Abstract
Proper brain function requires the precise localization of proteins and signaling molecules on a nanometer scale. The examination of molecular organization at this scale has been difficult in part because it is beyond the reach of conventional, diffraction-limited light microscopy. The recently developed method of superresolution, localization-based fluorescent microscopy (LBM), such as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), has demonstrated a resolving power at a 10 nm scale and is poised to become a vital tool in modern neuroscience research. Indeed, LBM has revealed previously unknown cellular architectures and organizational principles in neurons. Here, we discuss the principles of LBM, its current applications in neuroscience, and the challenges that must be met before its full potential is achieved. We also present the unpublished results of our own experiments to establish a sample preparation procedure for applying LBM to study brain tissue.
Keywords: superresolution imaging, LBM, PALM/STORM, light microscopy/EM image correlation, PSD-95
INTRODUCTION
The transmission, processing, and integration of information within neural cells rely on the precise orchestration of individual proteins and macromolecular complexes on a nanometer scale. For example, many synaptic proteins at the postsynaptic site of a neuronal synapse are organized within and around the postsynaptic density (PSD), which has a thickness on the order of a few tens of nanometers (Kennedy, 2000; Sheng and Kim, 2011). Changes in molecular organization at the nanoscale level can have profound impacts on cellular signaling and neuronal function (MacGillavry et al., 2011). Over the past several decades, neuroscience research has accumulated a wealth of knowledge about protein organization and interactions using indirect biochemical measurements, such as co-immunoprecipitation. However, to obtain an accurate picture of cellular function, it is necessary to directly measure the precise spatial organization of individual proteins and macromolecular complexes within neurons and glia.
Fluorescence light microscopy, such as confocal and two-photon microscopy, is widely used to study the cellular and subcellular organization of proteins. However, conventional fluorescence microscopy falls short when the relevant length scale approaches the size of individual macromolecular complexes (~10–100 nm). A fundamental limit called the “diffraction limit” prevents the resolution of details below approximately 200 nm. This diffraction limit results from the wave properties of light: a point source of light (e.g., a fluorescent molecule) in the sample plane of a microscope can no longer be re-focused to a single point in the image plane. Instead, diffraction creates an intensity distribution called the point-spread function (PSF). If two small objects are too close to each other, their PSFs will sum up and can no longer be resolved from each other (Figs. 1A and 1B). The smallest distance at which two PSF of the same brightness can be resolved defines the resolution limit of the microscope, which is described by the Rayleigh’s limit R ≈ 0.6λ/NA, where λ is the wavelength of light and NA is the numerical aperture of the objective. For a typical high-NA lens (e.g., 1.4) and the emission wavelength of GFP (509 nm), the resolution limit is approximately 220 nm. Please refer to recent reviews for in-depth discussions about the diffraction limit (Carlton, 2008; Schermelleh et al., 2010).
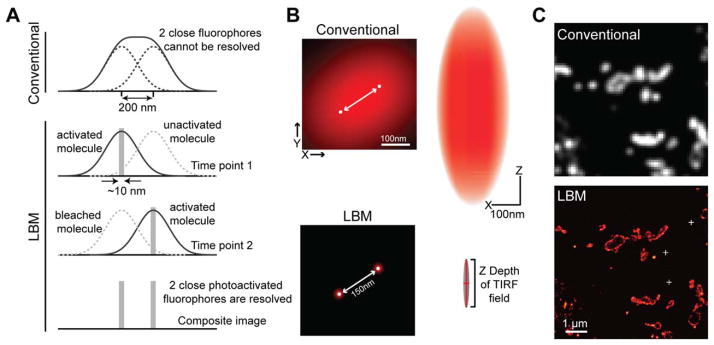
Fig. 1.
Mechanism and demonstration of LBM. A: Under a conventional microscope, two nearby fluorophores cannot be distinguished from each other because their PSFs overlap (top). LBM solves this problem using photoactivatable fluorophores (bottom). Each photoactivatable fluorophore is sequentially activated so that individual molecules can be imaged in isolation and precisely localized. In the composite image where the localization information of all molecules is overlaid, the two fluorophores are resolved [modified from Zhong (2010); with permission]. B: Schematic illustration of the resolving power and the PSF of LBM [modified from Long et al. (2014); with permission]. Left, the effective PSFs of two fluorophores shown in the x–y plane. Right, the PSF is shown in the x–z plane. C: Representative conventional microscopy and LBM images of membrane-tagged EosFPs expressed in fly brain. The sample was processed as described in the Focus Box. White crosses in the LBM image mark the position of gold fiducials for drift correction.
To meet the need for examining cellular processes at a level of resolution beyond the diffraction limit, several “superresolution” fluorescence imaging modalities have been developed over the past 15 years (Gould and Hess, 2008; Hell, 2007; Huang et al., 2010; Sengupta et al., 2012). These methods fall into three primary categories: stimulated emission depletion microscopy (STED) (Hell and Wichmann, 1994; Klar et al., 2000), structured illumination microscopy (SIM) (Gustafsson, 2000), and localization-based microscopy (LBM). LBM is the collective name used for several independently developed, yet conceptually similar methods, including photoactivated localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM) and fluorescence PALM (fPALM), as well as other related techniques (Betzig et al., 2006; Burnette et al., 2011; Egner et al., 2007; Folling et al., 2008; Heilemann et al., 2008; Hess et al., 2006; Rust et al., 2006; Sharonov and Hochstrasser, 2006; Simonson et al., 2011). Although each technique has its own advantages and disadvantages, LBM is being implemented in increasing numbers of biology laboratories due to its several strengths: (1) it routinely achieves a resolution of 10–20 nm, nearly 10-fold better than confocal microscopy, with biological samples. (2) It is the most efficient method with regard to signal photons utilization. This is particularly important for high resolution imaging where a limited photon budget often determines the achievable resolution. (3) It allows for the assessment of characteristics, such as diffusion, of single molecules, including intracellular protein molecules. (4) The design and optics of LBM are simple and can be set up relatively easily using an inverted microscope, allowing this technique to be implemented in many biological laboratories (Shroff et al., 2008b; Zhong, 2010).
