were cultured in RPMI 1640 medium supplemented with 100 units/mL penicillin–streptomycin and 10% fetal bovine serum (heat inactivated; Invitrogen).
Phosphoramidites
All of the DNA reagents needed for DNA synthesis were purchased from Glen Research or ChemGenes. All the DNA oligo sequences were chemically synthesized by attaching a fluorophore at the 3′ end using standard solid-phase phosphoramidite chemistry on an ABI394 DNA (Biolytics) synthesizer using a 0.2 μmol scale. The completed DNA sequences were de-protected and purified by using HPLC (Waters) that was equipped with a C-18 reversed-phase column (Phenomenex). All in vitro experiments were performed by using a binding buffer composed of Dulbecco's phosphate-buffered saline (DPBS) and 4.5 g/L glucose (Sigma-Aldrich), 5 mM MgCl2, 100 mg/L, tRNA (Sigma-Aldrich), and 1 g/L BSA (Sigma-Aldrich). The wash buffer was composed of DPBS with 5 mM MgCl2 and 4.5 g/L glucose (Sigma-Aldrich).
SELEX primers and library
Primers and SELEX library were obtained from Sefah et al. [11]. The SELEX library consisting of primers flanked by a 45-nucleotide randomized region was purchased from IDT DNA Technologies.
Cell-SELEX procedure
The PI staining of the cells and the flow cytometric analysis of expression of mIgM utilizing fluorescein isothiocyanate (FITC)-labeled anti-IgM antibody (1 μg, Goat anti human; Life Technologies) along with an isotype control (1 μg, Goat anti mouse IgG2a; Biolegend) were performed on a regular basis to maintain high-quality cells expressing mIgM before performing each round of SELEX.
The ss-SELEX DNA library binding buffer was heated at 95°C for 5 min and snapped cooled in ice for 30 min before selection. Cells were prepared for SELEX experiments by washing thrice with the wash buffer; subsequently, they were re-suspended in 100 μL of a cell suspension buffer (cell-binding buffer with 2 g/L BSA) before incubation with 100 μL of an ss-DNA library for 40 min on ice. The first round of selection was done with 10 × 106 cells and 100 nmol of the ss-DNA SELEX library.
The supernatant was collected as the unbound fraction. The cells that bound to the library were washed with wash buffer (10 mL) to remove weak or nonspecifically bound DNA strands. The bound DNA library was eluted by heating at 95°C for 10 min in 200 μL of DNAse/RNAse free water. A two-step PCR was employed for the optimization of the PCR conditions, and a large scale PCR was employed to expand the evolved library as reported elsewhere [12]. A double-stranded, PCR-amplified DNA library was made single stranded by using avidin agarose beads (Pierce) and desalted by using NAP-10 columns (GE) as described by Sefah et al. [11]. For subsequent SELEX rounds, 250 nM of the FITC-tagged ss-DNA library was used from round 2 to round 13.
Flow cytometric analysis
The progress of the selection was evaluated by utilizing flow cytometric analysis. The PCR-amplified DNA library is labeled with fluorescence tag FITC at the 5′ end and analyzed by a flow cytometric assay. A 250 nM FITC-tagged ss-DNA library (25 μL) was incubated with 2.5 × 105 Ramos cells in binding buffer for 40 min on ice. After washing twice with wash buffer (3 mL), the cells were suspended in 500 μL of wash buffer and were analyzed by an FACS Calibur flow cytometer (Cytek) by counting 10,000 events.
Cell-binding assays
The affinities of the aptamer sequences were evaluated by incubating Ramos cells (2.0 × 105) with a series of concentrations of FITC-labeled aptamer in 200 μL of binding buffer on ice for 60 min. The cells were then washed twice with 1 mL of wash buffer at 4°C and reconstituted in 400 μL of wash buffer. The binding of the constructs was analyzed using flow cytometry by counting 5000 events for each concentration. The calculation of Bmax/2 was done by using the same method as described in Sefah et al. [11].
