Abstract
Broad-spectrum disease resistance against two or more types of pathogen species is desirable for crop improvement. In rice, Xanthomonas oryzae pv. oryzae (Xoo), the causal bacteria of rice leaf blight, and Magnaporthe oryzae, the fungal pathogen causing rice blast, are two of the most devastating pathogens. We identified the rice BROAD-SPECTRUM RESISTANCE 1 (BSR1) gene for a BIK1-like receptor-like cytoplasmic kinase using the FOX hunting system, and demonstrated that BSR1-overexpressing (OX) rice showed strong resistance to the bacterial pathogen, Xoo and the fungal pathogen, M. oryzae. Here, we report that BSR1-OX rice showed extended resistance against two other different races of Xoo, and to at least one other race of M. oryzae. In addition, the rice showed resistance to another bacterial species, Burkholderia glumae, which causes bacterial seedling rot and bacterial grain rot, and to Cochliobolus miyabeanus, another fungal species causing brown spot. Furthermore, BSR1-OX rice showed slight resistance to rice stripe disease, a major viral disease caused by rice stripe virus. Thus, we demonstrated that BSR1-OX rice shows remarkable broad-spectrum resistance to at least two major bacterial species and two major fungal species, and slight resistance to one viral pathogen.
Keywords: rice, receptor-like cytoplasmic kinase, broad-spectrum disease resistance, Magnaporthe oryzae, Xanthomonas oryzae, Cochliobolus miyabeanus, Burkholderia glumae
Introduction
In the natural environment, plants encounter many species of pathogenic microorganisms, such as fungi, bacteria and viruses. The damage caused by microbial diseases is one of the most important limiting factors for crop production. To solve this problem, improvement of host resistance against these pathogens is the most economical and environmentally friendly approach. Rice (Oryza sativa L.) is one of the most important food crops and is a staple food for approximately 50% of the world’s population (Liu et al. 2014). Moreover, it is a model plant of monocotyledonous species. Bacterial leaf blight caused by bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo), blast by fungus Magnaporthe oryzae, brown (leaf) spot by fungus Cochliobolus miyabeanus, and stripe mosaic caused by viral pathogen rice stripe virus are the major rice diseases worldwide and result in serious losses of rice production.
To breed blast-resistant rice, efforts have aimed to introduce the resistance (R) genes into susceptible cultivars. The R gene is a key component of disease resistance to a particular pathogen and is often associated with a hypersensitive response (HR) (Flor 1971). In most cases, the resistant cultivars with R genes remain effective for only a few years in agricultural production (Dean et al. 2005) because new biotypes of the pathogen that can overcome the R gene often appear after release of the resistance cultivar. By contrast, although large numbers of quantitative trait loci (QTLs) (or quantitative genes) for bacterial leaf blight or blast resistance have been identified, these sources have not been used effectively in rice improvement because the genetic control of quantitative resistance is complex. Therefore, breeding for cultivars that exhibit broad-spectrum and durable disease resistance is a top priority in rice improvement programs around the world.
In addition to bacterial leaf blight and blast, there are several other important diseases in rice, such as brown spot and bacterial seedling rot. Brown spot disease is caused by the fungus C. miyabeanus, a representative necrotrophic pathogen, and is one of the most prevalent diseases in all rice-growing areas. C. miyabeanus infects plant tissues such as leaves and spikelets in all development stages. Fungicides, such as iprodione and propiconazole, are effective means to manage this disease (Moletti et al. 1997). However, the use of resistant varieties would be preferable because fungicides are expensive and not environmentally friendly. Meanwhile, rice cultivar ‘Tadukan’ offers quantitative resistance to brown spot disease. However, no major genes conferring immunity to this disease have been identified, although three QTLs for disease resistance have been identified (Sato et al. 2008). So far, genetic studies of resistance to brown spot disease have been superficial.
Bacterial seedling rot and bacterial grain rot (bacterial panicle blight) in rice are caused by bacterial pathogen Burkholderia glumae, which is also a necrotrophic pathogen (Iwai et al. 2002, Mizobuchi et al. 2013b). Recently, these diseases have become an increasingly serious problem in global rice production because of global warming and climate change; B. glumae prefers high temperature and humidity (Ham et al. 2011). Some studies reported partially resistant varieties for these diseases and several QTLs for resistance to bacterial seedling rot and bacterial grain rot have been identified (Mizobuchi et al. 2013a, 2013b, 2015, Pinson et al. 2010). However, these resistances are not strong. Meanwhile, Iwai et al. (2002) reported that transgenic rice lines overproducing Asthi1, an oat leaf thionin gene, showed enhanced resistance to B. glumae. However, it has not been applied to actual breeding.
Rice stripe disease, caused by rice stripe virus (RSV), is one of the major viral diseases in East Asia. The majority of japonica cultivars in East Asia are highly susceptible to RSV (Wang et al. 2014) although a few rice cultivars/lines that show resistance to RSV have been described (Noda et al. 1991, Zhang et al. 2011).
Breeding crops with broad-spectrum disease resistance using genetic resources is one of the principal goals of crop improvement. However, only a few genes have been identified as genetic resources for broad-spectrum disease resistance in rice. Hence, the transgenic approach could be a viable alternative. In recent years, several gain-of-function transgenic mutant populations have been developed in rice (Hsing et al. 2007, Jeong et al. 2002, Mori et al. 2007, Nakamura et al. 2007). Meanwhile, Kondou et al. (2009) produced more than 20,000 independent Arabidopsis transgenic lines overexpressing rice full-length cDNAs (rice-FOX Arabidopsis lines) to enable high-throughput screening for rice genes. We performed screening for pathogen resistance using these lines (Dubouzet et al. 2011). As a result, we identified several rice genes conferring resistance to both bacterial Pseudomonas syringae pv. tomato DC3000 and fungal Colletotrichum higginsianum in Arabidopsis. One of the genes named BROAD-SPECTRUM RESISTANCE1 (BSR1), encoding a receptor-like cytoplasmic kinase, conferred resistance to Xoo and M. oryzae when overexpressed in rice (Dubouzet et al. 2011).
In this paper, we report that overexpression of BSR1 not only conferred non-race-specific resistance to Xoo and M. oryzae, but also extended resistance to B. glumae and C. miyabeanus. Moreover, overexpression of BSR1 is likely to confer partial resistance to RSV.
Materials and Methods
Plant materials
Rice (Oryza sativa L.) wild-type (WT) cultivar ‘Nipponbare’, Xoo-resistant cultivar ‘Asominori’, RSV-resistant cultivar ‘Sainokagayaki’ and two transgenic plant lines (BSR1-OX-5 and -9) were grown under greenhouse conditions at 27°C to 30°C. BSR1-OX-5 and BSR1-OX-9 correspond to the previously reported AK070024:OX-5 and AK070024:OX-9 (Dubouzet et al. 2011), respectively.
For disease resistance tests, except for B. glumae, dehusked seeds were surface sterilized, sown on one-half-strength MS medium (Wako Pure Chemicals, Osaka, Japan), containing 3% (w/v) sucrose and 0.4% (w/v) Gelrite (Wako Pure Chemicals), in Agripots and grown in the growth chamber at 28°C in the dark for 3 days, then at 25°C under long-day conditions (16 h light [60–70 μmol m−2 s−1]/8 h dark) for 4–7 days. For transgenic seeds, Hygromycin B (30–50 μg/ml; Wako Pure Chemicals) was added to the medium. WT seedlings and hygromycin-resistant transgenic seedlings were transplanted into soil (Bonsol No. 2, Sumitomo Kagaku Kougyo, Osaka, Japan) and used for disease resistance tests.
Pathogens and pathogen cultures
The bacterial isolates used in this study were T7147 (MAFF311019, race II) and T7133 (MAFF311020, race III) of Xoo and AZ8204 (MAFF301682) of B. glumae, and the fungal isolates were Hoku1 (MAFF101512, race 007.0) of M. oryzae and H11-42-1 of C. miyabeanus.
