Abstract
The proto-oncogene PTPN11 encodes a cytoplasmic protein tyrosine phosphatase, SHP2, which is required for normal development and sustained activation of the Ras-MAPK signaling pathway. Germline mutations in SHP2 cause developmental disorders, and somatic mutations have been identified in childhood and adult cancers and drive leukemia in mice. Despite our knowledge of the PTPN11 variations associated with pathology, the structural and functional consequences of many disease-associated mutants remain poorly understood. Here, we combine X-ray crystallography, small angle X-ray scattering and biochemistry to elucidate structural and mechanistic features of three cancer-associated SHP2 variants harboring single point-mutations within the N-SH2:PTP interdomain autoinhibitory interface. Our findings directly compare the impact of each mutation on autoinhibition of the phosphatase and advance the development of structure-guided and mutation-specific SHP2 therapies.
The structural and functional impact of mutations frequently observed in oncogenes and tumor suppressors remain largely uncharacterized. One such oncogene, PTPN11, encodes a cytoplasmic protein tyrosine phosphatase, SHP2, which is required for normal development and sustained activation of the Ras-MAPK signaling pathway.1 Many germline mutations in SHP2 lead to developmental disorders,2 and somatic mutations have been identified in childhood and adult malignancies3 and cause leukemia in mice.4 Specifically, gain-of-function (GoF) PTPN11 mutations are found in 35% of juvenile myelomonocytic leukemias, 10% of myelodysplasic syndromes, 7% of B-precursor acute lymphoblastic leukemias and 5% of acute myeloid leukemias.5–9 SHP2 mutations have also been detected in multiple myeloma and solid tumors of the lungs, skin, nervous system and colon.5, 10, 11
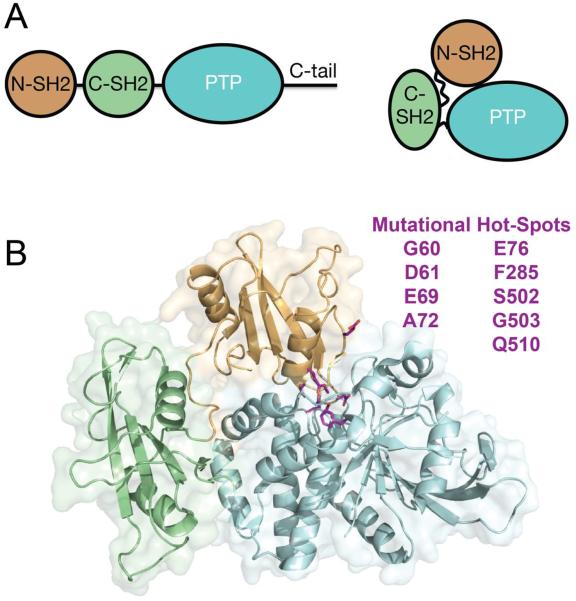
SHP2 is composed of two tandem Src homology 2 (SH2) domains, N-SH2 and C-SH2, followed by a catalytic protein tyrosine phosphatase (PTP) domain and a C-terminal tail that contains at least two tyrosyl phosphorylation sites and a proline-rich motif (Figure 1A).12 In the basal state, the N-SH2 domain packs against and sterically inhibits the catalytic activity of the PTP domain by inserting a loop into the active site cleft, which occludes substrate access (Figure 1A & B).13 Upon engagement of the N-SH2 and C-SH2 domains of SHP2 with tyrosine-phosphorylated signaling proteins, such as the epidermal growth factor receptor (EGFR),14 platelet derived growth factor receptor (PDGFR),15 or adaptor proteins,16, 17 SHP2 phosphatase activity is increased, presumably as a result of an induced conformational opening that alleviates N-SH2 autoinhibition of the PTP active site.18
Figure 1.
(A) Overview of SHP2 domain architecture and location of N-SH2, C-SH2 and PTP domains in the autoinhibited conformation. (B) Frequently occurring cancer mutations, shown in magenta, localize to the interaction interface between N-SH2 and PTP domains. Cancer-associated mutations identified from the COSMIC database. Structure PDB:2SHP.
Previous structural and functional analyses of pro-oncogenic SHP2 variants have revealed a correlation between position of amino acid mutation within the protein sequence and enhancement of phosphatase activity. Most cancer-associated mutations localize to the autoinhibition interface between the N-SH2 and PTP domains and, in general, activate the phosphatase.19 E76K, the most prevalent leukemia-associated mutation, enhances basal phosphatase activity by ~20-fold, induces aberrant growth of multiple hematopoetic lineages in cell culture20 and drives leukemia in murine models.21 Despite our knowledge of PTPN11 variations associated with pathology, the structural and functional consequences of many mutations are poorly understood, and quantitative comparison among various pro-oncogenic mutations remains largely incomplete.
Here we compare structural and mechanistic features of three SHP2 leukemia variants, SHP2E76Q, SHP2F285S and SHP2S502P, harboring single point mutations at the N-SH2:PTP interdomain autoinhibitory interface. The crystal structures of the mutated proteins show local differences in the inactive conformation of SHP2 associated with destabilization of the autoinhibited conformation. Analysis of the ensemble of solution structures, using small-angle X-ray scattering, shows evidence for conformational expansion and opening that varies among the different proteins, and correlates with the mutation-induced enhancement of phosphatase activity and sensitivity to activating ligands.
Materials & Methods
Materials
6,8-Difluoro-4-Methylumbelliferyl Phosphate (DiFMUP, D22065) and 6,8-difluoro-7-hydroxy-4-methylcoumarin (DiFMU, D6566) were purchased from LifeTechnologies, 2-mercaptoethanol from Aldrich (M6250), lysogeny broth from Invitrogen/LifeTechnologies (12795-084) and phosphorylated peptides from AnaSpec. Unless otherwise stated, all other reagents were obtained from commercial sources and used without further purification.
