Abstract
Natural killer/T-cell lymphoma (NKTCL) is a rare, aggressive form of non-Hodgkin lymphoma that is generally incurable at more advanced stages with systemic involvement. Clonal diagnostic markers (eg, unique T- or B-cell receptor rearrangements) are not available for NKTCLs. Killer cell immunoglobulin like receptors (KIRs) are a family of type I transmembrane glycoproteins involved in the inhibition or activation of NK cells. A restricted expression profile of KIRs has been proposed as clonal markers of NK-cell proliferations. Here we evaluated the transcription profile of all KIR family genes and C-type lectin receptor genes using RNA sequencing on NKTCL cases (n = 17) and NK-cell lines (n = 3). The expression of all KIRs tended to be markedly reduced or absent in NKTCL, except for the KIR family member killer Ig-like receptor 2DL4 (KIR2DL4; alias CD158D), which was selectively overexpressed in the majority (59%) of cases. No specific expression pattern was observed for C-type lectin receptors. KIR2DL4 is an unusual member of the KIR family that recognizes human leukocyte antigen G and mediates NK-cell activation through inducing proliferation and survival pathways such as AKT and NF-κB. Stable knockdown of KIR2DL4 in two malignant NK-cell lines with high KIR2DL4 expression significantly reduced cell growth. Selective overexpression of KIR2DL4 and down-regulation of inhibitory KIRs may contribute to NKTCL pathogenesis.
Natural killer/T-cell lymphoma (NKTCL) of the nasal type is a rare lymphoid malignancy with a poor prognosis.1 NKTCL prevalence varies geographically, with much higher prevalences in East Asia and Central and South America.2 NKTCL cases show good response to radiotherapy in stage I disease; however, the prognosis is markedly worse in patients with more advanced stages of the disease.3 Most NKTCLs originate from transformed NK-cells,4 and due to lack of NK-cell receptors with unique rearrangements, diagnostic markers of clonality have been challenging to identify.
NK-cell activation is regulated by the balance of opposing signals originating from activating and inhibitory receptors expressed on the NK-cell surface.5 Killer cell immunoglobulin like receptors (KIRs) are crucial plasma membrane receptors regulating NK-cell activation. KIRs are highly polymorphic and coevolve with their ligands, major histocompatibility complex class I molecules.6 KIRs with short cytoplasmic tails function as activation receptors, and KIR proteins with long cytoplasmic tails inhibit NK cells, with the exception of killer Ig-like receptor 2DL4 (KIR2DL4).7 The inhibitory KIRs transmit inhibitory signals when they are engaged with the cognate major histocompatibility complex class I molecules expressed on the host cells so that the normal cells of the body are protected from NK-cell–mediated cytotoxicity.8 The ligands for activating KIR family members are relatively poorly characterized7 compared with inhibitory KIR family proteins such as KIR2DL1, which binds human leukocyte antigen (HLA)-C2 allotypes,9 or KIR2DL2 and KIR2DL3 proteins, which bind HLA-C1 allotypes through the Asp residue at position 80.10 However, KIR2DL4 has been shown to interact with HLA-G. A distinct combination of KIRs is expressed on individual NK cells in the peripheral blood, giving a polyclonal pattern of KIR expression in the population.11 A clonal population of NK cells is expected to show a restricted pattern of KIR expression. Therefore, the identification of the KIR repertoire may provide information regarding the clonal nature of an NK-cell population.
Here, we applied unbiased RNA sequencing (seq), which has a wide dynamic range,12 to determine the KIR family gene expression pattern in NKTCL cases and NK-cell lines and compared their profile with resting and activated normal NK cells. This study demonstrated that KIR expression shows a clonal pattern and is also highly abnormal in malignant NK cells. We also demonstrated the frequent retention of KIR2DL4 expression, which may promote growth in NK cells.
Materials and Methods
Malignant NK-Cell Tumor Samples
The NKTCL cases, NK-cell lines, and normal NK-cell samples used in this study are historical samples already reported,13 and the characteristics of the NKTCL cases and NK-cell lines are available in Supplemental Tables S1 and S2.
NK-Cell Isolation and Activation
Resting NK cells were isolated from the peripheral blood of healthy donors using an NK-cell isolation kit (Miltenyi Biotec, Auburn, CA) as previously described.14 Resting NK cells were activated in the presence of 100 IU of IL-2 for 2 days to obtain IL-2–activated NK cells. Alternatively, NK cells were activated by co-culturing freshly isolated peripheral blood lymphocytes with K562-Clone9-mb21 cells (a gift from Dr. Dean Lee), which are K562 cells engineered to ectopically express 4-1BB ligand (4-1 BBL), CD86, and membrane-bound IL-21 on the cell surface to achieve higher levels of NK-cell activation,15 for 12 days as described previously.14 The purity of activated NK cells was evaluated with CD56-allophycocyanin and CD3-phycoerythrin staining, and >95% CD56+/CD3− cells were used for subsequent analysis.
RNA Isolation and Quality Control
RNA was isolated from the NKTCL cases using the AllPrep DNA/RNA Mini Kit (Qiagen Inc., Valencia, CA). Quality and integrity of the RNA was determined with the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). NKTCL cases with RNA having RNA integrity numbers >8 were used for library preparation and sequencing.
