Summary
Breviatea form a lineage of free living, unicellular protists, distantly related to animals and fungi1–3. This lineage emerged almost one billion years ago, when the oceanic oxygen content was low, and extant Breviatea have evolved or retained an anaerobic lifestyle4. Here we report the cultivation of Lenisia limosa, gen. et sp. nov., a newly discovered breviate colonized by relatives of animal-associated Arcobacter. Physiological experiments showed that the association of L. limosa with Arcobacter was driven by the transfer of hydrogen and was mutualistic, providing benefits to both partners. With whole genome sequencing and differential proteomics we show that an experimentally observed fitness gain of L. limosa could be explained by the activity of a so far unknown type of NAD(P)H accepting hydrogenase, which was expressed in the presence, but not in the absence of Arcobacter. Differential proteomics further revealed that the presence of Lenisia stimulated expression of known “virulence” factors by Arcobacter. These proteins typically enable colonization of animal cells during infection5, but may in the present case act for mutual benefit. Finally, re-investigation of two currently available transcriptomic datasets of other Breviatea4 revealed the presence and activity of related hydrogen-consuming Arcobacter, indicating that mutualistic interaction between these two groups of microbes might be pervasive. Our results support the notion that molecular mechanisms involved in virulence can also support mutualism6 as shown here for Arcobacter and Breviatea.
As a cause of genomic innovations and catalyst of diversification, close interactions between eukaryotes and prokaryotes are driving forces of evolution7. The importance of eukaryote-prokaryote interactions is clearly manifested in the remarkable diversity and abundance of bacteria that live in symbiosis with large multicellular eukaryotes such as animals. The basic adaptive requirements for symbiotic bacteria are the capability to recognize and colonize host tissue, evasion of defense mechanisms, replication and ultimately the transfer to new hosts. Bacteria may have first evolved the capability for symbiotic interactions with eukaryotes through associations with ancestral protists7. Today, several protist lineages are known that preserved ancestral eukaryotic features3,8,9. Characterizing these lineages and their molecular interactions with bacteria is vital to formulate evidence-based hypotheses for the origin and functions of ancestral bacterial-eukaryote symbioses.
By providing nitrous oxide as the only electron acceptor, and the bacterium Alteromonas macleodii as prey bacteria, we enriched an amoeboid flagellate colonized by spiral shaped bacteria from anoxic marine tidal flat sediment. DNA was extracted from the enrichment culture and used for metagenomic sequencing (see below). Phylogenetic analysis of a concatenated sequence alignment comprising 16 universal eukaryotic genes identified the flagellate as a new species within Breviatea (Fig. 1a), with Pygsuia biforma as its closest relative. We designated the protist as the novel genus and species Lenisia limosa (See Supplementary Notes for diagnosis, habitat description and etymology).
Figure 1. Relatives of animal-associated Arcobacter colonize Breviatea.
a, Maximum likelihood tree of breviates found in association with Arcobacter (in red). b, MrBayes tree of Arcobacter found in association with animals or breviates (in red). Dots indicate bootstrap support and posterior probabilities values, respectively. The scale bars represent substitution rate per site. c, Scanning electron micrograph (SEM) of L. limosa and associated bacteria. Pilus (1), pseudopodial extensions (2), prey bacteria (3), short anterior flagellum (4), Arcobacter (5), long posterior flagellum (6). The background of this image was removed and the bacteria were manually colored. See Extended Data Fig. 1 for an unmodified micrograph. d, e, Epifluorescence image of CARD-FISH labeled L. limosa (Euk516) and Epsilonproteobacteria (Epsy914). The nucleus was stained with DAPI. f, Transmission electron micrograph of L. limosa’s mitochondria-related organelles (mro), nucleus (nucl) and extracellular matrix (ex). g, Mitochondria-related organelles with inner (im) and outer membrane (om). h, Differential interference contrast (DIC) micrograph of L. limosa. i, Scanning electron micrograph of attached Arcobacter. Each specimen shown represents at least 10 specimens for which images were recorded.
L. limosa is a small amoeboid flagellate with a predatory lifestyle (Fig. 1c-i). Its morphology has both swimming and adherent gliding forms. For swimming, L. limosa beats its two flagella resulting in slow, wobbling locomotion. When it encounters a substrate, it attaches to it, wraps one flagellum around its lateral side, elongates in shape and starts gliding. While gliding, the second flagellum remains detached and assists in the acquisition of prey bacteria. These are captured with small pseudopodial extensions. Ultra-structural analysis showed the presence of several key features previously identified in other breviates (Extended Data Fig. 1). These features include a complex internal membrane system, centrioles, two basal bodies, a Golgi apparatus, digestive vacuoles as well as mitochondria-related organelles (“hydrogenosomes”).
Phylogenetic analysis of bacterial 16S rRNA gene sequences, combined with catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) identified L. limosa’s epibionts (typically 1-3 epibionts per cell) as a species of so far uncultivated Epsilonproteobacteria of the genus Arcobacter (Fig. 1b, d and e). Arcobacter sp. was closely related to several uncultivated species that were found in association with marine animals. CARD-FISH showed that Arcobacter bacteria successfully evaded ingestion, as they were never detected inside cells of L. limosa. Other denitrifying bacteria (related to Colwellia psychrerythracaea) were also detected, both by metagenomic sequencing and CARD-FISH microscopy, but these did not colonize cells of L. limosa.
We found that the symbiosis was facultative for both partners. Provided with dissolved organic carbon, hydrogen and nitrous oxide as the only electron acceptor, Arcobacter sp. grew independent of its host. However, in the absence of L. limosa, C. psychrerythracaea became much more abundant than Arcobacter (Extended Data Fig. 2). Further evidence for the facultative nature of the symbiosis was obtained by cultivating L. limosa without nitrous oxide, which resulted in the loss of its Arcobacter symbionts (Extended Data Fig. 2).
Why did Arcobacter sp. colonize L. limosa? To address this question, we investigated potential metabolic interactions between both partners with combined metagenomics approaches. We performed whole genome and transcriptome sequencing for the consortium as a whole and performed proteomics for the consortium and for both partners grown separately. Raw sequencing reads were assembled and binned, resulting in a 47 Mb (16x coverage) provisional whole genomes sequence for L. limosa and a 3 Mb genome (15x coverage) for its Arcobacter epibiont. For L. limosa we performed evidence driven gene prediction and repeat identification using the assembled contigs and a polyA-tail enriched transcriptome consisting of 48,530 assembled transcripts (22.5 Mb). We predicted 8,146 protein-coding genes covering 15.6% of the host genome. Both genomes were inferred to be nearly complete, with 95% of conserved eukaryotic and 99% of conserved Epsilonproteobacteria genes present (Extended Data Fig. 3).
L. limosa encoded a mosaic of genes enabling fermentative ATP production (Fig. 2a). Among genes typically supporting aerobic growth, we identified genes for a partial tricarboxylic acid cycle, an alternative oxidase, as well as a malate-aspartate shuttle. Despite the presence of these genes, L. limosa appeared to have lost the capability of oxidative phosphorylation because it lacked Complex IV and the capacity for the biosynthesis of ubiquinol and cytochrome c. Further, all subunits for the F-type ATP synthetase (complex V) were absent. Thus, L. limosa’s energy metabolism was inferred to depend strictly on fermentative ATP production.
Figure 2. Symbiotic metabolism of L. limosa and Arcobacter.
a, Symbiotic metabolism of L. limosa and Arcobacter sp. as inferred from genomics, transcriptomics and proteomics. We identified two main fermentation pathways in L. limosa of which one was coupled to the activity of a NAD(P)H-dependent Fe-hydrogenase (gene number 9, see also Extended Data Fig. 4). The latter pathway theoretically yields two times more ATP and was only expressed in presence of hydrogen oxidizing Arcobacter. Numbers correspond to gene names and expression values listed in figure panel b and c. Red circles indicate genes that are higher expressed under syntrophic conditions and blue circles indicate genes that are expressed higher at non syntrophic conditions. b, Expression levels of proteins involved in energy conservation in L. limosa in presence (red) and absence of Arcobacter (blue). c, Expression levels of proteins involved in energy conservation and organic carbon uptake expressed by Arcobacter in presence (red) and absence of L. limosa (blue). The error bars represent standard deviations from three independent experiments (see also Extended data Fig. 2). If a protein consisted of more than one subunit the average for all subunits is shown. Subcellular localization of proteins was inferred from the presence of N-terminal targeting signals. See Supplementary Table 1 for gene accession numbers.
