Abstract
Inter-specific hybrids are a useful source for increasing genetic diversity. Some reproductive barriers before and/or after fertilization prevent production of hybrid plants by inter-specific crossing. Therefore, techniques to overcome the reproductive barrier have been developed, and have contributed to hybridization breeding. In recent studies, identification of molecules involved in plant reproduction has been studied to understand the mechanisms of reproductive barriers. Revealing the molecular mechanisms of reproductive barriers may allow us to overcome reproductive barriers in inter-specific crossing, and to efficiently produce inter-specific hybrids in cross-combinations that cannot be produced through artificial techniques. Inter-specific hybrid plants can potentially serve as an elite material for plant breeding, produced through the merging of genomes of parental species by allopolyploidization. Allopolyploidization provides some benefits, such as heterosis, increased genetic diversity and phenotypic variability, which are caused by dynamic changes of the genome and epigenome. Understanding of allopolyploidization mechanisms is important for practical utilization of inter-specific hybrids as a breeding material. This review discusses the importance of reproductive barriers and the effect of allopolyploidization in crop breeding programs.
Keywords: inter-specific hybrid, pre-zygotic barrier, endosperm barrier, hybrid necrosis, heterosis, genomic rearrangements, non-additive expression
Introduction
Crop species have been improved for consumption by selecting for useful traits, and accumulating desirable genes from genetic resources. “Hybridization breeding” introduces agriculturally valuable traits into existing cultivars, such as biotic or abiotic stress tolerance, from breeding stocks or wild progenitors by intra-specific or inter-specific crossing. As genetic resources are limited in intra-species, inter-specific crossing is expected to contribute to further development of plant breeding programs.
In the process of hybridization breeding by inter-specific crossing, reproductive barriers, whose mechanisms maintain the genetic integrity of species or prevent gene flow with other species, challenge hybrid production. Reproductive barriers are roughly classified into two types, pre- (before fertilization) and post-zygotic (after fertilization) barriers. Pollination with other species is restricted by their natural habitat (geographic isolation), flower structure (i.e., shape or color), flowering time or pollen mediator, which are involved in pre-zygotic barriers (Lowry et al. 2008). Inter-specific incompatibility is the inhibition of pollen tube germination or growth. Inter-specific incompatibility is involved in post-zygotic barriers and occurs in the pistils when the pollen from other species pollinate the stigma (Dresselhaus et al. 2011). Even if pollen germination and fertilization are successful, embryo abortion or abnormal growth of endosperm (over- and under-proliferation of endosperm) causing disturbance of normal seed development are observed as post-zygotic barriers (Haig and Westoby 1991). Embryo abortion is thought to be caused by endosperm abnormality. Endosperm abnormality depends on the cross combination of species, direction of hybridization, and ploidy levels of parental species (Kinoshita 2007).
After hybrid seed formation, there are still reproductive barriers. Inter-specific hybrids sometimes show developmental defects compared to the parental lines, which is termed hybrid necrosis (or hybrid weakness). Furthermore, hybrid sterility caused by epistatic interactions between nuclear genes or between the nuclear genome and the maternally derived mitochondrial genome, termed cytoplasmic male sterility (CMS), remains as a post-zygotic barrier. However, male sterility is sometimes useful for breeding, and CMS is successfully applied to harvest the F1 hybrid seeds (Yamagishi and Bhat 2014). Several genes involved in hybrid sterility or CMS in rice have been isolated by a genetic approach (Fujii and Toriyama 2008). Overcoming reproductive barriers expands the possibility of hybridization breeding, and understanding the molecular mechanism of reproductive barriers is becoming more important.
There are polyploid species not only in wild species, but also in cultivated crops. Polyploids include important crops, such as maize, cotton, wheat, canola, tobacco, potato, and strawberry, indicating that polyploidization can potentially generate useful traits for cultivation. In plant speciation, whole-genome duplication that can lead to genetic redundancy and functional divergence of paralogous genes is considered as an important event related to the increase of genetic diversity (Innan and Kondrashov 2010). There are two types of polyploid species, autopolyploid and allopolyploid. Autopolyploid species are formed by multiplication of the genome from a single species. Genome size is increased by genome multiplication and the multiplied genes can provide useful traits for agriculture, such as enhanced photosynthesis or enlargement of fruit size by expansion of cell size (Lavania et al. 2012). Allopolyploid species are formed by merging genomes from two or more distantly related species. The merging genomes from divergent species not only leads to functional divergence of paralogous genes, but also heterozygosity (Chen 2010, Comai 2005, Osborn et al. 2003). Heterozygosity has advantages such as heterosis, an increase in genetic diversity, and phenotypic variability. Heterosis can give rise to growth vigour and an increase in crop yield. Genetic diversity and phenotypic variability are accompanied by various changes including genomic rearrangements by multivalent chromosome formation, homoeologous recombination, chromosome elimination, and transcriptome alteration (Adams et al. 2003, Gaeta and Chris Pires 2010, Otto 2007), some of which may play a role in generating desirable traits for cultivation.
This review focuses on the reproductive barriers in inter-specific hybridization and the dynamic genetic or epigenetic changes in inter-specific hybrids. We introduce the technical or genetic methods to overcome these reproductive barriers, molecular mechanism of reproductive barriers (especially pollen rejection, endosperm barriers and hybrid necrosis), and the dynamic changes caused by allopolyploidization.
1. Overcoming reproductive barriers
Producing inter-specific hybrids is an effective way to increase genetic diversity for breeding programs. However, it is necessary to overcome reproductive barriers. Artificial techniques to overcome the reproductive barriers have been developed for producing inter-specific hybrids. The use of genetic information to determine the crossability is also effective for hybridization breeding. In many plant species, different accessions of parental species have different crossability in inter-specific crossing, indicating that some genetic factors are important for the efficient production of inter-specific hybrids. Identification of key genes for inter-specific hybridization will allow rapid production of hybrids and contribute to accelerating the breeding program.
1-1. Artificial techniques for producing inter-specific hybrids
Techniques for producing inter-specific hybrids have been developed in many crops. There are several methods for overcoming inter-specific incompatibility as a pre-zygotic barrier. It has been suggested that these barriers are weaker at the early floral development stage; therefore, the bud-pollination method (pollinating stigmas of buds two to three days before anthesis) has been used in Brassicaceae (Hiscock and Dickinson 1993, Udagawa et al. 2010) and Solanaceae species (Gradziel and Robinson 1991, Kuboyama et al. 1994). Inter-specific incompatibility occurs at the stigma or style, and pollinating to a cut section of a stigma or style (cut-style pollination method) has been used in Liliaceae (Van Tuyl et al. 1988). Heat treatment of pistils or pollen has also been successful to overcome pre-zygotic barriers in Liliaceae (Matsubara 1981).
As a post-zygotic barrier, embryo abortion occurs during inter-specific crossing, especially between distantly related species. To overcome this barrier, embryo rescue techniques, including ovary, ovule, and embryo culture, have been developed (Van Tuyl et al. 1991). The timing of embryo abortion is different for each parental combination, and suitable method for embryo rescue in each inter-specific cross is required. If the embryo abortion occurs at the early stage, immediately after fertilization, ovary culture can be applied where young ovaries collected after pollination can be grown on tissue culture medium. Mature seeds can be obtained through ovary culture in some inter-specific crosses (Takahata 1990, Takahata and Takeda 1990). When mature seeds cannot be obtained by ovary culture or hybrid embryo because of abortion caused by the mismatch between embryo and endosperm development, ovule and embryo culture methods are suitable (Bajaj et al. 1986, Iwai et al. 1986). The embryo culture method rescues immature embryos by growing the embryo on the medium, and it is the most effective method applied in many inter-specific crosses (Sharma et al. 1996). The ovule culture method removes the immature ovules from the ovaries, and the ovules are germinated on the medium. This method is useful for plant species with small ovules when the dissection of immature embryos from the ovule is technically difficult.
Inter-specific hybrids between distantly related species are generally sterile or partially sterile. One of the reasons for hybrid sterility is reduced chromosome paring during meiosis. Hybrid sterility can often be restored by zygotic or somatic chromosome doubling through cell cycle disruption by antimitotic agents such as colchicine, oryzalin, or trifluralin (Dhooghe et al. 2011). Nitrous oxide (N2O) is also applied as a polyploidizing agent in lilies (Nukui et al. 2011).
1-2. Key genes for inter-specific hybridization
Key genes for crossability have been identified by genetic analysis of intra-specific genetic variation. In wheat (Triticum aestivum), two loci carrying dominant genes, Kr1 and Kr2, located on the long arm of chromosome 5B and 5A, respectively, affect the crossability in inter-specific crossing between wheat and rye (Secale cereale) (Lein 1943). Dominant alleles of Kr1 and Kr2 show poor crossability caused by inhibition of pollen germination and pollen tube growth before fertilization (Jalani and Moss 1980, Lange and Wojciechowska 1976, Riley and Chapman 1967). Most wheat cultivars with dominant alleles (Kr1 and Kr2) show low crossability in inter-specific crosses with rye; however, ‘Chinese spring’ and ‘Hope’, which have recessive alleles (kr1 and kr2), show high crossability (Riley and Chapman 1967). Other crossability loci, Kr3 located on chromosome 5D (Krolow 1970), Kr4 on chromosome 1A, and SKr on the short arm of chromosome 5B (Tixier et al. 1998) have been identified. These Kr genes have different effects on the crossability between wheat and rye; Kr1 has a stronger effect than other Kr genes (Krolow 1970, Riley and Chapman 1967). Moreover, wheat Kr genes affect the crossability with cultivated barley Hordeum vulgare (Koba and Shimada 1992), wild barley Hordeum bulbosum (Snape et al. 1979), sorghum Sorghum bicolor (Inagaki and Mujeeb-Kazi 1995) and Aegilops squarrosa (syn. tauschii) (Koba and Shimada 1993). Although fine mapping of Kr1 (Bertin et al. 2009) and the SKr locus have been carried out (Alfares et al. 2009), Kr genes have not been isolated and their molecular functions are not clear.
In Brassicaceae, the two lines of Brassica rapa, ‘Shogoin-kabu’ and ‘Chiifu’ show different crossability to Raphanus sativus; ‘Shogoin-kabu’ can produce several seeds when crossed with R. sativus, whereas ‘Chiifu’ does not produce any seeds. Three QTLs for crossability, qBrHFA-1, qBrHFA-2, and qBrHFA-3, which control embryo abortion, have been identified using a segregating population between ‘Shogoin-kabu’ and ‘Chiifu’ (Tonosaki et al. 2013). Moreover, the combination of qBrHFA-1 and qBrHFA-3 is important for increasing crossability with R. sativus.
Although most cultivars of wheat and B. rapa do not have high crossability for inter-specific crossing, a few cultivars such as “Chinese spring” of wheat and “Shogoin-kabu” of B. rapa show high crossability. This suggests that these cultivars gained high crossability by the mutation of the key genes during breeding steps. Thereby, varieties of other crops may also have a high crossability caused by mutation of these key genes. The generation of DNA markers based on this genetic information may be useful for effective production of inter-specific hybrids, and hybridization breeding.
