Abstract
Light, in a quality- and quantity-dependent fashion, induces nuclear import of the plant photoreceptors phytochrome, promotes interaction of phytochrome A (phyA) and phyB with transcription factors including phytochrome interacting factor 3 (PIF3), and is thought to trigger a transcriptional cascade to regulate the expression of ∼2500 genes in Arabidopsis thaliana. Here, we show that controlled degradation of the transcription factor PIF3 is a major regulatory step in light signaling. We demonstrate that accumulation of PIF3 in the nucleus in dark requires constitutive photomorphogenesis 1 (COP1), a negative regulator of photomorphogenesis, and show that red (R) and far-red light (FR) induce rapid degradation of the PIF3 protein. This process is controlled by the concerted action of the R/FR absorbing phyA, phyB, and phyD photoreceptors, and it is not affected by COP1. Rapid light-induced degradation of PIF3 indicates that interaction of PIF3 with these phytochrome species is transient. In addition, we provide evidence that the poc1 mutant, a postulated PIF3 overexpressor that displays hypersensitivity to R but not to FR, lacks detectable amounts of the PIF3 protein. Thus, we propose that PIF3 acts transiently, and its major function is to mediate phytochrome-induced signaling during the developmental switch from skotomorphogenesis to photomorphogenesis and/or dark to light transitions.
INTRODUCTION
To sense the environmental factor light, plants have evolved different sensory photoreceptors (Kendrick and Kronenberg, 1994). In Arabidopsis thaliana, five members of a small gene family (PHYTOCHROME A [PHYA] to PHYE) encode the photoreceptors phytochrome (Clack et al., 1994). Phytochromes are red (R)/far-red light (FR) photoreversible chromoproteins that form dimers with a molecular mass of ∼120 kD per monomer and in which an open-chain tetrapyrrole chromophore is autocatalytically attached to the apoprotein (Lagarias and Lagarias, 1989; Eichenberg et al., 2000). R-induced formation of the FR-absorbing active form of phytochrome (Pfr) initiates a signaling cascade, which results in modulated transcription of numerous genes controlling plant photomorphogenesis.
Of these phytochromes, phyA has a very specific mode of action by controlling very low fluence responses (VLFR) and far-red high irradiance responses (HIR) (Furuya and Schäfer, 1996). VLFR is initiated even by a few seconds of starlight and is saturated at ∼1 μmol/m2, whereas HIR requires prolonged irradiations with continuous far-red light (cFR). In contrast with phyA, phyB to phyE mediate responses to continuous red light (cR) and show the R/FR reversible induction responses.
In dark-grown seedlings, phyA is exclusively localized, whereas phyB to phyE are predominantly localized in the cytosol. After irradiation, phyA to phyE are translocated into the nucleus in a light quality- and quantity-dependent manner: phyA requires either VLFR or HIR treatments (Kircher et al., 1999; Kim et al., 2000), whereas phyB to phyE need irradiation with R (Gil et al., 2000; Kircher et al., 2002). Induction of the nuclear import of phyB is mediated by R and reverted by subsequent pulses of FR (Kircher et al., 1999). It has also been established that the majority of phy:GFP (green fluorescent protein) fusions in the nucleus are not distributed randomly but converge at subnuclear foci (Yamaguchi et al., 1999; Kircher et al., 2002).
Over the past few years, several genes potentially involved in phyA- and phyB-controlled signal transduction have been identified. Most of these genes also encode proteins, which are localized in the nucleus. One of the best characterized of these proteins is phytochrome interacting factor 3 (PIF3), a transcription factor interacting with phyA and phyB. PIF3 is a basic helix-loop-helix (bHLH) protein (Ni et al., 1998); it interacts in vitro with phyA and phyB in a conformation-specific manner (Ni et al., 1999). PIF3 binds specifically to a cis-acting regulatory element (G-box) in the promoters of a variety of phytochrome-responsive genes. Simultaneous binding of PIF3 to promoters of light-responsive genes and to the Pfr form of phyB described by Martinez-Garcia et al. (2000) indicates that PIF3 recruits phyB to the promoters of actively transcribed genes. Moreover, it was shown that PIF3 can heterodimerize with other transcription factors, such as HFR1 (Fairchild et al., 2000) and PIF4 (Huq and Quail, 2002), and manipulation of PIF3 expression levels in transgenic plants resulted in phenotypes indicating altered photomorphogenesis. By characterizing transgenic plants overexpressing the N-terminal truncated form of PIF3 or antisense PIF3, the physiological role of PIF3 has been classified as a positive regulator of PHYB-mediated signal transduction (Ni et al., 1998). Features of the poc1 mutant, which displays short hypocotyl phenotype and level of PIF3 mRNA higher than those in wild-type seedlings in cR, were then interpreted as a phenotype associated with overexpression of PIF3 (Halliday et al., 1999). These observations together with the microarray analyis of phytochrome-modulated gene transcription in Arabidopsis (Tepperman et al., 2001) led to a hypothesis that (1) phytochromes, notably phyA, through PIF3 and other yet unidentified factors, regulate transcription of a master set of regulators, such as CCA1 (Wang and Tobin, 1998), LHY1 (Schaffer et al., 1998), TOC1-L, RT2, DOF, and CO (Harmer et al., 2000; Tepperman et al., 2001), and (2) these regulators then control the transcription of genes encoding functions necessary for the terminal steps of the signaling cascade. A notable exception to this model is HY5, a nuclear basic domain/leucine zipper protein (Oyama et al., 1997; Chattopadhyay et al., 1998). Although HY5 transcript levels are rapidly induced by phyA, the HY5 promoter does not contain a G-box motif; thus, transcription of HY5 is likely to be regulated by phyA through a yet unidentified pathway independent of PIF3 (Quail, 2002).
The postulated positive regulatory role of PIF3 in phyB-mediated light signal transduction, however, has been challenged recently. Kim et al. (2003) reported that T-DNA insertion mutant lines lacking a detectable amount of PIF3 mRNA displayed hypersensitivity, whereas transgenic lines overexpressing PIF3 mRNA exhibited hyposensitivity to R regarding inhibition of hypocotyl growth. By analyzing in detail additional photomorphogenic responses of these lines, Kim et al. (2003) concluded that PIF3 acts mainly as a negative regulator of phyB-induced signaling. Matsushita et al. (2003) demonstrated that a fusion protein consisting of the N-terminal part of phyB fused to β-glucuronidase followed by the nuclear localization signal of the simian virus 40 signal is capable of complementing the phyB-5 mutant that lacks detectable amounts of phyB. This fusion protein does not contain the C-terminal domain of phyB, which has been shown to be important for interaction with PIF3 (Ni et al., 1998, 1999) and other regulatory proteins (for a review, see Gyula et al., 2003). In addition, we note that interaction of PIF3 with phyB is yet to be shown in vivo, and no data were available to assess the spatial and temporal distribution and level of the PIF3 protein during signaling. To analyze the mode of PIF3 action in planta at molecular level, we produced transgenic plants that either expressed PIF3 fused to the red-shifted green fluorescent protein (rsGFP) or coexpressed the PIF3:CFP (cyano fluorescent protein) with various phy species fused to the yellow fluorescent protein (YFP). We then monitored the level and nucleocytoplasmic partitioning of PIF3 in these transgenic lines and other mutants grown under various light conditions. Microscopic and protein gel blot hybridization studies indicate that the degradation of PIF3 is controlled by the concerted action of constitutive photomorphogenesis 1 (COP1), phyA, phyB, and phyD. Finally, we provide evidence that poc1, although it displays hypersensitive response to cR but not cFR, lacks a detectable amount of PIF3 protein.
