Abstract
The environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) elicits dose-dependent hepatotoxicity that includes fat accumulation, inflammation, and fibrosis that may progress to hepatocellular carcinoma. To further investigate these effects, RNA-Seq data were integrated with computationally identified putative dioxin response elements, and complementary targeted metabolomic and aryl hydrocarbon receptor (AhR) ChIP-Seq data from female C57BL/6 mice gavaged with TCDD every 4 days for 28 days. Data integration using CytoKEGG with manual curation identified dose-dependent alterations in central carbon and amino acid metabolism. More specifically, TCDD increased pyruvate kinase isoform M2 (PKM2) gene and protein expression. PKM2 has lower catalytic activity resulting in decreased glycolytic flux and the accumulation of upstream intermediates that were redirected to the pentose phosphate pathway and serine/folate biosynthesis, 2 important NADPH producing pathways stemming from glycolysis. In addition, the GAC:KGA glutaminase (GLS1) protein isoform ratio was increased, consistent with increases in glutaminolysis which serves an anaplerotic role for the TCA cycle and compensates for the reduced glycolytic flux. Collectively, gene expression, protein, and metabolite changes were indicative of increases in NADPH production in support of cytochrome P450 activity and ROS defenses. This AhR-mediated metabolic reprogramming is similar to the Warburg effect and represents a novel advantageous defense mechanism to increase anti-oxidant capacity in normal differentiated hepatocytes.
Keywords: TCDD, PKM2, glutaminolysis, oxidative stress, hepatotoxicity
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is the prototypical ligand for a series of structurally diverse environmental contaminants, natural products, and endogenous metabolites that bind and activate the aryl hydrocarbon receptor (AhR), a basic helix-loop-helix ligand activated transcription factor (Denison et al., 2011). Binding of TCDD and related compounds to the cytosolic AhR causes chaperone protein dissociation, followed by nuclear translocation and heterodimerization with the AhR nuclear translocator (ARNT). The TCDD-AhR-ARNT complex binds dioxin response elements (DREs, also known as AHREs and XREs) consisting of the invariant core sequence 5′-GCGTG-3′, eliciting changes in gene expression, although DRE-independent interactions have also been described (Beischlag et al., 2008; Dere et al., 2011b; Huang and Elferink, 2012).
TCDD and related compounds elicit a variety of adverse effects including teratogenesis, immunosuppression, and tumor promotion (Pohjanvirta and Tuomisto, 1994), and have also been implicated in the development of non-alcoholic fatty liver disease and type II diabetes (T2D) (Taylor et al., 2013). In mice, a single bolus dose of TCDD results in reversible hepatic fat accumulation (Boverhof et al., 2005; Lee et al., 2010) while repeated treatment sustains lipid accumulation with progression to steatohepatitis and fibrosis (Nault et al., 2015a; Pierre et al., 2014). TCDD-elicited metabolic disruption is not limited to lipid metabolism, but also disrupts glycolysis, the TCA cycle, and nucleoside metabolism (Forgacs et al., 2012; Lin et al., 2011; Matsubara et al., 2012). However, these were short term exposure studies involving a single bolus dose, or did not include complementary ‘omic’ data.
The objective of this study was to integrate complementary AhR ChIP-Seq, hepatic RNA-Seq, and hepatic and serum metabolomic data with computationally identified putative dioxin response elements (pDREs), to further elucidate the mechanisms involved in TCDD-elicited hepatotoxicity. Changes in central carbon and amino acid metabolism were reminiscent of the Warburg effect. This included pyruvate kinase isoform switching (Christofk et al., 2008), enhanced glutaminolysis (DeBerardinis et al., 2007), and increased serine (Ser)/folate metabolism (Locasale, 2013; Maddocks et al., 2013). This metabolic reprogramming is consistent with increased anti-oxidant capacity and may represent an advantageous, more general response, in normal differentiated tissue following toxic insult to support cell survival.
MATERIALS AND METHODS
Animal Treatment
Postnatal day 25 (PND25) female C57BL/6 mice (Charles River Laboratories, Portage, Michigan), housed in polycarbonate cages with cellulose fiber chips (Aspen Chip Laboratory Bedding, Warrensberg, New York) at 30–40% humidity and a 12-h light/dark cycle and acclimatized for 4 days, were fed ad libitum (Harlan Teklad 22/5 Rodent Diet 8940, Madison, Wisconsin) with free access to deionized water. On PND 28 and every following fourth day animals (n = 5) were orally gavaged with 0.1 ml sesame oil or 0.01, 0.03, 0.1, 0.3, 1, 3, 10, and 30 µg/kg TCDD (Dow Chemical Company, Midland, Michigan) for a total of 28 d. Blood was collected by submandibular vein puncture into BD microtainer tubes with serum separator and centrifuged at 10 000×g for 5 min and stored at −80°C. Liver samples were frozen in liquid nitrogen, and stored at −80°C. Only 30 µg/kg samples were analyzed by ChIP-Seq at 2 h. Livers from 92 days treated mice were collected using the same treatment and collection methods. Hepatic samples from female C57BL/6 mice treated with 100 µg/kg β-naphthoflavone (βNF) were collected following daily gavage for 28 days. TCDD time course, 3,3′,4,4′,5-pentachlorobiphenyl (PCB126), 2,3,7,8-tetrachlorodibenzofuran (TCDF), 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) were collected as described (Boverhof et al., 2005; Kopec et al., 2010a,b). All procedures were approved by the All-University Committee on Animal Use and Care.
pDRE Distribution
pDREs were updated using the mm10 GRCm38 genome build (Dere et al., 2011a). The position weight matrix used the same mouse and rat bona fide DRE’s (Dere et al., 2011a) with Gsta2 removed (ie, not present in the most recent rat genome build (rn5)). Four new functional DREs for human Bach2 and Cyp1b1 were added (Supplementary Table S1). pDREs UCSC genome browser tracks are available at http://dbzach.fst.msu.edu/index.php/supplementarydata.html.
Hepatic AHR ChIP-Seq
ChIP-Seq analysis was performed on liver samples collected at 2 h following a single oral gavage with 30 µg/kg TCDD. Cross-linked DNA was immunoprecipitated with either rabbit IgG or rabbit IgG and anti-AhR as previously described (Dere et al., 2011b; Lo and Matthews, 2012). Libraries prepared using the MicroPlex kit (Diagenode) were pooled and sequenced at a depth of approximately 30 M on an Illumina HiSeq 2500 at the MSU Research and Technology Support Facility Core. Read processing and analysis was performed using the MSU High Performance Computing Center. Quality was determined using FASTQC v0.11.2 and adaptor sequences removed using Cutadapt v1.4.1 while low-complexity reads were cleaned using FASTX v0.0.14. Reads were mapped to the mouse reference genome (GRCm38 release 76) using Bowtie 2.0.0 and alignments were converted to SAM format using SAMTools v0.1.19. Normalization and peak calling was performed using CisGenome (Ji et al., 2008) determined by comparison of IgG control and AhR enriched samples (n = 5) using a bin size (−b) of 25 and boundary refinement resolution (−bw) of 1 with default parameters.
Metabolite Extraction
Liver (approximately 100 mg) and serum (30 µl) metabolites were extracted using chloroform:water:methanol from animals orally gavaged every 4 days for 28 days with sesame oil vehicle, 3, 10, or 30 µg/kg TCDD. Briefly, frozen liver samples were homogenized (Polytron PT2100, Kinematica) or vortexed (serum) in HPLC grade methanol:water (5:3, 4.63 mL, −20°C) in Pyrex glass tubes. HPLC-grade chloroform was added following homogenization (methanol:water:chloroform, 5:3:5), vortexed, shaken on ice for 10 min, and centrifuged at 3000×g. The transferred aqueous phase was dried under nitrogen gas at room temperature and resuspended in HPLC-grade water prior to analysis. Hepatic extract protein layers were dried and a bicinchoninic acid assay (Sigma-Aldrich) on a Tecan Infinite 200 microplate reader (Männedorf, Switzerland) was used to normalize signal in liver.
Amino acids were carbobenzyloxy derivatized for analysis. Redissolved solutions (20 µl) were added to 80 µl of HPLC-grade methanol, and 2.5 µl of triethylamine. 0.5 µl benzylchloroformate was added and samples were vortexed then centrifuged, and the supernatant was transferred to HPLC vials for analysis.
Targeted Metabolomic Analysis
Hepatic extracts were first loaded onto a trapping column (C18, 4 × 2 mm, Phenomenex) and washed for 30 s with HPLC grade water containing 10 mM tributylamine and 15 mM acetic acid for desalting. Hepatic extracts were examined by liquid chromatography (LC) and tandem mass spectrometry (MS/MS) using a Paradigm MS4 HPLC (Michrom Bioresources, Auburn, California), and a Synergi Hydro column (4 µm particle size, 80 Å, 150 × 2 mm, from Phenomenex) (Lunt et al., 2015). HPLC was coupled with negative-mode electrospray ionization to a TSQ Vantage Triple Stage Quadrupole Mass Spectrometer (Thermo Scientific) operating in multiple reaction monitoring (MRM) mode. Serum extracts were analyzed using an Acquity UPLC system (Waters), Ascentis Express column (C18, 5 cm × 2.1 mm, 2.7 µm, Sigma-Aldrich) while MRM was performed using a Xevo TQ-S triple quadrupole mass spectrometer (Waters) (see Supplementary Materials). Raw data and MRM parameters are deposited in www.ebi.ac.uk/metabolights (MTBLS225).