We identified two potential pathways for pyruvate oxidation in the genome of L. limosa, and proteomics revealed that the expression of either pathway was dependent on the presence/absence of Arcobacter sp. In the absence of Arcobacter, L. limosa appeared to metabolize pyruvate mainly through the activity of pyruvate formate lyase and ethanol dehydrogenase (Fig. 2, #2,3). Inside the mitochondria-related organelles, reduced ferredoxin was oxidized by a ferredoxin-dependent hydrogen-evolving hydrogenase (Fig. 2, #8). This pathway does enable recycling of cytosolic NADH but is not coupled to the production of additional ATP.
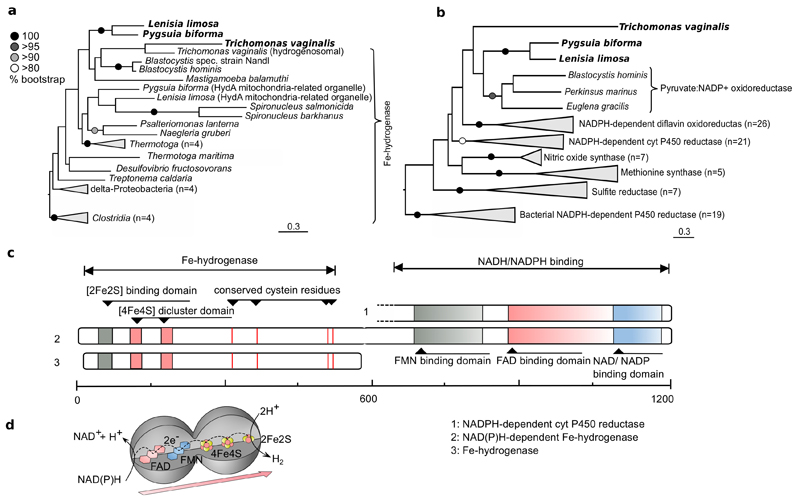
In the presence of Arcobacter, L. limosa was inferred to switch to a bio-energetically much more efficient metabolism that produces hydrogen not only from ferredoxin but also from NADH. Two distinct enzymes for NADH oxidation were identified and both were only expressed in the presence of the symbiont. A fusion enzyme with a NADH/NADPH accepting domain, homologous to P450 reductase, and a Fe-hydrogenase domain, was inferred to produce hydrogen from NADH in the cytosol (Fig. 2, #9, Extended Data Fig. 4). An enzyme with the same inferred domain structure is present in the breviate Pygsuia biforma10. Inside the mitochondria related organelles, a NADH dehydrogenase (Fig. 2, #10,11) might act together with the ferredoxin-dependent hydrogenase (#8) to produce hydrogen by electron confurcation10–12 (Fig. 2). In combination with pyruvate-ferredoxin-oxidoreductase, acetate:succinate CoA-transferase and succinyl-CoA synthetase, NADH-dependent hydrogen production would enable the oxidation of pyruvate to acetate and CO2 with the production of additional ATP. Expression of this entire metabolism was strongly stimulated in the presence of Arcobacter (Fig. 2).
While the production of hydrogen is thermodynamically not prone to product inhibition with electrons from ferredoxin, the production of molecular hydrogen from NAD(P)H only proceeds at low hydrogen partial pressure (<5.4 μM at NADH/NAD+=10)13. The activity of the pathways inferred above therefore requires an active hydrogen sink13. Our proteomic analysis suggested that Arcobacter acted as a hydrogen sink by expressing a high-affinity, hydrogen-oxidizing Ni/Fe-hydrogenase (Fig 2, #17, Extended Data Fig. 5). At a hydrogen turnover rate of 9·10-18 mol H2/s/cell of L. limosa, two Arcobacer epibionts should be able to maintain the hydrogen concentration in the L. limosa cytosol at ~5 μM. We also detected the expression of proteins that potentially catalyze the uptake and utilization of all other fermentation products inferred to be produced by L. limosa. This includes the anabolic uptake of acetate, as well as the catabolic oxidation of formate. The absence of essential genes supporting autotrophic growth (e.g. absence of succinyl-coenzyme-A synthetase and citrate lyase) indicates a general dependency of Arcobacter on organic substrates provided by its host. This is consistent with high expression levels observed for putative amino acid and fatty acid metabolizing enzymes. Our genomic analysis also indicated that hydrogen oxidation could in theory proceed in the presence of electron acceptors other than nitrous oxide. We found all key genes for denitrification, ammonification, as well as the respiration of fumarate and oxygen in the Arcobacter genome. Stimulation of growth of L. limosa/Arcobacter consortia was confirmed experimentally for both oxygen and nitrate (Extended Data Fig. 6g).
Cultivation of L. limosa in the absence and presence of its symbionts enabled us to directly measure the fitness effects resulting from the symbiosis. As fitness indicator we measured growth rates and growth yields of L. limosa in absence and in presence of Arcobacter (Fig 3). As expected, absence of a syntrophic partner resulted in an accumulation of hydrogen and in a significantly impaired fitness of L. limosa. This was apparent from reduced cell numbers, growth rates and growth yields. The growth yield of L. limosa was about two times higher during syntrophic growth. Negative effects on the fitness of L. limosa were also observed when hydrogen was added directly to the culture or when hydrogen oxidation was abolished through a respiration inhibitor (Extended Data Fig. 6). In combination, our physiological experiments demonstrated true metabolic advantage of L. limosa, provided by a hydrogen-scavenging partner, as inferred from combined genomics, transcriptomics and proteomics (Fig. 2).
Figure 3. L. limosa’s fitness depends on its symbiont.
Syntrophy was enabled by the presence of nitrous oxide acting as electron acceptor for bacterial hydrogen oxidation. a, Growth of L. limosa in the presence of nitrous oxide (syntrophy) compared to prey abundance (Alteromonas) and nitrous oxide concentration. b, Growth of L. limosa in the absence of nitrous oxide (no syntrophy) compared to prey abundance and hydrogen concentration. Cell numbers of L. limosa and concentrations were averaged from two independent experiments per treatment, with the error bars showing the full range between both measurements. For cell numbers of Alteromonas, determined with CARD-FISH, the error bars depict the standard deviation of the bacterial cell counts. Growth efficiency was calculated from the difference in bacterial and L. limosa’s cell numbers between two different time points two times during exponential growth. All four individual results are shown as circles, bar heights indicate averages and error bars indicate standard deviations. Error bars smaller than data points are not shown. See also Extended Data Fig. 6.
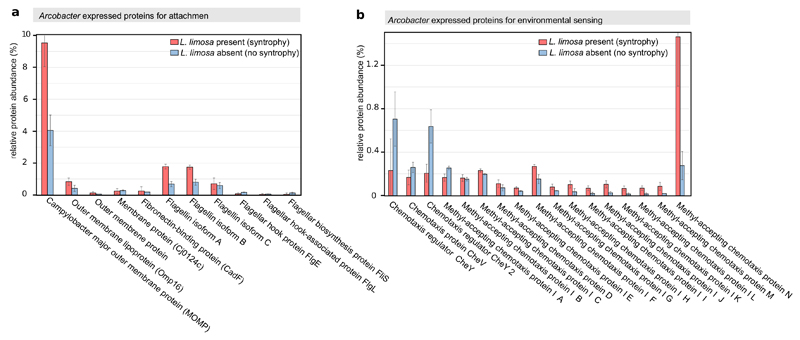
To understand how Arcobacter sp. managed to colonize cells of L. limosa, we inspected the genome of the epibiont for the presence of specific genes previously associated with host recognition and attachment. We found that the Arcobacter epibiont encoded an almost identical set of “virulence” genes previously identified in Arcobacter and Campylobacter pathogens of animals5. Among these gene products, the adhesion protein MOMP (Campylobacter jejuni major outer membrane protein), CadF and flagellin mediate binding to fibronectin type III, which is found in the extracellular matrix of animal cells14,15. The L. limosa epibiont was found to express these “virulence” genes and for MOMP, flagellin and a subset of chemotaxis proteins, expression was stimulated in the presence of L. limosa (Extended Data Fig. 7). Chemotaxis proteins were previously shown to enable pathogenic Campylobacter to move into the immediate proximity of their target16. Interestingly, we found that L. limosa expressed two fibronectin type III domain containing proteins, potential targets of Arcobacter MOMP, CadF and flagellin (Extended Data Fig. 8). Together, these findings suggest that the mechanism for colonization of L. limosa and animal cells by Arcobacter might be conserved at the molecular level. This adds to recent evidence that “virulence” genes, although initially described for pathogens, may in fact mediate both beneficial and pathogenic host-microbe interactions6.
