Abstract
The tripartite terminase complex of herpesviruses assembles in the cytoplasm of infected cells and exploits the host nuclear import machinery to gain access to the nucleus, where capsid assembly and genome-packaging occur. Here we analyzed the structure and conservation of nuclear localization signal (NLS) sequences previously identified in herpes simplex virus 1 (HSV-1) large terminase and human cytomegalovirus (HCMV) small terminase. We found a monopartite NLS at the N terminus of large terminase, flanking the ATPase domain, that is conserved only in α-herpesviruses. In contrast, small terminase exposes a classical NLS at the far C terminus of its helical structure that is conserved only in two genera of the β-subfamily and absent in α- and γ-herpesviruses. In addition, we predicted a classical NLS in the third terminase subunit that is partially conserved among herpesviruses. Bioinformatic analysis revealed that both location and potency of NLSs in terminase subunits evolved more rapidly than the rest of the amino acid sequence despite the selective pressure to keep terminase gene products active and localized in the nucleus. We propose that swapping NLSs among terminase subunits is a regulatory mechanism that allows different herpesviruses to regulate the kinetics of terminase nuclear import, reflecting a mechanism of virus:host adaptation.
Keywords: herpesvirus, nuclear translocation, nuclear transport, virus assembly, virus entry, genome packaging motor, importin α, terminase
Introduction
Herpesviruses are large double-stranded DNA (dsDNA) ubiquitously found in humans that replicate in the cell nucleus. They are broadly subdivided into three taxonomic subfamilies (α, β, and γ) thought to have diverged from a common ancestor around 400 million years ago (1). In total, there are 8 herpesvirus types that infect humans and present different cell and tissue tropism as well as genome size (between 120–240 kbp). The assembly of all herpesvirus proceeds via formation of an empty procapsid, which is filled with genetic material by the action of a powerful virus-encoded genome-packaging motor (for review, see Refs. 2–4). The icosahedral procapsid builds around a scaffolding protein not present in the mature virus and contains a dodecameric portal protein complex at a unique vertex (5, 6). Herpesvirus replication strategy is very similar to that of tailed bacteriophages (for review, see Refs. 7–10) and, to some extent, adenoviruses (11). In both bacterial viruses and herpesviruses, the terminase complex is formed by a small and a large terminase subunit (abbreviated as S-terminase2 and L-terminase) assembled in various stoichiometries (12) that dock at the portal vertex (13) and convert ATP hydrolysis into rotation of dsDNA, which is gradually encapsidated (14). L-terminase, known as pUL15 in HSV-1 (15) and pUL89 (16) in HCMV, is a bifunctional ATPase/nuclease that binds directly to portal protein (7, 17, 18). In contrast, the S-terminase subunit (e.g. pUL28 in HSV-1; Refs. 19 and 20) and pUL56 in HCMV (Refs. 21 and 22) binds packaging initiation sites on viral genome and, at least in bacterial viruses, regulates the ATPase activity of L-terminase (23–25). Herpesviruses also contain a third terminase subunit (herein referred to as “T-terminase), such as pUL33 in HSV1 or pUL51 in HMCV, of unknown function and structure, that can be isolated from infected cells in a complex with L- and S-terminase (26).
Herpesvirus L-terminase shares significant amino acid homology with L-terminase from bacteriophages (7), including an N-terminal ATPase domain with Walker A/B motifs and a C-terminal nuclease domain (27, 28) superimposable to that of most phages (13, 29–31). Although L-terminase is a bona fide ortholog of phage L-terminases, there is no sequence homology and predicted structural similarity between bacteriophage and herpesvirus S-terminases, suggesting the latter is an evolutionarily new protein in herpesviruses that plays a functionally similar role of DNA recognition subunit during genome packaging. In addition, herpesvirus S-terminase is larger in molecular mass than L-terminase (Fig. 1A), which prompted some authors to refer to this subunit as “large terminase” (22). In this paper we will refer to herpesvirus S-terminase as the functionally equivalent protein of phage S-terminase.
FIGURE 1.
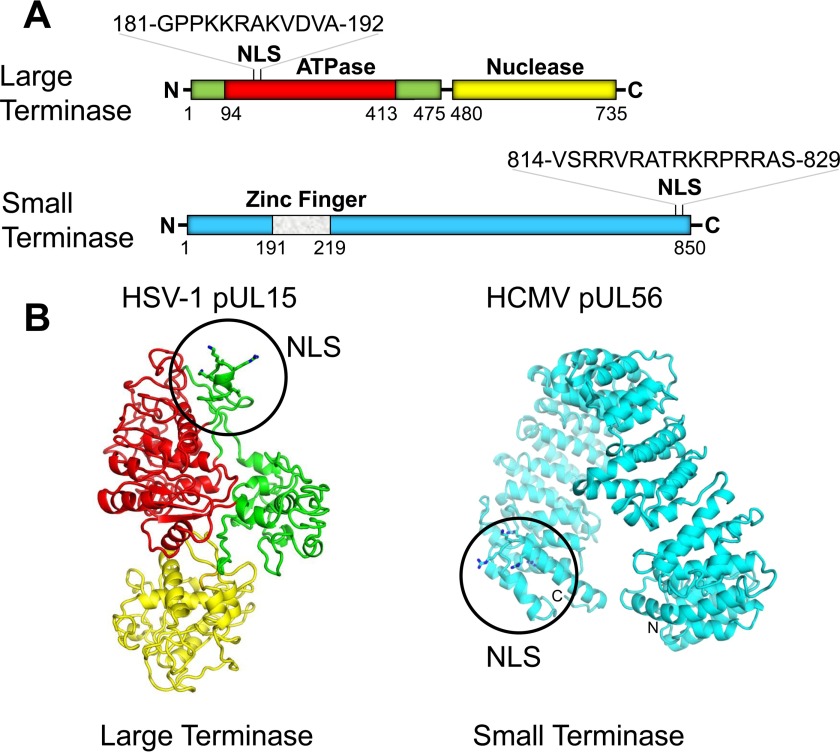
Mapping NLSs in herpesvirus terminase subunits. A, schematic diagram of L- and S-terminase subunits with predicted functional domains. The predicted zinc finger in S-terminase was described by Champier et al. (89). The amino acid sequence of pUL15-NLS (61) and pUL56-NLS (62) is shown in the magnified windows. B, ribbon diagrams of iTASSER (55) predicted three-dimensional-models of HSV-1 pUL15 (left panel) and HCMV S-terminase (right panel) subunits. NLS residues are shown as sticks with basic side chains colored in blue.
Trafficking of macromolecules between the nucleus and cytoplasm is typically an active, signal-mediated and highly regulated process that occurs through the nuclear pore complex (NPC) (for review, see Refs. 32–34). The majority of cellular and viral cargos moving through the NPC expose a transport signal on their surface, exemplified by the NLS (35) for import cargos and the nuclear export signal (36) for export cargos. NLS cargos are shuttled through the NPC by soluble transport factors of the importin β superfamily (also known as β-karyopherins) (37) in a process that requires the small GTPase Ran. Cargos bound to transport factors move bidirectionally through the NPC interior by making interactions with phenylalanine-glycine-rich repeats exposed by several nucleoporins mainly lining the NPC (38). Import complexes assemble in the cytoplasm upon interaction of NLS-cargos with the receptor importin β; this interaction can be direct (39) or mediated by transport adaptors such as importin α (40) and snurportin (41), which use an N-terminal Importin β binding (IBB) domain (42) to recruit importin β. Importin α is the universal adaptor that binds NLS-bearing import cargos (40). It exists in seven isoforms in humans, all structurally very similar (43), that generate a binding surface for NLSs, which can be monopartite like the SV40 T-large antigen NLS (44), or bipartite like nucleoplasmin NLS (45, 46). To obtain a quantitative description of the structure, potency, and conservation of NLSs found in herpesvirus terminase subunits, in this paper we carried out a structural and bioinformatic analysis of HSV-1 pUL15 and HCMV pUL56 NLS sequences.
Experimental Procedures
Biochemical Techniques
The gene encoding mouse importin α1 lacking the IBB domain was cloned in pET30a vector (Novagen) as previously described (43, 47). ΔIBB-importin α1 was expressed in Escherichia coli strain BL21 (DE3) by induction at 30 °C for 3 h with 1 mm isopropyl 1-thio-β-d-galactopyranoside and purified on nickel-agarose resin as described (43, 47). ΔIBB-Importin α1 was further purified over a Superdex 200 column (GE Healthcare) equilibrated in G.F. buffer (20 mm Tris pH 8.0, 150 mm NaCl, 5 mm β-mercaptoethanol, and 0.2 mm PMSF). Purified ΔIBB-Importin α1 was concentrated to 20 mg ml−1 using a Millipore concentrator (cutoff 10 kDa). Peptides encompassing HSV-1 pUL15 NLS (GPPKKRAKVDVA) and HCMV pUL56 NLS (VSRRVRATRKRPRRAS) were custom-synthesized (GenScript) and purified at 95% homogeneity by reverse phase chromatography. A 95% pure peptide comprising SV40 large T-antigen NLS (PKKKRKVEDPYC) was also purchased from GenScript.