RNA Sequencing
Aligned RNA-seq files generated in a previous study were used here for gene expression analysis.13 In this study, we concentrated on the expression of only KIR family members and C-type lectin receptors. Briefly, 100-bp paired-end libraries were prepared with the TruSeq RNA preparation kit (Illumina Inc., San Diego, CA), and high-throughput sequencing was performed in the University of Nebraska Medical Center Next Generation Sequencing Core Facility (Omaha, NE) and the Tufts University Genomics Core Facility (Boston, MA) using the Genome Analyzer IIX or the HiSeq 2000 sequencing system (Illumina). The raw files of the 17 NKTCL cases, KHYG1, NKYS, and NK92 cell lines, and normal NK-cell samples (resting, 48 hours IL-2 activated, and activated through co-culture with K562-Clone9-mb21 feeder cells14) are available from the Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/sra; accession number SRA200820).
RNA-Seq Data Analysis
FASTQC reports were evaluated for quality control after high-throughput sequencing. The sequencing reads were aligned to the February 2009 human reference sequence (GRCh37/hg19) with the TopHat software package version 1.3 (Cole Trapnell, University of Maryland, College Park, MD).16 Gene expression analysis was performed with the Cufflinks software package version 0.0.5 (http://cole-trapnell-lab.github.io/cufflinks, last accessed October 2012)17 in the Galaxy platform (https://usegalaxy.org, last accessed October 2012)18 using the following parameter settings: maximum intron length, 300,000; minimum isoform fraction, 0.1; pre mRNA fraction, 0.15; perform quartile normalization, no; use reference annotation, yes; reference annotation, USCS main on human; Refgene (genome)–perform bias correction, no; use multi read correct, no; and global model (for use in Trackster), no data set. The expression levels of KIR and C-type lectin receptor genes were determined by the fragment per kilobase in million reads estimate on the RNA-seq data. According to the algorithm of Cufflinks,17, 19 if a short read can be placed to multiple genomic positions, then the count of this read will be uniformly divided (using default settings) to these positions. The validity of the RNA-seq gene expression analysis pipeline was evaluated by determining the Pearson correlation of global or KIR family gene expression profile between GeneChip Human Genome U133 Plus version 2.0 (Affymetrix, Inc., Santa Clara, CA) chips and RNA-seq for two representative NKTCL cases (Supplemental Figure S1). For T-cell receptor gene expression analysis with Cufflinks, Gencode20 annotation version 23, which includes annotation of T-cell receptor genes, was used. The MiXCR software program version 1.1 (MiLaboratory LLC, Moscow, Russia)21 was used for searching for clonality in complementarity-determining region-3 regions of T-cell receptor α and β genes.
Design and Expression of Single or Double shRNAs for KIR2DL4 Knockdown in NK-Cell Lines
Two 19-mer KIR2DL4 siRNAs were designed using the siDesign Center (GE Dharmacon, Lafayette, CO). The siRNA nucleotide sequences were blasted to the human genome to ensure that they did not target other human genes. The siRNAs were converted to 97-mer shRNA-miRNAs and PCR-amplified with the high-fidelity PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies) to be cloned in the miR-30–adapted retroviral vector, microRNA-adapted retroviral vector MSCV-LTRmiR30-PIG (LMP) (Addgene; http://www.addgene.org; plasmid number 24071), which was digested at the XhoI and EcoRI restriction sites to remove the forkhead box-P3 shRNA according to the manufacturer's instructions (Thermo Fisher Scientific Inc, Rochester, NY) (Supplemental Figure S2A). The empty murine stem cell virus-LTRmiR30-PIG (control vector) was generated as follows. First, forkhead box-P3 shRNA was removed by gel extraction using QIAquick Gel Extraction Kit (Qiagen Inc.) after double-digestion with the XhoI and EcoRI restriction enzymes, which generated sticky overhangs. The sticky ends were then blunted with the Klenow Fragment (New England BioLabs Inc., Ipswich, MA), and empty LMP was circularized by ligating blunted ends with T4 DNA ligase (New England BioLabs Inc.). The sense nucleotide sequences of the siRNAs were as follows: KIR2DL4 1st siRNA, 5′-GATCATGGTCACAGGTCTA-3′; KIR2DL4 2nd siRNA, 5′-TCACAGGTCTATATGAGAA-3′.
KIR2DL4 first and second shRNAs were sequentially cloned into the LMP vector to generate the KIR2DL4–1st-2nd–double-shRNA using the following strategy: KIR2DL4–1st shRNA–LMP was digested with EcoRI and then treated with Antarctic Phosphatase (New England BioLabs Inc.) to prevent recircularization. After that, EcoRI-digested KIR2DL4–2nd shRNA insert, which was generated through amplification with PfuUltra II Fusion HS DNA polymerase (Agilent Technologies), was cloned into KIR2DL4–1st shRNA–LMP (Supplemental Figure S2C). Diagnostic restriction mapping and Sanger sequencing with forward and reverse primers were performed to validate each retroviral shRNA construct.
Gene Expression Profiling
KIR family gene expression data for NKTCL cases and NK-cell lines were obtained from our earlier study,4 which used the GeneChip Human Genome U133 Plus version 2.0 DNA microarray platform (Affymetrix, Inc.). The raw files of the microarray data are available from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE19067).