2. Molecular evidence of reproductive barriers
2-1. Pre-zygotic barriers
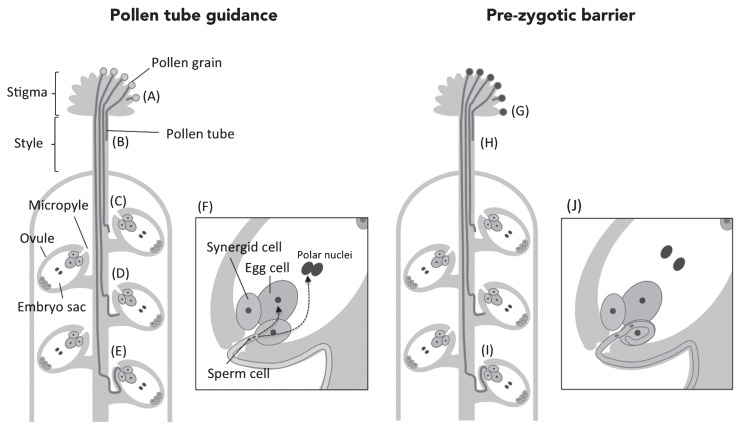
Pre-zygotic barriers in inter-specific crossing may be composed of multi-steps. Pollen tube guidance in angiosperms is a multi-stage mechanism (Okuda and Higashiyama 2010), which leads the pollen tube to the ovule from the stigma via the pistil by recognition of its own pollen (Fig. 1A–1F). It is thought that defective pollen tube guidance interferes with the plant reproductive mechanisms; therefore, all steps of pollen tube guidance potentially lead to a pre-zygotic barrier. Some studies on pollen tube guidance provide insights into pre-zygotic barriers by inter-specific pollen recognition (Fig. 1G–1J).
Fig. 1.
Schematic representations of multi-stage controlled pollen tube guidance and pre-zygotic barriers. Pollen tube guidance involves multistage controls (A–F). In pollen tube guidance, the pollen tube enters the stigma after pollen germination (A), and elongates the pollen tube through the stigma and style (B). The pollen tube penetrates the transmitting tract (C), grows on the inner surface of the ovule (D), and enters the micropyle toward the synergid cell (E). Finally, the pollen tube releases the sperm cells (F). Pre-zygotic barriers can also be a multi-stage mechanism (G–J). In the case of pollination with other species, pollen hydration and germination is inhibited on the stigma (G) and pollen tube elongation through stigma and style is prevented (H). In some inter-specific hybrids, these steps (G–H) may involve S-locus genes, which regulate self-incompatibility. The pollen tube is attracted by attractants, LURE peptide, in a species-preference manner (I). Pollen tubes entering the micropyle cannot burst to release the sperm cells, and overgrow inside the female gametophyte (J).
Pre-zygotic barriers and self-incompatibility
The first pre-zygotic barrier is pollen rejection; the inhibition of pollen hydration, germination, or pollen tube growth (Fig. 1G, 1H). These behaviors of pollen rejection are similar to that of self-incompatibility, which is regulated by a single multi-allelic locus, the S locus, in dicots (Dresselhaus et al. 2011, Murfett et al. 1996). In sporophytic self-incompatibility of Brassicaceae plants, the inhibition of pollen hydration or germination starts from an S haplotype specific interaction between the female S-determinant (S-locus receptor kinase (SRK)) and the male S-determinant (S-locus protein 11/S-locus-cysteine-rich protein (SP11/SCR)) in the stigma (Watanabe et al. 2003). In addition to genes, M-locus protein kinase (MLPK) was identified as a factor of self-incompatibility (Murase et al. 2004). In Solanaceae, Rosaceae, and Plantaginaceae plants with gametophytic self-incompatibility, the female S-determinant (S-RNase), which has ribonuclease activity, and the male S-determinant (S-locus F-box (SLF/SFB) protein), which is a component of the E3 ubiquitin ligase complexes, have been identified (Entani et al. 2003, Lai et al. 2002, Sijacic et al. 2004). Additionally, modifier genes, asparagine-rich HT proteins (HT-B) in tobacco and two HT proteins (HT-A and HT-B) in tomato, which play a role in pollen rejection and do not contribute to S specificity, were identified (Goldraij et al. 2006, O’Brien et al. 2002).
In inter-specific crosses, unilateral incompatibility, which occurs when pollination between species is successful in only one direction, is often observed. It often occurs in crosses between self-compatible and self-incompatible species in Solanaceae species. Pollen from self-compatible species is rejected by pistils of self-incompatible species, while pollen from self-incompatible species is not rejected by pistils of self-compatible species. This relationship is called the “SI × SC rule”. The molecular mechanism controlling self-incompatibility and inter-specific unilateral incompatibility may be linked in both Brassicaceae species with sporophytic self-incompatibility (Hiscock and Dickinson 1993) and Solanaceae species with gametophytic self-incompatibility (Lewis and Crowe 1958, Murfett et al. 1996).
Two lines of B. rapa stigmas pollinated with Brassica oleracea pollen grains show different pollen tube growth. Pollen from B. oleracea is rejected on pistils of self-incompatible lines of B. rapa, while pollen from B. oleracea is not rejected on pistils of self-compatible lines of B. rapa, which lack functional SP11, SRK, and MLPK (Fujimoto et al. 2006, Murase et al. 2004). However, this difference of inter-specific incompatibility in stigma may not involve the female determinants of self-incompatibility, SRK, and MLPK (Udagawa et al. 2010). Molecular genetic evidence that links the mechanisms of self-incompatibility and inter-specific unilateral incompatibility has not been clarified in Brassicaceae species.
Cultivated tomato (Solanum lycopersicum) is self-compatible and has lost the function of S-RNase, HT-A, and HT-B (Kondo et al. 2002). The introduction of functional S-RNase and HT genes from self-incompatible wild tomato (Solanum peruvianum) into self-compatible cultivated tomato causes inter-specific unilateral incompatibility, while a functional S-RNase alone did not (Tovar-Mendez et al. 2014). Similarly, in another Solanaceous genus, Nicotiana, transgenic plants with S-RNase and HT expression in Nicotiana tabacum show inter-specific unilateral incompatibility (Hancock et al. 2005, Murfett et al. 1996). These results suggested that the mechanism of pollen rejection of unilateral incompatibility overlaps with that of S-RNase dependent self-incompatibility. However, there are cases of an S-RNase-independent pollen rejection system.
In the genetic analysis of inter-specific unilateral incompatibility of the pollen side, two loci, ui1.1 and ui6.1, have been identified as factors of male-determinant for inter-specific unilateral incompatibility (Chetelat and Deverna 1991, Li et al. 2010). ui1.1 and ui6.1 encode SLF-23 and Cullin1 proteins, which are components of the SCF-type (Skp1, Cullin1, F-box) and ubiquitin ligase complexes (Li and Chetelat 2010, 2015), respectively. Cultivated tomato species and two wild relatives of cultivated tomato, which are self-compatible species, lack SLF-23 and have a loss-of-function mutation in Cullin1. Two transgenes, SLF-23 and Cullin1 from self-incompatible species, introduced into cultivated tomato, are sufficient to convert pollen rejection phenotype from unilateral incompatibility to compatible in inter-specific crossing (Li et al. 2010, Li and Chetelat 2015). Moreover, knockdown of Cullin1 loses self-incompatibility in Solanum arcanum (Li and Chetelat 2014), indicating that the SCF-type ubiquitin ligase complex may be involved not only in inter-specific unilateral incompatibility but also in self-incompatibility.
Species-preference of pollen tube attraction
Synergid cells play an important role in pollen tube guidance (Fig. 1E) (Higashiyama et al. 2006). In Torenia fournieri, LUREs, which are secreted cysteine-rich polypeptides derived from synergid cells, have been identified as attractants (Okuda et al. 2009). LUREs of T. fournieri do not attract the pollen tube of the closely related species, Lindernia micrantha, indicating pollen tube guidance by LUREs acts in a species-preferential manner (Fig. 1I) (Okuda et al. 2009). LUREs are also attractants in A. thaliana and A. lyrata. AtLURE1 peptide showed species-preference, while AlLURE1 peptide did not. In transgenic plants of T. fournieri expressing AtLURE1 in synergid cells, the pollen tube of A. thaliana reached the embryo sac of T. fournieri, suggesting that expression of AtLURE1 is sufficient to overcome inter-specific barriers in micropylar pollen tube guidance and penetration of the embryo sac (Takeuchi and Higashiyama 2012). Inter-specific hybrids between A. thaliana and A. lyrata can be obtained by pollinating A. thaliana stigma with pollen from A. lyrata by the cut-style pollination method; however, A. thaliana pollen cannot germinate on the A. lyrata stigma (Fujimoto et al. 2008). However, AlLURE1 has the ability to attract A. thaliana pollen, suggesting that the avoidance of pollen rejection on stigma by the cut-style pollination method may be applied to obtain reciprocal hybrids.
Species specificity of the pollen tube burst
The pollen tube bursts after entering into the micropyle in the last steps of pollen tube growth, and discharges two sperm cells into the female gametophyte for double fertilization (Fig. 1F) (Hamamura et al. 2011). In the A. thaliana feronia mutant, the pollen tube overgrows in the female gametophyte, and is unable to burst after entering the micropyle (Fig. 1J) (Huck et al. 2003, Rotman et al. 2003). The FERONIA gene encodes a receptor-like kinase, and is localized to the filiform apparatus of synergid cells. It has been suggested that FERONIA receives a male ligand, and triggers a signal cascade of feedback from the synergid cell to the pollen tube, causing an arrest of pollen tube growth, and release of sperm cells (Escobar-Restrepo et al. 2007). In inter-specific crossing of A. thaliana with pollen from a related Brassicacese species, the pollen tube showed overgrowth in the female gametophyte, similar to the phenotype observed in the feronia mutant (Fig. 1J). In the cross with A. lyrata, around half of the ovules received pollen tubes and were fertilized, but the others had pollen tubes that overgrew inside the female gametophyte. In the cross with a more distant relative, Cardamine flexuosa, around 70% of pollen tubes entered A. thaliana ovules, and displayed a feronia-like phenotype. This phenotype in inter-specific crossing correlated with high sequence divergence in the extracellular domain of FERONIA (Escobar-Restrepo et al. 2007), allowing us to speculate that the FERONIA pathway is involved in the pre-zygotic barrier just before fertilization in inter-specific crossing.
2-2. Reproductive barriers in endosperm
Embryo abortion in inter-specific crossing is thought to be caused by endosperm abnormality. In flowering plant, double fertilization gives rise to the embryo and endosperm (Berger et al. 2008). The endosperm is a triploid tissue with an unequal parental genomic contribution (the sperm nucleus (n) fuses with the two polar nuclei (2n) in the central cell), and it is a storage tissue for the control of nutrient transfer to the embryo during seed development and for seedling development after germination. In many cases of inter-specific crossing, embryo abortion phenotypes are observed together with endosperm abnormality, and can be overcome by embryo rescue techniques compensating with nutrients from the medium. This suggests that the major factor that is responsible for embryo abortion is the scarcity of nutrient supply during embryo development due to the failure of endosperm development.
The mechanism of endosperm abnormality
The abnormality of hybrid endosperm shows contrary phenotypes depending on the direction of the cross, especially when the parental lines have different ploidy levels. For example, when the ploidy level of the female parent is higher (maternal excess), under-proliferation of endosperm and the promotion of seed storage synthesis are observed. By contrast, when the ploidy level of the male parent is higher (paternal excess), over-proliferation of the endosperm and alteration of seed storage synthesis are observed (Scott et al. 1998, Sekine et al. 2013). Both under- and over-proliferation of hybrid endosperm are observed in many plant species, and it may be due to a general mechanism for preventing hybrid seed development. In classical genetic studies, models of ‘endosperm balance number’ in potatoes (Johnston et al. 1980) or ‘polar nuclei activation’ in oats (Nishiyama and Yabuno 1978) have suggested that the genome balance between male and female parents is important for hybrid endosperm development in inter-specific crossing. In inter-specific crossing, the balancing of parental genome dosages by manipulation of ploidy levels is required for successful hybrid seed development (Johnston and Hanneman 1982, Josefsson et al. 2006). These results support the importance of parental genome balance in hybrid endosperm.