Culture procedures for the various pathogens for inoculum were as follows. Xoo were cultured on PSA agar plates (1% proteose peptone, 1% sucrose, and 1.5% bacto agar) for 2 days at 28°C under dark conditions. B. glumae were cultured on King B agar plate (2% proteose peptone, 0.15% K2HPO4, 0.15% MgSO4, 1% glycerin and 1.5% agar, Eiken chemical, Tochigi, Japan) at 28°C for 2 days under dark conditions. M. oryzae was grown on oatmeal agar plates (3% oatmeal, 0.5% sucrose, and 1.6% bacto agar) at 25°C in the dark for 10 days, then under continuous illumination for 4 days to induce sporulation. C. miyabeanus was grown on V8 agar plates (20% V8 juice (Campbell soup company, Camden, NJ, USA), 0.3% CaCo3, and 1.5% bacto agar) at 25°C in the dark for 5 to 6 days, and then under a 12/12 h light/dark regime for 3 to 4 days to induce sporulation. The cultured pathogens were scraped and used to produce inocula.
Expression analysis of BSR1 by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated from rice leaves using Isogen (Wako Pure Chemicals) followed by further purification with the RNeasy mini kit (Qiagen, Valencia, CA, USA). First-strand cDNAs were synthesized from equal amounts of total RNA using a ReverTra Ace qPCR RT Master Mix with gDNA Remover Kit (Toyobo, Osaka, Japan) in a total volume of 10 μl, as described by the manufacturer. qRT-PCR was performed with the Thermal Cycler Dice TP800 system (Takara, Tokyo, Japan) using a Kapa SYBR FAST qPCR kit (Kapa Biosystems, Cape Town, South Africa) as described by the manufacturer. The primers used for qPCR were as follows: BSR1 5′-AGGTGAGGTTGCACTCTGCT-3′ and 5′-CCAAGAATCCACCAACTCGT-3′ as described Dubouzet et al. (2011); those for Rubq1 were 5′-GGAGCTGCTGCTGTTCTAGG-3′ and 5′-TTCAGACACCATCAAACCAGA-3′ as an internal control, as described Jiang et al. (2010). Transcript levels of BSR1 were normalized to the endogenous rice reference gene (Rubq1).
Test for resistance to Xoo
Rice seedlings transplanted in soil were grown in a growth chamber until the 6–8-leaf stage at 25°C under long-day conditions (16 h light/8 h dark) and then used for evaluation. The suspensions of Xoo (isolate T7147 or T7133) for inocula were adjusted to OD600 = 0.3 with sterile water. The top leaf blades of the tested plants were cut with scissors pre-wetted with inoculum at about 5 cm from the tip, and the cut ends (about 5 mm from the end) were dipped in inoculum for 10 s. The disease symptoms (lesion length) of inoculated plants were assessed 2 weeks after inoculation, as described previously (Dubouzet et al. 2011).
Test for resistance to M. oryzae
Rice seedlings transplanted in soil were grown in a greenhouse until the four-leaf stage at 28°C under a natural photoperiod. The plants were inoculated by spraying a spore suspension of M. oryzae (isolate Hoku1 (MAFF101512, race 007.0)). The detailed procedure for producing spore suspension is as follows: The mycelia of M. oryzae were scraped and the gel surface was flooded with sterile water containing 0.01% Tween 20. The suspension was filtered through a Kimwipe, and the resulted spore suspension was collected. The spore suspension was adjusted to a concentration of 6.7 × 105 spores/ml and used for inoculation. After inoculation, the plants were placed in a dark chamber at 26°C and 100% humidity for 20 h, and then further cultured in the greenhouse. Evaluation of resistance was based on the total number of compatible lesions that appeared on the 3rd and 4th leaf blades of each plant 5 days after inoculation.
Evaluation of bacterial pathogen B. glumae resistance
WT ‘Nipponbare’ rice seeds and T3–5 seeds of BSR1-OX-5 and -9 were sterilized by soaking in 70% ethanol for 30 s and Antiformin (available chlorine 5%) for 20 min. The seeds were then rinsed with sterilized water. The sterilized seeds were soaked in sterilized water at 28°C for 2 to 3 days in the dark and pre-germinated to 1–2 mm of sprout. The pre-germinated seeds were soaked in suspensions of B. glumae adjusted to OD520 = 0.004 and held under a vacuum for 1 min. The inoculated seeds were dried and sown in sterilized soil (Bonsol No. 2, Sumitomo Kagaku Kougyo). The inoculated seeds were incubated in a growth chamber at 28°C with 100% humidity under a 14-h photoperiod. Plant phenotypes were classified as ‘healthy’ or ‘diseased’ at 7–10 days after inoculation and the percentage of healthy plants among the total seeds used for inoculation was calculated as the survival ratio.
Evaluation of fungal pathogen C. miyabeanus resistance
Rice seedlings transplanted in soil were grown in a greenhouse until the six-leaf stage at 28°C under a natural photoperiod. The inoculation method of C. miyabeanus was the same as that of M. oryzae, except as follows. The concentration of the spore suspension was adjusted to 104 spores/ml. Evaluation of resistance was based on the total number of compatible lesions that appeared on the 5th and 6th leaf blades of each plant 5 days after inoculation.
Assessment of resistance to RSV
BSR1-OX seedlings selected by Hygromycin resistance (30–50 μg/ml), WT and cv. ‘Sainokagayaki’ seedlings were transplanted into soil at about 10 seedlings per pot, and grown in a greenhouse until the two-leaf stage at 28°C under a natural photoperiod. The rice seedlings were exposed to approximately 10 viruliferous small brown planthoppers (Laodelphax striatellus Fallén) per plant in an inoculation cage for 1 day to inoculate RSV, as described previously (Satoh et al. 2010). After the inoculation period, the insects were killed with insecticide and the seedlings were transferred to the same greenhouse. The virus infection was evaluated by enzyme-linked immunosorbent assay (ELISA) using antiserum against RSV nucleocapsid protein, as described previously (Shimizu et al. 2011). The pieces (about 1 cm) of leaf sheath plus stem tissue from inoculated seedlings were harvested for ELISA at 16 days after inoculation. Resistance to RSV was calculated by the ratio of diseased seedlings detected by ELISA among all inoculated seedlings.
