Abstract
A bioinformatics strategy was used to identify Scabin, a novel DNA-targeting enzyme from the plant pathogen 87.22 strain of Streptomyces scabies. Scabin shares nearly 40% sequence identity with the Pierisin family of mono-ADP-ribosyltransferase toxins. Scabin was purified to homogeneity as a 22-kDa single-domain enzyme and was shown to possess high NAD+-glycohydrolase (Km(NAD) = 68 ± 3 μm; kcat = 94 ± 2 min−1) activity with an RSQXE motif; it was also shown to target deoxyguanosine and showed sigmoidal enzyme kinetics (K0.5(deoxyguanosine) = 302 ± 12 μm; kcat = 14 min−1). Mass spectrometry analysis revealed that Scabin labels the exocyclic amino group on guanine bases in either single-stranded or double-stranded DNA. Several small molecule inhibitors were identified, and the most potent compounds were found to inhibit the enzyme activity with Ki values ranging from 3 to 24 μm. PJ34, a well known inhibitor of poly-ADP-ribosyltransferases, was shown to be the most potent inhibitor of Scabin. Scabin was crystallized, representing the first structure of a DNA-targeting mono-ADP-ribosyltransferase enzyme; the structures of the apo-form (1.45 Å) and with two inhibitors (P6-E, 1.4 Å; PJ34, 1.6 Å) were solved. These x-ray structures are also the first high resolution structures of the Pierisin subgroup of the mono-ADP-ribosyltransferase toxin family. A model of Scabin with its DNA substrate is also proposed.
Keywords: bacterial toxin, bioinformatics, crystallography, DNA binding protein, enzyme kinetics, fluorescence, molecular modeling
Introduction
Mono-ADP-ribosyltransferase (mART)3 toxins are a group of enzymes that catalyze the transfer of an ADP-ribose group from NAD+ to a target macromolecule within a host cell, which inactivates the target's function and leads to mutation or host cell apoptosis (1). Most mART toxins have NAD+-glycohydrolase (GH) activity in addition to transferase activity (2). GH activity occurs when the mART toxin has no target for transferase activity and thus utilizes a water molecule to accept the ADP-ribose moiety in a nucleophilic substitution reaction (2). Examples of mART toxins include ExoA from Pseudomonas aeruginosa, pertussis toxin from Bordetella pertussis, C3larvin from Paenibacillus larvae, and Pierisin-1 from Pieris rapae (3–5). Quite often, mART toxins target protein residues, such as Arg, Asn, and Cys (6). More specifically, mART toxins will label residues within target proteins that are often fundamental regulators of a cellular function (6). Target proteins can include RhoA, actin, elongation factor-2, and Ras. In rare cases, mART toxins may target DNA as the macromolecule substrate as in the case of Pierisin-1 (4). Those mART toxins that utilize DNA as a target macromolecule label the guanine base with an ADP-ribose moiety (7, 8).
Streptomyces scabies is a soil-dwelling, filamentous, Gram-positive bacterium that is known to cause the common scab disease in potatoes and other root and tuberous vegetables (9). The disease is characterized by deep-pitted and corky lesions found on the skin of the potato (9). Many varieties of potatoes are affected by the common scab disease, including the Yukon Gold potato (10). Notably, the mechanism used by S. scabies to infect potatoes is poorly understood. However, in silico analysis has revealed a putative mART toxin, herein named Scabin, identified within the genome of S. scabies strain 87.22.
Scabin is a 200-residue, 22-kDa, single-domain enzyme possessing a 29-residue N-terminal secretion signal peptide. Scabin was cloned, purified, and shown to possess both GH and ADP-ribosyltransferase activities. Five compounds were identified as good lead inhibitors against Scabin GH activity. The crystal structure of the apoenzyme has been resolved to 1.4 Å, and we have determined the structure of Scabin with two small molecule inhibitors bound to the active site. It also has sequence similarity to the Pierisin subgroup of mART toxins, as shown by multiple-sequence alignment (4, 7, 8). Using this information, we identified the Scabin transferase substrate as DNA and other small nucleotides possessing guanine (7). We have characterized the Michaelis-Menten kinetic parameters of Scabin for both β-NAD+ and deoxyguanosine substrates. Also, a catalytically less active variant of Scabin has been characterized. To our knowledge, this study presents the first reported inhibitors as well as the first crystal structure for a DNA-targeting enzyme within the mART toxin family.
Experimental Procedures
Unless otherwise noted, chemicals were purchased from Sigma-Aldrich.
Scabin Expression and Purification
The Scabin gene with a 29-residue N-terminal truncation (signal peptide removed) was overexpressed in Escherichia coli BL21, λDE3 cells and purified from the soluble fraction of the cell lysate. In brief, Scabin or a Q158A/E160A variant was cloned into a pET-TEV vector with an N-terminal His6 tag and tobacco etch virus protease cut site. Chemically competent E. coli BL21, λDE3 cells were transformed with plasmid and grown at 37 °C with shaking in 6 liters of 2× YT medium to an OD of 0.9 in the presence of kanamycin. Cells were subsequently induced with 1 mm isopropyl β-d-1-thiogalactopyranoside for 16 h at 16 °C. Cells were harvested by centrifugation at 4000 × g for 12 min. Pelleted cells were resuspended in lysis buffer containing 25 mm Tris-HCl, pH 8.2, 200 mm NaCl, 50 μg/ml CHAPS, 120 μm phenylmethylsulfonyl fluoride, 1 mm EDTA, and 100 μg/ml DNase. Cell lysis was performed using an Emulsiflex-C3 high pressure homogenizer (Avestin Inc., Ottawa, Canada). Lysate was subsequently centrifuged at 14,000 × g for 50 min. Supernatant was collected and incubated with 20 mm MgCl2 at 4 °C with stirring for 30 min.
Scabin and Q158A/E160A were purified by immobilized metal affinity chromatography by passing the supernatant over a HiTrap chelating Sepharose fast-flow column (GE Healthcare, Mississauga, Canada) charged with Ni2+ and equilibrated with Buffer A containing 50 mm TAPS, pH 8.5, 500 mm NaCl, and 5 mm imidazole. The column containing bound protein was subjected to a wash step with Buffer A containing 25 mm imidazole, and subsequently the column was developed with a linear gradient of imidazole from 25 mm to a final concentration of 250 mm. Fractions were analyzed by SDS-PAGE, and those found to contain the protein of interest were pooled and dialyzed overnight into 25 mm Tris-HCl, pH 8.2, and 50 mm NaCl.
Further purification was performed using a HiTrap Q-Sepharose HP column (GE Healthcare) equilibrated with dialysis buffer. The sample was passed over the column and washed with a linear NaCl gradient (50–500 mm) of dialysis buffer. Fractions containing pure protein were pooled and concentrated initially in a bed of PEG 20,000 at 4 °C until a volume of 5 ml was reached. The sample was then concentrated using Millipore 0.5-ml 10-kDa spin columns at 4250 rpm for 10-min intervals.
Mass Spectrometry of Scabin
The molecular weight of the recombinant Δ29 Scabin protein (herein called Scabin) was determined using an Agilent UHD 6530 Q-TOF mass spectrometer at the Mass Spectrometry Facility of the Advanced Analysis Centre, University of Guelph. The instrument was configured with the standard electrospray ionization source and operated in positive ion mode. Data analysis was performed using MassHunter Qualitative Analysis version B.06.00 (Agilent) software. Deconvolution of the m/z spectrum was achieved using the maximum entropy algorithm within the BioConfirm software (Agilent).
Circular Dichroism Spectra
CD spectra were acquired for both Scabin and the Q158A/E160A variant using a JASCO J-815 CD spectropolarimeter (250–190-nm scan, average of nine spectra). The protein was at 0.16 mg/ml in a buffer containing 20 mm Tris-HCl, pH 8.2, and 50 mm NaF in a 1-mm path length quartz CD cuvette.
Differential Scanning Fluorimetry
Differential scanning fluorimetry was performed on Scabin and the Q158A/E160A variant using an Applied Biosystems OneStepPlus Real-Time PCR light cycler. Samples were run in triplicate containing 14 μm protein in 25 mm Tris-HCl, pH 8.2, and 200 mm NaCl with the addition of 1× SYPRO® Orange stain. For Scabin, samples were incubated with and without 500 μm inhibitor for 30 min before the addition of 1× SYPRO Orange stain. Samples were subjected to a temperature gradient from 4 to 80 °C (scan rate 1.2 °C/min), and SYPRO fluorescence was monitored using the SYBR Green protocol in the StepOnePlusTM software.
Protein Crystallography
Crystal conditions for Scabin were screened using the PEG II suite (Qiagen) in 96-well screening trays using 1.5 mg/ml Scabin. Crystal hits were observed after 24 h in a condition containing 0.1 m MES, pH 6.5, and 15% PEG 400.
The crystal condition was scaled up using 18-mm hanging drop trays with 200 μl of well solution. One microliter of 1.5 mg/ml Scabin was mixed with 1 μl of well solution on a glass coverslide to form the crystal drop. Inhibitors were co-crystallized by preincubating Scabin with a 500 μm concentration of either PJ34 or P6-E for 30 min. After incubation at room temperature for 24–48 h, large single crystals appeared. Crystals were then routinely washed with mother liquor containing 30% PEG 400 to act as a cryoprotectant and subsequently were flash-frozen in liquid nitrogen. In the case of co-crystals containing inhibitor, cryoprotectant also contained 500 μm inhibitor. X-ray diffraction data were collected at the Canadian Light Source in the Canadian Macromolecular Crystallography Facility (beamline 08ID-1).
Scabin-apo Structure
A total of 266 frames of diffraction images were collected with a 0.75° oscillation range. Images were processed and scaled with XDS in the C2 space group (11). The Scabin-apo structure was solved using molecular replacement in MOLREP (12). For MOLREP, the template was obtained by generating a model of Scabin-apo from a homologous Bacillus sphaericus mosquitocidal toxin structure (Protein Data Bank code 2CB4) (13) using MODELLER (14). The model was further optimized by truncating loops. The molecular replacement solution was subjected to iterative model building and refinement using COOT (15) and Refmac5 in the CCP4 Suite (16). The final model was refined to a resolution of 1.4 Å with Rwork/Rfree (%) = 16.3/19.7.
Scabin PJ34 and Scabin P6-E Structures
The data collected were processed in XDS (11). Molecular replacement on the crystal data set was conducted using Phenix (17) with the Scabin-apo structure as the model. Iterative cycles of model building in COOT (15) and refinement in Phenix were performed. The Scabin-apo, Scabin PJ34, and Scabin P6-E structures have been deposited in the Protein Data Bank database, with codes 5DAZ, 5EWK, and 5EWY, respectively. Table 1 shows the refinement statistics for these three structures.
TABLE 1.