RT-qPCR
Real-time quantitative RT-PCR (RT-qPCR) was performed as previously described.13, 14 Briefly, 1 μg of RNA was reverse-transcribed with SuperScript II (Life Technologies, Carlsbad, CA). The DyNAmo HS SYBR Green qPCR Kit (Life Technologies) was used for amplifying and quantifying the cDNA with the CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories S.r.l., Segrate, Italy). RPL13A22 was used as the housekeeping gene for normalization. RT-qPCR primers used were: KIR2DL4-q-RT-PCR-F, 5′-CCTTCCCTTCTTTCTCCTTCATC-3′; KIR2DL4-q-RT-PCR-R, 5′-TGATCCAACTGTGCGTATGTC-3′.
Evaluation of Negative-Selection Pressure by Tracking GFP+ Cells
A FACSCalibur or BD LSRII flow cytometer (BD Biosciences, San Jose, CA) was used for quantifying the percentage of green fluorescent protein (GFP)-positive cells in regular time intervals after transduction with empty vector or KIR2DL4 shRNA vector. The results were analyzed with BD CellQuest Pro or BD FACSDiva software version 6.2 (BD Biosciences).
Apoptosis Assay
Apoptosis of KHYG1 cells retrovirally transduced with empty vector or KIR2DL4–1st-2nd–double-shRNA was evaluated on GFP+ gated cells using BD FACSCanto II (BD Biosciences) after staining the cells with annexin V–phycoerythrin (Apoptosis Detection Kit; BD Biosciences) according to the manufacturer's instructions.
Statistical Analysis
Statistical software package R language scripts version 2.15.2 (http://www.R-project.org, last accessed October 2012) were used for the identification of the Pearson correlations (R) for global gene expression values obtained using RNA-seq and GeneChip Human Genome U133 Plus version 2.0 (Affymetrix, Inc.) chips for two NKTCL cases. Excel 2007 software (Microsoft Corp., Redmond, WA) was used for t-tests whenever appropriate, and P < 0.05 was considered significant.
Results
KIR Expression Profile in NKTCLs
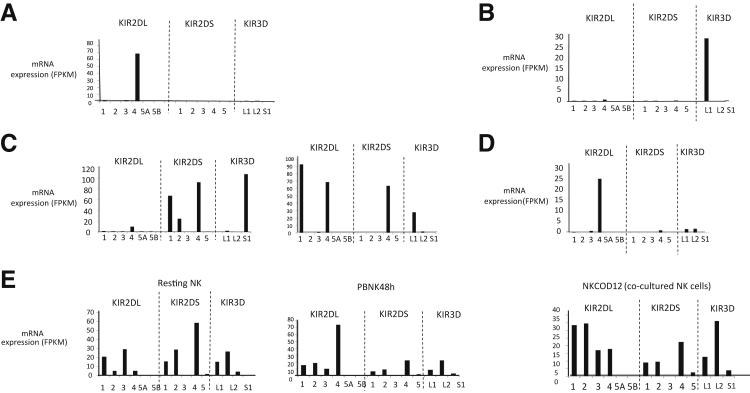
We determined the mRNA expression values for the KIR2 (n = 11; including KIR2DL1–5B, n = 6; and KIR2DS1–5, n = 5) and KIR3 (n = 3; including -L1, -L2, and -S1) genes. Most of the NKTCL cases (10 of 17; 59%) expressed only one KIR allele (Figure 1, A and B), nine of which exclusively expressed KIR2DL4 (Figure 1A). One NKTCL case expressed only KIR3DL1 (Figure 1B). We observed two NKTCL cases with expression of multiple KIRs (2 of 17; 12%), simultaneously at comparable levels (Figure 1C): KIR2DL1, KIR2DL4, KIR2DS4, and KIR3DL1 in one and KIR2DL4, KIR2DS1, KIR2DS2, KIR2DS4, and KIR3DS1 in the other case (Figure 1C). One NKTCL case expressed five KIR family genes at detectable levels; however, the expression of KIR2DL4 was much greater than that of the other four KIR genes (Figure 1D). In contrast to NKTCL cases, normal NK cells expressed multiple KIR genes simultaneously (Figure 1E). Interestingly, we did not observe KI2RDL5A or KIR2DL5B expression in normal resting or activated NK cells, and KIR2DS3 expression was very low or absent in normal NK cells (Figure 1E). In four NKTCL cases, of which two were T-cell receptor–αβ+ by RNA-seq, we did not observe a detectable level of mRNA expression (fragment per kilobase in million reads, <1) of any KIR genes. Collectively, these results indicate that malignant NK cells tend to down-regulate all KIR gene expression, with the exception of KIR2DL4. This profile could be a useful diagnostic biomarker of neoplastic NK-cell proliferations.
Figure 1.