Plasmid Construction
SHP2 (1-525) DNA was amplified from full-length SHP2 plasmid DNA and inserted into the pET19B vector between BamH1 restriction sites using InFusion cloning to generate pET19b-SHP2wt. Insertion was verified via DNA sequencing. Cancer associated SHP2 (1-525) mutants were generated using site directed mutagenesis (QuikChange II, Stratagene) with primers designed to install single or double point mutations at the desired codon. SHP2E76Q was designed using forward primer 5′-GCC ACT TTG GCT CAG TTG GTC CAG TAT TAC ATG G-3′ and reverse primer 5′-CCA TGT AAT ACT GGA CCA ACT GAG CCA AAG TGG C-3′, and SHP2S502P with forward primer 5′-GGT CTC AGA GGC CAG GGA TGG TCC AGA C-3′ and reverse primer 5′-GTC TGG ACC ATC CCT GGC CTC TGA GAC C-3″. The PTP domain of SHP2 (200–525) was amplified from pET19b-Shp2wt using forward primer 5′-ACC TGT ATT TTC AGG GAT CCG GAG GAC GTA TAA ATG CTG CTG AAA T-3′ and reverse 5′-GCT TTG TTA GCA GCC GGA TCC TTA TAG TGT TTC AAT ATA ATG-3′ and inserted into BamH1-digested and gel-excised empty pET19b using InFusion cloning. All plasmids reported herein were verified through DNA sequencing.
SHP2 Phosphatase Expression and His-tagged Protein Purification
SHP2-encoding plasmid DNA was transformed into BL21 DE3 E. coli, from which a single colony was selected, inoculated into a 50 mL starter culture of Lysogeny broth (LB) with 1 mg/mL ampicillin and allowed to grow overnight. The following morning, 5 mL of saturated culture was inoculated into 1 L of LB with 1 mg/mL ampicillin and allowed to grow until the cells reached an OD600 of 0.6–0.8, after which cells were placed at 4°C for 1 hour, and then allowed to express mutant or wild-type SHP2 proteins at 18°C in the presence of 0.5 mM ITPG overnight. The following morning, cells were pelleted at 4°C and 4,000 rpm for 20 min, resuspended in 45 mL of ice-cold 50 mM Tris pH 8.0, 200 mM NaCl, 5% glycerol, 10 mM β-mercaptoethanol, 0.1 % triton-X and 1 cOmplete, EDTA-free protease inhibitor cocktail tablet (Roche Diagnostics, #11873580001), and lysed via sonication. Total cell lysate was clarified at 20,000g for 30 minutes, after which the supernatant was added to Ni2+−NTA Agarose (QIAGEN, #1018240) and His-tagged SHP2 proteins were allowed to bind the resin at 4°C for 1 hour with gentle rocking. After binding, Ni2+−NTA resin was pelleted at 500g for 5 min and washed three times with 50 mL of ice-cold 50 mM Tris pH 8.0, 200 mM NaCl, 5% glycerol and 10 mM β-mercaptoethanol (Buffer 1) and three times with Buffer 1 containing 20 mL of 50 mM imidazole (pH 8.0). After washes, SHP2 proteins were eluted from the resin in 10 mL of Buffer 1 with 300 mM imidazole, pH 8.0, which was added to the resin in a 20 mL plastic column in 2 or 3 mL increments. To remove the His tag, eluted SHP2 proteins were incubated with 0.2 mg/mL TEV protease overnight at 4°C and tag removal was confirmed through polyacrylamide gel electrophoresis.
FPLC Purification of SHP2 Phosphatases
After His tag removal, SHP2 proteins were purified on a MonoQ™ 10/100 GL anion exchange column (GE Healthcare) using a linear gradient of 10 – 100% 1 M NaCl in 50 mM Tris pH 8.0, 5% glycerol, and 10 mM β-mercaptoethanol. Fractions were collected in 1 mL increments and assessed for purity with PAGE. Pure fractions were pooled, concentrated to 0.5 mL and purified further on a Superdex™ 200 10/300 GL size exclusion column (GE Healthcare) and equilibrated into 25 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol and 2 mM TCEP. Fractions were collected in 1 mL increments and assessed for purity via PAGE, and pure fractions were concentrated, dispensed into 5 μL single-use aliquots, and stored at −80°C until use. Purity was veryified through liquid chromatography-mass spectrometry.
Crystallization of Cancer-Associated SHP2 Mutants
Hanging drop vapor diffusion method was used for crystallization of SHP2 E76Q, S502P, and F285S proteins with the crystallization well containing 100 mM Tris pH 8.5, 20% PEG 3350 and 1 mM TCEP and a drop with a 1:2 volume of SHP2 protein and crystallization solution in 24-well VDX trays (Hampton Research, HR3-142). Crystals were formed within ten days and harvested, followed by cryoprotection using the crystallization solution with the addition of 20% glycerol, and flash freezing directly into liquid nitrogen.
X-Ray Data Collection, Phasing and Refinement
Diffraction data for the SHP2E76Q, SHP2S502P, and SHP2F285S crystals were collected on a Dectris Pilatus 6M Detector at beamline 17ID (IMCA-CAT) at the Advanced Photon Source at Argonne National Laboratories. The data were measured from a single crystal maintained at 100°K at a wavelength of 1Å, and the reflections were indexed, integrated, and scaled using XDS.22 The spacegroup of the crystals was P21 with 2 molecules in the asymmetric unit. The structure was determined with Fourier methods, using the SHP2 apo structure (PDB: 2SHP)13 with all waters removed. Structure determination was achieved through iterative rounds of positional and simulated annealing refinement using BUSTER,23 with model building using COOT.24 Individual B-factors were refined using an overall anisotropic B-factor refinement along with bulk solvent correction. The solvent was built into the density in later rounds of the refinement. Data collection and refinement statistics are shown in Table I. The coordinates for the structures of E76Q, F285S, and S502P have been deposited in the protein data bank with PDB ID codes 5IBS, 5I6V, and 5IBM, respectively.
Table I.
