NMR studies show at atomic resolution that the K-segment of a stress-induced dehydrin protein adapts α-helical structure when it binds membranes.
Abstract
Dehydrins are disordered proteins that are expressed in plants as a response to embryogenesis and water-related stress. The molecular function and structural action of the dehydrins are yet elusive, but increasing evidence points to a role in protecting the structure and functional dynamics of cell membranes. An intriguing example is the cold-induced dehydrin Lti30 that binds to membranes by its conserved K segments. Moreover, this binding can be regulated by pH and phosphorylation and shifts the membrane phase transition to lower temperatures, consistent with the protein’s postulated function in cold stress. In this study, we reveal how the Lti30-membrane interplay works structurally at atomic level resolution in Arabidopsis (Arabidopsis thaliana). Nuclear magnetic resonance analysis suggests that negatively charged lipid head groups electrostatically capture the protein’s disordered K segments, which locally fold up into α-helical segments on the membrane surface. Thus, Lti30 conforms to the general theme of structure-function relationships by folding upon binding, in spite of its disordered, atypically hydrophilic and repetitive sequence signatures. Moreover, the fixed and well-defined structure of the membrane-bound K segments suggests that dehydrins have the molecular prerequisites for higher level binding specificity and regulation, raising new questions about the complexity of their biological function.
Since rooted plants cannot flee environmental changes, their fitness and survival have come to rely on a series of molecular defense systems, reflected in genomes that are typically much larger than for animals (Rafalski, 2002). One of the most basal tasks of this defense is to manage cellular stress induced by desiccation, ionic strength, and thermal fluctuations. The molecular response includes the production of osmoprotectants, various sugars (Ingram and Bartels, 1996), and up-regulated expression of dedicated stress proteins like the intrinsically disordered dehydrins (Baker et al., 1988; Nylander et al., 2001; Bomal et al., 2002; Chakrabortee et al., 2007). Exactly how these stress proteins function and interfere with the cellular processes is yet unknown, but judging by their highly conserved sequence signatures, their action seems to be under precise molecular control (Close, 1996; Mouillon et al., 2006; Graether and Boddington, 2014). Accordingly, it is speculated that the dehydrins could have evolved to selectively target and modulate the properties of other proteins (Hara et al., 2001; Kovacs et al., 2008), DNA/RNA (Hara et al., 2009), or membranes (Dure, 1993; Danyluk et al., 1998; Koag et al., 2003).
Membrane targeting has come to stand out as particularly interesting, as both the structure and phase properties of lipid bilayers are known to depend critically on temperature and water content (Steponkus and Lynch, 1989; Crowe and Crowe, 1992). Also, some dehydrins are found to colocalize with membrane surfaces (Danyluk et al., 1998; Puhakainen et al., 2004) and bind to anionic phospholipids in vitro (Koag et al., 2003, 2009; Kovacs et al., 2008; Eriksson et al., 2011). Some of these interactions are driven by electrostatics and depend on positively charged residues in the dehydrin sequence pairing with negatively charged lipid head groups, following the same rules as other lipid-binding proteins (Lemmon, 2008; Moravcevic et al., 2010; Eriksson et al., 2011). A good example is the net positively charged dehydrin Lti30 from Arabidopsis (Arabidopsis thaliana), which binds membranes by recognizing the negatively charged head groups of phospholipids (Eriksson et al., 2011). The consequence of this membrane interaction is a decrease of the main lipid phase transition by 2.5°C in vitro (Eriksson et al., 2011), intriguingly matching the decreased survival temperature of 3°C observed upon Lti30 overexpression in Arabidopsis (Puhakainen et al., 2004). Consistent effects on the membrane phase transition are reported for the dehydrin K2 (Clarke et al., 2015). To achieve membrane binding, Lti30 comprises not less than six copies of the archetypical K segment (Fig. 1). However, the K segments are not alone accountable for the Lti30-membrane interaction, as binding also requires the protonation of flanking pairs of His residues. In essence, these colocalized pairs of His residues form a pH-controlled affinity switch, which can be further fine-tuned by protein phosphorylation (Eriksson et al., 2011). The complexity of the Lti30-membrane interplay naturally raises questions about the possible involvement of binding-induced folding (Arai et al., 2015; Wright and Dyson, 2015). Following general structure-function relationships, such three-dimensional organization of a specific structure would open up additional layers of target specificity and modes of function (Lee et al., 2010; Vuzman and Levy, 2012) analogous to those observed for other intrinsically disordered proteins (Kriwacki et al., 1996; Dunker and Obradovic, 2001; Tompa, 2002; Borg et al., 2007; Sigalov and Hendricks, 2009). Classical examples are found among the transcription factors (Wright and Dyson, 2015), but there also are cases where local folding is induced by membrane binding (Jao et al., 2004; Jarvet et al., 2007), resembling that of the dehydrins (Table I; Koag et al., 2003; Rahman et al., 2010).
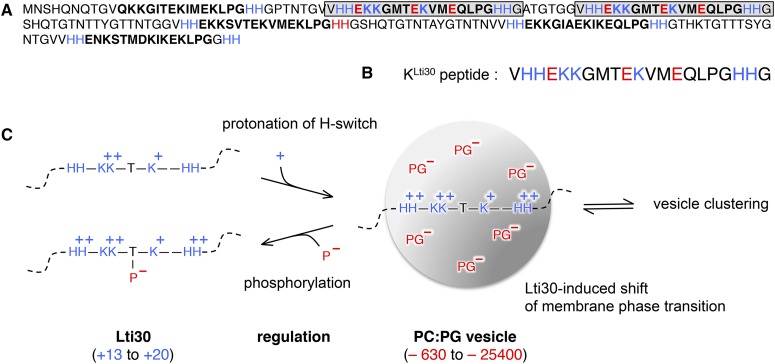
Figure 1.
Schematic image of disordered Lti30 binding to lipid (PC:PG) vesicles. Blue represents a positive charge and red represents a negative charge. A, Sequence of Lti30 with K segments (boldface) and the positions of the K segments matching the KLti30 peptide (gray boxes). B, Sequence of the KLti30 peptide. C, Binding equilibrium between Lti30 and lipid vesicles, where attraction is influenced by charge. Binding is promoted by protonation of the flanking His pairs (Eriksson et al., 2011) and down-regulated by phosphorylation of the K segment Thr residues (Eriksson et al., 2011). Upon Lti30 binding, the membrane phase decreases 2.5°C and the vesicles cluster into macroscopic aggregates (Eriksson et al., 2011). For size reference, Lti30 has an extended length of 20 nm and the diameter of the vesicles is 100 nm. PC, Phosphatidylcholine; PG, phosphatidylglycerol.
Table I. Summary of dehydrins observed previously to bind lipid vesicles and reported corresponding structural transitions.
| Protein | Plant | Type of Lipid | Gain of Structure | References |
|---|---|---|---|---|
| DHN1 | Zea mays | PA (100%) | α-Helix (CD) | Koag et al. (2003) |
| Lti30 | Arabidopsis | PC:PG (3:1) | α-Helix (NMR, FTIR) | – |
| ERD10 | Arabidopsis | PC:PS (1:1) | Disordered (CD) | Kovacs et al. (2008) |
| ERD14 | Arabidopsis | PC:PS (1:1) | Disordered (CD) | Kovacs et al. (2008) |
| TsDHN1 | Thellungiella salsuginea | PC:PS:PI (33:47:20) | β-Strand, α-helix (FTIR, CD) | Rahman et al. (2010) |
| TsDHN2 | Thellungiella salsuginea | PC:PS:PI (33:47:20) | β-Strand, α-helix (FTIR, CD) | Rahman et al. (2010) |
| K2 | Vitis riparia | PC:PA (1:1) | α-Helix (CD) | Clarke et al. (2015) |
In this study, we shed new light on this issue by identifying the generic physical-chemical features of the membranes that govern Lti30 binding, its dynamics, and, at atomic level resolution, the structural changes of the K segments that accompany this membrane binding (Fig. 1). The results show that, indeed, the anionic charges of the lipid head groups electrostatically capture the positively charged Lti30 K segments to establish a dynamic binding equilibrium. In the unbound state, the K segment remains fully disordered, whereas in the bound state, it turns into a fixed amphipathic helix that floats on the membrane surface. Such direct observation of binding-mediated folding of a K segment provides not only a strict structural base for elucidating stress-defense mechanisms but also points to the possibility that the functional repertoire of the dehydrin proteins is larger than previously anticipated.