LBM has found its way into neuroscience research and has already revealed neuronal organization at a new level of detail. However, it is worth noting that the application of LBM in neuroscience is often limited by practical factors. For example, the choice of fluorophore must be considered carefully and fluorophore options are often limited. In addition, with improved resolution comes the need for high quality sample preparation to adequately label and preserve native macromolecular structures. Here, we introduce the principle of LBM, its current applications in neuroscience, and the challenges that must be met to achieve the full potential of LBM in neuroscience studies. We also present our own efforts at addressing the need for proper sample preparation.
THE MECHANISM OF LBM
By definition, the diffraction limit only exists when the PSFs of multiple fluorescent molecules overlap with one another (Fig. 1A). If individual PSFs can be seen, such as in the case of very low labeling density (≪25 molecules/μm2), fitting of the isolated PSFs can pinpoint the centroid of each PSF, that is, the localization of each individual fluorophore, with high precision (Fig. 1A). The localization accuracy depends on the noise level and the number of signal photons collected for the molecule (Huang et al., 2013; Mortensen et al., 2010; Thompson et al., 2002). Under favorable conditions, this localization precision can approach ~10 nm [see also Pertsinidis et al. (2010) for a demonstration of ~1 nm localization precision]. However, a very low labeling density is insufficient to produce an image that reveals detailed spatial organization (Shroff et al., 2008a). The Nyquist–Shannon theorem requires that the mean distance between labeled molecules must be more than twice as fine as the desired resolution. Although a labeling density of 100 molecules/μm2 is sufficient to provide a Nyquist resolution of ~200 nm, 104 molecules/μm2 are required for a resolution of 20 nm. In other words, high resolution imaging requires high labeling density.
LBM breaks the diffraction limit by simultaneously achieving two seemingly paradoxical feats: low labeling density for highly precise localization of individual fluorophores, and high density labeling to reveal the spatial features (Fig. 1). The key to achieving both goals is to use fluorophores with a bright state and a dark state, such as photoactivatable or photoswitchable fluorescent proteins (PA-FPs); or organic caged dyes (Patterson and Lippincott-Schwartz, 2002; Shcherbakova et al., 2014; van de Linde et al., 2012), which do not fluoresce in the monitoring color channel until activated by short-wavelength light. In a typical LBM scheme, the sample is labeled at high density with photoactivatable/photoswitchable fluorophores, but these molecules are invisible as they are in the dark state. Using a very low dose of activation light, only a stochastic sparse subset of fluorophores is converted to the bright state. At a sufficient level of sparsity, the PSFs can be imaged and resolved individually, and their centroids can be precisely localized (Fig. 1A). After the activated molecules are bleached or driven back to the dark state via imaging, a second subset of fluorophores is then activated and imaged. Such iterations are repeated until all molecules have been imaged. The localization information of all the densely labeled molecules collected over time is then overlaid to produce a final LBM image at high Nyquist resolution (Fig. 1). In practice, LBM methods can routinely achieve a resolution of 10–20 nm. The z resolution of LBM can also be improved using the total internal reflection fluorescence (TIRF) imaging scheme or using a variety of 3D LBM methods (Huang et al., 2008b; Juette et al., 2008; Lew et al., 2010; Shtengel et al., 2009).
Since its inception, multicolor, 3D and live cell imaging variants of LBM have also been developed (Bates et al., 2007; Huang et al., 2008a,b; Jones et al., 2011; Juette et al., 2008; Lew et al., 2010, 2011; Shroff et al., 2007, 2008a; Shtengel et al., 2009). Furthermore, in single particle tracking PALM (sptPALM) and related techniques, individual protein molecules in living cells are tracked until each fluorophore bleaches to derive their mobile characteristics (Giannone et al., 2010; Lew et al., 2010; Manley et al., 2008). For more information regarding the mechanism of LBM, please refer to several detailed reviews on the topic (Huang et al., 2009; Patterson et al., 2010; Sengupta et al., 2012). LBM microscopes are commercially available from several companies, and custom systems can also be set up at a lower cost relatively easily using an inverted fluorescence microscope (Shroff et al., 2008b; Zhong, 2010).
CURRENT APPLICATIONS OF LBM IN NEUROSCIENCE
The great resolution gain provided by LBM has attracted researchers to apply this powerful method to examine biological architectures at a previously unattainable level of detail. Since its invention, many important discoveries have been made. The number of publications involving LBM increases rapidly every year (Long et al., 2014). Without attempting to be comprehensive, the summary below provides a representative set of examples of LBM-based neuroscience studies.
In neuroscience, a fascinating application of LBM is the examination of synaptic protein architectures. This is because synapses are organized at a level beyond the diffraction limit (Kennedy, 2000; Sheng and Kim, 2011), yet subtle changes in synaptic organization may profoundly impact synaptic function (MacGillavry et al., 2011). To this end, Dani et al. imaged pairs of pre- and post-synaptic proteins across many synapses in antibody-labeled thin sections of mouse cortex using two-color 3D STORM (Fig. 2A) (Dani et al., 2010). By systematically measuring 10 different proteins, they generated a schematic of protein localization relative to the synaptic cleft. Furthermore, by imaging antibodies targeted to different epitopes of certain proteins, they determined the orientation of the presynaptic proteins, piccolo, and bassoon. Such an ability to determine protein orientation, and thereby the layout of functional domains, may be important in revealing the precise functional roles of proteins.
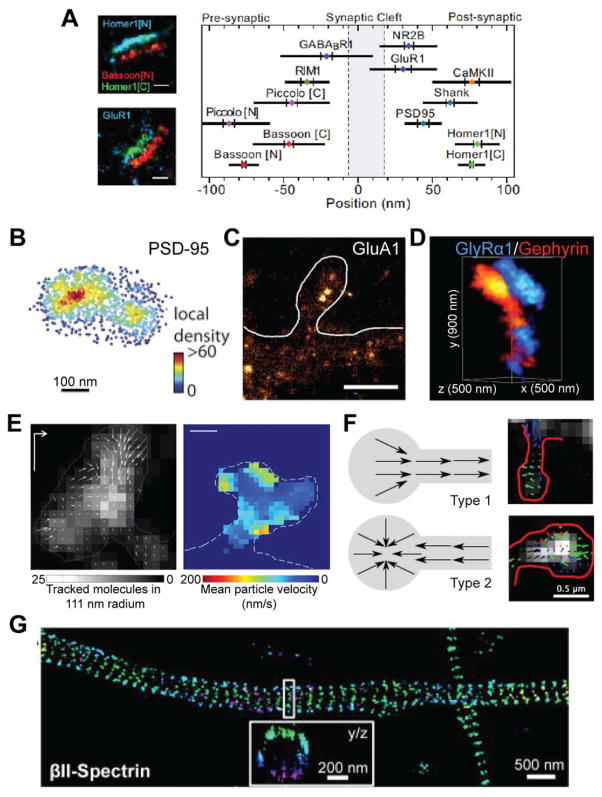
Fig. 2.