Results
Transcript level of BSR1 in BSR1-OX rice lines
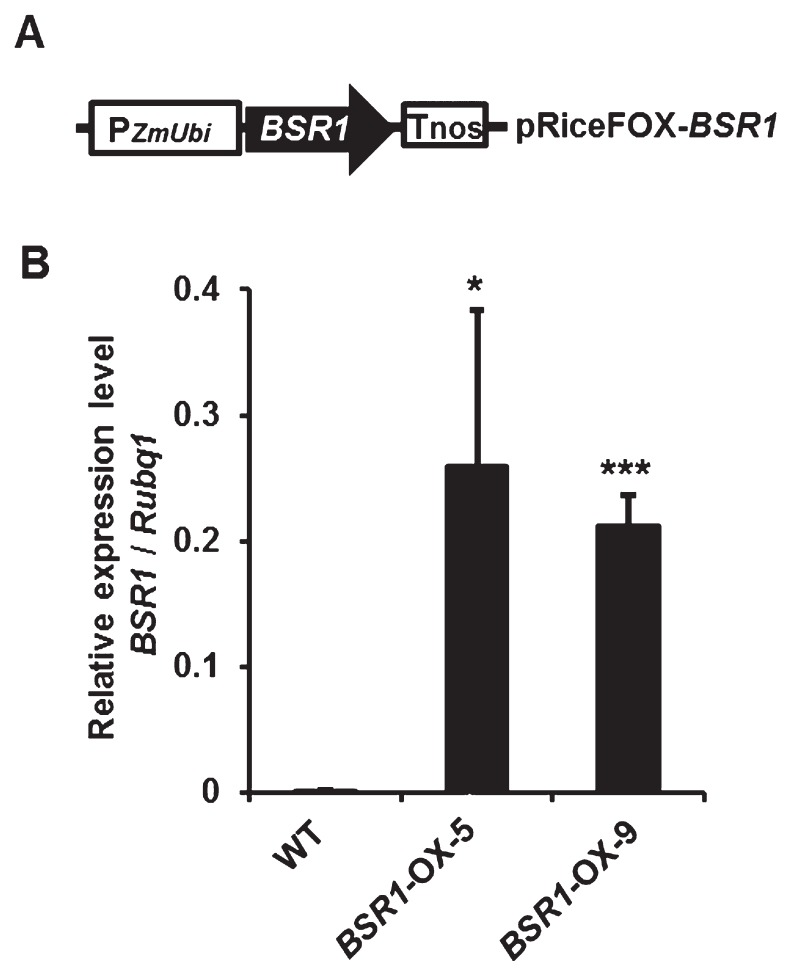
The cDNA of BSR1 was inserted downstream of the constitutive maize ubiquitin promoter (Fig. 1A), and the construct was used to generate transgenic rice lines overexpressing BSR1, as described previously (Dubouzet et al. 2011). The resulting two transgenic lines, BSR1-OX-5 (former name, AK070024:OX-5) and -9, were used for various disease resistance tests. To gain sufficient seeds for the disease resistance tests, the two transgenic lines were subjected to acceleration of advanced generations. To confirm overexpression of BSR1, we examined the transcript level of BSR1 by qRT-PCR in T3–4 generations of the BSR1-OX lines (Fig. 1B). Transcript levels of BSR1-OX-5 and -9 lines were 159- and 130-fold higher than that of ‘Nipponbare’ (WT), respectively. Thereafter, we used the plants of T3–5 lines for various disease resistance tests.
Fig. 1.
Schematic representation of pRiceFOX-BSR1 and the transcript level of BSR1 in BSR1-OX rice. (A) The pRiceFOX-BSR1 construct for overexpression of BSR1. (B) Transcript levels of BSR1 in T3–4 generations of BSR1-OX lines. Second youngest leaf blades of BSR1-OX and wild-type (WT) plants at the eight-leaf stage were used for measurement. Transcript levels of BSR1 were normalized to that of an endogenous Rubq1 housekeeping gene (Jiang et al. 2010). Values are means ± SD (n = 4).
Overexpression of BSR1 confers resistance to multiple races of Xoo and M. oryzae in rice
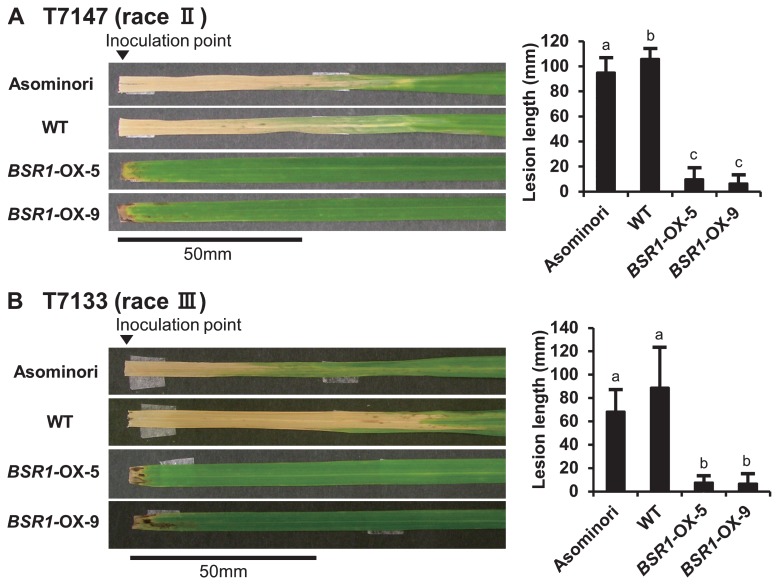
We have reported that BSR1-OX rice shows strong resistance to isolate T7174 (race I) of Xoo, a bacterial pathogen for rice bacterial leaf blight, and to isolate Kyu89-246 (race 003.0) of M. oryzae, a fungal pathogen for rice blast (Dubouzet et al. 2011). Hence, it would be plausible that BSR1 also confers resistance to other races of Xoo and M. oryzae. First, we examined whether BSR1-OX rice extended resistance to isolates T7147 (race II) and T7133 (race III) of Xoo. The WT and the resistant control, cv. ‘Asominori’, inoculated with isolate T7147 (race II) developed extended lesions from the cut (inoculated) end of the leaves, whereas BSR1-OX-5 and -9 plants showed restricted lesions (Fig. 2A). Lesion lengths in the inoculated BSR1-OX-5 and -9 were about 10 mm and 7 mm long, whereas those in WT and ‘Asominori’ were about 106 mm and 95 mm long (Fig. 2A). Two BSR1-OX lines showed 10- and 15-fold reductions in lesion length compared with the WT. Similarly, the results of inoculation of isolate T7133 (race III) are shown in Fig. 2B. The two BSR1-OX lines showed 10- and 12-fold reductions in lesion length compared with the WT. These results indicated that overexpression of BSR1 confers strong resistance to both T7147 (race II) and T7133 (race III), as well as to previously shown T7174 (race I). The results suggested that overexpression of BSR1 conferred non-race-specific resistance to Xoo. Although ‘Asominori’ has very strong resistance to the isolate T7174 (race I), the resistances to T7147 (race II) and T7133 (race III) were moderate (Kaku and Kimura 1989). Resistance levels of ‘Asominori’ shown here were similar to the report and ‘Asominori’ showed more race-specific resistance.
Fig. 2.
Disease resistance to multiple races of X. oryzae pv. oryzae. Isolates T7147 (race II; A) and T7133 (race III; B) of Xoo were used for infection. Arrowheads indicate the point of inoculation. Lesion lengths in BSR1-OX plants were significantly shorter than those in wild-type (WT) and ‘Asominori’, the resistance control cultivar. Values are means ± SD, n = 6–18. Different letters indicate significant differences (P < 0.05 by Tukey-Kramer’s test).