Crystallographic data and refinement statistics for Scabin structures from S. scabies
| Diffraction data and refinement statistics | Scabin-apo | Scabin-PJ34 | Scabin-P6-E |
|---|---|---|---|
| X-ray source | CLSI-08-ID-1 | CLSI-08-ID-1 | CLSI-08-ID-1 |
| Wavelength (Å) | 0.97949 | 0.97949 | 0.97949 |
| Unit cell parameters (Å) | a = 87.7, b = 61.0, c = 37.8, α = 90.0, β = 99.64, γ = 90.0 | a = 87.47, b = 61.18, c = 38.03, α = 90.0, β = 100.379, γ = 90.0 | a = 87.17, b = 60.53, c = 37.94, α = 90.0, β = 100.25, γ = 90.0 |
| Space group | C2 | C2 | C2 |
| Resolution range (Å)a | 50.0–1.45 (1.48–1.45) | 43.02–1.6 (1.667–1.6) | 42.89–1.40 (1.48–1.45) |
| Data completeness (%) | 94.4 | 99.7 | 99.5 |
| Rsym (%) | 4.8 (87.6) | 3.5 (60.27) | 3.5 (86.32) |
| Redundancy | 4.3 (4.1) | 3.8 (3.7) | 3.7 (3.5) |
| Average I/σ(I) | 18.81 (2.15) | 23.33 (2.56) | 20.44 (1.69) |
| Molecular replacement program | MolRep | Phenix | Phenix |
| Rwork (%)b | 16.3 | 16.6 | 16.3 |
| Rfree (%)c | 19.7 | 18.8 | 17.7 |
| No. of atoms in protein | 1319 | 1326 | 1326 |
| No. of waters | 183 | 143 | 195 |
| Root mean square deviation from ideal bond length (Å) | 0.024 | 0.009 | 0.008 |
| Root mean square deviation from ideal bond angles (degrees) | 2.21 | 1.05 | 1.16 |
| B-Factors (Å2) for Scabin | 20.8 | 34.76 | 30.83 |
| B-Factors (Å2) for water | 25.0 | 39.25 | 44.78 |
| Ramachandran plot favored (%) | 98.14 | 97.00 | 97.00 |
| Ramachandran plot outliers (%) | 1.86 | 0.00 | 0.61 |
a Values in parenthesis are for the highest resolution shell.
b |Σ‖Fo| − |Fc‖Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
c The Rfree value was calculated with a random 5% subset of all reflections excluded from refinement.
NAD+ Binding
The binding constant, Kd, for β-NAD+ and inhibitors was determined by measuring the quenching of intrinsic tryptophan fluorescence using a Cary Eclipse fluorescence spectrophotometer (Varian Instruments, Mississauga, Canada). An excitation wavelength of 295 nm, emission wavelength of 340 nm, and band passes of 5 nm were used. In quartz UV cuvettes (0.5 × 0.5 cm), 1.25 μm Scabin in 25 mm Tris-HCl, pH 8.2, and 200 mm NaCl was titrated with β-NAD+ (0–1000 μm) or inhibitor (0–1000 μm), and fluorescence intensity was monitored at 340 nm. Assays were performed in triplicate, and data were corrected to account for the increase in volume upon the addition of β-NAD+ or inhibitor. A blank titration with N-acetyl tryptophanamide was performed to correct for inner filter effects. Data were analyzed using OriginPro version 8 software (OriginLab, Northampton, MA) to determine the binding constants.
GH Activity
Glycohydrolase assays were performed on a Cary Eclipse fluorescence spectrophotometer using an excitation wavelength of 305 nm, emission wavelength of 405 nm, and band passes of 5 nm. Various concentrations of ϵ-NAD+ (0–450 μm) and 490 nm Scabin were added to NAD+ GH buffer containing 20 mm Tris-HCl, pH 7.9, and 50 mm NaCl. Triplicate reactions were monitored for 10-min intervals, and the initial slope of each reaction was recorded. An ϵ-AMP standard curve was derived to convert fluorescent units/min to [etheno-ADP-ribose] formed/min. A Michaelis-Menten curve was derived by fitting the data to the hyperbolic model in OriginPro version 8 software.
ADP-ribosyltransferase Reaction
ADP-ribosyltransferase assays with deoxyguanosine (dG) were performed as described under “GH Activity” with a few changes. ϵ-NAD+ was held at a concentration of 250 μm and was mixed with 10 nm Scabin in GH buffer containing 1% dimethyl sulfoxide and various concentrations of dG (0–1250 μm). The data were fit to the sigmoidal dose response model in OriginPro version 8.
Activity toward other small nucleotides was also tested using the method described above, with the exception of holding the small nucleotide substrate at a constant concentration of 1 mm. Substrates tested include GDP, cyclic guanosine monophosphate (cGMP), and deoxyinosine (dI).
Scabin activity toward genomic DNA was also tested using the method described above. The genomic DNA was extracted from various samples, as described previously (18), and was partially fragmented with 1 unit of DNase I/μg of DNA (375 ng/μl, final concentration) in Tris-EDTA buffer containing 5 mm MgCl2 for 10 min at room temperature.
Mass Spectrometry of mART Reaction Products
Liquid chromatography-mass spectrometry analyses were performed in the Mass Spectrometry Facility within the Advanced Analysis Centre at the University of Guelph. Reactions containing 1 mm nucleotide (GDP or cGMP) or 100 μm DNA (single-stranded pentamer of dG or double-stranded DNA with forward sequence 5′-ATTAAGTATT-3′ and reverse sequence 5′-TAATTCATAA-3′), 250 μm NAD+, and 50 nm Scabin in GH buffer were run overnight at room temperature with gentle shaking. Samples were diluted 1:100 in water before injection into a Dionex UHPLC UltiMate 3000 liquid chromatograph interfaced to an amaZon SL ion trap mass spectrometer (Bruker Daltonics, Billerica, MA). A Luna C18 column (5-μm particle size, 150 × 2 mm; Phenomenex) was used for chromatographic separation. The initial mobile phase conditions were 100% water (0.1% formic acid) isocratic for 5 min followed by a gradient to 60% acetonitrile (0.1% formic acid) in 20 min and then a single-step gradient to 100% acetonitrile in 10 min. The flow rate was maintained at 0.4 ml/min. The mass spectrometer electrospray capillary voltage was maintained at 4.5 kV, and the drying gas temperature was 280 °C with a flow rate of 10 liters/min. Nebulizer pressure was 40 p.s.i. Nitrogen was used as both nebulizing and drying gas, and helium was used as collision gas at 60 p.s.i. The mass spectrometer was set on enhanced resolution positive ion auto-MS/MS mode and scanned across 50–1500 m/z. The smart parameter setting was used to automatically optimize the trap drive level for selected precursor ions. The instrument was externally calibrated with the ESI TuneMix (Agilent).
Inhibitor Assays
Testing of potential inhibitors against Scabin GH activity was performed using compounds designed to act competitively with β-NAD+. P6 series inhibitors were obtained from Sinova Inc. (Bethesda, MD). Initial testing was performed on a Cary Eclipse fluorescence spectrophotometer using the same settings as described under “GH Activity.” Initial testing was performed in a reaction vessel containing 50 nm Scabin, 250 μm ϵ-NAD+, 100 μm inhibitor, and GH buffer. Because some of the inhibitors were dissolved in DMSO, a control (2% DMSO) was performed to take into account the effect of DMSO on enzyme activity. Triplicate reactions were monitored for 10-min intervals, and the initial slope of each reaction was normalized to take into account the effect of DMSO.
IC50 and Ki Determination
Inhibition assays were used to determine the IC50 values for selected inhibitors, and the experiments were performed on a Cary Eclipse fluorescence spectrophotometer with an excitation wavelength of 305 nm, emission wavelength of 405 nm, and band passes of 5 nm. Scabin (50 nm) was mixed with 250 μm ϵ-NAD+, various concentrations of inhibitors, and GH buffer. Triplicate reactions were monitored for 10-min intervals, and the initial slope of each reaction was normalized to a 2% DMSO control. An etheno-AMP standard curve was used to convert fluorescence units/min to [ϵ-ADP-ribose] formed/min. The data were fit using the dose-response function in OriginPro version 8 to determine the corresponding IC50 value. Experimental IC50 values were used to calculate Ki values using the Cheng-Prusoff equation (19), Ki = IC50/(1 + [S]/Km), where [S] is the concentration of ϵ-NAD+ and Km is for the β-NAD+ substrate. The Ki was determined experimentally for PJ34 from a Lineweaver-Burk plot of the inhibition data. This was developed by incubating 50 nm Scabin in GH buffer and varying the concentration of ϵ-NAD+ (12.5, 25, 50, and 100 μm) but holding the concentration of PJ34 at either 0, 6, 12, or 24 μm. Data were collected on a Cary Eclipse fluorescence spectrophotometer as described above.
Structure Preparation and Molecular Mechanics Calculations
Protein preparation, molecular mechanics calculations, and protein rendering were performed using the computational suite MOE (Molecular Operative Environment) release 2014.08 (Chemical Computing Group Inc., Montreal, Canada). The force field employed was Amber12:EHT, with the AMBER12 parameter set (ff12) for protein and parameters calculated from the extended Huckel theory for P6E, PJ34, NAD+, and dsDNA molecules. The MOE Protonate3D module was used to assign the ionization states and tautomers of protein side chains at T = 300 K, pH 7.4, and 0.1 m ionic strength, along with the GB-VI solvation model and MMFF94 partial charges. The molecular surfaces are solvent-excluded surfaces obtained by rolling a probe sphere of 1.4-Å diameter (water radius) and colored by several schemes. The van der Waals interaction surfaces correspond to zero-potential contours of the van der Waals potential, Evdw = 0, between the considered set of atoms and a water oxygen atom as mobile probe, using a standard 12-6 Lennard-Jones definition.
Modeling the Scabin·NAD+·DNA Complex
The Scabin·PJ34 complex (Protein Data Bank code 5EWK) was stripped from crystallographic waters, co-solvents, and ions, and the nicotinamide moiety of NAD+ was superposed to the fused amide of the PJ34 ring system. Then the PJ34 molecule was deleted, and the remaining Scabin·NAD+ structure was energy-minimized by fixing the coordinates of the nicotinamide moiety and Scabin atoms while allowing relaxation of the remaining NAD+ molecule into the binding cavity. This was followed by relaxing the side chains of the pocket residues with fixed NAD+ coordinates. This complex was named Scabinm·NAD+. To model the ternary Scabin·NAD+-DNA complex, a 10-mer dsDNA oligomer with the 5′-CAGTGCAGTG-3′ sequence in the B-helix conformation was constructed with a guanine base at the 5th position in both DNA strands. This dsDNA10 molecule was manually positioned around the Scabinm·NAD+ complex by a rigid body translation/rotation in order to locate the 5G bases near the bound NAD+ while avoiding direct clashes between their van der Waals surfaces. A distance-constrained energy term was set with limits of 10–12 Å between both reaction centers (the NAD+ C1′ atom of N-ribose and the exo-cyclic nitrogen atom of one of the 5-position guanines), and then a rigid-body optimization was conducted (root mean square gradient ≤0.01 kcal/mol/Å2). Next, a LowModeMD conformational search was performed (20) to optimize active site loop residues that contact the NAD+ and dsDNA molecules; this involved two steps: (i) 1000 iterations with fixed NAD+ and then (ii) 10,000 iterations with only the nicotinamide moiety fixed. In both cases, the coordinates of dsDNA and non-loop residues were constrained and set to inert protein atoms beyond 4.5 Å from any loop or NAD+ atoms. The decoy with lower energy was further energy-optimized for all side chains and loop atoms (with fixed NAD+) but allowing the dsDNA to approach as a rigid body into the distance range of 9–10 Å between reaction centers (root mean square gradient ≤0.001 kcal/mol/Å2). The final model was inspected for atom clashes and correct protein geometry (i.e. Ramachandran outliers, bond lengths, etc.). The model was herein called the Scabinm·NAD+·dsDNA10 complex.