Crystallographic Methods
Crystals of ΔIBB-Importin α1 bound to terminase NLSs were obtained using the hanging drop vapor diffusion methods. A droplet containing 2.3 μl of gel filtration-purified protein at 18 mg ml−1 and 0.7 μl of a 2-fold molar excess of peptide was mixed with an equal volume of 0.1 m sodium citrate tribasic dihydrate, pH 5.6, 0.7 m sodium citrate tribasic dihydrate, and 10 mm β-mercaptoethanol and equilibrated against 600 μl of the precipitant at 18 °C. Crystals were harvested in nylon cryo-loops, cryo-protected with 27% ethylene glycol, and flash-frozen in liquid nitrogen. Crystals were diffracted at LS-CAT Beamline 21-ID-F at Argonne Photon Source on a MARMOSAIC 225 CCD detector. Data were indexed, integrated, and scaled using HKL2000 (48). Initial phases were obtained by molecular replacement using Phaser (49) and PDB 3Q5U as a search model. Herpesvirus NLSs were built in Fo − Fc electron density difference maps using Coot (50), and complete atomic models were refined using phenix.refine (51) using cycles of positional and isotropic B-factor refinement with six distinct translation/libration/screw groups. Stereochemistry was checked using PROCHECK (52); the final models have excellent geometry, and the Ramachandran plot shows >96% of residues in the most favored regions of the Ramachandran plot and no outliers in disallowed regions. Data collection and refinement statistics are summarized in Table 1. All ribbon diagrams and surface representations were prepared using the program PyMOL (53). The structures were analyzed using the PISA server (54), and all structural superimpositions were carried out in Coot (50). Three-dimensional structural models of pUL15 and pUL56 were generated using I-TASSER (55).
TABLE 1.
Crystallographic data collection and refinement statistics
Values in parentheses are for highest resolution shells (2.02-1.95).
| ΔIBB- importin α1 |
||
|---|---|---|
| HSV-1 pUL15 NLS | HCMV pUL56 NLS | |
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 78.3, 90.7, 97.7 | 77.2, 90.8, 96.9 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.978 | 0.978 |
| Resolution (Å) | 50-1.95 | 50-1.95 |
| No. reflections (tot/unique) | 849,842/51,772 | 728,694/50,602 |
| Rsym | 8.7 (78.4) | 4.9 (74.9) |
| Rpin | 3.7 (48.1) | 2.4 (38.8) |
| I/σI | 33.1 (2.8) | 48.1 (3.8) |
| Completeness (%) | 99.7 (99.6) | 99.5 (99.5) |
| Redundancy | 3.9 (3.6) | 4.8 (4.5) |
| Refinement | ||
| Resolution (Å) | 15-1.95 | 15-1.95 |
| No. reflections | 51,068 | 47,920 |
| Rwork/Rfreea | 17.3/19.4 | 17.3/19.6 |
| No. protomers | 1 | 1 |
| No. protein atoms | 3,361 | 3,365 |
| B-factor (Å2) | ||
| Importin α/NLS major/NLS minor | 43.5/44.4/94.5 | 54.6/56.2/72.9 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.008 | 0.008 |
| Bond angles (°) | 0.887 | 1.041 |
a Rfree was calculated using ∼2000 randomly selected reflections.
Isothermal Titration Calorimetry (ITC)
ITC experiments were carried out at 25 °C using a nano-ITC calorimeter (TA Instruments). pUL15 NLS, pUL56 NLS, and SV40 NLS peptides were dissolved in G.F. buffer between 500 and 600 μm and injected in 1.5-μl increments into a calorimetric cell containing 195 μl of ΔIBB-importin α1 at 50 μm. The spacing between injections was 300 s. Titration data were analyzed using NanoAnalyze Data Analysis software version 3.5.0 (TA Instruments). Heats of dilution were determined from control experiments (NLS against buffer) and subtracted from heats of binding (NLS against importin α). Curve fitting using a two-independent-binding-site model gave a slightly better standard deviation around fit than a single binding site model (0.51 versus 0.54 for pUL15 NLS; 0.32 versus 0.45 for pUL56 NLS; 0.24 versus 0.34 for SV40 NLS).
Bioinformatic Analysis
Herpesvirus terminase subunits sequences were obtained from the NIAID Virus Pathogen Database and Analysis Resource (56) through the web site at Virus Pathogen Resource and with relevant website pages in the body of the paper. In addition, whole genome sequences accessed through NCBI (www.ncbi.nlm.nih.gov) were scanned for proteins related to HSV-1 large terminase and HCMV small terminase using either BLASTp (57) or manually. Global alignment of L- and S-terminase sequences was done using ClustalW at EMBL-EBI (58) using HSV-1 pUL15 and HCMV pUL56 as references to locate NLSs in orthologous terminases. Local alignment of NLSs was generated for every genus using the program WebLogo (59). Ab initio prediction of classical NLSs was done using NLS mapper (60).
Results
Topology of Herpesvirus L- and S-terminase Subunits
A functional NLS was previously identified and validated at the N terminus of HSV-1 L-terminase subunit (pUL15) (61) and at the C terminus of HCMV S-terminase (pUL56) (62) (Fig. 1A). To map the position of these import sequences in the three-dimensional structure of terminase subunits, we generated structural models of pUL15 and pUL56 using the prediction software iTASSER (55) (Fig. 1B). For L-terminase, iTASSER predicted a three-domain structure consisting of a highly conserved C-terminal nuclease domain (13, 27–31) and an amino-proximal ATPase domain containing Walker A and B motifs fused to an N-terminal insertion domain similar to that found in phage T4 L-terminase (13, 63). In this model the NLS is located at the surface of the insertion domain, adjacent to the ATPase module and readily accessible to importin α. In contrast, iTASSER predicted HCMV S-terminase to fold into a α-solenoid structure (Fig. 1B) built by tandemly repeated HEAT motifs, similar to importin β (37). This U-shaped structure is vastly different from phage S-terminases, which are typically smaller (∼20–25 versus ∼90 kDa), and oligomeric proteins assembled into ring-like oligomers (25, 64–67). Nonetheless, the iTASSER model is consistent with the four-prong structure of dimeric pUL56 obtained using negative stain electron microscopy (68). In this model pUL56 NLS (62) lays on the solvent-exposed loop connecting helix A with helix B of the last HEAT repeat and, in part, on the surface of helix B. It is not unusual for NLSs to be partially helical in the absence of importin α, as seen for the NLS of NF-κB p65 in the structure of NF-κB p65/p50 heterodimer bound to IkBa (69).
To shed light on the recognition of herpesvirus terminase NLSs by the host nuclear import machinery, we crystallized the adaptor importin α1 lacking the N-terminal IBB domain (42) (ΔIBB-importin α1) in complex with peptides encompassing HSV-1 pUL15 NLS (residues 180–191) and HCMV pUL56 NLS (residues 814–829) (Fig. 1A). For both complexes complete diffraction data to 1.95 Å resolution were measured, and the relative structures were solved by molecular replacement and refined to an Rwork/free of 17.3/19.4% and 17.3/19.6%, respectively (Table 1).