Determinants of the parental genome balance
Recent studies suggest that disturbance of the genome balance is caused by the epigenetic misregulation of imprinted genes (Kinoshita 2007). Genome imprinting is when the two alleles of a gene are expressed at different levels depending on their parent of origin, and is known to be regulated by epigenetic mechanisms. In flowering plant, genomic imprinting mainly occurs in the endosperm, and many imprinted genes have been identified by high-throughput sequencing techniques (Hsieh et al. 2011, Luo et al. 2011, Waters et al. 2011, Wolff et al. 2011). In A. thaliana, many mutants showing endosperm abnormality have been identified, some of which are imprinted genes. For example, the mutants of MEDEA (MEA), FERTILIZATION INDEPENDENT SEED2 (FIS2), which encode components of the Polycomb repressive complex 2 (PRC2), show extension of the syncytial phase, delay of cellularization of the endosperm (over-proliferation), and arrest of embryo development (Chaudhury et al. 1997, Grossniklaus et al. 1998, Kiyosue et al. 1999). In particular, the fis2 mutant not only delays the timing of endosperm cellularization, but also decreases the translocation rate of hexoses to the embryo (Hehenberger et al. 2012). MEA and FIS2 are imprinted genes in the endosperm, expressed only from the maternal allele (Jullien et al. 2006, Kinoshita et al. 1999). The PRC2, including MEA and FIS2, has an important role in the development of endosperm.
The PRC2 complex is conserved in mammals, Drosophila, and plants, and has methyltransferase activity, tri-methylation of lysine 27 on histone H3 (H3K27me3) at target genes. Type-I MADS transcription factors, AGAMOUS-LIKE GENE 62 (AGL62) and PHERES1 (PHE1), are downstream genes regulated by PRC2 (Kang et al. 2008, Köhler et al. 2005). The delay of cellularization of the endosperm and decrease of the translocation rate of hexoses to embryo observed in fis2 mutant are suppressed in double mutants of fis2 and agl62, suggesting that AGL62 regulates endosperm development, especially during cellularization (Hehenberger et al. 2012). PHE1 is an imprinted gene expressed only from the paternal allele and plays a role in endosperm development, because suppression of PHE1 in the mea mutant background prevents over-proliferation of the endosperm (Köhler et al. 2005).
Endosperm abnormality and PRC2 regulation
Endosperm abnormality is not only observed in inter-specific crosses, but also in crosses between parents of different ploidy (inter-ploidy cross). Increased paternal contribution by paternal excess during inter-ploidy crossing shows over-proliferation of the endosperm (Scott et al. 1998, Sekine et al. 2013). In the abnormal endosperm of paternal excess, expression of MEA is decreased, and expression of PRC2 target genes is increased; moreover, overexpression of MEA can restore normal endosperm development (Erilova et al. 2006). This indicates that reduced PRC2 function is likely to be responsible for endosperm abnormality in inter-ploidy crossing. On the other hand, in inter-specific crossing between A. thaliana and its close relative, A. arenosa, the hybrid endosperm shows over-proliferation and increased expression of PRC2 target genes such as PHE1 and AGL62 (Josefsson et al. 2006, Walia et al. 2009). Similarly, endosperm abnormality and the alteration of expression of OsMADS87, a PHE1 orthologue, are observed in inter-specific hybrids between rice and related wild species (Ishikawa et al. 2011). The maternal phe1 mutation improves the production of inter-specific hybrid seeds (Josefsson et al. 2006), suggesting that endosperm abnormality is attributed to misexpression of PRC2 target genes, which is caused by alteration of PRC2 regulation, in the hybrid endosperm. Some PRC2 components were identified as being involved in the post-zygotic barrier in the seeds of inter-specific hybrids in the inter-specific crossing of Arabidopsis (Burkart-Waco et al. 2013) or Brassica crops (Tonosaki et al. 2013). These results support PRC2 being related to endosperm abnormality in inter-specific crossing.
In inter-specific crossing, manipulation of ploidy levels was able to overcome endosperm abnormality (Johnston and Hanneman 1982), and moderate the misexpression of PRC2 target genes (Bushell et al. 2003, Johnston and Hanneman 1982). Disturbance of the genome balance may alter PRC2 regulation by a change in PRC2 dosage between parental species, and give rise to misexpression of PRC2 target genes (Fig. 2A). It has been suggested as a “quantitative difference” in inter-specific hybrids (Schatlowski and Köhler 2012). On the other hand, the endosperm abnormality and alteration of gene expression are observed in inter-specific crossing between cultivated rice and its close relatives, which have the same genome size and chromosome number (Ishikawa et al. 2011), indicating that endosperm abnormality occurred in inter-specific crossing without the quantitative difference between parental species. Due to the diversity of the amino acid sequences of the PRC2 component from different species in hybrid endosperm, the complex can be adversely affected. The formation of the functional PRC2 complex of maternally derived polycomb genes is limited in the hybrid endosperm (Ishikawa and Kinoshita 2009). Therefore, there is not enough functional PRC2 to regulate target genes of each allele from the two parental species leading to a “qualitative difference” in interspecies (Fig. 2B).
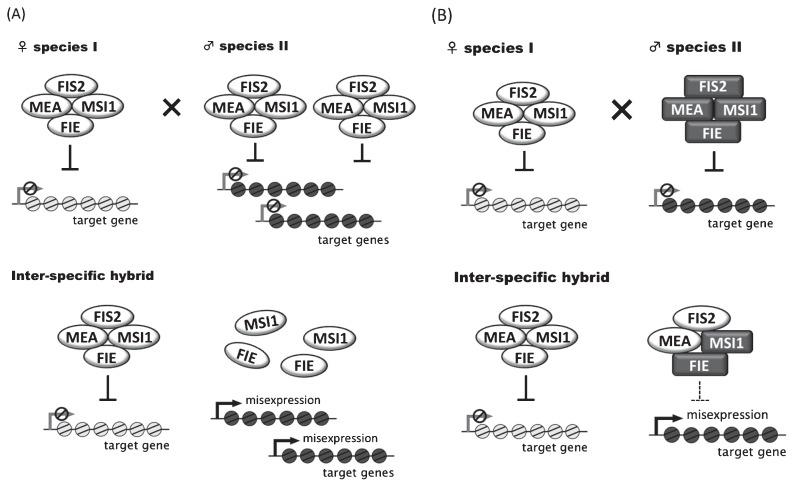
Fig. 2.
Regulation of target genes by PRC2 in inter-specific hybrid endosperm. (A) Alteration of PRC2 regulation by quantitative difference. The PRC2 proteins MEA, FIS2, FIE and MSI1 form a complex and repress target genes. The balance of PRC2 and target genes is different between parental species. In hybrid endosperm between maternal species I and paternal species II, misexpression of target genes is observed because maternally imprinted genes, MEA and FIS2, are not expressed from paternal species II. (B) Alteration of PRC2 regulation by qualitative difference. Two types of PRC2 complex, functional PRC2 and chimera complex, formed in hybrid endosperm. Chimera complex is assembled from MEA and FIS2, which are expressed only when they are derived from maternal species I allele, and FIE and MSI1 from species II allele. Chimera complex may be adversely affected by the diversity of the amino acid sequence of the PRC2 component.
Other factors in post-zygotic barriers
From the comparison of transcriptomes between hybrid seeds three days after pollination (DAP) in A. thaliana Columbia × A. arenosa (incompatible hybrid) and A. thaliana C24 × A. arenosa (compatible hybrid), misexpression of endosperm and seed regulator genes, and activation of ribosomal, photosynthesis, stress-related, and immune response genes have been identified (Burkart-Waco et al. 2013). The loss-of-function mutants of TRANSPARENT TESTA GLABRA2 (ttg2) and HAIKU1 (iku1) genes, which are endosperm growth factors, used as the maternal parent, showed partial improvement in seed survival when crossed to A. arenosa, suggesting that misregulation of genes involved in endosperm development causes hybrid incompatibility (Burkart-Waco et al. 2013). The loss-of-function of NON-EXPRESSOR OF PATHOGENESIS RELATED 1 (NPR1) and SALICYLIC ACID INDUCTION-DEFICIENT 2 (SID2), whose functions are in the defense response, also showed increased crossability, suggesting that a defense response dependent on the salicylic acid pathway is involved in the reproductive barrier.
2-3. Hybrid necrosis
Hybrid necrosis is one of the negative results of both intra- and inter-specific hybrids. Necrosis can be temperature dependent (Bomblies and Weigel 2007), and cause slow growth, wilting, discoloration, and lethality. Recent studies revealed that hybrid necrosis is caused by an autoimmune-like response through epistatic interaction between resistance (R) genes (Bomblies et al. 2007, Jeuken et al. 2009), and that the Dobzhansky-Muller model explains hybrid necrosis caused by the deleterious interaction between lineage-specific alleles at two or more loci (Dobzhansky 1937, Muller 1942).
In inter-specific hybrids in lettuce (Lactuca sativa and Lactuca saligna), hybrid necrosis is caused by the interaction between the L. sativa allele at the C6 locus and the L. saligna allele at the C9 locus. Segregation analysis found Rin4 at the C9 locus, which has the highest sequence similarity to RIN4 (RPM1 INTERACTING PROTEIN 4) of A. thaliana. Rin4 of L. saligna can cause severe necrosis in L. sativa when the C6 locus of L. sativa is present, but Rin4 of L. sativa and L. virosa does not lead to necrosis. There are six amino acid differences between Rin4 of L. sativa and L. saligna, and three of these changes in L. sativa Rin4 are required for hybrid necrosis (Jeuken et al. 2009).
Triploid hybrids (ABD) between tetraploid wheat (AABB) and diploid Aegilops tauschii (DD) showed abnormal growth phenotypes, some that are caused by two types of hybrid necrosis (type II and type III); type II necrosis occurs under low temperature conditions (4°C), while type III occurs gradually without temperature dependence (Mizuno et al. 2010). Segregation tests revealed that type II and type III necrosis are associated with a single genetic locus on the short arm of the D genome, chromosome 7D and 2D, respectively (Mizuno et al. 2010, 2011). Temperature-dependent hybrid weakness is also observed in inter-specific hybrids in rice (Chen et al. 2014, Fu et al. 2013). One high-temperature dependent hybrid weakness is caused by the interaction between Hwi1 in wild rice, Oryza furipogon, and Hwi2 in indica rice, Oryza sativa (Chen et al. 2014). Hwi1 and Hwi2 encode two LRR-RLK genes and putative subtilisin-like protease, respectively, and it has been considered that the activation of the autoimmune response by the interaction between these two genes causes hybrid weakness (Chen et al. 2014).
3. Effects of allopolyploidization
3-1. Heterosis
In the process of generating inter-specific hybrids, heterosis phenotype caused by heterozygosity of different species can be fixed through chromosome doubling, and may contribute to adaptation of allopolyploids and selection for breeding. Artificially synthesized Brassica juncea (AABB genome) sometimes show biomass heterosis relative to the parental species of B. rapa (AA genome) and B. nigra (BB genome), but are dependent on the hybrid combinations of parental lines (Bansal et al. 2012). Biomass heterosis is also observed in inter-specific hybrids between allopolyploid species B. napus (AACC genome) and one of its progenitor species, B. rapa (Liu et al. 2002, Qian et al. 2003). Intersubgenomic heterosis has been observed in B. napus introgressed with partial B. rapa genome, and introgression of the B. rapa genetic component into B. napus has been used for generating a new type of B. napus (Qian et al. 2005). In a study of B. rapa and B. napus, novel genomic alterations post-hybridization were associated with the greatest number (compared to allelic contribution by B. rapa) of QTLs for agronomic traits (Zou et al. 2011).