Representative studies of LBM in neuroscience. A: The localization of 10 synaptic proteins in thin sections of brain tissue [modified from Dani et al. (2010); with permission]. B: The postsynaptic scaffold protein PSD-95 forms nanometer clusters in cultured hippocampal neurons [modified from MacGillavry et al. (2013); with permission]. C: AMPA receptor GluA1 forms nanoclusters in cultured hippocampal neurons [modified from Nair et al. (2013); with permission]. D: Dual color imaging of the glycine receptor GlyRα1 and the scaffold protein Gephyrin in cultured spinal cord neurons [modified from Specht et al. (2013); with permission]. E: sptPALM tracking the flow of actin at the dendritic spine in cultured hippocampal neurons [modified from Frost et al. (2010); with permission]. Left shows the spatial distribution of actin movement directions in a spine, and right shows the mean actin movement velocity in a different spine. F: sptPALM tracking the flow of GluA1 movement in dendritic spines in hippocampal neurons, identifying two modes of GluA1 dynamics [modified from Hoze et al. (2012); with permission]. Schematics of the results (left) and representative images (right) are shown. G: The actin-associated cytoskeletal protein Spectrin forms a repeated structure in axons of cultured hippocampal neurons [modified from Xu et al. (2012); with permission].
LBM methods have also been used to reveal the organization and local heterogeneity within individual postsynaptic densities of both excitatory and inhibitory synapses. Macgillaery et al. and Nair et al. found that, in cultured hippocampal neurons, both postsynaptic scaffold proteins, such as PSD-95, and AMPA receptors formed non-random nanoclusters that were dynamically regulated (Figs. 2B and 2C) (MacGillavry et al., 2013; Nair et al., 2013, see also Fukata et al., 2013). The organization and dynamics of these nanoclusters were suggested to play important roles in controlling the amplitude and kinetic properties of synaptic currents. In addition to imaging the overall structure, LBM setups can also be used to count the number of a specific molecule since this method images each molecule individually. Specht et al. quantified the number of gephyrin molecules—the major scaffold protein of inhibitory synapses—in individual synapses in cultured spinal cord neurons (Fig. 2D) (Specht et al., 2013). On average, ~200 gephyrin molecules were present at a synapse, but this number increased to nearly 600 molecules if the synapse also expressed the glycine receptor. They also estimated the stoichiometry between gephyrin molecules and glycine receptor binding sites to be approximately 1:1.
LBM allows for the live visualization of the dynamics of individual protein molecules in cultured neurons. Frost et al. used sptPALM to identify discrete regions of actin polymerization in dendritic spines, which are the tiny protrusions on dendritic shafts that receive most excitatory synaptic inputs (Fig. 2E) (Frost et al., 2010). Similarly, sptPALM was used to directly measure the dynamics of glutamate receptors in cultured hippocampal neurons (Hoze et al., 2012; Nair et al., 2013), showing distinct modes of AMPA-type glutamate receptor dynamics across individual spines (Fig. 2F). Similar approaches have also been applied to the ATP-gated ion channel P2X7, and these channels were found to hardly move at the synapse (Shrivastava et al., 2013). However, a portion of extrasynaptic P2X7 was found to be highly mobile, suggesting the existence of two different populations of this channel that may serve distinct functions. Lu et al. also found that multiple populations of CaMKII exist in hippocampal neurons that display distinct movement characteristics and subcellular distributions (Lu et al., 2014).
The significant resolution gain achieved with LBM sometimes leads to exciting discoveries of entirely new phenomena and structures. One such example is the recent description of a periodic cytoskeletal structure in axons in the mammalian central nervous system (Fig. 2G) (Xu et al., 2012). Actin forms rings along the internal circumference of axons that repeat every ~200 nm. Spectrin connects these rings. Other actin-associated proteins were also found to form periodic structures colocalizing with the actin. Although the function of these periodic structures is not yet known, this result illustrates that unknown cellular structures remain to be discovered below the diffraction limit.
LIMITS AND CURRENT CHALLENGES OF LBM
LBM methods, while powerful, are not without limits. In addition to its intrinsic limits as a microscopy technique, the application of LBM is also constrained by several practical factors. Although microscopists continue to advance the instrumental aspects of LBM, biologists may also help to overcome some of the practical limitations, and in turn greatly facilitate the application of LBM in neuroscience. Below, I discuss several major challenges that my colleagues and I have encountered in our own experiments.
Two aspects of resolution limits: localization precision and labeling density
The localization precision of individual fluorophores is a major determinant of LBM resolution. Localization precision improves roughly in proportion to the square root of the number of photons collected from each molecule before bleaching. The collection of 1000 photons per molecule (mean photon counts from the widely used, UV-inducible green-to-red photocon-vertible fluorescence protein EosFP) provides a localization precision of ~10 nm (McKinney et al., 2009; Wiedenmann et al., 2004). Further improvement in localization precision beyond 10 nm is possible (Pertsinidis et al., 2010, 2013), but in additional to higher photon counts, it requires addressing other practical factors such the size of the fluorescent marker, variations among camera pixels, and drift of the stage and/or the sample.
In addition to localization precision, it is increasingly appreciated that the finite labeling density of the target protein is often the primary limit of imaging resolution in LBM (Nieuwenhuizen et al., 2013; Shroff et al., 2008a). As discussed above, the Nyquist–Shannon theorem states that the sampling frequency has to be more than twice as fine as the desired resolution. Therefore, a resolution of 20 nm would require a density of a labeled molecule every 10 nm and a resolution of 10 nm translates to a label every 5 nm. Regardless of the labeling method, labeling density at this very high level becomes increasingly difficult to achieve, because it begins to approach the size of individual probes (e.g., antibodies are ~12 nm and fluorescent proteins are ~2 nm in size), and sets a practical limit for LBM.