Results
Identification and Expression of Scabin
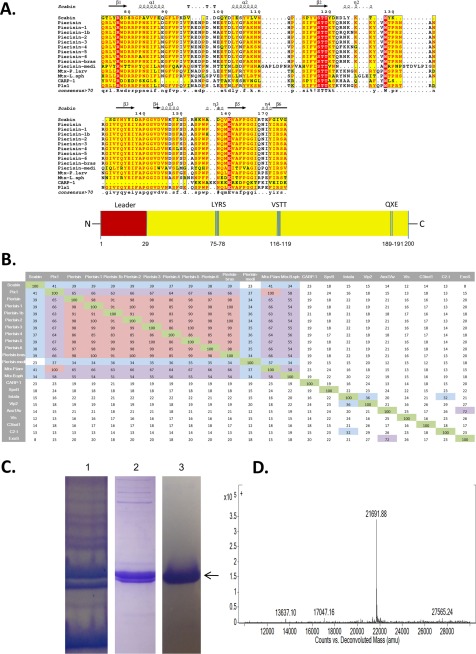
The Scabin gene, scab_27771, is found within the S. scabies pathogenic strain 87.22. The full-length protein consists of a single catalytic mART domain and a 29-residue N-terminal secretion signal. As shown from the multiple-sequence alignment depicted in Fig. 1A, Scabin shares common catalytic features with other mART toxins. This includes the hallmark Arg that is essential for NAD+ binding to the active site, the catalytic Gln-X-Glu motif, which is vital for the reaction mechanism, and the Ser-Thr-Thr motif that forms the scaffold of the NAD+ binding pocket (21) (Fig. 1A). The multiple-sequence alignment clearly shows that Scabin is closely related to the Pierisin mART toxin subgroup because there are large conserved regions shared by Scabin and the Pierisin members (Fig. 1A). A percentage identity matrix of several key members of the mART toxin family, including Scabin, revealed that Scabin shares nearly 40% sequence identity with Pierisin subgroup proteins, except for Pierisin-Medicago truncatula from a flowering plant (23% identity) (Fig. 1B). The Pierisin group of mART toxins is distinguished by its unique target specificity, where ADP-ribose is transferred to the guanine base in DNA, leading to apoptosis from P. rapae (4) (Fig. 1B).
FIGURE 1.
Multiple-sequence alignment of Scabin with various Pierisin-like mART toxins. A, sequence alignment of Scabin with some Pierisin-like toxins produced using the T-Coffee Web server to align the sequences and ESPript to generate the alignment figure (45). Key catalytic regions are highlighted. Identical residues are highlighted in red, and similar residues are printed in red type. B, identity matrix showing the amino acid identity between the 100 core catalytic residues of the known ExoS-like, C2-like toxins and Vis. Salmon, highly diverse sequences; light green, a large amount of conservation; yellow, an intermediate level of conservation between sequences. The identity matrix was generated using ClustalX2 (33) and colored using Microsoft Excel. C, purification and identification of Scabin from E. coli lysate. SDS-polyacrylamide gels showing the protein banding pattern for crude lysate (lane 1), immobilized metal affinity chromatography purification (lane 2), and FPLC ion exchange chromatography (lane 3). The arrow indicates the position of the Scabin protein. D, Q-TOF mass analysis of purified Scabin protein showing a single peak at 21,691.9 Da, corresponding to the expected mass of recombinant Scabin.
Before purification, an N-terminal 29-residue truncation of the Scabin gene was cloned for expression into E. coli BL21, λDE3 cells. Scabin did not express well with the N-terminal signal sequence, so removal of this region was required for good expression. Scabin was purified from whole-cell lysate via immobilized metal affinity chromatography and a subsequent anion exchange step. The purity level was assessed by inspection of the corresponding SDS-polyacrylamide gels after each purification step (Fig. 1C). Scabin protein yield of 1 mg/liter culture was obtained after purification to near homogeneity. Scabin was positively identified using MALDI-TOF mass spectrometry and gave the expected mass of 21,961.9 Da (Fig. 1D).
Folded Integrity of Scabin WT and Variant Toxins
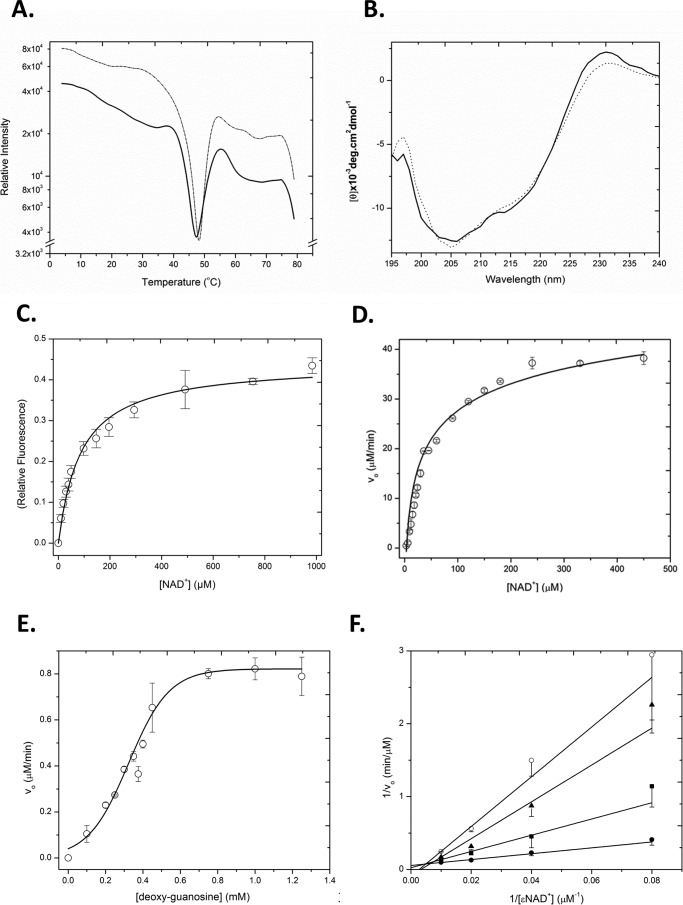
To assess the folded integrity of Scabin and the catalytic variant Q158A/E160A, both differential scanning fluorimetry and CD spectroscopy were used (Fig. 2, A and B). Differential scanning fluorimetry revealed a smooth transition between the folded and unfolded states, resulting in a melting temperature for Scabin and the catalytic variant Q158A/E160A of 46 ± 0.9 and 48 ± 0.4 °C, respectively (Fig. 2A). This revealed that the catalytic variant protein was slightly more stable than the WT protein. Scabin showed a typical CD spectrum for an α/β protein with a relatively higher β-strand composition than helical content (20.5% α-helix and 11.5% β-strand, respectively). There were no significant differences in the folding of the two proteins based on their CD spectra (Fig. 2B). This indicates that Scabin was properly folded into a stable and active conformation and that replacement of Gln158 and Glu160 residues did not significantly perturb the folded integrity of the enzyme (Fig. 2B).
FIGURE 2.
A, folded stability (Tm) of WT Scabin (thick line) and Scabin Q158A/E160A variant (thin dotted line) as measured by the SYPRO Orange thermal shift assay (derivative of the raw data traces). The traces are representative scans of three replicates for each sample, and the apex of the minima shows the position of the Tm, where the protein is half-unfolded. B, CD spectra of Scabin WT (thick line) and Q158A/E160A variant (thin dotted line) in 20 mm Tris, 50 mm NaF, pH 8.2, buffer. The concentrations of the proteins were both at 0.16 mg/ml, and each spectrum is the average of nine independent spectra. C, NAD+ binding by Scabin. The binding isotherm for NAD+ with 1.25 μm Scabin was determined by quenching of the intrinsic protein fluorescence. The raw fluorescence quenching data were converted to relative values and are plotted against the NAD+ concentration. The excitation was 295 nm, and the emission was 340 nm with excitation and emission band passes at 5 nm in 25 mm Tris-HCl, 50 mm NaCl, pH 8.2, buffer. D, GH activity of Scabin WT showing the hydrolysis rate of the NAD+ substrate by the protein (Scabin, 490 nm; ϵ-NAD+, 0–450 μm. Error bars, S.D. E, ADP-ribosyltransferase activity of WT Scabin. ϵ-NAD+ was held at a concentration of 250 μm and was mixed with 10 nm Scabin and buffer containing 1% dimethyl sulfoxide and various concentrations of dG (0–1250 μm). Error bars, S.D. F, inhibition plot of Scabin GH activity. Shown are Lineweaver-Burk plots for Scabin in the presence of various concentrations of inhibitor PJ34. The GH activity of Scabin was measured with 0 (filled circles), 6 μm (filled squares), 12 μm (filled triangles), and 24 μm (open circles). Vo indicates initial velocity in μm/min. Error bars, S.D.
NAD+ Glycohydrolase Activity and NAD+ Binding of Scabin
GH activity is characteristic of most mART toxins, whereby in the absence of a transferase substrate, an OH− molecule from aqueous solution acts as a nucleophile to accept the ADP-ribose group from NAD+ (3). The GH activity was characterized for Scabin in the absence of transferase substrate using a well established fluorescence-based assay (22). Scabin exhibited Michaelis-Menten behavior, and kinetic parameters (Table 2) were determined as Km = 68 ± 3 μm and kcat = 94 ± 2 min−1 (Fig. 2D). This is consistent with other mART toxins, such as Certhrax toxin, which has a Km of 42 ± 11 μm and a kcat of 8.7 ± 1.0 min−1 (23). Although some mART toxins do not possess GH activity, most have GH activity that is typically lower than their transferase activity (1). An active site variant of Scabin was prepared that involved replacing the QXE catalytic signature with AXA, which has been shown previously to render mART toxins catalytically impotent (5, 23). The catalytic Q158A/E160A variant was significantly less active, as anticipated for replacement of this key catalytic signature in mART toxins, and gave a kcat of 0.31 ± 0.02 min−1 (Table 2).
TABLE 2.
Kinetic parameters and NAD+ binding affinity of Scabin and Q158A/E160A variant
| Parametera | WT | Q158A/E160A |
|---|---|---|
| Km(GH) (μm) | 68 ± 3 | NDb |
| kcat(GH) (min−1) | 94 ± 2 | 0.31 ± 0.02 |
| KD(NAD+) (μm)c | 70 ± 3 | 86 ± 7 |
| K0.5(dG) (μm) | 302 ± 12 | ND |
| kcat(dG) (min−1) | 14 ± 1 | ND |
| kcat/Km(GH) (m−1 min−1) | 1.4 × 106 | ND |
| kcat/Km(dG) (m−1 min−1) | 4.6 × 104 | ND |
a Kinetic parameters were obtained as described under “Experimental Procedures” and represent the mean ± S.D. All results represent triplicate measurements for at least three separate experiments.
b ND, not determined.
c The KD value for the NAD+ substrate represents the binding affinity of NAD+ for the active site of Scabin and was measured by the Trp quenching caused by NAD+ docking within the active site of the enzyme (see “Experimental Procedures”). All results represent triplicate measurements for three separate experiments, mean ± S.D.
The dissociation constant, KD, of Scabin for NAD+ was determined by exploiting the intrinsic fluorescence of tryptophan residues that are located near the binding pocket (Scabin has 4 Trp residues). Scabin was titrated with NAD+, and the dissociation constant was determined to be 70 ± 3 μm (Table 2 and Fig. 2C). This is consistent with other mART toxins, such as Photox toxin, which has a KD for NAD+ of 11 ± 0.3 μm (24). The binding constant of the Q158A/E160A catalytic variant for NAD+ was determined to be 86 ± 7 μm, which is comparable with the NAD+ affinity of WT Scabin (Table 2). This suggests that the catalytic QXE motif in the enzyme is involved in the transferase mechanism and that it does not play a role in substrate binding, consistent with other mART toxins (21).