RESULTS
Lti30 Shows Electrostatic Recognition of Lipid Head Groups
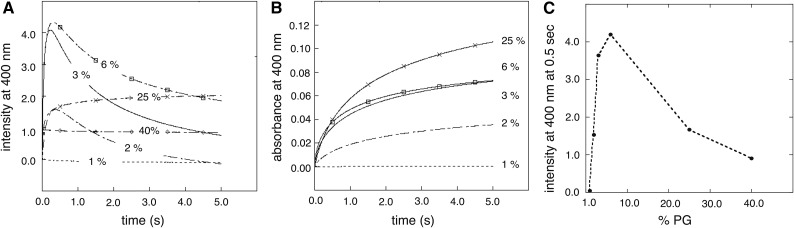
Previous data showed that Lti30 interacts with highly negatively charged vesicles (PC:PG ratio of 3:1; i.e. 25% negatively charged) and assemble these into large clusters. It was further shown that the binding is electrostatic and regulated by the protonation of His residues in a pH-dependent way (Eriksson et al., 2011). Full-length Lti30 binds and aggregates lipid vesicles upon His protonation, starting at pH below the pKA value of the bound state that is shifted to well above 8 (Eriksson et al., 2011). Accordingly, the pH titration of Lti30 gives a binding midpoint around pH 7.5, where 50% of the vesicle aggregation effect is manifested. Because of this flexibility in achieving suitable binding regimes in vitro, we opted here for pH 6.3 as the standard condition. The advantage is that pH 6.3 is close to the pKA of the His residues in unbound Lti30. The other pH of mechanistic interest is around the His pKA in the membrane-bound state (i.e. above pH 8), but this is too high to elucidate physiological action. In previous studies, we examined the Lti30-lipid binding from a His perspective by varying the pH (Eriksson et al., 2011), and here we expand the analysis by also varying the membrane charge. Large unilamellar vesicles (LUVs) were composed of DOPG (1%–40%) in the background of the neutral zwitter-ionic DOPC (99%–60%) lipids. The vesicles were rapidly mixed with Lti30 in a stopped-flow spectrophotometer to a final concentration of 0.5 mm lipids and 3.5 μm protein at pH 6.3, and the scattering effect of the vesicle clusters formed was monitored either as absorbance or fluorescence.
As seen in Figure 2, 2% negatively charged lipids are adequate for Lti30 to bind and cluster the vesicles. The intensity of the vesicle clustering increases when the negative net charge of the vesicles is elevated to 3% but seems to start to level off at 6% (Fig. 2). Hence, below binding saturation, the binding is proportional to the acidic charges of the vesicles. At a high negative net charge of the vesicles (above 6%; i.e. at 25% and 40% DOPG), the stopped-flow absorbance/fluorescence signal changes character. This can be explained by repulsion between the negative lipid head groups that interfere with the cluster reactions. The large clusters formed due to Lti30 binding are detectable with a light microscope, and images of the clusters further support the stopped-flow data (Fig. 3). The conclusion is that Lti30 has a strong affinity for low concentrations of negative lipids and needs only a few negative lipids to bind and cluster vesicles. This indicates that Lti30 also can bind to different cellular membranes in vivo, since most of these contain higher concentrations of negatively charged phospholipids than tested here (Uemura et al., 1995). The proposition seems even more plausible because the data presented here show vesicle clustering, a reaction secondary to lipid binding, and it is not unlikely that binding of Lti30 could take place at even lower vesicle charges.
Figure 2.
Stopped-flow data of Lti30 binding and clustering of PC:PG lipid vesicles (LUVs). Lti30 at 3.5 μm was rapidly mixed with 0.5 mm PC:PG vesicles at a 1:142 protein:lipid ratio. Binding of positively charged Lti30 promotes vesicle clustering, monitored by scattering at 400 nm. The process depends on the LUV net negative charge, which was varied between PG (1%–40%) and PC (99%–60%). A, Scattering at 400 nm. B, Absorbance at 400 nm. C, Scattering amplitude at 400 nm at 0.5 s, indicating two binding modes.
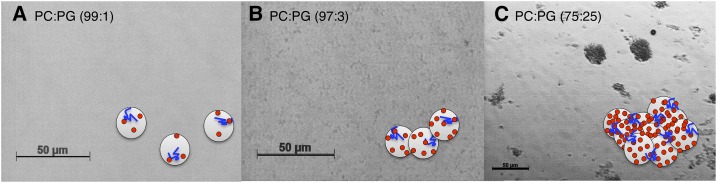
Figure 3.
Light microscopy analysis of the Lti30-induced clustering of PC:PG LUVs. Lti30 (14 μm) was used in the presence of LUVs (1.4 mm) with 1%, 3%, or 25% PG at pH 6.3. The protein:lipid ratio is 1:100. The schematic cartoons show a reductionist interpretation of the data: Lti30 (blue) and LUVs with increasing content of negatively charged PG (red). A, Lti30 and PC:PG (99:1): no visible clusters. B, Lti30 and PC:PG (97:3): small dispersed clusters less than 5 μm. C, Lti30 and PC:PG (75:25): clear macroscopic clusters larger than 5 μm (reduced magnification).
Identification and His-Flank Regulation of the Membrane-Binding Epitope
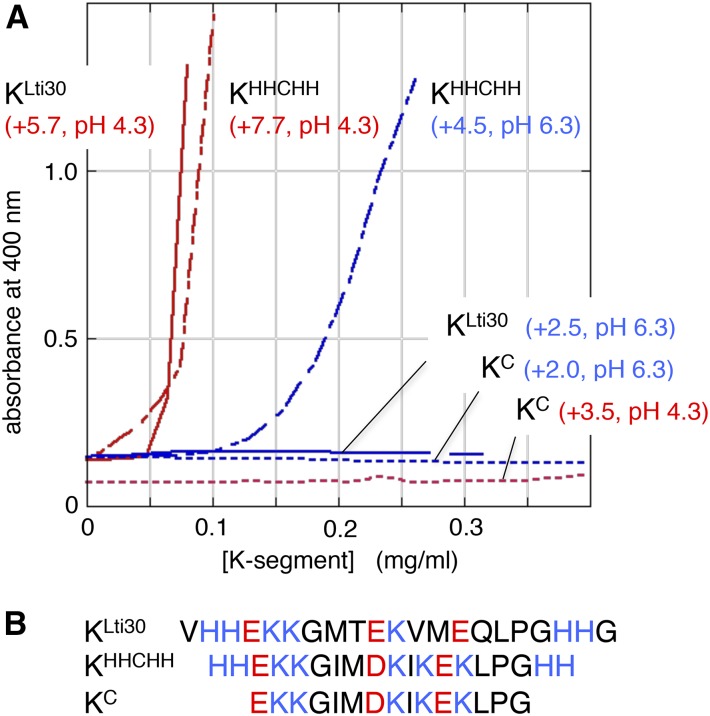
In previous studies (Eriksson et al., 2011; Petersen et al., 2012), it was demonstrated that the membrane binding of dehydrins is chiefly driven by the ubiquitous K segments (Figs. 1 and 4). The consensus sequence for these K segments is EKKGIMDKIKEKLPG, with slight variations across the protein family (Close, 1996; Graether and Boddington, 2014). We note here that the six K segments of Lti30 deviate from the consensus K segment by being less charged: the Lti30 segments are close to neutral at pH 7, whereas the consensus segment shows a net charge of +2 (Figs. 1 and 4). Moreover, the K segments of Lti30 are all flanked by His residues. These His flanks occur to varied extents across the dehydrin family and have been implicated as pH switches in the regulation of membrane binding (Eriksson et al., 2011). In essence, the increased positive charge from protonated His flanks activates binding, whereas deprotonation leads to dissociation (Eriksson et al., 2011). To examine the action of such regulation in Lti30, we constructed three peptide models: (1) the consensus K segment (KC); (2) the consensus K segment with flanking His residues (KHH-C-HH); and (3) the K segment of Lti30 with flanking His residues (KLti30; Fig. 4). The latter occurs twice in the Lti30 sequence and differs only marginally from the protein’s remaining four K segments (Fig. 1), whose charges vary in the same range as KLti30. Interestingly, these results show that both KHH-C-HH and KLti30 bind membranes in a pH-dependent manner, whereas KC that lacks the His flanks fails to bind in this model system (Fig. 4). Accordingly, the His flanks seem to be a functional feature of the K segment by providing extra positive charges to boost the interaction with the negatively charged membrane. Sequence comparison shows, consistently, that His flanks often are colocalized with the K segments across the dehydrin family, pointing to the possibility that the regulation of membrane binding by protonation is a widespread feature of the stress response (Eriksson et al., 2011). For further discussion of His flanks and the overall atypical positive net charge of dehydrins, see Eriksson et al. (2011). From the binding data in Figures 2 and 4, we conclude that the K segments of Lti30 play a key role in the vesicle binding of the full-length protein and that this binding is modulated both by the negative charge density of the membrane and the protonation state of the flanking His residues.
