LEC2, ABI3, and LEC1 synergistically interact to activate an oleosin promoter.
Abstract
In Arabidopsis (Arabidopsis thaliana), transcriptional control of seed maturation involves three related regulators with a B3 domain, namely LEAFY COTYLEDON2 (LEC2), ABSCISIC ACID INSENSITIVE3 (ABI3), and FUSCA3 (ABI3/FUS3/LEC2 [AFLs]). Although genetic analyses have demonstrated partially overlapping functions of these regulators, the underlying molecular mechanisms remained elusive. The results presented here confirmed that the three proteins bind RY DNA elements (with a 5′-CATG-3′ core sequence) but with different specificities for flanking nucleotides. In planta as in the moss Physcomitrella patens protoplasts, the presence of RY-like (RYL) elements is necessary but not sufficient for the regulation of the OLEOSIN1 (OLE1) promoter by the B3 AFLs. G box-like domains, located in the vicinity of the RYL elements, also are required for proper activation of the promoter, suggesting that several proteins are involved. Consistent with this idea, LEC2 and ABI3 showed synergistic effects on the activation of the OLE1 promoter. What is more, LEC1 (a homolog of the NF-YB subunit of the CCAAT-binding complex) further enhanced the activation of this target promoter in the presence of LEC2 and ABI3. Finally, recombinant LEC1 and LEC2 proteins produced in Arabidopsis protoplasts could form a ternary complex with NF-YC2 in vitro, providing a molecular explanation for their functional interactions. Taken together, these results allow us to propose a molecular model for the transcriptional regulation of seed genes by the L-AFL proteins, based on the formation of regulatory multiprotein complexes between NF-YBs, which carry a specific aspartate-55 residue, and B3 transcription factors.
Genetic and molecular analyses have delineated a complex network of transcriptional regulators controlling gene expression programs essential to accomplish seed maturation in Arabidopsis (Arabidopsis thaliana; for review, see Kagaya et al., 2005b; Braybrook et al., 2006; To et al., 2006; Braybrook and Harada, 2008; Santos-Mendoza et al., 2008; Suzuki and McCarty, 2008; Zhang and Ogas, 2009; Berger et al., 2011; Sreenivasulu and Wobus, 2013). This network comprises activators and repressors regulating phase transitions between embryogenesis, seed maturation, and vegetative development. Chromatin modifications also are involved, which repress the expression of the activators in late-maturing and germinating seeds as well as in vegetative organs (Suzuki and McCarty, 2008; Zhang and Ogas, 2009; Berger et al., 2011).
Among the key transcriptional activators of the maturation phase are three founding members of the B3 domain family, namely ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2; Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001). These AFL (ABI3/FUS3/LEC2) members act in concert with proteins homologous to the NF-YB subunit of the CCAAT box-binding protein called LEC1 and LEC1-LIKE (Lotan et al., 1998; Kwong et al., 2003; Cagliari et al., 2014; Hilioti et al., 2014). These genes will be named L-AFL (as proposed by Jia et al. [2013]). Additionally, basic leucine zippers (bZIPs) such as bZIP53 and bZIP67 (Alonso et al., 2009; Mendes et al., 2013), the MADS domain transcription factor AGAMOUS-LIKE15 (Thakare et al., 2008; Zheng et al., 2009), the MYB transcription factors MYB115 and MYB118 (Wang et al., 2009; Barthole et al., 2014), the WD repeat motif-containing EMB2757/TANMEI protein (Yamagishi et al., 2005), and the homeobox GLABRA2 (Shen et al., 2006) also were described as regulators of the maturation program.
LEC2, FUS3, and ABI3 share a B3 DNA-binding domain and play a major role in the transcriptional control of seed maturation (for review, see Koornneef et al., 1984; Giraudat et al., 1992; Meinke, 1992; Meinke et al., 1994; Parcy et al., 1997; Lotan et al., 1998; Luerssen et al., 1998; Harada, 2001). Complementary approaches have demonstrated the regulatory action exerted by these AFLs on embryo development, differentiation of zygotic tissues, accumulation of storage compounds (i.e. seed storage proteins [SSPs] and triacylglycerols [TAGs]), and acquisition of desiccation tolerance. Mutations affecting the AFL genes lead to partially overlapping and pleiotropic embryo phenotypes. Mutant embryos display abnormal suspensors, precocious cell cycle activation, growth of apical and root meristems, and cotyledons similar to young leaves accumulating high levels of chlorophyll and/or anthocyanins and lower amounts of storage compounds. Mutant seeds are less tolerant to desiccation and/or display precocious germination.
Several maturation-induced genes encode SSPs (e.g. At2S1-4 and CRA1) and proteins involved in the storage of TAGs (e.g. OLEOSIN1 [OLE1] and HSD1; Kagaya et al., 2005a; Santos Mendoza et al., 2005; Braybrook et al., 2006; Baud et al., 2009; Wang and Perry, 2013; Barthole et al., 2014). The B3 AFLs directly activate the expression of these genes through RY elements present in their promoter sequences (Fujiwara and Beachy, 1994; Ezcurra et al., 2000; Reidt et al., 2000; Reinders et al., 2002; Mönke et al., 2004). In agreement with this regulation, the core RY motif (5′-CATG-3′) was shown to be necessary for the correct expression of several other seed-specific genes in Arabidopsis (Bäumlein et al., 1986, 1992; Stålberg et al., 1993; Conceição and Krebbers, 1994; Ellerström et al., 1996). This motif also is found in the promoter sequence of legume and monocot SSP genes (Dickinson et al., 1988; Bobb et al., 1997; Suzuki et al., 1997).
If the B3 DNA-binding domains of the AFL transcription factors display some variability in terms of DNA recognition (Golovenko et al., 2014), both FUS3 and ABI3 interact with RY motifs in vitro (Reidt et al., 2000; Mönke et al., 2004), and in vivo analyses have allowed defining a consensus sequence recognized by the two transcription factors (Mönke et al., 2004, 2012; Wang and Perry, 2013). These results are consistent with the partially redundant phenotypes revealed both by loss- and gain-of-function analyses (To et al., 2006). In addition, yeast one-hybrid experiments and gel-shift assays have established that LEC2 and FUS3 can bind sequences containing RY elements separated by a G box (Kroj et al., 2003; Braybrook et al., 2006). Therefore, it was proposed that the B3 AFLs may act in concert with other transcription factors (e.g. bZIPs) interacting with these G boxes to confer proper expression to their common target genes during seed maturation (Sakata et al., 1997; Ezcurra et al., 1999; Hobo et al., 1999; Wobus and Weber, 1999; Kurup et al., 2000; Nakamura et al., 2001; Bensmihen et al., 2002; Brocard-Gifford et al., 2003; Lara et al., 2003; Zahn et al., 2005; Zhang et al., 2005; Nakashima et al., 2006).
Due to partial functional redundancy and the overlapping expression patterns of the B3 AFL genes (Roscoe et al., 2015), it is difficult to infer the specific in planta function of each AFL from genetic analyses. In the same line, ectopic overexpression of one of these proteins may trigger aspecific B3 effects, as observed previously with various transcriptional regulators (Xu et al., 2014), leading to possible misinterpretation. In order to decipher the specific role of each B3 AFL, we first conducted comprehensive analyses of the DNA-binding specificity of these proteins in vitro. In parallel, a functional analysis of the promoter of a known target of these AFLs, OLE1 (At4g25140), was carried out in planta. Interactions of the AFLs with the putative cis-regulatory elements identified in this promoter were then investigated both in planta and in the moss Physcomitrella patens protoplasts. Last, the molecular interactions between the B3 AFLs and other transcriptional regulators like LEC1 were analyzed in moss protoplasts and in vitro. The results obtained showed that a regulatory complex comprising LEC2, ABI3, LEC1, and NF-YC2 is responsible for the synergistic effects of the L-AFL regulators. These results provide new insights into our understanding of the molecular mechanisms underpinning the transcriptional control of maturation-related genes in Arabidopsis.
RESULTS
Comprehensive Characterization of the DNA-Binding Sites of LEC2, ABI3, and FUS3 in Vitro
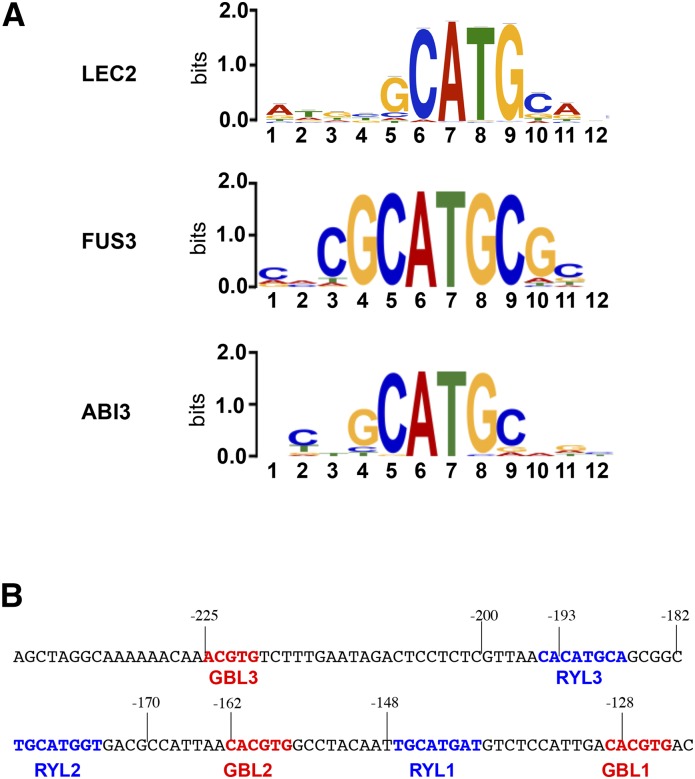
The DNA-binding sequences of the three AFL regulators were investigated in vitro using either high-throughput selection of ligand by exponential enrichment (SELEX) or protein-binding microarrays (Godoy et al., 2011; Franco-Zorrilla et al., 2014). The results confirmed that the three factors bind relatively similar elements made of a core 4-bp sequence (5′-CATG-3′) flanked by nucleotides whose relative importance differs depending on the protein considered (Fig. 1; Supplemental Fig. S1A). FUS3 has a strong requirement for the two positions flanking the core sequence (5′-GCATGC-3′), whereas the specificities of ABI3 and LEC2 are more relaxed, since these proteins only require the 4-bp core sequence. These analyses provided us with models (PWMs) for the DNA-binding specificities of the B3 AFLs that could be used to identify putative binding sites in the regulatory regions of their target genes. OLE1, which encodes an oleosin participating in TAG storage in maturing embryos, is a known target of the B3 AFLs (Braybrook et al., 2006; Mönke et al., 2012). Scanning of a 257-bp OLE1 promoter with these models resulted in the identification of three RY-like (RYL) elements (Fig. 1B; Supplemental Fig. S1B). The same sites were identified for ABI3, LEC2, and FUS3. In order to investigate the functions of these elements in vivo, a functional analysis of the OLE1 promoter was carried out.