To investigate whether Arcobacter might also engage beneficially with other Breviatea, we screened previously published3,4 transcriptomic data of Pygsuia biforma and Subulatomonas tetraspora for genes affiliated with Arcobacter. In both cases, we identified so far unreported Arcobacter 16S rRNA gene sequences (Fig. 1b), as well as transcription of high-affinity, hydrogen-oxidizing Ni/Fe-hydrogenases (Extended data Fig. 5). The phylogeny presented in Fig. 1b shows that the three Breviatea-associated Arcobacter do not form a monophyletic cluster, but are scattered phylogenetically among their animal associated relatives. This could be interpreted in at least two ways: The symbiosis might have originated once in the ancestor of either phylum. Subsequently, Arcobacter co-diversified together with its hosts, and radiated to the other phylum multiple times, potentially via intermediate, free-living forms. Alternatively, symbiotic Arcobacter may have evolved from free-living forms multiple times. In either case, this resulted in the use of a similar molecular mechanism for colonization (see above). To assess the co-occurrence of Breviatea and Arcobacter in present-day marine sediments, currently available shotgun metagenomes were screened for the presence or absence of L. limosa and other Breviatea, as well as for Breviatea-associated Arcobacter. Thirteen out of twenty-five samples obtained from sediment horizons favourable for growth of Breviatea/Arcobacter consortia potentially contained Breviatea (P<0.01), and seventeen potentially contained Breviatea-associated Arcobacter, including all samples positive for Breviatea (Extended Data Table 1). Although limited metagenomic data are available at present, they are consistent with the ecological interaction of Breviatea and Arcobacter observed for the enriched consortia. The evolutionary roots of this interaction can only be resolved by future investigation of more examples of Breviata/Arcobacter consortia.
In conclusion, we have shown that Lenisia limosa is a newly identified anaerobic protist, colonized by hydrogen-oxidizing Arcobacter. This colonization provided benefits to both partners via interspecies hydrogen transfer, which enables the activity of a newly identified NADH-dependent fusion hydrogenase in L. limosa, leading to increased ATP yield. The molecular mechanism of colonization may involve specific interactions between L. limosa fibronectin type III domain containing proteins and Arcobacter proteins similar to “virulence” factors previously shown to mediate infection of animal tissue by related bacteria. The detection of Arcobacter genes in transcriptomes of other Breviatea shows that these protists likely engage in similar symbioses.
Methods
Cultivation of L. limosa
Sediment samples for the initial enrichment of L. limosa were obtained from a tidal flat in the German Wadden Sea (53.73585 N 7.69905 E). For the enrichments 100 ml laboratory bottles (DURAN Glastechnik, Germany) were filled with 50 ml sediment from different depths (0.5-2 cm and 6-8 cm). The bottles were filled with cultivation medium and closed without creating a headspace. The cultivation medium was based on HEPES buffered seawater (34 g/L, 1 mM HEPES, pH=8) (Red Sea Deutschland, Germany) and contained prey bacteria for protists (final density approx. 109 cells/ml), as well as either nitrate, nitrite or nitrous oxide (0.2 mM final) as electron acceptor for bacterial denitrification. To allow for growth of μm-scaled protists, the small rod-shaped, strictly aerobic Gammaproteobacterium Alteromonas macleodii (strain ATCC 27126, 0.6 to 0.8 μm by 1.4 to 2 μm in size) was selected as prey bacterium. Prey bacteria were grown on solid marine broth medium (ATCC Medium 2216). The cells were washed three times by serial centrifugation in sterile seawater before they were added to the enrichment cultures. Enrichment cultures for protists were incubated in the dark at 22 °C and the nitrate as well as nitrite concentrations were regularly monitored. Depletion of electron acceptor was prevented by periodically adding small portions of anoxic concentrated stock solutions of nitrate, nitrite (from a 200 mM stock) or pure N2O to the cultures. Depletion of food was prevented by adding portions of anoxic suspensions of prey bacteria to the cultures. Growth of protists was regularly inspected by light microscopy. When enrichment was observed, a sub-sample of culture was transferred to sediment free anoxic medium for further investigations. To best preserve the microbial community naturally associated with L. limosa we did not isolate single protists. Instead we performed serial transfers into fresh medium until the presence of only one species could be confirmed via Sanger sequencing of 18S rDNA genes and light microscopy. For all results presented in this study we only used one strain (strain LL-12) obtained from an enrichment culture with nitrous oxide. This strain was maintained in medium containing 2 mM nitrous oxide and transferred weekly. For the cultivation of L. limosa in absence of Arcobacter we grew a subculture in medium containing no nitrous oxide. This culture was transferred once before proteomic analysis was performed (see below).
Cultivation of Arcobacter
For the cultivation of Arcobacter in absence of L. limosa we first incubated L. limosa/Arcobacter consortia in anoxic medium containing dissolved nutrients but no prey bacteria. The cultivation medium was based on HEPES buffered seawater (34 g/L, 1mM HEPES, pH=8) and supplemented with the following compounds: sodium acetate (10 mM), yeast extract (0.1 g/L), sodium phosphate (2 mM) and ammonium chloride (2 mM). The medium was filled in air tight cultivation bottles and made anoxic by flushing it with a mixture of 5% H2, 10% CO2 and 85% N2. Afterwards a part of the culture headspace was replaced with nitrous oxide to achieve a nitrous oxide concentration of 5 mM in the liquid. Under the given conditions no growth of L. limosa could be observed. The enrichment cultures were transferred twice before proteomic analysis was performed (See below).
Physiological experiments
Several growth experiments were performed to evaluate the importance of bacterial hydrogen oxidation for L. limosa’s fitness (Fig. 3 and Extended Data Fig. 6). As electron acceptor for hydrogen oxidation we used nitrous oxide which is the last intermediate in the serial denitrification pathway. Nitrous oxide is non-toxic and its reduction does not produce any intermediates. Thus, it allows for an easy and unambiguous determination of denitrification rates by measuring the consumption of nitrous oxide in the medium. To compare the efficiency of L. limosa’s growth under different conditions we determined cell numbers, growth yields as well as respiration rates. For the determination of L. limosa’s cell numbers, 50 µl subsamples of liquid culture were mixed with formaldehyde solution (0.02% final) which immobilized swimming cells. The cell abundance was determined by manually counting with an improved Neubauer counting chamber (BRAND® counting chamber, Neubauer improved, 0.1 mm depth). For bacterial cell counts, cells were fixed in 1.8% formaldehyde solution (prepared in 34 g/L sterile seawater, 1 mM HEPES, pH=8) for 2 h at room temperature, filtered onto 0.22 µm white polycarbonate filters (supported by 0.45 µm cellulose nitrate filter, Whatman) and stained with the DNA specific stain 4',6-diamidino-2-phenylindole (DAPI 1 µg/ml, 3 min, at 37°C). The filters were evaluated via epifluorescence microscopy (Leila DM: Osram centra mercury-vapor lamp) and the cells were manually counted at 1000x magnification.
Growth yields were calculated by measuring the bacterial cell numbers as well as the eukaryotic cell numbers between two different time points during exponential growth. The differences in bacterial abundance divided by the difference in eukaryotic cell numbers provides a proxy for the gross growth efficiency.
Respiration rates were determined by adding 13C enriched A. macleodii bacteria to an exponentially growing culture of L. limosa. Digestion of such labeled cells leads to the production of 13C bicarbonate, which was measured after conversion to 13CO2 (see chemical analysis). To investigate the growth of L. limosa in presence of antibacterial antibiotics ampicillin and streptomycin (0.1 mg/ ml each) were added to the medium. Since these antibiotics act on growing cells they did not affect the availability of food bacteria, which were provided in excess at the start of the incubation.
To inhibit denitrifying activity we added 5% acetylene final to the headspace of the cultures and dissolved it by gently shaking the cultures. Acetylene inhibits nitrous oxide reductase the enzyme complex that mediates nitrous oxide respiration.
To test the effect of nitrate on the growth of L. limosa we added 2 mM sodium nitrate to the cultivation medium. In addition, we incubated a culture in medium that contained 0.2 mM dissolved oxygen. L. limosa’s cell numbers in such treated cultures were compared to a culture provided with nitrous oxide (2.2 mM) and to a control culture that did not contain an electron acceptor for hydrogen oxidation.