Killer cell immunoglobulin like receptor (KIR) family gene expression profiles in cases of natural killer/T-cell lymphoma (NKTCL) and normal NK cells. The mRNA expression values for KIR2DL, KIR2DS, and KIR3D genes were quantified using RNA sequencing data on 17 NKTCL cases. The fragments per kilobase of transcript per million mapped reads (FPKM) values were calculated as explained in Materials and Methods. mRNA expression values of each KIR family member gene for NKTCL cases are shown separately. A: A representative NKTCL case (of nine) with exclusive KIR2DL4 expression. B: KIR expression pattern for an NKTCL case with only KIR3DL1. C: Two NKTCL cases with comparable levels of multiple KIR gene expression. D: KIR family gene expression profile for the NKTCL case with low but detectable expression of multiple KIR genes and high KIR2DL4 expression. E: KIR gene expression pattern in normal human NK-cell samples. n = 3 KIR3D; n = 5 KIR2DS; n = 6 KIR2DL; n = 14 KIR family member genes. NKCOD12, NK cells obtained by co-culturing peripheral blood lymphocytes with K562-Cl9-mb21 cells for 12 days; PBNK48h, 48-hour IL-2–activated peripheral blood NK cells; resting NK, freshly isolated peripheral blood NK cells.
C-Type Lectin Expression Profile in NKTCLs
Next, we evaluated the expression level of six C-type lectin family receptor genes—NKG2A, CD94, NKG2C, NKG2D, NKG2E, and NKG2F—in 17 NKTCL cases. Other than the NKG2F gene, whose expression was restricted to a subset of NKTCL cases, C-type lectin expression was detectable in the majority of the NKTCL cases. However, expression levels of these genes were highly variable (Supplemental Figure S3, A–F). Similarly, in normal NK-cell samples, except for NKG2F, we observed detectable expression of all C-type lectin receptors (Supplemental Figure S3, G and H). In general, we did not observe any marked difference of C-type lectin receptor gene expression pattern in NKTCL cases compared to normal NK-cell samples.
Stable Knockdown of KIR2DL4 Causes Negative-Selection Pressure in NK-Cell Lines
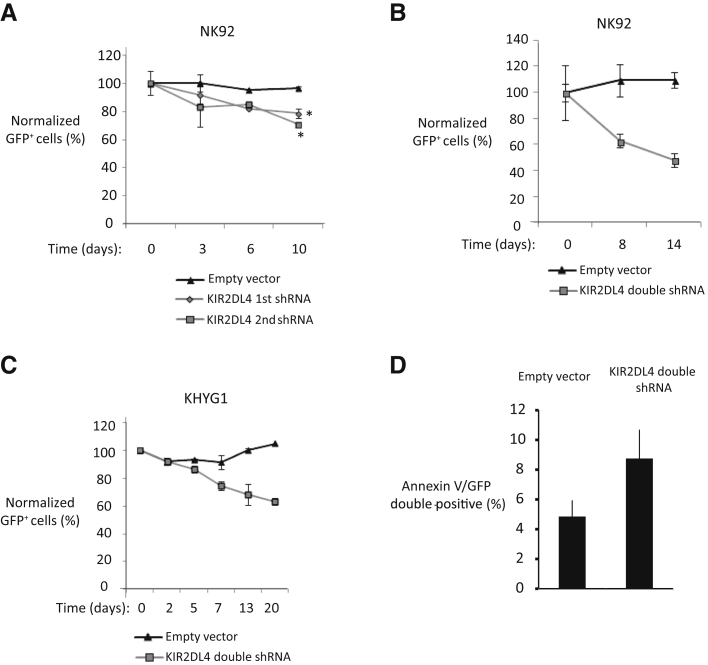
Next, we evaluated KIR2DL4 expression in three NK-cell lines with RNA-seq data (Supplemental Figure S4), and observed high selective KIR2DL4 expression, especially in the NK92 and KHYG1 cell lines, whereas other KIRs were not expressed, an observation consistent with the previous DNA microarray data (Supplemental Figure S4). The selectively retained expression of KIR2DL4 suggests that it may have a role in the neoplastic transformation of NK cells. To address this possibility, we stably knocked down KIR2DL4 expression using two different shRNAs with approximately 60% knockdown efficiency (Supplemental Figure S2B), and observed significant negative-selection pressure in KIR2DL4 shRNA–transduced NK92 cells (21.4% and 29.2% decreases on day 10 compared with day 0 in KIR2DL4 first or second shRNA–transduced cells, respectively) compared with empty vector–transduced cells (3.5% decrease on day 10 compared to day 0), as assessed by the reduction in the percentage of GFP+ cells (Figure 2A). We then generated a more effective retroviral shRNA construct (Supplemental Figure S2, C and D) by tandemly linking two KIR2DL4 shRNA–miRNAs as described previously.23 We observed a more robust decrease in the percentage of GFP+ cells in KIR2DL4 double-shRNA–transduced NK92 (Figure 2B) or KHYG1 (Figure 2C) cells compared to single-shRNA–transduced cells. Next, we evaluated apoptosis by quantifying the percentage of annexin V-phycoerythrin+/GFP+ KHYG1 cells transduced with the empty vector or KIR2DL4 double-shRNA, and observed moderately higher annexin V positivity in cells with KIR2DL4 knockdown, suggesting that an increased rate of apoptosis may be involved in the negative selection (Figure 2D). These results support a pro-oncogenic role of KIR2DL4 in NK-cell malignancies.
Figure 2.