Data analysis, including peak integration was performed using MAVEN (Clasquin et al., 2012; Melamud et al., 2010). Waters raw data were converted to mzxml format using msconvert in the ProteoWizard Tools (Kessner et al., 2008) prior to analysis by MAVEN. One-way ANOVA was performed in SAS 9.3 (SAS Institute, Inc., Cary, North Carolina).
Hepatic Gene Expression
Published RNA-Seq data for mice orally gavaged with sesame oil vehicle, 0, 0.01, 0.03, 0.1, 0.3, 1, 3, 10, or 30 µg/kg TCDD every 4 days for 28 days (GSE62902) were used for the integrative analysis (Nault et al., 2015b). Filtering criteria of |fold-change| ≥ 1.5 and P1(t) ≥ 0.8 were used for all data analysis. UCSC Genome browser tracks combine all reads from vehicle or 30 µg/kg doses (http://dbzach.fst.msu.edu/index.php/supplementarydata.html). Quantitative real-time PCR (QRTPCR) was performed as previously described (Boverhof et al., 2005, see Supplementary Materials). Relative expression was determined using the 2−ΔΔCT method and the geometric mean of reference genes ActB, Gapdh, and Hprt. Reference genes showed no evidence of treatment related mRNA effects. Statistical analyses of QRTPCR data were performed using a 1-way ANOVA for dose-response study designs followed by Dunnett’s post hoc test. For time-course designs a 2-way ANOVA with dose and time as factors was used followed by Tukey’s post hoc test. For chemical treatment, each study was considered independently and analyzed by Student’s t test. Statistical analyses were performed using SAS 9.3.
Protein Measurements
Liver samples from animals orally gavaged with either sesame oil or 30 µg/kg TCDD (approximately 25 mg, n = 5) were ground by mortar and pestle in radioimmunoprecipitation buffer supplemented with protease inhibitor cocktail (Sigma-Aldrich), sonicated on ice, and centrifuged. Total protein in the supernatant was quantitated by bicinchoninic acid assay (Sigma-Aldrich). Protein levels were determined using the Wes (ProteinSimple, San Jose, Califrnia) system. PKM1 (Sigma-Aldrich; 1:400), PKM2 (Cell Signaling, Danvers, Massachusetts; 1:65), PKM1/2 (Cell Signaling; 1:65), and GLS1 (ProteinTech, Chicago, Illinois; 1:30) antibodies were duplexed with β-actin (Cell Signaling; 1:65) as loading control and detected by goat anti-rabbit secondary antibody. Chemiluminescence signals were exported from Compass software (ProteinSimple) and used to calculate fold-change. Statistical analyses were performed using Student’s t test in SAS 9.3.
Enrichment Analyses and KEGG Pathway Integration
Over-represented metabolites within a pathway, as well as impact scores representing the connectivity to other metabolites or clusters were determined using MetPA (Xia and Wishart, 2010). Over-represented differentially expressed genes (DEGs) compared with all mouse Kyoto Encyclopedia of Genes and Genomes (KEGG; http://genome.jp/kegg/) pathway genes was performed using ClusterProfiler in R 3.2.0 (Yu et al., 2012). Pathways were manually curated to include only normal metabolism pathways which were considered relevant to the expression dataset (eg, signaling, tissue specific, and cancer pathways were removed).
The CytoKEGG 0.0.5 plugin (Cytoscape 3.2.0) was used to generate pathway templates. Through manual curation for completeness, and refinement, pathways were streamlined by combining reactions with the same compounds as a single node. pDRE, AhR enrichment, differential gene expression, and altered metabolite level data represent the most significant change for each node (eg, highest MSS score or largest |fold-change|). An expandable interactive versions can be viewed at http://dbzach.fst.msu.edu/index.php/supplementarydata.html.
RESULTS
Alterations in Central Carbon, Amino Acid, and Lipid Metabolism
TCDD elicits significant lipidomic changes consistent with the development of fatty liver and steatohepatitis in mice (Boverhof et al., 2005; Fernandez-Salguero et al., 1996). In this study, complementary targeted metabolomics was used to examine aqueous metabolite changes in hepatic extracts and serum from mice treated with 3, 10, and 30 µg/kg TCDD every 4 days for 28 days. Of the approximately 190 metabolites examined, only 124 and 84 were detected in hepatic and serum samples, respectively (Supplementary Table S2 and S3) with 48 hepatic and 41 serum metabolites altered (P ≤ .05) by treatment. This included an 11-fold increase in hepatic oxaloacetate (OAA) and >1000-fold decrease in serum xanthine. Altered metabolite levels were analyzed for pathway over-representation (enrichment) and connectivity within related metabolites (impact) using MetPA (Xia and Wishart, 2010). Central carbon (glycolysis/gluconeogenesis, the TCA cycle, and the pentose phosphate pathway [PPP]), and amino acid metabolism consistently exhibited higher enrichment (−log(p)) and/or impact scores (Figs. 1A and B, Supplementary Tables S4 and S5).
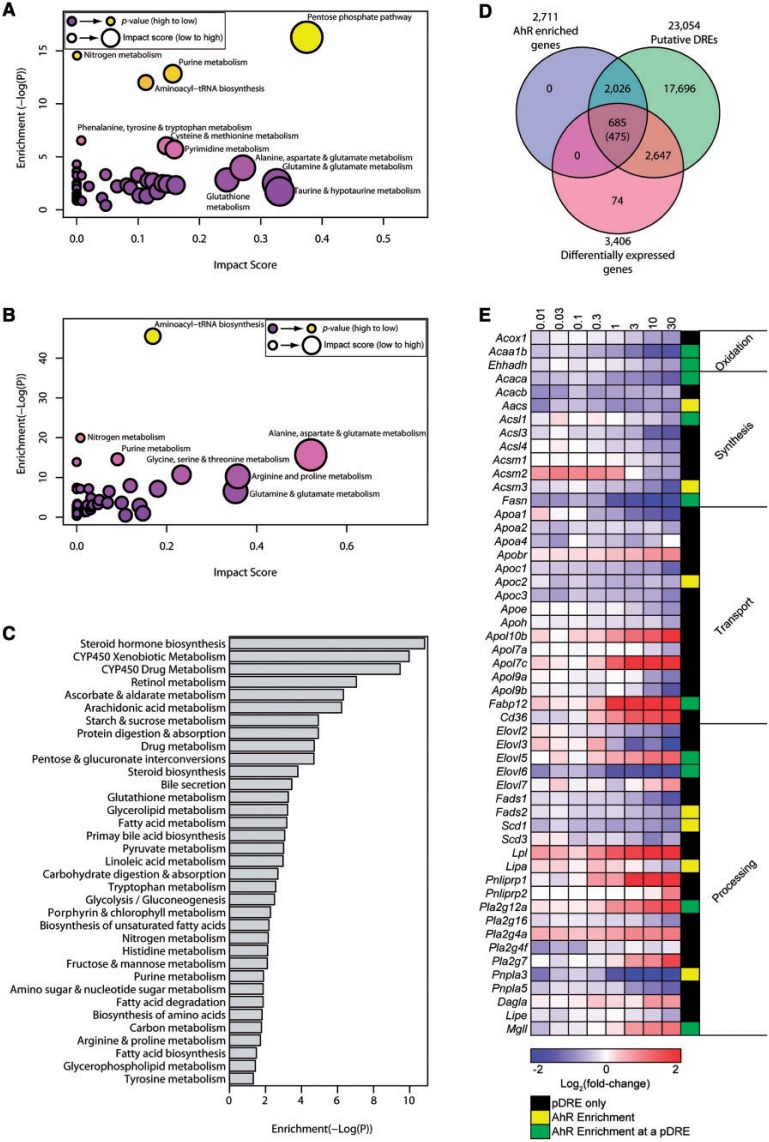
FIG. 1.
TCDD-elicited metabolomic disruption. Metabolic pathway enrichment analysis (MetPA) of altered metabolites (P ≤ .05) in (A) hepatic and (B) serum extracts. Enrichment values ≥ 4.0 were considered significant and impact scores were used to rank pathways. C, Significantly enriched (P ≤ .05) KEGG pathways for the 3406 DEGs (|fold-change| ≥ 1.5, P1(t) ≥ .8). D, Venn diagram of genes with AhR enrichment within its genomic region and 10 Kb upstream of the TSS determined by ChIP-Seq (FDR ≤ 0.05), putative dioxin response elements (pDREs), and DEGs. Genes possessing AhR enrichment, pDREs, and differential expression are shown. The number of genes meeting these criteria where the AhR enrichment peaks overlap a putative DRE is shown in parentheses. E, Heat map of DEGs associated with lipid transport, processing and metabolism. The presence of AhR enrichment at 2-h post-treatment and/or pDREs is indicated in last column on the right.