Atomic Structure of HSV-1 L-terminase NLS Bound to Importin α1
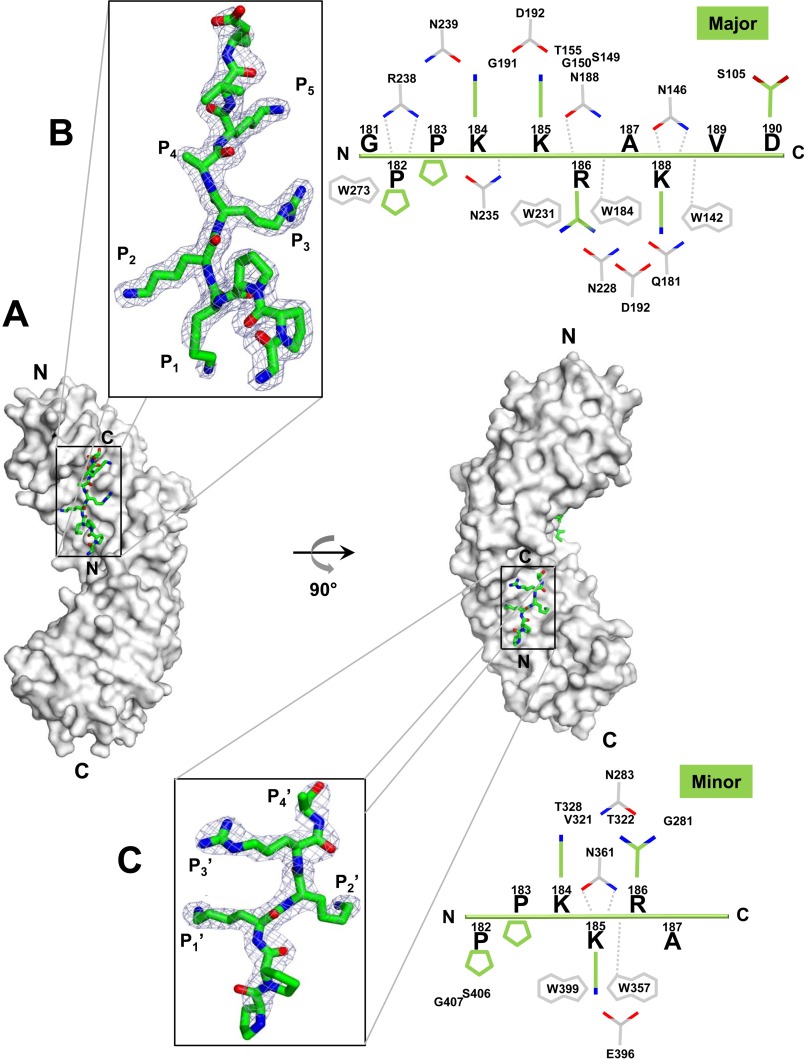
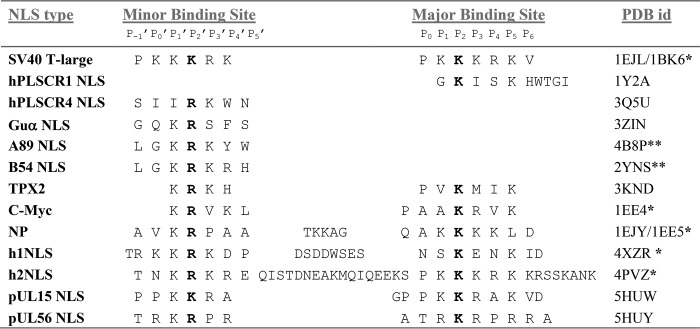
Importin α is made up of 10 stacked Armadillo (Arm) repeats, each formed by three α-helices (known as A, B, and C) (40, 70). Without the IBB-domain, the importin α Arm core generates a continuous α-helical surface that harbors a major and a minor NLS binding pocket. Two binding sites for NLSs have been identified and are usually referred to as major (Arm repeats 2–4) and minor (Arm repeats 7–8). At each site five main points of contact between NLS side chains and importin α are referred to as P1–P5 and P1′–P5′ at major and minor binding sites, respectively (35, 44–47, 71–76). HSV-1 L-terminase NLS binds at both sites of importin α (Fig. 2A) but is better ordered at the major pocket (Fig. 2B), where nine NLS residues (181GPPKKRAKV190) have excellent electron density, as opposed to only four residues at the minor binding site (184KKRA187) (Fig. 2C). Accordingly, the pUL15 NLS buries 733.9 and 542.3 Å2 of surface areas at major and minor binding sites, respectively, making a total of 17 hydrogen bonds, 1 salt bridge, and 153 non-bonded contacts at major and 8 hydrogen bonds, 2 salt bridges, and 65 non-bonded contacts at minor NLS site (Fig. 2, B and C). The different avidity for importin α core at the two sites is reflected by the peptide B-factor (Table 1), which is comparable to importin α average B-factor at the major NLS binding pocket (44.4 Å2 versus 43.5 Å2), but nearly twice as high (94.5 Å2) at the minor NLS site. Structural alignments with known classical and non-classical NLS peptides identified HSV-1 pUL15 NLS residues occupying conserved positions P1–P5 and P1′–P4′ (Table 2 and Fig. 2, B and C). A critical Lys (Lys-185) is invariantly conserved at position P2, as found in the majority of classical and non-classical NLSs (Table 2). Surprisingly, position P4 is occupied by an Ala, a residue not commonly found in the context of an NLS (35). Unlike NLSs that bind exclusively (or preferentially) to the importin α minor NLS site (47, 72, 73, 77), HSV-1 pUL15 NLS inserts a Lys at P2′ (Lys-185) as opposed to an Arg, possibly explaining the weak affinity for this site. The general features of pUL15 NLS recognition by importin α resemble those observed for the classical of SV40 NLS (44, 45). The NLS binds in an extended conformation, with the main chain running antiparallel to the direction of the importin α Arm core. The concave surface of importin α provides a continuous binding interface for the NLS, where conserved tryptophan and asparagine residues protruding at the surface of importin α make hydrophobic, cation-π (78), and polar contacts with NLS side chains and backbone, positioning the peptide in the helical groove of importin α (Fig. 2, B and C). Notably, importin α makes eight close contacts with pUL15 NLS backbone atoms at the major NLS pocket versus only three contacts at the minor NLS-box (dashed lines in Fig. 2, B and C), also explaining the reduced NLS avidity for this site.
FIGURE 2.
Crystal structure of HSV-1 l-terminase pUL15 NLS in complex with importin α1. A, structure of ΔIBB-importin α1 (gray surface) in complex with HSV-1 pUL15 NLS (green ribbon). B and C, left panel, zoom-in window showing the final 2Fo − Fc electron density map (displayed as a cyan mesh contoured at 1.25σ above background) calculated using all reflections between 15 and 1.95 Å resolution after omitting the NLS bound at the major (B) and minor (C) NLS-binding sites of importin α. In both cases, the density is overlaid to the NLS final model (the illustration was generated using PyMOL (53)). Right panel, schematic diagram of the interactions between pUL15 NLS (in green) and importin α1 (in gray) at the major (B) and minor (C) NLS-binding sites, in a distance range of 2.5–4.5 Å. Side chain nitrogen and oxygen atoms are color-colored in blue and red, respectively. Interactions between importin α side chains and the NLS main-chain atoms are shown by dashed gray lines.
TABLE 2.
Structural alignment of terminase NLSs with other NLSs visualized crystallographically in complex with Kap60/importinα
* denotes yeast importin α (Kap60). ** denotes rice importin α. In all other cases, mammalian importin α was co-crystallized with NLSs.
Atomic Structure of HCMV S-terminase NLS Bound to Importin α1
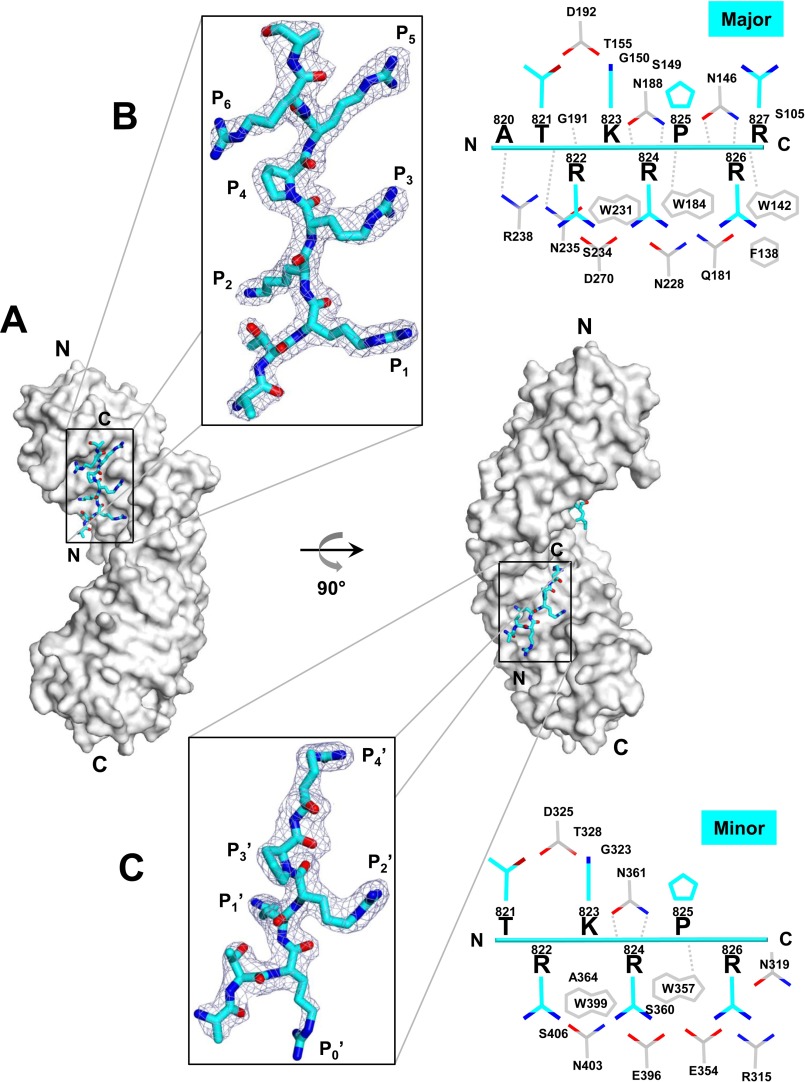
HCMV S-terminase (pUL56) NLS used for crystallization with importin α was longer than pUL15 NLS (16-mer versus 12-mer) and contained several basic residues clustered in three boxes (VSRRVRATRKRPRRAS) separated by a few residues (Fig. 1A). At 1.95 Å resolution, the crystal structure of importin α:pUL56 NLS complex revealed the peptide binds importin α like a monopartite NLS with different occupancy at major and minor binding pockets (Fig. 3A). Eight residues (821TRKRPRRA828) have clear electron density at the major site (Fig. 3B), whereas only six residues are seen clearly (821TRKRPR826) at the minor site (Fig. 3C). No density is observed for the N-terminal box 814VSRRVR819, which therefore is not part of this NLS. Structural alignment suggests pUL56 NLS occupies positions P1–P6 at the major binding pocket and P0′–P4′ at the minor binding pocket (Table 2). pUL56 NLS inserts a canonical Lys at position P2 (Lys-823) and an Arg (Arg-824) at position P2′. Position P4 is occupied by a Pro rather than a basic amino acid as seen in most NLSs (i.e. this residue was an alanine in HSV-1 pUL15) followed by two Arg residues at P5 and P6. The latter are not commonly found in classical NLSs. Interestingly, position P0 is occupied by a Thr (Table 2), a potential candidate for phosphorylation (34), which has been shown to affect the binding affinity for importin α depending on sequence context and importin α isoform specificity (40). At the minor NLS pocket (Fig. 3C), four basic side chains (RKRPR) contact sites P0′–P4′ as well as importin α makes three close contacts with pUL56 NLS backbone atoms (dashed lines in Fig. 3C), resulting in a stronger interaction than seen for HSV-1 pUL15 (Fig. 2C). Accordingly, the refined B-factor for pUL56 NLS at the major site is ∼56.2 Å2, comparable with importin α (54.6 Å2), and ∼72.9 Å2 at the minor site (Fig. 3B), lower than pUL15 NLS (Table 1). Overall, pUL56 NLS association with importin α is stabilized by 34 hydrogen bonds, 9 salt bridges, and total 224 non-bonded contacts (Fig. 3, B and C). The NLS peptide buries 773.4 Å2 and 511.5 Å2 of the surface areas at major and minor binding sites, respectively. All long chained basic residues of HCMV pUL56 NLS make very similar contacts with importin α backbone as reported in SV40 T-large antigen (45), except R122 at position P0′, which points in the opposite direction.