RESULTS
To analyze the intracellular localization of PIF3 in plants grown under various light conditions, we produced transgenic Arabidopsis lines expressing the PIF3:rsGFP or PIF3:CFP transgene under the control of the 35S promoter of the Cauliflower mosaic virus. Figure 1A illustrates that the selected 35S:PIF3 or 35S:PIF3:rsGFP transgenic lines (in wild-type background) showed a moderate hyposensitivity to R irradiation. This result is in contrast with the previously reported hypersensitive phenotype of transgenic lines overexpressing PIF3 (Ni et al., 1998; Halliday et al., 1999), but it is in good agreement with data reported by Kim et al. (2003). Expression levels of PIF3 and the PIF3:rsGFP fusion protein in the transgenic lines were then determined by protein gel blot hybridization using an antibody recognizing PIF3. Figures 1B and 1C show that the antibody raised against PIF3 cross-reacts with other proteins but is sufficiently specific to monitor accumulation levels of PIF3 (Figure 1B, lanes 2 and 3) and PIF3:rsGFP fusion protein (Figure 1C, lane 2). Figure 1C also illustrates that transgenic lines chosen for further studies overexpressed the PIF3 or PIF3:rsGFP proteins as compared with the wild type. We next monitored the nucleocytoplasmic distribution of PIF3:rsGFP in seedlings grown in constant dark or light after imbibition. Figure 2A demonstrates that the PIF3:rsGFP fusion protein is not detectable in seeds after imbibition. The expression of PIF3:rsGFP reaches detectable levels only when the radicle has already broken through the seed coat, mostly around day 2 after induction of germination. At later developmental stages the PIF3:rsGFP fusion protein is detected as diffuse staining exclusively in the nuclei. It is expressed in all cell types examined in etiolated seedlings (Figure 2A). By contrast, we had difficulties detecting any accumulation of the PIF3:rsGFP fusion protein in any cells of the same homozygous seedlings grown in constant light (Figure 2A). Similar data were obtained when the spatial and temporal distribution of the PIF3 protein fused to CFP was analyzed (data not shown). Although unexpected, these observations indicate that light may either regulate the expression of PIF3 at the transcriptional/posttranscriptional level or modulate the stability/turnover rate of the PIF3 protein itself.
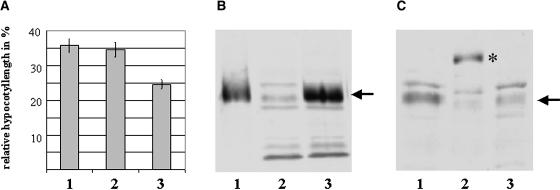
Figure 1.
Overexpression and Detection of PIF3 and PIF3:rsGFP Protein in Transgenic Plants.
(A) Transgenic seedlings overexpressing PIF3 or PIF3:rsGFP fusion protein are hyposensitive to cR. Transgenic Arabidopsis seedlings overexpressing PIF3 (1) or PIF3:rsGFP (2) and wild-type (ecotype Wassilewskija) (3) seedlings were grown for 4 d under 20 μmol/m2/s R, and inhibition of hypocotyl elongation was determined. Hypocotyl length as percentage of the corresponding dark control is shown. Each value represents an average of three independent experiments; error bars indicate the standard error of the means.
(B) The PIF3 antibody recognizes PIF3 in total plant cell extracts. The polyclonal antiserum raised against full-length recombinant PIF3 (rPIF3) cross-reacts with other proteins but recognizes the PIF3 in total plant protein extracts. Plant extracts were prepared from 6-d-old etiolated seedlings, and 2.0 ng rPIF3 and 0 μg plant protein extract (lane 1), 0 μg rPIF3 and 20 μg plant protein extract (lane 2), and 3.0 ng rPIF3 and 20 μg plant protein extract (lane 3) were subjected to protein gel blot hybridization after SDS-PAGE. The arrow indicates the position of rPIF3 and the endogenous plant PIF3 protein.
(C) Transgenic plants overexpress the PIF3 or PIF3:rsGFP fusion protein. Total protein extracts were prepared from etiolated wild-type (lane 3) and transgenic seedlings overexpressing PIF3 (lane 1) or PIF3:rsGFP fusion protein (lane 2). Accumulation of PIF3 or PIF3:rsGFP was determined by protein gel blot hybridization using the polyclonal antiserum described in Figure 1B. All lanes contain equal amounts of protein (20 μg). The arrow indicates the position of PIF3; the asterisk indicates the position of the PIF3:rsGFP fusion protein.
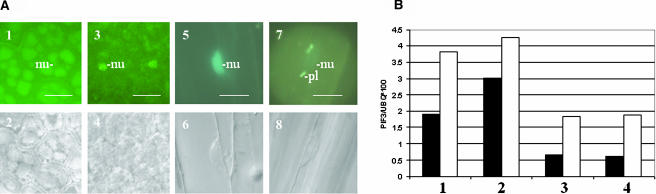
Figure 2.
Developmentally Regulated Expression of PIF3 Is Light Independent.
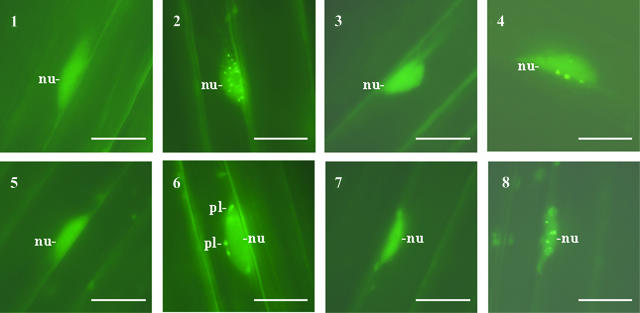
(A) Expression of 35S:PIF3:rsGFP is regulated developmentally. 35S:PIF3:rsGFP-expressing transgenic seedlings were germinated and grown in dark (1 to 6) or in 7 μmol/m2/s white light (7 and 8). Subcellular distribution of the PIF3:GFP fusion protein was monitored by fluorescence microscopy (1, 3, 5, and 7). Bright-field images of the cells analyzed are also shown (2, 4, 6, and 8). Accumulation level of PIF3:rsGFP in seeds 2 d after imbibition (1 and 2) and in seedlings grown 2 d in dark (3 and 4, cotyledon), for 4 d in dark (5 and 6, hypocotyl), or 4 d in light (7 and 8, hypocotyl) after germination was induced. Scale bars = 10 μm. Positions of nuclei (nu) and plastids (pl) are indicated.
(B) Expression of PIF3 is not regulated by light at the level of transcription. Accumulation of PIF3 and PIF3:rsGFP mRNA was determined by RNase protection experiments. Total RNA was extracted from seeds 1 d after imbibition (1), from 2-d-old (2) and 4-d-old seedlings (3) grown in dark, or from 4-d-old etiolated seedlings irradiated with 4 h of red light before harvesting (4). Closed bars represent PIF3, and open bars represent PIF3:rsGFP steady state mRNA levels normalized to the corresponding ubiquitin (UBQ) signals.