Figure 1.
Characterization of the B3 AFL-binding sites. A, Logos representing the DNA-binding specificities of ABI3, FUS3, and LEC2 as identified in vitro. Sequences presented for FUS3 and ABI3 correspond to the top-scoring 8-mers obtained for these proteins in protein-binding microarray assays; their corresponding position weight matrices (PWMs) were used for logo representations. The motif presented for LEC2 was derived from high-throughput SELEX experiments using MEME (see “Materials and Methods”). Scores are in arbitrary units (bits). B, Positions of the different cis-regulatory elements identified in the OLE1 promoter. The RYL elements identified using PWMs obtained in A are highlighted in blue, whereas GBL elements identified using PLACE (http://www.dna.affrc.go.jp/PLACE/index.html) are highlighted in red. Numbers placed above the DNA sequence indicate nucleotide positions from the translational start site.
Functional Dissection of the OLE1 Promoter in Planta
A deletion series of the OLE1 promoter was fused translationally to the uidA reporter gene and stably introduced into plants. For each construct, at least 14 (and up to 38) independent transformants were obtained and analyzed for GUS activity (Fig. 2A). Both 526- and 257-bp fragments yielded strong activation of the reporter gene in maturing embryos. Deletion to position −220 clearly decreased GUS activity. Further deletions to positions −187 and −140 resulted in a complete loss of activity. In light of these observations, the minimal 257-bp OLE1 promoter fragment was analyzed further. This promoter region contained the three RYL motifs, named RYL1 (5′-TGCATGAT-3′), RYL2 (5′-TGCATGGT-3′), and RYL3 (5′-CACATGCA-3′), that were identified previously as putative binding sites of the B3 AFLs (Fig. 1B). These elements share a common 5′-CATG-3′ sequence that fits well the consensus binding site determined for LEC2, ABI3, and FUS3 (Fig. 1A). This sequence is present in the four top-scoring motifs bound by FUS3 and ABI3 (Supplemental Fig. S1A). Interestingly, this promoter also contained three G box-like elements (GBL), two of those (GBL1 and GBL2) fitting perfectly the known consensus (5′-CACGTG-3′) and the third one (GBL3) harboring a different nucleotide composition (5′-AACGTG-3′; Fig. 1B).
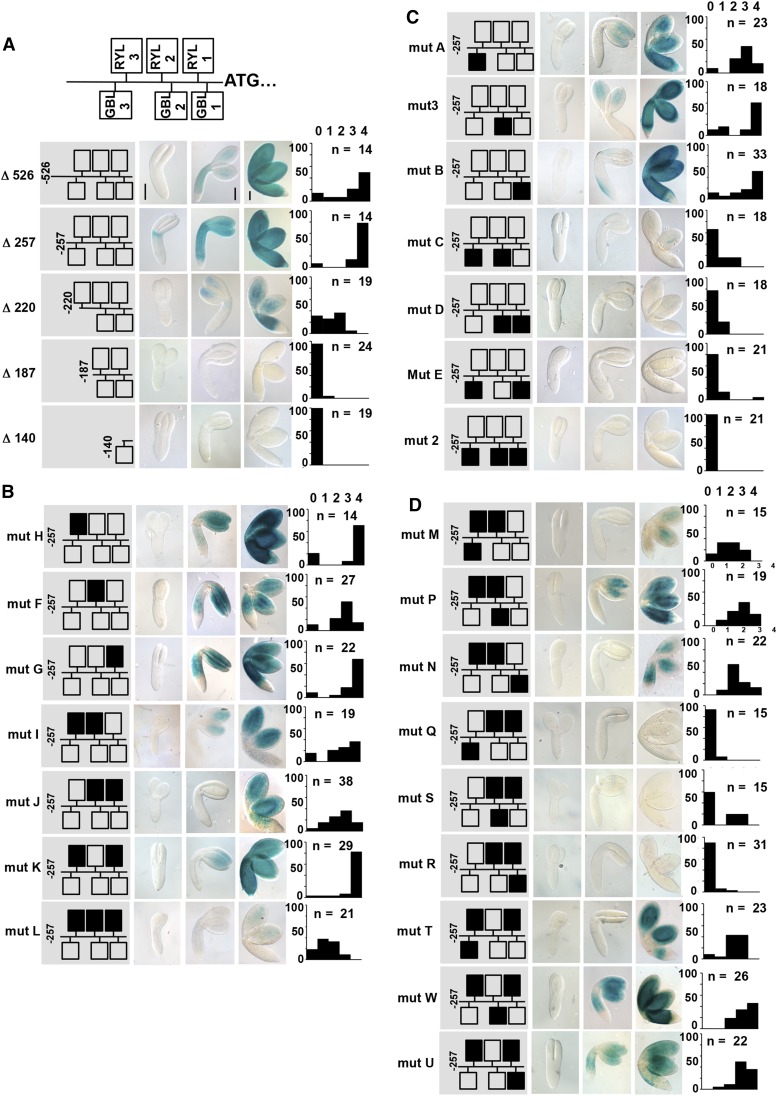
Figure 2.
Functional dissection of the OLE1 promoter in planta. A, At top is a schematic representation of the RYL and GBL elements identified within the OLE1 promoter sequence. A series of 5′ deletions was generated, and translational fusions to the uidA gene were prepared. The corresponding transgenic embryos were assayed for GUS activity at the torpedo, bent-cotyledon, and maturing stages. The length of the promoter tested is indicated at left. B and C, Mutations of the RYL (B) and GBL (C) elements were generated in the context of the 257-bp OLE1 promoter, and translational fusions to the uidA gene were prepared. The corresponding transgenic embryos were assayed for GUS activity. D, Combinations of mutations affecting GBL and RY elements were generated in the context of the 257-bp OLE1 promoter, and translational fusions to the uidA gene were prepared. The corresponding transgenic embryos were assayed for GUS activity. From left to right are the name of the mutagenized promoter under study, a schematic representation of the promoter with mutagenized elements in black, representative photographs of stained embryos at three different stages of the maturation process, and a bar graph showing the partition of staining intensities (from 0 = colorless to 4 = intense staining) among a population of n independent transformants.
Different mutated versions of the OLE1 minimal promoter (Supplemental Fig. S2) cloned upstream of the uidA reporter gene were stably introduced into Arabidopsis (Fig. 2, B and C). Mutation of one (mut H, F, and G) or two (mut I, J, and K) RY elements did not strongly affect the activity of the promoter, even though mutation of the RYL2 element led to slight and reproducible reduction of this activity (mut F, I, and J). Mutations of the three RYL elements (mut L) almost completely abolished the activity of the promoter. These results demonstrated a partial functional redundancy between the three elements in planta and confirmed the requirement for at least one RYL cis-regulatory element within the promoter of OLE1 for its activation in maturing embryos. Similar analyses for the GBL elements (mut A, 3, B, C, D, E, and 2) demonstrated that at least two GBLs are required for proper promoter activity. These results confirmed the importance of the GBL elements, which also display partial functional redundancy. Interestingly, combining the mutations of two RYL and one GBL elements yielded contrasting results ranging from no activation (mut Q, R, and S) to slightly decreased activation in comparison with the native promoter (mut M, N, P, T, U, and W), showing that not all combinations of RYL and GBL elements are equivalent in planta (Fig. 2D). Taken together, these results demonstrated both the partial redundancy and functional interactions of the RYL and GBL elements for the transcriptional activation of OLE1 in Arabidopsis embryos.
Synergistic Effect of the L-AFLs on the Activation of the OLE1 Promoter
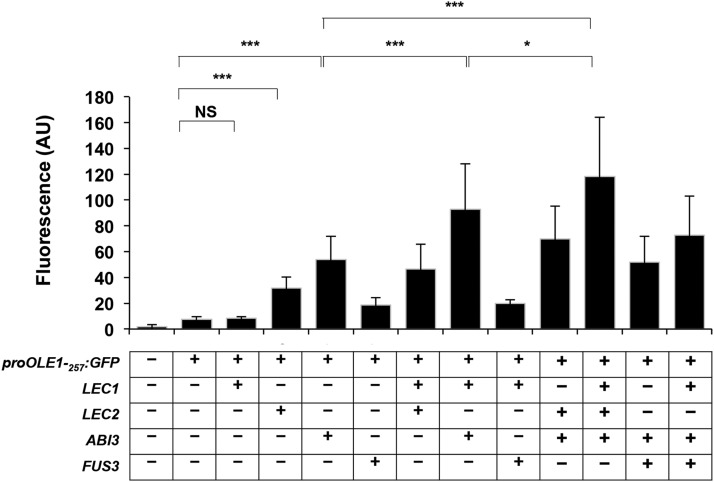
The ability of each transcriptional regulator to activate the OLE1 promoter was investigated in the moss P. patens following a procedure established by Thévenin et al. (2012). The regulators were transiently expressed in the presence of a reporter GFP placed under the control of a 257-bp-long OLE1 promoter that contained all regulatory elements (Fig. 3 and supplemental Fig S3.). The results obtained showed that LEC2, ABI3, and FUS3 are able to induce the activity of this promoter. The strongest activation was observed with ABI3 and the weakest with FUS3. When expressed alone, LEC1 had no effect on the activity of the OLE1 promoter. Combinations of transcription factors were then assayed. LEC2 and ABI3 synergistically activated the expression of the reporter gene. LEC1 further enhanced this transcriptional activity, whereas FUS3 did not (FUS3, therefore, was excluded from the next experiments).