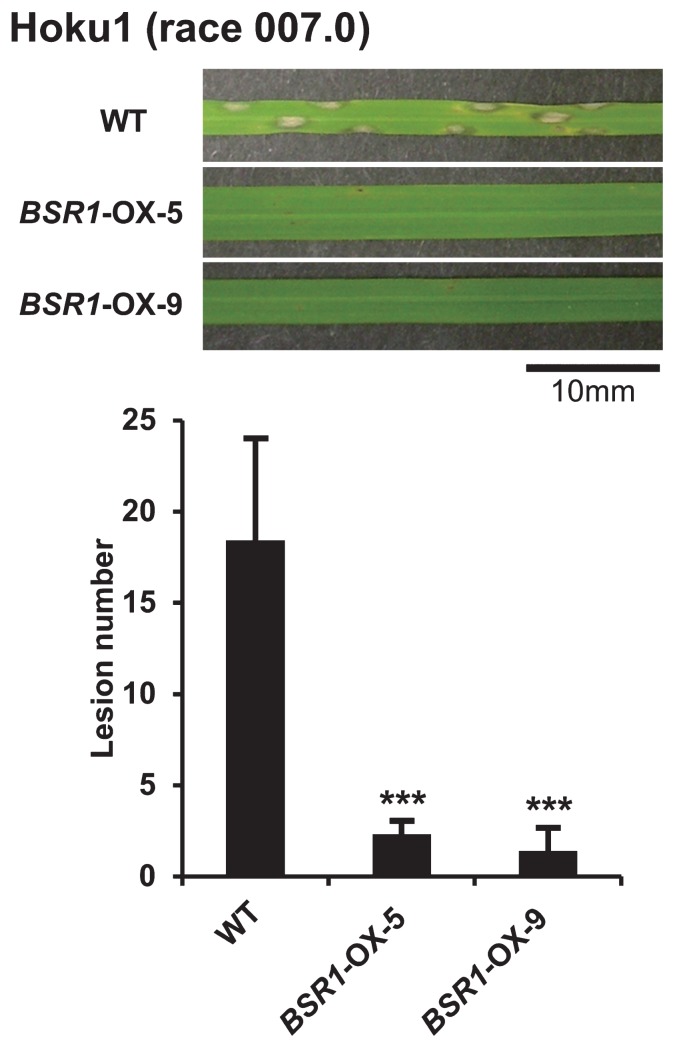
Next, to examine whether overexpression of BSR1 also confers resistance to another race of M. oryzae in rice, isolate Hoku1 (MAFF101512, race 007.0) was used for inoculation. Lesion numbers in the BSR1-OX-5 and -9 lines were significantly smaller than those in the WT plants (Fig. 3). Thus, because BSR1-OX lines conferred strong resistance to isolate Hoku1 (race 007.0) in addition to the previously shown isolate Kyu89-246 (race 003.0), we hypothesized that overexpression of BSR1 conferred non-race-specific resistance to M. oryzae.
Fig. 3.
Disease resistance to another race of M. oryzae. Isolate Hoku1 (race 007.0) of M. oryzae was used. Lesion numbers in BSR1-OX plants were significantly lower than those in wild-type (WT) plants (***P < 0.001 by t-test). Values are means ± SD, n = 3–7.
Extended resistance to another bacterial pathogen, Burkholderia glumae
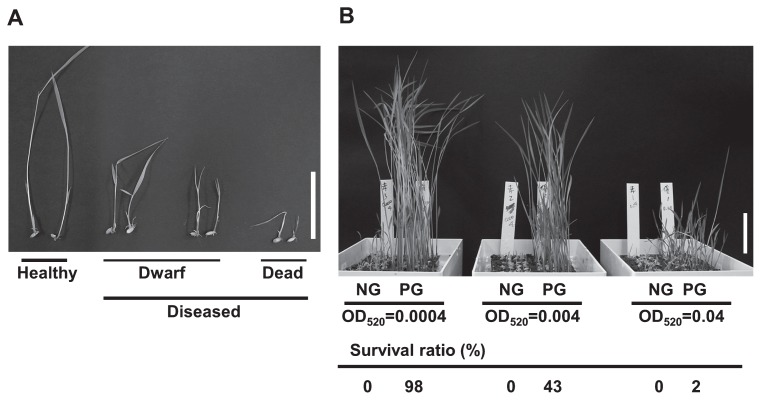
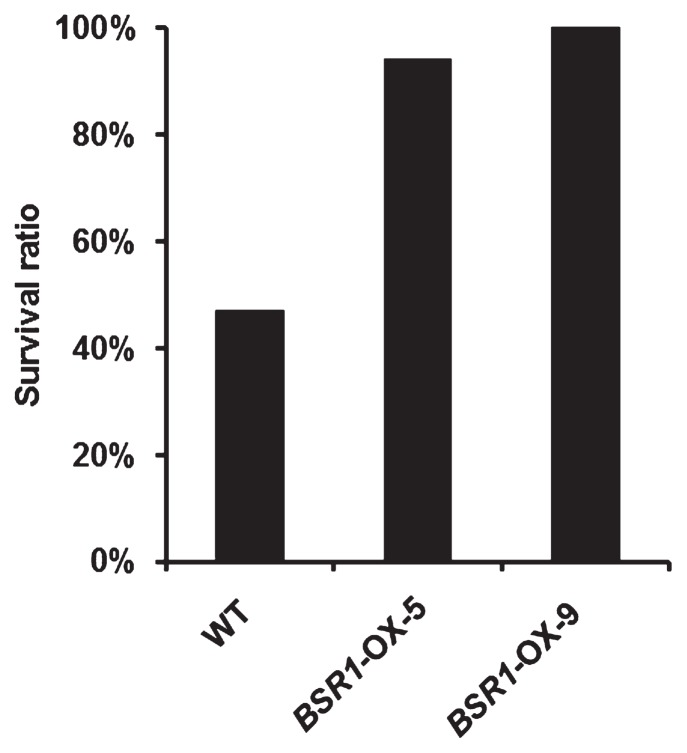
Bacterial seedling rot and bacterial grain rot (bacterial panicle blight) are caused by the bacterial pathogen B. glumae. The latter is an increasingly important disease problem in global rice production (Ham et al. 2011). Many genetic studies for resistance to bacterial grain rot have been reported (Mizobuchi et al. 2013a, 2015, Pinson et al. 2010, Sayler et al. 2006, Wasano and Okuda 1994). However, there are few reports on resistance to bacterial seedling rot, because such resistance is a complex characteristic influenced by environmental factors (Iwai et al. 2002, Mizobuchi et al. 2013b). We were interested in whether overexpression of BSR1 conferred resistance to bacterial seedling rot in rice. In the test for resistance to bacterial seedling rot, non-germinated seeds are usually used for inoculation by soaking. However, it was difficult to evaluate the resistance by this method because BSR1-OX rice displayed a decreased germination rate (Dubouzet et al. 2011). Therefore, we gathered only pre-germinated seeds for use in the disease resistance test. To determine the condition for inoculation of pre-germinated seeds, we performed a preliminary experiment using various concentrations of B. glumae suspension and WT seeds. The disease symptoms were classified as shown in Fig. 4A. Browning of the leaf sheath was usually detected together with a dwarf phenotype in diseased plants. Disease resistance was evaluated by the survival ratio, indicating the ratio of healthy plants to total seeds used for infection and shown as a percentage. When non-germinated (NG) seeds were inoculated by soaking in suspensions of three different concentrations of B. glumae, no healthy plant survived in all concentrations (Fig. 4B). In contrast, when pre-germinated seeds were used, 98%, 43% and 2% of healthy plants survived after soaking in suspensions of low (OD520 = 0.0004), medium (OD520 = 0.004) and high (OD520 = 0.04) concentrations of the bacteria, respectively. Thus, the optimal concentration range to evaluate resistance in this experiment was OD520 = 0.004–0.04. In subsequent experiments, we evaluated disease resistance by this method using pre-germinated seeds.
Fig. 4.
Preliminary experiments to determine the optimal concentration of bacterial pathogen, Burkholderia glumae, for inoculation. (A) The classification of disease symptoms at 7 days after inoculation. Healthy, normal phenotype; Diseased, dwarf or dead phenotype. Bar = 50 mm. (B) Results of the preliminary experiment. Photograph shows plants at 7 days after inoculation (n = 50–60). Non-germinated seeds (NG) and pre-germinated seeds (PG) were used for inoculation. Bar = 50 mm. Survival ratio (%) = (number of healthy plants/number of total seeds) × 100
Resistance to B. glumae was evaluated for BSR1-OX pre-germinated seeds. Survival ratios in BSR1-OX-5 and -9 lines were two times higher than that in the WT (Fig. 5). These results indicated that BSR1-OX lines displayed resistance to B. glumae in rice.
Fig. 5.
Disease resistance to another bacterial pathogenic species, Burkholderia glumae. Pre-germinated seeds of BSR1-OX and wild-type (WT) were inoculated with B. glumae. Concentration of inoculation was OD520 = 0.004. Survival ratio was calculated 8 days after inoculation (n = 17). Tests were performed three times with similar results.