Hybrids between A. thaliana and related species of A. arenosa, A. lyrata or A. halleri show a hybrid-vigour-like phenotype (Fujimoto et al. 2011, Ni et al. 2009). In the inter-specific hybrid between A. thaliana and A. arenosa, key genes of circadian rhythm, such as CCA1 (CIRCADIAN CLOCK ASSOCIATED 1), LHY1 (LATE ELONGATED HYPOCOTYL 1), TOC1 (TIMING OF CAB EXPRESSION 1), and GI (GIGANTEA), showed a differential expression pattern between the hybrid and its parental lines (Ni et al. 2009). Circadian rhythm plays a role in plant growth and development, and the transgenic plants down-regulated in CCA1 expression during the day increased its biomass. Thus, the authors considered that alteration of circadian rhythm genes in inter-specific hybrids results in the vigour phenotype (Ni et al. 2009).
3-2. Chromosome dynamics
Hybridization can lead to dynamic changes of the chromosomes in response to the “genomic shock” of the merging genomes. During this process, major (elimination, disjunction and doubling of chromosome) and minor (deletion or inclusion of DNA sequences, gene conversion and rDNA loci changes) chromosome changes are known to occur. Many of these chromosome changes occur after allopolyploidization events (hybridization of different species leading to doubling of the genome), and the effect of hybridization is thought to have greater influences on gene expression than the doubling of homologous chromosomes (Adams 2007). These chromosomal changes induced by hybridization can cause transcriptional changes that can ultimately affect the phenotype of the plants.
Homoeologous chromosome pairing
During meiosis, homologous/homoeologous chromosomes are paired to form so-called bivalents (association of three or more chromosomes is called multivalent). The formation of bivalents involves homologous recombination, which exchanges genetic information between paired homologous/homoeologous chromosomes, and results in segregation of a novel set of chromosomes into gametophytic cells. Understanding of the molecular mechanism of homologous/homoeologous pairing may be useful to control desired traits for breeding programs. For example, homoeologous recombination is well documented in B. napus (Cifuentes et al. 2010, Gaeta and Chris Pires 2010, Nicolas et al. 2008, 2009, Szadkowski et al. 2010), and homoeologous chromosome exchange in B. napus has been reported to affect seed yield (Osborn 2004) and Sclerotinia resistance (Zhao et al. 2006).
Chromosomally distant species hybrids generally show sterility due to chromosomal rearrangement on meiotic pairing. Generally, homologous chromosomes need to be associated for the allopolyploid hybrid plant to reduce chromosome number and recombine during meiosis. Pairing of chromosomes tend to favor homologs relative to homoeologs (Soltis and Soltis 2000), probably due to their similarity. The majority of chromosomes in wheat × barley hybrids was univalent, and an average of 0.7 bivalents per pollen mother cells was reported (Islam and Shepherd 1980). Similarly, bivalent formation was seen in allotriploid hybrids of Musa acuminata × Musa balbisiana, allotetraploid Festuca pratensis × Lolium perenne, and allohexaploid Lycopersicon esculentum × Lycopersicon peruvianum (Jeridi et al. 2011, Parokonny et al. 1997, Zwierzykowski et al. 2008). The occurrence of bivalent and multivalent pairings is naturally low, but is important for novel genetic exchanges between species.
In wheat, suppression of homoeologous chromosome pairing is known to be controlled by two key genes (Ph1 and Ph2) and a number of minor genes (Mello-sampayo 1971). Homoeologous pairing in allohexaploid wheat and Aegilops speltoides (wild relative of wheat) was shown to be controlled by the Ph1 locus, where deletion of Ph1 allowed homoeologous chromosomes to pair (Riley and Chapman 1958, Sears 1976). Increased (average of 5.03–6.63) bivalent formation per pollen mother cell, and a small number of trivalents and quadrivalents were seen in wheat × barley hybrids using the Ph mutant of wheat (Sethi et al. 1986). Homoeologous pairing between wheat and Haynaldia villosa chromosomes was also demonstrated by inhibiting Ph function (Bhullar et al. 2014, Chen et al. 1994), supporting Ph as a key player in homoeologous pairing. Another gene, PrBn, in B. napus has also been identified as an important gene controlling homoeologous pairing (Jenczewski et al. 2003).
Uniparental chromosomal elimination
Uniparental chromosomal elimination is another genomic change seen in hybrids, which biases the inheritance of chromosomes from one parent, and eliminates the chromosomes from the other parent. Complete chromosome elimination prevents an opportunity for genetic recombination between parental genomes, suggesting that this phenomenon can be regarded not only as allopolyploidization events, but also as a factor of post-zygotic barriers. Chromosome elimination can be temperature dependent (Pickering and Morgan 1985, Sanei et al. 2011), and genomic reorganization may be triggered when the merging parental genomes of the hybrid are genetically distant (Lim et al. 2004). However, complete chromosomal elimination has been reported in inter-specific hybrids between closely related species Hordeum vulgare or Hordeum parodii × H. bulbosum, and Hordeum marinum × H. vulgare; hybrids of Brassica species (Finch 1983, Kasha and Kao 1970, Li et al. 2004, Subrahmanyam 1977), between remotely related parental species (Aegilops spp, ryegrass (Lolium multiflorum), barley, oat, rye (S. cereal), and in crosses between wheat and Pennisetum glaucum, S. bicolor, Tripsacum dactyloides, H. bulbosum, Coix lacryma-jobi, Imperata cylindrical, pearl millet, or Zea mays (Barclay 1975, Chen et al. 1991, Gernand et al. 2005, Inagaki and Mujeeb-kazi 1995, Komeda et al. 2007, Laurie and Bennett 1988, 1989, Mochida and Tsujimoto 2001, Rines and Dahleen 1990, Zenkteler and Nitzsche 1984). Partial somatic elimination of chromosomes from one of the parent species was reported in Hordeum lechleri × H. vulgare (Linde-Laursen and von Bothmer 1999), Avena sativa × Z. mays (Riera-Lizarazu et al. 1996), or Triticum aestivum × H. vulgare (Barclay 1975). However, allopolyploids of Gossypium and Spartina did not show genomic rearrangements after hybridization (Ainouche et al. 2004, Liu et al. 2001). Chromosome elimination is seen not only in genetically distant hybrids, but also in closely related hybrids, suggesting that genetic distance is not the only factor determining elimination.
Observations in maize crosses and wheat × pearl millet crosses demonstrated that chromosome elimination occurs after pollination, during embryo development (Gernand et al. 2005, Zhao et al. 2013). Several hypotheses have been presented to explain uniparental chromosome elimination during hybrid embryo development in plants. Some of these are: differences in timing of essential mitotic processes due to asynchronous cell cycles (Gupta 1969), asynchrony in nucleoprotein synthesis leading to a loss of the most retarded chromosomes (Bennett et al. 1976, Laurie and Bennett 1989), degradation of alien chromosomes by host-specific nuclease activity (Davies 1974), spatial separation of genomes during interphase (Linde-Laursen and von Bothmer 1999) and metaphase (Schwarzacher-Robinson et al. 1987), formation of multipolar spindles (Subrahmanyam and Kasha 1973), and parent-specific inactivation of centromeres (Finch 1983, Jin et al. 2004, Kim et al. 2002, Mochida et al. 2004).
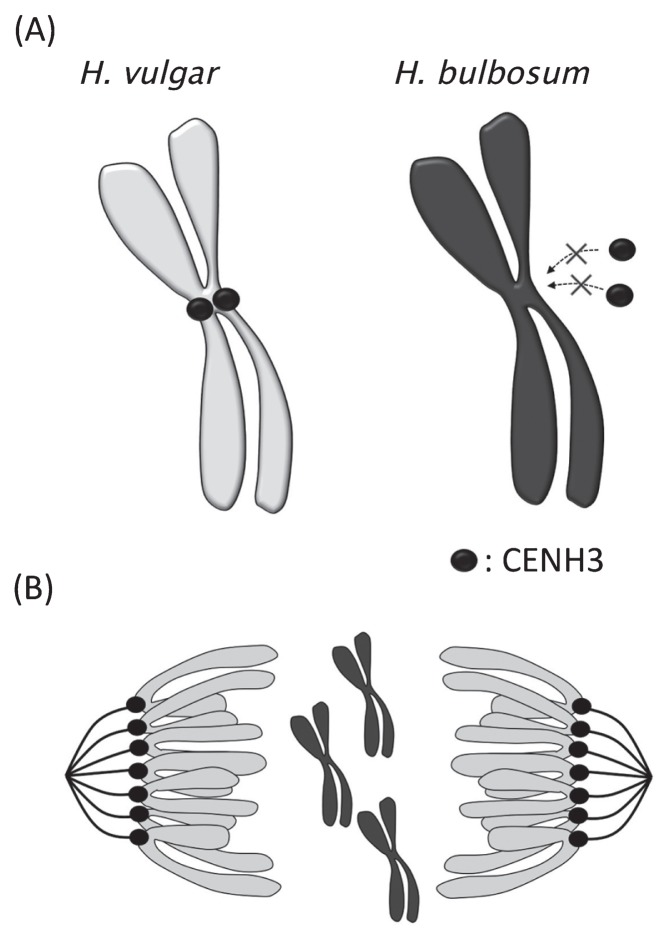
CENH3 was identified as a key player in uniparental chromosome elimination between H. vulgar × H. bulbosum inter-specific hybrids (Sanei et al. 2011). CENH3 encodes the Centromere-specific Histone H3 variant that functions in centromere formation and chromosome segregation, and is contained in all eukaryotic genomes. From this study, inactivity of the centromere caused the failure to assemble kinetochores that attach the microtubules for chromosome separation, leading to the loss of parental chromosome (Fig. 3). Modification of CENH3 has been demonstrated to efficiently produce homozygous diploid lines that may benefit breeding of hybrid lines by allowing rapid fixing of the homozygous locus linked to desired traits (Copenhaver and Preuss 2010, Ravi and Chan 2010).
Fig. 3.
Role of CENH3 in uniparental chromosome elimination in inter-specific hybrid embryos between H. vulgare × H. bulbosum. CENH3 (Filled circle) is loading into the centromere of H. vulgare but not of H. bulbosum, and incorporates only in the centromere of H. vulgare during the G2 phase (A). Sister chromatids of H. vulgare are separated and moved to opposite ends by attached microtubules in anaphase. However, chromosomes of H. bulbosum are abandoned, and are not inherited to daughter cells (B). As a result, chromosomes of H. bulbosum are eliminated in the inter-specific hybrid embryo.
Genomic rearrangement
In addition to major chromosomal changes, finer rearrangement of the genome can occur immediately after hybridization, and ultimately affect transcription. Mutation in simple sequence repeats (SSR), chromosomal rearrangements (translocation, duplications, inversions and deletions), and transposon activation were observed immediately after hybridization (ArArCnCn) between B. napus (AnAnCnCn) and B. rapa (ArAr) (Zou et al. 2011). Wheat allopolyploid showed DNA sequence elimination as an immediate response to allopolyploid formation (Feldman et al. 1997, Kashkush et al. 2002, Ozkan et al. 2001, Shaked et al. 2001). Loss, gain, and rearrangement of the nucleolus organizer region (NOR) and 5S rRNA gene were also seen immediately after hybridization in synthetic Arabidopsis allotetraploid (Pontes et al. 2004). Long-terminal-repeat-retrotransoposon (LTR-RTN) can be reactivated in allopolyploidization and inter-specific hybridization (Labrador et al. 1999, Liu and Wendel 2000), and it has been shown that transposable element (TE) activation can affect gene expression (Bennetzen 2005, Kashkush et al. 2003). An introgression of <0.1% of wild rice genomes into cultivated rice resulted in extensive genomic variation, and affected up to 30% of genomic loci including TE and protein-coding genes (Wang et al. 2005). In a study of B. rapa and B. napus hybrids, genomic changes that occurred post-hybridization were associated with a large number of QTLs (compared to the allelic contribution by B. rapa) for agronomic traits (Zou et al. 2011). This demonstrates that even minor rearrangements caused by hybridization can have a significant effect on gene expression and phenotype.