Figure 3.
Synergistic effects of LEC2, LEC1, and ABI3 on the activation of ProOLE1 in moss protoplasts. Transient expression assays were carried out in moss protoplasts. GFP activity was measured in protoplasts transfected with the ProOLE1-257:GFP reporter and constructs allowing the expression of B3 AFL (LEC2, ABI3, or FUS3) or LEC1, alone or in combination. Means of at least three replicates ± sd are presented. AU, Arbitrary units; NS, nonsignificant. Asterisks indicate significant differences. *P value 5%, ***P value 0.1%. Statistical analysis is provided in Supplemental Table S3.
The specific role of an amino acid residue (Asp-55) important for LEC1 function (Lee et al., 2003) and present in LEC1 but not in the homologous protein NF-YB7 (Asp being changed to Lys) was then investigated. LEC1, but neither NF-YB7 nor a mutated version of LEC1 (LEC1m D55K), could enhance the activity of the transcriptional complex, demonstrating the importance of residue Asp-55 in this activation (Fig. 4).
Figure 4.
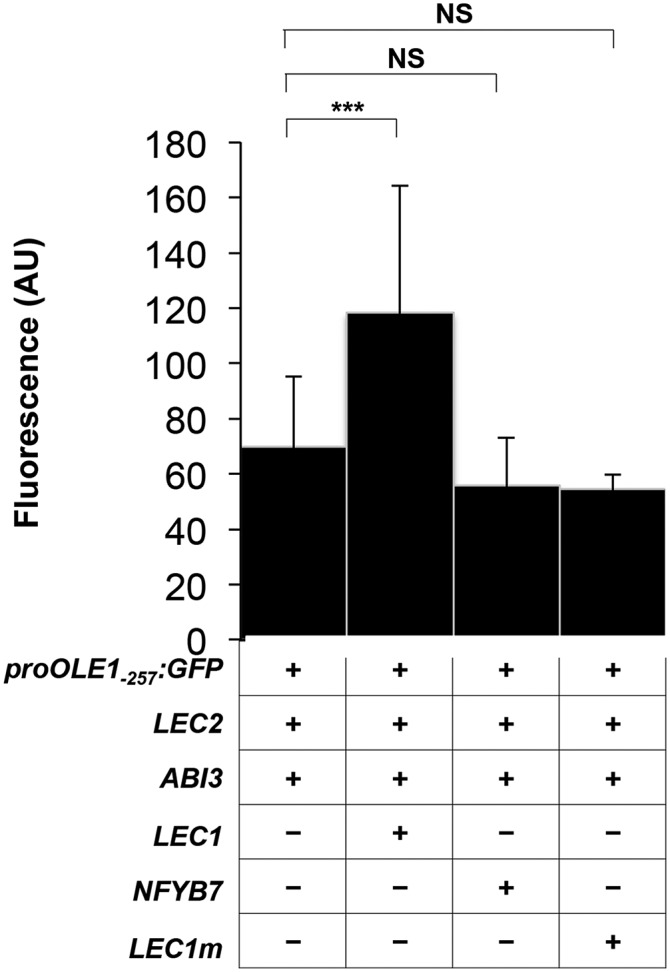
Enhanced activation of ProOLE1 by NF-YB proteins is specific to the LEC1 type. Transient expression assays were carried out in moss protoplasts. GFP activity was measured in protoplasts transfected with the ProOLE1-257:GFP reporter and constructs allowing the expression of LEC2, ABI3, and LEC2 and ABI3 together with different NF-YB proteins, namely LEC1, NF-YB7 (At2g13570), and LEC1m (LEC1 D55K). Means of at least three replicates ± sd are presented. AU, Arbitrary units; NS, nonsignificant. Asterisks indicate a significant difference. ***P value = 0.001. Statistical analysis is provided in Supplemental Table S3.
The synergistic effect observed between LEC1, LEC2, and ABI3 could result from an enhanced transcriptional activity of ABI3 or LEC2 or from changes in their DNA-binding properties within the complex. LEC1 also could affect promoter activity by providing, directly or indirectly, additional activation domains to the complex. To test the first hypothesis, ABI3 and LEC2 were fused to VP16, a strong transcriptional activation domain (Sadowski et al., 1988; Wilde et al., 1994; Parcy et al., 1998). Fusion proteins were expressed alone or in combination with LEC1 to test whether LEC1 still increased the transcriptional activity of complexes containing transcription factors fused to VP16 (Supplemental Fig. S4). VP16-LEC2 exhibited a stronger transcriptional activity than LEC2, demonstrating that the VP16 domain was fully active (Supplemental Fig. S4A). Yet, the addition of LEC1 further enhanced the activity of the VP16-LEC2-containing complex, suggesting that enhancement of the complex activity by LEC1 did not result from an increase in the transcriptional activity of LEC2 itself. A negative effect of the VP16 domain on ABI3 activity (lower activation of ProOLE expression, either alone or in combination with LEC1 or LEC2) prevented us from drawing any conclusion concerning ABI3 (Supplemental Fig. S4B).
Functional Interactions of the L-AFLs with the cis-Regulatory Elements of Oleosin Promoters
To study the interactions between the L-AFL complex and the cis-regulatory elements of its target promoters, we first performed activation assays in moss protoplasts using the promoters of two oleosin genes closely related to OLE1, namely OLE2 and OLE3. The promoter sequences of OLE1 and OLE3 both display repeats of RYL and GBL domains close to the translational start codon (Supplemental Fig. S5). On the contrary, RYL and GBL elements are scattered over a longer distance in ProOLE2. ProOLE2 was weakly induced by the L-AFL complex, whereas a strong and synergistic induction of ProOLE3 by the L-AFLs was observed, similar to the one reported previously for ProOLE1. These results showed that the relative positions of RYL and GBL elements within a promoter sequence dramatically affect the ability of the L-AFL complex to activate this promoter.
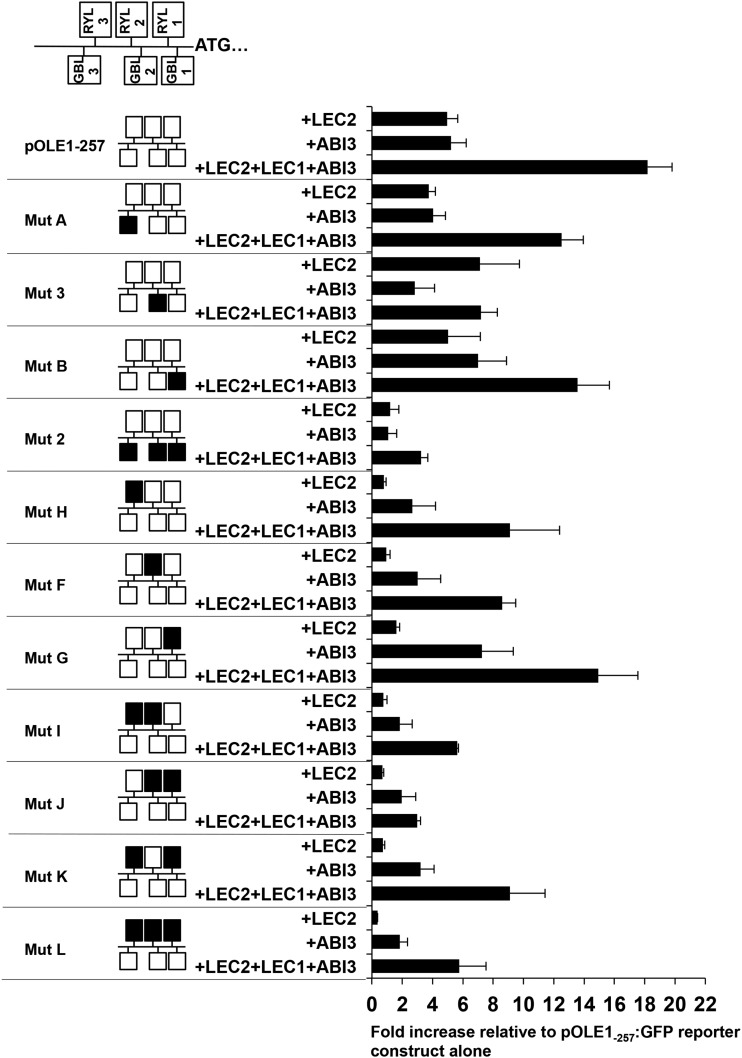
To further elucidate the interactions existing between the regulators under study and the cis-regulatory elements characterized, activation assays were then performed using mutagenized versions of the OLE1 promoter (Supplemental Fig. S2). The activation of mutagenized promoter versions by LEC2, ABI3, or a combination of the three regulators LEC1, LEC2, and ABI3 was tested (Fig. 5). Mutation of a single RYL element (mut H, F, or G) had limited impact on the activation by ABI3 or by the combination of the three proteins, whereas activation by LEC2 alone was strongly decreased. Mutation of two or three RYL elements (mut I, J, K, and L) dramatically affected the ability of the transcriptional regulators, alone or in combination, to activate the promoter. Mutations affecting the GBL1 or GBL3 element (mut A and B) did not affect the induction of the reporter gene by the regulators tested, whereas mutation of the GBL2 element (mut 3) affected its induction by the complex (Fig. 5). Consistent with the results observed in planta, deletion of the three GBL domains dramatically affected promoter activity (mut 2). Taken together, these results confirmed the functional interactions between the L-AFL complex and the RYL and GBL elements under study in the OLE1 promoter sequence. They also established the strong requirement of LEC2 for RYL elements to activate a promoter.