Figure 4.
Vesicle clustering by titration of various K-segments correlates to peptide net charge. Clustering of lipid vesicles was measured by absorbance at 400nm and peptide net charge was calculated at http://protcalc.sourceforge.net. A, Titration of different K-segments into 0.5 mM PC:PG vesicles (1:3) at pH 6.3 (blue lines) and pH 4.3 (red lines). The vesicle clustering follows peptide net charge. KLti30 represent the dominating type of K-segment in Lti30, KC is the consensus K-segment and KHHCHH is KC with the addition of histidine flanks. KC fail to bind vesicles regardless of pH, showing the regulatory impact of positively charged histidine flanks. B, Peptide sequences, with positively and negative residues in blue and red, respectively.
Lti30 Changes Structure upon Membrane Binding as Measured by Fourier Transform Infrared Spectroscopy
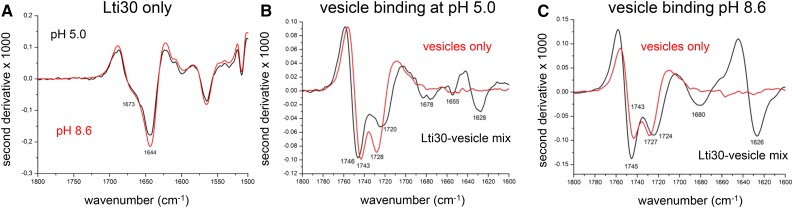
As vesicle clustering causes high background noise in the UV region, the structural response of Lti30 upon membrane association cannot be followed by conventional techniques like CD. Instead, we use here Fourier transform infrared (FTIR) spectroscopy, which targets conformation-sensitive vibrations associated with much longer wavelengths in the infrared regime. Following standard procedures, we use the second derivative of the absorption spectrum to identify peak positions, as this enables easier differentiation between overlapping bands. The negative bands in the second derivative spectra correspond to the component bands in the original absorption spectra. Figure 5 shows the second derivative spectra of the amide I′ vibrations of the peptide backbone, which report on secondary structure. A strong band at 1,644 cm−1 and a weak band at 1,673 cm−1 at both pH 5 and 8.6 indicate random coil structure (Fig. 5A). When lipid vesicles are added, the protein amide I′ spectrum remains largely unchanged, with both the main band near 1,644 cm−1 and the shoulder near 1,673 cm−1 being present. However, structural changes become obvious upon subtraction of the protein spectrum from the spectrum of the protein-vesicle mixture. Subtractions of the second derivative spectra show negative features near 1,680 and 1,630 cm−1, indicating additional absorption when Lti30 interacts with membranes (Fig. 5, B and C, black traces).
Figure 5.
FTIR spectra of Lti30 only and mixed with PC:PG vesicles. A, Second derivative infrared absorption spectra of Lti30 alone in the amide I region at pH 5 (black) and pH 8.6 (red). B and C, Corresponding spectra of vesicles alone (red line) and the Lti30-vesicle mix (black line) at pH 5 and 8.6, respectively.
These changes can be perceived in two ways: (1) as the appearance of distinct absorption bands near 1,680 and 1,630 cm−1; and (2) as a modification of the structures sampled in the random coil state of Lti30, leading to a broadening of the main band at 1,644 cm−1 and an enhanced shoulder at 1,673 cm−1. Both cases indicate that the ensemble of Lti30 structures, or the range of interaction strengths, alters upon interaction with vesicles. For example, interactions with protein carbonyls might be strengthened due to interactions with charges of the lipid head groups. Regarding the nature of the ensemble alteration, the additional absorption near 1,680 cm−1 is characteristic for turns, short helices, or the high-wave-number component of antiparallel β-sheets (Nevskaya and Chirgadze, 1976; Goormaghtigh et al., 1994; Barth and Zscherp, 2002) and that near 1,630 cm−1 is characteristic for β-sheets or solvated α-helices (Martinez and Millhauser, 1995; Reisdorf and Krimm, 1996; Williams et al., 1996; Yoder et al., 1997; Barth and Zscherp, 2002). Assignment of the spectral changes to β-sheet formation is disfavored by the generally low susceptibility of monomeric Lti30 to form antiparallel β-sheets (Battaglia et al., 2008; Koag et al., 2009; Thalhammer et al., 2010). At pH 5, we observe the appearance of a signal at 1,655 cm−1, which is characteristic of α-helices as found in globular proteins (Goormaghtigh et al., 1994; Barth and Zscherp, 2002). This 1,655 cm−1 feature is in agreement with the notion that Lti30 partially folds into regular, dehydrated helical segments upon interaction with vesicles.
Figure 5, B and C, also shows the second derivative spectra of the vesicles alone (red traces), which are distinctly different from those of the protein-vesicle mixtures. In particular, differences are observed at 1,750 to 1,700 cm−1, where the carbonyl-stretching vibrations of the lipids absorb. These vibrations are sensitive to hydrogen bonding and other parameters that affect the interfacial region of the lipids (Lewis and McElhaney, 2013). The band above 1,740 cm−1 stems from non-hydrogen-bonded C=O groups, while the band below 1,730 cm−1 comes from hydrogen-bonded C=O groups. When protein is present, the bands are farther apart. The downshift of the band below 1,730 cm−1 can be explained by stronger hydrogen-bonded C=O groups. Moreover, the Lti30 interaction decreases the amplitude of the band below 1,730 cm−1. Conclusions regarding the number of hydrogen-bonded C=O groups, however, cannot be based on amplitudes in second derivative spectra alone, as these also depend on the bandwidths of the component bands. Therefore, we analyzed baseline-corrected absorption spectra in two ways: (1) we integrated the two sides of the lipid C=O band (i.e. above 1,645 and below 1,625 cm−1 for non-hydrogen-bonded and hydrogen-bonded groups, respectively); and (2) we fitted the lipid band with three component bands near 1,723, 1,736, and 1,646 cm−1. The results show that the changes in band areas were only a few percent, indicating that there is no significant change in the number of hydrogen-bonded lipid carbonyls upon the addition of Lti30. The fits also confirm that the low-wave-number band of hydrogen-bonded C=O groups is downshifted in the presence of protein. Accordingly, in the absence of lipid vesicles, Lti30 displays the characteristic fingerprint of a fully disordered chain (Fig. 5A), consistent with previous reports (Mouillon et al., 2006, 2008). Then, upon interaction with DMPC:DMPG vesicles, Lti30 undergoes a conformational shift coupled to perturbation of the lipid bilayer (Fig. 5, B and C). The protein perturbs the lipid bilayer as demonstrated by spectral changes of the lipid carbonyls and causes stronger hydrogen bonding to these groups.