Stable knockdown of KIR2DL4 reduces growth of NK cell lines. A: Growth of NK92 cells was monitored by quantifying the percentage of green fluorescent protein (GFP)+ cells using flow cytometry at consecutive time points after transduction with each of two individual shRNAs. Empty vector–transduced cells were used as the negative control. Each data point is normalized to the percentage of GFP+ cells at day 0. Flow cytometry quantitation was initiated 6 days after transduction (day 0). The mean percentages of GFP+ cells at day 0 were 21%, 21.2%, and 15.5% for empty vector and KIR2DL4 1st or 2nd shRNA–transduced cells, respectively. B: NK92 cells were transduced with a retroviral KIR2DL4 shRNA construct in which 1st and 2nd KIR2DL4 shRNAs are tandemly linked as shown in Supplemental Figure S2C. The percentage of GFP+ cells was calculated as described in A. The mean percentages of GFP+ cells at day 0 were 14.1% and 4.9% for empty vector or KIR2DL4–1st-2nd–double-shRNA–transduced cells, respectively. Day 0 represents 2 days after transduction. C: The percentage of GFP+ population of KHYG1 cells transduced with empty vector or KIR2DL4–1st-2nd–double-shRNA were tracked with flow cytometry and normalized to the values at day 0. Day 0 represents 6 days after transduction. The mean percentages of GFP+ cells at day 0 were 25% and 13.8% for empty vector and KIR2DL4 double-shRNA–transduced cells, respectively. D: At 7 days after transduction, annexin V staining was evaluated on GFP-gated KHYG1 cells transduced with empty vector or KIR2DL4 double-shRNA. Data are expressed as means ± SD. n = 3 biological replicates (A); n = 2 biological replicates (B–D). ∗P < 0.05 versus empty vector.
Discussion
The role of the different KIR members in the inhibition or activation of NK cells has been relatively well-studied. However, there is little insight into the clonal expression of KIR family genes in malignant NK cells, in part due to the technical limitations of the DNA microarray technology in detecting the expression levels of highly similar genes and the experimental noise that may give false-positive signals. Recently, the application of the RNA-seq technology showed a better dynamic range compared to that of tiling arrays (Supplemental Figure S1A)12 and specificity in detecting gene expression values, which can provide improved results in clonality studies.
We observed a clear gene expression pattern in terms of the clone of origin of malignant NK cells after quantification of KIR family gene expression using RNA-seq. In addition, specific and exclusive expression of a single KIR family gene member, KIR2DL4, was observed, which suggests a possible oncogenic role for it during the transformation of NK cells. This observation is consistent but more extreme than previous reports showing dominant expression of KIR2DL4 in NKTCL cases using other methodologies such as DNA microarray,4 RT-PCR,24 or flow cytometry.25 This difference using different methodologies for quantification of expression may be due to the following reasons: KIR mRNA sequences are highly similar, and DNA microarray probe sets are not as specific as RNA-seq for the measurement of the expression of individual KIR genes. Based on the GeneAnnot database,26 most of the Affymetrix probe sets for KIR genes have many nonspecific targets, which can potentially influence the quantification of mRNA levels. Therefore, RNA-seq reads may have more unique coverage of the transcripts for regions that are different from other family members. KIR antibodies used for flow cytometry can be cross-reactive. However, additional studies may need to be conducted in the future to address these modest discrepancies. Several lines of evidence can potentially support a proto-oncogenic role of KIR2DL4, which is an unusual member of the KIR family receptors.
Interestingly, despite the presence of an immunoreceptor tyrosine–based inhibition motif in its cytoplasmic tail, KIR2DL4 activates NK cells.27 KIR2DL4 is an IL-2–induced NK-cell receptor28 with a strong potential for induction of interferon-γ secretion on engagement.29 Cross-linking of KIR2DL4 in an in vitro study showed activation of mitogen-activated protein kinases and phosphorylation of I κB kinase β, with consequent phosphorylation and degradation of inhibitor of κB α leading to the activation of NF-κB.30 In addition, KIR2DL4 was reported to be associated with DNA-dependent protein kinase, catalytic subunit,31 which activated AKT by phosphorylating it at Ser 473 in response to KIR2DL4 signaling.32 It is possible that KIR2DL4 may contribute to the neoplastic transformation of NK cells by constitutively activating these pro-survival or pro-proliferative pathways. In fact, the NF-κB and AKT pathways have been shown to be constitutively activated in NKTCLs.33
Our other observation is the frequent marked down-regulation of all other KIRs, which could be advantageous for the neoplastic NK cells by removing all inhibitory receptors. Although this process also removes the activating receptors, the balance may still be in favor of losing both receptors. The retention of KIR2DL4 may serve a unique role. KIR3DL2 expression has been reported in the CD4+ neoplastic T cells of Sézary syndrome, and it may serve to reduce activation-induced cell death in these T cells.34 Thus, certain KIRs may have functional significance in specific malignant conditions.
The ligand for KIR2DL4 in this situation is not clear. Soluble HLA-G has been determined to be a ligand for KIR2DL4.31 HLA-G has been reported to be associated with tumorigenesis and poor survival in carcinoma, mainly through immune suppression and tolerance.35, 36 It generally has a restricted pattern of expression, and whether HLA-G normally activates NK cells through KIR2DL4 is still a controversial topic.37 Thus, the ligand for KIR2DL4 in NKTCLs will need to be further defined.