Figure 5.
Importance of the RYL and GBL cis-regulatory elements in the activation of proOLE1 by the L-AFLs in moss protoplasts. At top is a schematic representation of the RYL (RYL1–RYL3) and GBL (GBL1–GBL3) elements identified in the minimal OLE1 promoter. At bottom are results from transient expression assays carried out in moss protoplasts with different mutagenized versions of the OLE1 promoter. GFP activity was measured in protoplasts transfected with the ProOLE1:GFP reporter and constructs allowing the expression of LEC2, ABI3, or a combination of LEC2, LEC1, and ABI3. From left to right are the name of the mutagenized promoter under study, a schematic representation of the promoter with mutagenized elements in black, and a bar graph showing fluorescence intensities in moss protoplasts expressed as a ratio of the intensity measured with and without expression of the L-AFLs. Statistical analysis is provided in Supplemental Table S3.
Molecular Interactions between the L-AFL Regulators and the OLE1 Promoter
The ability of L-AFL proteins to interact physically with the OLE1 promoter was tested in vitro by electrophoretic mobility-shift assay (EMSA; Supplemental Fig. S6). Both LEC2 and ABI3 proteins were able to interact with the ProOLE1 double-stranded probe (Supplemental Fig. S6, left). Surprisingly, LEC1 also was able to bind ProOLE1 in vitro, although LEC1 has never been reported to bind DNA before. A supershift (Supplemental Fig. S6, arrows) was detected when LEC1 and LEC2, or LEC1, LEC2, and ABI3, were tested in combination, but not when LEC1 and ABI3, or LEC2 and ABI3, were mixed together. The intensity of this supershift appeared to be well correlated with LEC1 abundance (Supplemental Fig. S6, right). Competition experiments with wild-type nonlabeled probe demonstrated the specificity of these interactions.
Molecular Interactions between the L-AFL Regulators
Interaction studies between the four regulators were first investigated using a yeast two-hybrid approach (Supplemental Fig. S8). Due to a strong autoactivation of the reporter genes by LEC2 and ABI3 fused to the GAL4 DNA-binding domain, only a limited subset of interactions could be tested. No interaction was detected (Supplemental Fig. S8A) except the homodimerization of LEC1 (Supplemental Fig. S8B). This homodimerization was not affected by the D55K mutation.
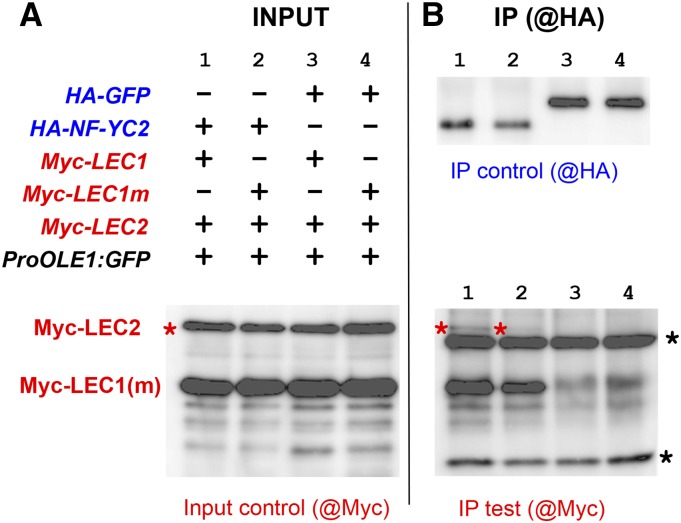
To test other interactions between the regulators, pull-down experiments were performed with recombinant hemagglutinin (HA)- or Myc-tagged proteins coexpressed in Arabidopsis protoplasts in the presence of a plasmid carrying the OLE1 promoter. Proteins were immunoprecipitated using anti-HA antibodies and protein G-agarose beads. The immunoprecipitated proteins were then analyzed by western blot using both HA antibodies (as a control for immunoprecipitation) and anti-Myc antibodies to discover coprecipitated partners (Fig. 6; Supplemental Fig. S7). No interaction was detected using LEC1, LEC2, or ABI3 (Supplemental Fig. S8). Because NF-YC, a member of the NF-Y complex, was shown previously to interact with LEC1 (Hackenberg et al., 2012), it was subsequently included in the experiments. Pairwise interactions were thus observed between NF-YC2 and LEC1 (or LEC1m D55K; Supplemental Fig. S6, A–C) but not with LEC2 (Supplemental Fig. S6, C and D). Then, NF-YC2, LEC2, and LEC1 (or LEC1m) were coexpressed simultaneously in the presence of the OLE1 promoter. A strong band and a faint band corresponding to Myc-LEC1 (or LEC1m) and Myc-LEC2, respectively, were reproducibly detected in the immunoprecipitated fraction (Fig. 6B, bottom, columns 1 and 2, respectively). This result demonstrated that the three proteins can form a ternary complex with the OLE1 promoter. Moreover, the detection of LEC2 was affected when replacing LEC1 by LEC1m (Fig. 6).
Figure 6.
Interaction of LEC1, LEC2, and NF-YC2 in pull-down experiments. Coimmunoprecipitation experiments were performed using native or mutated LEC1 (LEC1m D55K) as a bait and LEC2 as a prey in the presence (or absence) of NF-YC2 and of a proOLE1:GFP construct. HA- and Myc-tagged proteins were coexpressed in Arabidopsis protoplasts and unraveled by western blot using anti-Myc and anti-HA antibodies, respectively. INPUT, 2% of the full protein extract; IP, immunoprecipitated sample. Asterisks indicate heavy (black) and light (red) chains of the antibody used for immunoprecipitation.
DISCUSSION
Genetic analyses have unraveled functional interactions occurring in Arabidopsis seeds between the L-AFLs and the transcriptional regulation they exert, being mediated through RYL and GBL cis-regulatory elements present in their target promoters. Molecular analyses then established that the three B3 AFL regulators can bind RYL elements in vitro or in Saccharomyces cerevisiae. More recently, interactions of ABI3 and FUS3 with RYL and GBL domains were demonstrated in planta by chromatin immunoprecipitation (ChIP) approaches (Wang and Perry, 2013), although the binding to cis-regulatory elements reported in these experiments could be direct or indirect. Here, we bring new insights into the functional and molecular interactions between B3 AFLs and LEC1 (NF-YB9) on one side and between these regulatory proteins and the different RYL and GBL cis-regulatory elements present in their target promoters on the other side.
RY and G Boxes Are Involved in the Regulation of OLE1 Expression in Maturing Embryos
We have conducted comprehensive in vitro analyses of DNA sequences bound by the B3 AFLs LEC2, ABI3, and FUS3 and showed that they recognize a similar 6-bp consensus sequence (5′-gCATGc-3′), which encompasses the core RY motif 5′-CATG-3′. Specific binding of FUS3 to the RY element has been shown by band-shift assays (Reidt et al., 2000) or biacore (Mönke et al., 2004), and the 5′-CATGCA-3′ motif was found overrepresented within sequences bound by FUS3 (Wang and Perry, 2013). In our data, FUS3 exhibits a strong affinity for a longer consensus, namely 5′-cGCATGCg-3′, that fits well the ChIP data of Wang and Perry (2013). Overall, the core RY sequence is always identified for the three B3 domain-containing factors with few differences in the 5′ and 3′ ends of the core sequence. Scanning of the OLE1 minimal 257-bp promoter sequence with the PWMs led to the identification of three peaks, matching with RYL domains. Interestingly, a comparison of the promoter structures of the related OLE1, OLE2, and OLE3 genes, all induced in maturing seeds and regulated by ABI3 (Mönke et al., 2012), revealed contrasting organizations of their RYL and GBL motifs. The promoters of OLE1 and OLE3 displayed similar arrangements of GBL and RYL elements close to the translational start site and exhibited a similar synergistic induction by the L-AFLs in moss protoplasts. On the contrary, this synergistic activation was not observed with the OLE2 promoter, where GBL and RYL motifs are scattered over a long distance. These results and others (Che et al., 2009; Wang and Perry, 2013) emphasize the importance of the organization of RYL and GBL elements for the efficient activation of maturation-related genes by the L-AFLs. Further analyses with mutagenized versions of ProOLE1 carried out in vivo (both in moss protoplasts and in planta) confirmed the importance of RYL and GBL elements for mediating the induction by L-AFLs. At least one RYL and two GBL domains are required for proper ProOLE1 activity in planta. Nevertheless, it is still unknown whether the B3 AFLs can directly bind a GBL element. Although a direct interaction cannot be excluded, neither our in vitro experiments nor recent ChIP analyses carried out with FUS3 (Mönke et al., 2004; Wang and Perry, 2013) support this idea. Rather, one can hypothesize that additional factors such as bZIPs are necessary for direct binding of the GBL elements. In agreement with this, the involvement of bZIP proteins in the accumulation of reserves during seed maturation is well established (for review, see Kroj et al., 2003; Vicente-Carbajosa and Carbonero, 2005). What is more, interactions between bZIPs and ABI3 were reported (Lara et al., 2003; Alonso et al., 2009), and some bZIPs (e.g. bZIP67 and bZIP28) already have been demonstrated to interact with LEC1 to activate seed-specific promoters (Yamamoto et al., 2009; Liu and Howell, 2010). Consistent with this hypothesis, bZIP homologs have been described in P. patens (Yotsui et al., 2013), which may explain why the L-AFL system requires GBL elements for a stronger activation of the ProOLE1 promoter.