Most molecular and cytological studies of allopolyploids have been on synthesized polyploids, but chromosomal changes have been observed in a recently formed natural allopolyploid, Tragopogon (Chester et al. 2012). Chromosome rearrangement occurs in response to hybridization of different genomes, to adjust genome expression that may also lead to phenotypic novelty. Further understanding of the mechanisms may allow finer control and rapid introgression of desired traits after hybridization.
3-3. Non-additive expression in protein coding genes and micro RNAs
Allopolyploidization causes changes of gene expression, and these changes may ultimately affect the plant phenotype leading to heterosis (see 3-1 section) or stress tolerance, and ultimately to plant adaptation or domestication (Comai 2005). Following this concept, many studies have compared the transcriptome between artificially synthesized allopolyploids and their parental lines, or between natural allopolyploid species and their putative parental species (Adams and Wendel 2005, Chen 2007). These studies revealed that the majority of genes showed additive expression patterns (gene expression being equal to the average of the parental gene expression level) and some part of the genes showed non-additive gene expression (gene expression being different from the average of the parental gene expression level) in allopolyploids (Adams and Wendel 2005, Chen 2007).
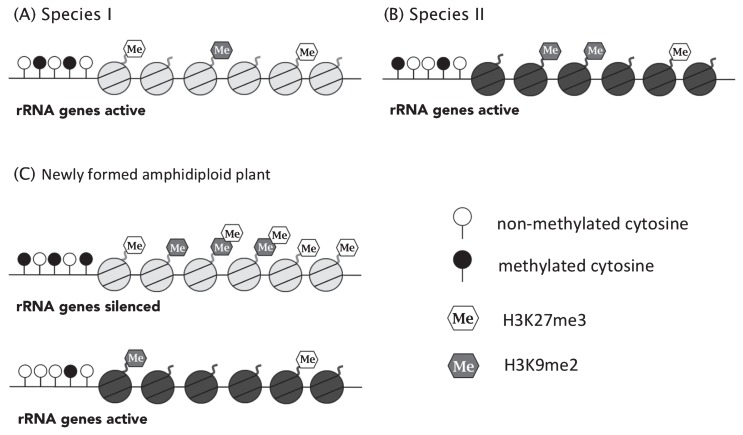
Genome dominance (genes in one parental genome tend to show higher expression levels than the other parent in allopolyploids) is reported in some allopolyploids (Cheng et al. 2012, Grover et al. 2012, Rapp et al. 2009, Wang et al. 2006). Nucleolar dominance (rRNA genes derived from one of the parental species inactivated in allopolyploids) is well known as an example of genome dominance, and silencing of one of the parental rRNA genes is epigenetically regulated (Earley et al. 2006, 2010, Lawrence et al. 2004, Preuss and Pikaard 2007). In inter-specific hybrids between A. arenosa and A. thaliana, between A. lyrata and A. thaliana, and between A. halleri and A. thaliana, A. thaliana-derived rRNA is silenced. Similarly, B. oleracea-derived rRNA is silenced in inter-specific hybrids between B. rapa and B. oleracea (Chang and Pikaard 2005, Fujimoto et al. 2011, Lewis et al. 2007). In A. arenosa × A. thaliana and B. rapa × B. oleracea hybrids, genes derived from A. thaliana and B. oleracea genomes tend to be silenced, respectively, suggesting that genome dominance in protein-coding genes is consistent with nucleolar dominance (Cheng et al. 2012, Wang et al. 2006). By contrast, genome dominance is not observed in protein coding genes in both A. thaliana × A. lyrata and A. thaliana × A. halleri hybrids (Fujimoto et al. 2011), suggesting that non-additive gene expression is independent from nucleolar dominance. In Aegilops longissima (SS) × Triticum uratu (AA) hybrids and T. uratu (AA) × Ae. tauschii (DD) hybrids, A-genome-derived rRNA is silenced, and A- and D-genome-derived rRNA are silenced in synthetic hexaploid wheat (BBAADD), indicating that the dominance order of rRNA is B > D > A genome. Silencing of rRNA is consistent with increased DNA methylation levels (especially CHH (where H = A, T, or C) and CHG methylation), and enrichment of H3K27me3 and H3K9me2 modifications, indicating that silencing of rRNA is epigenetically controlled (Fig. 4) (Guo and Han 2014).
Fig. 4.
Silencing of rRNA by epigenetic modification in newly formed amphidiploid plants. rRNA genes active in genomes of species I (A) and II (B). After formed amphidiploid plants, rRNA derived from species I is silenced by increased DNA methylation and enrichment of histone modification (H3K27me3 and H3K9me2) (C).
In allopolyploids, parental-biased gene expression change refers to biased expression of homoeologous genes (up- or down-regulated) depending on the parent-of-origin, which has been reported in wheat and cotton (Grover et al. 2012, Li et al. 2014, Li and Chetelat 2014, Shen et al. 2014, Yoo et al. 2013). In wheat, genes that have a differential expression between Ae. tauschii (DD) and Triticum turgidum (AABB), and an equal expression level between Ae. tauschii (DD) and T. aestivum (AABBDD), tend to include transcription factors such as key floral developmental genes. While genes that have differential expression between Ae. tauschii (DD) and T. turgidum (AABB), and equal expression levels between T. turgidum (AABB) and T. aestivum (AABBDD) tended to categorize into stress response. Thus, the authors suggested that merging of characteristic expression patterns for the two parental lines is important for adaptation in wheat (Li et al. 2014). Recent technical advances in next-generation sequencing have allowed not only large-scale analysis of transcriptome profiling (RNA-Sequencing), but also to distinguish the parental alleles of allopolyploids using nucleotide sequence differences between parental species (Akama et al. 2014).
Changes in transcription are not only observed in protein coding genes, but are also observed in small RNAs (Ha et al. 2009, Kenan-Eichler et al. 2011, Li et al. 2014). Micro RNAs (miRNAs) are a 21-nucleotide class of small RNAs, and they repress the target genes by mRNA cleavage or translational repression. Up to 25% of analyzed miRNAs displayed non-additive gene expression in synthetic hexaploid wheat, and negative correlations of expression levels between miRNA and their target genes among hybrids and their parental lines, suggesting that non-additive gene expression in some protein coding genes are due to non-additively expressed miRNAs (Li et al. 2014). Negative correlations between miRNA and target mRNA levels are also found in A. thaliana × A. arenosa hybrids, and the authors suggested that non-additive accumulation of miRNAs explains approximately 58% expression changes in their target mRNA in the A. thaliana × A. arenosa hybrids (Ha et al. 2009, Ng et al. 2012). Among A. thaliana, A. arenosa, and its synthetic F8 allotetraploids, miR163 expression in leaves and flowers is higher in A. thaliana than in A. arenosa and F8 allotetraploids. One of the target genes of miR163 encodes farnesoic acid methyltransferase, which converts farnesoic acid to methyl farcesoste, and changes in miR163 expression lead to changes in secondary metabolites, suggesting that miRNA regulation is also involved in adaptive evolution of allopolyploids (Ng et al. 2012).
Conclusions and perspectives
Reproductive barriers are challenges that must be overcome for developing efficient breeding programs by inter-specific crossing. Traditional techniques (medium culture methods) are effective methods, and have greatly contributed to producing inter-specific hybrids in various crops. However, inter-specific crossing requires different techniques depending on the cross-combinations. In many plant species, different accessions of parental species have different crossability in inter-specific crossing; therefore, cultivars that can be used by traditional techniques are limited.
Recent findings indicate that some reproductive barriers are controlled by the interaction between molecules from each species, suggesting that reproductive barriers may be overcome by modification of molecules related to the reproductive barrier. The pollen rejection behavior in inter-specific crossing is thought to be controlled by common factors related to the process during pollen recognition during pollen tube guidance. The loss of function mutation of either male or female S-determinants leads to the loss of the ability for self-incompatibility. It is possible that pre-zygotic barriers can also be overcome by the loss of function in male or female molecules related to inter-specific pollen recognition. One of the post-zygotic barriers, hybrid necrosis, is caused by an autoimmune-like response through epistatic interaction between two genes, indicating that the specific combination of causal genes is important. Hybrid necrosis can be efficiently avoided by crossing between species with a compatible combination of epistatic interaction. Endosperm abnormality may be caused by alteration of PRC2 regulation. In inter-specific hybrid endosperm, PRC2 regulation is affected by “quantitative difference” or “qualitative difference” in interspecies. In either case, it may be possible to overcome endosperm abnormality by modification of PRC2 dosage such as ploidy manipulation. The PRC2 genes, MEA and FIS2, are regulated epigenetically; therefore, if the expression levels of PRC2 genes are elevated by the modification of epigenetic gene regulation, PRC2 regulation in hybrid endosperm can be balanced with their target genes. As a result, endosperm abnormality of inter-specific crosses may be overcome without ploidy manipulation.
Allopolyploidization, caused by merging genomes, not only contributes to adaptation of allopolyploid species, but also for plant breeding programs. Heterozygosity, changes in chromosome and non-additive gene expression by allopolyploidization, will provide various advantages, and there is a potential to create novel agronomic traits. Indeed, some favorable traits, such as biomass heterosis (Bansal et al. 2012, Liu et al. 2002, Qian et al. 2003), seed yield (Osborn 2004), Sclerotinia resistance (Zhao et al. 2006) and loss of self-incompatibility (Okamoto et al. 2007), have been gained through allopolyploidization. Although the molecular mechanisms of allopolyploidization are not completely known, advances in new technologies enable new approaches, and will help elucidate the molecular mechanisms. Therefore, understanding the mechanisms of allopolyploidization may enable us to fine-tune the phenotype of inter-specific hybrids and to rapidly introduce desirable traits to cultivars from other species, and will provide further development for plant breeding of inter-specific hybrids.
Acknowledgement
We thank Dr. E.S. Dennis (CSIRO Agriculture, Australia), Dr. T. Kinoshita, Dr. T. Ohnishi and Dr. A. Ono (Kihara Institute for Biological Research, Yokohama City University) for helpful comments and discussions.