FIGURE 3.
Crystal structure of HCMV S-terminase pUL56 NLS in complex with importin α1. A, structure of ΔIBB-importin α1 (gray surface) in complex with HCMV pUL56 NLS (cyan ribbon). B and C, left panel, zoom-in window showing the final 2Fo − Fc electron density map (displayed as a cyan mesh contoured at 1.25σ above background) calculated using all reflections between 15 and 1.95 Å resolution after omitting the NLS bound at the major (B) and minor (C) NLS-binding sites of importin α. In both cases the density is overlaid to the NLS final model. Right panel, schematic diagram of the interactions between pUL56 NLS (in cyan) and importin α1 (in gray) at the major (B) and minor (C) NLS-binding sites in a distance range of 2.5–4.5 Å. Side chain nitrogen and oxygen atoms are color-colored in blue and red, respectively. Interactions between importin α side chains and the NLS main-chain atoms are shown by dashed gray lines.
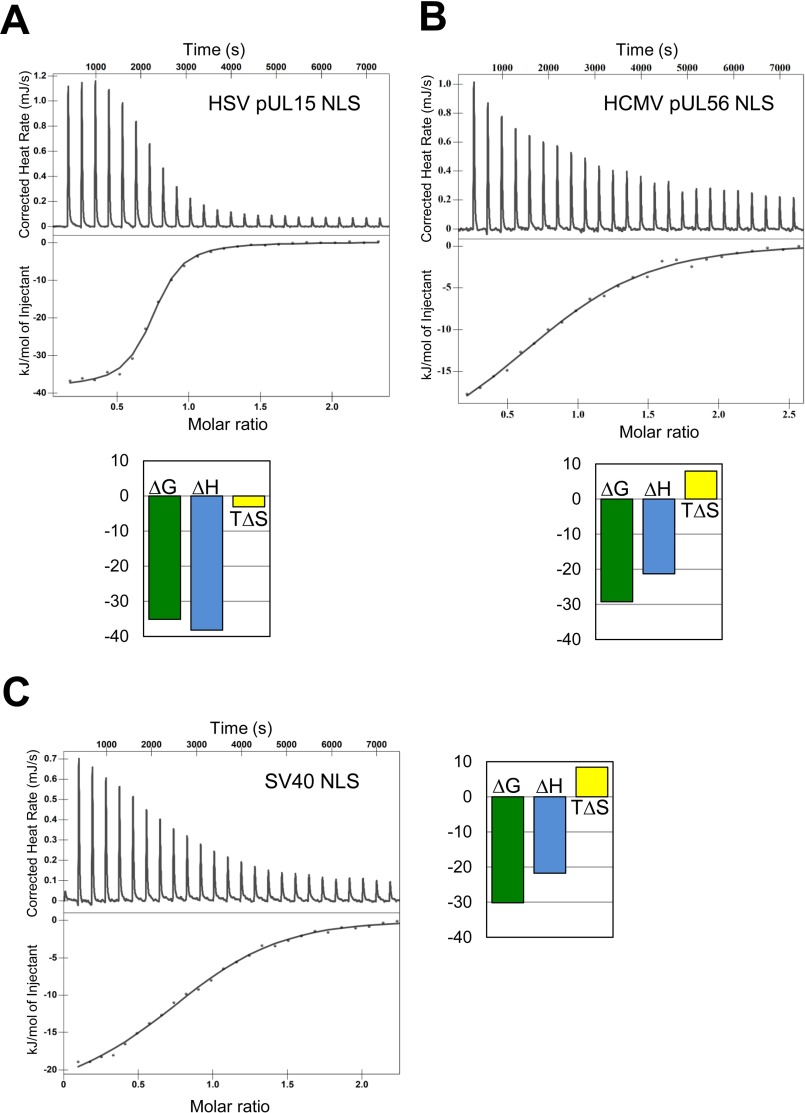
Binding Affinities of Terminase NLSs for Importin α
The intimate association of pUL15 and pUL56 NLSs with importin α1 observed crystallographically prompted us to measure their binding affinities in solution. Using nano-ITC, we measured the heat released upon titration of increasing concentrations of NLS peptide inside a cell containing purified ΔIBB-importin α. For pUL15 NLS (Fig. 4A), we observed a saturable endothermic reaction, which saturated within 11–12 injections after the NLS concentration in the cuvette was ∼50 μm. Binding data were fit using a two-independent binding site model yielding an equilibrium dissociation constant for the first site (Kd1) of 588.2 ± 10 nm (Fig. 4A) and a Kd2 ∼ 100 μm for the second site. We interpret the two binding events reflect the association of pUL15 NLS with major and minor NLS binding pockets, respectively. As suggested by the high B-factor at the minor pocket (Table 1), the contribution of this second NLS peptide to the overall heat released during titration is likely very small at 25 °C.
FIGURE 4.
Calorimetric analysis of the interaction of terminase NLSs with importin α. ITC analysis of the interaction of ΔIBB-importin α (in cell) with in syringe pUL15 NLS (A), pUL56 NLS (B), and as positive control SV40 T-large antigen NLS (C). Raw data are in the top panel, and the integrated enthalpy is plotted as a function of the NLS:ΔIBB-importin α molar ratio is shown in the bottom panel. The variation of enthalpy (ΔH), entropy (TΔS), and Gibbs (TΔG) energy associated to each binding event are shown next to each panel. Each ITC experiment was repeated three times, but only one representative run is shown.
For pUL56 NLS, ITC data could be fit unambiguously to a two-independent-binding-events model that yielded an equilibrium dissociation constant (Kd1) of 9.4 ± 1.4 μm for the first site (the major NLS binding pocket) and Kd2 = 66.9 ± 8.2 μm for the second binding site (the minor NLS binding pocket) (Fig. 4B). This model is consistent with the better occupied minor NLS binding site seen in the crystal structure, which suggests the enthalpy released during the experiment is a summation of two binding events. A similar enthalpy release was measured in a control experiment where the classical SV40-NLS was injected against ΔIBB-importin α under identical experimental conditions (Fig. 4C), which gave equilibrium dissociation constants Kd1 = 1.5 ± 0.3 μm and Kd2 = 9.7 ± 1.2 μm. It should be pointed out that the equilibrium binding constants reported here are significantly lower than those measured using fluorescence depolarization (79) or surface-immobilized NLSs (80) but nonetheless consistent with published ITC studies that employed short NLS peptides (47, 75, 76, 81). This is explained by the high solubility and charge of NLS peptides and their tendency to remain in solution as solvated ions. In light of this, pUL15 NLS appears to be a stronger binder than pUL56 NLS, which is similar to the classical SV40 NLS.
Using the two thermodynamic equations ΔG = ΔH − TΔS and ΔG = −RT ln(1/Kd), we compared the thermodynamic parameters associated to NLS binding to importin α to get further insight into the mechanisms of binding. Interestingly, HSV-1 NLS association to importin α1 (Fig. 4A) involves negative values of ΔH and ΔS at the experimental temperature, indicating a balanced binding affinity based on both favorable hydrogen and van der Waals interactions and hydrophobic interactions. The entropic contribution possibly reflects the involvement of six Trp in importin α1 and two Pro in pUL15 NLS (Fig. 2, B and C). In contrast, the binding affinity of HCMV pUL56 NLS, as well as the classical SV40 NLS for importin α (Fig. 4, B and C) is based exclusively on hydrogen and van der Waals interactions (ΔH < 0) but is accompanied by unfavorable entropy changes (ΔS > 0), possibly due to conformational effects.