Specific Roles of the B3 AFL Regulators
B3 proteins are classified into five subfamilies (Auxin Response Factor, Reproductive Meristem, High Level of Suc Inducible, Related to ABI3/VP1, and ABI3; Yamasaki et al., 2004), each subgroup exhibiting distinct DNA-binding characteristics (Boer et al., 2014; Franco-Zorrilla et al., 2014). They share a conserved B3 DNA-binding domain and can contain additional domains (e.g. B1 and B2) involved in protein-protein interactions (Romanel et al., 2009). The B3 AFLs, namely ABI3, LEC2, and FUS3, belong to the same subclass of ABI3-related transcription factors. Genetic and molecular analyses have demonstrated that these B3 AFL proteins have partially overlapping though nonidentical functions in planta (To et al., 2006; Mönke et al., 2012; Wang and Perry, 2013). Our results show that the three transcription factors can activate the OLE1 promoter at similar levels when expressed alone in moss protoplasts. Nevertheless, only ABI3 and LEC2 display clear synergistic effects. The differential ability of these actors to cooperate and/or to be part of common transcriptional complexes may explain why their functions are not identical. Different DNA-binding properties or protein-protein interaction capacities may constitute the molecular bases explaining these different behaviors. Nonoverlapping expression patterns of these transcriptional regulators also may play a part in specifying their functions in developing embryos.
LEC1 Acts Synergistically with LEC2 and ABI3
LEC1 (NF-YB9) is a member of the NF-YB protein family, which encodes components of an NF-Y regulatory complex (comprising NF-YA, NF-YB, and NF-YC subunits) involved in the recognition of the CCAAT box in yeast (Ronchi et al., 1995; Mantovani, 1999; Motta et al., 1999; Romier et al., 2003). With regard to the large number of NF-Y genes present in plant genomes (in comparison with mammals and fungi), other roles have been postulated for these proteins in plants (Gusmaroli et al., 2001, 2002; Petroni et al., 2012). Members of the NF-YB family have thus been shown to work through mechanisms independent from the CCAAT box and to interact with proteins other than the NF-Ys (Masiero et al., 2002; Wenkel et al., 2006; Liu and Howell, 2010; Li et al., 2011). Similarly, LEC1 interacts functionally with ABRE-binding bZIP67 and bZIP28 to directly activate seed-specific genes (Yamamoto et al., 2009; Liu and Howell, 2010; Mendes et al., 2013).
In a somehow similar manner, this study establishes that LEC1, although unable to activate the OLE1 promoter alone, enhances the transcriptional activity of LEC2 and ABI3. Interestingly, NF-YB7, a more distantly related protein used as a negative control, and a mutated version of LEC1 (D55K) did not. These results emphasized the importance of the amino acid Asp-55 in LEC1 for exerting a synergistic effect with LEC2 and ABI3. Consistent with these results is the role played by the Asp-55 residue in LEC1 function in planta (Lee et al., 2003).
To further elucidate the role of LEC1 in the synergistic activation of ProOLE1 by the L-AFLs, we tested the effect of LEC1 acting in concert with B3 AFL proteins fused to the strong VP16 activation domain. Consistent with previous negative results obtained with VP16-ABI3 in yeast one-hybrid experiments (Kroj et al., 2003), the lack of activity of the chimerical protein prevented us from drawing any conclusion concerning ABI3. On the contrary, VP16-LEC2 was highly functional, and its activity was still further enhanced by the addition of LEC1, suggesting that the role of LEC1 in the complex is not to stimulate the transcriptional activity of individual B3 AFLs. Instead, LEC1 may favor the stability of the multiprotein complex, enhance the DNA-binding efficiency of the B3 AFLs, or provide additional activation domains to the complex.
LEC1 and LEC2 Can Form a Ternary Complex in the Presence of NF-YC2
The tagged recombinant LEC1 and LEC2 proteins produced in Arabidopsis protoplasts can be coimmunoprecipitated when coexpressed with NF-YC2. Interestingly, the D55K mutation of LEC1 strongly impacted the coimmunoprecipitation of LEC2. This result is consistent with the lack of activity of the mutated LEC1m (D55K) and of NF-YB7 (Lys-55) in moss protoplasts. Then, homodimerization of LEC1 was demonstrated in yeast two-hybrid experiments. Taken together, these results provide some molecular bases for the key role played by LEC1 in structuring a complex (comprising NF-Y proteins and B3 AFLs) that is essential for the control of seed maturation (Lee et al., 2003). The Asp-55 residue found in LEC1 is of particular importance for the recruitment of LEC2 in the presence of NF-YC2. Interestingly, this amino acid is specific to LEC1 and L1L, which are evolutionarily closely related and functionally redundant proteins (Kwong et al., 2003; Xie et al., 2008; Laloum et al., 2013).
A Model for the Transcriptional Activation of Maturation-Related Genes by the L-AFLs
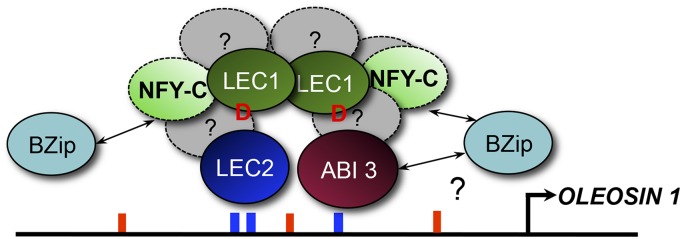
Taken together, the results of this study provide some important new insights into the molecular mechanisms underlying the partially overlapping functions of the B3 AFLs and the specific role of LEC1 in maturing seeds. They allow us to propose a model for the transcriptional activation of seed genes by LEC1, LEC2, ABI3, and NF-YC2 proteins through the formation of regulatory complexes (Fig. 7). In this model, LEC2 and ABI3 can bind RYL elements directly and independently. LEC1, which is able to form homodimers, also participates in the complex, and the amino acid Asp-55 of the protein is essential for LEC1 function through its synergistic action with LEC2 and ABI3. LEC1, as an NF-YB subunit, is expected neither to activate DNA transcription alone nor to directly bind DNA. In this respect, the shifted band obtained with LEC1 in gel-shift assays is surprising. LEC1 and L1L differ from the other NF-YB subunits and exhibit specific amino acid residues, some of which are essential for their activity. It might be that some of these residues may confer DNA-binding properties to LEC1. This aspect should be investigated further by performing a thorough structure-function analysis of the protein. Regardless of these DNA-binding capabilities, LEC1 remains unable to activate transcription by itself.
Figure 7.
Model for the transcriptional regulation of OLE1 expression by the L-AFL proteins. In this model, LEC2 and ABI3 transcription factors directly and independently bind DNA and have a synergistic effect, together with LEC1, on the activation of the ProOLE1 promoter. This may be due to their cooperating in a multiprotein complex, as shown by pull-down assays that unraveled the occurrence of LEC1/LEC2/NF-YC2 ternary complexes. bZIPs may play a role in this complex, although they were not tested in this study. D, Asp involved in the activity of LEC1; blue bars, RYL element; red bars, GBL element.
NF-YC2, which is also part of the complex, interacts with LEC1 (Calvenzani et al., 2012) and is essential for the formation of LEC1/LEC2/NF-YC2 ternary complexes in the presence of a target promoter in pull-down assays (Fig. 6). This role of NF-YC2 is consistent with its previously described involvement in the interactions between LEC1 and bZIP67 (Yamamoto et al., 2009). To explain the enhanced transcriptional activity of LEC2 and ABI3 by LEC1 observed in transient activation assays carried out in moss in the absence of AtNF-YC2, one has to consider that, whereas moss does not possess NF-YBs of the LEC1 type (containing the Asp-55 residue instead of a Lys; Kirkbride et al., 2013; Cagliari et al., 2014), NF-YA and NF-YC genes are present in moss and may substitute for their Arabidopsis counterparts. No direct evidence of direct or indirect binding between LEC1 and ABI3 could be obtained so far, despite the enhanced ABI3 activity observed in the presence of LEC1 in moss. Complementary analyses will be required to elucidate the molecular bases of this functional interaction.
A potential limit to the use of P. patens for performing activation studies may reside in the presence of endogenous transcription factors, such as B3s or bZIPs, that may be able to bind Arabidopsis promoter sequences and thus interfere with the candidates under study. Thirty-three B3 genes can be found in P. patens, seven of which belong to the ABI3-related or HIS families (Romanel et al., 2009). A gene homologous to ABI3 was even identified that partially complements the Arabidopsis abi3-6 mutant (Marella et al., 2006). PpABI3 was shown to interact cooperatively with PpNF-YC1 through the ACTT core element, which is different from the ATGC core of RY elements and the ACGT core of G boxes (Yotsui et al., 2013). As for bZIPs, the transcripts of which have been identified in P. patens (Richardt et al., 2010), it is difficult to speculate on their ability to interact with the system under study only on the basis of sequence similarities. Even if the possibility of PpB3s or PpbZIPs binding to ProOLE promoters cannot be ruled out (see above), the very low expression level of the reporter gene when the reporter construct was introduced alone in moss strongly suggests that the effect of endogenous P. patens factors alone is extremely limited.