To determine whether light rapidly downregulates PIF3 expression at the level of transcription, we analyzed steady state levels of PIF3 and PIF3:rsGFP mRNA using total RNA extracts prepared from seedlings grown under different light conditions. RNase protection experiments showed that neither the expression of the endogenous PIF3 nor that of the 35S:PIF3:rsGFP transgene is rapidly downregulated by light (Figure 2B). We note, however, that levels of PIF3 and PIF3:rsGFP mRNAs are significantly higher at early stages of development.
We next examined the accumulation and subcellular distribution of the PIF3:rsGFP fusion protein in seedlings irradiated with light of different wavelengths. Thus, we monitored expression and nucleocytoplasmic distribution of PIF3:rsGFP in 6-d-old etiolated seedlings irradiated with FR. Figures 3A and 3E show that before light treatment, PIF3:rsGFP accumulates in the nuclei. Irradiation with FR strongly affected the level and cellular localization of PIF3:rsGFP. FR treatment promoted rapid formation of PIF3:rsGFP-containing speckles reminiscent of phyA or phyB speckles described by Kircher et al. (2002) and dramatically changed the distribution of PIF3:rsGFP within the nuclei (Figures 3A and 3B). More interestingly, irradiation with FR induced rapid disappearance of PIF3:rsGFP fluorescence in the cells of the wild type (Figure 3C). However, the PIF3-rsGFP protein reaccumulated in the wild type when the seedlings were kept for 6 h in darkness after a short FR treatment. In seedlings lacking phyA, the accumulation level of PIF3:rsGFP was not affected either by pulse treatment or by irradiation with cFR for at least 20 h (data not shown). Furthermore, in wild-type but not in phyA-deficient seedlings, a second FR treatment, given at any time after reaccumulation of PIF3:rsGFP reached detectable levels in the dark, induced a new cycle of speckle formation followed by the disappearance of the fusion protein from the nuclei in ∼30 min (data not shown). These observations strongly suggest that FR induces rapid degradation of PIF3 and that this process is controlled by phyA.
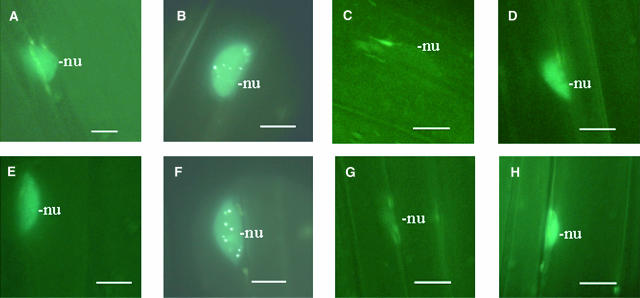
Figure 3.
Accumulation and Cellular Distribution of PIF3:CFP Is Affected by R and FR Light Treatment.
Epifluorescence images of hypocotyl cells expressing PIF3:CFP in 6-d-old dark-grown wild-type seedlings ([A] and [E]), irradiated with 2 min FR (B) or R (F) and returned afterwards to darkness for 30 min ([C] and [G]) and for 6 h ([D] and [H]). Scale bars = 10 μm. Positions of nuclei (nu) are indicated.
Short R irradiation also efficiently induced rapid formation of nuclear speckles containing PIF3:rsGFP (Figure 3F) and subsequent degradation of the fusion protein (Figure 3G). We found that within 30 min, PIF3:rsGFP disappeared from the nuclei of wild-type (Figure 3G) and phyB-deficient seedlings (data not shown), and it was also not detectable in the cytoplasm. PIF3:rsGFP, however, reaccumulated in darkness as diffuse staining in the nuclei of wild-type (Figure 3H) and phyB-deficient seedlings (data not shown) during an additional 6-h incubation. The fact that R treatment efficiently promotes degradation of PIF3 in mutant seedlings lacking phyA indicates that, besides phyA, other phytochrome species also control this response (data not shown).
This hypothesis was corroborated by the following experiments. First, a series of protein gel blot hybridization experiments was performed to determine the kinetics of the degradation of the PIF3 protein. To this end, total protein extracts were prepared from wild-type seedlings grown in the dark and treated by various light qualities. Figure 4A clearly demonstrates that PIF3 accumulates in the dark (lane 1), 2 min of R does not significantly affect the abundance of PIF3 (lane 2), and 10 min of R already decreases the level of the PIF3 protein (lane 3), whereas after 30 min, 45 min, and 6 h of R, the amount of PIF3 is below the level of detection (lanes 4, 5, and 6).
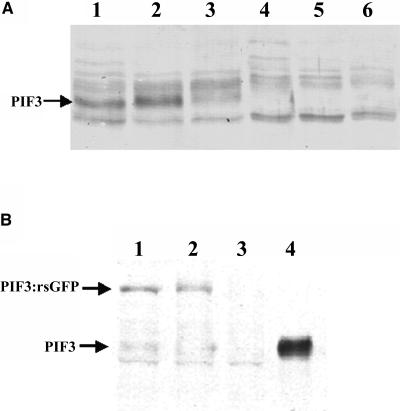
Figure 4.
Degradation of Both PIF3 and PIF3:rsGFP Is Induced by R Treatment.
Total protein extracts were prepared and the accumulation level of PIF3 was determined by protein gel blot hybridization using the antiserum raised against PIF3.
(A) Wild-type seedlings were grown for 6 d in dark (lane 1) and irradiated with R for 2 min (lane 2), 10 min (lane 3), 30 min (lane 4), 45 min (lane 5), and 6 h (lane 6). Each lane contains equal amounts of protein (30 μg). Position of the PIF3 is indicated by an arrow.
(B) Alternatively, transgenic seedlings expressing the PIF3:rsGFP were grown for 6 d in dark (lane 1) and were irradiated with R for 5 min (lane 2) or for 3 h (lane 3). Lane 4 contains 2.0 ng of recombinant PIF3 protein; lanes 1 to 3 contain equal amounts of plant total protein extract (20 μg). The bands representing PIF3 and PIF3:rsGFP are marked.
As expected, these light treatments also promoted degradation of the PIF3:rsGFP fusion protein. Figure 4B shows that accumulation of the endogenous PIF3 and PIF3:rsGFP fusion protein was readily detectable by the antibody used. Accumulation level of the PIF3:rsGFP fusion protein, similar to that of the endogenous PIF3, was the highest in etiolated transgenic seedlings (lane 1), and R treatment induced degradation of both the endogenous PIF3 and PIF3:rsGFP fusion protein. This figure also shows (lane 3) that 3 h of R irradiation was sufficient to decrease the amount even of the more highly expressed PIF3:rsGFP fusion protein below detection level. We obtained a similar degradation pattern for PIF3:CFP under these conditions (data not shown).
FR irradiation of etiolated seedlings, similarly to R treatment, also resulted in a gradual decrease of the abundance of PIF3, PIF3:rsGFP, and PIF3:CFP (data not shown).