X-Ray Data Collection, Phasing and Refinement Statistics.
| SHP2 E76Q | SHP2 F285S | SHP2 S502P | |
|---|---|---|---|
| Data Collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c, Å | 45.5, 214.8, 55.4 | 45.8, 214.3, 55.5 | 45.2, 214.2, 55.5 |
| α, β, γ, ° | 90.0, 95.6, 90.0 | 90.0, 96.1, 90.0 | 90.0, 95.6, 90.0 |
| Resolution, Å | 107.4–2.32 (2.33–2.32) | 55.16–1.87 (1.94–1.87) | 107.1–2.18 (2.19–2.18) |
| R merge | 13.6 (77.1) | 14.2 (64.4) | 8.6 (73.0) |
| l/σl | 7.7 (2.1) | 8.4 (1.7) | 9.7 (2.1) |
| Completeness (%) | 98.9 (98.2) | 99.0 (98.1) | 99.2 (99.7) |
| Multiplicity | 3.4 (3.5) | 2.5 (2.6) | 3.4 (3.4) |
| Refinement | |||
| Resolution, Å | 23.4 – 2.32 | 55.16 – 1.87 | 23.9 – 2.18 |
| No. reflections | 44,891 | 86,484 | 53,901 |
| Rwork /Rfree | 18.9 / 24.0 | 19.9 / 22.9 | 18.6 / 23.9 |
| No. atoms | 9,657 | 8,927 | 9,461 |
| Wilson B-factor | 44.86 | 17.02 | 43.4 |
| R.m.s deviations | |||
| Bond lengths, Å | 0.010 | 0.006 | 0.010 |
| Bond angles, ° | 1.13 | 0.86 | 1.09 |
Highest resolution shell is shown in parentheses.
SAXS Data Collection
The procedures for data collection, processing, and analysis are similar to those previously described.25 SAXS measurements were carried out at room temperature at the 12ID-B beamline of the Advanced Photon Source (APS), Argonne National Laboratory. A 14-KeV X-ray beam was used as the photon source with a sample-to-detector distance of 2 m for SAXS setup. Thirty two-dimensional (2D) images were recorded for each buffer and sample solution using a flow cell, with the exposure time of 1 second to minimize radiation damage and acquire adequate data statistics. The 2D images were reduced to 1D scattering profiles using the Matlab software package at the beamlines. The 1D SAXS profiles were grouped by sample and averaged, followed by buffer background subtraction. Concentration series measurements for each protein sample were carried out: 1.2, 2.4, 4.8 and 9.6 mg/ml for WT, 1.5, 3, 6, 9.6 mg/ml for F285S, and 1.0, 3.0, and 5.0 mg/ml for S502P and E76Q. in 50mM Hepes pH 7.4, 150mM NaCl, 5% glycerol and 2 mM TCEP for all samples. The SAXS intensity is extrapolated to infinite dilution and zero scattering angles to remove the scattering contribution due to interparticle interactions.
SAXS Data Analysis
SAXS data processing, scaling and calculations were carried out using the PRIMUS program of the ATSAS software suite.26, 27 The buffer solution scatter curve was subtracted from the sample scatter to generate a background-subtracted scatter curve. The Radius of gyration (Rg) was obtained from the Guinier plot of background-subtracted scatter data using the Radius of Gyration operation. The zero-intensity normalized Rg (Rg/I0) and maximum dimension of the particle (Dmax) were obtained from the p(r) function with the Distance Distribution operation. The zero-intensity normalized Rg (Rg/I0), P(r) function and maximum dimension of the particle (Dmax) were obtained using GNOM. The p(r)-derived radii or gyration were converted to real space values using GNOM.28 Theoretical X-ray scattering patterns of wild-type SHP2 (PDB:2SHP) and corresponding Rg values were calculated using the program Crysol.29
Measurement of Basal SHP2 Phosphatase Activity with the Small Molecule DiFMUP
To determine basal activity of SHP2 mutants, a ½ dilution series was performed for each phosphatase from 100 nM to 3 pM in the presence of 200 μM DiFMUP in 60 mM Hepes pH 7.2, 75 mM KCl, 75 mM NaCl, 1 mM EDTA and 0.05% Tween-20 (buffer 2), and changes in fluorescence emission resulting from DiFMUP dephosphorylation were measured at 450 nM with 358 nM excitation on a Spectramax M5. From each activity profile, an enzyme concentration was selected at the center of each linear response over 10 min. Specifically, 2.5 nM SHP2WT, 0.5 nM SHP2F285S, 0.05 nM SHP2E76Q, 0.025 nM SHP2S502P and 0.00625 nM SHP2PTP, were used in substrate titration experiments. To determine basal SHP2 enzyme kinetics, respective concentrations of each SHP2 phosphatase was prepared in buffer 2 and added to a linear titration of DiFMUP, from 4 mM to 1 fM and DiFMUP dephosphorylation was measured on a Spectramax M5 plate reader at 358 nm excitation and 450 nm emission. Raw velocity data was and converted to product (DIFMU)/time by preparing a standard curve of 6,8-difluoro-7-hydroxy-4-methylcoumarin (DIFMU)/DIFMUP and normalized to enzyme concentration to yield relative velocity, which was fit to the Michaelis-Menten equation (Equation 1) to extrapolate kinetic parameters:
| (Equation 1) |
Where V represents enzyme velocity, Vmax is the maximal enzyme activity, Km is the Michaelis-Menten constant, and [S] is substrate concentration.
Measurement of SHP2 Phosphatase Activity in the Presence of Synthetic Mono- and Bisphosphorylated Peptides Derived from IRS-1
Wild-type and mutant SHP2 proteins were diluted to 0.1 nM and treated with varying concentrations of either a monophosphorylated peptide derived from IRS-1 (pY1172, SLNY(p)IDLDLVK) or diphosphorylated peptide (pY1172-Peg-PY1222, SLNY(p)IDLDLVK-dPEG8-LSTY(p)ASINFQK) and phosphatase activity was measured through the dephosphorylation DiFMUP (200 μM, ex. 385, em. 450 nm) on a Spectramax M5 plate reader and converted to product (DiFMU)/time by preparing a standard curve of DiFMU/DiFMUP. Velocity data was fit to the following three-parameter dose response curve (Equation 2) to extrapolate EC50 values:
| (Equation 2) |
Where V represents enzyme velocity, basal is basal response, max is maximal response, [S] is substrate concentration, and EC50 is the substrate concentration that yields a response that represents 50% of the difference between basal and maximal.