In conclusion, this study reveals that the molecular mechanisms underpinning the transcriptional activation of maturation-related genes in seeds are much more complex than previously thought. They involve a set of protein partners assembled in a multiprotein complex, the structure of which may vary depending on the target promoter and the cell type considered. Aside from this modular organization, specific posttranslational modifications of the components also may contribute to regulate the activity of the complexes and remain to be investigated.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds of the accession Columbia were obtained from the Plant Observatory, Biological Resource Center (Institut National de la Recherche Agronomique). Plants were cultured as described previously (Baud et al., 2002). Material used for RNA extraction was frozen in liquid nitrogen immediately after harvest and then stored at −80°C.
Constructs for Plant and Moss Transformation
All the primers used in this work are described in Supplemental Table S1. For deletions of the OLE1 promoter (ProOLE1), genomic DNA was PCR amplified with Phusion High-Fidelity DNA Polymerase (Finnzymes) using primers carrying B1 and B2 Gateway recombination sites (Life Technologies). PCR products were cloned by BP recombination (according to the manufacturer’s recommendations) into the entry vector pDONR207 and then LR recombined into the destination vector pBS TPp-B for protoplast expression (Thévenin et al., 2012) or pBI101-R1R2-GUS for plant transformation. Mutations of the ProOLE1 promoter cloned in pDONR207 were performed with the QuickChange Site Directed Mutagenesis kit (Agilent) according to the manufacturer’s instructions. The mutations realized were as follows: GBL1*, ACGTG→AGTTG; GBL2*, ACGTG→AGTTG; GBL3*, ACGTG→TTGTG; RYL1*, TGCATGAT→TACAACAT; RYL2*, TGCATGGT→TACAACGT; and RYL3*, CACATGCA→CAGTTGTA (for oligonucleotide sequences, see Supplemental Table S2).
AFL transcription factors were PCR amplified from complementary DNAs (cDNAs) synthesized from RNAs extracted from developing seeds and BP recombined into pDONR207 (for oligonucleotide sequences, see Supplemental Table S1).
Expression in Plant and in Moss Protoplasts
For plant transformation, binary vectors were electroporated into Agrobacterium tumefaciens strain C58C1 (pMP90) and used for agroinfiltration of Arabidopsis inflorescences (Bechtold et al., 1993). Primary transformants were selected on Murashige and Skoog medium containing kanamycin (50 mg L−1) and then transferred to soil for further characterization. For each construct, several independent transgenic lines were analyzed (at least 14 and up to 38; Fig. 2). Transient expression in the moss Physcomitrella patens protoplasts was performed as described (Thévenin et al., 2012). Briefly, transcription factors were recombined into a specific vector (pBS TPp-A), allowing their expression in moss protoplasts. The promoters were recombined in a second vector (pBS TPp-B) upstream of a GFP reporter gene. Clones were selected with carbenicillin (100 µg mL−1). Moss was cultured on solid standard PpNH4 medium (Trouiller et al., 2006).
Cytometry Analysis
The activation studies were based on the quantification of the fluorescent signal in cotransformed protoplasts using a flow cytometer (Partec). Protoplasts were analyzed as described by Thévenin et al. (2012). The instrument was calibrated with calibration green beads (Sysmex-Partec; 05-4006). Transformed protoplasts in suspension were filtrated with a 50-µm cell strainer to allow the analysis of 20,000 to 250,000 events. All the graphs are presented as means of at least three replicates ± sd. The R stats package (https://www.r-project.org/) version 3.2.1 was used for statistical analysis (R Core Team, 2015).
Microscopy and Image Analysis
Microscopic observations of seeds and embryos were carried out as described previously (Baud et al., 2007). For histochemical detection of GUS activity, tissues were incubated in 0.1 m phosphate buffer, pH 7.2, containing 2 mm 5-bromo-3-indolyl-β-d-glucuronide (Duchefa), 0.1% (v/v) Triton X-100:water, 2 mm each potassium ferrocyanide and potassium ferricyanide, and 10 mm Na2-EDTA. A vacuum was applied for 1 h before incubating for 3 h at 37°C in the dark. After incubation, the staining solution was removed and samples were cleared by sequential changes of 75% and 95% (v/v) ethanol.
EMSA
The probe used comprised three GBL (underlined) and three RYL (boldface) domains: 5′-CAAACGTGTCTTTGAATAGACTCCTCTCGTTAACACATGCAGCGGCTGCATGGTGACGCCATTAACACGTGGCCTACAATTGCATGATGTCTCCATTGACACGTGACT-3′. It was obtained by annealing two 80-bp-long oligonucleotides exhibiting a 48-bp overlap that were 5′ biotinylated and synthesized by the Eurofins Company (5′-ggCAAACGTGTCTTTGAATAGACTCCTCTCGTTAACACATGCAGCGGCTGCATGGTGACGCCATTAACACGTGGCCTACA-3′ and 5′-ggAGTCACGTGTCAATGGAGACATCATGCAATTGTAGGCCACGTGTTAATGGCGTCACCATGCAGCCGCTGCATGTGTTA-3′). The oligonucleotides were diluted and annealed for 5 min at 95°C in a PCR thermal block. The sequences were then double stranded by the action of Klenow enzyme (Klenow fragment; Thermo Scientific) for 1 h at room temperature in order to obtain a 108-bp double-stranded biotinylated probe at a concentration of 20 fmol µL−1. Binding reactions were performed with the Shift Light Chemiluminescent EMSA kit (Thermo Scientific) and carried out with a binding buffer (10 mm Tris, 150 mm KCl, 1 mm dithiothreitol, 0.05% [v/v] Nonidet P-40, and 0.5 µg of poly [dI-dC], pH 7.5). One to 6 µg of proteins was incubated for 5 min on ice in binding buffer before adding 20 fmol of biotinylated probe. The binding reactions were then incubated at room temperature for 20 min in the dark. The competition reactions were performed with a 25-fold increase of unlabeled probe. The electrophoresis gel (5% acrylamide in 0.25% [v/v] glycerol and 0.5× Tris-borate/EDTA) was subjected to a 60-min prerun at 120 V, and the binding reactions were loaded onto the gel with 5 µL of loading buffer for 6 h of migration at 120 V and 4°C. The gel was transferred to a GeneScreen plus DNA membrane (Perkin-Elmer Life Sciences) for 1 h at 25 V with 0.5× Tris-borate/EDTA using a Transblot SD Semi Dry transfer cell system (Bio-Rad). DNA was cross-linked on the membrane with a transilluminator for 60 s at 254 nm with the autocrosslink function. The bands were revealed by chemiluminescence according to instructions of the Chemiluminescent Nucleic Acid Detection module kit (Thermo Scientific) using the Fujifilm Las 4000 imaging system.
Expression and Purification of Recombinant Proteins
LEC1 and LEC2 coding sequences were PCR amplified from AFLs in pDONR207 using primers carrying 5′ NcoI and 3′ SalI restriction sites (for primer sequences, see Supplemental Table S1). PCR products were ligated in the expression vector pETtrx-1a (Millipore) at the NcoI/XhoI sites with an N-terminal His-thioredoxin tag inserted in the oligonucleotide sequence. ABI3 in pDONR207 was directly LR recombined into direpDest17 (Life Technologies) with an N-terminal His tag inserted in the oligonucleotide sequence. Proteins were produced in Escherichia coli Rosetta [DE3, F− ompT hsdSB(rB− mB−) gal dcm (DE3) pRARE (CamR)] and induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside (IPTG) at 28°C for 3 h, 17°C for 3 h, and 17°C overnight. Bacterial pellets were sonicated in lysis buffer (20 mm Tris, 500 mm NaCl, 50 mm NaH2PO4, 10% [v/v] glycerol, and 5 mm imidazole, pH 8). Lysates were centrifuged at 21,000g for 45 min at 4°C. Soluble proteins were purified on nickel-nitrilotriacetic acid agarose (Ni-NTA) resin (Bio-Rad) in lysis buffer with increasing concentrations of imidazole (5–70 mm for washing and 300 mm for elution). Purified proteins were then dialyzed in buffer (0.05 m phosphate, pH 7.2, 0.15 m NaCl, and 20% [v/v] glycerol) at 4°C, concentrated on Amicon cell 10,000 MWCO (Millipore), and finally quantified by the Bradford method (Bradford, 1976). After purification, proteins were stored at −20°C before subsequent analyses.
Coimmunoprecipitation Experiments
The HindIII/StuI fragments of the pGWB15 and pGWB18 vectors (Nakagawa et al., 2007), containing promoter 35S/3× HA epitope/Gateway recombination cassette/terminator and promoter 35S/4× Myc epitope/Gateway recombination cassette/terminator sequence, respectively, were cloned into HindIII/SmaI sites of the pBluescript II KS + vector (Stratagene) and used as destination vectors (pBS 35SHA-R1R2 and pBS 35SMYC-R1R2) for coimmunoprecipitation experiments. LEC2, LEC1, and ABI3 in pDONR207 were recombined directly into these destination vectors. NF-YC2 coding sequence was PCR amplified using primers NF-YC2 F GW and NF-YC2 R GW (for primer sequences, see Supplemental Table S1) from an Arabidopsis Columbia cDNA library and recombined directly into pDONR207 using BP Clonase, according to the manufacturer’s instructions. The cDNA was then further transferred into the destination vector using LR Clonase.