Literature Cited
- Adams, K.L., Cronn, R., Percifield, R. and Wendel, J.F. (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L. and Wendel, J.F. (2005) Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Adams, K.L. (2007) Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 98: 136–141. [DOI] [PubMed] [Google Scholar]
- Ainouche, M.L., Baumel, A., Salmon, A. and Yannic, G. (2004) Hybridization, polyploidy and speciation in Spartina (Poaceae). New Phytol. 161: 165–172. [Google Scholar]
- Akama, S., Shimizu-Inatsugi, R., Shimizu, K.K. and Sese, J. (2014) Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Res. 42: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfares, W., Bouguennec, A., Balfourier, F., Gay, G., Berges, H., Vautrin, S., Sourdille, P., Bernard, M. and Feuillet, C. (2009) Fine mapping and marker development for the crossability gene SKr on chromosome 5BS of hexaploid wheat (Triticum aestivum L.). Genetics 183: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj, Y.P.S., Mahajan, S.K. and Labana, K.S. (1986) Interspecific hybridization of Brassica napus and B. juncea through ovary, ovule and embryo culture. Euphytica 35: 103–109. [Google Scholar]
- Bansal, P., Banga, S. and Banga, S.S. (2012) Heterosis as investigated in terms of polyploidy and genetic diversity using designed Brassica juncea amphiploid and its progenitor diploid species. PLoS ONE 7: e29607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay, I.R. (1975) High frequencies of haploid production in wheat (Triticum aestivum) by chromosome elimination. Nature 256: 410–411. [Google Scholar]
- Bennett, M.D., Finch, R.A. and Barclay, I.R. (1976) The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma 54: 175–200. [Google Scholar]
- Bennetzen, J.L. (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15: 621–627. [DOI] [PubMed] [Google Scholar]
- Berger, F., Hamamura, Y., Ingouff, M. and Higashiyama, T. (2008) Double fertilization-caught in the act. Trends Plant Sci. 13: 437–443. [DOI] [PubMed] [Google Scholar]
- Bertin, I., Fish, L., Foote, T.N., Knight, E., Snape, J. and Moore, G. (2009) Development of consistently crossable wheat genotypes for alien wheat gene transfer through fine-mapping of the Kr1 locus. Theor. Appl. Genet. 119: 1371–1381. [DOI] [PubMed] [Google Scholar]
- Bhullar, R., Nagarajan, R., Bennypaul, H., Sidhu, G.K., Sidhu, G., Rustgi, S., von Wettstein, D. and Gill, K.S. (2014) Silencing of a metaphase I-specific gene results in a phenotype similar to that of the Pairing homeologous 1 (Ph1) gene mutations. Proc. Natl. Acad. Sci. USA 111: 14187–14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J.L. and Weigel, D. (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e50236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K. and Weigel, D. (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8: 382–393. [DOI] [PubMed] [Google Scholar]
- Burkart-Waco, D., Ngo, K., Dilkes, B., Josefsson, C. and Comai, L. (2013) Early disruption of maternal-zygotic interaction and activation of defense-like responses in Arabidopsis interspecific crosses. Plant Cell 25: 2037–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell, C., Spielman, M. and Scott, R.J. (2003) The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15: 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S. and Pikaard, C.S. (2005) Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J. Biol. Chem. 280: 796–804. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S. and Peacock, W.J. (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Chen, H., Lin, Y.S., Shen, J.B., Shan, J.X., Qi, P., Shi, M., Zhu, M.Z., Huang, X.H., Feng, Q.et al. (2014) A two-locus interaction causes interspecific hybrid weakness in rice. Nat. Commun. 5: 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F.Q., Hayes, P.M. and Rivin, C.J. (1991) Wide hybridization of Hordeum vulgare × Zea mays. Genome 34: 603–605. [Google Scholar]
- Chen, P.D., Tsujimoto, H. and Gill, B.S. (1994) Transfer of PhI genes promoting homoeologous pairing from Triticum speltoides to common wheat. Theor. Appl. Genet. 88: 97–101. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J. (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J. (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15: 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, F., Wu, J., Fang, L., Sun, S., Liu, B., Lin, K., Bonnema, G. and Wang, X. (2012) Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS ONE 7: e36442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester, M., Gallagher, J.P., Symonds, V.V., Cruz da Silva, A.V., Mavrodiev, E.V., Leitch, A.R., Soltis, P.S. and Soltis, D.E. (2012) Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat, R.T. and DeVerna, J.W. (1991) Expression of unilateral incompatibility in pollen of Lycopersicon pennellii is determined by major loci on chromosomes 1, 6 and 10. Theor. Appl. Genet. 82: 704–712. [DOI] [PubMed] [Google Scholar]
- Cifuentes, M., Grandont, L., Moore, G., Chevre, A.M. and Jenczewski, E. (2010) Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 186: 29–36. [DOI] [PubMed] [Google Scholar]
- Comai, L. (2005) The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P. and Preuss, D. (2010) Haploidy with histones. Nat. Biotechnol. 28: 423–424. [DOI] [PubMed] [Google Scholar]
- Earley, K., Lawrence, R.J., Pontes, O., Reuther, R., Enciso, A.J., Silva, M., Neves, N., Gross, M., Viegas, W. and Pikaard, C.S. (2006) Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20: 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W., Pontvianne, F., Wierzbicki, A.T., Blevins, T., Tucker, S., Costa-Nunes, P., Pontes, O. and Pikaard, C.S. (2010) Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 24: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, D.R. (1974) Chromosome elimination in inter-specific hybrids. Heredity 32: 267–270. [Google Scholar]
- Dhooghe, E., Van Laere, K., Eeckhaut, T., Leus, L. and Van Huylenbroeck, J. (2011) Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tissue Organ Cult. 104: 359–373. [Google Scholar]
- Dobzhansky, T. (1937) Genetic and the origin of species. New York: Columbia University Press, pp. 364. [Google Scholar]
- Dresselhaus, T., Lausser, A. and Marton, M.L. (2011) Using maize as a model to study pollen tube growth and guidance, cross-incompatibility and sperm delivery in grasses. Ann. Bot. 108: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entani, T., Iwano, M., Shiba, H., Che, F.S., Isogai, A. and Takayama, S. (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213. [DOI] [PubMed] [Google Scholar]
- Erilova, A., Brownfield, L., Exner, V., Rosa, M., Twell, D., Scheid, O.M., Hennig, L. and Köhler, C. (2006) Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 5: e1000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo, J.M., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, J., Yang, W.C. and Grossniklaus, U. (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660. [DOI] [PubMed] [Google Scholar]
- Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A.A. and Vega, J.M. (1997) Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch, R.A. (1983) Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma 88: 386–393. [Google Scholar]
- Fu, C.Y., Wang, F., Sun, B.R., Liu, W.G., Li, J.H., Deng, R.F., Liu, D.L., Liu, Z.R., Zhu, M.S., Liao, Y.L.et al. (2013) Genetic and cytological analysis of a novel type of low temperature-dependent intrasubspecific hybrid weakness in rice. PLoS ONE 8: e73886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S. and Toriyama, K. (2008) Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility. Plant Cell Physiol. 49: 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, R., Sugimura, T., Fukai, E. and Nishio, T. (2006) Suppression of gene expression of a recessive SP11/SCR allele by an untranscribed SP11/SCR allele in Brassica self-incompatibility. Plant Mol. Biol. 61: 577–587. [DOI] [PubMed] [Google Scholar]
- Fujimoto, R., Kinoshita, Y., Kawabe, A., Kinoshita, T., Takashima, K., Nordborg, M., Nasrallah, M.E., Shimizu, K.K., Kudoh, H. and Kakutani, T. (2008) Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 4: e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, R., Taylor, J.M., Sasaki, T., Kawanabe, T. and Dennis, E.S. (2011) Genome wide gene expression in artificially synthesized amphidiploids of Arabidopsis. Plant Mol. Biol. 77: 419–431. [DOI] [PubMed] [Google Scholar]
- Gaeta, R.T. and Chris Pires, J. (2010) Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 186: 18–28. [DOI] [PubMed] [Google Scholar]
- Gernand, D., Rutten, T., Varshney, A., Rubtsova, M., Prodanovic, S., Brüß, C., Kumlehn, J., Matzk, F. and Houben, A. (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17: 2431–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij, A., Kondo, K., Lee, C.B., Hancock, C.N., Sivaguru, M., Vazquez-Santana, S., Kim, S., Phillips, T.E., Cruz-Garcia, F. and McClure, B. (2006) Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439: 805–810. [DOI] [PubMed] [Google Scholar]
- Gradziel, T.M. and Robinson, R.W. (1991) Overcoming unilateral breeding barriers between Lycopersicon peruvianum and cultivated tomato, Lycopersicon esculentum. Euphytica 54: 1–9. [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A. and Gagliano, W.B. (1998) Maternal control of embryogenesis by MEDEA, a Polycomb Group Gene in Arabidopsis. Science 280: 446–450. [DOI] [PubMed] [Google Scholar]
- Grover, C.E., Gallagher, J.P., Szadkowski, E.P., Yoo, M.J., Flagel, L.E. and Wendel, J.F. (2012) Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196: 966–971. [DOI] [PubMed] [Google Scholar]
- Guo, X. and Han, F. (2014) Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. Plant Cell 26: 4311–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S.B. (1969) Duration of mitotic cycle and regulation of DNA replication in Nicotiana plumbaginifolia and a hybrid derivative of N. tabacum showing chromosome instability. Can. J. Genet. Cytol. 11: 133–142. [Google Scholar]
- Ha, M., Lu, J., Tian, L., Ramachandran, V., Kasschau, K.D., Chapman, E.J., Carrington, J.C., Chen, X., Wang, X.J. and Chen, Z.J. (2009) Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106: 17835–17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig, D. and Westoby, M. (1991) Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. B Biol. Sci. 333: 1–13. [Google Scholar]
- Hamamura, Y., Saito, C., Awai, C., Kurihara, D., Miyawaki, A., Nakagawa, T., Kanaoka, M.M., Sasaki, N., Nakano, A., Berger, F.et al. (2011) Live-cell imaging reveals the dynamics of two sperm cells during double fertilization in Arabidopsis thaliana. Curr. Biol. 21: 497–502. [DOI] [PubMed] [Google Scholar]
- Hancock, C.N., Kent, L. and McClure, B.A. (2005) The stylar 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. Plant J. 43: 716–723. [DOI] [PubMed] [Google Scholar]
- Hehenberger, E., Kradolfer, D. and Köhler, C. (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Inatsugi, R., Sakamoto, S., Sasaki, N., Mori, T., Kuroiwa, H., Nakada, T., Nozaki, H., Kuroiwa, T. and Nakano, A. (2006) Species preferentiality of the pollen tube attractant derived from the synergid cell of Torenia fournieri. Plant Physiol. 142: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscock, S.J. and Dickinson, H.G. (1993) Unilateral incompatibility within the Brassicaceae: further evidence for the involvement of the self-incompatibility (S)-locus. Theor. Appl. Genet. 86: 744–753. [DOI] [PubMed] [Google Scholar]
- Hsieh, T., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M.J., Hashimoto, M., Kirkbride, R.C., Harada, J.J., Zilberman, D.et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck, N., Moore, J.M., Federer, M. and Grossniklaus, U. (2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159. [DOI] [PubMed] [Google Scholar]
- Inagaki, M. and Mujeeb-Kazi, A. (1995) Comparison of polyploid production frequencies in crosses of hexaploid wheat with maize, pearl millet and sorghum. Breed. Sci. 45: 157–161. [Google Scholar]