Results
X-Ray structural analyses of SHP2 and oncogenic mutants
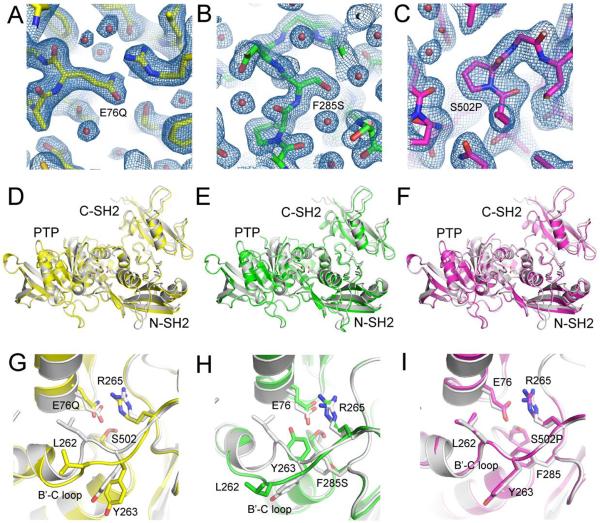
To investigate how cancer mutations impact the structure of SHP2, we prepared crystals of SHP2E76Q, SHP2S502P and SHP2F285S and solved the X-ray structure of each protein to 2.3 Å, 2.2 Å and 1.9 Å, respectively (Table I). The crystal structures capture the mutant proteins (see Figure 2A–C for electron density maps around the mutation sites) in closed, or autoinhibited conformations, in which the two SH2 domains flank the phosphatase domain, and the N-SH2 domain directly occludes access to the catalytic site (Figure 2D–F). The autoinhibited conformations of cancer-associated SHP2 proteins are similar to wild-type SHP2 (SHP2WT) (PDB:2SHP) with overall RMSD values for all protein atoms of 0.37 Å, 0.29 Å and 0.34 Å between SHP2WT and SHP2E76Q, SHP2S502P and SHP2F285S, respectively. Alignment of the SHP2 structures by superposition of the PTP domain (residues 220-525) reveal that in the closed conformation of each cancer variant, the N-SH2 and C-SH2 domains move away from the catalytic domain by 1–2 Å (Figure 2D–F), suggesting a destabilization of the autoinhibited conformation.
Figure 2. Mechanisms through which Cancer-Associated Mutations Destabilize the Autoinhibited Conformation of SHP2.
(A–C) Electron density maps (2Fo-Fc, contoured at 1σ) of E76Q (A), F285S (B), and S502P (C) shown around the sites of the mutations. (D–F) Overall structures of E76Q (D), F285S (E), and S502P (F) superimposed onto the structure of wild-type SHP2, aligned using the PTP (catalytic) domain. The wild-type structure is shown in white, and the side chains of the mutated residues are shown with sticks using CPK colors. Note the global shifts of the SH2 domains relative to the catalytic domain, which is most pronounced for the E76Q and F285S proteins. (G–I) Zoomed in views showing superposition of E76Q (G), F285S (H), and S502P (I) onto wild-type SHP2 around the mutation sites. Key residues at the interface described in the text are shown in sticks using CPK colors. Wild-type SHP2 PDB: 2SHP.
The SHP2 mutants display local structural deviations from SHP2WT within the interaction interface between the N-SH2 and PTP domains. In the basal conformation of SHP2WT, the N-SH2 domain is maintained in close proximity to the PTP domain through numerous direct and water-mediated interdomain hydrogen bonds, including a hydrogen-bond between the PTP active site S502 hydroxyl and N-SH2 E76 side-chain carboxylic acid and a salt bridge interaction between E76 and R265.13 Replacement of the E76 acid with a glutamine carboxamide yields a weaker, less electronegative, H-bond acceptor and a subtle displacement of the Q76 side-chain oxygen, so that the distance from the S502 hydroxyl shifts from 2.8 Å in the wild-type (E76) to 3.2 Å in the Q76 mutant. S502 also undergoes a side-chain rotamer change in the E76Q mutant, and the R265 guanidinium also adjusts relative to its position in SHP2WT (Figure 2G).
The X-ray structure of the leukemia-associated SHP2F285S mutation reveals a crankshaft movement of the hydrophobic B'C loop of the PTP domain away from residue 76 in the N-SH2 domain and an inward movement of Y263 to fill the region of the N-SH2:PTP domain interface occupied previously by F285 (Figure 2H). In the SHP2WT protein, L262 of the loop makes van der Waals interactions with the aliphatic portion of E76, whereas in the SHP2F285S mutant, the rotation of the loop moves L262 to a position more than 9 Å away from E76, and out of van der Waals contact (Figure 2H). A similar, though less extensive, rotation also occurs in the E76Q variant of SHP2 (Figure 2G). In both variant proteins, the movement of the B'C loop is accompanied by increased B-factors for L262 and other residues, suggesting increased mobility of this loop.