Chemical analysis
Nitrous oxide and hydrogen was measured from the gas headspace of the cultures using a GAM 400 mass spectrometer (In Process Instruments, Germany). Volatile fatty acids were measured with a Syham HPLC system (Fürstenfeldbruck, Germany) equipped with an Aminex HPX-87 H HPLC column (300 × 7.8 mm) and 5 mM H2SO4 as eluent. Separation was performed in isothermal mode at 40 °C and the eluted compounds were simultaneously detected with an UV and a RI detector at a detection limit of 0.1 mM. As calibration standard, a mixture of the fatty acids succinate, lactate, formate, acetate, propionate and butyrate was measured at different concentrations.
Respiration rates were measured as dissolved inorganic 13C-carbon (13C-DIC) released by the digestion of 13C-carbon enriched prey bacteria. Subsamples (1 ml) were poisoned with 0.2% zinc acetate solution at different time-points during the experiments. The isotopic component of DIC was determined after acidifying with hypo-phosphoric acid (1% final) and analyzed on a gas chromatography-isotope ratio monitoring mass spectrometry (Optima Micromass, Manchester, UK). To convert isotopic compositions to concentrations (mol/L), standard solutions of NaHCO3 with known DIC concentrations were measured.
Catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH)
CARD-FISH was performed on polycarbonate filters as described elsewhere18. For the hybridization of ribosomal RNA we used the following probes. For eukaryotes, Euk516, ACCAGACTTGCCCTCC (5’-3’, 0% formamide), for Epsilonproteobacteria, Epsi914, GGTCCCCGTCTATTCCTT (5’-3’, 35% formamide) and for Alteromonas, Alt184, CCCGTTTGGTCCGAAGAC (5’-3’, 25% formamide). Before microscopic evaluation all samples were counter-stained with DAPI and embedded in a 3 to 1 mixture of Citifluor-VectaShield (Citiflour, Leicester, UK/ Vector Labs, Burlingame, CA, USA). Cells were imaged at 1000x magnification with an epifluoresecene microscope (AxioSkop 2 mot plus, Carl Zeiss, Goettingen, Germany) connected to an AxioCam MRm camera (Carl Zeiss).
Electron microscopy
For transmission electron microscopy cells were harvested at 2000 rpm using a Stat Spin Microprep 2 table top centrifuge. After centrifugation the pellet was vitrified in a BAL-TEC HPM-010 high-pressure freezer. The samples were substituted at -90 °C in a solution containing 0.1% tannic acid and 0.5% glutaraldehyde in anhydrous acetone for 72 hours, and for additional 8 hours in 2% OsO4 in anhydrous acetone. After a further incubation over 20 hours at -2° C samples were warmed up to +4° C and washed with anhydrous acetone. The samples were embedded at room temperature in Agar 100 (Epon 812 equivalent) at 60 °C over 24 hours. After ultrathin sectioning (60 nm), section were counter-stained with lead citrate. Samples were analyzed with a Philips CM 120 transmission electron microscope (Philips Inc. Eindhoven, The Netherlands) and images were taken with a TemCam F416 CMOS camera (TVIPS, Gauting, Germany).
For scanning electron microscopy cells were harvested at 400 rpm for 6 minutes using a table top centrifuge. After centrifugation the pellet was placed on teflon slides. We then waited 5 minutes to allow attachment of cells to the slide and fixed them with 2% glutaraldehyde solution (in 34 g/L sterile seawater, 1 mM HEPES, pH=8) for 60 minutes at room temperature. Fixation was followed by a washing step in milliQ-water and an ethanol dehydration series in 30%, 50%, 70%, 90% and 100% ethanol (20 minutes each). Finally the specimens were subjected to critical point drying with CO2 to remove any volatile solvents. The objects were stored in a silica filled desiccator until microscopic evaluation. Imaging was performed with a nova NanoLab 600 scanning electron microscope (FEI Company, The Netherlands). For a better identification of bacteria (in Fig 1c), colors were manually added and the background was removed using the imaging processing software GNU Image Manipulation Program (The GIMP team, v-2.8.14).
Percoll density gradient centrifugation
Enrichment of L. limosa/Arcobacter consortia from suspended bacterial cells was done via Percoll® (Sigma-Aldrich, Germany) based density gradient centrifugation. To form a density gradient, 4.5 ml percoll was mixed with 3.5 ml buffered sterile seawater (1 mM HEPES), filled in 10 ml centrifugation tubes and centrifuged for 30 minutes at 10,000 g and 15 °C. Afterward 1ml of culture was carefully loaded on top of the gradient and immediately centrifuged for another 10 min at 8,000 g at 15 °C. Cells of L. limosa were enriched in a dim white band in the upper third of the gradient, approximately 0.5 cm above the bacteria. The L. limosa fraction was diluted in 50 ml sterile seawater and washed two times by serial centrifugation at 800 g for 5 minutes (15 °C) to remove percoll and suspended bacteria. The enriched L. limosa fraction was pelleted, frozen and stored at -80 °C until DNA extraction.
Next generation sequencing and in silico procedures
Shotgun metagenomic Illumina libraries were constructed from genomic DNA of enriched L. limosa/Arcobacter consortia (See Percoll density gradient centrifugation). Sequencing yielded ~7.5 million raw 2x250 bp paired-end MiSeq reads. Reads were Q10 quality trimmed and filtered to a minimum length of 99 nucleotides using nesoni (https://github.com/Victorian-Bioinformatics-Consortium/nesoni). Initial assembly of the quality filtered reads was performed with velvet19. Bacterial genomes were then binned based on %GC, tetratnucleotide composition and sequence coverage using metawatt20. The binning yielded provisional whole genome sequences for L. limosa, Arcobacter as well as for co-enriched Lacinutrix and Colwellia. For a refined assembly of the L. limosa genome, we filtered out all reads that mapped to the bacterial bins using bowtie221. The remaining reads were then used to generate a final assembly for L. limosa, using velvet and gapfiller22.This assembly was 48.1 Mb in size and had an N50 value of 9.5 kb. The length of the longest scaffold was 135.5 kb. Structural annotations and gene predictions for L. limosa were performed with the MAKER pipeline23. After an initial ab-initio gene prediction with GeneMark-ES24 we refined the obtained gene models with evidence driven gene predictions using snap25 and 48.530 assembled transcripts of a polyA-tail enriched transcriptome (see below). Repeat identification, annotation and masking were implemented by RepeatMasker26. Genome completeness was estimated by representation of 159 universal eukaryotic genes previously identified in P. biforma27 using hidden markov models based searches with hmmer and blast homology searches. Functional annotation was performed with the KEGG automatic annotation server28. For L. limosa’s core metabolism, we manually validated the correct prediction and annotation of each gene model by blastx and blastp homology searches against the UniProtKB/Swiss-Prot protein database. Translocation of gene products to mitochondria-related organelles was predicted based on the presence of N-terminal target peptides using TargetP29 and MitoPROT30. Transcriptional activity of genes was evaluated by mapping the reads of a polyA+ tail enriched transcriptome to the predicted nucleotide gene models. Protein architectures and conserved protein domains were identified using HMMER31, the Pfam32 database and the SMART33 protein domain detection tools.
For the Arcobacter genome gene predictions and functional annotations were performed using the online annotation-pipeline RAST17. Completeness of the Arcobacter genome was evaluated with CheckM34 using the implemented set of conserved Epsilonproteobacteria reference genes. For the core metabolism and virulence related genes we manually validated the correct prediction and annotation of each gene model with blastx and blastp homology searches against the UniProtKB/Swiss-Prot protein database.
For mRNA sequencing, RNA was preserved in RNA-later and was extracted as previously described35. Extracted RNA was treated with RQ1 DNase (Promega), purified with RNeasy MinElute columns (Qiagen) and stored in TE buffer at -80° C. Approximataly 1 µg of total RNA was used for preparation of an mRNA sequencing library following the Illumina TruSeq RNA Sample Preparation v2 Guide, using poly-T oligo-attached magnetic beads to enrich eukaryotic mRNA. The RNA library was sequenced on a MiSeq instrument in a 2x250 bp paired-end run.
Phylogenetic analysis
Eukaryotic phylogenies were determined with an alignment consisting of 16 universal eukaryotic genes from 88 different taxa covering all major eukaryotic groups as previously published36. The alignment was complemented with genes from Lenisia limosa, Pygusia biforma, Breviata anathema as well as Subulatomonas tetraspora. Alignments were calculated with MAFFT37 and phylogenies were constructed with RaxML38 using the GTR+GAMMA model for the SSU-rDNA partition and the WAG replacement matrix with Maximum likelihood estimated base frequencies for the amino acid partitions. We performed 400 rapid bootstrap iterations followed by a search for the best scoring Maximum Likelihood tree.