RNA-Seq (Nault et al., 2015b) and metabolomic data were integrated using KEGG metabolic pathways. Analysis of 3406 DEGs (|fold-change| ≥ 1.5 and P1(t) ≥ .8) using clusterProfiler (Yu et al., 2012) identified over-represented (P ≤ .05) pathways associated with altered glucose, amino acid, and fatty acid metabolism (Figure 1C). KEGG enrichment also identified drug, starch and sucrose, and pyruvate metabolism enrichment. Steroid hormone biosynthesis was the most enriched pathway, consistent with previous reports (Angrish et al., 2013; Tanos et al., 2012b) although our targeted metabolomic data did not include steroid metabolites. Overall, metabolite changes were consistent with the identified DEGs.
To further evaluate AhR-mediated effects, ChIP-Seq was performed 2 h post dose with 30 µg/kg TCDD. Approximately 13 936 enrichment peaks were identified, of which 5766 satisfied the FDR ≤ 0.05 criteria, with 2711 located within the coding region or 10 kb upstream region (Supplementary Table S6). Comparing the 3406 DEGs with 2711 AhR enriched regions with 23 054 pDREs identified 685 genes at the intersection. Only 475 had a pDRE within an AhR enriched region (Figure 1D) suggesting AhR-mediated DRE-dependent regulation, while 210 genes with AhR enrichment lacked an overlapping pDRE.
Increased Hepatic Lipid Uptake but Inhibited Fatty Acid Synthesis and Oxidation
KEGG enrichment analysis identified DEGs associated with lipid transport, processing, and metabolism (Figure 1E). This included the down-regulation of apolipoproteins (ie, 3.3-fold Apoa1) involved in lipid packaging and transport. Conversely, Apol7c, Apol10d, and Fabp12 were induced 3.6-, 14-, and 46-fold, respectively. Cd36 involved in lipid uptake was also induced 3.2-fold as previously reported (Lee et al., 2010). However, AhR enrichment with a pDRE was only observed for Fabp12. Similarly, lipid processing genes such as elongases (Elovl2, 3, and 6) and desaturases (Fads, Scd) were repressed >2-fold. Lipases Lpl, Mgll, Pnliprp1, and Pnliprp2 were induced 4.8-, 2.1-, 48-, and 1.9-fold, respectively, while Lipa, Pnpla3, and Pnpla5 were repressed 1.5-, 8.0-, and 2.4-fold, respectively. Overall, de novo fatty acid (FA) synthesis and β-oxidation genes were repressed (Figure 1E and Supplementary Figure S1) including a 5.3-fold repression of fatty acid synthase (Fasn) with AhR enrichment that included a pDRE. β-Oxidation was marked by repression of acetyl-CoA acyltransferase (Acaa1b 3.1-fold), carboxylase (Acaca 2.5-fold and Acacb 1.5-fold), and enoyl-CoA hydratase (Ehhadh 1.6-fold). Acsl1, Acsm3, Acaa1b, Acaca, and Ehhadh exhibited AhR enrichment with a pDRE, except for Acsm3. The inhibition of β-oxidation is consistent with short acyl-CoA reductions, particularly decreases in butanoyl-CoA (2.9-fold).
Induction of Oxidative Stress and Anti-Oxidant Defenses
CytoKEGG analysis with manual curation identified the enrichment of oxidative stress (eg, P450 induction, glutathione metabolism), central carbon metabolism, and amino acid metabolism pathways (Figs. 2 and 3). These pathways were found to converge on NADP+/NADPH, indicating altered production and/or consumption of reducing equivalents.
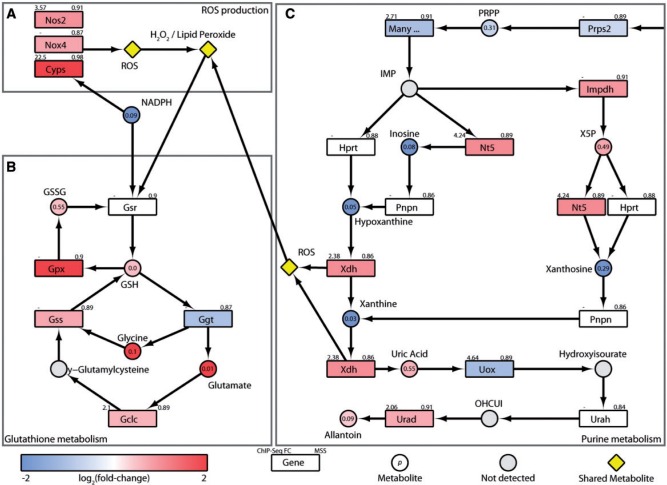
FIG. 2.
Induction of oxidative stress and defense pathways. Integration of transcriptomic, hepatic and serum metabolomic, and KEGG pathway data for (A) ROS producing xenobiotic metabolizing enzymes, (B) Glutathione metabolism, and (C) Purine metabolism. Color scale represents the log2(fold-change) for genes and metabolites. Yellow symbols indicate converging metabolites. Gray symbols indicate metabolites not measured or detected. Genes are identified as rectangles and metabolites as circles. The upper left corner indicates maximum AhR enrichment fold-change and upper right corner indicates the highest scoring pDRE matrix similarity score (MSS). Circles contain the P-value for metabolite changes. An expandable and interactive version of this figure with further information is available at http://dbzach.fst.msu.edu/index.php/supplementarydata.html.
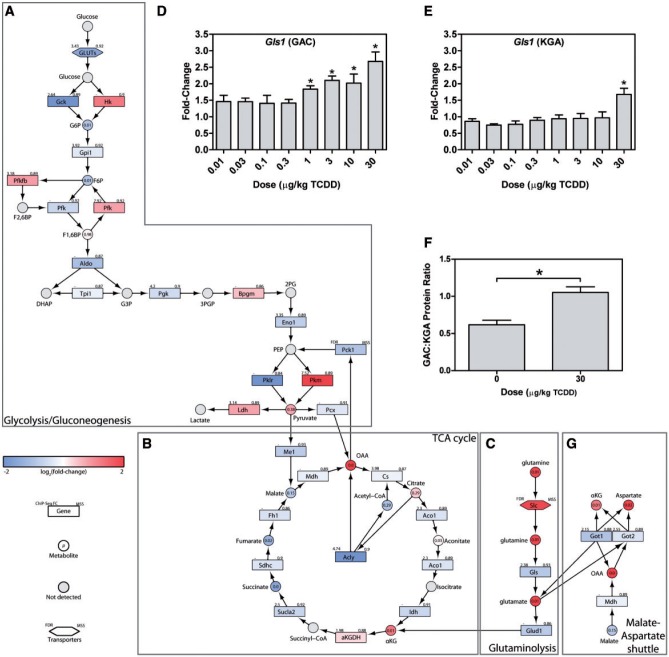
FIG. 3.
Reprogramming of central carbon and amino acid metabolism. Integration of transcriptomic, hepatic and serum metabolomic, and KEGG pathway data for (A) glycolysis/gluconeogenesis, (B) the TCA cycle, (C) glutaminolysis. QRTPCR confirmation of hepatic Gls1 (D) GAC, and (E) KGA isoforms, as well as (F) the GAC:KGA protein ratio are shown as barplots. (G) Transcriptomic, metabolomic, and KEGG pathway data were integrated for the malate-aspartate shuttle. In integrative pathways, the color scale represents the log2(fold-change) for genes and metabolites. Gray indicates metabolites not measured or detected. Genes are identified as rectangles, metabolites as circles, and transporters as hexagons. The upper left corner provides the maximum AhR enrichment fold-change and upper right corner indicates the highest pDRE matrix similarity score (MSS). Circles contain the P-value for detected metabolites. An expandable and interactive version of this figure which provides additional information can be viewed at http://dbzach.fst.msu.edu/index.php/supplementarydata.html. Bars in barplots represent mean + SEM. Asterisks (*) indicate P ≤ .05 compared with vehicle determined by 1-way ANOVA followed by Dunnett’s post hoc test.
The 1250-, 807-, and 16-fold induction of classical AhR battery genes Cyp1a1, Cyp1b1, and Cyp1a2 as well as the 1.6- and 1.9-fold increase in NADPH oxidase (Nox4) and nitric oxide synthase (Nos2) represent major NADPH consumers as well as producers of reactive oxygen species (ROS; Figure 2A). In response, glutathione peroxidase (Gpx2) was induced 11-fold, an important defense mechanism that uses glutathione (GSH) to reduce lipid peroxide and hydrogen peroxide oxidative stress products to alcohols and water, respectively (Figure 2B). The resulting oxidized glutathione (GSSG) is then recycled to GSH (increased 1.4-fold) using NADPH by glutathione reductase (Gsr), which was induced 1.4-fold, to preserve cellular anti-oxidant capacity. TCDD also induced a 1.7-fold increase in glutathione synthetase (Gss), which catalyzes the condensation of glycine (Gly) with γ-glutamylcysteine to produce more GSH (Figure 2B). Accordingly, these processes increased total hepatic glutathione levels 1.3-fold (Supplementary Figure S2). The absence of AhR enrichment within Gpx2, Gsr, and Gss loci suggest these responses were mediated by oxidative stress as opposed to direct AhR regulation, and likely involve Nrf2 signaling which is known to be induced following AhR activation (Lu et al., 2011; Wang et al., 2011b; Yeager et al., 2009).