Interestingly, conversion of the E76 interaction partner, S502, to proline results in loss of the S502-E76 H-bond but does not detectably alter the distance between E76 and the PTP catalytic site or the distance between E76 and R265 (Figure 2I). The minimal interface disruption in the closed conformation of SHP2S502P is likely due to maintenance of the water-mediated hydrogen-bonding network between E76 and localized regions of the PTP domain.13
SAXS-determination of SHP2 particle sizes
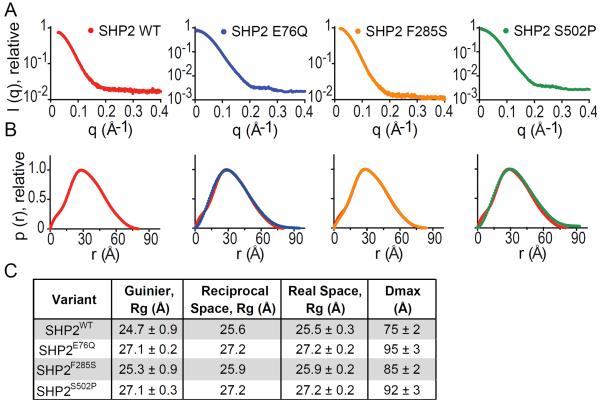
To determine whether the structural changes observed in the SHP2 crystals correlate with changes in the distribution of conformations populated in solution, we analyzed the small angle X-ray scattering (SAXS) behavior of each variant.30,31 Indirect Fourier transformation of 2-dimensional vector-averaged scattering profiles (Figure 3A) allowed the data to be fit to the pair-distance distribution p(r) function, which describes distances between all electron pairs within a macromolecule and is useful for detecting conformational changes in solution.32 p(r)-analysis of the SAXS profile obtained for SHP2WT revealed that the phosphatase adopts a compact conformation, with a P(r)-derived radius of gyration (Rg) of 25.5 ± 0.3 Å, in agreement with Rg calculated from a Guinier-fit of the scattering intensity (24.7 ± 0.9 Å) and theoretical Rg derived from the X-ray structure of SHP2 (PDB:2SHP, 25.2 Å) (Figure 3B & C).
Figure 3. Small Angle X-Ray Scattering (SAXS) Analysis of Cancer-Associated SHP2 Proteins.
(A) SHP2 protein SAXS intensity profiles across radially-averaged scattering vector, q. (B) Distance distribution analysis of SHP2 proteins, calculated through the p(r) function with indirect Fourier transformation of the SAXS intensity profiles. For each mutant, the p(r) function of SHP2WT (red) is displayed for comparison. (C) Radii of gyration (Rg) and maximal end-to-end distance (Dmax) values shown with standard deviations, calculated as described in methods.
To identify how cancer-associated mutations impact the size and shape distribution of SHP2 conformations, we analyzed SHP2E76Q, SHP2F285S and SHP2S502P by SAXS, and compared them with SHP2WT (Figure 3A). SHP2F285S exhibited a small 0.6 Å Rg enlargement relative to wt (Figure 3B & C), which is within error for the Rg of SHP2WT and consistent with the more modest increase in basal catalytic activity of F285S (see below). Both SHP2E76Q and SHP2S502P displayed statistically significant Rg increases over SHP2WT (Figure 3B & C). While both the Guinier and P(r)-calculated Rg values of the SHP2 cancer mutants were similar to those of wild-type, larger differences were observed in the tails of the distance-distribution function. Specifically, the p(r) function of SHP2WT tailed to a maximal end-to-end distance (Dmax) value of 75 ± 2 Å, while those of SHP2E76Q, SHP2F285S, and SHP2S502P tailed to Dmax values of 95 ± 3 Å, 85 ± 2 Å, 92 ± 3 Å and 87 ± 3 Å, respectively, arguing that these variants populate “open” conformations in solution that are not as substantially populated for SHP2WT in the basal state.33
Basal activity of SHP2 cancer variants
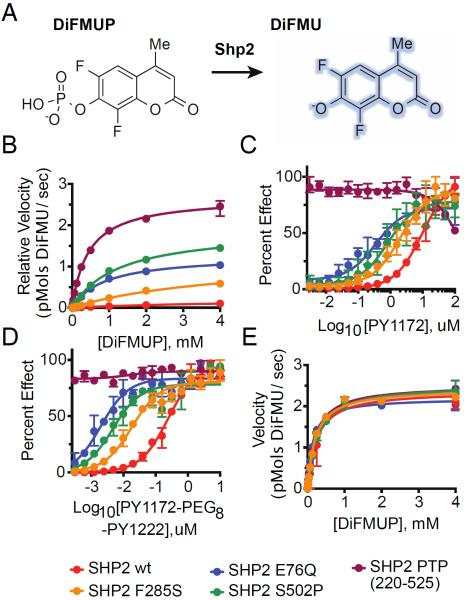
To better understand how cancer mutations impact SHP2 function, we assayed the phosphatase activity of SHP2 proteins using the fluorogenic substrate DiFMUP (Figure 4A).34 In agreement with Michaelis-Menten parameters reported by others for similar substrates,35, 36 SHP2WT displayed a kcat of 12.2 ± 1.0 s−1 and kcat/Km of 2.1·103 M−1s−1 with DiFMUP as substrate (Figure 4B & Table I), and the isolated Shp2 PTP domain (residues 220-525) revealed a kcat of 135 ± 3 s−1 and kcat/Km of 2.98·105 M−1s−1. Some of the SHP2 cancer mutants also displayed enhanced basal activity, and similar levels of activation were observed for SHP2S502P (kcat 96 ± 3 s−1, kcat/Km 7.7·104 M−1s−1) and SHP2E76Q (kcat 64 ± 1 s−1, kcat/Km 7.1·104 M−1s−1), while SHP2F285S displayed a more modest level of activation (kcat 61 ± 5 s−1, kcat/Km 1.4·104 M−1s−1).
Figure 4. Cancer-Associated SHP2 Mutations Increase Basal Phosphatase Activity and Sensitivity to IRS-1 Phosphopeptide ligand Stimulation.
(A) Schematic illustrating DiFMUP dephosphorylation by SHP2. (B) Basal enzymatic trajectories of wild-type and cancer-associated SHP2 proteins across varying concentrations of DiFMUP. (C) Enzymatic activity of SHP2 proteins (0.05 nM) across varying concentrations of an IRS-1-derived monophosphorylated peptide. (D) Identical experiment to C but with synthetic bisphosphorylated peptide derived from IRS-1. (E) Phosphatase activity of SHP2 proteins (0.01 nM) at saturating concentrations of a double-phosphorylated peptide derived from IRS-1 (6 μM) across varying concentrations of DiFMUP. Velocity data is shown as mean ± standard deviation and was fit to the standard Michaelis-Menten equation to extrapolate kcat and KM values.