NMR-Monitored Folding of the Conserved K Segment Flanked by His Residues
To map out at atomic resolution the structural changes accompanying the membrane binding of Lti30, we used NMR. As a means to optimize structural resolution, we focused on the interaction between the KLti30 segment peptide with flanking His residues (VHHEKKGMTEKVMEQLPGHHG; Eriksson et al., 2011), which has been shown previously to drive the Lti30-membrane association, and bicelles (i.e. mixed micelles of DMPC/DMPG lipids and the detergent 1,2-dihexanoyl-sn-glycero-3-phosphocoline [DHPC]). Under the conditions in these experiments, these bicelles form disc-shaped ellipsoids with a relatively flat lipid bilayer surrounded peripherally by DHPC molecules (Andersson and Mäler, 2005; Lind et al., 2008). The free KLti30 is soluble in buffer and gives a good one-dimensional 1H-NMR spectrum, typical for a fully disordered peptide (Fig. 6). Upon the addition of bicelles, however, the chemical shift signatures of the KLti30 segment spectra undergo distinct alterations consistent with binding (Fig. 6). Notably, these spectra show no peaks corresponding to those of the isolated K segment, meaning either that 100% of the peptides are bound to the bicelles or that the peptides are in fast, dynamic exchange between free and bound conformations where the observed chemical shifts are the population-weighted averages. In support of a scenario with dynamic exchange between free and bound peptide, the NMR spectra in bicelle solution show quite narrow line width. In contrast, a homogenous population of tightly bound species would display significant line broadening, as the protein would be slaved to the slower motions of the large bicelles. To clarify this issue, we determined the translational diffusion coefficient (Dt) of the isolated KLti30 segment in buffer as well as in bicellar solution. As the magnitude of Dt is linked directly to the size of the diffusing object, small unbound KLti30 segments are expected to diffuse faster ( ) than the much larger bicelles (
) than the much larger bicelles (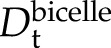 ). Thus, if all KLti30 segments are firmly bound to the bicelles (i.e. codiffuse), Dt should be the same for both (i.e. approximately
). Thus, if all KLti30 segments are firmly bound to the bicelles (i.e. codiffuse), Dt should be the same for both (i.e. approximately 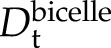 ). In the case of a dynamic exchange, on the other hand, the observed Dt for the KLti30 segment (
). In the case of a dynamic exchange, on the other hand, the observed Dt for the KLti30 segment (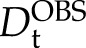 ) is the population-weighted mean of the diffusion of free peptide and the bicelle. The population of bound K segments (pbound) at every given time point then can be calculated from
) is the population-weighted mean of the diffusion of free peptide and the bicelle. The population of bound K segments (pbound) at every given time point then can be calculated from
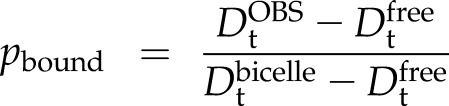 |
In 100% D2O at 25°C, the unbound KLti30 segment yields 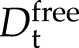 = 1.42 ± 0.01 10−10 m2 s−1 and the bicelles yield
= 1.42 ± 0.01 10−10 m2 s−1 and the bicelles yield 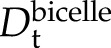 = 0.48 ± 0.01 10−10 m2 s−1, as determined with PFG NMR diffusion experiments (Fig. 5). The unbound KLti30 segment shows a diffusion coefficient that is in good agreement with the expected value for an unfolded peptide in D2O (Danielsson et al., 2002). In bicelle solution, however, the KLti30 segment yields
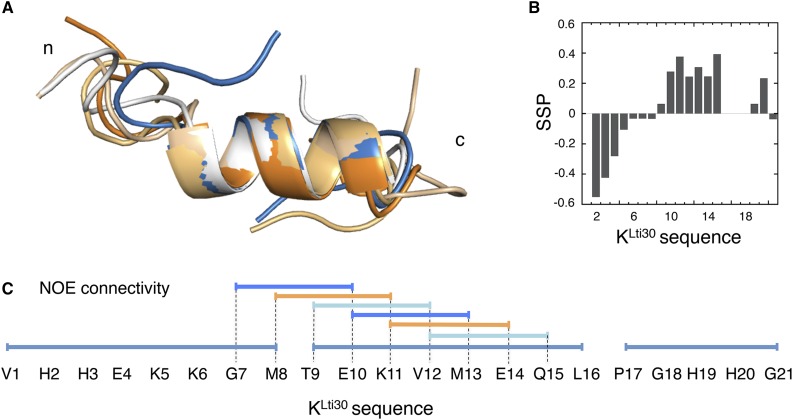
= 0.48 ± 0.01 10−10 m2 s−1, as determined with PFG NMR diffusion experiments (Fig. 5). The unbound KLti30 segment shows a diffusion coefficient that is in good agreement with the expected value for an unfolded peptide in D2O (Danielsson et al., 2002). In bicelle solution, however, the KLti30 segment yields 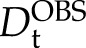 = 0.97 ± 0.03 10−10 m2 s−1, suggesting a dynamic equilibrium where, at each moment, 48% of the peptides are bound to the bicelle. Thus, the peptide is in unremitting exchange between the bound and free states, which is fast on the NMR time scale, both on the chemical shift time scale and the diffusion time scale. Although the chemical shifts induced upon bicelle binding are moderate at the level of population average (Fig. 6), we conclude from the diffusion data that the chemical shift perturbations of the bound population are twice the observed values and, consequently, in line with the folding of the KLti30 segment into an ordered structure. To determine at atomic resolution the structure of the membrane-bound KLti30, we used deuterated bicelles and two-dimensional NMR. The results reveal six αHi-NHi+3 nuclear Overhauser effect (NOE) couplings in the central part of the KLti30 segment (Fig. 6C), constraining residues Gly-7 to Gln-15 into a well-ordered α-helix, when bound to bicelles (Figs. 6 and 7).
= 0.97 ± 0.03 10−10 m2 s−1, suggesting a dynamic equilibrium where, at each moment, 48% of the peptides are bound to the bicelle. Thus, the peptide is in unremitting exchange between the bound and free states, which is fast on the NMR time scale, both on the chemical shift time scale and the diffusion time scale. Although the chemical shifts induced upon bicelle binding are moderate at the level of population average (Fig. 6), we conclude from the diffusion data that the chemical shift perturbations of the bound population are twice the observed values and, consequently, in line with the folding of the KLti30 segment into an ordered structure. To determine at atomic resolution the structure of the membrane-bound KLti30, we used deuterated bicelles and two-dimensional NMR. The results reveal six αHi-NHi+3 nuclear Overhauser effect (NOE) couplings in the central part of the KLti30 segment (Fig. 6C), constraining residues Gly-7 to Gln-15 into a well-ordered α-helix, when bound to bicelles (Figs. 6 and 7).
Figure 6.
Structural analysis of membrane-bound KLti30 peptide by NMR. A, The five best structures of bicelle-bound KLti30 peptide from 1H-NMR constraints, where the central nine residues of the peptide adopt a fixed α-helix, whereas the N and C termini are more disordered. B, Secondary structure propensity (SSP) from 1Ha, 1HN, and 1Hb chemical shifts, where positive values show the induced α-helix in the central part of the peptide. Negative secondary structure propensity values indicate extended conformation in the N-terminal region of the peptide. C, Determined NOE connectivity between peptide residues along the KLti30. The ordered n+3 couplings within the residue segment Gly-7 to Gln-15 are the hallmark of the α-helical structure.
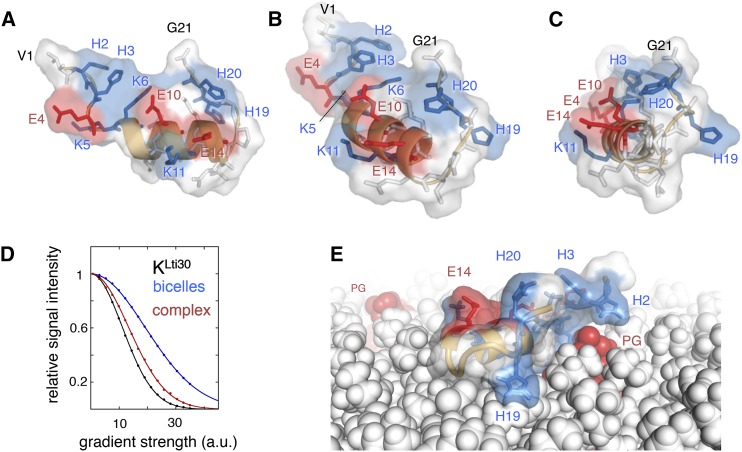
Figure 7.
Atomic resolution structure of the membrane-binding domain of Lti30. A to C, Atomic model of the membrane-binding KLti30 segment of Lti30, in side, diagonal, and front views, respectively. Positively charged residues are in blue, negatively charged residues are in red, and the hydrophobic side of the amphipathic α-helix faces downward. D, NMR diffusion data of KLti30 alone, bicelles alone, and KLti30 mixed with bicelles, verifying KLti30 micelle binding. a.u., Arbitrary units. E, Model of the positioning of the His-flanked KLti30 segment into a lipid bilayer (PC:PG 98:2 mix), using periodic boundary conditions. Consistent with the binding data in Figures 2 to 5, the positive side chains H2, H3, K5, and K6 coordinate the negative charge of a PG head group.
The secondary chemical shifts that report on secondary structure formation (Marsh et al., 2006) show approximately 40% fully formed helix at any given time point, in line with a dynamic equilibrium between the bound and free states, in good agreement with diffusion data. Interestingly, this helical structure shows archetypical amphipathic characteristics, with one polar and one hydrophobic side stretching along its length. The amphipathic properties of the short helix together with the fast-exchanging dynamic equilibrium between bound and free peptides are consistent with the structured peptide positioned on the surface of the bicelle. Manually positioning the KLti30 segment structure into a lipid bilayer positions the α-helix parallel to the membrane plane, with the hydrophobic side chains anchoring into the apolar regions of the lipids and the polar/charged side chains facing the solvent and the hydrophilic environment of the lipid head groups (Fig. 7). The His residues of KLti30 are not part of the helix but are in good position to form electrostatic contacts with the lipids. The induced folding of the conserved KLti30 segment suggests a mechanism for reversible binding to negatively charged membranes. This reversible binding corresponds well to that observed for several other intrinsically disordered proteins upon binding specific protein targets (Jao et al., 2004; Ulmer and Bax, 2005; Ulmer et al., 2005). Moreover, a seemingly analogous helix induction was observed recently by NMR for the dehydrin K2 upon binding to SDS micelles (Clarke et al., 2015), pointing to the possibility that this mode of membrane interaction is a general feature of the conserved K segments.