The fluorophores
LBM methods rely on photoactivatable fluorophores. Many varieties of such fluorophores are available, and the choice for individual experiments requires careful considerations (Bates et al., 2013a,b; Shcherbakova et al., 2014). Notably, most currently available PA-FPs emit a median of less than 400 photons before bleaching or switching to a dark state (Shcherbakova et al., 2014). New proteins with increased photon counts may improve localization precision. In sptPALM and similar applications, where repeated tracking of a molecule is desired, an increased photon count also allows for longer single molecule trajectories and more precise determination of the molecule’s mobile characteristics. Furthermore, EosFP variants remain one of the most popular fluorophores in LBM applications where genetic tagging is used to label the sample. Despite their advantages, EosFPs can potentially blink multiple times and thus be counted as multiple molecules before they finally bleach (Annibale et al., 2011). In one of our own tests (see Focus Box Fig. 1C), ~70% of activated mEos2 molecules gave only one molecular count in an imaging period of 100 s. However, the remaining ~30% of activated mEos2 molecules blinked, resulting in multiple molecular counts, with sometimes as many as 15 counts (Focus Box Fig. 1C). Such blinking behavior makes it difficult to quantify the exact number of molecules. A few molecules that blink many times may also create artifacts that appear as local clusters of molecules.
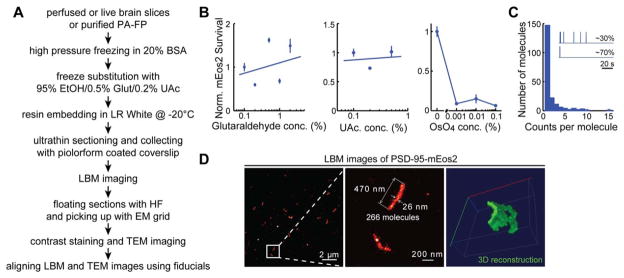
Focus Box Fig. 1.
Example of a sample preparation procedure for applying LBM to brain tissue. A: Sample preparation procedure flow chart. B: Characterization of the survivability of mEos2 in the presence of glutaraldehyde, uranyl acetate (UAc), or osmium tetroxide (OsO4). C: Characterization of the blinking behavior of mEos2. The inset shows the signal intensity collected from putative individual activated mEos2 molecules. While most activated mEos2 molecules (~70%) exhibited a single bright state and were counted only once, the remaining ~30% of activated mEos2 molecules exhibited multiple bright states due to blinking and were therefore counted several times. The histogram shows the distribution of mEos2 molecules that exhibited different blinking behaviors. The mean over-counting factor was calculated to be 1.72 ±0.04, n =5. D: Representative LBM images of PSD-95-mEos2 in mouse layer 2/3 cortical pyramidal neurons. The sections were 70-nm thick. The indicated molecule count was determined after correcting for the overcounting factor.
LBM also requires photoactivatable fluorophores with a very high bright-dark contrast ratio because high-resolution images require high labeling density. At high labeling density, the weak dark-state fluorescence of molecules with a low contrast ratio can add up to create unacceptable levels of fluorescence background, preventing the detection of single molecules. The contrast ratio of the fluorophore therefore limits the highest possible labeling density and in turn limits the achievable Nyquist resolution. Currently, both fluorescent proteins and organic dyes with high contrast ratios exist, but the selection is limited and not all photoactivatable organic dyes are commercially available. However, it was recently discovered that many types of conventional organic dyes can become photoswitchable under certain imaging conditions (Dempsey et al., 2011; Folling et al., 2008; Heilemann et al., 2008; Testa et al., 2010), expanding the repertoire of fluorophore selection. LBM applications would most certainly benefit from further development efforts to expand the currently limited choices of fluorophores for multi-color imaging. The development of FPs suitable for strong sample preparation conditions, which are required to appropriately preserve the sample structure at the nanometer scale, is also desired (see Focus Box).
The labeling method
The labeling of samples remains another major challenge for LBM. There are two main classes of fluorescent labeling methods: affinity labeling (e.g., with antibodies) and genetic tagging (e.g., with FP tagging). Affinity labeling is widely used to visualize endogenous proteins. However, although antibodies exist for many targets, whether their affinity and specificity are suitable for superresolution imaging needs to be examined carefully [see reference Luby-Phelps et al. (2003) for an example of unreliable antibody labeling]. Furthermore, the bulky size of antibodies (~12 nm for a single antibody molecule and ~25 nm if both primary and secondary antibodies are used) often limits their penetration into brain tissue and sets an upper boundary for the labeling density, and in turn the achievable Nyquist resolution. The bulky antibody size also introduces significant uncertainty in the localization of a protein target. Finally, antibody labeling is not usually compatible with live imaging of intracellular molecules. Therefore, there is a need for highly specific antibodies and smaller alternatives such as single-chain antibodies (Huston et al., 1988) and nanobodies (Ries et al., 2012). These alternatives have further advantages in that they are single polypeptides that can be expressed intracellularly, making them compatible with live cell imaging by tagging them with an FP and expressing them within a cell (Fukata et al., 2013; Gross et al., 2013; Nizak et al., 2003). However, achieving stoichiometric labeling remains difficult, and excess labels can be mislocalized.
Conversely, FPs can be specifically attached to most proteins stoichiometrically by molecular cloning methods, and their small sizes (~2 nm) allow for high packing density and localization precision. Importantly, data from our lab (Focus Box Fig. 1B) and others (Brown et al., 2010; Shtengel et al., 2014) have shown that many FPs are resistant to glutaraldehyde fixation, which is important for the preservation of molecular structures at the nanometer level. In most experiments, an FP-tagged protein is overexpressed, but overexpression can lead to undesired perturbations in cell function and artifacts in protein localization [see Schnell et al. (2002) for an example]. In principle, gene-replacement techniques can be used to express the FP-tagged protein at endogenous levels, but this is rarely done because the associated cost is high and typical knock-in strategies lead to the expression of the FP tagged protein in all cells where the target protein is typically expressed, resulting in a loss of cell-specific contrast [e.g., see Herzog et al. (2011)]. We have recently developed a conditional knock-in strategy that allows the FP-tagged protein to be expressed at endogenous levels in a sparse subset of cells or in specific cell types (Fortin et al., 2014). The recent development of mouse transgenic techniques, such as the CRISPR technology, may also reduce the time and cost required to generate knock-in mice (Sander and Joung, 2014).