Extended resistance to another major fungal pathogen, Cochliobolus miyabeanus
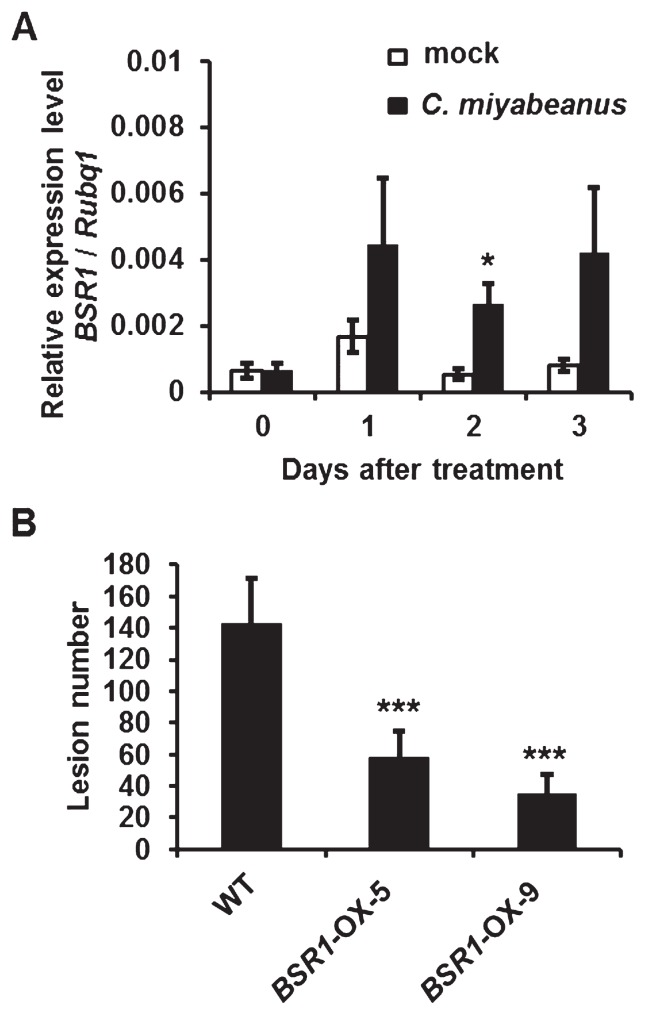
Brown spot disease is caused by the fungal pathogen, C. miyabeanus. We hypothesized that BSR1 conferred resistance to brown spot disease as well as to rice blast in rice. First, we investigated whether the expression of BSR1 changed by inoculation with C. miyabeanus in WT plants (Fig. 6A). C. miyabeanus was spray-inoculated onto WT plants, and the transcript level of BSR1 was measured by qRT-PCR. As a result, inoculated plants showed inducible expression of BSR1 compared with mock control after inoculation (Fig. 6A), although the transcript levels of BSR1 in the inoculated plants were much lower than those in BRS1-OX lines. This result suggested that BSR1 is involved in innate immunity against brown spot in rice. Hence, we examined whether overexpression of BSR1 conferred resistance to C. miyabeanus (Fig. 6B). Lesion numbers in BSR1-OX-5 and -9 plants were significantly lower than those in the WT plants (***P < 0.001 by t-test, Fig. 6B). Thus, overexpression of BSR1 conferred significant resistance to C. miyabeanus.
Fig. 6.
Disease resistance to another fungal pathogenic species, Cochliobolus miyabeanus. (A) Relative expression levels of BSR1 in wild-type (WT) plants on inoculation with mock or C. miyabeanus. Seventh-leaf blades were used for inoculation. Total RNAs at 0 to 3 days after inoculation were extracted. Transcript levels of BSR1 were normalized to that of Rubiq1. Expression levels of BSR1 in inoculated leaves were up-regulated compared with mock at 1 to 3 days. Values are means ± SD, n = 3. An asterisk indicates a statistically significant difference from the mock at 2 days (P < 0.05 by t-test). (B) Resistance to C. miyabeanus in BSR1-OX rice. Lesion numbers in BSR1-OX plants were significantly lower than those in WT plants (***P < 0.001 by t-test). Values are means ± SD, n = 4–12.
BSR1-OX rice were slightly resistant to rice stripe virus (RSV)
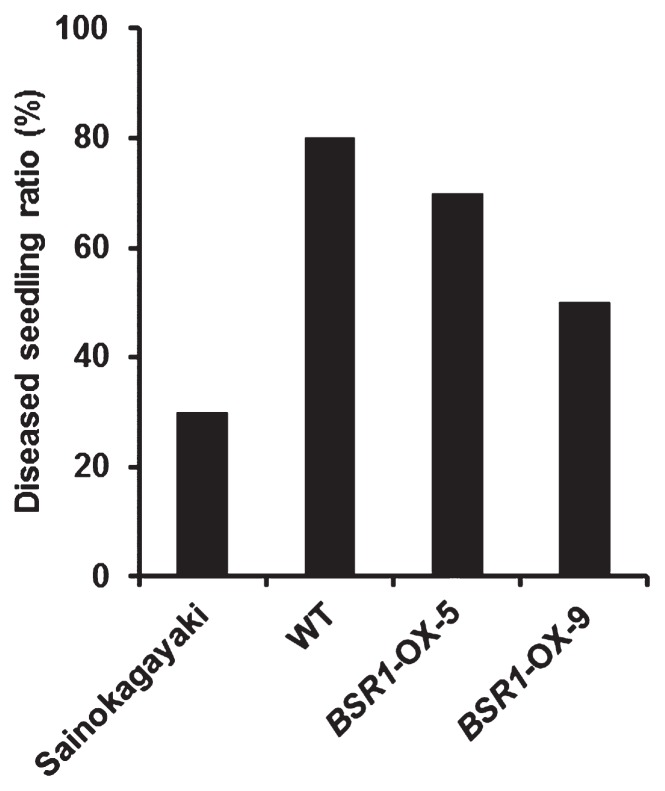
We examined whether overexpression of BSR1 could confer resistance against a viral pathogen, RSV, in rice, because the majority of japonica cultivars, including cv. ‘Nipponbare’, are susceptible to RSV. The results are shown in Fig. 7. After inoculation of RSV, the percentages of diseased seedlings detected by ELISA in BSR1-OX-5 and -9 seedlings were slightly lower than those in WT seedlings, but were higher than that in cv. ‘Sainokagayaki’, the RSV-resistant control cultivar (Fig. 7). Thus, overexpression of BSR1 could confer slight resistance to RSV, although its resistance level was weaker than that of ‘Sainokagayaki’, which possesses the highly resistant Stvb-i gene to RSV (Shimizu et al. 2011).
Fig. 7.
Disease resistance to a viral pathogen, rice stripe virus. Diseased seedling ratio, percentage of diseased seedlings detected by enzyme-linked immunosorbent assay (ELISA) in all inoculated seedlings at 16 days after inoculation. n = 8 to 10. Tests were performed three times with similar results.
Discussion
From the point of view of breeding, the quality of broad-spectrum resistance against two or more different pathogen species is an agronomically important trait. We previously reported that overexpression of BSR1, encoding a receptor-like cytoplasmic kinase, confers remarkable resistance to both bacterial and fungal pathogens in Arabidopsis, and that it confers resistance to the bacterial pathogen, Xoo, and fungal pathogen, M. oryzae, in rice (Dubouzet et al. 2011). In this study, we first showed that BSR1 confers resistance to multiple races of Xoo and M. oryzae in overexpressing rice. In addition, BSR1-OX rice showed extended resistance to another bacterial pathogen, B. glumae, and another fungal pathogen, C. miyabeanus. Thus, overexpression of BSR1 conferred broad-spectrum disease resistance to at least two bacterial and two fungal pathogenic species in rice. Therefore, it would be natural to consider that the resistance to bacterial and fungal pathogens by BSR1 is non-race specific. Furthermore, BSR1-OX rice showed slight resistance to RSV. To the best of our knowledge, there are no other genes that confer such multi-disease resistance.