DISCUSSION
Forces Driving Lti30-Membrane Binding
To maintain in-cell solubility and freedom of movement, biomolecules like proteins, membranes, and DNA/RNA as a rule carry a net negative charge (Kurnik et al., 2012). In the few cases where proteins violate this principle of general negative repulsion by being net positive, this has been linked to functional binding. Typical examples are the positively charged proteins associating with DNA/RNA (Ellenberger et al., 1992; Niessing et al., 2004) and also local distributions of positive side chains controlling transmembrane protein topology (von Heijne, 2006), membrane binding (McLaughlin and Murray, 2005; Moravcevic et al., 2010), molecular dyes (Lira-De León et al., 2013), and protein-protein interaction (Sinha and Smith-Gill, 2002; Müller et al., 2003; Bartels et al., 2011). In this context, it is intriguing that Lti30, as well as some other members of the dehydrin family, stand out not only as intrinsically disordered but also as net positively charged (Supplemental Table S1; Eriksson et al., 2011). The positive charges of these proteins are clustered in repetitive and highly conserved sequence segments (Fig. 1) that sometimes have the benefit of flanking His residues, allowing local charge tuning (Supplemental Table S1; Eriksson et al., 2011). Together with the regulation of membrane binding/net charge by phosphorylation (Eriksson et al., 2011) and metal binding (Svensson et al., 2000; Alsheikh et al., 2003), this sets the stage for an intricate physiological function. In this study, we focus on the interaction between Lti30 and membranes, addressing the protein’s putative function in membrane protection during environmental stress (Steponkus et al., 1998). The results show that the Lti30-membrane binding and macroscopic clustering of vesicles respond to the density of negatively charged DOPG lipids (Figs. 2 and 3). Accordingly, the interaction between Lti30 and membranes is partly controlled by electrostatics, where the affinity can be regulated by either protonation of the His residues flanking the protein’s K segments (Eriksson et al., 2011) or by changing the negative potential of the membrane. An interesting detail here is that Lti30 manages to cluster LUVs even upon increasing their DOPG content to 40%, and the intervesicular repulsion must be substantial (Fig. 2). Even though the spatial arrangement between protein and vesicles under these conditions is yet unclear, the vesicular integrity seems to be maintained in the clusters, as they do not leak calcein (Eriksson et al., 2011) and are readily redispersed upon deprotonation of Lti30 His residues at pH 9 (Supplemental Fig. S1). Also, Lti30-induced clustering of purified thylakoids is reversed by increased ionic strength (Supplemental Fig. S1). Taken together, these findings point to an efficient and responsive mode of operation, as would be expected if membrane binding is part of Lti30’s physiological action.
Structure of Membrane-Bound Lti30: The Full-Length Protein
Following general structure-function relationships, the question is, to what extent does the interaction between Lti30 and membranes involve specificity in terms of uniquely ordered protein structure? To shed light on this issue, we used FTIR spectroscopy to probe for the induction of secondary structure. An advantage of this technique is that it can resolve subtle rearrangements of the protein backbone in dense vesicle solutions where light scattering renders conventional CD unsuitable. The results show that the Lti30 structure changes from overall random coil to partly helical upon vesicle binding (Fig. 5). Coil-to-helix transitions, as a phenomenon, have been observed previously for dehydrins when dehydration was simulated with glycerol and polyethylene glycol (Mouillon et al., 2008) and Late Embryogenesis Abundant (LEA) proteins (Battaglia et al., 2008; Thalhammer et al., 2010) in desiccated samples, reflecting the generic way any protein would satisfy backbone hydrogen-bonding constraints in the absence of water (Tanford, 1970). Similarly, dehydrins and LEA proteins tend to increase their helical content when lowering the water potential by osmolytic titration (Battaglia et al., 2008; Mouillon et al., 2008). In our case, however, the situation seems different, as Lti30 forms helices upon binding to a putatively physiological target in aqueous solution. This indicates that the structural response of Lti30 yields functional specificity above that exerted by the electrostatics alone.
Structure and Dynamics of Membrane-Bound Lti30: KLti30
The molecular details of the membrane-induced folding of Lti30 are revealed by the NMR structure of the bicelle-associated KLti30 segment (Figs. 6 and 7). Corroborating FTIR data of the full-length protein, the NMR structure of the bicelle-bound peptide shows a well-ordered α-helix stretching locally along residues of the KLti30 segment (i.e. Gly-7 to Gln-15). Flanking this helical unit are the segments VHHEKK at the N-terminal end and LPGHH at the C-terminal end, neither of which indicates a fixed structural order (Fig. 6). The helical structure also comprises clear amphipathic sidedness, guiding the positioning of the K segment to the membrane surface (Figs. 6 and 7). Judging by the disordered character of the unbound KLti30 segment, this docking could then occur either by conformational selection of preformed helical segments or by induced folding upon binding, according to the fly-casting mechanism (Trizac et al., 2010). In either case, the NMR data show that the membrane-associated state is in rapid equilibrium (relaxation time faster than approximately 300 s−1) with the unbound disordered state of the KLti30 segment, indicating a swift dynamic response. Even if the NMR analysis was done under conditions where the KLti30 segment does not promote bicelle clustering, it is reasonable to assume that such sandwiching of the α-helical structure between membrane surfaces will significantly reduce its binding dynamics. Thus, the membrane clustering in Figure 3 and Supplemental Figure S2 may be promoted by the amphipathic KLti30 segment helix binding hydrophobically to one vesicle and electrostatically to the other, possibly with the aid of the pH-tunable, flanking His residues (Lee et al., 2005; Eriksson et al., 2011). Although this idea remains to be tested, it is clear from the ordered structure of the KLti30 segment in Figures 6 and 7 that Lti30 has potential for a higher degree of binding specificity and diversity of targets than were explored in this study. How such specificity can be achieved is exemplified by the zinc fingers (Klug, 2010), protein-protein interaction (Matthews et al., 2009), and the YscU protein (Weise et al., 2014).
Implications for Biological Function
In addition to the ubiquitous K segment, some dehydrins also contain conserved segments such as the Y and S segments; the final segment composition for each dehydrin also gives the subgroup (i.e. K, SK, YSK, KS, and YK; Close, 1996). In line with a broad functional repertoire of the dehydrin proteins, it has been observed that different combinations of conserved segments are recruited at different types of physiological stresses: lowered temperature triggers the expression of K, SK, and KS subfamily dehydrins (Nylander et al., 2001; Graether and Boddington, 2014), whereas drought or increased salt exposure triggers the expression of YSK dehydrins (Nylander et al., 2001; Graether and Boddington, 2014). Considering also the ability to regulate the action of individual segments, the dehydrins may be seen functionally as a permuted set of Swiss Army knives (Alexander et al., 2013), expressed as needed and adjusted to their specific tasks by protonation (Eriksson et al., 2011), posttranslational phosphorylation (Alsheikh et al., 2003; Jiang and Wang, 2004), and metal binding (Svensson et al., 2000; Hara et al., 2005). When it comes to the molecular mechanism, it is suggested in this study that the K segment fulfills a role in attaching to membranes (Fig. 6). Regarding the role of the other types of segments, there is yet no structural information at hand. Even so, the S segment has been identified as a key phosphorylation site (Plana et al., 1991; Graether and Boddington, 2014) that not only modulates the protein global charge but also enables Ca2+ binding (Alsheikh et al., 2005). Phosphorylation of the S segment is further found to spatially relocate the dehydrin action by moving the proteins out of the cell nucleus (Riera et al., 2004). The role of the Y segment, on the other hand, is currently more difficult to envisage. From sequence BLAST, it is notable that the Y segment shows similarity to the ATP-binding motifs of the chaperones GroEL and GroES (Martin et al., 1993), hinting at a role in molecular recognition. The dehydrin MtCAs31, which comprises two Y and four K segments, likewise has been observed to interact with the stress-associated ICE transcription factor, yielding decreased numbers of stomata in leaves (Xie et al., 2012; Peng et al., 2014). In addition to the K, S, and Y segments, some dehydrins also contain H repeats found to control homodimerization (Hernández-Sánchez et al., 2014) as well as charge segments implicated in nuclear targeting and DNA binding (Hara et al., 2005; Rosales et al., 2014). Interestingly, these charge segments show further sequence similarity to a linker region in HSP90 (Mouillon et al., 2006).