The sample preparation method
It is sometimes underappreciated that regardless of the resolution achieved by a superresolution imaging method, any microscopic image can only be as good as what the sample allows. LBM experiments are intrinsically a slow technique (~1–30 min per image) due to the need for many imaging iterations, and are associated with severe photobleaching. Therefore, most LBM experiments examine fixed samples. However, the preparation of fixed samples is not a trivial task. Milder fixation conditions are usually preferred for conventional fluorescence microscopy because strong fixatives, such as glutaraldehyde, can increase background fluorescence and directly crosslink with proteins to reduce the antigenicity of antibody epitopes. At the same time, mild fixation conditions may result in insufficient levels of fixation and cause artifacts that are visible at the resolution level of LBM. Fortunately, because many FPs used in LBM, such as the EosFP, are resistant to even high concentrations of glutaraldehyde (e.g., 2%) (Focus Box Fig. 1B), this fixative can be used as part of sample preparation for LBM when genetic tagging is used as the labeling strategy. Overall, optimization of sample preparation conditions is required to achieve a balance between the preservation of cellular architecture and the labeling density required for superresolution imaging.
To date, most LBM applications in neuroscience are on single layer, dissociated neuronal cultures. This is because LBM relies on single molecule imaging, which is very sensitive to the light scattering and aberration associated with thick brain samples. For brain tissue, very thin sections under fixed conditions have to be prepared (Dani et al., 2010). An active area of research is to adapt EM sample preparation methods to produce resin-embedded ultrathin sections for LBM imaging (Brown et al., 2010; Micheva and Smith, 2007; Watanabe et al., 2011, 2014; Yao et al., 2011). Such an approach has many advantages: it improves the z resolution, as section thicknesses are typically 100 nm or less, it eliminates most background fluorescence and light aberration that are associated with thicker samples and it usually achieves a high degree of structural preservation. In the Focus Box, we illustrate our sample preparation procedure and example application to embed mouse brain tissues expressing PSD-95-mEos2 in LR White resin (Focus Box Fig. 1), and show that mEos2 readily survives this procedure. The samples were then sectioned to 50–100 nm and imaged with LBM. Serial reconstruction of the images revealed that PSD-95 exists as a sheet-like structure, consistent with the notion that it is the major constituent of the PSD in excitatory synapses.
The big leap in resolution provided by LBM may sometimes uncover a level of detail that is difficult to interpret on its own. Using resin-embedded or other types of ultrathin sections, it may be possible to first image a section with LBM and then with EM. The LBM and EM images can then be aligned to interpret the protein localization information from LBM images in the context of EM-visible ultrastructures [Focus Box Fig. 2 and Betzig et al. (2006); Sochacki et al. (2014); Watanabe et al. (2011)]. Such LBM and EM correlation has significant advantages over immuno-EM, because LBM/EM correlation can achieve much higher labeling density, and thus better Nyquist resolution. In our preliminary studies, we tested mEos2 survivability under the treatment of common EM staining reagents. mEos2 survives uranyl acetate treatments, but not osmium treatments (Focus Box Fig. 1B). We further demonstrated that LBM/EM correlation in brain samples is possible (Focus Box Fig. 2), although the reproducibility of the sample preparation procedure remains to be improved. When fully developed, the correlation of EM with LBM has the potential to revolutionize our understanding of protein organization at the ultrastructural level.
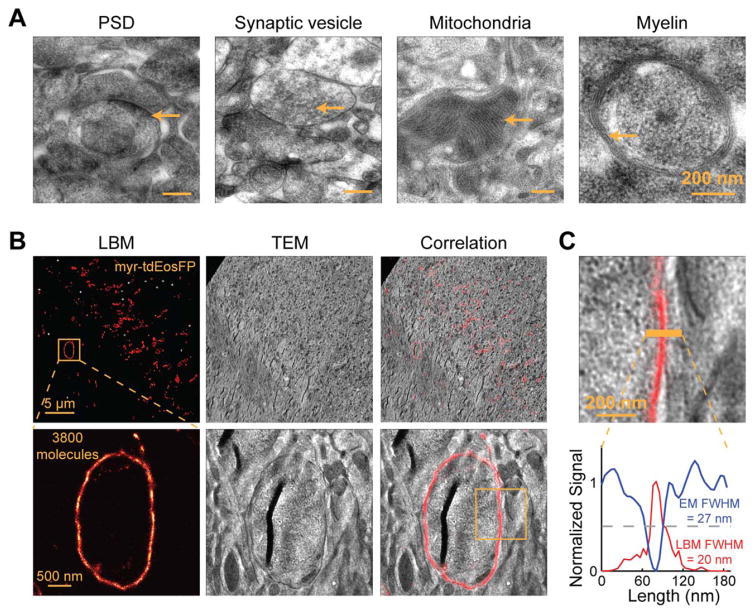
Focus Box Fig. 2.
EM characterization of our sample procedure and LBM/EM correlations. A: Representative TEM images showing ultrastructures in mouse brain tissue preserved using our sample preparation procedure. B: Representative LBM/EM correlative images. Myristoylated tdEosFP was expressed in a subset of neurons in the fly optic lobe to label the plasma membrane, and imaged by LBM (left). The same section was later imaged with TEM (middle), and the LBM and EM images were then correlated with each other (right). The yellow box in the lower right panel corresponds to the zoom-in image in panel (C). C: A zoom-in view of the LBM/EM correlative image from the yellow box in panel (B) (top) with the LBM and EM intensity profiles shown as red and blue lines, respectively (bottom). Note that stronger EM intensity is darker and has lower values. The overlapping peaks and troughs, and comparable full width half maximums (FWHMs) from LBM and EM demonstrate the alignment accuracy and confirm the resolution of the LBM method.
SUMMARY
Recent progress in LBM has started to make it possible for neuroscientists to investigate subcellular protein architecture and molecular dynamics in neurons at 10–20 nm resolution. LBM imaging experiments have already described previously unknown structures, clarified the organization of macromolecular complexes, and revealed the dynamics of critical synaptic proteins. However, the improvements in microscopic resolution also present demands and challenges in associated areas, such as fluorophore development, labeling methods, and sample preparation. Although challenges and limitations remain, LBM is rapidly becoming a fundamental tool for neuroscience studies.