Investigating how cancer-mutations tune SHP2 activation by phosphotyrosine ligands
Engagement of SHP2 SH2 domains with phosphotyrosine ligands has been observed to increase phosphatase activity,37, 38 and previous research has revealed that many leukemia-associated SHP2 mutations enhance sensitivity to phospholigand-activation, relative to wild-type enzyme.19 To investigate how SHP2 cancer variants respond to phosphopeptide stimulation, we incubated wt and mutant SHP2 proteins in the presence of either a monophosphorylated peptide derived from insulin receptor substrate-1 (IRS-1), which preferentially engages the N-SH2 domain (PY1172),37 or a bisphosporylated peptide that occupies both SH2 domains and induces full activation of the phosphatase (PY1172-PEG-PY1222),38 and quantified activity via DiFMUP dephosphorylation.
SHP2WT displayed a dose-dependent increase in DiFMUP dephosphorylation upon titration of increasing amounts of PY1172, with half-maximal stimulation (EC50) at 10 μM (Figure 4C). The monophosphopeptide activation-profiles of SHP2E76Q, SHP2F285S and SHP2S502P were left-shifted, relative to SHP2WT, with corresponding EC50 values of 350 nM, 2 μM, 830 nM and 4 μM, respectively, and of the four proteins, only SHP2S502P achieved complete stimulation of phosphatase activity under saturating concentrations of PY1172.
Similar differences were observed when SHP2 variants were incubated with bisphosphopeptide PY1172-PEG-PY1222 (Figure 4D), however, rather than the partial activation observed in the presence of monophosphopeptide for most of the variants, the double-phosphorylated IRS-1 peptide resulted in complete activation of SHP2WT at low-micromolar concentrations (Figure 4D) with an EC50 value of 250 nM. The dose-response curves of SHP2E76Q, SHP2F285S and SHP2S502P shifted left, relative to wt enzyme, with EC50 values of 2 nM, 18 nM, 5 nM and 49 nM, respectively (Figure 4D). When SHP2 variants were incubated with saturating concentrations of bisphosphopeptide, all constructs displayed near-identical enzyme activity (Figure 4E & Table II).
Table II.
Michaelis-Menten Parameters of DiFMUP Dephosphorylation by wt and mutant SHP2 under basal conditions.
| Variant | KM (mM) | kcat (s−1) | kcat /KM (M−1 s−1) |
|---|---|---|---|
| wt | 5.8 ± 0.7 | 12.2 ± 1.0 | 2.10 · 103 |
| E76Q | 0.89 ± 0.05 | 63.7 ± 1.4 | 7.12 · 104 |
| F285S | 4.28 ± 0.56 | 61.4 ± 4.9 | 1.43 · 104 |
| S502P | 1.25 ± 0.09 | 95.6 ± 2.8 | 7.65 · 104 |
| PTP | 0.45 ± 0.03 | 134.5 ± 3.0 | 2.98 · 105 |
Discussion
To better understand the effect of GoF SHP2 cancer mutations on the regulation of its phosphatase activity, we investigated the structure and function of three cancer variants, and uncovered a spectrum of differences resulting from single amino-acid substitutions within the 1176 Å2 N-SH2:PTP autoinhibitory interface (Supplementary Figure 1). Using X-ray crystallography, we visualized the closed conformation of each protein and identified local changes that lead to destabilization of the autoinhibited conformation, which we further assessed via SAXS solution studies and enzymatic characterization using a small molecule phosphotyrosine mimetic.
The X-ray structures of SHP2E76Q and SHP2S502P revealed a weakening and loss, respectively, of the critical S502-E76 interdomain H-bond that favors N-SH2 inhibition of substrate access to the active site of the catalytic domain. While the E76Q substitution led to a distinct movement of the N-SH2 domain away from the PTP domain, the distance between the two domains remained essentially unperturbed in SHP2S502P. This difference may be accounted for by the extensive water-mediated hydrogen-bonding network that links E76 to distant residues throughout the PTP catalytic pocket. Upon perturbation of E76, this interaction network is likely destabilized more substantially than when E76 is maintained and S502 is mutated.
The X-ray structure of SHP2F285S revealed an unanticipated movement of the B'C loop of the PTP domain, which was also seen to a lesser extent in SHP2E76Q but not in SHP2S502P. F285 likely maintains van der Waals contacts with hydrophobic residues of the B'C loop and maintains proximity of this loop to the N-SH2:PTP interface. We suggest that the B'C loop of SHP2 partially shields interfacial residues from solvent and thus strengthens interdomain interactions. Destabilization of the B'C loop, via mutation of F285 induces a subtle conformational change that modestly destabilizes autoinhibition.
Our SAXS experiments suggest that the local differences observed in SHP2 crystal structures are accompanied by an increase in the fraction of molecules in an “open” and active conformation in solution. In particular, the increased values of Rg and the tails of the distance distribution functions for SHP2E76Q and SHP2S502P suggest that, at any give time in solution, a detectable fraction of these variants are in a more open and extended conformation than that favored by the crystalline lattice. While the Rg of SHP2F285S is within error of the Rg determined for SHP2WT, the Dmax value of SHP2F285S is significantly elevated, which suggests that although the average particle size of SHP2F285S is similar to that of SHP2WT, a larger population of SHP2F285S molecules adopt conformations in solution that are more open, consistent with the slightly increased basal activity of SHP2F285S observed in vitro. The modest expansion of Rg values for the other two mutant proteins is in agreement with the 10 Å N-SH2:PTP interdomain opening trajectory suggested by molecular dynamics simulations39 and argues that only a subtle conformational change in SHP2 is necessary to expose the active site and enhance activity. The further exploration of SAXS as a means of assessing activation, and reporting on the conformation of “open”, phosphopeptide-bound SHP2 is underway.
To determine how local and global differences in cancer-associated SHP2 proteins translate to changes in phosphatase activity, we analyzed the ability of wild-type and mutant SHP2 proteins to dephosphorylate a synthetic, quenched-umbelliferone surrogate substrate in the presence and absence of activating phosphopeptide ligands derived from IRS-1. Previous research has found that many cancer-associated SHP2 mutations enhance basal phosphatase activity and sensitivity to stimulation, and in accordance with these findings, we discovered that SHP2F285S, SHP2E76Q and SHP2S502P activate basal enzymatic activity to varying degrees.