Scabin Acts on Substrates Containing Guanine
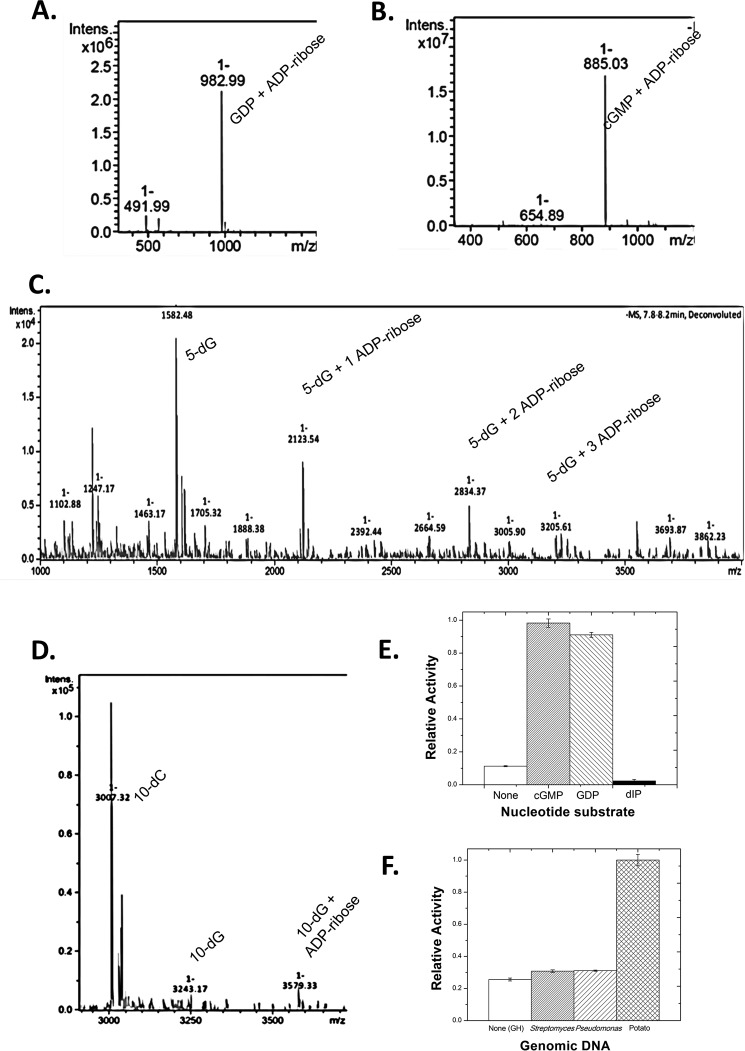
mART activity occurs when a transferase substrate is present to act as a nucleophile in the reaction to replace the endogenous OH− nucleophile from water. Using the fluorescence-based assay for measuring mART activity, several potential substrates were tested. Initial studies involved testing whole cell lysates from potato, yeast, and mammalian cells for Scabin transferase activity against target proteins. However, there was no activity or labeling of any proteins from these lysates (data not shown). As demonstrated previously by Wakabayashi and co-workers (7), Pierisin-1 labels guanine-containing substrates, such as small nucleotides or DNA strands. Consequently, potential substrates that were tested for Scabin activity include GDP and cGMP for small nucleotides as well as a single-stranded oligonucleotide consisting of 5 dG bases and a double-stranded oligonucleotide containing a single guanine base at the fifth position (from the 5′-end) of the forward strand. A sharp increase in activity was observed for all substrates as compared with the baseline GH activity. To provide solid evidence that Scabin is indeed labeling these substrates, mass spectrometry was performed on reactions that were allowed to run overnight. This showed that Scabin was able to label GDP and cGMP with an ADP-ribose moiety, giving products with masses of 983.0 and 885.0 Da, respectively (Fig. 3, A and B). Scabin was also able to label the single-stranded 5-nucleotide poly(dG) substrate with one, two, and three ADP-ribose moieties at unique positions, as shown by the masses of 2123.5, 2834.3, and 3205.6 Da, respectively (Fig. 3C).
FIGURE 3.
Mass spectrometry of Scabin with oligonucleotide substrates. A, product ion spectra (singly charged, positive mode) after liquid chromatographic separation of the reaction products from the incubation of Scabin with 0.5 mm GDP. B, product ion spectra (singly charged, positive mode) after liquid chromatographic separation of the reaction products from the incubation of Scabin with 0.5 mm cGMP. C, product ion spectra (singly charged, positive mode) after liquid chromatographic separation of the reaction products from the incubation of Scabin with annealed poly(5)-deoxyguanidine/deoxycytidine oligonucleotide. Peaks corresponding to unlabeled (1582.5 Da), singly labeled (2123.5 Da), doubly labeled (2834.4 Da), and triply labeled (3205.6 Da) oligonucleotide were clearly resolved. D, product ion spectra (singly charged, positive mode) after liquid chromatographic separation of the reaction products from the reaction of Scabin with annealed poly(10)-deoxyguanidine/deoxycytidine oligonucleotide. Peaks corresponding to unlabeled (3038.6 Da) and singly labeled (3579.3 Da) oligonucleotide are shown. E, histogram showing the relative activity of Scabin against the following substrates: none (baseline GH activity only), cGMP, GDP, and 2′-deoxyinosine-5′-monophosphate at 1 mm concentrations in GH buffer. F, histogram showing the relative transferase activity of Scabin against genomic DNA from the following organisms: none (GH background activity), S. scabies, P. aeruginosa, and Solanum tuberosum (potato).
In addition, Scabin labeled the DNA strand containing a single deoxyguanosine base with an ADP-ribose group, yielding a total mass of 3579.3 Da (Fig. 3D). Importantly, Scabin did not label oligonucleotides that did not possess guanine bases (data not shown).
Pierisin-1 was previously shown to label the exocyclic N2 substituent on guanine bases of DNA substrates (7). To test whether this was true for Scabin, dI, a nucleotide that is identical to dG with the exception that it is missing the exocyclic N2 substituent, was tested for activity with Scabin. Scabin showed no significant activity toward dI above the baseline GH as compared with cGMP and GDP substrates (Fig. 3E), demonstrating that Scabin labels the same position on deoxyguanosine as Pierisin-1.
Further studies were performed with dG to determine kinetic parameters for this substrate. Notably, Scabin exhibited sigmoidal kinetic behavior in the presence of the dG substrate, with K0.5 = 302 ± 12 μm and kcat = 83 ± 1 min−1 (Fig. 2E and Table 2).
Scabin Labels Genomic DNA
Scabin was also tested for its ability to modify genomic DNA of the producing organism, S. scabies, and of a Gram-negative bacterium, P. aeruginosa, and the target plant, potato (Fig. 3F). Scabin showed weak activity against the genomic DNA of both S. scabies and P. aeruginosa. However, it showed much higher activity against potato genomic DNA, suggesting the presence of target genes or a preferred DNA conformation in the plant species.
Inhibition of Scabin
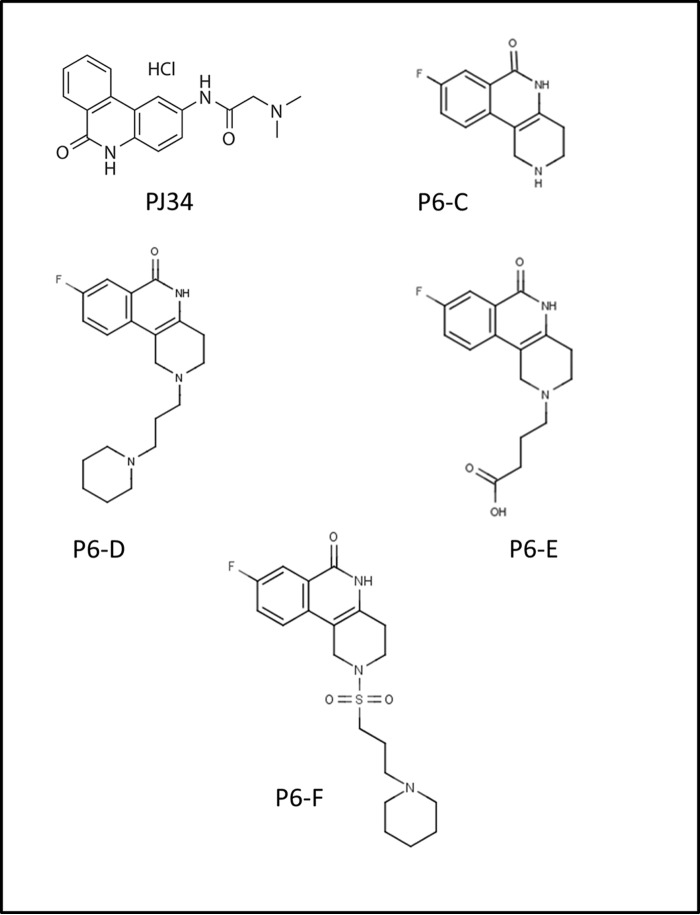
Several small molecules were shown to inhibit Scabin GH activity. These compounds, PJ34, P6-C, P6-D, P6-E, and P6-F (Fig. 4), are known inhibitors of other mART toxins (23, 25). Lineweaver-Burk analysis was performed for kinetic inhibition data of Scabin with PJ34 inhibitor to validate the mechanism of inhibition for the P-series inhibitors. As expected, it was clear that PJ34 acts as a competitive inhibitor against Scabin GH activity (Fig. 2F). Another small in-house library of known mART inhibitors were also tested against Scabin, but only those that exhibited significant levels of inhibition were further characterized by determining their respective IC50, pIC50, Ki, and KD values (Table 3). All of these compounds bind tighter than NAD+ with affinity in the range of low micromolar (KD < 50 μm), although the dissociation constant of P6-F was not reported due to technical difficulties inherent to the estimation of the binding affinity by means of the quenching of the Trp fluorescence emission. Nevertheless, based on a competition model, the affinities (KD values) of a series of inhibitors must correlate with their inhibition constants (Ki values). Because Ki is the thermodynamic parameter that characterizes the inhibition, and due to the lower relative uncertainties with respect to the nominal values, the Ki values were considered as the representative parameter of the binding/activity of the inhibitors. Thus, PJ34 showed the most promise as a potential lead compound for inhibition of Scabin (Ki = 3 ± 0.2 μm). For the P6 series, the activity is limited between P6-F with the highest activity (Ki = 7 ± 0.2 μm) and P6-E with the lowest one (Ki = 24 ± 0.3 μm). Evidently, the chemical and topological constitution of the inhibitor “tail” is a determining factor for the inhibition of Scabin GH activity. In this sense, the hydrophobic character of the terminal piperidine of P6-D and P6-F favors the inhibition in comparison with the “tail-less” benzo-naphthyridinone ring system of the P6-C compound as the reference structure. Notably, the anionic terminal carboxylate of P6-E is detrimental for the inhibitor activity (Table 3 and Fig. 6).
FIGURE 4.
P-series inhibitors effective against Scabin GH activity. PJ34, 2-[[3-(dimethylamino)-2-oxopropyl]amino]-5,6-dihydrophenanthridin-6-one; P6-C, 8-fluoro-1H,2H,3H,4H,5H,6H-benzo[c]1,6-naphthyridin-6-one; P6-D, 8-fluoro-2-[3-(piperidin-1-yl)propyl]-1H,2H,3H,4H,5H,6H-benzo[c]1,6-naphthyridin-6-one; P6-E, 4-[8-fluoro-6-oxo-1H,2H,3H,4H,5H,6H-benzo[c]1,6-naphthyridin-2-yl]butanoic acid; P6-F, 8-fluoro-2-[3-(piperidin-1-yl)propanesulfonyl]-1H,2H,3H,4H,5H,6H-benzo[c]1,6-naphthyridin-6-one.