Focus box: our efforts at applying LBM and LBM/EM correlation to brain tissue
The application of LBM has been largely limited to samples appropriate for single molecule imaging such as single layer cell cultures. Severe background fluorescence, light scattering and light aberration have limited the ability of these techniques to be applied directly to image deep into thick tissues, such as the brain. Sectioning is required to study protein organization deep within thick samples (Betzig et al., 2006; Dani et al., 2010; Sochacki et al., 2014; Vaziri and Shank, 2010; Watanabe et al., 2011; York et al., 2011).
Inspired by the array tomography sample preparation method (Micheva and Smith, 2007; Punge et al., 2008; Watanabe et al., 2011, 2014), we have developed a procedure to section intact brain samples embedded in the LR White acrylic resin into ultra-thin (50–500 μm) sections, which allows for LBM imaging. To achieve a high level of Nyquist resolution, we typically label our samples using genetic tagging methods with PA-FPs, such as mEos2 (McKinney et al., 2009). The key is to appropriately preserve protein structures for high resolution imaging while maintaining the function of PA-FPs. After comparing multiple procedures, we favored a procedure in which the sample was high pressure frozen in the presence of 20% BSA, followed by freeze substitution in a medium containing 95% ethanol and fixatives for preserving structure (Focus Box Fig. 1A). To test whether our current PA-FP of choice, mEos2, could survive the strong fixation and processing conditions for resin embedding, we froze a known concentration of purified recombinant mEos2 and freeze substituted the sample under a variety of conditions, such as different glutaraldehyde concentrations. The sample was then infiltrated and polymerized in LR White resin at −20°C using a chemical catalyst and an accelerator, sectioned to 100 nm and examined with LBM. In contrast to the relatively poor preservation of antigenicity of antibody epitopes, mEos2 survived well even when high concentrations of glutaraldehyde (2%) were added to the freeze substitution medium (Focus Box Fig. 1B). Similarly, mEos2 also survived well in the presence of uranyl acetate (UAc, up to 0.5%). However, as little as 0.001% osmium tetroxide (OsO4) led to a significant decrease in the protein counts. Finally, few mEos2 molecules survived if the LR White resin was polymerized at an elevated temperature (50°C overnight; data not shown). Based on these fundings, we typically perform freeze substitution in the presence of 1–2% glutaraldehyde and 0.2–0.5% uranyl acetate, and embed in LR White resin at −20°C using a chemical catalyst and an accelerator.
Photo-switching of EosFPs is irreversible. In principle, such proteins allow for quantification of absolute molecular counts. However, EosFPs are known to blink (Annibale et al., 2011), which can lead to the overcounting of molecules. We examined the blinking behavior of individual mEos2 molecules embedded in LR White resin after a single pulse of light activation and found that ~70% of molecules exhibited a single bright peak (Focus Box Fig. 1C). However, the remaining ~30% of the activated mEos2 molecules blinked on and off multiple times before they finally bleached. Some blinked as many as 15 times. Overall, blinking of mEos2 in our preparation resulted in an overestimation of mEos2 by a factor of 1.7. Given our finite observation time (100 s) after activation, this likely is an underestimate of the overcounting factor.
We then tested whether this procedure allows for the examination of synaptic protein organization in brain tissue. We expressed mEos2-tagged PSD-95 (PSD-95-mEos2) in vivo in mouse layer 2/3 cortical pyramidal neurons using in utero electroporation [see Kim and Sheng (2004) for a review of PSD-95]. To minimize sample damage during dissection, adult mice (P30–140) were perfused with 4% paraformaldehyde and 0.2% glutaraldehyde before the brains were dissected. The brain tissue was sectioned to 100-μm thick using a vibratome and then high pressure frozen and freeze substituted. The samples were then embedded in LR White resin at −20°C. For many brain samples, the resin polymerized poorly inside the tissue, presumably because the viscosity of the unpolymerized resin at −20°C made the penetration of the accelerator into the brain tissue very slow. At the same time, the resin outside of the tissue started to polymerize soon after the accelerator was added. As a result, the sample block often had a soft “yolk” inside a hard shell. Despite this limitation, we could still sometimes obtain samples with adequate resin polymerization throughout the brain tissue. A representative LBM image of a 70 nm section of a well polymerized sample is shown in Focus Box Figure 1D. Most of the PSD-95-mEos2 structures exist as thin lines with characteristics of the PSD, consistent with the established role of PSD-95 as one of the major constituents of the PSD. By producing serial sections, we were able to image the same set of PSD-95 structures over many consecutive sections and perform 3D reconstruction (Focus Box Fig. 1D, right panel). To evaluate the structural preservation, we examined processed mouse brain samples by transmission EM (TEM). Although the contrast was poor overall due to a lack of osmium staining, we were able to recognize many ultrastructures, including the PSD, presynaptic vesicles, mitochondria, and myelin, indicating adequate structural preservation of the sample (Focus Box Fig. 2A). This procedure was also compatible with other types of samples, including the fly brain (Focus Box Fig. 2B).
We measured labeling density, which defines the achievable Nyquist resolution, in our brain tissue samples. We scored 266 molecules in the PSD-95-mEos2 structure in a mouse layer 2/3 cortical pyramidal neuron shown in the middle panel of Focus Box Figure 1D (after correcting for overcounting due to blinking). We estimated the volume of the PSD-95-mEos2 structure to be 470 nm × 26 nm × 70 nm =8.6 × 105 nm3. We calculated the intermolecular distance between two neighboring PSD-95 molecules to be 15 nm. In the fly brain tissue example shown in the lower left panel of Focus Box Figure 2B, we scored ~3800 membrane bounded, tandem-dimer EosFP (myr-tdEosFP) molecules (after correction for overcounting due to blinking and the tdEosFP used here) in a ring-like structure of a fly optic lobe neuron, presumably corresponding to a position close to the edge of a soma. By measuring the circumference (3 μm) and knowing that the thickness of the section was 50 nm, we estimate that the molecular density along the membrane was around 1/ 100 nm2 with an intermolecular distance of 10 nm.