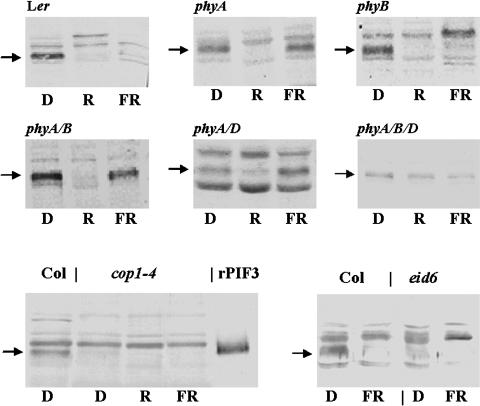
Second, another series of protein gel blot hybridizations was performed to identify the photoreceptors controlling R- and FR-induced turnover of the PIF3 protein. To this end, wild-type and mutant seedlings lacking various phytochrome species were grown in the dark and subjected to 1 h of FR or R illumination. Total protein extracts prepared from dark-grown and illuminated plant material were then used to determine the amount of PIF3 protein in these samples. Figure 5 shows that PIF3 accumulated to the same level in etiolated wild-type and various phytochrome-deficient seedlings. This figure also demonstrates that FR treatment induced rapid degradation of PIF3 in the wild type and phyB but not phyA, phyA phyB, phyA phyD, and phyA phyB phyD mutant seedlings. Thus, we concluded that the FR-induced degradation of PIF3 is controlled by phyA. In contrast with FR, R treatment induces rapid degradation of PIF3 in phyA, phyB, and phyA phyB mutant seedlings, indicating that other phy species are also involved in regulating turnover of PIF3. Analysis of the phyA phyD double mutant showed that phyD plays a role in regulating this process. Figure 5 shows that R-induced degradation of PIF3 is slower in the phyA phyD mutants than it is in the phyA phyB mutants or the wild type because a residual amount of PIF3 is still readily detectable after 1 h of R treatment. By contrast, neither R nor FR treatment affected accumulation level of PIF3 in the phyA phyB phyD triple mutant background. In this case, the amount of PIF3 detected in the total protein extract isolated from etiolated and R-illuminated seedlings did not differ after 1 h (Figure 5) or several hours of illumination (data not shown). These data demonstrate that different signaling pathways initiated by these phytochrome species mediate FR- and R-induced degradation of PIF3.
Figure 5.
FR- and R-Induced Degradation of PIF3 Is Regulated by the Concerted Action of phyA, phyB, and phyD, whereas COP1 Is Required for the Accumulation of PIF3 in Dark.
Wild-type (Landsberg erecta and Columbia), phyA-201, phyB-5, phyA-201 phyB-5, phyA-201 phyD-1, phyA-201 phyB-5 phyD-1, cop1-4, and eid6 mutant seedlings were grown for 6 d in dark and then exposed to 1 h of R or FR. Total protein extracts were prepared and PIF3 abundance was determined by protein gel blot hybridization. All lanes contain identical amounts of protein extract (20 μg). D, dark; rPIF3, recombinant PIF3. Arrows mark the positions of PIF3.
COP1 was shown to be a negative regulator of photomorphogenesis. COP1 mutants display a constitutive photomorphogenetic phenotype in the dark and show a characteristic hypersensitive phenotype in light. Genetic and biochemical evidence indicates that COP1 controls light signaling by regulating the degradation of several key transcription factors (Ma et al., 2003). This conclusion is supported by recent reports showing that COP1, acting as an E3 ligase, directs HY5 (Hardtke et al., 2000; Osterlund et al., 2000) and, in the presence of the SPA1 protein, LAF1 (Seo et al., 2003) to the proteasome. Similarly to HY5 and LAF1, PIF3 was also postulated to act as a positive regulator of photomorphogenesis. However, we found that PIF3, in contrast with HY5, accumulated efficiently in the nuclei of dark-grown seedlings and turned over rapidly upon illumination by light. These findings prompted us to examine the possible role of COP1 in mediating the light-induced proteolysis of PIF3. Quite unexpectedly we found that (1) the accumulation level of PIF3 in dark-grown cop1-4 seedlings was significantly lower than in the wild type and (2) the residual amount of PIF3 detected in dark-grown cop1-4 seedlings degraded quickly, similarly to the wild type, upon illumination by R or FR. These data, shown in Figure 5, indicate that COP1 does not play a significant role in mediating the light-induced turnover of PIF3 protein but is required for its high-level accumulation in the dark. To corroborate these findings, we monitored the accumulation of PIF3 protein in a new COP1 allele designated eid-6. In contrast with cop1-4, which displays constitutive photomorphogenic phenotype, eid-6 has no dark phenotype, yet it shows a phyB-dependent hypersensitive light phenotype (Dieterle et al., 2003). Figure 5 demonstrates that the accumulation of PIF3 in dark-grown eid-6 seedlings was significantly inhibited as compared with dark-grown wild-type seedlings, whereas the light-induced degradation of PIF3 was as rapid and complete as in wild-type seedlings. These data provide evidence that (1) COP1 promotes the accumulation of PIF3 protein in dark-grown seedlings independently of the manifestation of the characteristic COP1 constitutive photomorphogenic phenotype and (2) COP1 does not affect the light-induced turnover of the PIF3 protein. Furthermore, we conclude that a low level of PIF3 is not sufficient for the manifestation of the constitutive photomorphogenic phenotype.
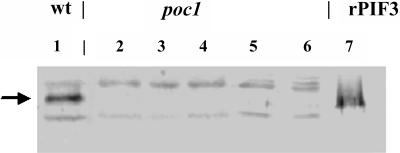
These findings prompted us to characterize the accumulation and light-induced degradation of PIF3 in the poc1 mutant. According to Halliday et al. (1999), the poc1 mutant contains a T-DNA insertion at the 5′ untranslated region of the PIF3 gene. The authors showed that poc1 seedlings display a hypersensitive hypocotyl elongation inhibition response in cR but not in cFR. Furthermore, the authors reported that poc1 seedlings grown in cR have elevated levels of PIF3 mRNA as compared with the wild type; thus, they classified the poc1 mutant as a PIF3 overexpressor. However, Kim et al. (2003) and we found that transgenic seedlings overexpressing the PIF3 mRNA exhibited hyposensitivity to R. Moreover, Kim et al. (2003) reported that PIF3 mutants lacking a detectable amount of PIF3 mRNA displayed hypersensitivity to cR concerning inhibition of hypocotyl elongation, a phenotype conspicuously similar to that of the poc1. To solve the discrepancy between these reports, we determined the accumulation level of PIF3 protein in dark- and cR-grown poc1 and wild-type seedlings. We found that total protein extracts prepared from poc1 seedlings grown in dark or cR did not contain detectable amounts of the PIF3 protein (Figure 6). These data provide evidence that the poc1 mutant is a PIF3 null mutant and that PIF3 acts negatively in signaling cascades regulating hypocotyl elongation of Arabidopsis seedlings in cR. Halliday et al. (1999) and we found that the poc1 mutation does not affect the rate of hypocotyl growth in cFR (data not shown). Thus, we conclude that PIF3, although FR can also induce its degradation, is not involved in signaling that mediates phyA-controlled hypocotyl growth.
Figure 6.
poc1 Mutant Seedlings Do Not Contain Detectable Amounts of PIF3.
Abundance of PIF3 in total protein extracts prepared from wild-type and poc1 seedlings was determined by protein gel blot hybridization. Wassilewskija wild-type (lane 1) and poc1 seedlings were germinated and grown in dark for 6 d. The 6-d-old etiolated poc1 seedlings (lane 2) were then irradiated with R for 1 h (lane 3) and 2 h (lane 4). Alternatively, 6-d-old etiolated poc1 seedlings were grown for an additional 4 d in dark (lane 5) or irradiated for 4 d with cR (lane 6). Lanes 1 to 6 contain identical amounts of plant total protein extract (20 μg), and lane 7 contains 2 ng of recombinant PIF3 (rPIF3) protein isolated from Escherichia coli. An arrow indicates the position of PIF3 and rPIF3.