TABLE 3.
Comparison of KD, IC50, and solubility parameter values for inhibitors of Scabin
| Inhibitor | KDa | IC50b | Kic | pIC50d | ΔTme |
|---|---|---|---|---|---|
| μm | μm | μm | °C | ||
| PJ34 | 14 ± 0.5 | 12 ± 1 | 3 ± 0.2 | 4.9 | 2.4 ± 0.3 |
| P6-C | 25 ± 1 | 89 ± 4 | 19 ± 1 | 4.1 | 2.2 ± 0.6 |
| P6-D | 42 ± 5 | 97 ± 7 | 18 ± 1 | 4.0 | 1.8 ± 0.4 |
| P6-E | 50 ± 6 | 119 ± 2 | 24 ± 0.3 | 3.9 | 2.6 ± 0.7 |
| P6-F | NDf | 38 ± 2 | 7 ± 0.2 | 4.4 | 1.5 ± 0.5 |
a The binding affinity of inhibitors to Scabin was measured by the quenching of the intrinsic Trp fluorescence caused by the binding of the ligand to the enzyme active site.
b The IC50 values were determined by fitting each dose-response curve to a Boltzmann sigmoidal function in MicroCal Origin version 8.0.
c The inhibition constant (Ki) was calculated from the experimentally determined IC50 values according to the relationship, Ki = IC50/(1 + ([SNAD]/Km(NAD)) using fixed values for [Scabin] = 10 μm, [ϵ-NAD+] = 400 μm, and Km(ϵ-NAD+) = 276 μm.
d The pIC50 values were calculated from the IC50 values as follows, pIC50 = −log IC50. The higher the pIC50 value, the lower dose that is required for 50% inhibition of Scabin toxin activity.
e The ΔTm value (°C) was determined by the expression, ΔTm°C(Scabin-inhibitor) − ΔTm°C (Scabin-apo).
f Not determined.
FIGURE 6.
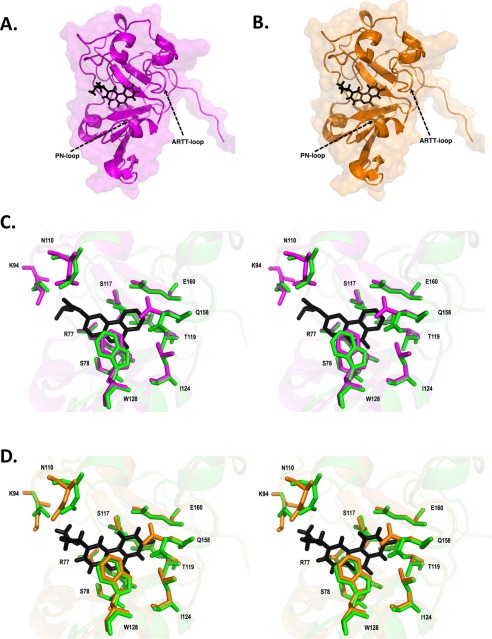
Scabin inhibitor crystal structures. A, Scabin·PJ34 complex structure shown as a ribbon diagram. PJ34 is colored black and represented in stick format. B, Scabin·P6-E complex structure shown as a ribbon diagram. P6-E is colored black and represented in stick format. C, stereo view of active site of Scabin·PJ34 complex structure (magenta) and Scabin-apo structure (green). PJ34 is colored black and represented in stick format. Structural differences among important catalytic residues (Arg77 Ser78, Lys94, Asn110, Ser117, Thr119, Leu124, Tyr128, Gln158, and Glu160) are highlighted. D, stereo view of active site of Scabin·P6-E complex structure (magenta) and Scabin-apo structure (green). P6-E is colored black and represented in stick format. Structural differences among important catalytic residues (Arg77 Ser78, Lys94, Asn110, Ser117, Thr119, Leu124, Tyr128, Gln158, and Glu160) are highlighted.
Using differential scanning fluorimetry, the change in Tm when inhibitors were bound to the active site was studied. A higher Tm than the Scabin apoenzyme is characteristic of inhibitor binding to the active site and stabilizing the folded protein (26). However, the change in Tm does not necessarily correlate with the affinity/activity for a dissimilar set of compounds (Table 3). Indeed, among the compounds tested, PJ34 exhibited the highest affinity but was not the best stabilizer of Scabin folded structure (Table 3, last column). Moreover, for the P6 series, there is a direct relationship with respect to the changes in affinity (or its equivalent Ki) compared against a reference compound (ΔKi = Ki − Ki,ref) and the changes in the variation of Tm (ΔΔTm = ΔTm − ΔTm,ref). In effect, the inhibitory activity and effect induced by the backbone ring structure of P6-C, P6-D, and P6-F compounds showed greater activity yet reduced stability compared with the P6-E inhibitor.
Scabin Crystal Structure
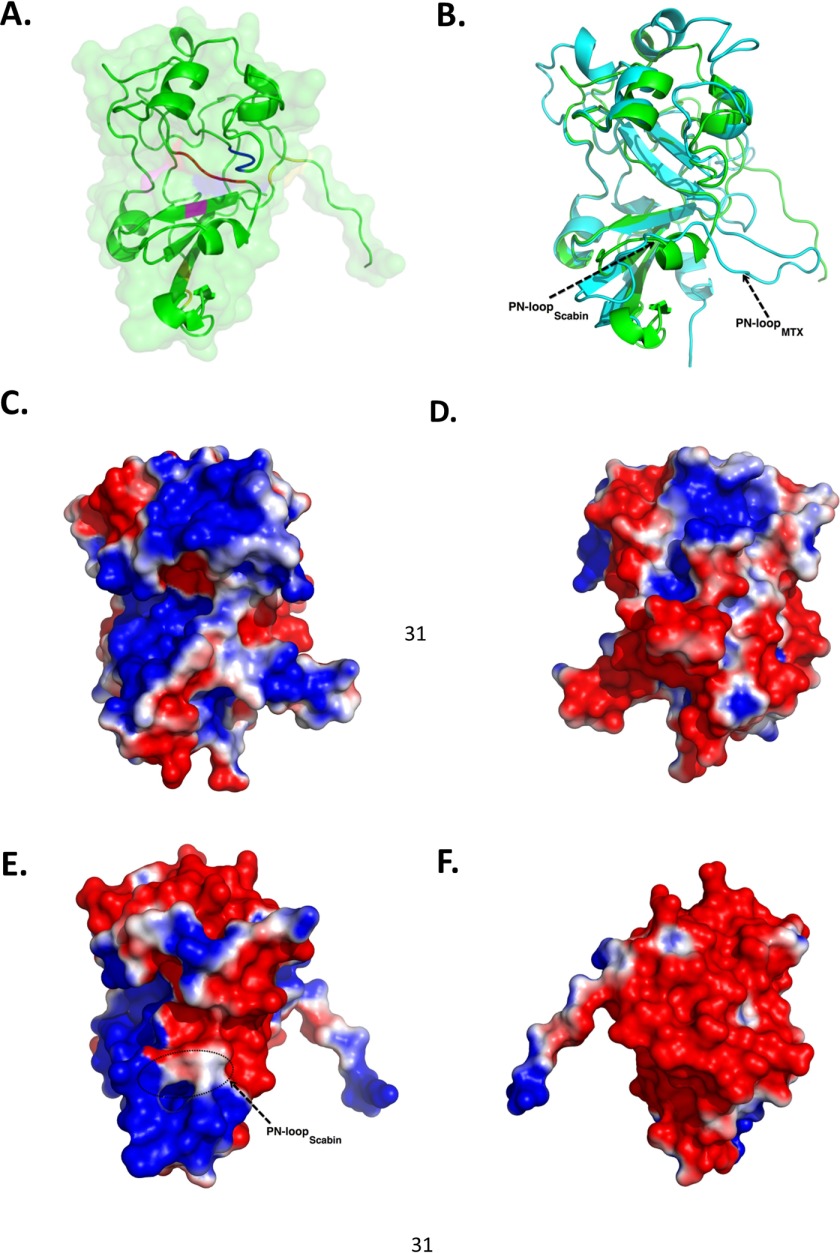
The structure of recombinant Scabin was refined to a resolution of 1.50 Å in the apo-form (substrate-free) (Fig. 5A). The Scabin structure displays a characteristic mART fold and contains the conserved RSQXE motif. Most of the mART enzymes studied to date have predominant α/β structure (e.g. ι-toxin catalytic, ∼52% (27); C3bot1, ∼55% (28); Vis, ∼55% (29); and Spvb, ∼59% (30)). Furthermore, Scabin has low sequence identity with most mART toxins except for the Pierisin subgroup (Fig. 1, A and B) and significantly differs in topology from well characterized mARTs like ι-toxin and C3-group toxins, which are dominated by high helical content at the N terminus (31). Scabin shares sequence homology with mosquitocidal toxin (MTX) from B. sphaericus (13) and the apoptosis-inducing Pierisin-1 from the cabbage butterfly P. rapae (no structure available). Superposition of the backbone Cα atoms of Scabin with the catalytic subunit of MTX toxin (Protein Data Bank entry 2CB6) reveals a low root mean square deviation of 0.671 Å (for 436 atoms). However, upon closer observation of the backbone traces, some structural differences in certain regions are observed (Fig. 5B). The PN loop is longer and more extended compared with Scabin (indicated by an arrow in Fig. 5B). Notably, the PN loop motif in Scabin begins as a helical segment and later becomes coil structure. The characteristic EXE catalytic motif found in MTX and in other Arg-modifying ADP-ribosyltransferases is replaced by QXE in Scabin. The QXE motif is also found in the C3 group of toxins and in pertussis toxin, which modify Asn (5, 32) and Cys residues (34), respectively. In the case of Scabin, it modifies deoxyguanosine bases on a DNA substrate like Pierisin group members (7). The Scabin structure is also uniquely stabilized by two disulfide bridges (Cys42-Cys72 and Cys176-Cys190).
FIGURE 5.
Scabin-apo crystal structures. A, close-up of the Scabin-apo crystal structure shown as a ribbon diagram. Catalytic residues Gln158 and Glu160 are colored blue. Other important residues in the reaction mechanism, Arg77 (pink) and STS motif (red), are also highlighted. Disulfide bridges are colored yellow. B, structural comparison of Scabin-apo (green) and the catalytic domain of the MTX toxin structure (cyan) based on an iterative, three-dimensional alignment of protein backbone Cα atoms using PyMOL. C, surface potential of the catalytic subunit of MTX (front view). Molecular surfaces are colored by the relative electrostatic potential (red, negative or acidic; blue, basic or positive). Surface potentials were calculated using PyMOL APBS software. D, surface potential of the catalytic subunit of MTX toxin (back view). E, surface potential of Scabin-apo (side view). F, surface potential of Scabin-apo (opposite side view).
Further analyses of these crystal structures of MTX and Scabin using electrostatic potential data from their corresponding solvent-accessible surface areas show distinct patterns from one another (Fig. 5, C–F), suggesting that these two toxins do not share a similar substrate. MTX toxin shows patches of both electropositive and electronegative potential throughout its structure (Fig. 5, C and D), whereas Scabin is predominantly electronegative, with patches of electropositive surface potential near the PN loop (Fig. 5, E and F). Given the propensity of DNA to associate with its cognate binding proteins via electropositive surfaces and the lack of these positively charged surface patches in other regions of the Scabin structure, the structural data suggest that the PN loop and nearby areas probably play a crucial role in DNA substrate binding.