In addition, purine metabolism, producing the anti-oxidant, uric acid, was increased 1.5-fold in the liver (Figure 2). Inosine monophosphate (IMP) is converted to inosine by 5′ nucleotidase (Nt5e), and then to hypoxanthine by purine nucleoside phosphorylase (Pnp) and subsequently to xanthine and uric acid by xanthine dehydrogenase (Xdh). Although decreases in serum inosine (33-fold), hypoxanthine (170-fold), and xanthine (>1000-fold), are consistent with the 2.1- and 1.9-fold AhR-mediated induction of Nt5e and Xdh (Figure 2C), increased enzymatic activity are likely the primary factors. Interestingly, the oxidase form (XO) of Xdh irreversibly produced under low energy conditions is also a significant ROS contributor.
Interaction of Glucose, Amino Acid, and Glutathione Metabolism
Alterations in the catabolic oxidation of glucose were also revealed by enrichment analysis (Figs. 3A and B). Although hexose transporters with different kinetic properties and substrate preferences were differentially expressed (Slc2a3, 2a6, 2a10, and 2a3 were induced 1.6-, 2.0-, 1.7-, and 1.6-fold, respectively while Slc2a2, 2a4, and 2a5 were repressed 2.1-, 3.0-, and 1.8-fold, respectively), only Slc2a2 was associated with AhR enrichment. Hexokinases (Hk) 1, 2, and 3 were induced 2.3-, 2.5-, and 2.2-fold, respectively, while glucokinase (Gck) was repressed 4.2-fold. HK2 induction is consistent with the Warburg effect. Glucose-6-phosphate (G6P) levels in liver extracts were reduced 1.9-fold, consistent with a approximately 30% decrease at 168 h after a single bolus dose of TCDD (Forgacs et al., 2012). There was a negligible decrease in glucose-6-phosphate isomerase (Gpi) mRNA with a 1.9-fold decrease in fructose-6-phosphate levels (F6P). Liver phosphofructokinase (Pfkl), which catalyzes the production of fructose 1,6-bisphophate (F1,6BP) was also repressed 1.5-fold while fructose 1,6 bisphosphatase (Fbp1) catalyzing the reverse reaction was induced 1.7-fold. Moreover, phosphofructokinase-2/fructose-2,6-bisphosphatase (Pfkfb3) was induced 1.8 fold, which could decrease fructose-2,6-bisphosphate (F2,6BP) levels, a potent phosphofructokinase allosteric activator (Chesney, 2006). Fructose bisphosphate aldolase B (Aldob) expression was also repressed 2.1-fold. Although cytosolic glyceraldehyde-3-phosphate dehydrogenase (Gpd1) which converts DHAP to glycerol-3-phosphate to support triglyceride synthesis was unaffected, mitochondrial Gpd2 was repressed 2.8-fold which may compromise glycerol-3-phosphate re-oxidation to DHAP and FADH2 production, limiting cytosolic NADH electron transfer to support oxidative phosphorylation. TCDD did not affect glyceraldehyde-3-phosphate dehydrogenase (Gapdh) or phosphoglycerate mutase (Pgam1 and 5) expression while modestly inducing phosphoglycerate kinase (Pgk1, 1.3-fold), and repressing enolase (Eno1, 1.6-fold) which catalyzes the transformation of 2-phosphoglycerate to phosphoenolpyruvate (PEP). Both Pgam1 and Eno1 showed AhR enrichment.
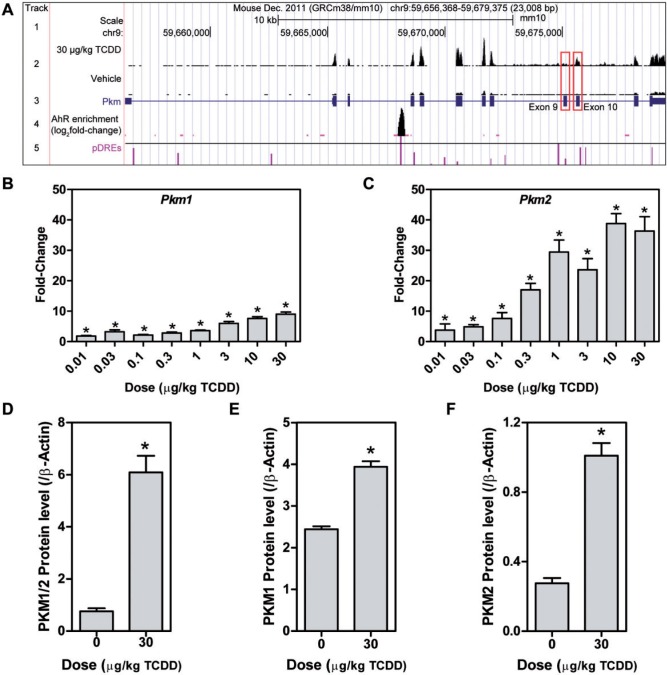
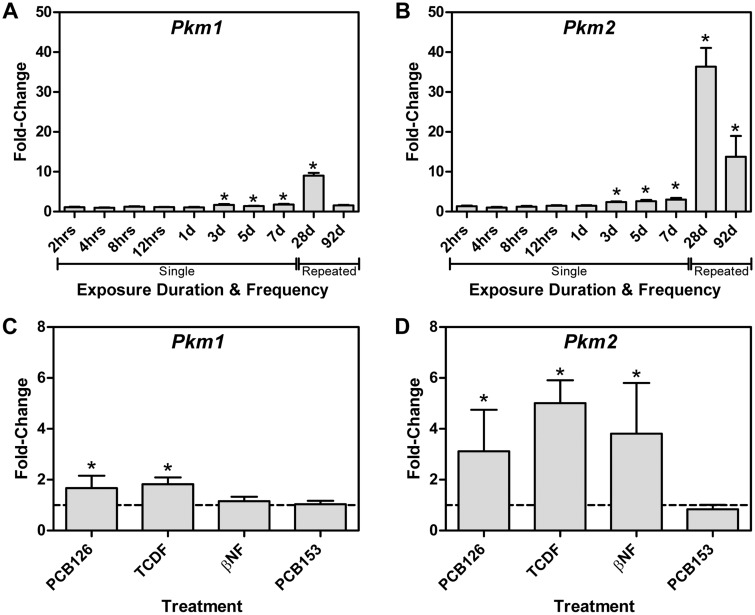
PEP conversion to pyruvate by pyruvate kinase is a rate limiting step in glycolysis. Liver/red blood cell pyruvate kinase (Pklr) was repressed 6.7-fold, consistent with a reported 3.2-fold decrease (Forgacs et al., 2012) while muscle pyruvate kinase (Pkm) was induced 4.6-fold with AhR enrichment, as previously reported (Forgacs et al., 2012). Further analysis of Pkm mapped RNA-Seq reads to exon 10 as opposed to exon 9 indicating isoform switching from Pkm1 to Pkm2 (Figure 4A). Pkm2 is typically expressed in rapidly proliferating cancer cells and is characteristic of the Warburg effect, with a lower catalytic rate in the dimeric or monomeric form, leading to the accumulation of upstream glycolytic intermediates. QRTPCR confirmed a 9-fold induction of Pkm1 compared with 40-fold for Pkm2 (Figs. 4B and C). Protein measurements found a 13.4-fold increase in total PKM, with a 1.6-fold increase in PKM1, and a 3-fold increase in PKM2 using 3 different antibodies (Figs. 4D and F). Although Pkm2 was only induced 2.4-fold compared with 1.7-fold for Pkm1 at 72 h, it was induced 13.8-fold compared with 1.6-fold for Pkm1 after treatment every 4 days for 92 days (Figs. 5A and B). Other AhR agonists, including 3,3′,4,4′,5-pentachlorobiphenyl (PCB126), 2,3,7,8-TCDF, and βNF also induced Pkm2, while 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153), which does not bind to the AhR, had no effect on Pkm2 mRNA levels (Figs. 5C and D). ChIP-Seq analysis at 2 h after treatment with 30 µg/kg TCDD identified a 7.5-fold increase in AhR enrichment within the Pkm coding region that contained a pDRE suggesting DRE-dependent AhR-mediated regulation (Figure 4A). This same peak was previously identified by ChIP-chip and confirmed by ChIP-PCR (Dere et al., 2011b).
FIG. 4.