In IL-2–activated NK cells (Figure 1E), other KIR genes still retain mRNA expression to a comparable level. Of note, KIR2DL4 expression is not the most remarkable one in the activated NK cell sample obtained by co-culturing peripheral blood lymphocytes with the engineered K562 cells (NKCOD12 cells). It is possible that KIR2DL4 expression is augmented by IL-2; however, NKTCL cases, which were not cultured in the presence of exogenous IL-2, also show unique expression of KIR2DL4, whereas no other KIR mRNA was present in most NKTCL cases. This observation supports the idea that KIR2DL4 expression is a unique feature of clonal NKTCL cases and not the artifact of a high IL-2 concentration. However, we cannot rule out the possibility that IL-2 present in the tumor microenvironment may contribute to higher levels of KIR2DL4 expression.
Global down-regulation of KIRs with selective retention of KIR2DL4 is a common profile and is not only a useful diagnostic marker of malignant NK-cell proliferation but may also shed light on the role of KIR in the pathogenesis of NKTCLs.
Acknowledgments
We thank the University of Nebraska DNA Sequencing Core Facility and Tufts University Genomic Core Facility for RNA-seq library preparation and sequencing, and Dr. Dean Lee for the K562-Clone9-mbIL21 cells for the NK cell activation experiments.
C.K. designed and performed the experiments, analyzed the data, and wrote the manuscript; X.H. and B.J. performed experiments; Q.G., A.C., and T.M. analyzed RNA-seq data; P.G. provided the patient material for the study; and W.C.C. conceived and supervised the project, and edited the manuscript.
Footnotes
Supported by NIH Lymphoma Specialized Programs of Research Excellence grant P50CA136411-01 (W.C.C.); Science Academy’s Young Scientist Awards Program (BAGEP) (C.K.); The Scientific and Technological Research Council of Turkey (TÜBİTAK) 2232, project 115C006 (C.K.); the National Cancer Institute of the NIH award P30CA033572 and P50CA107399 (W.C.C.). The University of Nebraska DNA Sequencing Core receives partial support from National Center for Research Resources grants 1S10RR027754-01, 5P20RR016469, and RR018788-08, and from National Institute for General Medical Science grants 8P20GM103427 and GM103471-09.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.02.011.
Contributor Information
Can Küçük, Email: can.kucuk@deu.edu.tr.
Wing C. Chan, Email: jochan@coh.org.
Supplemental Data
Box plots of pairwise comparisons of the correlation of Human Genome U133 Plus version 2.0 (Affymetrix, Inc., Santa Clara, CA) and RNA sequencing (seq) data from two representative cases of natural killer/T-cell lymphoma. A: Log2-transformed Affymetrix or RNA-seq expression values were compared, and box plots generated for two representative cases are shown. B: Killer cell immunoglobulin-like receptor (KIR) family gene expression profile based on RNA-seq or Affymetrix has been compared for the same representative cases shown in A.
The shRNA constructs used to knock down expression of KIR2DL4 in natural killer (NK)-cell lines. A: The KIR2DL4 shRNA–LMP construct used in our study. B: KIR2DL4 knockdown efficiency, as tested with real-time quantitative RT-PCR in KIR2DL4 first or second shRNA–transduced Green fluorescent protein (GFP)-sorted cells. C: The KIR2DL4 double-shRNA–LMP construct used in our study. D: Knockdown efficiency of GFP+-sorted NK92 cells transduced with KIR2DL4–1st-2nd–double-shRNA is displayed. Data are expressed as means ± SD. n = 2 replicates. eGFP, enhanced green fluorescent protein; IRES, internal ribosomal entry site; LTR, long terminal repeat; KIR2DL4, KIR, killer Ig-like receptor 2DL4; LMP, LTRmiR30-PIG; MSCV, murine stem cell virus; Ppgk, phosphoglycerate kinase (PGK) promoter.
C-type lectin family receptor gene expression pattern in natural killer/T-cell lymphoma (NKTCL) cases. The mRNA expression profile of the C-type lectin family receptor genes—NKG2A (A), CD94 (B), NKG2C (C), NKG2D (D), NKG2E (E), and NKG2F (F)—across 17 NKTCL cases or three normal NK samples (G and H) are shown based on RNA sequencing values. FPKM, fragments per kilobase of transcript per million mapped reads; NKCOD12, NK cells obtained by co-culturing peripheral blood lymphocytes with K562-Cl9-mb21 cells for 12 days; PBNK48h, 48-hour IL-2–activated peripheral blood NK cells; resting NK, freshly isolated peripheral blood NK cells.
KIR2DL4 expression in malignant NK cell lines. A–C: KIR2DL4 mRNA is exclusively expressed in PMIG-NK92, KHYG1, and NKYS cell lines, as determined by RNA sequencing (top) or DNA microarray (bottom). The order of the KIR family genes is shown at the top of each plot. For DNA microarray, data previously analyzed and reported in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE19067) was used for KIR expression analysis. Data from nine of the KIR genes that have both RNA-seq and gene expression profiling data available are shown. KIR family gene mRNA values are shown as gray columns apart from KIR2DL4, which was shown in black. RNA-seq data from NK92 cells were not available, so PMIG-NK92 cells were used. PMIG-NK92, RNA-seq data from PMIG-transduced NK92 cells enriched for green fluorescent protein (GFP)-positive fraction with fluorescence-activated cell sorting that was originally prepared for another project.