Scabin in Complex with Its Inhibitor PJ34
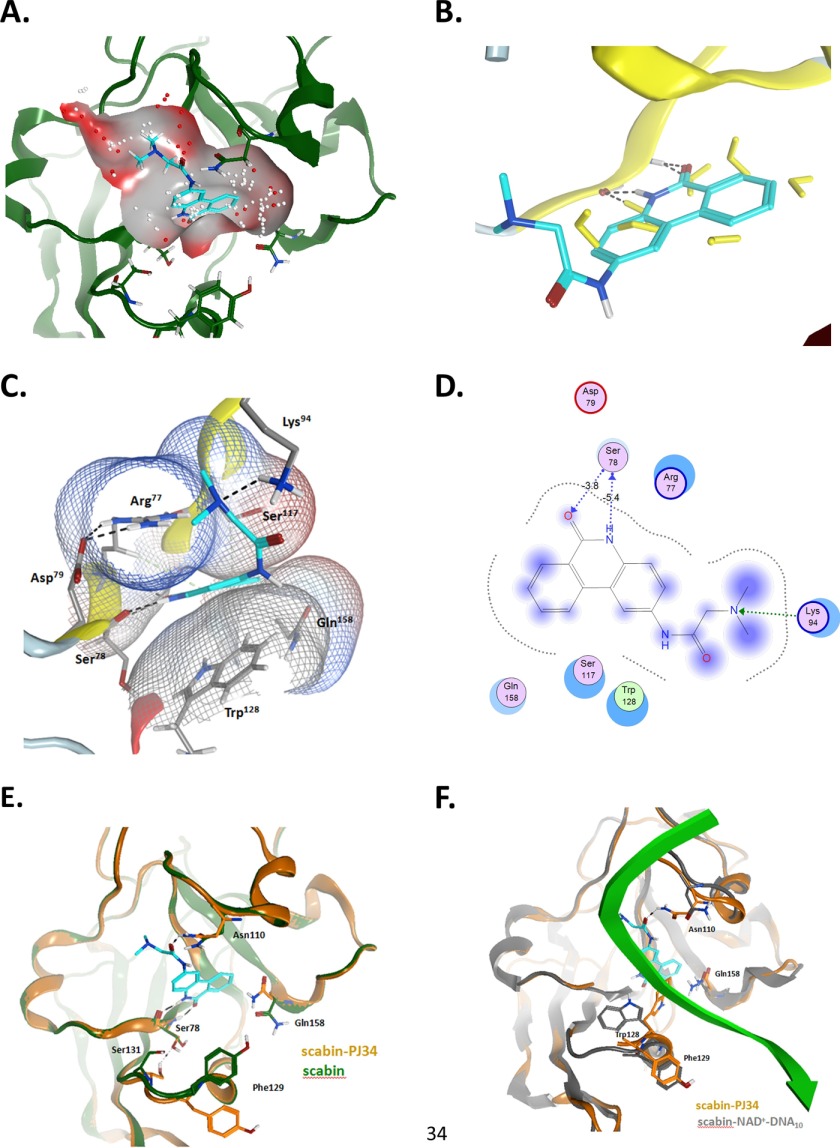
The PJ34-bound structure of Scabin was refined to a resolution of 1.60 Å (Fig. 6A). It shows a low root mean square deviation with the Scabin-apo structure (0.127 Å for 1053 atoms). As expected, the catalytic residues within the ARTT loop orient toward the inhibitor (Fig. 6C) with the catalytic Gln158 shifting nearly 2.5 Å. The PJ34 molecule is stabilized within the active site of Scabin through hydrophobic interactions and H-bonds. PJ34 forms two H-bonds with Ser78. One H-bond forms between the main-chain nitrogen of Ser78 (2.8 Å), and the other forms with the main-chain oxygen of Ser78 (2.8 Å (Figs. 6C and 8E). Additionally, Trp128 from the PN loop appears to play a key role in inhibitor binding and stabilization. The side-chain oxygen of Asn110 is situated 2.9 Å away from the tertiary amine of the R-group of PJ34, which forms another important H-bond. As reported earlier in other mART toxin structures with PJ34 (25, 35, 36), the electron density of the hetero-ring system of the inhibitor is clearly defined; however, the electron density is weaker for the R-group (tertiary amine) (supplemental Fig. S1), suggesting some flexibility in the “tail” of the inhibitor. Despite this, the electron density for the PJ34 R-group shows orientation toward the loop containing Asn110 (supplemental Fig. S1).
FIGURE 8.
The Scabin·PJ34 complex. Shown are details of the interaction between Scabin and the PJ34 inhibitor in the context of the apo-form and the modeled complexed with NAD+ and dDNA10. A, binding pocket on the Scabin-apo. The binding pocket was calculated based upon the x-ray coordinates of the Scabin-apo. The small dots correspond to the center of hydrophilic (red) and hydrophobic (white) α spheres into the N-site of the binding pocket. The PJ34 molecule was superposed for reference. B, binding of PJ34. Shown is the binding pose of PJ34 on the Scabin·PJ34 complex. The crystallographic waters are depicted in yellow from the Scabin-apo structure for reference. C, Scabin-PJ34 ligand interactions; van der Waals interaction surfaces (vdw-interaction surfaces) around pocket residues, colored by the electrostatic potential. D, two-dimensional ligand interactions. Shown is a two-dimensional depiction of the bound PJ34 and interacting residues. The arrows represent the H-bonds, with backbone atoms (blue) and side-chain atoms (green). The degree of exposure is shown by the purple sphere. E, pocket variation upon binding. Shown are pocket residues with major side-chain variation upon PJ34 binding. Depicted are ribbons and carbon atoms for Scabin (in green) and for Scabin·PJ34 complex (in orange). F, comparison between Scabin-bound forms. Shown is superposition of the Scabin·PJ34 complex (in orange ribbons and carbon atoms) and the modeled Scabinm·NAD+·dDNA10 complex (in gray ribbons and carbon atoms), with NAD+ not shown. The backbone of the inward facing DNA strand is shown in green. In all panels, PJ34 is shown as cyan carbon atoms. Figures were rendered by MOE.
Scabin in Complex with Its Inhibitor P6-E
The structure of Scabin in complex with the P6-E inhibitor was also determined at 1.50 Å resolution (Fig. 6B). P6-E is a member of the P6 series of inhibitors (25), and this is the first report of its crystal complex bound to a mART enzyme. The P6-E-bound structure shows a low root mean square deviation from Scabin-apo (0.114 Å for 1053 atoms). As observed in the Scabin·PJ34 structure, the catalytic Gln158 is shifted nearly 2.5 Å upon inhibitor binding, whereas the catalytic Glu160 and other conserved residues that interact with P6-E (Arg77,Ser78, Lys94, Asn110, Ser117, Thr119, Leu124, and Tyr128) do not significantly change position upon inhibitor binding (Fig. 6D). P6E is stabilized by hydrophobic interactions and two H-bonds. The first H-bond is between the main-chain guanidinium group of Arg77 and the oxygen of P6-E (3.4 Å). The second is between the main-chain nitrogen of Ser78 and the oxygen of P6-E (2.8 Å). P6-E bound in the active site closely mimics the PJ34 structural position and has a well defined electron density throughout the molecule (supplemental Fig. S1).
Scabin Modeled with DNA
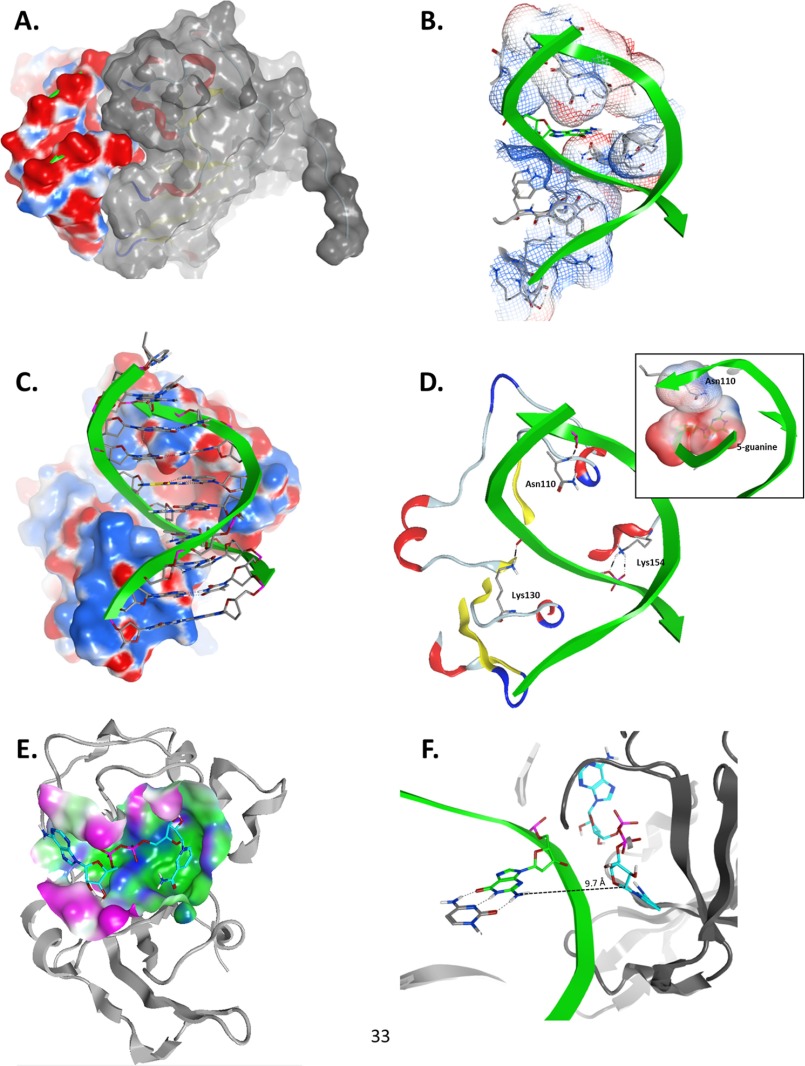
A model of Scabin in complex with DNA was constructed to provide a working basis for understanding and testing the interaction of Scabin with its DNA substrate(s) (Fig. 7). Based upon molecular modeling results, we propose a ternary complex between Scabin, NAD+, and the optimal target DNA substrate tested so far, a double-stranded 10-base pair DNA oligonucleotide with a guanine base at the 5th position from the 5′-end (Fig. 7, A–F). In this model, because the guanine NH2 group faces the minor groove of the dsDNA molecule in a B-helix conformation, this groove must face the NAD+-binding pocket. Based on this constraint, the backbone of a DNA strand was docked into a protein cleft that runs diagonally with respect to the vertical and main molecular axis of Scabin (Fig. 7B). The embedding of the DNA strand fulfilled steric or shape complementarity between both molecular surfaces (Fig. 7A). Electrostatic complementarity is observed between the negative DNA ribose-phosphate backbone (red surfaces in Fig. 7A) and the exposed regions of Scabin (blue surfaces in Fig. 7C). In addition, specific H-bond interactions are predicted to occur between the backbone of the inward facing DNA strand with the side chains of Lys130 and Lys154 (Fig. 7D). In summary, the model suggests that Scabin clamps the inward facing DNA strand with two helix-turn motifs, one located in the minor groove and the other at the major groove (supplemental Fig. S2).
FIGURE 7.