TCDD elicited Pkm isoform switching. A, Pkm UCSC genome tracks representing (1) the scale, (2) raw number of aligned RNA-Seq reads (scaled to a maximum of 403 for both treatments - red boxes highlight location of exon 9 (Pkm1) and exon 10 (Pkm2)), (3) Pkm exons and introns, (4) AhR enrichment sites, and (5) pDREs with horizontal line indicating matrix similarity score of 0.874. QRTPCR verification of hepatic (B) Pkm1 and (C) Pkm2 mRNA expression, and confirmation of changes in hepatic protein levels of (D) PKM1/2, (E) PKM1, and (F) PKM2. Bars represent mean + SEM (n = 4–5). Asterisks (*) indicate P ≤ .05 compared with vehicle determined by 1-way ANOVA followed by Dunnett’s post hoc test or Student’s t test.
FIG. 5.
AhR-mediated Pkm isoform expression. Time-dependent expression of (A) Pkm1 and (B) Pkm2 mRNA induced by 30 µg/kg TCDD at 2 h to 7 days following a single bolus dose, or dosing every 4 days for 28 or 92 days and ligand dependent expression of (C) Pkm1 and (D) Pkm2 following treatment with PCB126, TCDF, or βNF, or the non-dioxin like PCB153. Bars represent mean + SEM (n = 4–5). Asterisks (*) indicate P ≤ .05 compared with vehicle determined by 2-way ANOVA followed by Tukey’s post hoc test or Student’s t test.
Reduction of glycolytic flux and the inhibition of β-oxidation not only reduce hepatic acetyl-CoA levels, but also impact the TCA cycle and reducing equivalent production. Less pyruvate from glycolysis, inhibition of pyruvate dehydrogenase by pyruvate dehydrogenase kinase (Pdk3 and 4 induced 1.9 and 1.7-fold, respectively), and the inhibition of β-oxidation reduce acetyl-CoA availability for condensation with OAA. In cancer cells and the Warburg effect, central carbon metabolism reprogramming also involves compensatory changes in glutamine metabolism (Gln) to maintain oxidative phosphorylation and ATP production (van den Heuvel et al., 2012). TCDD increased glutamate (Glu) levels in the serum, altered its metabolism (Figure 1) and induced Slc1a5 (5.1-fold), Slc6a19 (3.6-fold), and Slc38a1 (1.5-fold) transporters while repressing the expression of antiporters Slc38a3, 4 and 7 (repressed 1.8 -, 1.7 -, and 1.3-fold respectively; Figure 3C). A concomitant 1.4-fold increase in glutaminase (Gls1), which catalyzes the transformation of Gln to Glu and ammonia, and a 1.9-fold decrease in glutaminase 2 (Gls2) are consistent with increased glutaminolysis, as reported in some cancer cell lines (Wise et al., 2008). Glutaminolysis may also provide Glu to support GSH biosynthesis.
Coincidentally, mRNA for the cystine (Cyss) transporters, xCT (Slc7a11 induced 1.8-fold), Slc3a2 (induced 1.9-fold), and Slc7a6 (induced 1.8-fold) are increased. Slc7a11 is regulated by Nfe2l2 (aka Nrf2) (Conrad and Sato, 2012) which was also induced 2.4-fold by TCDD. Once inside the cell, Cyss is reduced to Cys, the rate limiting precursor for de novo GSH biosynthesis (Yin et al., 2015). Although Cys may be synthesized from Met by the transsulfuration pathway, the primary source of intracellular Cys is more likely transport as hepatic Met levels were decreased 5.9-fold in the liver.
Two splice variants associated with tumor environments exist for Gls1; kidney-type glutaminase (KGA) consisting of exons 1–14 and 16–19, and the GAC isoform with an alternate carboxy-terminus consisting of exon 15. The GAC:KGA mRNA and protein ratio is increased in tumors (van den Heuvel et al., 2012), and also increased by TCDD, with GAC and KGA mRNA induced 2.3 - and 1.6-fold, respectively (Figs. 3D and E). Accordingly, the GAC:KGA protein ratio was increased 1.7-fold (Figure 3F).
Higher hepatic Glu levels (2.5-fold) would be expected to be converted to α-ketoglutarate (αKG) by glutamate dehydrogenase (Glud1) to replenish the TCA cycle (Figure 3C), and may explain the 11-fold increase in OAA in the treated liver (Figure 3C). The 2-fold repression of PEP carboxykinase (Pck1), the rate-limiting step of gluconeogenesis, is also consistent with gluconeogenesis inhibition (Ahmed et al., 2015; Diani-Moore et al., 2010; Weber et al., 1995), further contributing to OAA accumulation. However, Glud1 is repressed 1.8-fold as are succinyl-CoA synthetase (1.6 - and 1.5-fold repression of Sucla2 and Suclg2 1.5-fold), succinate dehydrogenase (1.3-fold repression of Sdhd), and fumarase (1.4-fold repression of Fh1) which may reduce glutaminolysis flux towards OAA.
Alternatively, Glu could be used in the malate-aspartate shuttle to produce pyruvate, which is reported not to contribute to lactate production (Son et al., 2013). In this scenario, mitochondrial OAA and Glu are converted to αKG and aspartate (Asp) by glutamate oxaloacetate transaminase (Got2) which is modestly repressed 1.3-fold (Figure 3F). The 1.9-fold repression of cytosolic Got1 would limit the reconversion of cytosolic Asp, and may be the source of the 4.1-fold increase in hepatic Asp levels.
Redirecting Central Carbon and Amino Acid Metabolism to NADPH Production
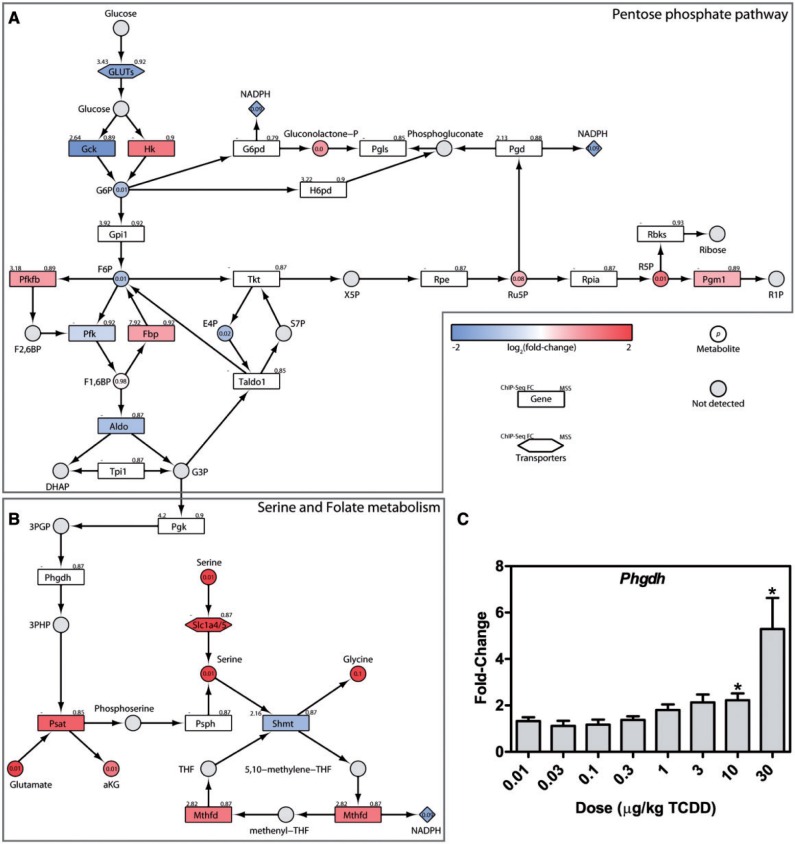
Increased ROS production and lipid peroxidation create an oxidative environment requiring NADPH-dependent counter-measures for cell survival. In cancer cells, NADPH demands are satisfied by the PPP with smaller contributions from NADP+-dependent malic enzyme (Me1) and NADP+-dependent isocitrate dehydrogenase (Idh) (Jiang et al., 2014). With increased PKM2 expression, accumulating G6P levels are redirected to the PPP (Figure 6A) consistent with decreases in hepatic G6P (1.9-fold), F6P (1.9-fold), 6-phospho-D-gluconate (6PG; 4.5-fold), erythrose-4-phosphate (E4P; 2.1-fold), and 5-phosphoribose 1-diphosphate (1.6-fold). Increases in hepatic ribulose 5-phosphate (Ru5P; 1.5-fold), and ribose 5-phosphate (R5P; 2-fold) are consistent with increased PPP flux. In mammalian cells, oxidative stress is reported to induce glucose-6-phosphate dehydrogenase (G6pdh) activity so that this enzyme is not rate limiting (Cosentino et al., 2011; Wang et al., 2014). Phosphoglucomutase (Pgm1), involved in the catalysis of R5P to ribose 1-phosphate (R1P) was also induced 1.6-fold further supported suggestions of increased flux through the PPP that would produce NADPH.
FIG. 6.