Cell suspension culture of Arabidopsis ecotype Columbia, subcultured weekly in JPL medium (Takahashi et al., 2001), was used for the preparation of protoplasts and transformations. Protoplasts were isolated and transfected by the polyethylene glycol-mediated method (Kiegerl et al., 2000). After a 16-h incubation, cells were harvested by centrifugation, and proteins were extracted in an extraction buffer containing 25 mm Tris HCl, pH 7.7, 10 mm MgCl2, 15 mm EGTA, 75 mm NaCl, 15 mm β-glycerophosphate, 0.1% (v/v) Tween 20, 10% (v/v) glycerol, 1 mm dithiothreitol, 1 mm NaF, 0.5 mm NaVO3, 0.5 mm phenylmethylsulfonyl fluoride, and protease inhibitors (Complete; Roche). From 500 µg of total protein extract of transfected Arabidopsis protoplasts, HA-tagged proteins were immunoprecipitated with 500 ng of 3F10 anti-HA monoclonal antibody (Roche) and washed three times with the extraction buffer and once with 20 mm Tris-HCl, pH 7.5. Proteins bound to the beads were eluted with 50 µL of SDS loading buffer. Coimmunoprecipitation of the Myc-tagged proteins was revealed by SDS-PAGE followed by western blot using the 9E10 anti-Myc monoclonal antibody (Roche Life Sciences) and ECL detection (Amersham). The efficiency of the immunoprecipitation was tested using HA-probe (Y-11) polyclonal antibody (Santa Cruz Biotech).
Yeast Experiments
LEC2, LEC1, and ABI3 in pDONR207 were directly LR recombined into pDEST22 and pDEST32 vectors (Life Technologies) and stably cotransformed into yeast. The yeast AH109 strain (Clontech) was grown on YPDA plates (Clontech) at 28°C. A 2-d-old culture was collected and resuspended in 1 mL of sterile water. The yeast was pelleted by centrifugation (at maximum speed for 15 s) and resuspended in 1 mL of 0.1 m lithium acetate in 1× Tris-EDTA solution. Five milligrams of each plasmid construct was mixed with 5 μL of carrier DNA (Finnzymes, Fisher Scientific), 0.1 mL of yeast cell solution, and 0.6 mL of 40% [v/v] polyethylene glycol 4000/0.1 m lithium acetate/1× Tris-EDTA solution. After a 30-min incubation at 28°C, the transformation mixture was heat shocked at 42°C for 25 min. After centrifugation (at maximum speed for 15 s), the pellet was resuspended in 250 μL of sterile water and plated onto synthetic dextrose-Trp-Leu medium (Clontech). To analyze interactions, cotransformed yeast cells were transferred to 100 μL of sterile water. Growth of a 5-μL yeast solution was tested on selective medium. Colonies growing on medium lacking His or both His and Ala, and in the presence of various concentrations of 3-aminotriazole, were considered as positive interactions.
Systematic Evolution of Ligands by Exponential Enrichment Experiments (SELEX) for the LEC2 Protein
LEC2 coding sequence was cloned in pET-trx 1a. LEC2 protein was expressed using E. coli strain Rosetta Blu (DE3). After induction by 0.5 mm IPTG, cells were grown overnight at 17°C under shaking. For cell lysis, the pellet of 1 L of culture was sonicated in 35 mL of loading buffer (500 mm NaCl, 20 mm Tris‐HCl, pH 8, 5 mm imidazole, 5% [v/v] glycerol, and 5 mm tris(2-carboxyethyl)phosphine TCEP) with one protease inhibitor cocktail tablet (Complete EDTA‐free; Roche) and centrifuged for 40 min at 16,000g. The clear supernatant was incubated for 1 h with 1 mL of Ni-NTA resin (Qiagen). The resin was transferred into a column, washed with 15 mL of loading buffer and then 15 mL of washing buffer (loading buffer with 20 mm imidazole), and eluted with 10 mL of elution buffer (loading buffer with 300 mm imidazole). The protein concentration of each fraction was estimated using the Bradford assay (Bradford, 1976).
For SELEX, a random library was first synthesized using the 73-mer 5′-TGGAGAAGAGGAGAGATCTAGC(N)30CTTGTTCTTCTTCGATTCCGG-3′ as a template with a fluorescent TAMRA-labeled forward primer (SElex-F) and a nonlabeled reverse primer (SElex-R), as described previously (Moyroud et al., 2011).
For each selection cycle, 1 µm protein was mixed with 10 nm fluorescent double-stranded DNA in 225 µL of SELEX buffer (25 mm HEPES, pH 8, 1 mg mL−1 bovine serum albumin, 75 nm KCl, 10% glycerol, and 5 mm tris(2-carboxyethyl)phosphine [TCEP]) with 60 μg mL−1 fish sperm DNA added. After a 15-min incubation under shaking at 4°C, 25-µL Ni-NTA magnetic beads (Qiagen), previously equilibrated in SELEX buffer, were added to the reaction mix to immobilize DNA/protein complexes via the His tag of the protein. After 30 min of incubation under shaking at 4°C, the reaction mix was placed on a tube magnet for 1 min, and the supernatant was removed from the separated beads to eliminate the unbound DNA. Six washes were performed subsequently at 4°C, each of them consisting of first adding 50 µL of SELEX buffer and then 250 µL of SELEX buffer containing 20 μg mL−1 fish sperm DNA. Between the two buffer additions, an aliquot was taken to perform PCR (see below). Each wash was followed by a 2-min incubation and 1 min on the tube magnet to discard the supernatant, and finally, the magnetic beads were resuspended in 50 µL of SELEX buffer. Selected 73-mers were amplified by PCR as described previously (Moyroud et al., 2011) using 1 µL of the magnetic beads solution as a template. PCR products were quantified as described previously (Moyroud et al., 2011), and the selection cycle was repeated two times, each time using the newly synthesized fluorescent DNA as a library. The 73-mer libraries obtained after three cycles of selection were used subsequently for barcoding and high-throughput sequencing as described previously (Chahtane et al., 2013). Sequences were finally analyzed with MEME software (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) to produce a PWM.
Protein-Binding Microarrays on ABI3-B3 and FUS3
The FUS3 coding sequence was PCR amplified using Phusion High Fidelity DNA Polymerase (Thermo Scientific) and the oSB110R/oETH1132 primers. The amplified PCR product was subcloned into Zero Blunt (Life Technologies) and further transferred into pETM-40 (Dümmler et al., 2005) at NcoI-XhoI restriction sites to yield pETH221 expression vector, allowing the expression of FUS3 fused to a maltose-binding protein tag followed by a tobacco etch virus cleavage site.
The ABI3 B3 domain coding sequence was PCR amplified using Phusion High Fidelity DNA Polymerase (Thermo Scientific) and the oSB109F/oSB109R primers. The amplified PCR product was subcloned into Zero Blunt (Life Technologies) and further transferred into pMAL-C5x (New England Biolabs) at NdeI-NotI restriction sites to yield pETH214. This expression vector allows the expression of the ABI3 B3 domain fused to a maltose-binding protein tag followed by a Factor-Xa cleavage site.
These two proteins were expressed using E. coli strain Rosetta 2 (DE3). After induction by 0.4 mm IPTG, cells were grown overnight at 17°C, and soluble protein extracts were used for protein-binding microarray studies as described previously (Godoy et al., 2011).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. E-score distribution of top four 6mers recognized by FUS3 and ABI3.
Supplemental Figure S2. Schematic representation of the mutagenized versions of the OLE1 promoter used in this study.
Supplemental Figure S3. Activation of the native or mutated OLE1 promoter versions by LEC2 in moss protoplasts.
Supplemental Figure S4. Complementary analyses related to activation studies in moss protoplasts.
Supplemental Figure S5. Test of synergistic effect of LEC2, LEC1, and ABI3 on the activation of several oleosin promoters in moss protoplasts.
Supplemental Figure S6. In vitro binding of the L-AFLs to the OLE1 promoter.
Supplemental Figure S7. Complementary results to pull-down experiments.
Supplemental Figure S8. Study of protein-protein interactions in yeast.
Supplemental Table S1. Primers and genes used in this study.
Supplemental Table S2. List of primers used for site-directed mutagenesis of ProOLE1.
Supplemental Table S3. Statistical analyses.
Supplementary Material
Acknowledgments
We thank the Observatoire du Végétal for plant culture, access to the imaging facility, and assistance.
Glossary
- SSP
seed storage protein
- TAG
triacylglycerol
- SELEX
selection of ligand by exponential enrichment
- PWM
position-weight matrix
- EMSA
electrophoretic mobility-shift assay
- HA
hemagglutinin
- ChIP
chromatin immunoprecipitation
- cDNA
complementary DNA
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- Ni-NTA
nickel-nitrilotriacetic acid agarose
Footnotes
This work was supported by ANR projects Plant TF Code (grant no. ANR–07–BLAN–0211), CERES (grant no. ANR–BLAN–1238), and ERA-CAPS ABCEEDS and by Labex Saclay Plant Sciences (grant no. ANR–10–LABX–0040–SPS to the Institut Jean-Pierre Bourgin).