PIF3 is a bHLH transcription factor, which has been shown to interact with phyA and phyB in vitro and postulated to play a key role in mediating phytochrome-controlled light signaling. This working model ascertains a sustained interaction of PIF3 with phyB and other signaling molecules. Here, however, we show that the dynamics of PIF3 degradation are astonishingly fast: the half-life for degradation is ∼10 min in R (Figure 4A). This finding is in conflict with the proposed model, and to overcome this apparent discrepancy, we analyzed (1) the level and subcellular localization of PIF3 and (2) its postulated interacting partners phyA and phyB as well as (3) that of phyD, which has been shown to be involved in R-induced degradation of PIF3 in planta. To this end, we produced transgenic Arabidopsis lines expressing YFP fusion proteins of PHYA, PHYB, or PHYD together with PIF3:CFP fusion proteins. The expression of each chimeric gene was under the control of the viral 35S promoter. Expression levels of the fusion proteins were determined by protein gel blot hybridization (data not shown).
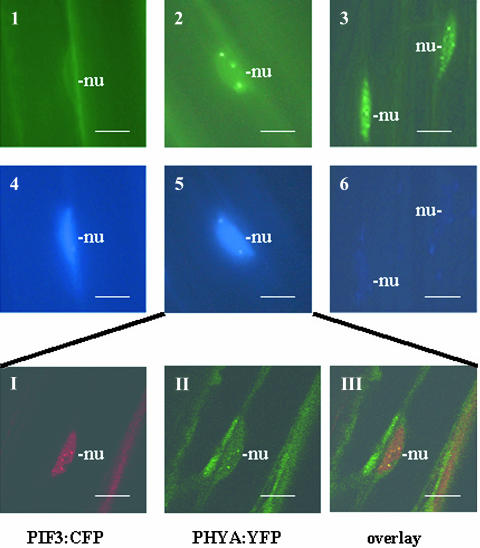
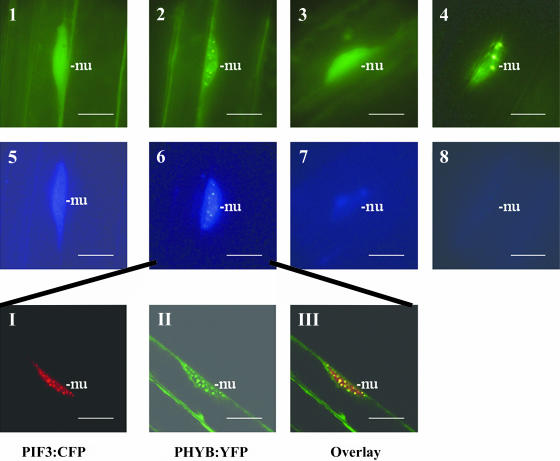
Figure 7 shows that PHYA:YFP localizes exclusively to the cytoplasm, whereas PIF3:CFP accumulates in the nucleus of etiolated seedlings. A short FR treatment induces translocation of PHYA:YFP and formation of speckles containing PHYA:YFP and PIF3:CFP (Figures 2, 5, and 7). Overlay of the microscopic images indicates that the PHYA:YFP and PIF3:CFP fusion proteins are colocalized in the nucleus (Figure 7, insert). An extended FR treatment leads to the disappearance of PIF3:CFP staining, whereas the PHYA:YFP staining first becomes diffuse before newly formed PHYA speckles appear. A short R treatment also induces translocation of PHYA to the nucleus, speckle formation, and colocalization of PHYA and PIF3 therein. Extended R treatment, however, results in the complete loss of both PHYA:YFP and PIF3:CFP fluorescence (data not shown). Analysis of transgenic plants coexpressing PHYB:YFP or PHYD:YFP with the PIF3:CFP fusion protein confirmed FR-induced degradation of PIF3. A short FR pulse induced formation of PIF3:CFP but not PHYB:YFP and PHYD:YFP containing speckles, whereas extended FR treatment led to the disappearance of PIF3:CFP without affecting the abundance of PHYB:YFP and PHYD:YFP fusion proteins (data not shown). A short R illumination of these seedlings, however, efficiently promoted the formation of PHYB:YFP, PHYD:YFP, as well as PIF3:CFP speckles. Overlay pictures of the speckles indicate that PIF3:CFP colocalizes transiently with both PHYB:YFP (Figure 8, insert) and PHYD:YFP (data not shown). An extended R treatment again triggered rapid loss of PIF3:CFP, whereas PHYB:YFP and PHYD:YFP staining first became diffuse, and then newly formed PHYB:YFP and PHYD:YFP containing speckles appeared. Figure 8 illustrates studies performed on seedlings expressing PHYB:YFP and PIF3:CFP. We note that the number and size of the PHYB (Figure 8) and PHYD:YFP speckles (data not shown) formed immediately after a short R treatment (early speckles) is characteristically different from those appearing after extended R illumination (late speckles).
Figure 7.
PIF3:CFP Is Colocalized Transiently with PHYA:YFP in Nuclei of FR Light–Irradiated Transgenic Seedlings.
Transgenic lines expressing both PHYA:YFP and PIF3:CFP in phyA-211 background were used to determine cellular distribution of the fusion proteins. Epifluorescent images of PhyA:YFP (1 to 3) and PIF3-CFP (4 to 6) are shown in 6-d-old etiolated seedlings (1 and 4) irradiated with 2 min of FR (2 and 5) and 20 h of cFR (3 and 6). The insert presents confocal microscopic images of cells expressing both fusion proteins after 2 min of FR treatment. PIF3:CFP is displayed in red (I), PHYA:YFP is shown by green (II), and overlay of the two signals is indicated by yellow (III). Positions of nuclei (nu) are indicated. Scale bars = 10 μm.
Figure 8.
PHYB:YFP Forms Early and Late Speckles.
Early PHYB:YFP speckles are colocalized transiently with PIF3:CFP speckles in the nuclei of R-irradiated transgenic seedlings. Transgenic lines expressing both PHYB:YFP and PIF3:CFP in phyB-9 background were used to determine cellular distribution of these fusion proteins. Epifluorescent images of PHYB:YFP (1 to 4) and PIF3:CFP (5 to 8) are shown in 6-d-old etiolated seedlings (1 and 5) irradiated with 2 min (2 and 6), 1 h (3 and 7), and 16 h (4 and 8) of red light. The insert shows confocal microscopic images of cells expressing both fusion proteins after 2 min of R treatment. PIF3:CFP is displayed in red (I), PHYB:YFP is shown by green (II), and overlay of the two signals is indicated by yellow (III). Positions of nuclei (nu) are indicated. Scale bars = 10 μm.