Metabolic reprogramming for NADPH production. Integration of transcriptomic, hepatic and serum metabolomic, and KEGG pathway data for (A) the PPP, and (B) serine and folate metabolism. The color scale represents the log2(fold-change) for genes and metabolites. Yellow identifies converging metabolites and gray indicates metabolites not measured or detected. Genes are identified as rectangles and metabolites as circles. The upper left corner provides the maximum AhR enrichment fold-change and upper right corner indicates the highest pDRE MSS. Circles contain the P-value for detected metabolites. An expandable and interactive version that provides additional complementary information can be viewed at http://dbzach.fst.msu.edu/index.php/supplementarydata.html. C, Expression of hepatic Phgdh was confirmed by QRTPCR. Bars represent mean + SEM (n = 5). Asterisks (*) indicates P ≤ .05 compared with vehicle determined by 1-way ANOVA followed by Dunnett’s post hoc test.
Recently, folate-dependent reactions have been estimated to account for 10–40% of NADPH production, compared with approximately 30% from the PPP (Fan et al., 2014). PKM2 reduced glycolytic flux is consistent with the shunting of 3-phosphoglycerate (3PG) towards Ser biosynthesis and folate-dependent NADPH production (Figure 6B). 3PG is transformed to 3-phosphohydroxypyruvate by phosphoglycerate dehydrogenase (Phgdh, 5.8-fold) and subsequently to Ser by phosphoserine aminotransferase (Psat1, 3-fold), which simultaneously converts Glu to αKG, another potential source of OAA accumulation (Figure 3C). Ser and tetrahydrofolate (THF) can then be converted to Gly and 5,10-methylene-tetrahydrofolate (5,10-CH2-THF) by cytosolic and mitochondrial serine hydroxymethyl transferase 1 and 2 (Shmt1/2). TCDD did not affect Shmt2 expression but repressed Shmt1 2.3-fold. 5,10-CH2-THF is then used by methylenetetrahydrofolate dehydrogenase (Mthfd1/2) to produce NADPH. Mitochondrial Mthfd2 was induced 2.4-fold while cytosolic Mthfd1 was modestly repressed suggesting greater NADPH production to support mitochondrial oxidative defenses. Besides de novo synthesis, Ser is available from protein and Ser-containing phospholipid catabolism, as well as transporter-mediated uptake. The 4.2-fold increase in serum Ser levels may be made available via the 3.4 - and 5.1-fold induction of Slc1a4 and 1a5, respectively, 2 high affinity transporters. In addition to producing NADPH, this pathway also provides Gly, another precursor required for GSH biosynthesis, thus serving dual antioxidant roles.
Cell Cycle Arrest by TCDD
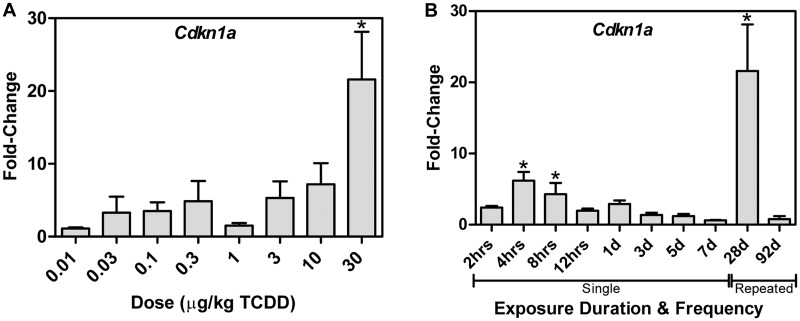
PKM2 lies at the intersection of cell proliferation and cell survival (Cairns et al., 2011; Gorrini et al., 2013; Harris et al., 2012). The AhR has also been linked to anti-oxidant defenses and cell cycle regulation (Jackson et al., 2014; Puga et al., 2002). Jackson et al. (2014) report that p21 gene expression regulation (Cdkn1a) is critical to TCDD-induced G1 cell cycle arrest. Similarly, we confirm Cdkn1a was induced 7.6-fold at 28 d with 3 AhR enriched regions, but was repressed 1.2-fold following treatment with TCDD every 4 days for 92 days (Figure 7), while Pkm2 was induced 13.8-fold. Other cell cycle regulators also demonstrated altered expression by TCDD, although their role in TCDD-induced in cell cycle arrest is not as well understood including the 8.6-fold induction of Fhit, the 1.7-fold repression of Gadd45α and the 3.1-fold induction of Gadd45β.
FIG. 7.
Dose- and time-dependent Cdkn1a expression. A, Dose-dependent hepatic Cdkn1a mRNA expression following gavage every 4 days for 28 days, and (B) time-dependent expression following a single bolus dose of 30 µg/kg TCDD. Bars represent mean + SEM (n = 4–5). Asterisks (*) indicate P ≤ 0.05 compared with vehicle determined by 1-way ANOVA followed by Dunnett’s post hoc test or 2-way ANOVA following by Tukey’s post hoc test.
DISCUSSION
Results from previous TCDD ‘omic’ studies in various in vivo and in vitro models have used different designs, doses, treatment protocols, and models that limit extrapolations between species, and confound integration and interpretation. Consequently, we examined the metabolomic and transcriptomic effects of TCDD in the same animals, and used KEGG pathways to integrate and manually curate the data while incorporating complementary pDRE and AhR-binding (ChIP-Seq) data to further investigate metabolic reprogramming in the mouse liver. Oral treatment every 4 days for 28 days simulated continuous exposure that increased the severity of the hepatic effects but was well tolerated (Nault et al., 2015a). Our results demonstrate that TCDD elicited a dose-dependent reorganization of hepatic metabolism characterized by the reprogramming of central carbon and amino acid metabolism, reminiscent of the Warburg effect, which supported NADPH demand created by increased anti-oxidant defense activities.
TCDD induced hepatic steatosis by altering gene expression consistent with lipid accumulation. This also included the inhibition of de novo fatty acid synthesis and β-oxidation (Boverhof et al., 2005; Fernandez-Salguero et al., 1996; Lee et al., 2010; Sato et al., 2008; Tanos et al., 2012a). Although other studies have suggested that AhR-mediated inhibition of β-oxidation may involve suppression of PPARα activity (Shaban et al., 2004; Wang et al., 2011a), the presence of AhR enrichment within the loci of several de novo fatty acid synthesis and β-oxidation genes suggest AhR regulation involving tethering mechanism (Beischlag et al., 2008; Dere et al., 2011b). Hepatic fat accumulation and the induction of xenobiotic (eg, cytochrome P450 activity) and purine metabolism (ie, Xdh/XO) would not only increase ROS levels but also lipid peroxidation (Bansal et al., 2014; Sugihara et al., 2001), creating a sustained oxidative burden requiring counter-measures to minimize cell damage. In response, AhR and Nrf2 cooperate to induce antioxidant counter-measures (Lu et al., 2011; Wang et al., 2011b; Yeager et al., 2009). Furthermore, redirection of glycolytic flux and the inhibition of β-oxidation would also compromise NADH and ATP production further compounding cell damage (Lu et al., 2011).
Notably, TCDD repressed Pklr expression while inducing Pkm2, the isoform typically associated with the Warburg effect and cell proliferation (Lunt et al., 2015). This is the first report of PKM isoform switching elicited by an exogenous chemical in normal differentiated tissue, although switching has been shown in PTEN-null mice fatty liver tissue (Panasyuk et al., 2012). PKM2 sits at the intersection of cell growth and cell survival (Cairns et al., 2011; Gorrini et al., 2013; Harris et al., 2012), and its induction with subsequent metabolic changes highlights PKM2’s putative role in TCDD toxicity. Although we did not determine whether Pkm2 was present in monomeric, dimeric, and tetrameric forms, under oxidative stress conditions such as those elicited by TCDD, PKM2 is oxidized at Cys358 which further reduces its activity resulting in the accumulation of upstream intermediates and shunting towards NADPH producing pathways (Anastasiou et al., 2011). Interestingly, AhR enrichment has been repeatedly demonstrated within the Pkm loci indicating direct regulation. Furthermore, PCB126, TCDF, and βNF also induced Pkm2 while PCB153, which does not bind to the AhR, had no effect on expression. Collectively, these results indicate AhR-mediated regulation of PKM2 expression is a sustained response as long as TCDD exposure is maintained.
In cancer cells, PKM2 is tightly regulated, both transcriptionally and post-translationally (Cairns et al., 2011; Gorrini et al., 2013; Harris et al., 2012). In response to TCDD, our data are consistent with the shunting of accumulating upstream intermediates towards anti-oxidant pathways as opposed to cell proliferation, especially given the induction of the cell cycle inhibitor p21 (Jackson et al., 2014). More specifically, G6P and 3PG are redirected to the PPP and Ser/folate biosynthesis, respectively, to satisfy increasing NADPH demand in order to maintain redox balance that include GSH biosynthesis and GSSG reduction. Similar metabolic reprogramming is reported in cancer cells (Amelio et al., 2014; Cairns et al., 2011; Gorrini et al., 2013; Wise et al., 2008), lipopolysaccharide-activated macrophages (Palsson-McDermott et al., 2015), and normal cells experiencing oxidative stress (Kuehne et al., 2015).