Arcobacter phylogenies were calculated using 16S rRNA sequences as phylogenetic marker genes. Alignments were constructed with MAFFT. Phylogenetic tree calculation was performed by bayesian inference using the software MrBayes39 with the GTR substitution model and Gamma rate variation.
Phylogenetic analysis for the putative NAD(P)H-dependent Fe-hydrogenase was done individually for the NAD(P)H accepting domain and the Fe-hydrogenase domain. For this, closely related sequences were obtained from the NCBI non-redundant protein database and used to construct a sequence alignment with MAFFT. Phylogenies were inferred using RAxML and the WAG replacement matrix with fixed base frequencies. We ran 600 rapid bootstrap iterations followed by a search for the best scoring maximum likelihood tree.
Protein extraction and peptide preparation
For proteomics three parallel cultures of L. limosa were grown with nitrous oxide in presence of Arcobacter and three cultures were grown without nitrous oxide and in absence of Arcobacter. In addition three cultures of Arcobacter were grown with nitrous oxide and in absence of L. limosa (See Cultivation for details on medium composition). The cultures were harvested during the early exponential growth phase and immediately frozen at -80 °C.
For each of the three treatments we prepared tryptic digests from three biological replicates following the filter-aided sample preparation (FASP) protocol described by Wisniewski et al.40. In brief, SDT-lysis buffer (4% (w/v) SDS, 100 mM Tris-HCl pH 7.6, 0.1 M DTT) was added in a 1:10 sample/buffer ratio to the sample pellets. Samples were heated to 95° C for 10 minutes followed by pelleting of debris for 5 min at 21,000 x g. 30 µl of the cleared lysate were mixed with 200 µl of UA solution (8 M urea in 0.1 M Tris/HCl pH 8.5) in a 10 kDa MWCO 500 µl centrifugal filter unit (VWR International) and centrifuged at 14,000 x g for 40 min. 200 µl of UA solution were added again and centrifugal filter spun at 14,000 x g for 40 min. 100 µl of IAA solution (0.05 M iodoacetamide in UA solution) were added to the filter and incubated at 22° C for 20 min. The IAA solution was removed by centrifugation and the filter was washed three times by adding 100 µl of UA solution and then centrifuging. The buffer on the filter was then changed to ABC (50 mM Ammonium Bicarbonate), by washing the filter three times with 100 µl of ABC. 2 µg of MS grade trypsin (Thermo Scientific Pierce, Rockford, IL, USA) in 40 µl of ABC were added to the filter and filters incubated over night in a wet chamber at 37° C. The next day, peptides were eluted by centrifugation at 14,000 x g for 20 min, followed by addition of 50 µl of 0.5 M NaCl and again centrifugation. Peptides were desalted using C18 spin columns (Thermo Scientific Pierce, Rockford, IL, USA) according to the manufacturer’s instructions. Approximate peptide concentrations were determined using the Pierce Micro BCA assay (Thermo Scientific Pierce, Rockford, IL, USA).
1D-LC-MS/MS
Samples were analyzed by 1D-LC-MS/MS using a block-randomized design as previously described41. Two blank runs were done between samples to reduce carry over. For each sample a technical replicate was run. For each run 800 ng of peptide were loaded onto a 2 cm, 75 µm ID C18 Acclaim® PepMap 100 pre-column (Thermo Fisher Scientific) using an EASY-nLC 1000 Liquid Chromatograph (Thermo Fisher Scientific) set up in 2-column mode. The pre-column was connected to a 50 cm x 75 µm analytical EASY-Spray column packed with PepMap RSLC C18, 2µm material (Thermo Fisher Scientific), which was heated to 35° C via the integrated heating module. The analytical column was connected via an Easy-Spray source to a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were separated on the analytical column at a flow rate of 225 nl/min using a 260 min gradient going from buffer A (0.2% formic acid, 5% acetonitrile) to 20% buffer B (0.2% formic acid in acetonitrile) in 200 min, then from 20 to 35% B in 40 min and ending with 20 min at 100% B. Eluting peptides were ionized with electrospray ionization (ESI) and analyzed in the Q Exactive Plus. Full scans were acquired in the Orbitrap at 70,000 resolution. MS/MS scans of the 15 most abundant precursor ions were acquired in the Orbitrap at 17,500 resolution. The mass (m/z) 445.12003 was used as lock mass as described by Olsen et al.42 with the modification that lock mass was detected in the full scan rather than by separate SIM scan injection. Lock mass use was set to “best”. Ions with charge state +1 were excluded from MS/MS analysis. Dynamic exclusion was set to 30 sec. Roughly 160,000 MS/MS spectra were acquired per sample run.
Protein identification, quantification and statistics
For protein identification a custom protein sequence database containing 28,966 proteins, predicted from the L. limosa, Arcobacter sp., Alteromonas, Colwellia and Lacinutrix provisional whole genome sequences, was used. The database was submitted to the PRIDE repository (see below). For protein identification technical replicates were combined and MS/MS spectra were searched against the database using the Sequest HT node in Proteome Discoverer version 2.0.0.802 (Thermo Fisher Scientific) with the following parameters: Trypsin (Full), max. 2 missed cleavages, 10 ppm precursor mass tolerance, 0.6 Da fragment mass tolerance and max. 3 equal dynamic modifications per peptide. The following three dynamic modifications were considered: oxidation on M (+15.995 Da), carbamidomethyl on C (+57.021 Da) and acetyl on protein N-terminus (+42.011 Da). False discovery rates (FDRs) for peptide spectral matches (PSMs) were calculated and filtered using the Percolator Node in Proteome Discoverer. The Percolator algorithm43 “uses semi-supervised learning and a decoy database search strategy to learn to distinguish between correct and incorrect PSMs”. Percolator was run with the following settings: Maximum Delta Cn 0.05, a strict target FDR of 0.01, a relaxed target FDR of 0.05 and validation based on q-value.
Search results for all samples were combined into a multiconsensus report with Proteome Discoverer and additional filtering criteria applied on the protein level, which were at least 6 PSMs per protein, at least one unique peptide and only PSMs with a concatenated rank of 1. This resulted in the following overall FDRs: 0.6% for PSMs, 2.6% for peptides and 5% for proteins. The multiconsensus report was then exported as a tab-delimited file for further processing.
For protein quantification normalized spectral abundance factors (NSAFs) were calculated based on PSMs using the method described previously44 and multiplied by 10,000. The NSAFx10,000 gives the relative abundance of a protein in a sample as a fraction of 10,000. For statistical analyses of differences between treatments L. limosa and Arcrobacter proteins were analyzed separately i.e. separate tables were generated. For both organisms the set of proteins was reduced by only including proteins that had at least three NSAF values greater than 0 across all nine samples (3 treatments x 3 biological replicates) and at least one NSAF value greater than 0 in each of the treatments that were compared in the respective statistical test. The NSAFx10,000 values in these reduced tables were then again normalized to 10,000 for each sample to generate organism specific NSAF values (orgNSAFs) as described by Mueller et al.45. This normalization procedure ensures that differences in organism abundance across samples do not lead to the false detection of differentially expressed proteins. The tables with orgNSAFs were loaded into the Perseus software (version 1.5.1.6, http://www.perseus-framework.org/doku.php) and log2 transformed. Missing values produced by log2(0) were replaced by sampling from a normal distribution assuming that the missing values are on the lower end of abundance (normal distribution parameters in Perseus: width 0.3, down shift 1.8, do separately for each column). A t-test with permutation based FDR calculation was used to detect proteins that differed significantly in their expression level between two treatments. The statistical method implemented in Perseus that we used is based on the “significance analysis of microarrays” described by Tusher et al.46 which by using a permutation based FDR accounts for the multiple-testing problem inherent in testing for significant expression differences for a large number of genes. The following parameters were used for the test: groupings were not preserved for randomizations, both sides, 250 randomizations, FDR of 1% and s0 of 0.1.