Several physical dimensions of the cell will be affected by desiccation, such as the shape and proximity of membrane structures. As a consequence, desiccation also is expected to alter the dynamics of membrane fusion and budding, distort phase transitions, and contribute to topological frustration. By binding to membranes, the role of Lti30 may be to interfere in the progression of any of these processes in order to keep the membranes biologically viable. Regardless of functional mode, the elements of recognition with the cellular target are likely to be the conserved segments, such as the K segment of Lti30 (Fig. 7). Taken together, the diversity of conserved sequence segments among the dehydrins points to a complex and versatile biological function, evolvable in a simple modular fashion to meet new needs. A pressing task now is to establish how the intriguing behavior of dehydrins in vitro translates to the crowded conditions in vivo, where other putative interaction partners will likely compete with, and modulate, membrane binding. Also, it remains to be determined the extent to which the observed behavior in vitro reflects true functional traits or simply reports on the generic propensities of unstructured proteins (Thalhammer et al., 2014; Popova et al., 2015). In any case, the molecular clues emerging from this and other studies stand out as a solid base for mapping out in detail how this unique class of disordered proteins works, and can be rationally interfered with, in stressed plant cells.
MATERIALS AND METHODS
Protein Production
Expression, purification, and identification of the recombinant Arabidopsis (Arabidopsis thaliana) dehydrin Lti30 were performed as described by Svensson et al. (2000) with minor changes. Glycerol stocks of the Escherichia coli strain were made, and 150 μL was spread on Luria agar plates with 150 μg of ampicillin and grown at 37°C overnight. The cells were suspended and added to 2 L of Luria-Bertani medium containing 50 μg mL−1 ampicillin and kept at 37°C. Expression was induced at an optical density at 600 nm of 0.6 to 0.7 by adding 1 mm isopropyl-β-d-thiogalactopyranoside and kept at 23°C overnight. Cells were harvested by centrifugation at 6,000 rpm for 15 min. The pellet from 1-L cultures was suspended in 25 mL of 20 mm Na2HPO4, pH 7.2, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and one tablet of Complete (Roche). Cells were sonicated for five 1-min periods on ice followed by centrifugation at 18,000 rpm for 30 min. The supernatant was placed in an 80°C water bath for 30 min to precipitate heat-denatured proteins and then centrifuged at 18,000 rpm for 30 min. Lti30 was purified by metal ion affinity chromatography. The supernatant from heat precipitation was diluted 1:2 with 20 mm Na2HPO4, pH 7.2, 1.88 m NaCl, and 1 mm phenylmethylsulfonyl fluoride. The sample was loaded on a 5-mL HiTrap IDA-Sepharos column (GE Healthcare) charged with 7 mL of 3 mg mL−1 CuSO4. The column was equilibrated with 5 volumes of 20 mm Na2HPO4, pH 7.2, and 1 m NaCl. The same buffer (40 volumes) was used to wash off unbound sample from the column. Fractions of 5 mL were collected for analysis during the whole run. Elution was performed with 2 m NH4Cl in 20 mm Na2HPO4, pH 7.2, and 1 m NaCl in one step. The column was then equilibrated with 10 volumes of 20 mm Na2HPO4, pH 7.2, followed by elution of the copper with 10 mm EDTA in 20 mm Na2HPO4, pH 7.2. Precipitation of protein was performed with 80% (NH4)2SO4 overnight, and protein was collected by centrifugation at 18,000 rpm for 45 min. Lti30 pellet was suspended with 2.5 mL of 5 mm MES, pH 6.3, and, in the case of FTIR spectroscopy, 2.5 mL of D2O. The protein was desalted on two PD-10 columns (GE Healthcare). Purity was analyzed by the Ready Gel SDS-PAGE system (Bio-Rad). Protein quantification was measured with the bicinchoninic acid assay (Sigma-Aldrich).
Preparation of LUVs and Bicelles
All lipids were purchased from Avanti Polar Lipids. LUVs of DOPC:DOPG were prepared with an extrusion method. The lipids were dissolved in chloroform, and lipid mixtures were dried under a gentle flow of liquid nitrogen for 3 to 4 h. The lipid film was solved in 5 mm MES, pH 6.3, and then vortexed for 10 min; for FTIR experiments, the lipids were solved in 10 mm potassium phosphate in D2O at pH 5 and 8.6. Five freeze-thaw cycles in liquid nitrogen to reduce lamellarity followed, and then the lipid solution was extruded 20 times through a hand-driven extruder with 0.1-μm pore-size polycarbonate filter. The measurements of size distribution of the LUVs produced were done on a ALV/CGS-3 compact goniometer system (ALV) with a helium-neon laser (632.8 nm), with a scattering angle of 150°. Bicelles for NMR measurements were prepared with DHPC as a detergent. DMPC:DMPG (4:1) lipids were used to provide the bilayer of the bicelles. Bicelles with q = 0.3 (q = {[DMPC]+[DMPG]}/[DHPC], where q equals the molar ratio of lipids and detergents) were prepared by mixing DMPC:DMPG (4:1) with 20 mm potassium phosphate, pH 6.3. The mixture was vortexed followed by centrifugation at 13,300 rpm for 1 min, and this cycle was repeated until the solution looked homogenous. DHPC was added, and the centrifugation cycle was repeated until a clear nonviscous lipid solution was obtained.
Thylakoid Membrane Preparation
Forty grams of spinach (Spinacia oleracea) and 100 mL of 0.3 m Suc, 5 mm MgCl2, and 50 mm sodium phosphate, pH 7.4, were placed in a cold mixer and mixed for 5 × 10 s. The solution was filtered and centrifuged at 3,000 rpm for 3 min at 4°C. The pellet was suspended in 30 mL of 0.3 m Suc, 5 mm MgCl2, and 50 mm sodium phosphate, pH 7.4, and centrifuged at 4,500 for 5 min at 4°C. The solution was homogenized in 30 mL of 5 mm MgCl2, 5 mm NaCl, and 10 mm sodium phosphate, pH 7.4, and centrifuged at 4,500 rpm for 5 min at 4°C. The pellet was homogenized in 12 mL of 0.1 m Suc, 5 mm MgCl2, 5 mm NaCl, and 10 mm phosphate, pH 7.4.
Stopped-Flow Measurements
Scattering of protein-lipid cluster formation was monitored on an SX.18-MV stopped-flow fluorimeter (Applied Photophysics). Excitation was at 400 nm, and all measurements were done at room temperature in 5 mm MES, pH 6.3. Protein concentration after mixing was 3.5 μm, and lipid concentration was 0.5 mm.
Absorbance Measurements
Absorbance measurements were performed on an Ultrospec 3300 pro (Amersham) at 400 nm and 25°C.
Light Microscopy Images
To examine light microscopy images, an inverted Zeiss Axiovert 40 CFL microscope equipped with a digital Aciocam ICc1 camera at 10× magnification was used. A solution of Lti30 (14 μm) together with vesicles (1.4 mm) was placed on a glass slide, and three images per droplet were examined.
FTIR Spectroscopy
All samples were prepared with 10 mm potassium phosphate in D2O at pH 5 or 8.6. Samples containing Lti30 had a concentration of 0.2 mm, and samples containing lipids of DOPC:DOPG (3:1) had a concentration of 8 mm. Samples of 3 μL were deposited on a CaF2 window with a trough of 55 μm, covered with a flat CaF2 window, and placed in the spectrometer. Infrared spectra were recorded with a Tensor 37 Fourier transform spectrometer (Bruker Optics) equipped with a liquid N2-cooled HgCdTe detector and continuously purged with CO2-free, dry air. Samples were mounted on a two-position sample shuttle, which allowed for interleaved acquisition of sample and background spectra. A wait time of 15 min was allowed after inserting the sample to ensure complete purging. All measurements were made at room temperature. Interferograms were recorded at a resolution of 2 cm−1, apodized using a three-term Blackman-Harris apodization function, and Fourier transformed with a zero-filling factor of 2. Sixteen consecutive interferograms were averaged to obtain a single sample or background spectrum. Infrared spectra were recorded and analyzed using the OPUS software from the instrument manufacturer. Second derivative spectra were calculated using a smoothing length of 13 data points (approximately 13 cm−1).
NMR
In all NMR experiments, we used an unlabeled synthesized peptide, and the experiments where performed at 298 K on a Bruker Avance 600-MHz spectrometer equipped with a room-temperature probe with z-gradients. To obtain proton assignments, we recorded two homonuclear TOCSY experiments with 80- and 100-ms Hartman-Hahn mixing times (Bax and Davis, 1985a) and two homonuclear NOESY experiments with 150- and 250-ms mixing times (Bax and Davis, 1985b; Hwang and Shaka, 1995). The NOESY experiments also were used for the identification of long-range contacts. All two-dimensional experiments were recorded with 512 increments in the indirect detected dimension and used excitation sculpting to reduce the water signal. Processing was performed in NMRPipe (Delaglio et al., 1995), and analysis was performed using Sparky (T.D. Goddard and D.G. Kneller, SPARKY 3, University of California, San Francisco).