Model of the Scabinm·NAD+·dsDNA10 complex. Panels show details of the interaction between Scabin, NAD+, and the dsDNA10 molecules from the ternary complex generated by using the Scabin-apo structure as the template. A, Scabin-DNA shape complementarity; side view of the molecular surfaces of the modeled complex of Scabin (colored with gray surface) and the 10-mer dsDNA (colored according to the electrostatic potential; red, negative; blue, positive). B, DNA-binding cleft. Front view of the van der Waals interaction surface of the active conformation of Scabin, colored by the electrostatic potential. The 5-position guanine base is depicted in green carbon atoms, whereas the protein residues making DNA contact are shown in gray carbon atoms. C, Scabin-DNA electrostatic complementarity. Shown is a front view of the complex, depicting the DNA ribose and base atoms. The molecular surface of the Scabin is colored by the electrostatic potential. D, specific Scabin-DNA interactions. Details of the H-bond interactions between three active site loop residues (Asn110, Lys130, and Lys154) in the Scabin active conformation and the phosphate backbone of the dsDNA molecule are shown. Inset, contact between the van der Waals interaction surfaces of Asn110 and the 5-position guanine. E, NAD+-binding pocket. The molecular surface of Scabin around the binding pocket of NAD+ is colored according to its polar (blue), hydrophobic (green), or exposed (fuchsia) character. The bound NAD+ substrate is shown in cyan carbon atoms, and the backbone trace of Scabin is shown in light gray ribbons. F, distance and configuration of the reactive centers. A side view of the ternary complex showing the 5-position guanine (in green carbon atoms) of the dsDNA10 and the active conformation of NAD+ (in cyan carbon atoms) are shown in the ternary complex. The drawn segmented line connects the exo-cyclic amino nitrogen atom of the 5-position guanine with the C1′ atom of the NAD+ N-ribose with a separation distance of ∼9.7 Å. Figures were rendered by MOE.
Furthermore, the outward facing DNA strand contacts the protein by an H-bond with the backbone of Asn110. Importantly, this residue is conserved in all of the protein sequences shown in Fig. 1A except for Pierisin-medi and CARP-1, implying a specific side-chain interaction with the DNA substrate. Indeed, this residue is the only one with its side chain protruding into the DNA minor groove, making van der Waals contact with the target 5-position guanine (Fig. 7D, inset). Accordingly, its putative role might be to assist the destabilization of the GC pair, to expose the 5-position guanine as the nucleophile for the reaction. Alternatively, the active conformation of the NAD+ substrate was also modeled with emphasis on the binding mode of the NMN moiety, to locate the C1′ atom of the N-ribose as the electrophilic center for the nucleophilic reaction (Fig. 7E). Thus, based on the location of the electrophilic atom in the Scabin·NAD+ complex, the closest approach of the nucleophilic center (the nitrogen atom of the exo-cyclic amino group of 5-guanine) achieved by our modeling strategy corresponded to a distance of 9.7 Å (Fig. 7F). Notably, the guanine NH2 group faces the minor groove of the dsDNA molecule, which limits the closest approach between both macromolecules. Nevertheless, this distance is comparable with previously reported values for the reactive centers in other Michaelis-Menten complexes of mART toxins (36–38); however, this distance is certainly much greater than the expected separation of 3–3.5 Å between reactive centers as a ground state complex for an SN1 nucleophilic reaction (39). Hence, further conformational changes or “reaction steps” are required, such as electrophile migration and/or “strain alleviation” of the electrophile by dihedral rotation around the O3–NP and NP–NO5 bonds (38).
Scabin Pocket Analysis with PJ34
The x-ray structure of the Scabin·PJ34 complex reveals that the N-site of the binding pocket is well formed in the Scabin apo-structure (Fig. 8A), exhibiting a cavity with principal hydrophobic character (white spheres) and a deeper hydrophilic one (red spheres). The PJ34 ring system fits into the subpocket with no significant variation in the pocket topology. Nevertheless, the binding of PJ34 induces dehydration of the N-locus, with a major impact on the displacement of two water molecules bound to the backbone of Ser78 (Fig. 8B), due to its negative effect on the binding energy. The backbone of Ser78 and the side chains of Arg77 and Asp79 (both conserved), Lys94 and Trp128 (conserved), and Glu158 (conserved QXE motif) are the main interacting residues, with Arg77 and Trp128 constituting the flat volume of the N-subpocket (Fig. 8C). The charged side chain of Lys94 prevents the protonation of the terminal dimethyl-amino group of the PJ34 “tail,” in opposition to the protonated state for the soluble PJ34 at neutral pH. Fig. 8D depicts a two-dimensional representation of the pocket interactions. The reciprocal H-bonds between the fused amide of PJ34 and the backbone atoms of Ser78 is a key feature of the binding mode for the P series of inhibitors in several mART toxins (40, 41). However, some side chains of neighboring residues reorient significantly upon binding of PJ34 (Fig. 8E). The side chain of Asn110 is shifted to bind the O-carbonyl of the PJ34 “tail,” whereas the conformational variation in the side chain of Ser78 and Ser131 may align with changes in the water structure and/or may be an “artifact” of the modeled coordinates of Ser131 because it is not well resolved in the x-ray structure. Remarkably, the conformations of Phe129 and Gln158 in the Scabin·PJ34 complex resemble their conformations in the modeled Scabin·NAD+·DNA ternary complex (Fig. 8F). This is not the case for Trp128 and Asn110, which adapt their conformations to interact with the inward facing NAD+ strand (Figs. 7D and 8F).
Discussion
In this work, a bioinformatics strategy was used to identify Scabin as a novel mART enzyme with potential as a putative virulence factor from S. scabies, a well known and persistent pathogen of tuberous plants, such as potatoes (9). Scabin is clearly a member of the Pierisin subgroup of mART toxins based upon its high sequence identity and common core enzyme pattern with that group. Scabin shares common enzymatic function with the Pierisin subgroup in its ability to target the guanine base within DNA (8). Specifically, Scabin labels nucleosides, mononucleotides, and both dsDNA and ssDNA by transferring ADP-ribose to the exocyclic amino group on the guanine base. It also labels genomic DNA with an apparent preference for potato (target) DNA. However, it is not known whether this represents the presence of target genes or a preferred DNA conformation in the plant species.
The Scabin gene (scab_27771) encodes a signal peptide (residues 1–29), indicating that it is a factor secreted by S. scabies and a putative virulence factor. Previously, it was shown that a number of proteins in S. scabies are secreted by the twin Arg protein transport (Tat) pathway (42). Scabin was shown to be secreted into the extracellular medium and was proposed to use the Tat secretion pathway based on prediction models utilizing the signal peptide sequence. The Scabin signal peptide sequence contains a twin Arg motif, which is indicative of Tat-mediated secretion (42).
Interestingly, there is a gene in S. scabies (scab_27781) that is adjacent to the Scabin gene (scab_27771), and the former gene encodes for a putative toxin B domain that features a YVTN β-propeller that may be involved in Scabin toxicity. This B-domain also possesses a signal peptide, implying secretion as an exported protein along with Scabin.
The Scabin catalytic core region shares common features with the mART CT toxin family (6, 43), including the conserved Arg β-strand residue, an STT motif, and the catalytic Glu motif (QXE). The enzyme showed elevated GH activity compared with most mART toxins, nearly 100 min−1 compared with 0.2 min−1 for ExoA (1). The Scabin Q158A/E160A variant exhibits the classic mART effect of mutation on the enzyme activity with a 300-fold reduction in GH activity.
Scabin was shown to react with and label mononucleotides, such as GDP and cGMP; mononucleosides, such as deoxyguanosine; and both dsDNA and ssDNA but not DNA substrates void of guanine bases. When tested for activity toward dI, no catalytic activity was detected above baseline GH activity. This further supports the proposal that Scabin shares a similar reaction mechanism with the Pierisin subgroup by labeling the exocyclic NH2 substituent on the guanine base.
Scabin exhibited Michaelis-Menten behavior for its GH activity as expected; however, the kinetic profile for Scabin modification of dG was sigmoidal. The molecular basis for the non-Michaelis-Menten kinetic behavior of Scabin is not known; however, one possibility is that Scabin may oligomerize to form a dimer or higher order aggregate that acts cooperatively to label macromolecule substrates (nucleosides, nucleotides, DNA). Another possibility is that the sigmoidal kinetic behavior of Scabin toward the dG substrate may be due to a complex interplay between the dG guanine base and another binding site on the Scabin enzyme. Hence, there may be two binding sites for the dG substrate that show a cooperative effect as a function of concentration. Further studies are required in order to determine the molecular basis for Scabin sigmoidal kinetic behavior toward the dG substrate.
In our Scabin-DNA model, the main contact between both molecules is by the docking of a DNA strand into a small cleft of the protein rather than by defined protein motifs (e.g. helix-turn-helix) that protrude and dock into the major groove of the dsDNA molecule. In the model, only two symmetric and small protein segments (i.e. two short helix-loop motifs (see supplemental Fig. S2)) “clamp” onto the dsDNA molecule. The steric (van der Waals) and charge (electrostatic) complementarity between the protein cleft and the ribose-phosphate DNA backbone are the main interacting forces. Only three H-bonds offer some degree of directionality to the interaction, although with only modest specificity, since the interactions mostly involve protein-DNA backbone atoms. The specific interactions with the guanine base are currently not known; however, we are pursuing structural data to probe the nature of these interactions.
The crystal structure of Scabin was determined in the absence of substrate or ligand (Scabin-apo, 1.45 Å) and complexed with two active site competitive inhibitors of Scabin GH activity, PJ34 and P6-E (1.6 and 1.4 Å, respectively). The structures reveal that Scabin is a single-domain mART toxin that possesses a mART fold, which features a mixed α/β structure. However, the Scabin core contains less α/β structure than is usually observed in mART crystal structures; it features more random coil, with some rather large loop regions. It is believed that this added flexibility in the mART core of Scabin may be necessary to adapt/conform to the DNA substrate. Additionally, the abundance of electropositive patches on the surface of Scabin, especially in the region around the PN loop, implies that Scabin interacts with the phosphate backbone of DNA in these regions.
Scabin was tested against several in-house mART toxin libraries of inhibitors, which include the P6 series (25). Several potent inhibitors were identified, including a previously characterized inhibitor of mART toxins known as PJ34 (25, 35, 36). The inhibitor structures are stabilized within the active site of Scabin, largely through hydrophobic interactions combined with a few critical H-bonds. Upon inhibitor binding, rather small changes were observed overall within the Scabin active site, with the major structural movement being the reorientation of the ARTT loop and shifting of the catalytic Gln158 residue by 2.5 Å. This side-chain conformer of Gln158 is also observed in the Scabin P6-E complex and in the modeled Scabin·NAD+ complex. Thus, the binding of a ligand that occupies the N-subsite is enough to trigger the conformational change of Gln158 side chain toward an inward facing conformation.
The lower affinity/activity of P6-E is remarkable when the cationic character and/or H-bond capability of residues flanking the “tail” of P6-E is considered, in particular Arg77, Lys94, and Asn110. These interactions may arise from desolvation penalties associated with water displacement in the vicinity of these residues and/or an entropic component of the P6-E “tail.” In summary, neutral and hydrophobic terminal “tails” (e.g. the dimethylamino group of PJ34 or the piperidine ring of P6-D and P6-F) are preferred over negatively charged substituents (e.g. the carboxylate of P6-E) and are also preferred over positively charged terminal groups (e.g. the cationic PJ34 tail). Nevertheless, the polar and H-bond capability of the proximal section to the inhibitor “core” is also a determinant, as observed in the enhanced affinity/activity of PJ34 and P6-F compared with the other P6 compounds.