Screening of sediment metagenomes
A SRAdb R/Bioconductor47 query of NCBI’s Short Read Archive (SRA) with search term “marine sediment” yielded 130 shotgun metagenomes. After downloading and converting the SRA files to FASTQ format, the FASTQ short, <300bp reads were mapped onto the assembled contigs (only those > 5kbp) of L. limosa and Arcobacter, using BBMap with parameters “ambiguous=random qtrim=lr trimq=10 minid=0.90”. The remaining long, >300bp reads were quality trimmed using BBMap with parameters “qtrim=lr trimq=10” before mapping onto the same contigs using BWASW48 with default settings. For robustness of phylogenetic classification, the low complexity reads identified using sga49 with parameters “preprocess--dust--dust-threshold=4” were filtered out from the mapped reads; with diamond blastx50 the remaining mapped reads were searched against a database containing amino acid sequences of genus-level (Bacteria) or family-level (Eukarya) representatives of all forms of life, including L. limosa and other Breviatea, as well as L. limosa-affiliated Arcobacter, A. nitrofigilis and seven other Arcobacter. Only reads with hits to Breviatea, L. limosa-affiliated Arcobacter and A. nitrofigilis and only if the hits within 10% of the best bit-score with e-value lower than 0.01 and min bit-score at least 50 were considered. Distribution of positive hits over multiple loci of L. limosa and Arcobacter sp. genome sequences was verified manually. Of the 130 available metagenomes, 15 had to be rejected because they were not obtained from marine sediments, and 65 were rejected because they contained too few reads to enable detection of L. limosa. Of the remaining 50 metagenomes, 25 were obtained from the top of the sediment, a potentially favourable habitat for L. limosa. The remaining 25 were obtained from deeper sediment horizons with probably unfavourable conditions. For the latter we calculated an average false positive rate (FPR) for Breviatea detection of (0.004 reads/1 million reads). This FPR is likely overestimated, because we cannot completely exclude that the metagenomes from unfavourable conditions did not contain Breviatea or were contaminated during sampling. The FPR was used to calculate, for each sample, using a binomial distribution, the probability that the actual number of reads assigned to Breviatea was coincidental and not related to the potential presence of Breviatea. We did not apply the same statistical procedure to the read maps for Arcobacter, because classifying samples as unfavourable for Arcrobacter was not possible based on the fact that Arcobacter can occupy many different ecological niches. Extended Data Tab. 1 presents the results and SRA accession numbers for the analysed metagenomes.
Bioenergetics and hydrogen fluxes
Per-cell Hydrogen production rates were calculated from nitrous oxide consumption rates and cell numbers presented in Fig. 3, assuming a stoichiometry of 1/3 mol H2/mol N2O (Fig. 2). The hydrogen concentration sustained in L. limosa’s mro by a single Arcobacter epibiont was calculated to be 10 μM with the equation F (mol H2/s) = D/d * C * A, with: F, the hydrogen flux (~9·10-18 mol/s); D, the diffusivity of H2 in water (~4.6·10-9 m2/s); d, the distance between the H2 source and the H2 sink (2.1·10-6 m); C, the H2 concentration in the mro (mM or mol/m3); A, the area of diffusion (0.4·10-12 m2). The maximum hydrogen concentration enabling the reaction NADH + H+ = NAD+ + H2 (ΔG +18 kJ/mol51) was calculated to be 5.4 μM (with Henry’s Law constant for H2 6.33·109 Pa and an NADH/NAD+ ratio of ten52). The calculations are presented in Supplementary Table 2.
Extended Data
Extended Data Figure 1. Micrographs for L. limosa and epibiotic Arcobacter.
a, Scanning electron micrograph (SEM) showing L. limosa and associated bacteria. Pilus (1) connecting Arcobacter (5) with L. limosa. Pseudopodial extensions (2) are used for the acquisition of prey bacteria (3) (Alteromonas). Short anterior flagellum (4). Long posterior flagellum (6). b–e, CARD-FISH labeling with probes targeting the SSU rRNA of L. limosa (Euk516 in red) and Arcobacter (Epsy914 in green). The scale bar applies to all figures. f-i, CARD-FISH labeling of L. limosa with probes targeting the SSU rRNA of Alteromonas macleodii (Alt184 in green). The scale bar applies to all figures. j-r, Transmission electron micrographs showing different structural features of L. limosa. Mitochondria-related organelle (mro), nucleus (nucl), digestive vacuoles (dv), double basal body (bb), endoplasmic reticulum (er), inner (im) and outer membrane (om), tubular cristae (cristae), extracellular matrix (ex), bacterium (bac), membrane (mem), flagellum (flag), multivesicular body (mb). For a-i: Each specimen shown represents at least 10 specimens for which images were recorded.
Extended Data Figure 2. Relative abundance of L. limosa and co-enriched bacteria under different growth conditions.
The abundance of L. limosa and its associated microbiota was determined at three different conditions (treatments) with three independent experiments per treatment: 1-3, Presence of nitrous oxide and prey bacteria. 4-6, Absence of nitrous oxide and presence of prey bacteria. 7-9, Presence of nitrous oxide, dissolved organic nutrients and hydrogen and absence of prey bacteria. Relative abundances were determined via proteomics and estimated based on the total normalized spectrum count per population.
Extended Data Figure 3. Genome statistics for L. limosa and epibiotic Arcobacter.
The pie chart represents the classifications of gene models into functional categories for Arcobacter. Gene classifications were performed with the RAST functional annotations and the SEED subsystem database17.
Extended Data Figure 4. A new type of NAD(P)H dependent Fe-hydrogenase.
L. limosa’s genome encoded a so far undescribed NAD(P)H dependent Fe-hydrogenase. Genes with identical domain architecture were also identified in P. biforma and T. vaginalis (shown in bold). The scale bars represent substitution rate per site. a, Phylogeny of the Fe-hydrogenases domain. b, Phylogeny of the NAD/NADP binding domain. Phylogenies were inferred by RAxML using the WAG amino acid replacement matrix. c, Domain architecture of the NAD(P)H-dependent Fe-hydrogenase (2) in comparison with the domain architecture of Fe-hydrogenase (3) and the NADPH accepting domain of the cyt P450 reductase (1). The scale bar shows approximate amino acid positions. d, Predicted electron flow within the NAD(P)H-dependent Fe-hydrogenase indicates the capability for a proton dependent recycling of NAD(P)H. Note: the shape of the model does not intent to depict the actual three-dimensional structure of the protein.
Extended Data Figure 5. Maximum likelihood tree of quinone reactive Ni/Fe-hydrogenases (Subunit hydB).
The tree shows the phylogenetic relation of quinone reactive Ni/Fe-hydrogenases from Arcobacter associated with S. tetraspora, P. biforma and L. limosa (indicated in red). Circles represent bootstrap support values for each node. The scale bar represents substitution rate per site.
Extended Data Figure 6. The fitness of L. limosa’s depends on its symbiont.
Syntrophy was enabled by the presence of nitrous oxide acting as electron acceptor for bacterial hydrogen oxidation. a, Inhibition of nitrous oxide reduction (addition of the competitive inhibitor acetylene, see arrow) leads to a reduced growth of L. limosa and reduced respiration rates. To monitor respiration rates, 13C-carbon enriched Alteromonas were added together with acetylene. Digestion of 13C-carbon labeled bacteria by L. limosa led to the production of 13C-bicarbonate which was measured after conversion to 13CO2 (right panel). Similar effects on the growth and respiration rates were observed after adding hydrogen (b) or hydrogen and acetate (c) to a culture. d, Growth of L. limosa and production of hydrogen and fatty acids while growing syntrophically (nitrous oxide present). e, Growth of L. limosa in presence of antibacterial antibiotics (nitrous oxide absent) f, Growth of L. limosa in presence of antibacterial antibiotics (nitrous oxide present). g, Growth of L. limosa in presence of nitrate (2 mM) and oxygen (0.2 mM). Growth of L. limosa was compared to a culture that contained nitrous oxide (2.2 mM) and to a control culture that did not contain an electron acceptor for hydrogen oxidation. Each panel shows the results for at least five independent experiments, with cell numbers depicted as averages of seven cell counts per experiment and the error bars indicate the standard deviation.
Extended Data Figure 7. Expression levels for Arcobacter proteins involved in attachement and chemotaxis.
a. Expression level of proteins involved in attachment in presence (red) and absence of L. limosa (blue). b, Expression level of proteins involved in chemotaxis. Expression levels were measured and averaged for three independent experiments per treatment (see also Extended data Fig. 2). The error bars indicate the standard deviations. See Supplementary Table 1 for gene accession numbers and statistical tests.
Extended Data Figure 8. Domain architecture of L. limosa’s fibronectin type III domain containing proteins.
Protein architectures and conserved protein domains were identified using the SMART protein domain detection tools. See Supplementary Table 1 for gene accession numbers and expression levels.