In earlier studies, Kircher et al. (2002) monitored only the formation of late PHYB:GFP and PHYD:GFP containing speckles in seedlings exposed to light. We therefore tested whether the appearance of early PHYB and PHYD speckles in the double transgenic lines was because of the elevated levels of PHYB, PHYD, and PIF3 proteins as compared with previous experiments. To this end, we generated PHYB:GFP-expressing transgenic plants in the poc1 mutant background. The kinetics of the appearance of PHYB:GFP speckles after various R treatments was then analyzed in the poc1 and the wild-type backgrounds. Plants expressing PHYB:GFP in wild-type and poc1 backgrounds at the same levels were chosen for further studies. Figure 9 demonstrates that a short R treatment, similar to the early appearance of PHYB:YFP speckles in the PIF3:CFP expressing lines, promotes formation of early and transient PHYB:GFP speckles in the wild-type background. This observation indicates that different expression levels of the PHYB:GFP or PHYB:YFP transgene cannot be accounted for in the appearance of early phyB-containing speckles in the wild-type background. By contrast, however, we failed to detect early PHYB:GFP speckles in the poc1 mutant lacking detectable amounts of PIF3 protein (Figure 9). Furthermore, we found that after extended R treatment, late PHYB:GFP speckles appeared in a similar fashion in all these transgenic plants, independently of their genetic backgrounds (Figure 9). These data strongly suggest that (1) the presence of PIF3 with phyB is critical for the formation of early PHYB:GFP but not of late PHYB:GFP speckles and (2) these two types of PHYB:GFP speckles consist of different proteins.
Figure 9.
PHYB:GFP Does Not Form Early Speckles in poc1 Seedlings.
Epifluorescent analysis of the cellular distribution of PHYB:GFP is shown in 6-d-old etiolated wild-type (1 to 4) and poc1 (5 to 8) seedlings irradiated with 2 min (2 and 6), 1 h (3 and 7), and 6 h of R (4 and 8). Positions of nuclei (nu) and plastids (pl) are indicated. Scale bars = 10 μm.
DISCUSSION
Photoperception by the five known phytochrome photoreceptors PHYA to PHYE triggers partly overlapping, partly specific intracellular pathways that culminate in modulated transcription of ∼10% of examined genes in Arabidopsis after FR treatment (Tepperman et al., 2001). Attempts to identify primary phytochrome signaling partners led to the discovery of new molecules, such as PIF3 (Ni et al., 1998), PKS1 (Fankhauser et al., 1999), NPDK2 (Choi et al., 1999), and ARR4 (Sweere et al., 2001) and also showed that phytochromes can interact with proteins required for the functional plant circadian system, such as ELF3 (Liu et al., 2001) and ADO/ZTL (Somers et al., 2000; Jarillo et al., 2001). DNA microarray analysis of the transcription of phytochrome-regulated genes provided strong support for a model that postulates that light directly regulates transcription of a master set of regulators. According to this model, G-box–bound PIF3 interacts with the Pfr conformer of phyA and/or phyB in the nucleus and directly regulates transcription of other transcription factors, such as LHY and CCA1 (Martinez-Garcia et al., 2000). Different binding affinities of PIF3 to phyA and phyB, heterodimerization of PIF3 with other bHLH-type transcription factors, and the markedly different kinetics of nuclear import and stability of the Pfr conformer of phyA and phyB were enumerated to explain the light quality- and quantity-dependent transcription of phytochrome-responsive genes (Quail, 2002).
Here, we report that accumulation of both PIF3 mRNA and protein is regulated developmentally. The level of the PIF3 mRNA gradually increases up to 2 to 3 d after germination and then declines. The PIF3 protein is not detectable in imbibed seeds; however, its accumulation increases after imbibition up to 4 d. The molecular mechanism responsible for the delayed accumulation of PIF3 protein as compared with that of PIF3 mRNA remains to be elucidated. We show that independent of its level, PIF3, when it is detectable, is localized constitutively in the nuclei of etiolated seedlings, and both the endogenous PIF3 as well as the PIF3:rsGFP and PIF3:CFP fusion proteins degrade after exposure to short pulses or continuous illumination. PIF3 degradation induced by R or FR is rapid; the half-life of PIF3 is ∼10 min in R and controlled by the concerted action of phyA, phyB, and phyD. The PIF3 protein is not detectable in the nuclei of cells exposed to light longer than 1 h. Degradation of PIF3 takes place equally fast in dark and light after the inductive light treatment, and PIF3 readily reaccumulates again to high levels in the dark. Taken together, these data indicate that the expression of PIF3 is negatively regulated by light at the level of protein degradation. Thus, we propose that PIF3 is mainly required for phytochrome signaling during the developmental transition from etiolated growth to photomorphogenesis or during the transition from dark to light.
There is little evidence that regulated proteolysis plays a role in phyB- or phyD-initiated signaling. By contrast, isolation of the EID1 gene encoding an F-box protein (Büche et al., 2000; Dieterle et al., 2001) as well as the observation that SPA1 acts as a cofactor in COP1-mediated degradation of the transcription factor LAF1 (Seo et al., 2003) provided evidence that phyA signaling is mediated, at least partly, by proteasome-related pathways. Light-induced rapid degradation of PIF3 and its reaccumulation in dark suggests that the function of PIF3, similarly to those of HY5 and LAF1, is regulated by proteolysis. There is, however, a significant difference between the modes of action of these transcriptional regulators. HY5 is targeted by COP1 to the COP9 signalosome and is degraded in dark, whereas FR induces transcription of the HY5 gene and accumulation of the HY5 protein in the nucleus. LAF1, similarly to HY5, also acts as a positive regulator of phy-controlled signaling. Signaling by LAF1 in light is attenuated by the concerted action of SPA1 and COP1. By contrast, PIF3 accumulates in dark only in the presence of COP1, and both FR and R treatments promote its degradation in a COP1-independent fashion.
These data prompted us to speculate that PIF3, although originally postulated to act as a positive regulator (Halliday et al., 1999; Martinez-Garcia et al., 2000), might regulate phyB signaling negatively, similarly to PIF4 (Huq and Quail, 2002). Here, we show that the poc1 mutant, reported to overexpress the PIF3 mRNA (Halliday et al., 1999), in fact does not contain detectable amounts of PIF3 protein either in dark or even after such extended cR irradiation that is sufficient for the manifestation of the characteristic poc1 phenotype (Figure 6, lane 6). Moreover, we found that transgenic plants overexpressing the PIF3 or PIF3:rsGFP protein (Figure 1) are moderately hyposensitive to cR. poc1 seedlings display hypersensitivity to cR but not to cFR. Halliday et al. (1999) showed by the analysis of the poc1 phyB double mutant that poc1 is epistatic to phyB. Moreover, Kim et al. (2003) reported that mutants lacking a detectable amount of PIF3 mRNA display a phenotype very similar to that of poc1. Considering these data, we conclude that (1) poc1 is a PIF3 null mutant, (2) PIF3 is a negative regulator of hypocotyl growth inhibition in cR, and (3) light-induced degradation of PIF3 represents a key regulatory step in phyB-controlled signaling.
While our work was in progress, Matsushita et al. (2003) reported that a chimeric gene containing the N-terminal domain of PHYB fused to β-glucuronidase and that the nuclear localization signal of the simian virus 40 is capable of complementing the phyB-5 mutant lacking a functional phyB photoreceptor. These authors postulated that (1) the N-terminal domain of phyB positively regulates signaling, (2) the C-terminal domain regulates translocation of phyB into the nucleus, and (3) it probably mediates the interaction of the photoreceptor with negative regulatory factors. These results together with the data presented here radically changed our view about phyB-mediated signaling. We show here that PIF3 is a negative regulator of phyB signaling, whereas all other interacting proteins except ARR4 (for a review, see Nagy and Schäfer, 2002) bind to the C-terminal domain of the photoreceptor. Thus, we should conclude that, despite recent advances, the molecular nature of phyB-initiated signaling regulating photomorphogenesis still remains elusive.