- Innan, H. and Kondrashov, F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108. [DOI] [PubMed] [Google Scholar]
- Ishikawa, R. and Kinoshita, T. (2009) Epigenetic programming: the challenge to species hybridization. Mol. Plant 2: 589–599. [DOI] [PubMed] [Google Scholar]
- Ishikawa, R., Ohnishi, T., Kinoshita, Y., Eiguchi, M., Kurata, N. and Kinoshita, T. (2011) Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J. 65: 798–806. [DOI] [PubMed] [Google Scholar]
- Islam, A. and Shepherd, K.W. (1980) Meiotic restitution in wheat-barley hybrids. Chromosoma 79: 363–372. [Google Scholar]
- Iwai, S., Kishi, C., Nakata, K. and Kawashima, N. (1986) Production of Nicotiana tabacum × Nicotiana acuminata hybrid by ovule culture. Plant Cell Rep. 5: 403–404. [DOI] [PubMed] [Google Scholar]
- Jalani, B.S. and Moss, J.P. (1980) The site of action of the crossability genes (Kr1, Kr2) between Triticum and Secale. I. Pollen germination, pollen tube growth, and number of pollen tubes. Euphytica 29: 571–579. [Google Scholar]
- Jenczewski, E., Eber, F., Grimaud, A., Huet, S., Lucas, M.O., Monod, H. and Chevre, A.M. (2003) PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeridi, M., Bakry, F., Escoute, J., Fondi, E., Carreel, F., Ferchichi, A., D’Hont, A. and Rodier-Goud, M. (2011) Homoeologous chromosome pairing between the A and B genomes of Musa spp. revealed by genomic in situ hybridization. Ann. Bot. 108: 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken, M.J., Zhang, N.W., McHale, L.K., Pelgrom, K., den Boer, E., Lindhout, P., Michelmore, R.W., Visser, R.G. and Niks, R.E. (2009) Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21: 3368–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W.W., Melo, J.R., Nagaki, K., Talbert, P.B., Henikoff, S., Dawe, R.K. and Jiang, J.M. (2004) Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell 16: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, S.A., den Nijs, T.P., Peloquin, S.J. and Hanneman, R.E.Jr. (1980) The significance of genic balance to endosperm development in interspecific crosses. Theor. Appl. Genet. 57: 5–9. [DOI] [PubMed] [Google Scholar]
- Johnston, S.A. and Hanneman, R.E.Jr. (1982) Manipulations of endosperm balance number overcome crossing barriers between diploid Solanum species. Science 217: 446–448. [DOI] [PubMed] [Google Scholar]
- Josefsson, C., Dilkes, B. and Comai, L. (2006) Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol. 16: 1322–1328. [DOI] [PubMed] [Google Scholar]
- Jullien, P.E., Kinoshita, T., Ohad, N. and Berger, F. (2006) Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, I.H., Steffen, J.G., Portereiko, M.F., Lloyd, A. and Drews, G.N. (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasha, K.J. and Kao, K.N. (1970) High frequency haploid production in barley (Hordeum vulgare L.). Nature 225: 874–876. [DOI] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M. and Levy, A.A. (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M. and Levy, A.A. (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33: 102–106. [DOI] [PubMed] [Google Scholar]
- Kenan-Eichler, M., Leshkowitz, D., Tal, L., Noor, E., Melamed-Bessudo, C., Feldman, M. and Levy, A.A. (2011) Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188: 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N.S., Armstrong, K.C., Fedak, G., Ho, K. and Park, N.I. (2002) A microsatellite sequence from the rice blast fungus (Magnaporthe grisea) distinguishes between the centromeres of Hordeum vulgare and H. bulbosum in hybrid plants. Genome 45: 165–174. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B. and Fischer, R.L. (1999) Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T. (2007) Reproductive barrier and genomic imprinting in the endosperm of flowering plants. Genes Genet. Syst. 82: 177–186. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Mardossian, L., Harada, J.J., Goldberg, R.B.et al. (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koba, T. and Shimada, T. (1992) Variations in the crossability of common wheat cultivars with cultivated barley. Hereditas 116: 187–192. [Google Scholar]
- Koba, T. and Shimada, T. (1993) Crossability of common wheat with Aegilops squarrosa. Wheat Inf. Serv. 77: 7–12. [Google Scholar]
- Köhler, C., Page, D.R., Gagliardini, V. and Grossniklaus, U. (2005) The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37: 28–30. [DOI] [PubMed] [Google Scholar]
- Komeda, N., Chaudhar, H.K., Suzuki, G. and Mukai, Y. (2007) Cytological evidence for chromosome elimination in wheat × Imperata cylindrica hybrids. Genes Genet. Syst. 82: 241–248. [DOI] [PubMed] [Google Scholar]
- Kondo, K., Yamamoto, M., Matton, D.P., Sato, T., Hirai, M., Norioka, S., Hattori, T. and Kowyama, Y. (2002) Cultivated tomato has defects in both S-RNase and HT genes required for stylar function of self-incompatibility. Plant J. 29: 627–636. [DOI] [PubMed] [Google Scholar]
- Krolow, K.D. (1970) Investigations on compatibility between wheat and rye. Z. PXanzenzüchtg 64: 44–72. [Google Scholar]
- Kuboyama, T., Chung, C.S. and Takeda, G. (1994) The diversity of interspecific pollen-pistil incongruity in Nicotiana. Sex. Plant Reprod. 7: 250–258. [Google Scholar]
- Labrador, M., Farré, M., Utzet, F. and Fontdevilla, A. (1999) Interspecific hybridization increases transposition rates of Osvaldo. Mol. Biol. Evol. 16: 931–937. [DOI] [PubMed] [Google Scholar]
- Lai, Z., Ma, W., Han, B., Liang, L., Zhang, Y., Hong, G. and Xue, Y. (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50: 29–42. [DOI] [PubMed] [Google Scholar]
- Lange, W. and Wojciechowska, B. (1976) The crossing of common wheat (Triticum aestivum L.) with cultivated rye (Secale cereale L.) I. Crossability, pollen grain germination and pollen tube growth. Euphytica 25: 609–620. [Google Scholar]
- Laurie, D.A. and Bennett, M.D. (1988) Cytological evidence for fertilization in hexaploid wheat × sorghum crosses. Plant Breed. 100: 73–82. [Google Scholar]
- Laurie, D.A. and Bennett, M.D. (1989) The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome 32: 953–961. [Google Scholar]
- Lavania, U.C., Srivastava, S., Lavania, S., Basu, S., Misra, N.K. and Mukai, Y. (2012) Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant J. 71: 539–549. [DOI] [PubMed] [Google Scholar]
- Lawrence, R.J., Earley, K., Pontes, O., Silva, M., Chen, Z.J., Neves, N., Viegas, W. and Pikaard, C.S. (2004) A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Lein, A. (1943) The genetical basis of the crossability between wheat and rye. Z. Indukt. Abstamm. Vererbungsl. 81: 28–59. [Google Scholar]
- Lewis, D. and Crown, L.K. (1958) Unilateral interspecific incompatibility in flowering plants. Heredity 12: 233–256. [Google Scholar]
- Lewis, M.S., Pikaard, D.J., Nasrallah, M., Doelling, J.H. and Pikaard, C.S. (2007) Locus-specific ribosomal RNA gene silencing in nucleolar dominance. PLoS ONE 2: e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A., Liu, D., Wu, J., Zhao, X., Hao, M., Geng, S., Yan, J., Jiang, X., Zhang, L., Wu, J.et al. (2014) mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: 1878–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.T., Qian, W., Meng, J.L. and Li, Z.Y. (2004) Construction of novel Brassica napus genotypes through chromosomal substitution and elimination using interploid species hybridization. Chromosome Res. 12: 417–426. [DOI] [PubMed] [Google Scholar]
- Li, W. and Chetelat, R.T. (2010) A pollen factor linking inter- and intraspecific pollen rejection in tomato. Science 330: 1827–1830. [DOI] [PubMed] [Google Scholar]
- Li, W., Royer, S. and Chetelat, R.T. (2010) Fine mapping of ui6.1, a gametophytic factor controlling pollen-side unilateral incompatibility in interspecific Solanum hybrids. Genetics 185: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. and Chetelat, R.T. (2014) The role of a pollen-expressed Cullin1 protein in gametophytic self-incompatibility in Solanum. Genetics 196: 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. and Chetelat, R.T. (2015) Unilateral incompatibility gene ui1.1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc. Natl. Acad. Sci. USA 112: 4417–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, K.Y., Matyasek, R., Kovarik, A. and Leitch, A.R. (2004) Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 82: 599–606. [Google Scholar]
- Linde-Laursen, I. and von Bothmer, R. (1999) Orderly arrangement of the chromosomes within barley genomes of chromosome-eliminating Hordeum lechleri × barley hybrids. Genome 42: 225–236. [Google Scholar]
- Liu, B. and Wendel, J.F. (2000) Retrotransposon activation followed by rapid repression in introgressed rice plant. Genome 43: 874–880. [PubMed] [Google Scholar]
- Liu, B., Brubaker, C.L., Mergeai, G., Cronn, R.C. and Wendel, J.F. (2001) Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44: 321–330. [PubMed] [Google Scholar]
- Liu, R., Qian, W. and Meng, J. (2002) Association of RFLP markers and biomass heterosis in trigenomic hybrids of oilseed rape (Brassica napus × B. campestris). Theor. Appl. Genet. 105: 1050–1057. [DOI] [PubMed] [Google Scholar]
- Lowry, D.B., Modliszewski, J.L., Wright, K.M., Wu, C.A. and Willis, J.H. (2008) Review. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 363: 3009–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Taylor, J.M., Spriggs, A., Zhang, H., Wu, X., Russell, S., Singh, M. and Koltunow, A. (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, S. (1981) Overcoming the self-incompatibility of Lilium longiflorum Thunb. by application of flower-organ extract or temperature treatment of pollen. Euphytica 30: 97–103. [Google Scholar]
- Mello-Sampayo, T. (1971) Genetic regulation of meiotic chromosome pairing by chromosome 3D of Triticum aestivum. Nature New Biol. 230: 22–23. [DOI] [PubMed] [Google Scholar]
- Mizuno, N., Hosogi, N., Park, P. and Takumi, S. (2010) Hypersensitive response-like reaction is associated with hybrid necrosis in interspecific crosses between tetraploid wheat and Aegilops tauschii coss. PLoS ONE 5: e11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, N., Shitsukawa, N., Hosogi, N., Park, P. and Takumi, S. (2011) Autoimmune response and repression of mitotic cell division occur in inter-specific crosses between tetraploid wheat and Aegilops tauschii Coss. that show low temperature-induced hybrid necrosis. Plant J. 68: 114–128. [DOI] [PubMed] [Google Scholar]
- Mochida, K. and Tsujimoto, H. (2001) Production of wheat doubled haploids by pollination with Job’s tears (Coix lachryma-jobi L.). J. Hered. 92: 81–83. [DOI] [PubMed] [Google Scholar]
- Mochida, K., Tsujimoto, H. and Sasakuma, T. (2004) Confocal analysis of chromosome behavior in wheat × maize zygotes. Genome 47: 199–205. [DOI] [PubMed] [Google Scholar]
- Muller, H.J. (1942) Isolating mechanisms, evolution and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Murase, K., Shiba, H., Iwano, M., Che, F.S., Watanabe, M., Isogai, A. and Takayama, S. (2004) A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303: 1516–1519. [DOI] [PubMed] [Google Scholar]
- Murfett, J., Strabala, T.J., Zurek, D.M., Mou, B., Beecher, B. and McClure, B.A. (1996) S RNase and interspecific pollen rejection in the genus Nícotíana: Multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. Plant Cell 8: 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D.W., Lu, J. and Chen, Z.J. (2012) Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr. Opin. Plant Biol. 15: 154–161. [DOI] [PubMed] [Google Scholar]
- Ni, Z., Kim, E.D., Ha, M., Lackey, E., Liu, J., Zhang, Y., Sun, Q. and Chen, Z.J. (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, S.D., Leflon, M., Liu, Z., Eber, F., Chelysheva, L., Coriton, O., Chevre, A.M. and Jenczewski, E. (2008) Chromosome ‘speed dating’ during meiosis of polyploid Brassica hybrids and haploids. Cytogenet. Genome Res. 120: 331–338. [DOI] [PubMed] [Google Scholar]