It was also observed that the binding of a small ligand occupying the N-subpocket of Scabin (e.g. P6-C) is able to stabilize the toxin structure, which is reflected in the ΔTm of 2.2 °C. Accordingly, it has been described previously that the dynamics of the central β-core that characterizes the scaffold of the mART toxins fluctuate with several correlated and anti-correlated modes of movement with exposed subdomains and motifs (44). Therefore, inhibitor/ligand substituents may differentially affect the overall dynamics and stability of the protein, despite interactions with exposed protein loops and motifs only. The binding of pocket residues with various ligands might produce a new mode of coupling between “ligand-bridged” protein subdomains and may not be observed in complex with a simple ligand (i.e. in the absence of the ligand motif(s)) or for the apoprotein. Incidentally, P6 derivatives with higher affinity for Scabin than P6-C, due to favorable interactions with pocket loops and motifs (e.g. P6-D and P6-F), induce lower Tm changes in Scabin, whereas derivatives with lower affinity (e.g. P6-E) exhibit a higher Tm shift.
In summary, our structural and biochemical data herein suggest that Scabin is a member of the Pierisin subgroup within the mART toxin family. The members within this subgroup target DNA by attaching ADP-ribose to the exocyclic amino group of the guanine base (4, 7, 8). We are pursuing further studies with Scabin to determine its role in S. scabies as a putative virulence factor. We are also working to determine the structure of Scabin in complex with various substrates, including DNA oligonucleotides. Future studies will also involve a search for the DNA base sequence specificity for Scabin as a DNA-modifying enzyme and its biological DNA/gene target(s).
Author Contributions
B. L., R. R., J. L., M. R. L., D. D., and S. C. performed the experiments. B. L., R. R., M. R. L., and A. R. M. wrote the paper.
Acknowledgments
We thank Tom Keeling for excellent technical assistance in the cloning and related molecular biology pertaining to this work. We thank Dr. Sherif Sherif for providing the S. scabies genomic DNA. We also thank Dr. Robert Fieldhouse for the initial bioinformatics research to identify the Scabin gene as a putative virulence factor from S. scabies 87.22 strain. X-ray data for this research were collected at Beamline 08ID-1 at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan.
This work was supported by the Canadian Institutes of Health Research (Grant MOP-106661 to A. R. M.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 5DAZ, 5EWK, and 5EWY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- mART
- mono-ADP-ribosyltransferase
- ϵ-NAD+
- etheno-NAD+
- GH
- glycohydrolase
- TAPS
- 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid
- cGMP
- cyclic GMP
- MTX
- mosquitocidal toxin
- H-bond
- hydrogen bond.
References
- 1. Yates S. P., Jørgensen R., Andersen G. R., and Merrill A. R. (2006) Stealth and mimicry by deadly bacterial toxins. Trends Biochem. Sci. 31, 123–133 [DOI] [PubMed] [Google Scholar]
- 2. Holbourn K. P., Shone C. C., and Acharya K. R. (2006) A family of killer toxins: exploring the mechanism of ADP-ribosylating toxins. FEBS J. 273, 4579–4593 [DOI] [PubMed] [Google Scholar]
- 3. Simon N. C., Aktories K., and Barbieri J. T. (2014) Novel bacterial ADP-ribosylating toxins: structure and function. Nat. Rev. Microbiol. 12, 599–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Watanabe M., Kono T., Matsushima-Hibiya Y., Kanazawa T., Nishisaka N., Kishimoto T., Koyama K., Sugimura T., and Wakabayashi K. (1999) Molecular cloning of an apoptosis-inducing protein, pierisin, from cabbage butterfly: possible involvement of ADP-ribosylation in its activity. Proc. Natl. Acad. Sci. U.S.A. 96, 10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krska D., Ravulapalli R., Fieldhouse R. J., Lugo M. R., and Merrill A. R. (2015) C3larvin toxin, an ADP-ribosyltransferase from Paenibacillus larvae. J. Biol. Chem. 290, 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fieldhouse R. J., and Merrill A. R. (2008) Needle in the haystack: structure-based toxin discovery. Trends Biochem. Sci. 33, 546–556 [DOI] [PubMed] [Google Scholar]
- 7. Takamura-Enya T., Watanabe M., Totsuka Y., Kanazawa T., Matsushima-Hibiya Y., Koyama K., Sugimura T., and Wakabayashi K. (2001) Mono(ADP-ribosyl)ation of 2′-deoxyguanosine residue in DNA by an apoptosis-inducing protein, pierisin-1, from cabbage butterfly. Proc. Natl. Acad. Sci. U.S.A. 98, 12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakano T., Takahashi-Nakaguchi A., Yamamoto M., and Watanabe M. (2015) Pierisins and CARP-1: ADP-ribosylation of DNA by ARTCs in butterflies and shellfish. Curr. Top. Microbiol. Immunol. 384, 127–149 [DOI] [PubMed] [Google Scholar]
- 9. Lerat S., Simao-Beaunoir A. M., and Beaulieu C. (2009) Genetic and physiological determinants of Streptomyces scabies pathogenicity. Mol. Plant. Pathol. 10, 579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goyer C., and Beaulieu C. (1997) Host range of streptomycete strains causing common scab. Plant Dis. 81, 901–904 [DOI] [PubMed] [Google Scholar]
- 11. Kabsch W. (2010) XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 13. Reinert D. J., Carpusca I., Aktories K., and Schulz G. E. (2006) Structure of the mosquitocidal toxin from Bacillus sphaericus. J. Mol. Biol. 357, 1226–1236 [DOI] [PubMed] [Google Scholar]
- 14. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 15. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 16. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Healey A., Furtado A., Cooper T., and Henry R. J. (2014) Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng Y., and Prusoff W. H. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 20. Labute P. (2010) LowModeMD: implicit low-mode velocity filtering applied to conformational search of macrocycles and protein loops. J. Chem. Inf. Model. 50, 792–800 [DOI] [PubMed] [Google Scholar]
- 21. Masignani V., Balducci E., Serruto D., Veggi D., Aricò B., Comanducci M., Pizza M., and Rappuoli R. (2004) In silico identification of novel bacterial ADP-ribosyltransferases. Int. J. Med. Microbiol. 293, 471–478 [DOI] [PubMed] [Google Scholar]
- 22. Armstrong S., and Merrill A. R. (2001) Application of a fluorometric assay for characterization of the catalytic competency of a domain III fragment of Pseudomonas aeruginosa exotoxin A. Anal. Biochem. 292, 26–33 [DOI] [PubMed] [Google Scholar]
- 23. Visschedyk D., Rochon A., Tempel W., Dimov S., Park H. W., and Merrill A. R. (2012) Certhrax toxin, an anthrax-related ADP-ribosyltransferase from Bacillus cereus. J. Biol. Chem. 287, 41089–41102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Visschedyk D. D., Perieteanu A. A., Turgeon Z. J., Fieldhouse R. J., Dawson J. F., and Merrill A. R. (2010) Photox, a novel actin-targeting mono-ADP-ribosyltransferase from Photorhabdus luminescens. J. Biol. Chem. 285, 13525–13534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turgeon Z., Jørgensen R., Visschedyk D., Edwards P. R., Legree S., McGregor C., Fieldhouse R. J., Mangroo D., Schapira M., and Merrill A. R. (2011) Newly discovered and characterized antivirulence compounds inhibit bacterial mono-ADP-ribosyltransferase toxins. Antimicrob. Agents Chemother. 55, 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niesen F. H., Berglund H., and Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 27. Sakurai J., Nagahama M., Hisatsune J., Katunuma N., and Tsuge H. (2003) Clostridium perfringens ι-toxin, ADP-ribosyltransferase: structure and mechanism of action. Adv. Enzyme Regul. 43, 361–377 [DOI] [PubMed] [Google Scholar]
- 28. Han S., Arvai A. S., Clancy S. B., and Tainer J. A. (2001) Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. J. Mol. Biol. 305, 95–107 [DOI] [PubMed] [Google Scholar]
- 29. Ravulapalli R., Lugo M. R., Pfoh R., Visschedyk D., Poole A., Fieldhouse R. J., Pai E. F., and Merrill A. R. (2015) Characterization of vis toxin, a novel ADP-ribosyltransferase from Vibrio splendidus. Biochemistry 54, 5920–5936 [DOI] [PubMed] [Google Scholar]
- 30. Lesnick M. L., Reiner N. E., Fierer J., and Guiney D. G. (2001) The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol. Microbiol. 39, 1464–1470 [DOI] [PubMed] [Google Scholar]
- 31. Vogelsgesang M., Pautsch A., and Aktories K. (2007) C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn Schmiedebergs Arch. Pharmacol. 374, 347–360 [DOI] [PubMed] [Google Scholar]
- 32. Aktories K., Rösener S., Blaschke U., and Chhatwal G. S. (1988) Botulinum ADP-ribosyltransferase C3: purification of the enzyme and characterization of the ADP-ribosylation reaction in platelet membranes. Eur. J. Biochem. 172, 445–450 [DOI] [PubMed] [Google Scholar]
- 33. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 34. Barbieri J. T., Mende-Mueller L. M., Rappuoli R., and Collier R. J. (1989) Photolabeling of Glu-129 of the S-1 subunit of pertussis toxin with NAD. Infect. Immun. 57, 3549–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yates S. P., Taylor P. L., Jørgensen R., Ferraris D., Zhang J., Andersen G. R., and Merrill A. R. (2005) Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa. Biochem. J. 385, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jørgensen R., Purdy A. E., Fieldhouse R. J., Kimber M. S., Bartlett D. H., and Merrill A. R. (2008) Cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. J. Biol. Chem. 283, 10671–10678 [DOI] [PubMed] [Google Scholar]
- 37. Jørgensen R., Merrill A. R., Yates S. P., Marquez V. E., Schwan A. L., Boesen T., and Andersen G. R. (2005) Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature 436, 979–984 [DOI] [PubMed] [Google Scholar]
- 38. Tsurumura T., Tsumori Y., Qiu H., Oda M., Sakurai J., Nagahama M., and Tsuge H. (2013) Arginine ADP-ribosylation mechanism based on structural snapshots of ι-toxin and actin complex. Proc. Natl. Acad. Sci. U.S.A. 110, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parikh S. L., and Schramm V. L. (2004) Transition state structure for ADP-ribosylation of eukaryotic elongation factor 2 catalyzed by diphtheria toxin. Biochemistry 43, 1204–1212 [DOI] [PubMed] [Google Scholar]
- 40. Lugo M. R., and Merrill A. R. (2015) A comparative structure-function analysis of active-site inhibitors of Vibrio cholerae cholix toxin. J. Mol. Recognit. 28, 539–552 [DOI] [PubMed] [Google Scholar]
- 41. Lugo M. R., and Merrill A. R. (2015) Pocket analysis of the full-length cholix toxin: an assessment of the structure-dynamics of the apo catalytic domain. J. Biomol. Struct. Dyn. 33, 2452–2468 [DOI] [PubMed] [Google Scholar]
- 42. Joshi M. V., Mann S. G., Antelmann H., Widdick D. A., Fyans J. K., Chandra G., Hutchings M. I., Toth I., Hecker M., Loria R., and Palmer T. (2010) The twin arginine protein transport pathway exports multiple virulence proteins in the plant pathogen Streptomyces scabies. Mol. Microbiol. 77, 252–271 [DOI] [PubMed] [Google Scholar]
- 43. Fieldhouse R. J., Turgeon Z., White D., and Merrill A. R. (2010) Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLoS Comput. Biol. 6, e1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lugo M. R., Ravulapalli R., Dutta D., and Merrill A. R. (2016) Structural variability of C3larvin toxin: intrinsic dynamics of the α/β fold of the C3-like group of mono-ADP-ribosyltransferase toxins. J. Biomol. Struct. Dyn. 26, 1–62 [DOI] [PubMed] [Google Scholar]
- 45. Gouet P., Courcelle E., Stuart D. I., and Metoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]