To test whether our sample preparation procedure allowed for LBM/EM correlation, we first used LBM to image a section of fly optic lobe expressing myr-tdEosFP on a Pioloform-coated coverslip. We then floated the sections together with the Pioloform film on a water surface using 0.1% hydrofluoric acid to etch the coverslip, and collected the section onto an EM grid (Watanabe et al., 2014). The sections were contrast stained with uranyl acetate and lead, and imaged with TEM. Gold fiducials that were observable both by LBM and by TEM were used to align and correlate the LBM image with the corresponding TEM image using an affine transformation algorithm (Focus Box Fig. 2B). At high magnification, it can be seen that the membrane structure observed by LBM correlated well with the ultrastructures shown by TEM (Focus Box Fig. 2B, lower panels). Across our samples based on fiducial alignments, we estimated that we achieve an alignment accuracy of ~50 nm. This LBM/EM correlation also provided further validation of the resolution for LBM: the full width half maximum (FWHM) of structures measured by LBM approached that measured by EM at many places (Focus Box Fig. 2C).
In summary, our sample preparation procedure can be used to embed thick tissues such as brain samples, in LR White resin, which allows the sample to be processed into ultrathin sections. This sample preparation procedure allows for the application of LBM and LBM/EM correlative studies to brain tissue. A major challenge in the current procedure is that the resin polymerization of brain tissue is variable. In addition, not being able to use osmium during fixation results in low contrast EM ultrastructures. Future development of PA-FPs with increased heat stability and osmium resistance will likely overcome these obstacles.
Acknowledgments
Contract grant sponsor: NIH Director’s New Innovator Award (H.Z.); Contract grant number: DP2OD008425; Contract grant sponsor: NIH R21 grant (H.Z.); Contract grant number: R21NS084315; Contract grant sponsor: HHMI.
Unpublished experiments were performed with the support of Dr. Eric Betzig at the HHMI Janelia Research Campus. Dr. Richard Fetter contributed to sample preparation and performed all TEM imaging. Myristoylated tdEosFP-expressing flies were a gift from Barret Pfeiffer in Dr. Gerry Rubin’s Lab at the HHMI Janelia Research Campus. 3D reconstruction of PSD-95-mEos2 on ultrathin sections was performed by Dr. Dmitri Chklovskii at the HHMI Janelia Research Campus. The authors thank Drs. Richard Fetter (HHMI Janelia Research Campus) and Tianyi Mao (Oregon Health & Science University) for their critical comments on this manuscript, and Teresa Newton (Oregon Health & Science University) for proof-reading the manuscript.
References
- Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Quantitative photo activated localization microscopy: Unraveling the effects of photoblinking. PLoS One. 2011;6:e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photoswitchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M, Jones SA, Zhuang X. Preparation of photoswitchable labeled antibodies for STORM imaging. Cold Spring Harb Protoc. 2013a;2013:540–541. doi: 10.1101/pdb.prot075168. [DOI] [PubMed] [Google Scholar]
- Bates M, Jones SA, Zhuang X. Transfection of genetically encoded photoswitchable probes for STORM imaging. Cold Spring Harb Protoc. 2013b;2013:537–539. doi: 10.1101/pdb.prot075150. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Brown TA, Fetter RD, Tkachuk AN, Clayton DA. Approaches toward super-resolution fluorescence imaging of mitochondrial proteins using PALM. Methods. 2010;51:458–463. doi: 10.1016/j.ymeth.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Sengupta P, Dai Y, Lippincott-Schwartz J, Kachar B. Bleaching/blinking assisted localization microscopy for superresolution imaging using standard fluorescent molecules. Proc Natl Acad Sci USA. 2011;108:21081–21086. doi: 10.1073/pnas.1117430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton PM. Three-dimensional structured illumination microscopy and its application to chromosome structure. Chromosome Res. 2008;16:351–365. doi: 10.1007/s10577-008-1231-9. [DOI] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based superresolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner A, Geisler C, von Middendorff C, Bock H, Wenzel D, Medda R, Andresen M, Stiel AC, Jakobs S, Eggeling C, Schonle A, Hell SW. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folling J, Bossi M, Bock H, Medda R, Wurm CA, Hein B, Jakobs S, Eggeling C, Hell SW. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- Fortin DA, Tillo SE, Yang G, Rah JC, Melander JB, Bai S, Soler-Cedeno O, Qin M, Zemelman BV, Guo C, Mao T, Zhong H. Live Imaging of endogenous PSD-95 using ENABLED: A conditional strategy to fluorescently label endogenous proteins. J Neurosci. 2014;34:16698–16712. doi: 10.1523/JNEUROSCI.3888-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Dimitrov A, Boncompain G, Vielemeyer O, Perez F, Fukata M. Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J Cell Biol. 2013;202:145–161. doi: 10.1083/jcb.201302071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Hosy E, Levet F, Constals A, Schulze K, Sobolevsky AI, Rosconi MP, Gouaux E, Tampe R, Choquet D, Cognet L. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys J. 2010;99:1303–1310. doi: 10.1016/j.bpj.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TJ, Hess ST. Chapter 12: Nanoscale biological fluorescence imaging: Breaking the diffraction barrier. Methods Cell Biol. 2008;89:329–358. doi: 10.1016/S0091-679X(08)00612-2. [DOI] [PubMed] [Google Scholar]
- Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GC, McGee AW, Sabatini BL, Roberts RW, Arnold DB. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- Herzog E, Nadrigny F, Silm K, Biesemann C, Helling I, Bersot T, Steffens H, Schwartzmann R, Nagerl UV, El Mestikawy S, Rhee J, Kirchhoff F, Brose N. In vivo imaging of intersynaptic vesicle exchange using VGLUT1 Venus knock-in mice. J Neurosci. 2011;31:15544–15559. doi: 10.1523/JNEUROSCI.2073-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoze N, Nair D, Hosy E, Sieben C, Manley S, Herrmann A, Sibarita JB, Choquet D, Holcman D. Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proc Natl Acad Sci USA. 2012;109:17052–17057. doi: 10.1073/pnas.1204589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008a;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008b;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Hartwich TM, Rivera-Molina FE, Lin Y, Duim WC, Long JJ, Uchil PD, Myers JR, Baird MA, Mothes W, Davidson MW, Toomre D, Bewersdorf J. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat Methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- Kennedy MB. Signal-processing machines at the postsynaptic density. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew MD, Thompson MA, Badieirostami M, Moerner WE. In vivo three-dimensional superresolution fluorescence tracking using a double-helix point spread function. Proc Soc Photo Opt Instrum Eng. 2010;7571:75710Z. doi: 10.1117/12.842608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew MD, Lee SF, Ptacin JL, Lee MK, Twieg RJ, Shapiro L, Moerner WE. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus. Proc Natl Acad Sci USA. 2011;108:E1102–1110. doi: 10.1073/pnas.1114444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BR, Robinson DC, Zhong H. Subdiffractive microscopy: Techniques, applications, and challenges. Wiley Interdiscip Rev Syst Biol Med. 2014;6:151–168. doi: 10.1002/wsbm.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HE, MacGillavry HD, Frost NA, Blanpied TA. Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J Neurosci. 2014;34:7600–7610. doi: 10.1523/JNEUROSCI.4364-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K, Ning G, Fogerty J, Besharse JC. Visualization of identified GFP-expressing cells by light and electron microscopy. J Histochem Cytochem. 2003;51:271–274. doi: 10.1177/002215540305100301. [DOI] [PubMed] [Google Scholar]