- Nicolas, S.D., Leflon, M., Monod, H., Eber, F., Coriton, O., Huteau, V., Chevre, A.-M. and Jenczewski, E. (2009) Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. Plant Cell 21: 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, I. and Yabuno, T. (1978) Causal relationships between the polar nuclei in double fertilization and interspecific cross-incompatibility in Avena. Cytologia 43: 453–466. [Google Scholar]
- Nukui, S., Kitamura, S., Hioki, T., Ootsuka, H., Miyoshi, K., Satou, T., Takatori, Y., Oomiya, T. and Okazaki, K. (2011) N2O induces mitotic polyploidization in anther somatic cells and restores fertility in sterile interspecific hybrid lilies. Breed. Sci. 61: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien, M., Kapfer, C., Major, G., Laurin, M., Bertrand, C., Kondo, K., Kowyama, Y. and Matton, D.P. (2002) Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J. 32: 985–996. [DOI] [PubMed] [Google Scholar]
- Okamoto, S., Odashima, M., Fujimoto, R., Sato, Y., Kitashiba, H. and Nishio, T. (2007) Self-compatibility in Brassica napus is caused by independent mutations in S-locus genes. Plant J. 50: 391–400. [DOI] [PubMed] [Google Scholar]
- Okuda, S., Tsutsui, H., Shiina, K., Sprunck, S., Takeuchi, H., Yui, R., Kasahara, R.D., Hamamura, Y., Mizukami, A., Susaki, D.et al. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Okuda, S. and Higashiyama, T. (2010) Pollen tube guidance by attractant molecules: LUREs. Cell Struct. Funct. 35: 45–52. [DOI] [PubMed] [Google Scholar]
- Osborn, T.C., Pires, J.C., Birchler, J.A., Auger, D.L., Chen, Z.J., Lee, H.S., Comai, L., Madlung, A., Doerge, R.W., Colot, V.et al. (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Osborn, T.C. (2004) The contribution of polyploidy to variation in Brassica species. Physiol. Plant. 121: 531–536. [Google Scholar]
- Otto, S.P. (2007) The evolutionary consequences of polyploidy. Cell 131: 452–462. [DOI] [PubMed] [Google Scholar]
- Ozkan, H., Levy, A.A. and Feldman, M. (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parokonny, A.S., Marshall, J.A., Bennett, M.D., Cocking, E.C., Davey, M.R. and Power, J.B. (1997) Homoeologous pairing and recombination in backcross derivatives of tomato somatic hybrids [Lycopersicon esculentum (+) L. peruvianum]. Theor. Appl. Genet. 94: 713–723. [Google Scholar]
- Pickering, R.A. and Morgan, P.W. (1985) The influence of temperature on chromosome elimination during embryo development in crosses involving Hordeum spp., wheat (Triticum aestivum L.) and rye (Secale cereale L.). Theor. Appl. Genet. 70: 199–206. [DOI] [PubMed] [Google Scholar]
- Pontes, O., Neves, N., Silva, M., Lewis, M.S., Madlung, A., Comai, L., Viegas, W. and Pikaard, C.S. (2004) Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 101: 18240–18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, S. and Pikaard, C.S. (2007) rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim. Biophys. Acta 1769: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W., Liu, R. and Meng, J. (2003) Genetic effects on biomass yield in interspecific hybrids between Brassica napus and B. rapa. Euphytica 134: 9–15. [Google Scholar]
- Qian, W., Chen, X., Fu, D., Zou, J. and Meng, J. (2005) Intersubgenomic heterosis in seed yield potential observed in a new type of Brassica napus introgressed with partial Brassica rapa genome. Theor. Appl. Genet. 110: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Rapp, R.A., Udall, J.A. and Wendel, J.F. (2009) Genomic expression dominance in allopolyploids. BMC Biol. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi, M. and Chan, S.W.L. (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu, O., Rines, H.W. and Phillips, R.L. (1996) Cytological and molecular characterization of oat × maize partial hybrids. Theor. Appl. Genet. 93: 123–135. [DOI] [PubMed] [Google Scholar]
- Riley, R. and Chapman, V. (1958) Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182: 713–715. [Google Scholar]
- Riley, R. and Chapman, V. (1967) The inheritance in wheat of crossability with rye. Genet. Res. 9: 259–267. [Google Scholar]
- Rines, H.W. and Dahleen, L.S. (1990) Haploid oat plants produced by application of maize pollen to emasculated oat florets. Crop Sci. 30: 1073–1078. [Google Scholar]
- Rotman, N., Rozier, F., Boavida, L., Dumas, C., Berger, F. and Faure, J.E. (2003) Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13: 432–436. [DOI] [PubMed] [Google Scholar]
- Sanei, M., Pickering, R., Kumke, K., Nasuda, S. and Houben, A. (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. USA 108: E498–E505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatlowski, N. and Köhler, C. (2012) Tearning down barriers: understanding the molecular mechanisms of interploidy hybridizations. J. Exp. Bot. 63: 6059–6067. [DOI] [PubMed] [Google Scholar]
- Schwarzacher-Robinson, T., Finch, R.A., Smith, J.B. and Bennett, M.D. (1987) Genotypic control of centromere positions of parental genomes in Hordeum × Secale hybrid metaphases. J. Cell Sci. 87: 291–304. [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J. and Dickinson, H.G. (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- Sears, E.R. (1976) Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10: 31–51. [DOI] [PubMed] [Google Scholar]
- Sekine, D., Ohnishi, T., Furuumi, H., Ono, A., Yamada, T., Kurata, N. and Kinoshita, T. (2013) Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J. 76: 792–799. [DOI] [PubMed] [Google Scholar]
- Sethi, G.S., Finch, R.A. and Miller, T.E. (1986) A bread wheat (Triticum aestivum) × cultivated barley (Hordeum vulgare) hybrid with homoeologous chromosome pairing Can. J. Genet. Cytol. 28: 777–782. [Google Scholar]
- Shaked, H., Kashkush, K., Ozkan, H., Feldman, M. and Levy, A.A. (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, D.R., Kaur, R. and Kumar, K. (1996) Embryo rescue in Plants—a review. Euphytica 89: 325–337. [Google Scholar]
- Shen, Y., Zhao, Q., Zou, J., Wang, W., Gao, Y., Meng, J. and Wang, J. (2014) Characterization and expression patterns of small RNAs in synthesized Brassica hexaploids. Plant Mol. Biol. 85: 287–299. [DOI] [PubMed] [Google Scholar]
- Sijacic, P., Wang, X., Skirpan, A.L., Wang, Y., Dowd, P.E., McCubbin, A.G., Huang, S. and Kao, T.H. (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Snape, J.W., Chapman, V., Moss, J., Blanchard, C.E. and Miller, T.E. (1979) The crossabilities of wheat varieties with Hordeum Bulbosum. Heredity 42: 291–298. [Google Scholar]
- Soltis, P.S. and Soltis, D.E. (2000) The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA 97: 7051–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyam, N.C. and Kasha, K.J. (1973) Selective chromosomal elimination during haploid formation in barley following interspecific hybridization. Chromosoma 42: 111–125. [Google Scholar]
- Subrahmanyam, N.C. (1977) Haploidy from Hordeum interspecific crosses: I. polyhaploids of H. parodii and H. procerum. Theor. Appl. Genet. 49: 209–217. [DOI] [PubMed] [Google Scholar]
- Szadkowski, E., Eber, F., Huteau, V., Lode, M., Huneau, C., Belcram, H., Coriton, O., Manzanares-Dauleux, M.J., Delourme, R., King, G.J.et al. (2010) The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186: 102–112. [DOI] [PubMed] [Google Scholar]
- Takahata, Y. (1990) Production of intergeneric hybrids between a C3–C4 intermediate species Moricandia arvensis and a C3 species Brassica oleracea through ovary culture. Euphytica 46: 259–264. [Google Scholar]
- Takahata, Y. and Takeda, T. (1990) Intergeneric (intersubtribe) hybridization between Moricandia arvensis and Brassica A and B genome species by ovary culture. Theor. Appl. Genet. 80: 38–42. [DOI] [PubMed] [Google Scholar]
- Takeuchi, H. and Higashiyama, T. (2012) A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 10: e1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixier, M.H., Sourdille, P., Charmet, G., Gay, G., Jaby, C., Cadalen, T., Bernard, S., Nicolas, P. and Bernard, M. (1998) Detection of QTLs for crossability in wheat using a doubled-haploid population. Theor. Appl. Genet. 97: 1076–1082. [Google Scholar]
- Tonosaki, K., Michiba, K., Bang, S.W., Kitashiba, H., Kaneko, Y. and Nishio, T. (2013) Genetic analysis of hybrid seed formation ability of Brassica rapa in intergeneric crossings with Raphanus sativus. Theor. Appl. Genet. 126: 837–846. [DOI] [PubMed] [Google Scholar]
- Tovar-Mendez, A., Kumar, A., Kondo, K., Ashford, A., Baek, Y.S., Welch, L., Bedinger, P.A. and McClure, B.A. (2014) Restoring pistil-side self-incompatibility factors recapitulates an interspecific reproductive barrier between tomato species. Plant J. 77: 727–736. [DOI] [PubMed] [Google Scholar]
- Udagawa, H., Ishimaru, Y., Li, F., Sato, Y., Kitashiba, H. and Nishio, T. (2010) Genetic analysis of interspecific incompatibility in Brassica rapa. Theor. Appl. Genet. 121: 689–696. [DOI] [PubMed] [Google Scholar]
- Van Tuyl, J.M., Straathof, T.P., Bino, R.J. and Kwakkenbos, A.A.M. (1988) Effect of three pollination methods on embryo development and seedset in intra- and interspecific crosses between seven Lilium species. Sex. Plant Reprod. 1: 119–123. [Google Scholar]
- Van Tuyl, J.M., Van Dien, M.P., Van Creij, M.G.M., Kleinwee, T.C.M., Franken, J. and Bino, R.J. (1991) Application of in vitro pollination, ovary culture, ovule culture and embryo rescue for overcoming incongruity barriers in interspecific Lilium crosses. Plant Sci. 74: 115–126. [Google Scholar]
- Walia, H., Josefsson, C., Dilkes, B., Kirkbride, R., Harada, J. and Comai, L. (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol. 19: 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Tian, L., Lee, H.S., Wei, N.E., Jiang, H., Watson, B., Madlung, A., Osborn, T.C., Doerge, R.W., Comai, L.et al. (2006) Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.M., Dong, Z.Y., Zhang, Z.J., Lin, X.Y., Shen, Y., Zhou, D.W. and Liu, B. (2005) Extensive de novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.). Genetics 170: 1945–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M., Takayama, S., Isogai, A. and Hinata, K. (2003) Recent progresses on self-incompatibility research in Brassica species. Breed. Sci. 53: 199–208. [Google Scholar]
- Waters, A.J., Makarevitch, I., Eichten, S.R., Swanson-Wagner, R.A., Yeh, C.T., Xu, W., Schnable, P.S., Vaughn, M.W., Gehring, M. and Springer, N.M. (2011) Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, P., Weinhofer, I., Seguin, J., Roszak, P., Beisel, C., Donoghue, M.T., Spillane, C., Nordborg, M., Rehmsmeier, M. and Köhler, C. (2011) High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, H. and Bhat, S.R. (2014) Cytoplasmic male sterility in Brassicaceae crops. Breed. Sci. 64: 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, M.J., Szadkowski, E. and Wendel, J.F. (2013) Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenkteler, M. and Nitzsche, W. (1984) Wide hybridization experiments in cereals. Theor. Appl. Genet. 68: 311–315. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Udall, J.A., Quijada, P.A., Grau, C.R., Meng, J. and Osborn, T.C. (2006) Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor. Appl. Genet. 112: 509–516. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Xu, X.W., Xie, H.X., Chen, S.J. and Jin, W.W. (2013) Fertilization and uniparental chromosome elimination during crosses with maize haploid inducers. Plant Physiol. 163: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J., Fu, D.H., Gong, H.H., Qian, W., Xia, W., Pires, J.C., Li, R.Y., Long, Y., Mason, A.S., Yang, T.J.et al. (2011) De novo genetic variation associated with retrotransposon activation, genomic rearrangements and trait variation in a recombinant inbred line population of Brassica napus derived from interspecific hybridization with Brassica rapa. Plant J. 68: 212–224. [DOI] [PubMed] [Google Scholar]
- Zwierzykowski, Z., Zwierzykowska, E., Taciak, M., Jones, N., Kosmala, A. and Krajewski, P. (2008) Chromosome pairing in allotetraploid hybrids of Festuca pratensis × Lolium perenne revealed by genomic in situ hybridization (GISH). Chromosome Res. 16: 575–585. [DOI] [PubMed] [Google Scholar]