The specific binding of each aptamer was evaluated by incubating Ramos cells (0.5 or 1.0 × 105) and Jurkat.E6 cells (0.5 or 1.0 × 105) with FITC-labeled aptamers of concentrations of 0.5 or 1 μM in 100 μL of cell suspension buffer on ice for 60 min. The cells were then washed twice with 1 mL of wash buffer at 4°C and reconstituted in 250 μL of wash buffer. Aptamer binding was analyzed using flow cytometry by counting 5000 events for each concentration. As a positive control, a similar assay was performed by using an Alexa Fluor 647 labeled anti-IgM antibody (1 μg, Goat anti human μ-chain; Life Technologies) along with an isotype control (1 μg; Biolegend).
Ligand-guided cell-selection protocol
Ligand competition
The enriched 13th pool FITC-tagged ss-DNA pool or control zero cycle ss-DNA pool was heated at 95°C for 5 min and cooled on ice for 20 min. 2.5 × 105 cells were incubated with 250 nM 13th SELEX-pool and 25 μL ss-DNA pool for 40 min on ice and washed once with 3 mL of wash buffer. The pretreated Ramos cells with the 13th SELEX pool were suspended in 50 μL of binding buffer and then incubated with an Alexa Fluor 647 goat anti-human IgM antibody (1 μg) for 30 min on ice to compete and elute the potential aptamer candidates. After incubation, the eluted 13th pool was obtained through competition, which in the supernatant was collected and amplified by PCR. To ensure the presence of mIgM expressed on Ramos cells, 2.5 × 105 cells were incubated in parallel with an Alexa Fluor® 647 goat anti-human IgM or Alexa Fluor 647 goat IgG Isotype antibody for 30 min. After incubation, all the samples were washed and the sample was analyzed by FACS Calibur flow cytometry (cytek) by counting 10,000 events.
Two different SELEX libraries generated from (1) the DNA pool from the SELEX 13th round specifically enriched against Ramos cells, (2) the competitively eluted fraction of the SELEX 13th round by using antibody competition specific for epitopes on the mIgM were cloned into bacteria by using a TOPO TA cloning kit (Invitrogen), and positive colonies were subsequently sequenced by the DNA sequencing core facility at Albert Einstein College of Medicine.
Antigen specificity
Antigen specificity is determined by the competition between anti-IgM antibody and aptamers. To investigate the competition between anti-IgM (mu) antibody and aptamers, first, 0.5 μg/mL of APC anti-human CD20 antibody and 0.25 μg/mL of Alexa Fluor 647 goat anti-human IgM antibody were incubated with 4 × 105 Ramos cells on ice for 30 min. Then, the free antibody was washed with 3 mL of wash buffer, and cells were reconstituted with 400 μL of cell suspension buffer. A final concentration of 0.4–0.5 μM of FITC-labeled aptamer or corresponding random control was incubated in 125 μL of cell suspension buffer for another 60 min on ice. Then, the cells were washed with 1 mL of wash buffer and binding events were monitored in FL1 for the aptamer and in FL4 for the antibody counting 5000 events using flowcytometry.
We also conducted blocking experiments with aptamers that had been preincubated with antibody. First, 10 × 104 of Ramos cells were incubated with 1 μM of corresponding aptamer or random control on ice for 45 min. Then, the preincubated cells with the aptamer or random were added to serially diluted concentrations from 20 ng/μL to 0.2 g/μL of anti-IgM solution and allowed free competition for an additional 35 min on ice. Next, the cells were washed twice with 1 and 0.5 mL of wash buffer and re-suspended in 300 μL of wash buffer, and the antibody binding was analyzed with flow cytometry.
Results and Discussion
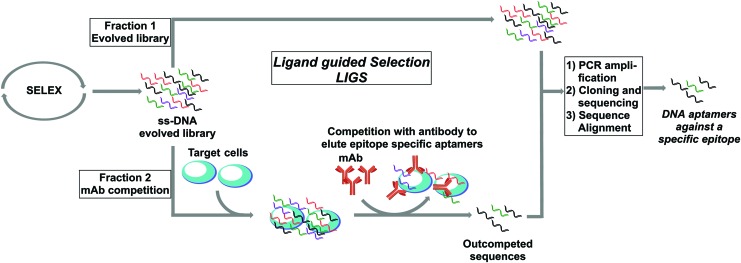
The selection method is outlined in Figure 1. Briefly, the conventional cell-SELEX method was first employed against Ramos cells that naturally express high levels of the desired epitope (mIgM). Cell-SELEX was continued until a partial enrichment of the evolved SELEX library against the target cells was observed [11]. Next, the partially enriched library was divided into fractions. The first fraction was PCR amplified, cloned, and sequenced. These sequences are specific toward target cells (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat). An excess of Ab was introduced on the second fraction, which was preincubated with Ramos cells that were subsequently washed to remove nonbinding sequences, to selectively outcompete and elute potential aptamers that bind to the cognate epitope less strongly when the anti-IgM Ab is present (Fig. 1). The sequences outcompeted by Ab were PCR amplified, cloned, and sequenced. Finally, sequences obtained from DNA sequencing of two fractions of the SELEX pool were aligned using the ClustalX.2 program and based on set criteria; specific aptamer candidates against mIgM in target cells were screened and identified [13,14].