Extended Data Table 1. Potential presence of Breviatea and Breviatea-associated Arcobacter detected in presently available shotgun metagenomes from marine sediments.
| SRA accession | F* | Description | #reads | Breviatea (including L. limosa) | Breviatea-associated Arcobacter | |
|---|---|---|---|---|---|---|
|
| ||||||
| Abundance | P-value | Abundance | ||||
|
| ||||||
| (per 1 milion reads) | (per 1 milion reads) | |||||
| SRR2638107 | + | sediment, upper 2cm, Barataria Bay (USA) | 869558078 | 0.043 | 0.0000 | 0.64 |
| SRR2657558 | + | sediment, upper 2cm, Bay jimmy (USA) | 943961688 | 0.036 | 0.0000 | 0.86 |
| SRR2636951 | + | sediment, upper 2cm, Barataria Bay (USA) | 738401828 | 0.037 | 0.0000 | 0.79 |
| SRR2637699 | + | sediment, upper 2cm, Barataria Bay (USA) | 986158388 | 0.028 | 0.0000 | 0.13 |
| SRR2657575 | + | sediment, upper 2cm, Bay jimmy (USA) | 642092762 | 0.028 | 0.0000 | 3.09 |
| SRR1793930 | - | igneous basalt, Louisville Seamounts | 65221092 | 0.123 | 0.0000 | 0.02 |
| SRR577221** | + | tidal flat sediment upper 2cm | 577090 | 5.198 | 0.0000 | 3.47 |
| SRR577224** | + | tidal flat sediment upper 2cm | 682998 | 4.392 | 0.0000 | 10.25 |
| SRR2657076 | + | sediment, upper 2cm, Barataria Bay (USA) | 164252226 | 0.055 | 0.0000 | 0.15 |
| SRR2657962 | + | sediment, upper 2cm, Barataria Bay (USA) | 595816470 | 0.015 | 0.0006 | 3.65 |
| SRR577220** | + | tidal flat sediment upper 2cm | 581409 | 1.720 | 0.0023 | 6.88 |
| SRR2637322 | + | sediment, upper 2cm, Barataria Bay (USA) | 144887606 | 0.028 | 0.0025 | 0.22 |
| SRR2656923 | + | sediment, upper 2cm, Barataria Bay (USA) | 69560055 4 | 0.012 | 0.0052 | 0.14 |
| SRR2657585 | + | sediment, upper 2cm, Barataria Bay (USA) | 607701178 | 0.012 | 0.0083 | 0.02 |
| SRR2637706 | - | Sediment, 8-12cm, Barataria Bay (USA) | 140402750 | 0.021 | 0.0164 | 0.14 |
| SRR2657208 | + | sediment, upper 2cm, Terrebonne (USA) | 402338618 | 0.012 | 0.0173 | 0.01 |
| SRR2657207 | + | sediment, upper 2cm, Barataria Bay (USA) | 137181860 | 0.015 | 0.0855 | 0.04 |
| SRR2658004 | + | sediment, upper 2cm, Barataria Bay (USA) | 162613058 | 0.012 | 0.1087 | 2.64 |
| SRR1627906 | - | ocean sediment CostaRica, 32m deep | 35468880 | 0.028 | 0.1219 | 0.00 |
| SRR2637708 | - | Sediment, 8-12cm, Barataria Bay (USA) | 943467314 | 0.002 | 0.1668 | 0.06 |
| SRR2637690 | - | Sediment, 8-12cm, Barataria Bay (USA) | 596039480 | 0.005 | 0.2067 | 0.22 |
| SRR2658026 | - | Sediment, 8-12cm, Barataria Bay (USA) | 604747158 | 0.005 | 0.2086 | 0.24 |
| SRR2657594 | - | Sediment, 8-12cm, Barataria Bay (USA) | 619813316 | 0.002 | 0.2113 | 0.84 |
| SRR2657566 | - | Sediment, 8-12cm, Barataria Bay (USA) | 607030206 | 0.002 | 0.2176 | 0.09 |
| SRR2657582 | + | sediment, upper 2cm, Barataria Bay (USA) | 803328968 | 0.004 | 0.2229 | 0.10 |
| SRR2657625 | + | sediment, upper 2cm, Barataria Bay (USA) | 50533 6664 | 0.004 | 0.2707 | 4.71 |
| SRR2656927 | - | Sediment, 8-12cm, Barataria Bay (USA) | 120576078 | 0.008 | 0.2960 | 0.15 |
| SRR2657579 | + | sediment, upper 2cm, Barataria Bay (USA) | 135244394 | 0.007 | 0.3133 | 0.13 |
| SRR2656924 | + | sediment, upper 2cm, Barataria Bay (USA) | 143593200 | 0.007 | 0.3218 | 0.10 |
| SRR2656926 | + | sediment, upper 2cm, Barataria Bay (USA) | 157234396 | 0.006 | 0.3339 | 0.43 |
| SRR2656925 | - | Sediment, 8-12cm, Barataria Bay (USA) | 618546874 | 0.000 | - | 0.01 |
| SRR2657909 | - | Sediment, 8-12cm, Barataria Bay (USA) | 484269622 | 0.000 | - | 0.21 |
| SRR1179191 | - | Mahoney Lake (euxinic) | 199487463 | 0.000 | - | 0.00 |
| SRR2638077 | - | Sediment, 8-12cm, Barataria Bay (USA) | 196861140 | 0.000 | - | 0.03 |
| SRR2657627 | + | Sediment, 0-2cm, Barataria Bay (USA) | 157919092 | 0.000 | - | 2.29 |
| SRR2657590 | + | Sediment, 0-2cm, Barataria Bay (USA) | 149050036 | 0.000 | - | 0.02 |
| SRR2657155 | - | Sediment, 8-12cm, Barataria Bay (USA) | 121323126 | 0.000 | - | 0.17 |
| SRR1627905 | - | ocean sediment Costa Rica margin, 2.9m | 86658932 | 0.000 | - | 0.02 |
| SRR1628696 | - | pacific ocean 280m deep, igneous rock | 84164942 | 0.000 | - | 0.00 |
| SRR1971620 | - | Haakon Mosby mud volcano, 3.7m deep | 46060794 | 0.000 | - | 0.00 |
| SRR1628698 | - | pacific ocean 280m deep, igneous rock | 45206218 | 0.000 | - | 1.26 |
| SRR1628697 | - | pacific ocean 280m deep, igneous rock | 42072910 | 0.000 | - | 0.00 |
| SRR1971621 | - | Haakon Mosby mud volcano, 3.7m deep | 41183206 | 0.000 | - | 0.15 |
| SRR1627907 | - | ocean sediment Costa Rica margin, 94m deep | 34240066 | 0.000 | - | 0.00 |
| SRR1793929 | - | pacific ocean, basalt | 30470478 | 0.000 | - | 0.00 |
| SRR1793931 | - | pacific ocean 130m deep, igneous rock | 30177380 | 0.000 | - | 0.00 |
| SRR1022349 | + | marine fish farm sediment | 2656105 6 | 0.000 | - | 0.15 |
| SRR1971622 | - | Haakon Mosby mud volcano, 2.8m deep | 20696408 | 0.000 | - | 0.00 |
| SRR1793928 | - | pacific ocean, drill fluid | 1537452 | 0.000 | - | 0.00 |
| SRR577219** | + | tidal flat sediment, upper 2cm | 667625 | 0.000 | - | 1.50 |
Sediments were grouped into two habitat types, (+) indicates habitats favourable for growth of Breviatea, (-) indicates habitats unfavourable for growth of Breviatea. The P-values (calculated from a binomial distribution with a false-positive rate obtained from the unfavourable habitats) are the probabilities that the actual number of reads assigned to Breviatea was coincidental and not related to the potential presence of Breviatea.
These metagenomes were obtained from the same site as the inocula for the enrichment of L. limosa in the present study.
Supplementary Material
Acknowledgment
We thank Theresa Hargesheimer, Dan Liu, Gabi Klockgether, Phillip Hach, Ramona Appel and Ines Kattelmann for technical assistance, Alastair Simpson, Ger Strous and Sergio for comments on electron micrographs, Casey Hubert, Emil Ruff, Soeren Ahmerkamp and Nicole Dubilier for stimulating discussions. This study was supported by the European Research Council (ERC) starting grant MASEM 242635 (M.S., E.H., J.C.), the Campus Alberta Innovation Chair Program (M.S., E.H., X.D.), the Canadian Foundation for Innovation (M.S), the Alberta Small Equipment Grant Program (M.S.) the German Federal State Nordrhein-Westfalen (M.S.), the Max Planck Society, and NSERC for both a Banting fellowship to M.K. and a Discovery Grant to M.S.