The structure was generated with PEP-fold (Maupetit et al., 2009) using six experimentally determined (i, i+3) HαHN distances as constraints when generating the structure. The obtained coarse-grained structure was further energy minimized using the Yasara force field (Krieger et al., 2009), and the best five structures were selected as representative models for KLti30 peptide structure in membrane mimetica. The PDB representation of the double-layered lipid membrane was generated using the CHARMM-GUI membrane builder (Jo et al., 2007, 2008, 2009; Wu et al., 2014).
Diffusion experiments were performed in 100% D2O at 298 K. A list of 16 linearly spaced gradient strengths was used. Gradient pulse length was set to 5 ms, and diffusion time was set to 100 ms. Diffusion coefficients were obtained by integration of the aromatic side chain protons.
Estimation of the End-to-End Distance of the Vesicle-Bound Lti30
The size dimension of Lti30 located on a vesicle surface was estimated as the end-to-end distance of a self-avoiding random walk of a bead-on-a-string Cα model on a two-dimensional surface. The overall dimensions (e.g. radius of gyration) scales similarly to the end-to-end distance. Here, we model the protein by a chain of n = 193 spheres with 3.5-Å diameter, linked by 3.88-Å-long bonds. No overlap of the spheres is allowed. As Lti30 is an intrinsically disordered protein, the size is a distribution of sizes rather than a well-defined value. To estimate the distribution, we calculated 104 chains, using an in-house script, and found that the end-to-end distance, as expected, follow a skewed Gaussian distribution where the mean end-to-end distance is 7.5 ± 3.5 nm. In the vesicle-bound state, Lti30 coordinates the charged lipid head groups, and as these are sparsely and evenly distributed over the vesicle surface, we assume Lti30 to be significantly more extended than a random Gaussian chain. Therefore, to estimate the length distribution on such vesicles, we only included the 2% most extended conformations of the ensemble. This subensemble has a mean end-to end-distance of 20.4 ± 2.5 nm.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number Lti30: NP_190666.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effects of NaCl on thylakoid and LUV clustering by Lti30 and KLti30.
Supplemental Figure S2. Light microscopy analysis of the K segment-induced clustering of PC:PG LUVs.
Supplemental Figure S3. FTIR spectra of Lti30, lipid vesicles, or a mix of Lti30 and vesicles (raw data).
Supplemental Table S1. Summary of positive net charge among LEA proteins.
Supplementary Material
Acknowledgments
We thank Mikael Oliveberg for helpful discussions.
Glossary
- LUV
large unilamellar vesicle
- FTIR
Fourier transform infrared
- DHPC
1,2-dihexanoyl-sn-glycero-3-phosphocoline
Footnotes
This work was supported by the Knut and Alice Wallenberg Foundation (to J.D. and A.B.) and the Magnus Bergwall Foundation (to P.H. and J.D.).
Articles can be viewed without a subscription.
References
- Alexander CG, Jürgens MC, Shepherd DA, Freund SM, Ashcroft AE, Ferguson N (2013) Thermodynamic origins of protein folding, allostery, and capsid formation in the human hepatitis B virus core protein. Proc Natl Acad Sci USA 110: E2782–E2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsheikh MK, Heyen BJ, Randall SK (2003) Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J Biol Chem 278: 40882–40889 [DOI] [PubMed] [Google Scholar]
- Alsheikh MK, Svensson JT, Randall SK (2005) Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant Cell Environ 28: 1114–1122 [Google Scholar]
- Andersson A, Mäler L (2005) Magnetic resonance investigations of lipid motion in isotropic bicelles. Langmuir 21: 7702–7709 [DOI] [PubMed] [Google Scholar]
- Arai M, Sugase K, Dyson HJ, Wright PE (2015) Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci USA 112: 9614–9619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J, Van Dennsteele C, Dure L III (1988) Sequence and characterization of 6 Lea proteins and their genes from cotton. Plant Mol Biol 11: 277–291 [DOI] [PubMed] [Google Scholar]
- Bartels T, Choi JG, Selkoe DJ (2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477: 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A, Zscherp C (2002) What vibrations tell us about proteins. Q Rev Biophys 35: 369–430 [DOI] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A, Davis DG (1985a) Mlev-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson Imaging 65: 355–360 [Google Scholar]
- Bax A, Davis DG (1985b) Practical aspects of two-dimensional transverse noe spectroscopy. J Magn Reson Imaging 63: 207–213 [Google Scholar]
- Bomal C, Le VQ, Tremblay FM (2002) Induction of tolerance to fast desiccation in black spruce (Picea mariana) somatic embryos: relationship between partial water loss, sugars, and dehydrins. Physiol Plant 115: 523–530 [DOI] [PubMed] [Google Scholar]
- Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS (2007) Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci USA 104: 9650–9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Boschetti C, Walton LJ, Sarkar S, Rubinsztein DC, Tunnacliffe A (2007) Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc Natl Acad Sci USA 104: 18073–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MW, Boddington KF, Warnica JM, Atkinson J, McKenna S, Madge J, Barker CH, Graether SP (2015) Structural and functional insights into the cryoprotection of membranes by the intrinsically disordered dehydrins. J Biol Chem 290: 26900–26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ. (1996) Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant 97: 795–803 [Google Scholar]
- Crowe LM, Crowe JH (1992) Anhydrobiosis: a strategy for survival. Adv Space Res 12: 239–247 [DOI] [PubMed] [Google Scholar]
- Danielsson J, Jarvet J, Damberg P, Graslund A (2002) Translational diffusion measured by PFG-NMR on full length and fragments of the Alzheimer A beta(1-40) peptide: determination of hydrodynamic radii of random coil peptides of varying length. Magn Reson Chem 40: S89–S97 [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F (1998) Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 10: 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z (2001) The protein trinity: linking function and disorder. Nat Biotechnol 19: 805–806 [DOI] [PubMed] [Google Scholar]
- Dure L., III (1993) A repeating 11-mer amino acid motif and plant desiccation. Plant J 3: 363–369 [DOI] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 71: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Eriksson SK, Kutzer M, Procek J, Gröbner G, Harryson P (2011) Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 23: 2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormaghtigh E, Cabiaux V, Ruysschaert JM (1994) Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. I. Assignments and model compounds. Subcell Biochem 23: 329–362 [DOI] [PubMed] [Google Scholar]
- Graether SP, Boddington KF (2014) Disorder and function: a review of the dehydrin protein family. Front Plant Sci 5: 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Fujinaga M, Kuboi T (2005) Metal binding by citrus dehydrin with histidine-rich domains. J Exp Bot 56: 2695–2703 [DOI] [PubMed] [Google Scholar]
- Hara M, Shinoda Y, Tanaka Y, Kuboi T (2009) DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ 32: 532–541 [DOI] [PubMed] [Google Scholar]
- Hara M, Terashima S, Kuboi T (2001) Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J Plant Physiol 158: 1333–1339 [Google Scholar]
- Hernández-Sánchez IE, Martynowicz DM, Rodríguez-Hernández AA, Pérez-Morales MB, Graether SP, Jiménez-Bremont JF (2014) A dehydrin-dehydrin interaction: the case of SK3 from Opuntia streptacantha. Front Plant Sci 5: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang TL, Shaka AJ (1995) Water suppression that works: Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson A 112: 275–279 [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Jao CC, Der-Sarkissian A, Chen J, Langen R (2004) Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc Natl Acad Sci USA 101: 8331–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvet J, Danielsson J, Damberg P, Oleszczuk M, Gräslund A (2007) Positioning of the Alzheimer Abeta(1-40) peptide in SDS micelles using NMR and paramagnetic probes. J Biomol NMR 39: 63–72 [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang Y (2004) Beta-elimination coupled with tandem mass spectrometry for the identification of in vivo and in vitro phosphorylation sites in maize dehydrin DHN1 protein. Biochemistry 43: 15567–15576 [DOI] [PubMed] [Google Scholar]
- Jo S, Kim T, Im W (2007) Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2: e880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29: 1859–1865 [DOI] [PubMed] [Google Scholar]
- Jo S, Lim JB, Klauda JB, Im W (2009) CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys J 97: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. (2010) The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys 43: 1–21 [DOI] [PubMed] [Google Scholar]