As a typical example of interactions with host plants and pathogens, the plant defense against pathogens, such as bacteria and fungi, is provided through cell-surface-localized pattern recognition receptors that detect pathogen-associated molecular patterns (PAMPs), and result in pattern-triggered immunity (PTI). To counteract this innate immunity, pathogens deliver effector proteins into the host cell to suppress PTI. In some cases, plants use intracellular resistance (R) proteins to detect race-specific effectors, which results in effector-triggered immunity, often associated with hypersensitive response and programmed cell death. The BSR1 gene, OsRLCK278, is one of 379 RLCK genes encoding receptor-like cytoplasmic kinases in rice (Dubouzet et al. 2011). BSR1 is classified into the RLCK-VII protein family, which belongs to the same family as BIK1 and PBS, the well-characterized Arabidopsis RLCKs involved in plant defense (Dubouzet et al. 2011). Arabidopsis BIK1 was originally isolated as a gene involved in the defense against necrotrophic fungal pathogens (Veronese et al. 2006). BIK1 also associates with a flagellin receptor complex, FLS2/BAK1, which is rapidly phosphorylated upon perception of flagellin. In addition, BIK1 is also phosphorylated by another PAMP, translation elongation factor (EF-Tu). Hence, BIK1 is considered to mediate PTI signal transduction, such as production of a reactive oxygen species (ROS) burst, activation of mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CPKs or CDPKs), transcriptional reprogramming, and ultimately immunity, from multiple PAMP receptor complexes (Chinchilla et al. 2007, Heese et al. 2007, Lu et al. 2010, Roux et al. 2011, Schulze et al. 2010, Schwessinger et al. 2011, Sun et al. 2013). Furthermore, BIK1 regulates calcium influx, phosphorylates and activates NADPH oxidase RBOHD, and not only induces the defense response (including the ROS burst that is a poisonous factor directly attacking pathogens), but also prevents invasion of pathogenic bacteria by closing stoma (Kadota et al. 2014, Li et al. 2014). Thus, BIK1 plays pivotal roles in recognition of PAMPs and subsequent signal transductions. In rice, OsRLCK185, which belongs to the RLCK-VII family, was reported recently (Yamaguchi et al. 2013). OsRLCK158 interacts with a pattern recognition receptor OsCERK1, which recognizes chitin and peptidoglycan at the plasma membrane, and regulates a MAP kinase cascade that leads to a PTI defense response. Taken together, it is likely that the BSR1 protein interacts with various transmembrane receptors that recognize PAMPs of Xoo, M. oryzae, B. glumae and/or C. miyabeanus, which functions in linking multiple PAMP receptor complexes to downstream intracellular signaling and enhances a part of PAMP-mediated defense.
From the viewpoint of the pathogen character, the lifestyles of pathogens used in this study are classified as hemibiotrophs or necrotrophs. The blast fungus, M. oryzae, and bacterial leaf blight bacteria pathogen, Xoo, are considered hemibiotrophs (Van Bockhaven et al. 2013). Hemibiotrophs are characterized by an initial infectious period of the biotrophic stage in which the pathogens grow within host cells before switching to a necrotrophic growth stage when lesions become apparent (Wilson and Talbot 2009). Xoo has been considered mostly as a biotroph but is probably best classified as a hemibiotroph (De Vleesschauwer et al. 2013). Furthermore, the pathogens of bacterial seedling rot, B. glumae, and of brown spot, C. miyabeanus, have been classified as necrotrophs (Iwai et al. 2002, Su’udi et al. 2012). Thus, we demonstrated that BSR1-OX rice conferred resistance not only to hemibiotrophs but also necrotrophs.
Salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) are typical defense hormones, and the regulation of their signaling networks against pathogen infection is well established, especially in the dicotyledonous model plant Arabidopsis (Van Bockhaven et al. 2013). In Arabidopsis, SA-dependent defenses are generally associated with resistance to biotrophs, whereas JA/ET-dependent defenses are generally associated with necrotrophs (Van Bockhaven et al. 2013, Thomma et al. 2001, De Vos et al. 2006). By contrast, the defense system in rice is considered to be controlled by a more complicated signaling network (De Vleesschauwer et al. 2013). SA-, JA- and ET-dependent defenses all enhanced resistance to hemibiotrophic M. oryzae (Ahn et al. 2005, Helliwell et al. 2013, Iwai et al. 2006, Mei et al. 2006, Peng et al. 2012, Qiu et al. 2007, Schweizer et al. 1998, Shimono et al. 2007, 2012). Meanwhile, SA- and JA-dependent defenses were involved in disease resistance against hemibiotrophic Xoo (Babu et al. 2003, Qiu et al. 2007, Shimono et al. 2012, Tanaka et al. 2014, Taniguchi et al. 2014), whereas ET-dependent defense played a negative role (Shen et al. 2011). Among the hemibiotrophs, the role of ET is the opposite of Xoo and M. oryzae. Similarly, ET-dependent defense is involved negatively in the resistance against C. miyabeanus, a necrotrophic pathogen (De Vleesschauwer et al. 2010), while no reports have implicated SA- and JA-dependent defenses against C. miyabeanus (Van Bockhaven et al. 2013). More interestingly, ABA, which antagonizes the SA pathway, is involved in the resistance to C. miyabeanus (De Vleesschauwer et al. 2010), although ABA promotes susceptibility against Xoo and M. oryzae (Jiang et al. 2010, Xu et al. 2013). The plant hormone defense network against B. glumae is unknown in rice. RSV resistance is implicated in enhancing the SA signal (Wang et al. 2014). Taken together, understanding the mechanism of broad-spectrum resistance of BSR1 from the point of view of plant hormones is complex.
Currently, several genes have been reported to confer broad-spectrum disease resistance in rice. For example, overexpression of OsWRKY13 or OsWRKY45 enhanced resistance to Xoo and M. oryzae in rice by mediating SA signaling (Qiu et al. 2007, Shimono et al. 2007, 2012). Unlike BSR1-OX rice, however, there are few reports that overexpressed genes enhanced resistance to necrotrophic pathogens, B. glumae and C. miyabeanus in rice. Meanwhile, overexpression of OsWRKY30 or OsACS2 encoding a key enzyme of ET biosynthesis enhanced resistance to fungal pathogens M. oryzae and necrotrophic Rhizoctonia solani in rice. The resistance conferred by OsWRKY30 was associated with the activation of JA synthesis-related genes and the increased accumulation of endogenous JA (Peng et al. 2012), and that by OsACS2 was associated with the increased level of endogenous ET (Helliwell et al. 2013). Hence, we speculate that the broad-spectrum disease resistance against two hemibiotrophs (Xoo and M. oryzae) and two necrotrophs (B. glumae and C. miyabeanus) in BSR1-OX rice is based on an SA and JA/ET combined pathway.
Here, BSR1-OX rice displayed strong resistance to three tested races (races I to III) of Xoo. In contrast, ‘Asominori’ showed more race-specific resistance. Although the resistance to isolate T7174 (race I) on ‘Asominori’ was very strong (Dubouzet et al. 2011), the resistances to T7147 (race II) and T7133 (race III) were moderate (Fig. 2), almost similar to the report of Kaku and Kimura (1989). ‘Asominori’ has been reported to have an Xa1-as(t) gene at the Xa1 locus (Ise et al. 1998). Xa1-as(t) is implicated in the strong resistance to T7174 (race I). ‘Asominori’ has also been suggested to have minor-affected loci that are involved in the quantitative resistance to T7133 (race III) (Yoshimura et al. 1996). BSR1-OX rice showed strong resistance to both T7174 (race I) and T7133 (race III); therefore, the defense mechanism of BSR1-OX rice would be different from that of ‘Asominori’. From the viewpoint of breeding, overexpression of BSR1 could confer more useful non-race-specific resistance to Xoo in rice compared with using the resistance genes of ‘Asominori’.