Comparative Analysis of Herpesvirus NLSs in Terminase Subunits
Next, we explored the evolutionary conservation of terminase NLSs in members of the herpesvirus superfamily. Sixty-eight herpesvirus genomes have been sequenced and annotated and are available for analysis through the NIAID Virus Pathogen Database and Analysis Resource (56). We identified the genes encoding L-, S-, and T-terminase in each available genome and generated a global alignment of all sequences grouped in α, β, and γ subfamilies and further divided in genera according to established taxonomic criteria (82). Strikingly, all three terminase subunits are remarkably conserved among subfamilies with sequence identity exceeding 60% for L-terminase and close to 50% for T-terminase, which is the most divergent (Table 3).
TABLE 3.
Evolutionary conservation of terminase subunits expressed as degree of aminoacid identity and similarity in different genera of the herpesvirus-superfamily
Human terminase subunits from HSV-1, HHV-5 and HHV-4 were used as reference sequences for the α-, β- and γ-subfamilies and to calculate percent identity/similarity among different genera of each subfamily.
| Herpesvirus subfamilies | Genera | Large terminase subunit | Small terminase subunit | Third terminase subunit |
|---|---|---|---|---|
| α Subfamily | Iltovirus | 48.9/57.7 | 36.7/51.3 | 41.1/74.3 |
| Mardivirus | 59.1/70.0 | 48.0/62.7 | 41.6/74.8 | |
| Simplexvirus | 89.2/92.1 | 80.0/85.1 | 70.8/86.6 | |
| Vericellovirus | 64.2/72.3 | 50.9/64.6 | 43.0/71.6 | |
| Scutavirus | 57.2/63.2 | 36.4/54.6 | 47.0/76.0 | |
| Average identity/similarity | 63.7/71.0 | 50.4/63.6 | 48.7/76.7 | |
| β Subfamily | Cytomegalovirus | 88.3/92.2 | 76.2/78.3 | 67.2/72.4 |
| Muromegalovirus | 65.8/75.6 | 53.4/62.2 | 41.1/54.2 | |
| Proboscivirus | 48.1/60.9 | 36.0/46.4 | 36.8/55.8 | |
| Roseolovirus | 55.6/67.3 | 42.7/52.9 | 42.4/59.1 | |
| Average identity/similarity | 64.5/74.0 | 52.1/60.0 | 46.9/60.4 | |
| γ Subfamily | Lymphocryptovirus | 76.28/85.5 | 78.9/83.6 | 100/100 |
| Macavirus | 50.1/60.7 | 36.7/44.6 | 30.3/34.6 | |
| Rhadinovirus | 49.5/62.2 | 38.6/43.3 | 29.9/33.1 | |
| Percavirus | 52.3/63.9 | 37.5/45.0 | 31.2/35.6 | |
| Average identity/similarity | 57.1/68.1 | 47.9/54.1 | 47.9/67.8 |
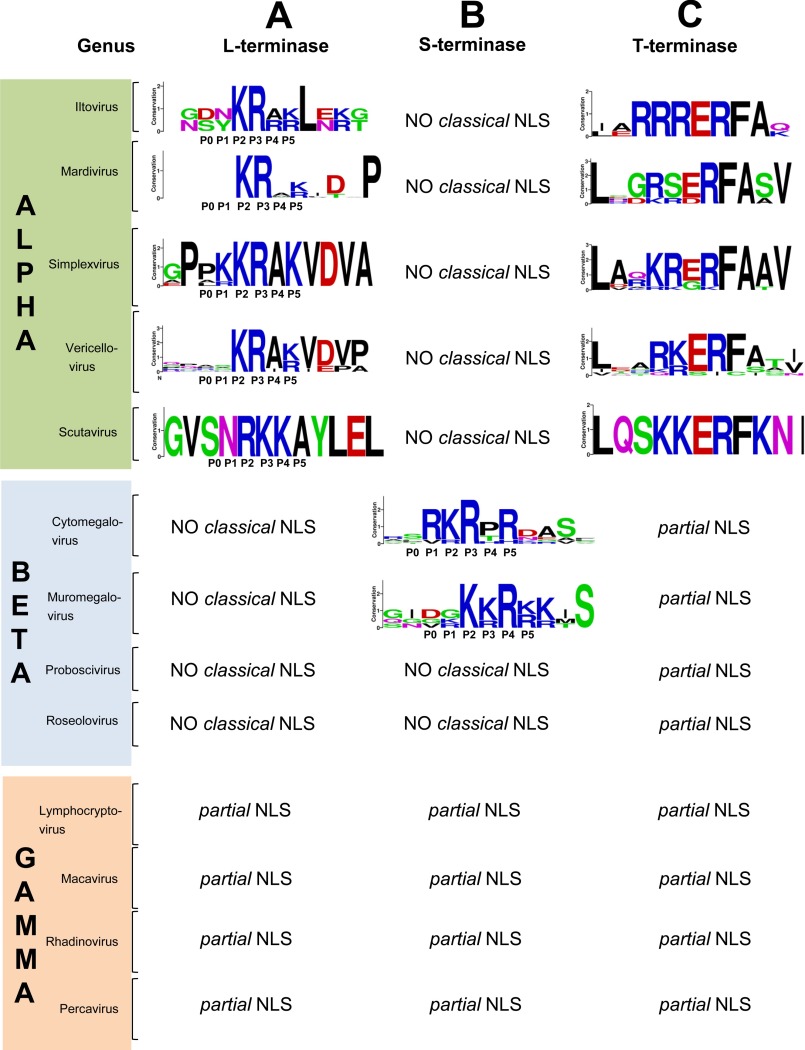
To investigate the conservation of NLS sequences in terminase subunits, we focused on the regions of L- and S-terminase that contain a functional NLS and generated a local alignment around pUL15 and pUL56 NLSs, which was displayed with WebLogo (59) for each genus of the three subfamilies. For L-terminase (Fig. 5A), we found HSV-1 pUL15 NLS is conserved not only in viruses of the Simplexvirus genus, which have very high sequence identity (76–95%), but also in genera with relatively lower percentages of sequence identity/similarity such as Varicellovirus (Table 3), supporting the idea of a strong evolutionary conservation among α-herpesviruses. Interestingly, critical Lys and Arg at positions P2/P3 are conserved in all four major genera of α-herpesviruses. The only exception is perhaps the Scutavirus genus that includes only two species, the testudinid herpesvirus 3 and chelonid herpesvirus 5. Testudinid herpesvirus 3 has an inverted RK motif, whereas a putative L-terminase was not found in chelonid herpesvirus 5. Likewise, a systematic search for pUL15-like NLS in the L-terminase subunit of β- and γ-herpesviruses failed to identify a sequence consistent with a classical import signal (Fig. 5A).
FIGURE 5.
Conservation of NLS sequences in herpesvirus terminase subunits. Conserved NLS sequences for L-terminase (A), S-terminase (B), and T-terminase (C) displayed using WebLogo (59). For each genus conserved amino acids in the NLS are represented by a stack of letters. The height of the stack (measured in bits) reflects the degree of sequence conservation at the corresponding position, and the height of each letter represents the relative frequency of the amino acids at the corresponding location.
For S-terminase (Fig. 5B), global alignment of β-herpesvirus sequences revealed a pUL56-like NLS is well conserved in Cytomegalovirus and Muromegalovirus genera but surprisingly absent in Roseolovirus and Proboscivirus despite high sequence identity (>48%) and similarity >60% (Table 3). The latter viruses have truncated S-terminases that lack the C-terminal moiety harboring the NLS. Finally, a pUL56-like NLS was not found in members of the α and γ subfamilies (Fig. 5B), consistent with the cytoplasmic localization of S-terminase from HSV-1 (61, 83) and Kaposi's sarcoma-associated herpesvirus (84), a prototypical γ-herpesvirus. Thus, the NLS sequence of herpesvirus L- and S-terminase subunits has diverged more rapidly than the rest of the amino acid sequence of these conserved gene products.
Prediction of Classical NLSs in Terminase Subunits
To determine if loss of pUL15-like and pUL56-like NLSs could be compensated by new NLSs somewhere else in the tripartite terminase complex, we probed the amino acid sequence of L- and S-terminases from γ-herpesviruses using NLS mapper (60). We predicted two putative NLSs spanning residues 278–284 of L-terminase and 429–446 of S-terminase (using Kaposi's sarcoma-associated herpesvirus terminases as a reference). Weblogo representation revealed conservation of two basic residues in L-terminase, possibly indicative of a partial (or weak) monopartite NLS (Fig. 6A). In contrast, the predicted NLS of γ-herpesvirus S-terminase contains two small basic patches separated by 10–12 residues similar to a bipartite NLS (45, 46) (Fig. 6B). Structural modeling suggests these putative NLSs fall in the ATPase domain of L-terminase (named ORF29), close to pUL15-like NLS, and on the outer surface of pUL56 paralog (named ORF7).
FIGURE 6.