- Koag MC, Fenton RD, Wilkens S, Close TJ (2003) The binding of maize DHN1 to lipid vesicles: gain of structure and lipid specificity. Plant Physiol 131: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koag MC, Wilkens S, Fenton RD, Resnik J, Vo E, Close TJ (2009) The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol 150: 1503–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs D, Kalmar E, Torok Z, Tompa P (2008) Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol 147: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins (Suppl 9) 77: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE (1996) Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci USA 93: 11504–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnik M, Hedberg L, Danielsson J, Oliveberg M (2012) Folding without charges. Proc Natl Acad Sci USA 109: 5705–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Martinez-Yamout MA, Dyson HJ, Wright PE (2010) Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry 49: 9964–9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Eyeson R, Cheever ML, Geng J, Verkhusha VV, Burd C, Overduin M, Kutateladze TG (2005) Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc Natl Acad Sci USA 102: 13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. (2008) Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9: 99–111 [DOI] [PubMed] [Google Scholar]
- Lewis RN, McElhaney RN (2013) Membrane lipid phase transitions and phase organization studied by Fourier transform infrared spectroscopy. Biochim Biophys Acta 1828: 2347–2358 [DOI] [PubMed] [Google Scholar]
- Lind J, Nordin J, Mäler L (2008) Lipid dynamics in fast-tumbling bicelles with varying bilayer thickness: effect of model transmembrane peptides. Biochim Biophys Acta 1778: 2526–2534 [DOI] [PubMed] [Google Scholar]
- Lira-De León KI, García-Gutiérrez P, Serratos IN, Palomera-Cárdenas M, Figueroa-Corona MdelP, Campos-Peña V, Meraz-Ríos MA (2013) Molecular mechanism of tau aggregation induced by anionic and cationic dyes. J Alzheimers Dis 35: 319–334 [DOI] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD (2006) Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci 15: 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Geromanos S, Tempst P, Hartl FU (1993) Identification of nucleotide-binding regions in the chaperonin proteins GroEL and GroES. Nature 366: 279–282 [DOI] [PubMed] [Google Scholar]
- Martinez G, Millhauser G (1995) FTIR spectroscopy of alanine-based peptides: assignment of the amide I′ modes for random coil and helix. J Struct Biol 114: 23–27 [DOI] [PubMed] [Google Scholar]
- Matthews JM, Bhati M, Lehtomaki E, Mansfield RE, Cubeddu L, Mackay JP (2009) It takes two to tango: the structure and function of LIM, RING, PHD and MYND domains. Curr Pharm Des 15: 3681–3696 [DOI] [PubMed] [Google Scholar]
- Maupetit J, Derreumaux P, Tuffery P (2009) PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res 37: W498–W503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Murray D (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438: 605–611 [DOI] [PubMed] [Google Scholar]
- Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA (2010) Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell 143: 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon JM, Eriksson SK, Harryson P (2008) Mimicking the plant cell interior under water stress by macromolecular crowding: disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol 148: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon JM, Gustafsson P, Harryson P (2006) Structural investigation of disordered stress proteins: comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol 141: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JJ, Lapko A, Ruckpaul K, Heinemann U (2003) Modeling of electrostatic recognition processes in the mammalian mitochondrial steroid hydroxylase system. Biophys Chem 100: 281–292 [DOI] [PubMed] [Google Scholar]
- Nevskaya NA, Chirgadze YN (1976) Infrared spectra and resonance interactions of amide-I and II vibration of alpha-helix. Biopolymers 15: 637–648 [DOI] [PubMed] [Google Scholar]
- Niessing D, Hüttelmaier S, Zenklusen D, Singer RH, Burley SK (2004) She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell 119: 491–502 [DOI] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45: 263–279 [DOI] [PubMed] [Google Scholar]
- Peng PH, Lin CH, Tsai HW, Lin TY (2014) Cold response in Phalaenopsis aphrodite and characterization of PaCBF1 and PaICE1. Plant Cell Physiol 55: 1623–1635 [DOI] [PubMed] [Google Scholar]
- Petersen J, Eriksson SK, Harryson P, Pierog S, Colby T, Bartels D, Röhrig H (2012) The lysine-rich motif of intrinsically disordered stress protein CDeT11-24 from Craterostigma plantagineum is responsible for phosphatidic acid binding and protection of enzymes from damaging effects caused by desiccation. J Exp Bot 63: 4919–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plana M, Itarte E, Eritja R, Goday A, Pagès M, Martínez MC (1991) Phosphorylation of maize RAB-17 protein by casein kinase 2. J Biol Chem 266: 22510–22514 [PubMed] [Google Scholar]
- Popova AV, Rausch S, Hundertmark M, Gibon Y, Hincha DK (2015) The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim Biophys Acta 1854: 1517–1525 [DOI] [PubMed] [Google Scholar]
- Puhakainen T, Hess MW, Mäkelä P, Svensson J, Heino P, Palva ET (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol 54: 743–753 [DOI] [PubMed] [Google Scholar]
- Rafalski JA. (2002) Plant genomics: present state and a perspective on future developments. Brief Funct Genomics Proteomics 1: 80–94 [DOI] [PubMed] [Google Scholar]
- Rahman LN, Chen L, Nazim S, Bamm VV, Yaish MW, Moffatt BA, Dutcher JR, Harauz G (2010) Interactions of intrinsically disordered Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 with membranes: synergistic effects of lipid composition and temperature on secondary structure. Biochem Cell Biol 88: 791–807 [DOI] [PubMed] [Google Scholar]
- Reisdorf WC Jr, Krimm S (1996) Infrared amide I′ band of the coiled coil. Biochemistry 35: 1383–1386 [DOI] [PubMed] [Google Scholar]
- Riera M, Figueras M, López C, Goday A, Pagès M (2004) Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc Natl Acad Sci USA 101: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales R, Romero I, Escribano MI, Merodio C, Sanchez-Ballesta MT (2014) The crucial role of Φ- and K-segments in the in vitro functionality of Vitis vinifera dehydrin DHN1a. Phytochemistry 108: 17–25 [DOI] [PubMed] [Google Scholar]
- Sigalov AB, Hendricks GM (2009) Membrane binding mode of intrinsically disordered cytoplasmic domains of T cell receptor signaling subunits depends on lipid composition. Biochem Biophys Res Commun 389: 388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N, Smith-Gill SJ (2002) Electrostatics in protein binding and function. Curr Protein Pept Sci 3: 601–614 [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Lynch DV (1989) Freeze/thaw-induced destabilization of the plasma membrane and the effects of cold acclimation. J Bioenerg Biomembr 21: 21–41 [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson J, Palva ET, Welin B (2000) Purification of recombinant Arabidopsis thaliana dehydrins by metal ion affinity chromatography. Protein Expr Purif 20: 169–178 [DOI] [PubMed] [Google Scholar]
- Tanford C. (1970) Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem 24: 1–95 [PubMed] [Google Scholar]
- Thalhammer A, Bryant G, Sulpice R, Hincha DK (2014) Disordered cold regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol 166: 190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A, Hundertmark M, Popova AV, Seckler R, Hincha DK (2010) Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. Biochim Biophys Acta 1798: 1812–1820 [DOI] [PubMed] [Google Scholar]
- Tompa P. (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27: 527–533 [DOI] [PubMed] [Google Scholar]
- Trizac E, Levy Y, Wolynes PG (2010) Capillarity theory for the fly-casting mechanism. Proc Natl Acad Sci USA 107: 2746–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL (1995) Cold acclimation of Arabidopsis thaliana: effect on plasma membrane lipid composition and freeze-induced lesions. Plant Physiol 109: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer TS, Bax A (2005) Comparison of structure and dynamics of micelle-bound human alpha-synuclein and Parkinson disease variants. J Biol Chem 280: 43179–43187 [DOI] [PubMed] [Google Scholar]
- Ulmer TS, Bax A, Cole NB, Nussbaum RL (2005) Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem 280: 9595–9603 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (2006) Membrane-protein topology. Nat Rev Mol Cell Biol 7: 909–918 [DOI] [PubMed] [Google Scholar]
- Vuzman D, Levy Y (2012) Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol Biosyst 8: 47–57 [DOI] [PubMed] [Google Scholar]
- Weise CF, Login FH, Ho O, Gröbner G, Wolf-Watz H, Wolf-Watz M (2014) Negatively charged lipid membranes promote a disorder-order transition in the Yersinia YscU protein. Biophys J 107: 1950–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S, Causgrove TP, Gilmanshin R, Fang KS, Callender RH, Woodruff WH, Dyer RB (1996) Fast events in protein folding: helix melting and formation in a small peptide. Biochemistry 35: 691–697 [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu EL, Cheng X, Jo S, Rui H, Song KC, Dávila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, et al. (2014) CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem 35: 1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang R, Qu Y, Miao Z, Zhang Y, Shen X, Wang T, Dong J (2012) Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol 195: 124–135 [DOI] [PubMed] [Google Scholar]
- Yoder G, Pancoska P, Keiderling TA (1997) Characterization of alanine-rich peptides, Ac-(AAKAA)n-GY-NH2 (n = 1-4), using vibrational circular dichroism and Fourier transform infrared: conformational determination and thermal unfolding. Biochemistry 36: 15123–15133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.