In the previous report, BSR1-OX rice displayed strong resistance to race 003.0 (isolate Kyu89-246) and the resistance level was stronger than that in cv. ‘Sensho’ (Dubouzet et al. 2011), which has a strong non-race-specific resistance to M. oryzae associated with pi21 (Fukuoka et al. 2009). In this paper, BSR1-OX rice displayed extended strong resistance to race 007.0 (isolate Hoku1; Fig. 3). Hence, non-race-specific resistance or field resistance against M. oryzae is also promising in BSR1-OX rice. Taken together, overexpression of BSR1 could confer more promising leaf blight and blast resistances compared with the resistant cultivars ‘Asominori’ and ‘Sensho’, respectively, in many useful O. sativa varieties. Furthermore, it is plausible that BSR1-OX rice also shows non-race-specific resistance to B. glumae and C. miyabeanus, and exhibits resistance to other pathogen species, because BSR1-OX rice showed resistance to all pathogens tested.
In conclusion, BSR1, when overexpressed in rice, conferred broad-spectrum disease resistance against at least four diseases: bacterial leaf blight, blast, bacterial seedling rot and brown spot, and slight resistance against rice stripe disease by RSV. BSR1 represents a highly valuable and convenient genetic resource because it confers resistance to various diseases by a single gene. In the future, the defense mechanism conferred by BSR1 will be clarified to use the BSR1 gene effectively.
Acknowledgements
This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement, GMO-1006). We thank Dr. Hiroaki Ichikawa (NIAS, Japan) for providing the pRiceFOX vector, Dr. Hirokazu Ochiai (NIAS, Japan) for providing Xoo and ‘Asominori’ rice seeds, Dr. Koji Azegami (NARC, Japan) for providing B. glumae and Dr. Ritsuko Mizobuchi (Institute of Crop Science, NARO, Japan) for advice on the B. glumae assay. We also thank Lois Ishizaki, Tomiko Senba and Chiyoko Umeda (NIAS, Japan) for their support during the rice transformation experiments and for overall technical assistance.
Literature Cited
- Ahn, I.P., Kim, S., Kang, S., Suh, S.C. and Lee, Y.H. (2005) Rice defense mechanisms against Cochliobolus miyabeanus and Magnaporthe grisea are distinct. Phytopathology 95: 1248–1255. [DOI] [PubMed] [Google Scholar]
- Babu, R.M., Sajeena, A., Samundeeswari, A.V., Sreedhar, A., Vidhyasekaran, P., Seetharaman, K. and Reddy, M.S. (2003) Induction of systemic resistance to Xanthomonas oryzae pv. oryzae by salicylic acid in Oryza sativa (L.). J. Plant Dis. Prot. 110: 419–431. [Google Scholar]
- Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J.D., Felix, G. and Boller, T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer, D., Yang, Y., Cruz, C.V. and Höfte, M. (2010) Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 152: 2036–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer, D., Gheysen, G. and Höfte, M. (2013) Hormone defense networking in rice: tales from a different world. Trends Plant Sci. 18: 555–565. [DOI] [PubMed] [Google Scholar]
- De Vos, M., Van Zaanen, W., Koornneef, A., Korzelius, J.P., Dicke, M., Van Loon, L.C. and Pieterse, C.M. (2006) Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiol. 142: 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A., Talbot, N.J., Ebbole, D.J., Farman, M.L., Mitchell, T.K., Orbach, M.J., Thon, M., Kulkarni, R., Xu, J.R., Pan, H.et al. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986. [DOI] [PubMed] [Google Scholar]
- Dubouzet, J.G., Maeda, S., Sugano, S., Ohtake, M., Hayashi, N., Ichikawa, T., Kondou, Y., Kuroda, H., Horii, Y., Matsui, M.et al. (2011) Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 9: 466–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9: 275–296. [Google Scholar]
- Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., Hayashi, N., Takahashi, A., Hirochika, H., Okuno, K.et al. (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325: 998–1001. [DOI] [PubMed] [Google Scholar]
- Ham, J.H., Melanson, R.A. and Rush, M.C. (2011) Burkholderia glumae: next major pathogen of rice? Mol. Plant Pathol. 12: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese, A., Hann, D.R., Gimenez-Ibanez, S., Jones, A.M., He, K., Li, J., Schroeder, J.I., Peck, S.C. and Rathjen, J.P. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, E.E., Wang, Q. and Yang, Y. (2013) Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 11: 33–42. [DOI] [PubMed] [Google Scholar]
- Hsing, Y.I., Chern, C.G., Fan, M.J., Lu, P.C., Chen, K.T., Lo, S.F., Sun, P.K., Ho, S.L., Lee, K.W., Wang, Y.C.et al. (2007) A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 63: 351–364. [DOI] [PubMed] [Google Scholar]
- Ise, K., Li, C.Y., Sun, Y.Q. and Ye, C.R. (1998) Inheritance of resistance to bacterial leaf blight in differential rice variety Asominori. IRRN 23: 13–14. [Google Scholar]
- Iwai, T., Kaku, H., Honkura, R., Nakamura, S., Ochiai, H., Sasaki, T. and Ohashi, Y. (2002) Enhanced resistance to seed-transmitted bacterial diseases in transgenic rice plants overproducing an oat cell-wall-bound thionin. Mol. Plant Microbe Interact. 15: 515–521. [DOI] [PubMed] [Google Scholar]
- Iwai, T., Miyasaka, A., Seo, S. and Ohashi, Y. (2006) Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice rice plants. Plant Physiol. 142: 1202–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, D.H., An, S., Kang, H.G., Moon, S., Han, J.J., Park, S., Lee, H.S., An, K. and An, G. (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130: 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.J., Shimono, M., Sugano, S., Kojima, M., Yazawa, K., Yoshida, R., Inoue, H., Hayashi, N., Sakakibara, H. and Takatsuji, H. (2010) Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 23: 791–798. [DOI] [PubMed] [Google Scholar]
- Kadota, Y., Sklenar, J., Derbyshire, P., Stransfeld, L., Asai, S., Ntoukakis, V., Jones, J.D., Shirasu, K., Menke, F., Jones, A.et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kaku, H. and Kimura, T. (1989) Qualitative resistance reaction of rice cultivar Asominori to certain race II strains of Xanthomonas campestris pv. oryzae. Ann. Phytopathol. Soc. Jpn. 55: 657–659. [Google Scholar]
- Kondou, Y., Higuchi, M., Takahashi, S., Sakurai, T., Ichikawa, T., Kuroda, H., Yoshizumi, T., Tsumoto, Y., Horii, Y., Kawashima, M.et al. (2009) Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 57: 883–894. [DOI] [PubMed] [Google Scholar]
- Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., Cai, G., Gao, L., Zhang, X., Wang, Y.et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Liu, W., Liu, J., Triplett, L., Leach, J.E. and Wang, G.L. (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52: 213–241. [DOI] [PubMed] [Google Scholar]
- Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L. and He, P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, C., Qi, M., Sheng, G. and Yang, Y. (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 19: 1127–1137. [DOI] [PubMed] [Google Scholar]