Metabolic reprogramming, including increased intermediate transport, also explains increased hepatic Gly, Cys, OAA and Asp levels. 3PG accumulation due to PKM2 expression is shunted to Ser biosynthesis indicated by Phgdh and Psat1 induction. Ser is then converted to Gly to produce 5,10-CH2-THF which is oxidized to produce NADPH. Ser/folate metabolism is estimated to account for 10–40% of NADPH production, compared with approximately 30% from the PPP (Fan et al., 2014). Moreover, Ser biosynthesis involves the conversion of Glu to αKG, another potential source of OAA, in addition to glutaminolysis and the malate-aspartate shuttle. Decreases in acetyl-CoA, and the inhibition of β-oxidation and gluconeogenesis further contribute to OAA accumulation. Pck1 repression also inhibits OAA utilization in gluconeogenesis and metabolism to lactate. Increased OAA levels likely drive Asp biosynthesis despite Got repression. Nevertheless, more precise tracer studies are required to further characterize TCDD-elicited metabolic reprogramming since metabolic regulation primarily occurs via post-translational mechanisms (Stincone et al., 2014).
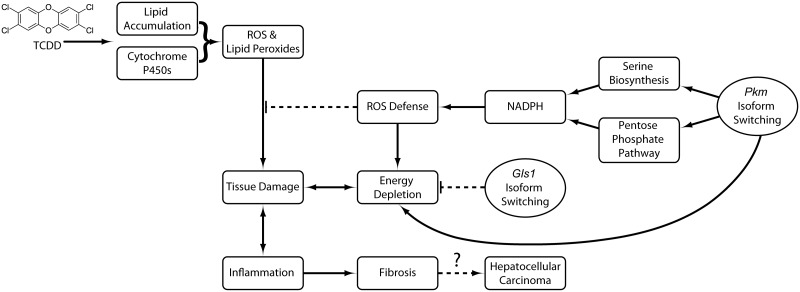
In summary, compromised energy production, increasing lipid peroxidation, and the resulting oxidative environment cause cell damage that triggers not only immune cell infiltration but also the reprogramming of central carbon and amino acid metabolism towards defensive responses (Figure 8). Induction of p21 and its known role in TCDD mediated cell cycle arrest (Jackson et al., 2014) further supports cell survival by ensuring accumulating intermediates are shunted towards oxidative stress counter-measures and cell survival, as opposed to biomass production and cell proliferation. Resistance to cell death, energy metabolism reprogramming, and the initiation of inflammation are important hallmarks of cancer (Hanahan and Weinberg, 2011). TCDD is a known carcinogen in rodents, but its human carcinogenicity is debated (Boffetta et al., 2011). The continued induction of PKM2, and return to basal Cdkn1a expression at 92 days suggests possible support of proliferation of surviving cells with accumulated DNA damage that have evaded repair, leading to tumor development. This balance between survival and proliferation regulated by metabolic reprogramming could also have roles in AhR-mediated species- and sex-specific sensitivity and ligand toxicity differences. Consequently, regulating metabolic reprogramming in normal tissue, reminiscent of the Warburg effect, may be an advantageous defensive strategy used by normal cells to promote survival in response to other toxicities that is later co-opted by transformed cells to support proliferation and tumor development.
FIG. 8.
Proposed consequences of TCDD-elicited metabolic reprogramming in the mouse liver. TCDD induces pro-oxidant systems such as xenobiotic and purine metabolism with the accumulation of hepatic lipids that generate ROS and lipid peroxides. Pkm isoform switching redirects accumulating glycolytic intermediates to NADPH producing pathways to support cell defenses, leading to energy depletion. Compensatory Gls1 isoform switching and glutaminolysis provide intermediates for the TCA cycle in order to maintain oxidative phosphorylation. Together, this advantageous metabolic reorganization protects the liver from further ROS damage. However, with higher doses or prolonged exposure, the system is overwhelmed, and steatosis progresses to steatohepatitis with fibrosis, and potentially hepatocellular carcinoma.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Ed Dere for his help with computational pDRE identification, and Dr. John Froehlich and Brendan Johnson for ProteinSimple Wes assay assistance.
FUNDING
This work was supported by the National Institute of Environmental Health Sciences Superfund Research Program (NIEHS SRP P42ES04911). T.R.Z. is supported by AgBioResearch at MSU. RN is supported by the MSU Barnett Rosenberg Endowed Assistantship and Integrative Training in the Pharmacological Sciences grant (the National Institutes of Health 5T32GM092715). K.A.F. is supported by the Canadian Institutes of Health Research (CIHR) Doctoral Foreign Study Award (DFS-140386). S.Y.L. is supported by the Department of Defense (W81XWH-12-1-0466). JM is supported by a CIHR operating grant (MOP-125919), an unrestricted research grant from DOW Chemical, and an Early Researcher Award from the Ontario Ministry of Innovation.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Ahmed S., Bott D., Gomez A., Tamblyn L., Rasheed A., Cho T., MacPherson L., Sugamori K. S., Yang Y., Grant D. M., et al. (2015). Loss of the Mono-ADP-ribosyltransferase, tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J. Biol. Chem. 290, 16824–16840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I., Cutruzzola F., Antonov A., Agostini M., Melino G. (2014). Serine and glycine metabolism in cancer. Trends Biochem. Sci. 39, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J. K., Shen M., Bellinger G., Sasaki A. T., Locasale J. W., Auld D. S., et al. (2011). Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrish M. M., Dominici C. Y., Zacharewski T. R. (2013). TCDD-elicited effects on liver, serum, and adipose lipid composition in C57BL/6 mice. Toxicol. Sci. 131, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S., Leu A. N., Gonzalez F. J., Guengerich F. P., Chowdhury A. R., Anandatheerthavarada H. K., Avadhani N. G. (2014). Mitochondrial targeting of cytochrome P450 (CYP) 1B1 and its role in polycyclic aromatic hydrocarbon-induced mitochondrial dysfunction. J. Biol. Chem. 289, 9936–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag T. V., Luis Morales J., Hollingshead B. D., Perdew G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18, 207–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffetta P., Mundt K. A., Adami H. O., Cole P., Mandel J. S. (2011). TCDD and cancer: A critical review of epidemiologic studies. Crit. Rev. Toxicol. 41, 622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof D. R., Burgoon L. D., Tashiro C., Chittim B., Harkema J. R., Jump D. B., Zacharewski T. R. (2005). Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol. Sci. 85, 1048–1063. [DOI] [PubMed] [Google Scholar]
- Cairns R. A., Harris I. S., Mak T. W. (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Chesney J. (2006). 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase and tumor cell glycolysis. Curr. Opin. Clin. Nutr. Metab. Care 9, 535–539. [DOI] [PubMed] [Google Scholar]
- Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008). The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233. [DOI] [PubMed] [Google Scholar]
- Clasquin M. F., Melamud E., Rabinowitz J. D. (2012). LC-MS data processing with MAVEN: A metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics Chapter 14, Unit14 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M., Sato H. (2012). The oxidative stress-inducible cystine/glutamate antiporter, system x (c) (-) : Cystine supplier and beyond. Amino acids 42, 231–246. [DOI] [PubMed] [Google Scholar]
- Cosentino C., Grieco D., Costanzo V. (2011). ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. Embo. J. 30, 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C. B. (2007). Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U. S. A. 104, 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Soshilov A. A., He G., DeGroot D. E., Zhao B. (2011). Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 124, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Forgacs A. L., Zacharewski T. R., Burgoon L. D. (2011a). Genome-wide computational analysis of dioxin response element location and distribution in the human, mouse, and rat genomes. Chem. Res. Toxicol. 24, 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Lo R., Celius T., Matthews J., Zacharewski T. R. (2011b). Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics 12, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diani-Moore S., Ram P., Li X., Mondal P., Youn D. Y., Sauve A. A., Rifkind A. B. (2010). Identification of the aryl hydrocarbon receptor target gene TiPARP as a mediator of suppression of hepatic gluconeogenesis by 2,3,7,8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J. Biol. Chem. 285, 38801–38810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Ye J., Kamphorst J. J., Shlomi T., Thompson C. B., Rabinowitz J. D. (2014). Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., Gonzalez F. J. (1996). Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 140, 173–179. [DOI] [PubMed] [Google Scholar]