FIG. 1.
Outline of ligand-guided selection. Color images available online at www.liebertpub.com/nat
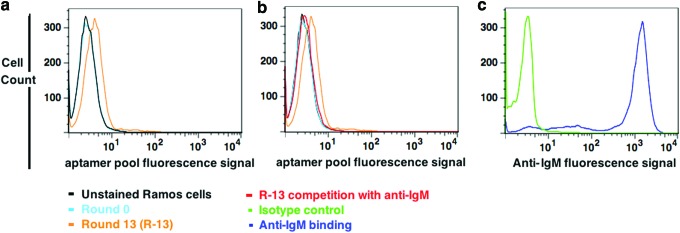
The SELEX-library used in the selection has been published elsewhere [15]. Cell-SELEX was carried out without incorporating a negative selection, because we hypothesized that if potential aptamer candidates could be partially enriched toward the desired epitope, that is, mIgM, applying an antibody against the aptamer would elute these sequences, despite the existence of unrelated off-target sequences in the partially evolved pool. We first validated the expression of mIgM on Ramos cell lines utilizing anti-IgM antibody (Supplementary Fig. S2). Ten million cells and a high concentration of the initial DNA library were employed during the first round of selection to increase the probability of capturing potential “binders.” We detected a partial enrichment of the evolved pool starting at round 13 of the cell-SELEX pool, compared with the unselected pool (Fig. 2a). At this point, the remaining round 13 was used in LIGS. To elute mIgM-specific sequences, we introduced an excess amount of Ab (1 μg) to compete with the aptamer from the fraction of round 13 of the cell-SELEX pool that was preincubated with Ramos cells followed by a wash to remove nonbinding DNA molecules. The supernatant containing sequences outcompeted by Ab were then collected. To confirm that the Ab had, indeed, interacted with mIgM, and to investigate Ab's effect on aptamer pool 13 fraction-2 binding to Ramos cells, cells after Ab competition were analyzed by flow cytometry (Fig. 2b red histogram for binding of SELEX pool round 13 after adding Ab and Fig. 2c blue histogram for binding of anti-IgM Ab). As shown in Figure 2b and c, the binding of the anti-IgM Ab to its epitope on Ramos cells replaces the binding of some aptamer sequences that are enriched in the evolved pool. This observation suggests that at least few sequences that are enriched in the round 13 fraction-2 are eluted by anti-mIgM Ab. Based on the PCR of eluted pool at this step, we observed that a low number of sequences was eluted during this step, mainly because: (1) the DNA pool was only partially evolved, with a low number of aptamer copies; (2) a low number of cells was employed in the LIGS; and (3) Ab competition, which is designed to selectively elute specific sequences, only generates a low number of specific sequences for one target epitope.
FIG. 2.