- Mizobuchi, R., Sato, H., Fukuoka, S., Tanabata, T., Tsushima, S., Imbe, T. and Yano, M. (2013a) Mapping a quantitative trait locus for resistance to bacterial grain rot in rice. Rice (N Y) 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizobuchi, R., Sato, H., Fukuoka, S., Tsushima, S., Imbe, T. and Yano, M. (2013b) Identification of qRBS1, a QTL involved in resistance to bacterial seedling rot in rice. Theor. Appl. Genet. 126: 2417–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizobuchi, R., Sato, H., Fukuoka, S., Tsushima, S. and Yano, M. (2015) Fine mapping of RBG2, a quantitative trait locus for resistance to Burkholderia glumae, on rice chromosome 1. Mol. Breed. 35: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moletti, M., Giudici, M.L. and Villa, B. (1997) Rice Akiochi-brown spot disease in Italy: Agronomic and chemical control. In: Chataigner, J. (ed.) Maladies du riz en région méditerranéenne et les possibilités d’ amélioration de sa résistance, CIHEAM, Montpellier, pp. 79–85. [Google Scholar]
- Mori, M., Tomita, C., Sugimoto, K., Hasegawa, M., Hayashi, N., Dubouzet, J.G., Ochiai, H., Sekimoto, H., Hirochika, H. and Kikuchi, S. (2007) Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol. Biol. 63: 847–860. [DOI] [PubMed] [Google Scholar]
- Nakamura, H., Hakata, M., Amano, K., Miyao, A., Toki, N., Kajikawa, M., Pang, J., Higashi, N., Ando, S., Toki, S.et al. (2007) A genome-wide gain-of function analysis of rice genes using the FOX-hunting system. Plant Mol. Biol. 65: 357–371. [DOI] [PubMed] [Google Scholar]
- Noda, S., Omura, T., Murakami, M. and Tsuchizaki, T. (1991) Infectivity of rice viruses to the varieties resistant to rice stripe virus. Ann. Phytopathol. Soc. Jpn. 57: 259–262. [Google Scholar]
- Peng, X., Hu, Y., Tang, X., Zhou, P., Deng, X., Wang, H. and Guo, Z. (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236: 1485–1498. [DOI] [PubMed] [Google Scholar]
- Pinson, S.R., Shahjahan, A.K., Rush, M.C. and Groth, D.E. (2010) Bacterial panicle blight resistance QTLs in rice and their association with other disease resistance loci and heading date. Crop Sci. 50: 1287–1297. [Google Scholar]
- Qiu, D., Xiao, J., Ding, X., Xiong, M., Cai, M., Cao, Y., Li, X., Xu, C. and Wang, S. (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20: 492–499. [DOI] [PubMed] [Google Scholar]
- Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., Malinovsky, F.G., Tör, M., de Vries, S. and Zipfel, C. (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, H., Ando, I., Hirabayashi, H., Takeuchi, Y., Arase, S., Kihara, J., Kato, H., Imbe, T. and Nemoto, H. (2008) QTL analysis of brown spot resistance in rice (Oryza sativa L.). Breed. Sci. 58: 93–96. [Google Scholar]
- Satoh, K., Kondoh, H., Sasaya, T., Shimizu, T., Choi, I.R., Omura, T. and Kikuchi, S. (2010) Selective modification of rice (Oryza sativa) gene expression by rice stripe virus infection. J. Gen. Virol. 91: 294–305. [DOI] [PubMed] [Google Scholar]
- Sayler, R.J., Cartwright, R.D. and Yang, Y. (2006) Genetic characterization and real-time PCR detection of Burkholderia glumae, a newly emerging bacterial pathogen of rice in the United States. Plant Dis. 90: 603–610. [DOI] [PubMed] [Google Scholar]
- Schulze, B., Mentzel, T., Jehle, A.K., Mueller, K., Beeler, S., Boller, T., Felix, G. and Chinchilla, D. (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, P., Buchala, A., Dudler, R. and Métraux, J.P. (1998) Induced systemic resistance in wounded rice plants. Plant J. 14: 475–481. [Google Scholar]
- Schwessinger, B., Roux, M., Kadota, Y., Ntoukakis, V., Sklenar, J., Jones, A. and Zipfel, C. (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., Liu, H., Yuan, B., Li, X., Xu, C. and Wang, S. (2011) OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 34: 179–191. [DOI] [PubMed] [Google Scholar]
- Shimizu, T., Nakazono-Nagaoka, E., Uehara-Ichiki, T., Sasaya, T. and Omura, T. (2011) Targeting specific genes for RNA interference is crucial to the development of strong resistance to rice stripe virus. Plant Biotechnol. J. 9: 503–512. [DOI] [PubMed] [Google Scholar]
- Shimono, M., Sugano, S., Nakayama, A., Jiang, C.J., Ono, K., Toki, S. and Takatsuji, H. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19: 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, M., Koga, H., Akagi, A., Hayashi, N., Goto, S., Sawada, M., Kurihara, T., Matsushita, A., Sugano, S., Jiang, C.J.et al. (2012) Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Li, L., Macho, A.P., Han, Z., Hu, Z., Zipfel, C., Zhou, J.M. and Chai, J. (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628. [DOI] [PubMed] [Google Scholar]
- Su’udi, M., Park, J.M., Kang, W.R., Park, S.R., Hwang, D.J. and Ahn, I.P. (2012) Quantification of rice brown leaf spot through Taqman real-time PCR specific to the unigene encoding Cochliobolus miyabeanus SCYTALONE DEHYDRATASE1 involved in fungal melanin biosynthesis. J. Microbiol. 50: 947–954. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Taniguchi, S., Tamaoki, D., Yoshitomi, K., Akimitsu, K. and Gomi, K. (2014) Multiple roles of plant volatiles in jasmonate-induced defense response in rice. Plant Signal. Behav. 9: e29247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, S., Hosokawa-Shinonaga, Y., Tamaoki, D., Yamada, S., Akimitsu, K. and Gomi, K. (2014) Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 37: 451–461. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Penninckx, I.A., Broekaert, W.F. and Cammue, B.P. (2001) The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13: 63–68. [DOI] [PubMed] [Google Scholar]
- Van Bockhaven, J., De Vleesschauwer, D. and Höfte, M. (2013) Towards establishing broad-spectrum disease resistance in plants: silicon leads the way. J. Exp. Bot. 64: 1281–1293. [DOI] [PubMed] [Google Scholar]
- Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., Dietrich, R.A., Hirt, H. and Mengiste, T. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Liu, Y., He, J., Zheng, X., Hu, J., Liu, Y., Dai, H., Zhang, Y., Wang, B., Wu, W.et al. (2014) STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 5: 4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasano, K. and Okuda, S. (1994) Evaluation of resistance of rice cultivars to bacterial grain rot by the syringe inoculation method. Breed. Sci. 44: 1–6. [Google Scholar]
- Wilson, R.A. and Talbot, N.J. (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7: 185–195. [DOI] [PubMed] [Google Scholar]
- Xu, J., Audenaert, K., Hofte, M. and De Vleesschauwer, D. (2013) Abscisic acid promotes susceptibility to the rice leaf blight pathogen Xanthomonas oryzae pv oryzae by suppressing salicylic acid-mediated defenses. PLoS ONE 8: e67413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., Yamada, K., Ishikawa, K., Yoshimura, S., Hayashi, N., Uchihashi, K., Ishihama, N., Kishi-Kaboshi, M., Takahashi, A., Tsuge, S.et al. (2013) A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13: 347–357. [DOI] [PubMed] [Google Scholar]
- Yoshimura, A., Lei, J.X., Matsumoto, T., Tsunematsu, H., Yoshimura, S., Iwata, N., Baraoidan, M.R., Mew, T.W. and Nelson, R.J. (1996) Analysis of pyramiding of bacterial blight resistance genes in rice by using DNA markers. In: Khush, G.S. (ed.) Rice Genetics III: Proceedings of the Third International Rice Genetics Symposium, IRRI, Manila, pp. 577–581. [Google Scholar]
- Zhang, Y.X., Wang, Q., Jiang, L., Liu, L.L., Wang, B.X., Shen, Y.Y., Cheng, X.N. and Wan, J.M. (2011) Fine mapping of qSTV11KAS, a major QTL for rice stripe disease resistance. Theor. Appl. Genet. 122: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]