In addition, although PIF3 has been shown to interact with phyA in a conformation-dependent fashion (Ni et al., 1999), Halliday et al. (1999) and we demonstrated that inhibition of hypocotyl elongation in poc1 and wild-type seedlings is identical in cFR. By contrast, degradation of PIF3 is rapidly induced by FR and requires signaling by phyA, but FR induced degradation of PIF3 is independent of EID1, FHY3, and SPA1 proteins (data not shown). Taken together, these observations suggest that (1) phyA controls PIF3 degradation and seedling growth in FR by different signaling cascades, (2) PIF3 does not play a major role in the FR light–induced transcription cascade controlling hypocotyl elongation, and (3) phyA may affect phyB signaling by modulating the level of PIF3.
Interestingly, we found that the PIF3 protein accumulates to significantly lower levels in the dark in cop1-4 and eid6 mutants as compared with the wild type and that poc1 seedlings similar to the pif3 null mutant seedlings (Kim et al., 2003) do not exhibit a dark phenotype. These data suggest that (1) PIF3 does not play a role in establishing the characteristic cop1 phenotype, and (2) it is not required for the elevated level of transcription of light-responsive genes in the dark. Thus, we suggest that COP1 directly promotes the degradation of positive regulators such as HY5 and probably indirectly promotes the buildup of negative regulators such as PIF3 during skotomorphogenesis. Independent of the mechanism by which COP1 protects PIF3 from degradation in darkness (COP1, for example, can promote destruction of factors required for degradation of PIF3), we postulate that dual function of COP1 could be important to determine the ratio of regulatory proteins required during the early stage of photomorphogenesis. This hypothesis is in good agreement with recent genome-wide analysis of cop/det/fus mutants (Ma et al., 2003).
PIF3 was shown to interact in vitro with the full-length phyA and phyB photoreceptors in a conformation-dependent fashion (Ni et al., 1999). We observed that R but not FR pulses induced transient colocalization of PIF3 with phyB in nuclear speckles. Extended R treatment led to the disintegration of early speckles and the appearance of late phyB speckles, which differed in size and number and did not contain PIF3. Similar data were obtained by analyzing the formation of phyA and phyD containing speckles after FR and R treatments, respectively. Experiments to determine colocalization of PIF3 and phyC and phyE are in progress.
By contrast, Matsushita et al. (2003) did not observe the formation of any phyB-containing speckles in transgenic plants exhibiting active phyB phototransduction and concluded that speckle formation may not be required for phyB signaling. We found that detection of speckles is affected by a variety of factors, including light conditions, the level of tagged proteins, and the size of speckles. The fact that formation of at least two types of phyB speckles was induced by R treatment in the wild-type background and that the early ones were absent in seedlings lacking PIF3 indicates that the presence of PIF3 is essential for the detection of early phyB speckles. These data, together with observations showing that mutant versions of phyA and phyB also fail to form speckles or display aberrant speckles (Kircher et al., 2002; Yanovsky et al., 2002), lend credible support to the hypothesis that these subnuclear structures are required for or are characteristic of phytochrome signaling. Sorting out conclusively the functional relevance of the possibly many types of phyA to phyE related speckles and determining the factors influencing their appearance, however, will remain a challenging task.
METHODS
Light Sources, Plant Material, and Growth Conditions
Handling of irradiated and dark-grown seedlings under a dim-green safe light and the white, R, and FR light sources used in these studies have been described previously by Kircher et al. (2002). Transgenic plants were generated in Arabidopsis thaliana Wassilewskija lacking functional phyD (Aukerman et al., 1997), in Columbia ecotypes, and in poc1 (Halliday et al., 1999), phyA-211 (Reed et al., 1994), and phyB-9 (Reed et al., 1993) mutant backgrounds. Transgenic seeds were germinated as follows. Seeds were sown on four-layer filter paper and imbibed in water in the dark for 48 h at 4°C. Cold-treated seeds were then irradiated with 18 h of white light to induce homogeneous germination, transferred to 25°C, and grown for additional days in dark. Six-day-old dark-grown seedlings were then subjected to various light treatments as described in the text. In addition to mutants described above, the following mutant lines were used: phyA-201 (Nagatani et al., 1993), phyB-5 (Reed et al., 1993), phyA-201 phyB-5 (Reed et al., 1993), phyA201 phyD-1 and phyA-201 phyB-5 phyD-1 (kind gift from Garry Whitelam), cop1-4 (McNellis et al., 1994), and eid6 (Dieterle et al., 2003).
Recombinant DNA Technology, Construction of Chimeric Genes, and RNase Protection
The pPCV812 binary vector was modified, resulting in pPCVB812 (details available on request) in which the hygromycin cassette was replaced with the BASTA cassette. The 35S:PHYA:YFP, 35S:PHYB:YFP, and 35S:PHYD:YFP chimeric genes were constructed as described previously by Kircher et al., 2002 using the coding region of YFP (Clontech, Palo Alto, CA) instead of the GFP reporter gene.
The full-length PIF3 cDNA was reconstituted from two partial cDNA fragments isolated from a cDNA library and modified by PCR to replace its stop codon by a unique XhoI site to facilitate the construction of PIF3:rsGFP and PIF3:CFP (Clontech) chimeric genes. These chimeric genes were inserted as BamHI-SacI fragments into the pPCVB812 binary vector containing the 35S promoter and the nopalin synthase (NOS3′) terminator.
The pPCVB812 binary vector containing 35S:PHYA:YFP:NOS3′, 35S:PHYB:YFP:NOS3′, or 35S:PHYD:YFP:NOS3′ was linearized, and the fragment carrying the 35S:PIF3:CFP:NOS3′ construct was inserted to yield a new plasmid that contains the PIF3 and PHY genes fused to different reporter genes. All constructs were verified by automated sequencing. The 35S:PHYB:GFP construct was described earlier (Kircher et al., 2002). Total RNA isolation and RNase protection assays (the fragment used as a probe consisted of the 198-bp-long 3′ fragment of the PIF3 cDNA and the 165-bp-long 5′ part of the rsGFP) were performed according to Tóth et al. (2001). All DNA manipulations were performed as described by Sambrook and Russell (2001). PCR reactions were done using the ProofSprinter polymerase system (AGS, Heidelberg, Germany).
Plant Transformation Regeneration of Transgenic Arabidopsis Lines and Measurement of Hypocotyl Length
The pPCV812 and pPCVB812 binary vectors containing the selected chimeric gene(s) were transferred from Escherichia coli to Agrobacterium tumefaciens GV3101. Arabidopsis plants were transformed via the inflorescence infiltration method (Clough and Bent, 1998), and transgenic plants were selected on sterile medium containing hygromycin (15 μg/mL) or 0.005% BASTA (active constituent 20% glufosinate ammonium; Hoechst Schering AgrEvo, Berlin, Germany) and transferred to the greenhouse after 2 weeks. Selected plants were grown to maturation, and at least 20 independent lines were generated for each construct.
Hypocotyl length measurements were performed as follows: seeds were sown on four-layer filter paper and imbibed in water in dark for 48 h at 4°C. Cold-treated seeds were then irradiated with 10 h of white light and then transferred to 25°C dark for an additional 16 h. After this treatment, seedlings were grown in 20 μmol/m2/s R for 4 d. Measurement of the length of hypocotyls was performed using MetaMorph Software (Universal Imaging, Downingtown, PA).