Flow cytometric analysis of evolved library and LIGS. Fluorescence intensity on x-axis is indicative of binding of fluorescently labeled R10 that evolved round 13 pool or anti-IgM. (a) Analysis of binding of evolved pool from round 13 against Ramos cells. At the 13th round, an increase in fluorescence intensity was observed, indicating that the library was partially evolved with Ramos-specific DNA aptamer sequences. (b) Pool from round 13 during LIGS, introduction of anti-IgM Ab to round 13 bound Ramos cells, resulted in decreased fluorescence intensity. (c) Binding of anti-IgM mAb during LIGS showing Ab binding to Ramos cells (blue line). LIGS, ligand-guided selection. Color images available online at www.liebertpub.com/nat
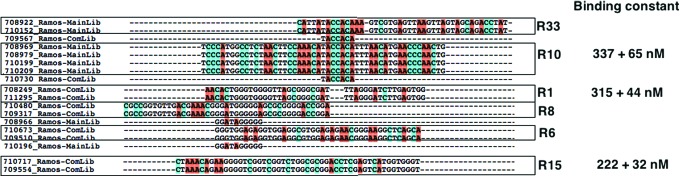
Since the SELEX pool is partially enriched, we cloned and sequenced multiple fractions of round 13 of the cell-SELEX pool and competitively eluted a fraction of round 13 of the cell-SELEX pool. About 500 sequences were obtained from all fractions, which could be categorized into families based on their sequence homology. We hypothesized that enriched sequences toward the cell line (Ramos) predominate in the library and have a higher probability in “surviving” the pool. Therefore, the sequences resulting from sequencing of round 13 of the cell-SELEX pool would contain all of the sequences that were enriched toward Ramos cells. On the other hand, the sequences obtained from LIGS would favor the set of sequences selectively eluted by the ligand. Analysis of the sequences obtained from sequencing of the competitively eluted pool or the cell-SELEX round 13 pool showed two types of sequences: (1) Sequences share motifs that are common to sequences in round 13 of the cell-SELEX pool and the competitively eluted pool. (2) Sequences are repeated within the competitively eluted library. We focused on both types of sequences that repeatedly appeared within a family with common motifs from both pools or within the competitively eluted pool. For example, as shown in Figure 3, sequence R10 only appeared on cell-SELEX round 13; however, a shorter version of a common motif was identified to appear in the competitively eluted pool. Also, R6, R8, and R1 share a common GGG motif, differing only in few bases within the motif (Fig. 3). We also observed the same sequence repeated within the competitively eluted library (Supplementary Table S1) and within the main cell-SELEX round 13 pool. Since the scope of this research is to identify sequences that are specific toward mIgM, even though sequences repeated within the main library might be potential aptamer candidates, we did not investigate the sequences that did not show any common motifs with competitively eluted sequences. Based on these criteria, we synthesized and tested 33 different sequences either from the competitively eluted library or from the main SELEX-library of round 13. Out of 33 sequences, 27 sequences are from the competitively eluted library and 6 sequences share the same motif as that of the competitively eluted library but from the cell-SELEX round 13 main pool.
FIG. 3.