Specifically, SHP2F285S, which destabilizes the hydrophobic B'C loop of the PTP domain and leads to a modest 2 Å opening of the SHP2 closed conformation, evokes a 5-fold increase in kcat under basal conditions. SHP2E76Q and SHP2S502P, which perturb the S502-E76 interdomain hydrogen bond and yield similar radii of gyration, increase the value of kcat from 5- to 8-fold. In general, we also observe a correlation between the enhanced levels of basal phosphatase activity and SAXS-derived radii of gyration.
SHP2E76Q, SHP2F285S and SHP2S502P also increase sensitivity of the phosphatase to stimulation by phosphorylated ligands derived from IRS-1. PY1172, a monophosphorylated IRS-1 peptide that preferentially interacts with the N-SH2 domain,37 partially stimulated SHP2E76Q and SHP2F285S, and stimulated SHP2S502P to a specific activity comparable to that of the isolated phosphatase domain. The three SHP2 cancer mutants also displayed elevated responses to a bisphosphorylated peptide ligand peptide derived from IRS-1, which stimulated full activity (PY1172-PEG-PY1222),38 with greatest sensitivity observed for SHP2E76Q and SHP2S502P and intermediate for SHP2F285S, relative to that observed for SHP2WT.
Our analysis of the influence of cancer mutations on the distribution of solution conformations and catalytic efficiency of SHP2 indicates that SHP2WT exists predominantly in a closed, autoinhibited conformation. The oncogenic mutations investigated herein destabilize autoinhibition and reduce the energetic barrier to adopting an open, catalytically competent conformation. Such mutations do not fully abolish the kinetic barrier to achieving an open state, since the values of kcat are still influenced by the rate of opening of the structure to allow substrate access.
While our quantification of SHP2 variants has allowed us to elucidate the consequences of specific pro-oncogenic mutations, we plan to expand our analysis to build a more comprehensive understanding of PTPN11 mutations observed in human cancer and investigate their corresponding effects on the structure, function and oncogenic signaling properties of SHP2 and the progression of leukemia. We aim to complement our molecular analysis with cellular assays that report on the activation and localization of SHP2 within cultured cancer cells. Ultimately, our results hold the promise to improve understanding of how specific SHP2 point mutations influence oncogenic signaling and advance the development of structure-guided and mutation-specific SHP2 targeted therapies.
Supplementary Material
Table III.
Michaelis-Menten Parameters DiFMUP dephosphorylation by wt and mutant SHP2 under full phosphopeptide stimulation.
| Variant | KM (mM) | kcat (s−1) | kcat /KM (M−1 s−1) |
|---|---|---|---|
| wt | 0.24 ± 0.04 | 119 ± 6.0 | 4.96 · 105 |
| E76Q | 0.14 ± 0.01 | 121 ± 2.0 | 8.64 · 105 |
| F285S | 0.21 ± 0.02 | 121 ± 3.0 | 5.76 · 105 |
| S502P | 0.26 ± 0.02 | 126 ± 3.2 | 4.85 · 105 |
| PTP | 0.22 ± 0.02 | 126 ± 2.4 | 5.73 · 105 |
ACKNOWLEDGMENT
For SAXS experiments, we gratefully acknowledge use of SAXS Core facility of Center for Cancer Research, National Cancer Institute (NCI). The SAXS data were collected at beamline 12-ID-B. The shared scattering beamline 12-ID-B resource is allocated under the PUP-24152 agreement between the National Cancer Institute and Argonne National Laboratory (ANL). We thank Dr. Lixin Fan (NCI) and Dr. Xiaobing Zuo (ANL) for their expert support. Use of the IMCA-CAT beamline 17-ID (or 17-BM) at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences by Argonne National Laboratory, under Contract No. DE-AC02-06CH11357.
Funding Sources This work was funded in part by the Novartis - Dana Farber Cancer Institute Drug Development Program, P50 GM107618-01A1 (XX and SCB) and by the William Lawrence-Blanche Hughes Foundation (to SCB). MF, TS, HMC, MJL, RC, PW, PDF and MGA are employees of Novartis. JRL gratefully acknowledges postdoctoral fellowship funding from the American Cancer Society®, Award ID 128126.
Footnotes
Supporting Information. The following files are available free of charge. Supplementary Information, consisting of one figure.