Prediction of partial NLS sequences in herpesvirus terminase subunits. Predicted basic sequences similar to partial NLSs in L-terminase (A), S-terminase (B), and T-terminase (C) displayed using WebLogo (59).
We also investigated the existence and conservation of a classical NLS in the T-terminase subunit that forms a cytoplasmic complex with S- and L-terminase (26). Using HSV1 pUL33 as prototypical T-terminase sequence, NLS mapper (60) identified a classical monopartite NLS at the C terminus of the protein spanning residues 107–117. Global alignment revealed this NLS is well conserved in α-herpesviruses (Fig. 5C) with 3–4 conserved Arg/Lys clustered together as in classical NLSs. Although NLS mapper (60) failed to identify this same NLS in T-terminase from β- and γ-herpesviruses, a global sequence alignment against pUL33 revealed partial conservation of pUL33 NLS in other herpesviruses (Fig. 6C). This putative NLS is only “partial” in β- and γ-herpesviruses, where it contains two basic residues scattered >3–5 amino acids (Fig. 6C) similar to the partial NLSs found in γ-herpesvirus L- and S-terminase subunits (Fig. 6, A and B).
Discussion
In this paper we investigated the structure and conservation of import signals in herpesvirus terminase subunits. Structural analysis of pUL15 and pUL56 NLSs in complex with importin α revealed both terminases use a classical monopartite NLS, bound predominantly to the major NLS-binding site of importin α. The higher B-factor of the NLS peptide bound at the minor NLS binding pocket, also observed for SV40 T-large antigen NLS (45, 46), argues against the physiological significance of this binding site that is not likely occupied when the NLS is in the context of a full-length cargo. In analogy to classical NLS-cargos, terminase NLSs present a conserved Lys at position P2, and their nuclear import is disrupted by a point mutation at this position (85, 86). This was validated by introducing Ala substitutions at P1 and P2 of pUL56 that abolished nuclear import of a reporter protein consisting of GFP-β-galactosidase fused to pUL56 NLS (62).
Conservation of NLSs in Terminase Subunits
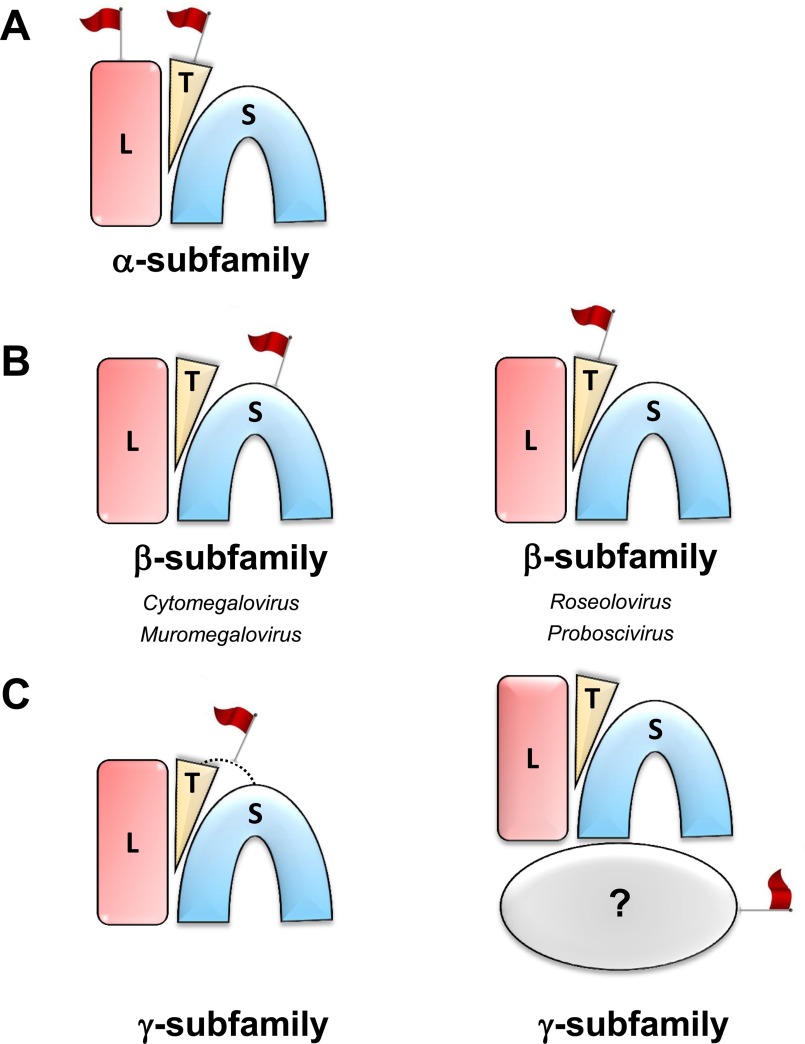
All herpesviruses contain a set of 41 core ortholog proteins (87) that include L-, S-, and T-terminase. Together these three factors form a complex that was purified from HSV-1-infected cells (26) and identified using immunoblot analysis in cells infected with HCMV (88). Growing evidence supports a model where the terminase complex assembles in the cytoplasm, and its translocation to the nucleus is dependent on L-terminase NLS (61, 83). Our bioinformatic analysis of pUL15 NLS suggests this scenario is certainly possible for α-herpesviruses (Fig. 7A), where pUL15 harbors a strong NLS but is unlikely for β- and γ-herpesviruses that lack an NLS in L-terminase. We also predicted a putative classical NLS in T-terminase, which has not been functionally validated. We speculate a second NLS in the tripartite terminase complex could synergize with pUL15 NLS to provide a kinetic advantage to the nuclear import reaction. Unlike α-herpesviruses, two genera of the β-subfamily (Cytomegalovirus and Muromegalovirus) have a strong NLS in the S-terminase subunit that is expected to substitute for the lack of a pUL15-like NLS in L-terminase (Fig. 7B). The conundrum is how the tripartite terminase complex of Proboscivirus and Roseolovirus (both β-herpesviruses) and γ-herpesviruses that lack both pUL15-like and pUL56-like NLSs can be imported in the nucleus without a “proper” import signal. None of the putative NLSs predicted in L-, S-, and T-terminase (Fig. 6) appeared strong enough to promote nuclear import of the terminase complex, which exceeds 200 kDa assuming a 1:1:1 stoichiometry (26). We envision three ways by which these herpesviruses could import the terminase complex (Fig. 7C). First, γ-herpesviruses have a partial pUL56-like NLS in S-terminase (Fig. 6B) and a weak NLS-like sequence in T-terminase (Fig. 6C). Two partial NLSs could generate a functional NLS in trans that becomes exposed and signals only in the quaternary structure of the terminase assembly. Trans-NLSs generated by complementation of partially basic sequences have been explored and validated for the dimeric transcription factor STAT1, which is imported only upon Tyr-phosphorylation (81). Second, the terminase complex of γ-herpesviruses may interact in the cytoplasm of infected cells with a yet unknown factor harboring a strong NLS, sufficient to translocate the entire complex in the nucleus. Finally, we cannot rule out these viruses contain other non-classical NLSs in one or more subunits of the terminase complex, impossible to predict with current algorithms, that are necessary and sufficient for nuclear import of the terminase complex.
FIGURE 7.
Divergent evolution of NLS sequences in herpesvirus terminase subunits. Schematic depiction of the tripartite terminase complex assuming a 1:1:1 stoichiometry of L-, S-, and T-terminase subunits. The NLS is shown as a red flag. A, α-herpesviruses use a strong NLS in L-terminase, possibly aided by an NLS in T-terminase. B, a subset of the β-subfamily uses a strong NLS in S-terminase (left panel) and possibly an NLS in T-terminase (right panel). C, γ-herpesviruses may use three strategies: either (left panel) partial NLSs in T- and S-terminase complement in trans to generate a functional NLS or a fourth factor carrying a functional NLS binds the terminase complex in the cytoplasm and promotes its nuclear translocation. Finally (not shown) one or more non-classical NLSs may be present somewhere else in the terminase complex.