Sequence alignment of the major family of LIGS using ClustalX2. Sequences repeated in the competitively eluted library were synthesized and screened against Ramos cells. Also, sequences from the main library share common motifs with competitively eluted sequences that are synthesized and screened against Ramos cells. MainLib, sequences obtained from SELEX-13 round; ComLib, sequences obtained from competitively eluted SELEX-13 round. Three specific aptamer candidates R1, R10, and R15 were evaluated and their Bmax/2 was calculated. Color images available online at www.liebertpub.com/nat
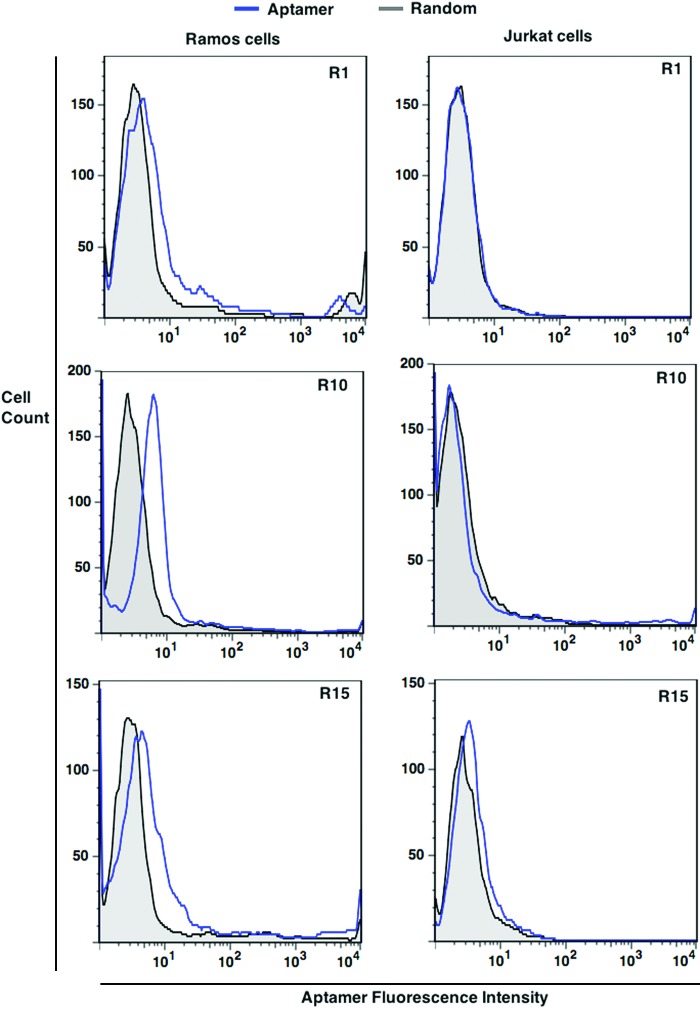
Since the LIGS is predominantly aimed at increasing specificity, individual chemically synthesized sequences based on set criteria of sequence selection were first tested for specificity. We used target Ramos cells, which express high levels of mIgM and nontarget Jurkat.E6 cells. Since Jurkat.E6 cells are human T-cell leukemia that are mainly designed to investigate the T-cell receptor complex, by definition, these cells do not express mIgM; thus, Jurkat.E6 cells are comparable to a cell line that does not express the antigen that validates its use as a nonspecific cell line [16]. All specificity assays were done by using FITC-labeled aptamers, and a randomized DNA sequence was used as a control. Tested aptamer candidates are listed in Supplementary Table S2, and corresponding histograms of specificity analysis are in Supplementary Figure S3a and b. Interestingly, we observed three types of binding patterns within tested 33 sequences: (1) Sequences do not bind to either Ramos cells or Jurkat.E6 cells, which might be nonspecific amplicons in the library. (2) Sequences bind to both Ramos and Jurkat.E6 cells, and these sequences might be binding to commonly present Fcγ receptors that are present in both types of cells that are eluted by the Ab competition. (3) Sequences bind to only Ramos cells but not to Jurkat.E6 cells, which might be specific sequences toward Ramos cells. This suggests that the pool resulting from competitive elution does not necessarily contain only specific sequences, and screening of individual aptamer candidates for specificity is needed to identify epitope-specific aptamer candidates. Out of the tested 33 sequences, we identified three unique sequences that show specificity toward Ramos cells (Fig. 4).
FIG. 4.
Analysis of specificity of R1, R10, and R15. Specificity of aptamers was analyzed against mIgM-expressing Ramos cells and mIgM-negative Jurkat.E6. Color images available online at www.liebertpub.com/nat
The sequences that showed specificity toward mIgM-positive Ramos cells were further investigated for binding affinity. We evaluated Bmax/2 for the sequences that showed specific binding. The calculated Bmax/2 for R1, R10, and R15 are in the sub-micro molar range (see affinity curves in Supplementary Fig. S4) against Ramos cells, suggesting that sequences generated using LIGS show lower affinities. The issue of lower affinities of the identified aptamers could be predominantly because LIGS was applied to a partially evolved SELEX pool, and the evolution of sequences was interrupted. Therefore, a partially evolved SELEX pool might contain sequences with lower to moderate affinities. However, the affinity of these aptamers could be further enhanced given their high specificity by post-SELEX modification followed by linear multimerization approaches, as described earlier [17]. We are currently investigating the effect of the degree of enrichment of an SELEX pool against whole cells on LIGS to improve LIGS technique.