References
- 1.Oshimi K. Progress in understanding and managing natural killer-cell malignancies. Br J Haematol. 2007;139:532–544. doi: 10.1111/j.1365-2141.2007.06835.x. [DOI] [PubMed] [Google Scholar]
- 2.Kwong Y.L. Natural killer-cell malignancies: diagnosis and treatment. Leukemia. 2005;19:2186–2194. doi: 10.1038/sj.leu.2403955. [DOI] [PubMed] [Google Scholar]
- 3.Ishida F., Kwong Y.L. Diagnosis and management of natural killer-cell malignancies. Expert Rev Hematol. 2010;3:593–602. doi: 10.1586/ehm.10.51. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal J., Weisenburger D.D., Chowdhury A., Tsai M.Y., Srivastava G., Greiner T.C., Kucuk C., Deffenbacher K., Vose J., Smith L., Au W.Y., Nakamura S., Seto M., Delabie J., Berger F., Loong F., Ko Y.H., Sng I., Liu X., Loughran T.P., Armitage J., Chan W.C., International Peripheral T-cell Lymphoma Project Natural killer cell lymphoma shares strikingly similar molecular features with a group of non-hepatosplenic gammadelta T-cell lymphoma and is highly sensitive to a novel aurora kinase A inhibitor in vitro. Leukemia. 2011;25:348–358. doi: 10.1038/leu.2010.255. [DOI] [PubMed] [Google Scholar]
- 5.Lanier L.L. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Single R.M., Martin M.P., Gao X., Meyer D., Yeager M., Kidd J.R., Kidd K.K., Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 7.Purdy A.K., Campbell K.S. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR) Cancer Biol Ther. 2009;8:2211–2220. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karre K. Natural killer cell recognition of missing self. Nat Immunol. 2008;9:477–480. doi: 10.1038/ni0508-477. [DOI] [PubMed] [Google Scholar]
- 9.Hilton H.G., Vago L., Older Aguilar A.M., Moesta A.K., Graef T., Abi-Rached L., Norman P.J., Guethlein L.A., Fleischhauer K., Parham P. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol. 2012;189:1418–1430. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandelboim O., Reyburn H.T., Vales-Gomez M., Pazmany L., Colonna M., Borsellino G., Strominger J.L. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shilling H.G., Young N., Guethlein L.A., Cheng N.W., Gardiner C.M., Tyan D., Parham P. Genetic control of human NK cell repertoire. J Immunol. 2002;169:239–247. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kucuk C., Jiang B., Hu X., Zhang W., Chan J.K., Xiao W., Lack N., Alkan C., Williams J.C., Avery K.N., Kavak P., Scuto A., Sen E., Gaulard P., Staudt L., Iqbal J., Zhang W., Cornish A., Gong Q., Yang Q., Sun H., d'Amore F., Leppa S., Liu W., Fu K., de Leval L., McKeithan T., Chan W.C. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun. 2015;6:6025. doi: 10.1038/ncomms7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kucuk C., Hu X., Iqbal J., Gaulard P., Klinkebiel D., Cornish A., Dave B.J., Chan W.C. HACE1 is a tumor suppressor gene candidate in natural killer cell neoplasms. Am J Pathol. 2013;182:49–55. doi: 10.1016/j.ajpath.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somanchi S.S., Senyukov V.V., Denman C.J., Lee D.A. Expansion, purification, and functional assessment of human peripheral blood NK cells. J Vis Exp. 2011;48:2540. doi: 10.3791/2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–568. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goecks J., Nekrutenko A., Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 22.Kucuk C., Iqbal J., Hu X., Gaulard P., De Leval L., Srivastava G., Au W.Y., McKeithan T.W., Chan W.C. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Natl Acad Sci U S A. 2011;108:20119–20124. doi: 10.1073/pnas.1115128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun D., Melegari M., Sridhar S., Rogler C.E., Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 24.Lin C.W., Lee W.H., Chang C.L., Yang J.Y., Hsu S.M. Restricted killer cell immunoglobulin-like receptor repertoire without T-cell receptor gamma rearrangement supports a true natural killer-cell lineage in a subset of sinonasal lymphomas. Am J Pathol. 2001;159:1671–1679. doi: 10.1016/s0002-9440(10)63014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuo Y., Drexler H.G. Immunoprofiling of cell lines derived from natural killer-cell and natural killer-like T-cell leukemia-lymphoma. Leuk Res. 2003;27:935–945. doi: 10.1016/s0145-2126(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 26.Chalifa-Caspi V., Yanai I., Ophir R., Rosen N., Shmoish M., Benjamin-Rodrig H., Shklar M., Stein T.I., Shmueli O., Safran M., Lancet D. GeneAnnot: comprehensive two-way linking between oligonucleotide array probesets and GeneCards genes. Bioinformatics. 2004;20:1457–1458. doi: 10.1093/bioinformatics/bth081. [DOI] [PubMed] [Google Scholar]