Footnotes
Author contributions
E. H., J. C. performed sampling, cultivation and physiological experiments. M.K. performed proteomics and data analysis. D.R. performed transmission electron microscopy. S.L. performed scanning electron microscopy. E.H. performed CARD-FISH imaging. H.T. performed next generation sequencing, H.G.-V. performed read processing, assembly and binning. E.H. performed in silico processing of next generation sequencing data with assistance from H.G.-V., X.D. and M.S. M.W.B. C.W.S. and A.J.R. analyzed sequences for Arcobacter associated with S. tetraspora . E.H., B.V. and K.B performed chemical analysis with input from J. M, K.-U.H. and M.S.. The experimental design was developed jointly by M.S., J.M. K.-U.H. and E.H. E. H. wrote the manuscript with input from all co-authors.
The authors declare no conflict of interest
Author information
Reprints and permissions information is available at www.nature.com/reprints
Reference Material and Data availability: The culture was deposited in the cryo-preserved state in liquid nitrogen at the American Type Culture Collection and has been accessioned as Lenisia limosa (Strain LL-12) AcqID-00721. Sequence data are available for download from the NCBI Sequence Read Archive (SRA) database and the Whole Genome Shotgun (WGS) database and are grouped as NCBI BioProject PRJNA277740. The SSU rRNA gene for L. limosa is available in Genbank under the accession number KT023596. The mass spectrometry proteomics data and the protein sequence database have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD003275.
Contributor Information
Emmo Hamann, Microbial Fitness Group, Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany; Department of Geoscience, University of Calgary, Calgary, 2500 University Dr NW, Alberta, T2N 1N4, Canada.
Harald Gruber-Vodicka, Symbiosis Department, Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany.
Manuel Kleiner, Department of Geoscience, University of Calgary, Calgary, 2500 University Dr NW, Alberta, T2N 1N4, Canada.
Halina E. Tegetmeyer, Microbial Fitness Group, Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany; Institute for Genome Research and Systems Biology, Center for Biotechnology, University of Bielefeld, Universitätsstraße 25, 3615 Bielefeld, Germany
Dietmar Riedel, Max Planck Institute for Biophysical Chemistry, Am Faßberg 11, 37077 Göttingen. Germany.
Sten Littmann, Biogeochemistry Department, Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany.
Jianwei Chen, Microbial Fitness Group, Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany; Department of Geoscience, University of Calgary, Calgary, 2500 University Dr NW, Alberta, T2N 1N4, Canada.
Jana Milucka, Biogeochemistry Department, Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany.
Bernhard Viehweger, MARUM Centre for Marine Environmental Sciences, Bibliothekstraße 1, University of Bremen, 28359 Bremen, Germany.
Kevin W. Becker, MARUM Centre for Marine Environmental Sciences, Bibliothekstraße 1, University of Bremen, 28359 Bremen, Germany
Xiaoli Dong, Department of Geoscience, University of Calgary, Calgary, 2500 University Dr NW, Alberta, T2N 1N4, Canada.
Courtney W. Stairs, Centre for Comparative Genomics and Evolutionary Bioinformatics, Department of Biochemistry and Molecular Biology, 6299 South St Dalhousie University, Halifax, Nova Scotia, B3H 4R2, Canada
Kai-Uwe Hinrichs, MARUM Centre for Marine Environmental Sciences, Bibliothekstraße 1, University of Bremen, 28359 Bremen, Germany.
Matthew W. Brown, Department of Biological Sciences, Mississippi State University, Mississippi State, MS USA 39762
Andrew J. Roger, Centre for Comparative Genomics and Evolutionary Bioinformatics, Department of Biochemistry and Molecular Biology, 6299 South St Dalhousie University, Halifax, Nova Scotia, B3H 4R2, Canada
Marc Strous, Microbial Fitness Group, Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany; Department of Geoscience, University of Calgary, Calgary, 2500 University Dr NW, Alberta, T2N 1N4, Canada; Institute for Genome Research and Systems Biology, Center for Biotechnology, University of Bielefeld, Universitätsstraße 25, 3615 Bielefeld, Germany.
References
- 1.Steinberg R, et al. Low Mid-Proterozoic atmospheric oxygen levels and the delayed rise of animals. Science. 2014;346:635–638. doi: 10.1126/science.1258410. [DOI] [PubMed] [Google Scholar]
- 2.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MW, et al. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc R Soc B. 2013;280:1769. doi: 10.1098/rspb.2013.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stairs CW, et al. A SUF Fe-S cluster biogenesis system in the mitochondrion-related organelles of the anaerobic protist. Pygsuia Curr Biol. 2014;24:1176–1186. doi: 10.1016/j.cub.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira S, Queiroz JA, Oleastro M, Domingues FC. Insights in the pathogenesis and resistance of Arcobacter: A review. Crit Rev Microbiol. 2015;7828:1–20. doi: 10.3109/1040841X.2014.954523. [DOI] [PubMed] [Google Scholar]
- 6.Sayavedra L, et al. Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels. Elife. 2015;4:e07966. doi: 10.7554/eLife.07966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suga H, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 2013;4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stairs CW, Leger MM, Roger AJ. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140326. doi: 10.1098/rstb.2014.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schut GJ, Adams MWW. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol. 2009;191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrdy I, Hirt RP, Dolezal P, Bardonová L, Foster PG, Tachezy J, Embley TM. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 13.Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nature Rev Microbiol. 2009;7(8):568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 14.Moser I, Schroeder W, Salnikow J. Campylobacter jejuni major outer membrane protein and a 59-kDa protein are involved in binding to fibronectin and INT 407 cell membranes. FEMS Microbiol Lett. 1997;157:233–238. doi: 10.1111/j.1574-6968.1997.tb12778.x. [DOI] [PubMed] [Google Scholar]
- 15.Monteville MR. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology. 2003;149:153–165. doi: 10.1099/mic.0.25820-0. [DOI] [PubMed] [Google Scholar]
- 16.van Alphen LB, et al. Active migration into the subcellular space precedes Campylobacter jejuni invasion of epithelial cells. Cell Microbiol. 2008;10:53–66. doi: 10.1111/j.1462-5822.2007.01014.x. [DOI] [PubMed] [Google Scholar]
- 17.Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010;5 doi: 10.1101/pdb.prot5368. pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 18.Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol. 2002;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strous M, Kraft B, Bisdorf R, Tegetmeyer HE. The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol. 2012;3:410. doi: 10.3389/fmicb.2012.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadalin F, Vezzi F, Policriti A. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics. 2012;13 doi: 10.1186/1471-2105-13-S14-S8. S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2004;25:4.10.1–4.10.14. doi: 10.1002/0471250953.bi0410s05. [DOI] [PubMed] [Google Scholar]
- 27.Brown MW, et al. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc R Soc B. 2013;280:1769. doi: 10.1098/rspb.2013.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 30.Claros MG. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci. 1995;11:441–447. doi: 10.1093/bioinformatics/11.4.441. [DOI] [PubMed] [Google Scholar]
- 31.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucl Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, et al. The Pfam protein families database. Nucl Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2014;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donovan H Parks, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes Donovan. PeerJ. 2014;3:1–4. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CJ, Nedwell DB, Dong LF, Osborn AM. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol. 2007;73:3612–3622. doi: 10.1128/AEM.02894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parfrey LW, et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 41.Oberg AL, Vitek O. Statistical design of quantitative mass spectrometry-based proteomic experiments. J Proteome Res. 2009;8:2144–2156. doi: 10.1021/pr8010099. [DOI] [PubMed] [Google Scholar]
- 42.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Spivak M, et al. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J Proteome Res. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Florens L, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller RS, et al. Ecological distribution and population physiology defined by proteomics in a natural microbial community. Mol Syst Biol. 2010;6:374. doi: 10.1038/msb.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Y, Stephens RM, Meltzer PS, Davis SR. Sradb: query and use public next-generation sequencing data from within R. BMC Bioinformatics. 2013;14(1):19. doi: 10.1186/1471-2105-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012 doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature Meth. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 51.Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nature Rev Microbiol. 2009;7(8):568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 52.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging Cytosolic NADH-NAD+ Redox State with a Genetically Encoded Fluorescent Biosensor. Cell Metabolism. 14(4):545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.