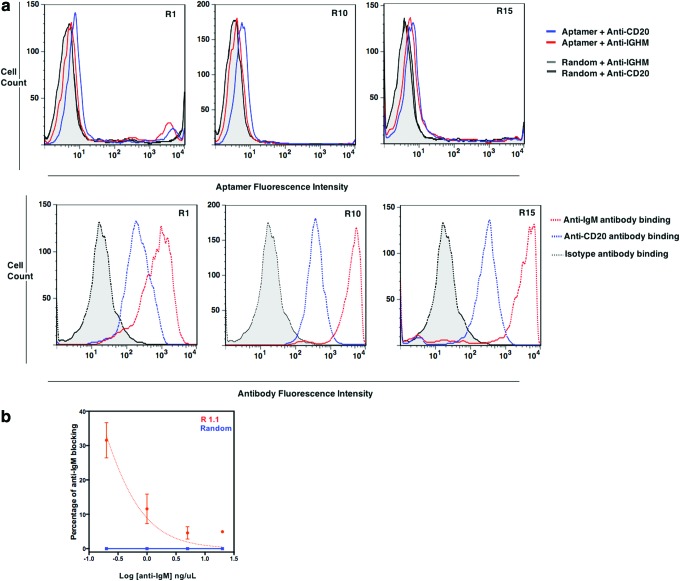
We further investigated whether the identified specific aptamer candidates compete with an anti-IgM Ab for the binding site. The validation of the target using competition against the corresponding antibody has been used earlier [18,19]. We performed competition by first preincubating Ramos cells with anti-IgM Ab or anti-CD20 Ab. CD20 is uniquely expressed in mature, normal B-cells in early developmental stages. CD-20-positive B-cells are the source of a variety of B-cell neoplasms, including Ramos cells, which is a B-cell Non-Hodgkin's Lymphoma. Therefore, the use of anti-CD20 antibody as a control to investigate the antigen specificity further confirms the specificity of the aptamers toward mIgM [20]. Ramos cells pretreated with Ab were then incubated with individual aptamer sequences. After washing, the binding of each aptamer was analyzed using flow cytometry. As expected, the introduction of anti-IgM Ab diminished the binding of the aptamer for R1, R10, and R15 (Fig. 5a red line), which is indicated by the diminished binding of each aptamer in the presence of anti-IgM but not when anti-CD20 is present in the corresponding histograms (Fig. 5a blue line). R1 and R10 showed substantial competition with anti-IgM based on the diminished aptamer fluorescence intensity compared with Ramos cells preincubated with anti-CD20 antibody, whereas R15 did not show substantial competition, suggesting that the binding of R15 might be stabilized by a secondary epitope that is specific for Ramos cells. The competition with only anti-IgM but not with anti-CD20 demonstrates that R10 and R1 are specific for mIgM. Also, we have investigated whether each aptamer can block the binding of the anti-IgM. Since post-SELEX modification of aptamers is essential to increase the homogenous fold and to obtain better yields in chemical synthesis, we optimized the structure of R1 and R10 by systematically truncating bases from 3′ and 5′ ends. We used a truncated and improved version of R1.1 (Bmax/2 ∼82 nM) and R10.T1 (Bmax/2 ∼160 nM) for blocking experiments (see Supplementary Table S2 for sequences of R1.1 and R10.T1). Interestingly, we observed that R1.1 blocked at lower concentrations of anti-IgM, but no significant blockage of the binding of the anti-IgM to Ramos cells was observed at higher concentrations of anti-IgM (Fig. 5b). We did not observe any significant difference in the binding of anti-IgM when R10.T1 was present compared with the randomized control. Antibodies are bivalent in nature; therefore, the avidity of an antibody is higher than monovalent aptamers, and, thus, antibody binding is kinetically more favored than the aptamers.
FIG. 5.
Investigation of epitope identity. Flow cytometric competitive binding analysis of R1, R10, and R15 in the presence of IgM (a) and competitive blocking of anti-IgM binding by R 1.1 (b). Each FITC-labeled library (0.5 μM for R10 and 0.4 μM for R1, and R15) was incubated for 60 min on ice with 1 × 105 Ramos cells that were preincubated with either anti-IgM or anti-CD20 followed by washing with 3 mL of wash buffer; they were subsequently analyzed by flow cytometry. Aptamer fluorescence intensity on the x-axis is indicative of binding of each aptamer. Thus, an increment of fluorescence intensity directly translates into aptamer binding to pretreated Ramos cells. When the cells are preincubated with anti-CD20, all three aptamers show an increase in fluorescence intensity (blue line). Aptamer fluorescence intensity on the x-axis shifts to a lower value in the presence of the anti-IgM antibody (red line), indicating that the anti-IgM substantially blocks R1, R10 and effects R15 binding to its target. No difference in fluorescence intensity was observed for the random control (gray–black), when Ramos cells were preincubated with anti-IgM antibody or anti-CD20, compared with an un-evolved pool from round 0 (black line). Binding of the corresponding antibody indicated in the lower panel. (b) 10 × 104 Ramos cells were incubated with either the random control or R1.1 for 45 min on ice and added to a serially diluted anti-IgM solution. The competitive blocking was allowed for an additional 35 min. followed by wash and were analyzed by flow cytometry. FITC, fluorescein isothiocyanate. Color images available online at www.liebertpub.com/nat
Since we observed a similar pattern of molecular recognition of R10 and R1, we investigated whether these two aptamers are competing for binding to the same epitope. In doing so, we incubated fivefold excess of unlabeled R1.1 and fluorescently labeled R10 with Ramos cells. Washing of unbound aptamer followed by flowcytometric analysis revealed that R1.1 replaces R10, suggesting that the aptamers are binding to the same epitope (Supplementary Fig. S5). This evidence further confirms that the aptamers can be identified by utilizing LIGS binding to the same epitope on Ramos cells.