References
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthole G, To A, Marchive C, Brunaud V, Soubigou-Taconnat L, Berger N, Dubreucq B, Lepiniec L, Baud S (2014) MYB118 represses endosperm maturation in seeds of Arabidopsis. Plant Cell 26: 3519–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype Ws. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Dichow NR, Kelemen Z, d’Andréa S, To A, Berger N, Canonge M, Kronenberger J, Viterbo D, Dubreucq B, et al. (2009) Regulation of HSD1 in seeds of Arabidopsis thaliana. Plant Cell Physiol 50: 1463–1478 [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Bäumlein H, Nagy I, Villarroel R, Inzé D, Wobus U (1992) Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J 2: 233–239 [PubMed] [Google Scholar]
- Bäumlein H, Pustell J, Wobus U, Case ST, Kafatos FC (1986) The 3′ ends of two genes in the Balbiani ring c locus of Chironomus thummi. J Mol Evol 24: 72–82 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N, Dubreucq B, Roudier F, Dubos C, Lepiniec L (2011) Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23: 4065–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobb AJ, Chern MS, Bustos MM (1997) Conserved RY-repeats mediate transactivation of seed-specific promoters by the developmental regulator PvALF. Nucleic Acids Res 25: 641–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliari A, Turchetto-Zolet AC, Korbes AP, Maraschin FdS, Margis R, Margis-Pinheiro M (2014) New insights on the evolution of Leafy cotyledon1 (LEC1) type genes in vascular plants. Genomics 103: 380–387 [DOI] [PubMed] [Google Scholar]
- Calvenzani V, Testoni B, Gusmaroli G, Lorenzo M, Gnesutta N, Petroni K, Mantovani R, Tonelli C (2012) Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS ONE 7: e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahtane H, Vachon G, Le Masson M, Thévenon E, Périgon S, Mihajlovic N, Kalinina A, Michard R, Moyroud E, Monniaux M, et al. (2013) A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J 74: 678–689 [DOI] [PubMed] [Google Scholar]
- Che N, Yang Y, Li Y, Wang L, Huang P, Gao Y, An C (2009) Efficient LEC2 activation of OLEOSIN expression requires two neighboring RY elements on its promoter. Sci China C Life Sci 52: 854–863 [DOI] [PubMed] [Google Scholar]
- Conceição AS, Krebbers E (1994) A cotyledon regulatory region is responsible for the different spatial expression patterns of Arabidopsis 2S albumin genes. Plant J 5: 493–505 [DOI] [PubMed] [Google Scholar]
- Dickinson CD, Evans RP, Nielsen NC (1988) RY repeats are conserved in the 5′-flanking regions of legume seed-protein genes. Nucleic Acids Res 16: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dümmler A, Lawrence AM, de Marco A (2005) Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb Cell Fact 4: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerström M, Stålberg K, Ezcurra I, Rask L (1996) Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol Biol 32: 1019–1027 [DOI] [PubMed] [Google Scholar]
- Ezcurra I, Ellerström M, Wycliffe P, Stålberg K, Rask L (1999) Interaction between composite elements in the napA promoter: both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol Biol 40: 699–709 [DOI] [PubMed] [Google Scholar]
- Ezcurra I, Wycliffe P, Nehlin L, Ellerström M, Rask L (2000) Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J 24: 57–66 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Beachy RN (1994) Tissue-specific and temporal regulation of a beta-conglycinin gene: roles of the RY repeat and other cis-acting elements. Plant Mol Biol 24: 261–272 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy M, Franco-Zorrilla JM, Pérez-Pérez J, Oliveros JC, Lorenzo O, Solano R (2011) Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J 66: 700–711 [DOI] [PubMed] [Google Scholar]
- Golovenko D, Manakova E, Zakrys L, Zaremba M, Sasnauskas G, Grazulis S, Siksnys V (2014) Structural insight into the specificity of the B3 DNA-binding domains provided by the co-crystal structure of the C-terminal fragment of BfiI restriction enzyme. Nucleic Acids Res 42: 4113–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G, Tonelli C, Mantovani R (2001) Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene 264: 173–185 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G, Tonelli C, Mantovani R (2002) Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283: 41–48 [DOI] [PubMed] [Google Scholar]
- Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B (2012) Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant 5: 876–888 [DOI] [PubMed] [Google Scholar]
- Harada JJ. (2001) Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J Plant Physiol 158: 405–409 [Google Scholar]
- Hilioti Z, Ganopoulos I, Bossis I, Tsaftaris A (2014) LEC1-LIKE paralog transcription factor: how to survive extinction and fit in NF-Y protein complex. Gene 543: 220–233 [DOI] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T (1999) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19: 679–689 [DOI] [PubMed] [Google Scholar]
- Jia H, McCarty DR, Suzuki M (2013) Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol 163: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T (2005a) Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol 46: 300–311 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005b) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, Liwosz A, Eklöf S, Till S, Bögre L, Hirt H, et al. (2000) SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkbride RC, Fischer RL, Harada JJ (2013) LEAFY COTYLEDON1, a key regulator of seed development, is expressed in vegetative and sexual propagules of Selaginella moellendorffii. PLoS ONE 8: e67971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013) CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci 18: 157–166 [DOI] [PubMed] [Google Scholar]
- Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Lee H, Fischer RL, Goldberg RB, Harada JJ (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA 100: 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Distelfeld A, Comis A, Dubcovsky J (2011) Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J 67: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010) bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Miséra S (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1999) The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27 [DOI] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS (2006) Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J 46: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Masiero S, Imbriano C, Ravasio F, Favaro R, Pelucchi N, Gorla MS, Mantovani R, Colombo L, Kater MM (2002) Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J Biol Chem 277: 26429–26435 [DOI] [PubMed] [Google Scholar]
- Meinke DW. (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes A, Kelly AA, van Erp H, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013) bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke G, Altschmied L, Tewes A, Reidt W, Mock HP, Bäumlein H, Conrad U (2004) Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219: 158–166 [DOI] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Caretti G, Badaracco GF, Mantovani R (1999) Interactions of the CCAAT-binding trimer NF-Y with nucleosomes. J Biol Chem 274: 1326–1333 [DOI] [PubMed] [Google Scholar]
- Moyroud E, Minguet EG, Ott F, Yant L, Posé D, Monniaux M, Blanchet S, Bastien O, Thévenon E, Weigel D, et al. (2011) Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 23: 1293–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Miséra S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF III, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Reidt W, Wohlfarth T, Ellerström M, Czihal A, Tewes A, Ezcurra I, Rask L, Bäumlein H (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21: 401–408 [DOI] [PubMed] [Google Scholar]
- Reinders A, Schulze W, Kühn C, Barker L, Schulz A, Ward JM, Frommer WB (2002) Protein-protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell 14: 1567–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardt S, Timmerhaus G, Lang D, Qudeimat E, Corrêa LG, Reski R, Rensing SA, Frank W (2010) Microarray analysis of the moss Physcomitrella patens reveals evolutionarily conserved transcriptional regulation of salt stress and abscisic acid signalling. Plant Mol Biol 72: 27–45 [DOI] [PubMed] [Google Scholar]
- Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M (2009) Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE 4: e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D (2003) The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem 278: 1336–1345 [DOI] [PubMed] [Google Scholar]
- Ronchi A, Bellorini M, Mongelli N, Mantovani R (1995) CCAAT-box binding protein NF-Y (CBF, CP1) recognizes the minor groove and distorts DNA. Nucleic Acids Res 23: 4565–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol 56: 1215–1228 [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Sakata Y, Chiba Y, Fukushima H, Matsubara N, Habu Y, Naito S, Ohno T (1997) The RY sequence is necessary but not sufficient for the transcription activation of a winged bean chymotrypsin inhibitor gene in developing seeds. Plant Mol Biol 34: 191–197 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Shen B, Sinkevicius KW, Selinger DA, Tarczynski MC (2006) The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol Biol 60: 377–387 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Wobus U (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol 64: 189–217 [DOI] [PubMed] [Google Scholar]
- Stålberg K, Ellerström M, Josefsson LG, Rask L (1993) Deletion analysis of a 2S seed storage protein promoter of Brassica napus in transgenic tobacco. Plant Mol Biol 23: 671–683 [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Katagiri T, Hirayama T, Yamaguchi-Shinozaki K, Shinozaki K (2001) Hyperosmotic stress induces a rapid and transient increase in inositol 1,4,5-trisphosphate independent of abscisic acid in Arabidopsis cell culture. Plant Cell Physiol 42: 214–222 [DOI] [PubMed] [Google Scholar]
- Thakare D, Tang W, Hill K, Perry SE (2008) The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol 146: 1663–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin J, Dubos C, Xu W, Le Gourrierec J, Kelemen Z, Charlot F, Nogué F, Lepiniec L, Dubreucq B (2012) A new system for fast and quantitative analysis of heterologous gene expression in plants. New Phytol 193: 504–512 [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouiller B, Schaefer DG, Charlot F, Nogué F (2006) MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res 34: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Niu QW, Teng C, Li C, Mu J, Chua NH, Zuo J (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19: 224–235 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde RJ, Cooke SE, Brammar WJ, Schuch W (1994) Control of gene expression in plant cells using a 434:VP16 chimeric protein. Plant Mol Biol 24: 381–388 [DOI] [PubMed] [Google Scholar]
- Wobus U, Weber H (1999) Seed maturation: genetic programmes and control signals. Curr Opin Plant Biol 2: 33–38 [DOI] [PubMed] [Google Scholar]
- Xie Z, Li X, Glover BJ, Bai S, Rao GY, Luo J, Yang J (2008) Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol Biol Evol 25: 1581–1592 [DOI] [PubMed] [Google Scholar]
- Xu W, Lepiniec L, Dubos C (2014) New insights toward the transcriptional engineering of proanthocyanidin biosynthesis. Plant Signal Behav 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi K, Nagata N, Yee KM, Braybrook SA, Pelletier J, Fujioka S, Yoshida S, Fischer RL, Goldberg RB, Harada JJ (2005) TANMEI/EMB2757 encodes a WD repeat protein required for embryo development in Arabidopsis. Plant Physiol 139: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58: 843–856 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, et al. (2004) Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. Plant Cell 16: 3448–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsui I, Saruhashi M, Kawato T, Taji T, Hayashi T, Quatrano RS, Sakata Y (2013) ABSCISIC ACID INSENSITIVE3 regulates abscisic acid-responsive gene expression with the nuclear factor Y complex through the ACTT-core element in Physcomitrella patens. New Phytol 199: 101–109 [DOI] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack J, DePamphilis CW, Ma H, Theissen G (2005) To B or not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J Hered 96: 225–240 [DOI] [PubMed] [Google Scholar]
- Zhang H, Ogas J (2009) An epigenetic perspective on developmental regulation of seed genes. Mol Plant 2: 610–627 [DOI] [PubMed] [Google Scholar]
- Zhang W, Ruan J, Ho TD, You Y, Yu T, Quatrano RS (2005) Cis-regulatory element based targeted gene finding: genome-wide identification of ABA- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 21: 3074–3081 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.