- Forgacs A. L., Kent M. N., Makley M. K., Mets B., DelRaso N., Jahns G. L., Burgoon L. D., Zacharewski T. R., Reo N. V. (2012). Comparative metabolomic and genomic analyses of TCDD-elicited metabolic disruption in mouse and rat liver. Toxicol. Sci. 125, 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C., Harris I. S., Mak T. W. (2013). Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12, 931–947. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: The next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Harris I., McCracken S., Mak T. W. (2012). PKM2: A gatekeeper between growth and survival. Cell Res. 22, 447–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Elferink C. J. (2012). A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol. Pharmacol. 81, 338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. P., Li H., Mitchell K. A., Joshi A. D., Elferink C. J. (2014). Ah receptor-mediated suppression of liver regeneration through NC-XRE-driven p21Cip1 expression. Mol. Pharmacol. 85, 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Jiang H., Ma W., Wong W. H. (2008). Using cisgenome to analyze ChIP-chip and ChIP-seq data. In Curr Protoc Bioinformatics. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- Jiang P., Du W., Wu M. (2014). Regulation of the pentose phosphate pathway in cancer. Protein Cell 5, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008). ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec A. K., Burgoon L. D., Ibrahim-Aibo D., Burg A. R., Lee A. W., Tashiro C., Potter D., Sharratt B., Harkema J. R., Rowlands J. C., et al. (2010a). Automated dose-response analysis and comparative toxicogenomic evaluation of the hepatic effects elicited by TCDD, TCDF, and PCB126 in C57BL/6 mice. Toxicol. Sci. 118, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec A. K., Burgoon L. D., Ibrahim-Aibo D., Mets B. D., Tashiro C., Potter D., Sharratt B., Harkema J. R., Zacharewski T. R. (2010b). PCB153-elicited hepatic responses in the immature, ovariectomized C57BL/6 mice: comparative toxicogenomic effects of dioxin and non-dioxin-like ligands. Toxicol. Appl. Pharmacol. 243, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne A., Emmert H., Soehle J., Winnefeld M., Fischer F., Wenck H., Gallinat S., Terstegen L., Lucius R., Hildebrand J., et al. (2015). Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell 59, 359–371. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Wada T., Febbraio M., He J., Matsubara T., Lee M. J., Gonzalez F. J., Xie W. (2010). A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology 139, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Yang Z., Liu H., Cai Z. (2011). Metabolomic analysis of liver and skeletal muscle tissues in C57BL/6J and DBA/2J mice exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol. Biosyst. 7, 1956–1965. [DOI] [PubMed] [Google Scholar]
- Lo R., Matthews J. (2012). High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol. Sci. 130, 349–361. [DOI] [PubMed] [Google Scholar]
- Locasale J. W. (2013). Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Cui W., Klaassen C. D. (2011). Nrf2 protects against 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced oxidative injury and steatohepatitis. Toxicol. Appl. Pharmacol. 256, 122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt S. Y., Muralidhar V., Hosios A. M., Israelsen W. J., Gui D. Y., Newhouse L., Ogrodzinski M., Hecht V., Xu K., Acevedo P. N., et al. (2015). Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell 57, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O. D., Berkers C. R., Mason S. M., Zheng L., Blyth K., Gottlieb E., Vousden K. H. (2013). Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T., Tanaka N., Krausz K. W., Manna S. K., Kang D. W., Anderson E. R., Luecke H., Patterson A. D., Shah Y. M., Gonzalez F. J. (2012). Metabolomics identifies an inflammatory cascade involved in dioxin- and diet-induced steatohepatitis. Cell Metab. 16, 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E., Vastag L., Rabinowitz J. D. (2010). Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault R., Colbry D., Brandenberger C., Harkema J. R., Zacharewski T. R. (2015a). Development of a computational high-throughput tool for the quantitative examination of dose-dependent histological features. Toxicol. Pathol. 43, 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault R., Fader K. A., Zacharewski T. (2015b). RNA-Seq versus oligonucleotide array assessment of dose-dependent TCDD-elicited hepatic gene expression in mice. BMC Genomics 16, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott E. M., Curtis A. M., Goel G., Lauterbach M. A., Sheedy F. J., Gleeson L. E., van den Bosch M. W., Quinn S. R., Domingo-Fernandez R., Johnston D. G., et al. (2015). Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasyuk G., Espeillac C., Chauvin C., Pradelli L. A., Horie Y., Suzuki A., Annicotte J. S., Fajas L., Foretz M., Verdeguer F., et al. (2012). PPARgamma contributes to PKM2 and HK2 expression in fatty liver. Nat. Commun. 3, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre S., Chevallier A., Teixeira-Clerc F., Ambolet-Camoit A., Bui L. C., Bats A. S., Fournet J. C., Fernandez-Salguero P., Aggerbeck M., Lotersztajn S., et al. (2014). Aryl hydrocarbon receptor-dependent induction of liver fibrosis by dioxin. Toxicol. Sci. 137, 114–124. [DOI] [PubMed] [Google Scholar]
- Pohjanvirta R., Tuomisto J. (1994). Short-term toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in laboratory animals: effects, mechanisms, and animal models. Pharmacol. Rev. 46, 483–549. [PubMed] [Google Scholar]
- Puga A., Xia Y., Elferink C. (2002). Role of the aryl hydrocarbon receptor in cell cycle regulation. Chem. Biol. Interact. 141, 117–130. [DOI] [PubMed] [Google Scholar]
- Sato S., Shirakawa H., Tomita S., Ohsaki Y., Haketa K., Tooi O., Santo N., Tohkin M., Furukawa Y., Gonzalez F. J., et al. (2008). Low-dose dioxins alter gene expression related to cholesterol biosynthesis, lipogenesis, and glucose metabolism through the aryl hydrocarbon receptor-mediated pathway in mouse liver. Toxicol. Appl. Pharmacol. 229, 10–19. [DOI] [PubMed] [Google Scholar]
- Shaban Z., El-Shazly S., Abdelhady S., Fattouh I., Muzandu K., Ishizuka M., Kimura K., Kazusaka A., Fujita S. (2004). Down regulation of hepatic PPARalpha function by AhR ligand. J. Vet. Med. Sci. 66, 1377–1386. [DOI] [PubMed] [Google Scholar]
- Son J., Lyssiotis C. A., Ying H., Wang X., Hua S., Ligorio M., Perera R. M., Ferrone C. R., Mullarky E., Shyh-Chang N., et al. (2013). Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stincone A., Prigione A., Cramer T., Wamelink M. M., Campbell K., Cheung E., Olin-Sandoval V., Gruning N., Kruger A., Tauqeer Alam M., et al. (2014). The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 90, 927–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K., Kitamura S., Yamada T., Ohta S., Yamashita K., Yasuda M., Fujii-Kuriyama Y. (2001). Aryl hydrocarbon receptor (AhR)-mediated induction of xanthine oxidase/xanthine dehydrogenase activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 281, 1093–1099. [DOI] [PubMed] [Google Scholar]
- Tanos R., Murray I. A., Smith P. B., Patterson A., Perdew G. H. (2012a). Role of the Ah receptor in homeostatic control of fatty acid synthesis in the liver. Toxicol. Sci. 129, 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos R., Patel R. D., Murray I. A., Smith P. B., Patterson A. D., Perdew G. H. (2012b). Aryl hydrocarbon receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology 55, 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. W., Novak R. F., Anderson H. A., Birnbaum L. S., Blystone C., Devito M., Jacobs D., Kohrle J., Lee D. H., Rylander L., et al. (2013). Evaluation of the association between persistent organic pollutants (POPs) and diabetes in epidemiological studies: A national toxicology program workshop review. Environ. Health Perspect. 121, 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel A. P., Jing J., Wooster R. F., Bachman K. E. (2012). Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol. Ther. 13, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Xu C. X., Krager S. L., Bottum K. M., Liao D. F., Tischkau S. A. (2011a). Aryl hydrocarbon receptor deficiency enhances insulin sensitivity and reduces PPAR-alpha pathway activity in mice. Environ. Health Perspect. 119, 1739–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. P., He X. Q., Bi Y. Y., Szklarz G., Ma Q. (2011b). Ah receptor interacts with Nrf2 to mediate induction of NQO1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin and benzo[a]pyrene. Faseb J. 25,1014.3. [Google Scholar]
- Wang Y. P., Zhou L. S., Zhao Y. Z., Wang S. W., Chen L. L., Liu L. X., Ling Z. Q., Hu F. J., Sun Y. P., Zhang J. Y., et al. (2014). Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. Embo. J. 33, 1304–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L. W., Lebofsky M., Stahl B. U., Smith S., Rozman K. K. (1995). Correlation between toxicity and effects on intermediary metabolism in 2,3,7,8-tetrachlorodibenzo-p-dioxin-treated male C57BL/6J and DBA/2J mice. Toxicol. Appl. Pharmacol. 131, 155–162. [DOI] [PubMed] [Google Scholar]
- Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., et al. (2008). Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U. S. A. 105, 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Wishart D. S. (2010). MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26, 2342–2344. [DOI] [PubMed] [Google Scholar]
- Yeager R. L., Reisman S. A., Aleksunes L. M., Klaassen C. D. (2009). Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol. Sci. 111, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Ren W., Yang G., Duan J., Huang X., Fang R., Li C., Li T., Yin Y., Hou Y., et al. (2015). l-Cysteine metabolism and its nutritional implications. Mol. Nutr. Food Res. 60, 134–146. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L. G., Han Y., He Q. Y. (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.