Theoretically, aptamer interactions are specific toward one target; therefore, the set of aptamers generated in the end of the cell-based selection are expected to correlate to the altered levels of molecules in the positive cell line. Using this approach, a number of aptamers had been selected. We and others have shown that aptamers selected using cell-SELEX compete with the cognate antibody for binding to its target epitope. For example, the aptamer TD05, selected using the cell-SELEX method targeting Burkitt's lymphoma, binds to the heavy chain of membrane-bound IgM (mIgM) and competes with the anti-IgM antibody, permitting the bound aptamer from the target to be eluted into the solution [17]. Similarly, an RNA aptamer selected against CD71-expressing cells using the hybrid-SELEX method competes with the anti-transferrin antibody [19]. A cell-SELEX selected aptamer against myeloid leukemia binds to the sialic acid-binding Ig-like lectin protein and competes with its respective antibody [21]. These reported observations suggest that the aptamers can bind to a region of the receptor that is close to an Ab-binding site. The decrease in aptamer binding when the respective cognate ligand is present can be due to steric hindrance resulting from the large size or the high affinity of the ligand, eluting the aptamer, or the structural changes in the receptor protein induced by ligand (Ab) binding. Also, the bivalent nature of an antibody with favorable kinetic parameters enables antibody binding compared with monovalent aptamers. It has been already shown that aptamers usually bind to ligand-binding sites on receptors or to active sites of proteins [22,23]. Therefore, a partially evolved cell-SELEX aptamer library can also be utilized to identify epitope-specific aptamers, by simply using a ligand against the desired target to elute the respective aptamer sequences.
In this study, we selected aptamers against mIgM. The mIgM molecule is considered the hallmark of B-cells, plays a major role in B-cell development, and is a major player in transformation of B-cells into malignant B-cells [24–26]. Also, mIgM plays a major role in autoimmune disorders and 95% of human lymphomas originate from B-cells [24–26]. There is evidence of activated protein kinase stimulated downstream of B-cell receptor (BCR), demonstrating the significance of developing therapeutics against BCR [24–26]. Currently, there are no successful targeting agents available against mIgM. We have identified three different aptamer candidates with specificity toward mIgM.
The LIGS is a simple approach for generating aptamers toward predetermined epitopes expressed on the cell membranes. Previously, it was not possible to select aptamers against predetermined epitopes of target proteins without post- or pre-SELEX sample manipulation, demonstrating the novelty and significance of this method. Determination of ligands to outcompete the specific aptamers could be done based on the application of the generated aptamers. For example, either ligands could be growth factors interacting with their cognate growth factor receptors expressed on the cell surface, or the interaction of ligands such as hormones and neurotransmitters to activate signaling of G-protein-coupled-receptors could also be chosen as a secondary ligand [27]. Enzyme substrates binding to enzymes, any pre-existing ligand's interaction with its cognate receptor could be exploited in LIGS to generate specific aptamers.
In conclusion, we report on a novel strategy, that is, LIGS, for selecting aptamers against desired epitopes on extracellular receptors using Ab binding as a model ligand. This simple method could also be modified and utilized as a selective screening platform not only to select aptamers but also in phage-display libraries, peptide libraries, and small-molecular libraries to identify artificial molecules toward active sites of macromolecules utilizing pre-existing molecular and cellular interactions as a guide.