- MacGillavry HD, Kerr JM, Blanpied TA. Lateral organization of the postsynaptic density. Mol Cell Neurosci. 2011;48:321–331. doi: 10.1016/j.mcn.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78:615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: A new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen KI, Churchman LS, Spudich JA, Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat Methods. 2010;7:377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D, Hosy E, Petersen JD, Constals A, Giannone G, Choquet D, Sibarita JB. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J Neurosci. 2013;33:13204–13224. doi: 10.1523/JNEUROSCI.2381-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen RP, Lidke KA, Bates M, Puig DL, Grunwald D, Stallinga S, Rieger B. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–562. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizak C, Monier S, del Nery E, Moutel S, Goud B, Perez F. Recombinant antibodies to the small GTPase Rab6 as conformation sensors. Science. 2003;300:984–987. doi: 10.1126/science.1083911. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Superresolution imaging using single-molecule localization. Annu Rev Phys Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertsinidis A, Zhang Y, Chu S. Subnanometre single-molecule localization, registration and distance measurements. Nature. 2010;466:647–651. doi: 10.1038/nature09163. [DOI] [PubMed] [Google Scholar]
- Pertsinidis A, Mukherjee K, Sharma M, Pang ZP, Park SR, Zhang Y, Brunger AT, Sudhof TC, Chu S. Ultrahigh-resolution imaging reveals formation of neuronal SNARE/Munc18 complexes in situ. Proc Natl Acad Sci USA. 2013;110:E2812–2820. doi: 10.1073/pnas.1310654110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punge A, Rizzoli SO, Jahn R, Wildanger JD, Meyer L, Schonle A, Kastrup L, Hell SW. 3D reconstruction of high-resolution STED microscope images. Microsc Res Tech. 2008;71:644–650. doi: 10.1002/jemt.20602. [DOI] [PubMed] [Google Scholar]
- Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci USA. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Van Engelenburg S, Lippincott-Schwartz J. Visualizing cell structure and function with point-localization superresolution imaging. Dev Cell. 2012;23:1092–1102. doi: 10.1016/j.devcel.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Sengupta P, Lippincott-Schwartz J, Verkhusha VV. Photocontrollable fluorescent proteins for superresolution imaging. Annu Rev Biophys. 2014;43:303–329. doi: 10.1146/annurev-biophys-051013-022836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E. The postsynaptic organization of synapses. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava AN, Rodriguez PC, Triller A, Renner M. Dynamic micro-organization of P2X7 receptors revealed by PALM based single particle tracking. Front Cell Neurosci. 2013;7:232. doi: 10.3389/fncel.2013.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci USA. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008a;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, White H, Betzig E. Photoactivated localization microscopy (PALM) of adhesion complexes. Curr Protoc Cell Biol. 2008b;Chapter 4(Unit 4):21. doi: 10.1002/0471143030.cb0421s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, Hess HF. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultra-structure. Proc Natl Acad Sci USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtengel G, Wang Y, Zhang Z, Goh WI, Hess HF, Kanchanawong P. Imaging cellular ultrastructure by PALM, iPALM, and correlative iPALM-EM. Methods Cell Biol. 2014;123:273–294. doi: 10.1016/B978-0-12-420138-5.00015-X. [DOI] [PubMed] [Google Scholar]
- Simonson PD, Rothenberg E, Selvin PR. Single-molecule-based super-resolution images in the presence of multiple fluorophores. Nano Lett. 2011;11:5090–5096. doi: 10.1021/nl203560r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochacki KA, Shtengel G, van Engelenburg SB, Hess HF, Taraska JW. Correlative super-resolution fluorescence and metal-replica transmission electron microscopy. Nat Methods. 2014;11:305–308. doi: 10.1038/nmeth.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht CG, Izeddin I, Rodriguez PC, El Beheiry M, Rostaing P, Darzacq X, Dahan M, Triller A. Quantitative nanoscopy of inhibitory synapses: Counting gephyrin molecules and receptor binding sites. Neuron. 2013;79:308–321. doi: 10.1016/j.neuron.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Testa I, Wurm CA, Medda R, Rothermel E, von Middendorf C, Folling J, Jakobs S, Schonle A, Hell SW, Eggeling C. Multi-color fluorescence nanoscopy in fixed and living cells by exciting conventional fluorophores with a single wavelength. Biophys J. 2010;99:2686–2694. doi: 10.1016/j.bpj.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Linde S, Heilemann M, Sauer M. Live-cell super-resolution imaging with synthetic fluorophores. Annu Rev Phys Chem. 2012;63:519–540. doi: 10.1146/annurev-physchem-032811-112012. [DOI] [PubMed] [Google Scholar]
- Vaziri A, Shank CV. Ultrafast widefield optical sectioning microscopy by multifocal temporal focusing. Opt Express. 2010;18:19645–19655. doi: 10.1364/OE.18.019645. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, Davis MW, Hell SW, Jorgensen EM. Protein localization in electron micrographs using fluorescence nanoscopy. Nat Methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Lehmann M, Hujber E, Fetter RD, Richards J, Sohl-Kielczynski B, Felies A, Rosenmund C, Schmoranzer J, Jorgensen EM. Nanometer-resolution fluorescence electron microscopy (nano-EM) in cultured cells. Methods Mol Biol. 2014;1117:503–526. doi: 10.1007/978-1-62703-776-1_22. [DOI] [PubMed] [Google Scholar]
- Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2012;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Fetter RD, Hu P, Betzig E, Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011;25:569–580. doi: 10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H. Photoactivated localization microscopy (PALM): An optical technique for achieving ~10-nm resolution. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.top91. pdb top91. [DOI] [PubMed] [Google Scholar]