Author Contributions JRL, PDF, MGA, and SCB designed experiments. JLR, MF, XX, PW and ID performed experiments. All authors interpreted data. JLR and SCB wrote the manuscript with input from all co-authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- [1].Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Role of SHPTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol Cell Biol. 1994;14:6674–6682. doi: 10.1128/mcb.14.10.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- [3].Xu R, Yu Y, Zheng S, Zhao X, Dong Q, He Z, Liang Y, Lu Q, Fang Y, Gan X, Xu X, Zhang S, Dong Q, Zhang X, Feng GS. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood. 2005;106:3142–3149. doi: 10.1182/blood-2004-10-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mohi MG, Williams IR, Dearolf CR, Chan G, Kutok JL, Cohen S, Morgan K, Boulton C, Shigematsu H, Keilhack H, Akashi K, Gilliland DG, Neel BG. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 2005;7:179–191. doi: 10.1016/j.ccr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- [5].Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hahlen K, Hasle H, Licht JD, Gelb BD. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- [6].Loh ML, Reynolds MG, Vattikuti S, Gerbing RB, Alonzo TA, Carlson E, Cheng JW, Lee CM, Lange BJ, Meshinchi S, Children's Cancer G. PTPN11 mutations in pediatric patients with acute myeloid leukemia: results from the Children's Cancer Group. Leukemia. 2004;18:1831–1834. doi: 10.1038/sj.leu.2403492. [DOI] [PubMed] [Google Scholar]
- [7].Tartaglia M, Martinelli S, Cazzaniga G, Cordeddu V, Iavarone I, Spinelli M, Palmi C, Carta C, Pession A, Arico M, Masera G, Basso G, Sorcini M, Gelb BD, Biondi A. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104:307–313. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- [8].Yamamoto T, Isomura M, Xu Y, Liang J, Yagasaki H, Kamachi Y, Kudo K, Kiyoi H, Naoe T, Kojma S. PTPN11, RAS and FLT3 mutations in childhood acute lymphoblastic leukemia. Leuk Res. 2006;30:1085–1089. doi: 10.1016/j.leukres.2006.02.004. [DOI] [PubMed] [Google Scholar]
- [9].Yoshida N, Omoto Y, Inoue A, Eguchi H, Kobayashi Y, Kurosumi M, Saji S, Suemasu K, Okazaki T, Nakachi K, Fujita T, Hayashi S. Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci. 2004;95:496–502. doi: 10.1111/j.1349-7006.2004.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bentires-Alj M, Paez JG, David FS, Keilhack H, Halmos B, Naoki K, Maris JM, Richardson A, Bardelli A, Sugarbaker DJ, Richards WG, Du J, Girard L, Minna JD, Loh ML, Fisher DE, Velculescu VE, Vogelstein B, Meyerson M, Sellers WR, Neel BG. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- [11].Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Neel BG, Gu H, Pao L. The `Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends in biochemical sciences. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- [13].Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- [14].Holgado-Madruga M, Emlet DR, Moscatello DK, Godwin AK, Wong AJ. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- [15].Kazlauskas A, Feng GS, Pawson T, Valius M. The 64-kDa protein that associates with the platelet-derived growth factor receptor beta subunit via Tyr-1009 is the SH2-containing phosphotyrosine phosphatase Syp. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6939–6943. doi: 10.1073/pnas.90.15.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kuhne MR, Pawson T, Lienhard GE, Feng GS. The insulin receptor substrate 1 associates with the SH2-containing phosphotyrosine phosphatase Syp. The Journal of biological chemistry. 1993;268:11479–11481. [PubMed] [Google Scholar]
- [17].Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Molecular and cellular biology. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cunnick JM, Meng S, Ren Y, Desponts C, Wang HG, Djeu JY, Wu J. Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. The Journal of biological chemistry. 2002;277:9498–9504. doi: 10.1074/jbc.M110547200. [DOI] [PubMed] [Google Scholar]
- [19].Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. The Journal of biological chemistry. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- [20].Schubbert S, Lieuw K, Rowe SL, Lee CM, Li X, Loh ML, Clapp DW, Shannon KM. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. 2005;106:311–317. doi: 10.1182/blood-2004-11-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, Neel BG. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nature medicine. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- [22].Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER version x.y.z. Global Phasing Ltd.; Cambridge, United Kingdom: 2011. [Google Scholar]
- [24].Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang J, Zuo X, Yu P, Xu H, Starich MR, Tiede DM, Shapiro BA, Schwieters CD, Wang YX. A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J Mol Biol. 2009;393:717–734. doi: 10.1016/j.jmb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, Svergun DI. New developments in the program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS - a windows-pc based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- [28].Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- [29].Svergun D, Barberato C, Koch MHJ. CRYSOL – a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768. [Google Scholar]
- [30].Doniach S, Lipfert J. Comprehensive Biophysics, Vol 1: Biophysical Techniques for Structural Characterization of Macromolecules. 2011. Small and Wide Angle X-ray Scattering from Biological Macromolecules and their Complexes in Solution; pp. 376–397. [Google Scholar]
- [31].Graewert MA, Svergun DI. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS) Curr Opin Struc Biol. 2013;23:748–754. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- [32].Hong XG, Hao Q. High resolution pair-distance distribution function P(r) of protein solutions. Appl Phys Lett. 2009;94 [Google Scholar]
- [33].Hirai M, Arai S, Iwase H, Takizawa T. Small-angle X-ray scattering and calorimetric studies of thermal conformational change of lysozyme depending on pH. J Phys Chem B. 1998;102:1308–1313. [Google Scholar]
- [34].Gee KR, Sun WC, Bhalgat MK, Upson RH, Klaubert DH, Latham KA, Haugland RP. Fluorogenic substrates based on fluorinated umbelliferones for continuous assays of phosphatases and beta-galactosidases. Analytical biochemistry. 1999;273:41–48. doi: 10.1006/abio.1999.4202. [DOI] [PubMed] [Google Scholar]
- [35].Qiu W, Wang X, Romanov V, Hutchinson A, Lin A, Ruzanov M, Battaile KP, Pai EF, Neel BG, Chirgadze NY. Structural insights into Noonan/LEOPARD syndrome-related mutants of protein-tyrosine phosphatase SHP2 (PTPN11) BMC structural biology. 2014;14:10. doi: 10.1186/1472-6807-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yu ZH, Zhang RY, Walls CD, Chen L, Zhang S, Wu L, Liu S, Zhang ZY. Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations. Biochemistry. 2014;53:4136–4151. doi: 10.1021/bi5002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sugimoto S, Wandless TJ, Shoelson SE, Neel BG, Walsh CT. Activation of the SH2-containing protein tyrosine phosphatase, SH-PTP2, by phosphotyrosine-containing peptides derived from insulin receptor substrate-1. The Journal of biological chemistry. 1994;269:13614–13622. [PubMed] [Google Scholar]
- [38].Pluskey S, Wandless TJ, Walsh CT, Shoelson SE. Potent Stimulation of Sh-Ptp2 Phosphatase-Activity by Simultaneous Occupancy of Both Sh2 Domains. Journal of Biological Chemistry. 1995;270:2897–2900. doi: 10.1074/jbc.270.7.2897. [DOI] [PubMed] [Google Scholar]
- [39].Darian E, Guvench O, Yu B, Qu CK, MacKerell AD. Structural mechanism associated with domain opening in gain-of-function mutations in SHP2 phosphatase. Proteins-Structure Function and Bioinformatics. 2011;79:1573–1588. doi: 10.1002/prot.22984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.