Supplementary Material
Acknowledgments
The authors are grateful to the National Institute for General Medical Sciences (NIGMS) grant SC3 GM105578 and Lehman start-up funds for financial support.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Keefe AD, Pai S. and Ellington A. (2010). Aptamers as therapeutics. Nat Rev Drug Discov 9:537–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayasena SD. (1999). Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45:1628–1650 [PubMed] [Google Scholar]
- 3.Tuerk C. and Gold L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510 [DOI] [PubMed] [Google Scholar]
- 4.Ellington AD. and Szostak JW. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822 [DOI] [PubMed] [Google Scholar]
- 5.Gold L. (1995). The SELEX process: a surprising source of therapeutic and diagnostic compounds. Harvey Lect 91:47–57 [PubMed] [Google Scholar]
- 6.Shangguan D, Cao ZC, Li Y. and Tan W. (2007). Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin Chem 53:1153–1155 [DOI] [PubMed] [Google Scholar]
- 7.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ. and Tan W. (2006). Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA 103:11838–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Gong Q, Maheshwari N, Eisenstein M, Arcila ML, Kosik KS. and Soh HT. (2014). Particle display: a quantitative screening method for generating high-affinity aptamers. Angew Chem Int Ed Engl 53:4796–4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White RR, Sullenger BA. and Rusconi CP. (2000). Developing aptamers into therapeutics. J Clin Invest 106:929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He W, Elizondo-Riojas MA, Li X, Lokesh GL, Somasunderam A, Thiviyanathan V, Volk DE, Durland RH, Englehardt J, Cavasotto CN. and Gorenstein DG. (2012). X-aptamers: a bead-based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry 51:8321–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sefah K, Shangguan D, Xiong X, O'Donoghue MB. and Tan W. (2010). Development of DNA aptamers using Cell-SELEX. Nat Protoc 5:1169–1185 [DOI] [PubMed] [Google Scholar]
- 12.Thiel WH, Thiel KW, Flenker KS, Bair T, Dupuy AJ, McNamara JO, II, Miller FJ. and Giangrande PH. (2015). Cell-internalization SELEX: method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol Biol 1218:187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD, Higgins DG. and Gibson TJ. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y. and Tan W. (2007). Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem 79:4900–4907 [DOI] [PubMed] [Google Scholar]
- 16.Gillis S. and Watson J. (1980). Biochemical and biological characterization of lymphocyte regulatory molecules. V. Identification of an interleukin 2-producing human leukemia T cell line. J Exp Med 152:1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, McDevitt MR, Patel DJ. and Scheinberg DA. (2011). A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res 39:2458–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D. and Tan W. (2007). Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's lymphoma cells. Mol Cell Proteomics 6:2230–2238 [DOI] [PubMed] [Google Scholar]
- 19.Wilner SE, Wengerter B, Maier K, de Lourdes Borba Magalhaes M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A. and Levy M. (2012). An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids 1:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Meerten T. and Hagenbeek A. (2010). CD20-targeted therapy: the next generation of antibodies. Semin Hematol 47:199–210 [DOI] [PubMed] [Google Scholar]
- 21.Yang M, Jiang G, Li W, Qiu K, Zhang M, Carter CM, Al-Quran SZ. and Li Y. (2014). Developing aptamer probes for acute myelogenous leukemia detection and surface protein biomarker discovery. J Hematol Oncol 7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell SD, Denu JM, Dixon JE. and Ellington AD. (1998). RNA molecules that bind to and inhibit the active site of a tyrosine phosphatase. J Biol Chem 273:14309–14314 [DOI] [PubMed] [Google Scholar]
- 23.Tian Y, Adya N, Wagner S, Giam CZ, Green MR. and Ellington AD. (1995). Dissecting protein:protein interactions between transcription factors with an RNA aptamer. RNA 1:317–326 [PMC free article] [PubMed] [Google Scholar]
- 24.Bojarczuk K, Bobrowicz M, Dwojak M, Miazek N, Zapala P, Bunes A, Siernicka M, Rozanska M. and Winiarska M. (2015). B-cell receptor signaling in the pathogenesis of lymphoid malignancies. Blood Cells Mol Dis 55:255–265 [DOI] [PubMed] [Google Scholar]
- 25.Danilov AV. (2013). Targeted therapy in chronic lymphocytic leukemia: past, present, and future. Clin Ther 35:1258–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matutes E, Wotherspoon A. and Catovsky D. (2007). Differential diagnosis in chronic lymphocytic leukaemia. Best Pract Res Clin Haematol 20:367–384 [DOI] [PubMed] [Google Scholar]
- 27.Kobilka BK. and Deupi X. (2007). Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci 28:397–406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.