Evolution of Import Signals in Herpesviridae
Genome packaging is essential to the herpesvirus replications cycle, and all herpesviruses replicate in the cell nucleus. Together, these two functional constraints on herpesvirus biology have likely driven the evolution of these viruses to maintain the tripartite terminase complex both active and localized in the nucleus. This idea is supported by the strong conservation of terminase subunits in diverse members of the herpesvirus superfamily that exceeded 50% sequence identity (Table 3). Likewise, critical motifs associated with essential enzymatic function such as the Walker A (258VPRRHGKT265) and Walker B (352LLFVDE357) motifs in L-terminase (essential for ATP binding/hydrolysis) and a putative zinc finger motif at the N terminus of S-terminase likely implicated in DNA binding (89) (Fig. 1A) are 100% conserved in all herpesviruses, underscoring the strong evolutionary pressure to maintain these gene products active in DNA packaging. Unexpectedly, although the terminase complex functions strictly in the cell nucleus, in this paper we demonstrate that terminase subunit NLSs are not conserved among herpesviruses but vary significantly even in relatively similar genera of the same subfamily (Fig. 5B). There appear to have been significant loss of NLS-function during evolution, possibly compensated by gain of NLSs in other subunits, a process that we will refer to as “NLS-swapping.” How does this observation correlate with what is known about conservation of genes in the herpesvirus superfamily? In general terms, there are three ways by which herpesvirus-common proteins evolve and diversify (90). Certain factors preserve common function as well as sequence homology. This is perhaps the case of the Walker A/B motifs in L-terminase and the putative zinc finger in S-terminase (Fig. 1A), which are absolutely invariant in all 68 herpesvirus genomes analyzed here. In other cases the function has been retained yet with only limited sequence homology. This is especially true for structural components that have fewer constraints on their structure than enzymes and tend to diverge in sequence conserving a similar three-dimensional organization (91). Finally, there are examples of genes that retain high sequence homology while acquiring distinct function. For instance, the large subunit of ribonucleotide reductase (e.g. HSV-1 pUL39), which forms an active enzyme with a small subunit in α- and γ-herpesviruses, lacks enzymatic activity in β-herpesviruses despite being conserved. Our analysis of NLS conservation in terminase subunits suggests a fourth way to maintain a critical function in herpesviruses, by swapping NLSs among different subunits of the terminase complex. The divergent evolution of NLSs in terminase subunits likely followed the evolution of the host import machinery. With two structurally distinct and functionally independent NLS-binding sites, importin α is perfectly suited to accommodate partial NLSs exposed at the surface of a terminase oligomer. In this scenario, the cumulative avidity of a terminase tripartite complex NLS for importin α would be the product of the Kd values of each NLS for importin α (92), suggesting that even weak NLSs, not functional on their own, could become active if simultaneously bound to one equivalent of importin α. The same would not hold true if importin α could associate with only one NLS at a time. Thus, the divergent evolution of herpesvirus terminase NLSs may reflect a highly specialized mode of virus:host adaptation meant at regulating the affinity of the terminase complex for importin α and the kinetics of terminase translocation into the cell nucleus. We propose that by adjusting the number, strength, and synergy of NLSs in different subunits of the terminase complex, herpesviruses regulate the kinetic at which the terminase complex is translocated into the cell nucleus and becomes available for genome-packaging.
Author Contributions
R. S. S. purified and crystallized importin α with NLS-peptides, determined x-ray structures in Figs. 2 and 3, and carried out all bioinformatics in Fig. 5. R. K. L. carried out all ITC studies in Fig. 4. G. C. conceived and coordinated the study, collected x-ray data, and wrote the paper with the help of other authors. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We are grateful to the beamline staff at APS LS-CAT 21-ID-G/F stations and SSRL 14-1 station for beamtime and assistance in data collection.
This work was supported by National Institutes of Health Grant R01GM100888 (to G. C.). Research in this publication includes work carried out at the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interaction Facility, which is supported in part by National Institutes of Health Grant P30 CA56036 (NCI). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5HUW and 5HUY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- S-terminase
- small terminase
- L-terminase
- large terminase
- T-terminase
- third terminase
- HCMV
- human cytomegalovirus
- ITC
- isothermal titration calorimetry
- NLS
- nuclear localization signal
- NPC
- nuclear pore complex
- IBB
- importin β binding
- Arm
- Armadillo.
References
- 1. McGeoch D. J., Rixon F. J., and Davison A. J. (2006) Topics in herpesvirus genomics and evolution. Virus Res. 117, 90–104 [DOI] [PubMed] [Google Scholar]
- 2. Brown J. C., McVoy M. A., and Homa F. L. (2002) Packaging DNA into herpesvirus. In Structure-Function Relationships of Human Pathogenic Viruses (Holzenburg A., and Bogner E., eds) pp. 111–153, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 3. Brown J. C., and Newcomb W. W. (2011) Herpesvirus capsid assembly: insights from structural analysis. Curr. Opin. Virol. 1, 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baines J. D., and Weller S. K. (2005) Cleavage and packaging of herpes simplex virus 1 DNA. In Viral Genome Packaging Machines: Genetics, Structure, and Mechanism (Catalano C. E., ed.) pp. 135–150, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 5. Trus B. L., Cheng N., Newcomb W. W., Homa F. L., Brown J. C., and Steven A. C. (2004) Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78, 12668–12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olia A. S., Prevelige P. E. Jr., Johnson J. E., and Cingolani G. (2011) Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 18, 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rao V. B., and Feiss M. (2008) The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42, 647–681 [DOI] [PubMed] [Google Scholar]
- 8. Casjens S. R. (2011) The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 9, 647–657 [DOI] [PubMed] [Google Scholar]
- 9. Guo P., Zhao Z., Haak J., Wang S., Wu D., Meng B., and Weitao T. (2014) Common mechanisms of DNA translocation motors in bacteria and viruses using one-way revolution mechanism without rotation. Biotechnol. Adv. 32, 853–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhardwaj A., Olia A. S., and Cingolani G. (2014) Architecture of viral genome-delivery molecular machines. Curr. Opin. Struct. Biol. 25, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ostapchuk P., and Hearing P. (2005) Control of adenovirus packaging. J. Cell Biochem. 96, 25–35 [DOI] [PubMed] [Google Scholar]
- 12. McNulty R., Lokareddy R. K., Roy A., Yang Y., Lander G. C., Heck A. J., Johnson J. E., and Cingolani G. (2015) Architecture of the complex formed by large and small terminase subunits from bacteriophage P22. J. Mol. Biol. 427, 3285–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun S., Kondabagil K., Draper B., Alam T. I., Bowman V. D., Zhang Z., Hegde S., Fokine A., Rossmann M. G., and Rao V. B. (2008) The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 14. Liu S., Chistol G., Hetherington C. L., Tafoya S., Aathavan K., Schnitzbauer J., Grimes S., Jardine P. J., and Bustamante C. (2014) A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 157, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu D., and Weller S. K. (1998) Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72, 7428–7439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Champier G., Hantz S., Couvreux A., Stuppfler S., Mazeron M. C., Bouaziz S., Denis F., and Alain S. (2007) New functional domains of human cytomegalovirus pUL89 predicted by sequence analysis and three-dimensional modelling of the catalytic site DEXDc. Antivir. Ther. 12, 217–232 [PubMed] [Google Scholar]
- 17. White C. A., Stow N. D., Patel A. H., Hughes M., and Preston V. G. (2003) Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77, 6351–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dittmer A., Drach J. C., Townsend L. B., Fischer A., and Bogner E. (2005) Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-d-ribonucleosides. J. Virol. 79, 14660–14667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Addison C., Rixon F. J., and Preston V. G. (1990) Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71, 2377–2384 [DOI] [PubMed] [Google Scholar]
- 20. Adelman K., Salmon B., and Baines J. D. (2001) Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. U.S.A. 98, 3086–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bogner E., Reschke M., Reis B., Mockenhaupt T., and Radsak K. (1993) Identification of the gene product encoded by ORF UL56 of the human cytomegalovirus genome. Virology 196, 290–293 [DOI] [PubMed] [Google Scholar]
- 22. Bogner E., Radsak K., and Stinski M. F. (1998) The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72, 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baumann R. G., and Black L. W. (2003) Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J. Biol. Chem. 278, 4618–4627 [DOI] [PubMed] [Google Scholar]
- 24. Leffers G., and Rao V. B. (2000) Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 275, 37127–37136 [DOI] [PubMed] [Google Scholar]
- 25. Roy A., Bhardwaj A., Datta P., Lander G. C., and Cingolani G. (2012) Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure 20, 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heming J. D., Huffman J. B., Jones L. M., and Homa F. L. (2014) Isolation and characterization of the herpes simplex virus 1 terminase complex. J. Virol. 88, 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nadal M., Mas P. J., Mas P. J., Blanco A. G., Arnan C., Solà M., Hart D. J., and Coll M. (2010) Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc. Natl. Acad. Sci. U.S.A. 107, 16078–16083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selvarajan Sigamani S., Zhao H., Kamau Y. N., Baines J. D., and Tang L. (2013) The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J. Virol. 87, 7140–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roy A., and Cingolani G. (2012) Structure of p22 headful packaging nuclease. J. Biol. Chem. 287, 28196–28205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smits C., Chechik M., Kovalevskiy O. V., Shevtsov M. B., Foster A. W., Alonso J. C., and Antson A. A. (2009) Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 10, 592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao H., Christensen T. E., Kamau Y. N., and Tang L. (2013) Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc. Natl. Acad. Sci. U.S.A. 110, 8075–8080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bednenko J., Cingolani G., and Gerace L. (2003) Nucleocytoplasmic transport: navigating the channel. Traffic 4, 127–135 [DOI] [PubMed] [Google Scholar]