- 27.Faure M., Long E.O. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002;168:6208–6214. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 28.Kikuchi-Maki A., Yusa S., Catina T.L., Campbell K.S. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 29.Rajagopalan S., Fu J., Long E.O. Cutting edge: induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. 2001;167:1877–1881. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 30.Miah S.M., Hughes T.L., Campbell K.S. KIR2DL4 differentially signals downstream functions in human NK cells through distinct structural modules. J Immunol. 2008;180:2922–2932. doi: 10.4049/jimmunol.180.5.2922. [DOI] [PubMed] [Google Scholar]
- 31.Rajagopalan S., Long E.O. KIR2DL4 (CD158d): an activation receptor for HLA-G. Front Immunol. 2012;3:258. doi: 10.3389/fimmu.2012.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajagopalan S. Endosomal signaling and a novel pathway defined by the natural killer receptor KIR2DL4 (CD158d) Traffic. 2010;11:1381–1390. doi: 10.1111/j.1600-0854.2010.01112.x. [DOI] [PubMed] [Google Scholar]
- 33.Ng S.B., Selvarajan V., Huang G., Zhou J., Feldman A.L., Law M., Kwong Y.L., Shimizu N., Kagami Y., Aozasa K., Salto-Tellez M., Chng W.J. Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J Pathol. 2011;223:496–510. doi: 10.1002/path.2823. [DOI] [PubMed] [Google Scholar]
- 34.Thonnart N., Caudron A., Legaz I., Bagot M., Bensussan A., Marie-Cardine A. KIR3DL2 is a coinhibitory receptor on Sezary syndrome malignant T cells that promotes resistance to activation-induced cell death. Blood. 2014;124:3330–3332. doi: 10.1182/blood-2014-09-598995. [DOI] [PubMed] [Google Scholar]
- 35.Fukushima Y., Oshika Y., Nakamura M., Tokunaga T., Hatanaka H., Abe Y., Yamazaki H., Kijima H., Ueyama Y., Tamaoki N. Increased expression of human histocompatibility leukocyte antigen-G in colorectal cancer cells. Int J Mol Med. 1998;2:349–351. doi: 10.3892/ijmm.2.3.349. [DOI] [PubMed] [Google Scholar]
- 36.Tripathi P., Agrawal S. Non-classical HLA-G antigen and its role in the cancer progression. Cancer Invest. 2006;24:178–186. doi: 10.1080/07357900500524579. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopalan S., Long E.O. Comment on “Killer Ig-like receptor 2DL4 does not mediate NK Cell IFN-gamma responses to soluble HLA-G preparations”. J Immunol. 2014;192:4003. doi: 10.4049/jimmunol.1400430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Box plots of pairwise comparisons of the correlation of Human Genome U133 Plus version 2.0 (Affymetrix, Inc., Santa Clara, CA) and RNA sequencing (seq) data from two representative cases of natural killer/T-cell lymphoma. A: Log2-transformed Affymetrix or RNA-seq expression values were compared, and box plots generated for two representative cases are shown. B: Killer cell immunoglobulin-like receptor (KIR) family gene expression profile based on RNA-seq or Affymetrix has been compared for the same representative cases shown in A.
The shRNA constructs used to knock down expression of KIR2DL4 in natural killer (NK)-cell lines. A: The KIR2DL4 shRNA–LMP construct used in our study. B: KIR2DL4 knockdown efficiency, as tested with real-time quantitative RT-PCR in KIR2DL4 first or second shRNA–transduced Green fluorescent protein (GFP)-sorted cells. C: The KIR2DL4 double-shRNA–LMP construct used in our study. D: Knockdown efficiency of GFP+-sorted NK92 cells transduced with KIR2DL4–1st-2nd–double-shRNA is displayed. Data are expressed as means ± SD. n = 2 replicates. eGFP, enhanced green fluorescent protein; IRES, internal ribosomal entry site; LTR, long terminal repeat; KIR2DL4, KIR, killer Ig-like receptor 2DL4; LMP, LTRmiR30-PIG; MSCV, murine stem cell virus; Ppgk, phosphoglycerate kinase (PGK) promoter.
C-type lectin family receptor gene expression pattern in natural killer/T-cell lymphoma (NKTCL) cases. The mRNA expression profile of the C-type lectin family receptor genes—NKG2A (A), CD94 (B), NKG2C (C), NKG2D (D), NKG2E (E), and NKG2F (F)—across 17 NKTCL cases or three normal NK samples (G and H) are shown based on RNA sequencing values. FPKM, fragments per kilobase of transcript per million mapped reads; NKCOD12, NK cells obtained by co-culturing peripheral blood lymphocytes with K562-Cl9-mb21 cells for 12 days; PBNK48h, 48-hour IL-2–activated peripheral blood NK cells; resting NK, freshly isolated peripheral blood NK cells.
KIR2DL4 expression in malignant NK cell lines. A–C: KIR2DL4 mRNA is exclusively expressed in PMIG-NK92, KHYG1, and NKYS cell lines, as determined by RNA sequencing (top) or DNA microarray (bottom). The order of the KIR family genes is shown at the top of each plot. For DNA microarray, data previously analyzed and reported in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE19067) was used for KIR expression analysis. Data from nine of the KIR genes that have both RNA-seq and gene expression profiling data available are shown. KIR family gene mRNA values are shown as gray columns apart from KIR2DL4, which was shown in black. RNA-seq data from NK92 cells were not available, so PMIG-NK92 cells were used. PMIG-NK92, RNA-seq data from PMIG-transduced NK92 cells enriched for green fluorescent protein (GFP)-positive fraction with fluorescence-activated cell sorting that was originally prepared for another project.