Epifluorescent, Light, and Confocal Laser Scanning Microscopy
For epifluorescence and light microscopy, seedlings were transferred to glass slides under dim-green safelight and analyzed with an Axioskop microscope (Zeiss, Oberkochem, Germany). Excitation and detection of the different fluorophors (GFP, CFP, and YFP) were performed with GFP, CFP, and YFP filter sets (AHF Analysentechnik, Tübingen, Germany). Each experiment was repeated three times using at least five seedlings. Representative cells were documented by photography with a digital Axiocam camera system (Zeiss). Documentation of cells was performed during the first 30 s of microscopic analysis.
For laser scanning microscopy, the seedlings were also transferred to glass slides under dim-green safelight and analyzed with the LSM 510 laser scanning microscope (Zeiss). Excitation of the fluorophores was performed with argon laser lines; 458 nm for CFP and 514 nm for YFP. For simultaneous two-channel detection of CFP and YFP signals, band pass 535 to 590 IR and band pass 480 to 520 IR filters, respectively, were used. Photographs were processed for optimal presentation using the Photoshop 5.0 (Adobe Systems Europe, Edinburgh, UK) and MS Office 97 (Microsoft, Redmond, WA) software packages.
Protein Extraction, Protein Gel Blotting, Immunodetection, and Antiserum Production
Two hundred milligrams of 7-d-old dark-grown Arabidopsis seedlings were homogenized in a potter using hot extraction buffer containing 65 mM Tris-HCl, pH 7.8, 4 M urea, 5% (w/v) SDS, 14 mM 2-mercaptoethanol, 15% (v/v) glycerol, and 0.05% (w/v) bromophenol blue. The homogenate was heated for 5 min at 95°C, the resulting suspension was cleared by centrifugation (15 min at 15000g and 25°C), and the supernatant was used for further experiments. Protein assays were performed as described by Kircher et al. (1999). Twenty micrograms of crude protein extract was separated on an SDS-PAGE gel and blotted to a polyvinylidene difluoride membrane. Immunodetection of PIF3 was performed using a polyclonal antiserum as primary antibody and alkaline phosphatase–coupled anti-rabbit antiserum (Bio-Rad, Hercules, CA) as a secondary antibody. The PIF3 antiserum was produced (Eurogentec, Seraing, Belgium) against recombinant full-length PIF3 protein expressed with the pET system (Novagen, Bad Soden, Germany) and purified as described by Kircher et al. (1998).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AT1G09570 (PhyA), AT2G18790 (PhyB), AT4G16250 (PhyD), and AT1G09530 (PIF3).
Acknowledgments
Work in Freiburg, Germany was supported by the Wolfgang Paul Award to F.N. and Sonderforschungsbereich 592 and Fonds Chemical Industry grants to E.S. Work in Szeged, Hungary was supported by Országos Tudományos Kutatási Alapprogramok Grant T046710 and Howard Hughes Medical Institute Grant 55000325 to F.N.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eberhard Schäfer (eberhard.schaefer@biologie.uni-freiburg.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021568.
References
- Aukerman, M.J., Hirschfeld, M., Wester, L., Weaver, M., Clack, T., Amasino, R.M., and Sharrock, R.A. (1997). A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büche, C., Poppe, C., Schäfer, E., and Kretsch, T. (2000). eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12, 547–558. [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.H., Puente, P., Deng, X.W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.K., Soh, M.S., Shin, B., Luka, Z., Hahn, T.R., and Song, P.S. (1999). Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613. [DOI] [PubMed] [Google Scholar]
- Clack, T., Matthews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequence and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–417. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dieterle, M., Buche, C., Schafer, E., and Kretsch, T. (2003). Characterization of a novel non-constitutive photomorphogenic cop1 allele. Plant Physiol. 133, 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle, M., Zhou, Y.C., Schäfer, E., Funk, M., and Kretsch, T. (2001). EID1, an F-box protein involved in phytochrome A-specific light signalling. Genes Dev. 15, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberg, K., Baurle, I., Paulo, N., Sharrock, R.A., Rüdiger, W., and Schäfer, E. (2000). Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 470, 107–112. [DOI] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 28, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Furuya, M., and Schäfer, E. (1996). Photoperception and signalling of induction reactions by different phytochromes. Trends Plant Sci. 1, 301–307. [Google Scholar]
- Gil, P., Kircher, S., Adam, E., Bury, E., Kozma-Bognar, L., Schäfer, E., and Nagy, F. (2000). Photocontrol of subcellular partitioning of phytochrome-B:GFP fusion protein in tobacco seedlings. Plant J. 22, 135–145. [DOI] [PubMed] [Google Scholar]
- Gyula, P., Schaefer, E., and Nagy, F. (2003). Light perception and signalling in higher plants. Curr. Opin. Plant Biol. 6, 446–452. [DOI] [PubMed] [Google Scholar]
- Halliday, K.J., Hudson, M., Ni, M., Qin, M., and Quail, P.H. (1999). poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T-DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl. Acad. Sci. USA 96, 5832–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19, 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hoegenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signalling in Arabidopsis. EMBO J. 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo, J.A., Capel, J., Tang, R.H., Yang, H.Q., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410, 487–490. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.M.H., eds (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Kim, J., Yi, H., Choi, G., Shin, B., Song, P.S., and Choi, G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, L., Kircher, S., Toth, R., Adam, E., Schäfer, E., and Nagy, F. (2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22, 125–134. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Adam, E., Schäfer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is differentially regulated by light and exhibits a diurnal rhythm. Plant Cell 25, 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schäfer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Ledger, S., Hayashi, H., Weisshaar, B., Schafer, E., and Frohnmeyer, H. (1998). CPRF4a, a novel plant bZIP protein of the CPRF family: Comparative analyses of light-dependent expression, post-transcriptional regulation, nuclear import and heterodimerization. Mol. Gen. Genet. 257, 595–605. [DOI] [PubMed] [Google Scholar]
- Lagarias, J.C., and Lagarias, D.M. (1989). Selfassembly of synthetic phytochrome holoprotein in vivo. Proc. Natl. Acad. Sci. USA 86, 5778–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Fankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Zhao, H., and Deng, X.W. (2003). Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130, 969–981. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Matsushita, T., Mochizuki, N., and Nagatani, A. (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571–574. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.-W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggest functional roles for the multiple protein domains. Plant Cell 6, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, F., and Schäfer, E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53, 329–355. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wie, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Photosensory perception and signalling in plant cells: New paradigms? Curr. Opin. Cell Biol. 14, 180–188. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis thaliana disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballasteros, M.L., and Chua, N.-H. (2003). LAF1 ubiquination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 424, 995–999. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Baurle, I., Kudla, J., Nagy, F., Schäfer, E., and Harter, K. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 942, 1108–1111. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.-S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth, R., Kevei, É., Hall, A., Millar, A.J., Nagy, F., and Kozma-Bognár, L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.-Y., and Tobin, E.M. (1998). Constitutive expression of the circadian clock associated 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B:GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., Luppi, J.P., Kirchenbauer, D., Ogorodnikova, O.B., Sineshchekov, V.A., Adam, E., Kircher, S., Staneloni, R.J., Schäfer, E., Nagy, F., and Casal, J.J. (2002). Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. Plant Cell 14, 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]