- 33. Cook A., Bono F., Jinek M., and Conti E. (2007) Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 [DOI] [PubMed] [Google Scholar]
- 34. Nardozzi J. D., Lott K., and Cingolani G. (2010) Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., and Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 36. Güttler T., Madl T., Neumann P., Deichsel D., Corsini L., Monecke T., Ficner R., Sattler M., and Görlich D. (2010) NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol. 17, 1367–1376 [DOI] [PubMed] [Google Scholar]
- 37. Cingolani G., Petosa C., Weis K., and Müller C. W. (1999) Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 [DOI] [PubMed] [Google Scholar]
- 38. Milles S., Mercadante D., Aramburu I. V., Jensen M. R., Banterle N., Koehler C., Tyagi S., Clarke J., Shammas S. L., Blackledge M., Gräter F., and Lemke E. A. (2015) Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 163, 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cingolani G., Bednenko J., Gillespie M. T., and Gerace L. (2002) Molecular basis for the recognition of a nonclassical nuclear localization signal by importin β. Mol. Cell 10, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 40. Pumroy R. A., and Cingolani G. (2015) Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem. J. 466, 13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lott K., Bhardwaj A., Mitrousis G., Pante N., and Cingolani G. (2010) The importin β binding domain modulates the avidity of importin β for the nuclear pore complex. J. Biol. Chem. 285, 13769–13780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lott K., and Cingolani G. (2011) The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813, 1578–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pumroy R. A., Ke S., Hart D. J., Zachariae U., and Cingolani G. (2015) Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7. Structure 23, 374–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Conti E., Uy M., Leighton L., Blobel G., and Kuriyan J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 45. Fontes M. R., Teh T., and Kobe B. (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 46. Conti E., and Kuriyan J. (2000) Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure 8, 329–338 [DOI] [PubMed] [Google Scholar]
- 47. Lott K., Bhardwaj A., Sims P. J., and Cingolani G. (2011) A minimal nuclear localization signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin α1. J. Biol. Chem. 286, 28160–28169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 49. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 51. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 52. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 53. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.8, Schrodinger, LLC, New York [Google Scholar]
- 54. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 55. Roy A., Kucukural A., and Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pickett B. E., Sadat E. L., Zhang Y., Noronha J. M., Squires R. B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., Zhou L., Larson C. N., Dietrich J., Klem E. B., and Scheuermann R. H. (2012) ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 40, D593–D598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M., Buso N., Cowley A. P., and Lopez R. (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kosugi S., Hasebe M., Tomita M., and Yanagawa H. (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang K., Homa F., and Baines J. D. (2007) Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 81, 6419–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Giesen K., Radsak K., and Bogner E. (2000) The potential terminase subunit of human cytomegalovirus, pUL56, is translocated into the nucleus by its own nuclear localization signal and interacts with importin α. J. Gen. Virol. 81, 2231–2244 [DOI] [PubMed] [Google Scholar]
- 63. Sun S., Kondabagil K., Gentz P. M., Rossmann M. G., and Rao V. B. (2007) The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell 25, 943–949 [DOI] [PubMed] [Google Scholar]
- 64. de Beer T., Fang J., Ortega M., Yang Q., Maes L., Duffy C., Berton N., Sippy J., Overduin M., Feiss M., and Catalano C. E. (2002) Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell 9, 981–991 [DOI] [PubMed] [Google Scholar]
- 65. Zhao H., Finch C. J., Sequeira R. D., Johnson B. A., Johnson J. E., Casjens S. R., and Tang L. (2010) Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc. Natl. Acad. Sci. U.S.A. 107, 1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sun S., Gao S., Kondabagil K., Xiang Y., Rossmann M. G., and Rao V. B. (2012) Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 109, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Büttner C. R., Chechik M., Ortiz-Lombardía M., Smits C., Ebong I. O., Chechik V., Jeschke G., Dykeman E., Benini S., Robinson C. V., Alonso J. C., and Antson A. A. (2012) Structural basis for DNA recognition and loading into a viral packaging motor. Proc. Natl. Acad. Sci. U.S.A. 109, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Savva C. G., Holzenburg A., and Bogner E. (2004) Insights into the structure of human cytomegalovirus large terminase subunit pUL56. FEBS Lett. 563, 135–140 [DOI] [PubMed] [Google Scholar]
- 69. Jacobs M. D., and Harrison S. C. (1998) Structure of an IκBα/NF-κB complex. Cell 95, 749–758 [DOI] [PubMed] [Google Scholar]
- 70. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., and Adam S. A. (2004) Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 71. Chen M. H., Ben-Efraim I., Mitrousis G., Walker-Kopp N., Sims P. J., and Cingolani G. (2005) Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin α. J. Biol. Chem. 280, 10599–10606 [DOI] [PubMed] [Google Scholar]
- 72. Chang C. W., Couñago R. L., Williams S. J., Bodén M., and Kobe B. (2012) Crystal structure of rice importin-α and structural basis of its interaction with plant-specific nuclear localization signals. Plant Cell 24, 5074–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chang C. W., Couñago R. M., Williams S. J., Bodén M., and Kobe B. (2013) Distinctive conformation of minor site-specific nuclear localization signals bound to importin-α. Traffic 14, 1144–1154 [DOI] [PubMed] [Google Scholar]
- 74. Roman N., Christie M., Swarbrick C. M., Kobe B., and Forwood J. K. (2013) Structural characterisation of the nuclear import receptor importin α in complex with the bipartite NLS of Prp20. PLoS ONE 8, e82038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lokareddy R. K., Hapsari R. A., van Rheenen M., Pumroy R. A., Bhardwaj A., Steen A., Veenhoff L. M., and Cingolani G. (2015) Distinctive properties of the nuclear localization signals of inner nuclear membrane proteins Heh1 and Heh2. Structure 23, 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Barros A. C., Takeda A. A., Dreyer T. R., Velazquez-Campoy A., Kobe B., and Fontes M. R. (2016) Structural and calorimetric studies demonstrate that xeroderma pigmentosum type G (XPG) can be imported to the nucleus by a classical nuclear import pathway via a monopartite NLS sequence. J. Mol. Biol. 10.1016/j.jmb.2016.01.019 [DOI] [PubMed] [Google Scholar]
- 77. Giesecke A., and Stewart M. (2010) Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-α. J. Biol. Chem. 285, 17628–17635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Koerner C., Guan T., Gerace L., and Cingolani G. (2003) Synergy of silent and hot spot mutations in importin β reveals a dynamic mechanism for recognition of a nuclear localization signal. J. Biol. Chem. 278, 16216–16221 [DOI] [PubMed] [Google Scholar]
- 79. Fanara P., Hodel M. R., Corbett A. H., and Hodel A. E. (2000) Quantitative analysis of nuclear localization signal (NLS)-importin α interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J. Biol. Chem. 275, 21218–21223 [DOI] [PubMed] [Google Scholar]
- 80. Catimel B., Teh T., Fontes M. R., Jennings I. G., Jans D. A., Howlett G. J., Nice E. C., and Kobe B. (2001) Biophysical characterization of interactions involving importin-α during nuclear import. J. Biol. Chem. 276, 34189–34198 [DOI] [PubMed] [Google Scholar]
- 81. Nardozzi J., Wenta N., Yasuhara N., Vinkemeier U., and Cingolani G. (2010) Molecular basis for the recognition of phosphorylated STAT1 by importin α5. J. Mol. Biol. 402, 83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Davison A. J. (2007) Overview of classification. In Source Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. (Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., Yamanishi K., eds) pp. 1–8, Cambridge University Press, Cambridge, UK: [PubMed] [Google Scholar]
- 83. Higgs M. R., Preston V. G., and Stow N. D. (2008) The UL15 protein of herpes simplex virus type 1 is necessary for the localization of the UL28 and UL33 proteins to viral DNA replication centres. J. Gen. Virol. 89, 1709–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Koslowski K. M., Shaver P. R., Wang X. Y., Tenney D. J., and Pederson N. E. (1997) The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71, 9118–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kalderon D., Richardson W. D., Markham A. F., and Smith A. E. (1984) Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38 [DOI] [PubMed] [Google Scholar]
- 86. Kalderon D., Roberts B. L., Richardson W. D., and Smith A. E. (1984) A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 [DOI] [PubMed] [Google Scholar]
- 87. McGeoch D. J., and Gatherer D. (2005) Integrating reptilian herpesviruses into the family herpesviridae. J. Virol. 79, 725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Borst E. M., Kleine-Albers J., Gabaev I., Babic M., Wagner K., Binz A., Degenhardt I., Kalesse M., Jonjic S., Bauerfeind R., and Messerle M. (2013) The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J. Virol. 87, 1720–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Champier G., Couvreux A., Hantz S., Rametti A., Mazeron M. C., Bouaziz S., Denis F., and Alain S. (2008) Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole d-ribonucleoside activity. Antivir. Ther. 13, 643–654 [PubMed] [Google Scholar]
- 90. Mocarski E. S., Jr. (2007) Comparative analysis of herpesvirus-common proteins. In Source Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. (Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., and Yamanishi K., eds) pp. 44–58, Cambridge University Press, Cambridge, UK: [PubMed] [Google Scholar]
- 91. Sankhala R. S., Lokareddy R. K., and Cingolani G. (2015) A greasy aid to capsid assembly: lessons from a salty virus. Structure 23, 1777–1779 [DOI] [PubMed] [Google Scholar]
- 92. Rao J., Lahiri J., Isaacs L., Weis R. M., and Whitesides G. M. (1998) A trivalent system from vancomycin d-Ala-d-Ala with higher affinity than avidin.biotin. Science 280, 708–711 [DOI] [PubMed] [Google Scholar]