Control of female flower opening in cucumber by hormone and nutritional cues, and genes related to cell wall, photosynthesis, protein degradation, and signaling.
Abstract
Flower opening is essential for pollination and thus successful sexual reproduction; however, the underlying mechanisms of its timing control remain largely elusive. We identify a unique cucumber (Cucumis sativus) line ‘6457’ that produces normal ovaries when nutrients are under-supplied, and super ovaries (87%) with delayed corolla opening when nutrients are oversupplied. Corolla opening in both normal and super ovaries is divided into four distinct phases, namely the green bud, green-yellow bud, yellow bud, and flowering stages, along with progressive color transition, cytological tuning, and differential expression of 14,282 genes. In the super ovary, cell division and cell expansion persisted for a significantly longer period of time; the expressions of genes related to photosynthesis, protein degradation, and signaling kinases were dramatically up-regulated, whereas the activities of most transcription factors and stress-related genes were significantly down-regulated; concentrations of cytokinins (CKs) and gibberellins were higher in accordance with reduced cytokinin conjugation and degradation and increased expression of gibberellin biosynthesis genes. Exogenous CK application was sufficient for the genesis of super ovaries, suggesting a decisive role of CKs in controlling the timing of corolla opening. Furthermore, 194 out of 11,127 differentially expressed genes identified in pairwise comparisons, including critical developmental, signaling, and cytological regulators, contained all three types of cis-elements for CK, nitrate, and phosphorus responses in their promoter regions, indicating that the integration of hormone modulation and nutritional regulation orchestrated the precise control of corolla opening in cucumber. Our findings provide a valuable framework for dissecting the regulatory pathways for flower opening in plants.
The flower is the most distinguishing organ in higher plants, and artists and scientists have been attracted to explore the mystery of flower structure and origin for decades. The evolution of flowering plants was greatly accelerated approximately 100 million years ago with the development of flowers, which were essential for recruiting animals to help distribute pollen and seeds (Danielson and Frommer, 2013). Flowers are produced from a specialized structure in the shoot tip called the shoot apical meristem, which consists of a pool of stem cells that continuously divide and replenish themselves (Fletcher et al., 1999). Morphologically, flowers are comprised of four basic structures arranged in concentric whorls: sepals in the outermost whorl 1, petals in whorl 2, stamens in whorl 3, and carpels in the innermost whorl 4. Bisexual flowers possess all four basic structures, while unisexual flowers lack one or more structures, usually the male organ stamen or the female organ carpel (Dellaporta and Calderon-Urrea, 1993). In flowering plants, approximately 90% species produce bisexual flowers, 6% species are dioecious with male and female flowers on separate plants, and 4% species are monoecious with male and female flowers on the same plant (Yampolsky and Yampolsky, 1922; Renner and Ricklefs, 1995). Arabidopsis (Arabidopsis thaliana) is a model species for bisexual plant, in which the patterning of floral organs is controlled by the combinatorial actions of three classes of homeotic genes, i.e. the A, B, and C class of genes (Bowman et al., 1991; Coen and Meyerowitz, 1991). In contrast, cucumber (Cucumis sativus) is a monoecious vegetable that has served as a model species for sex determination and differentiation (Malepszy and Niemirowicz-Szczytt, 1991). The unisexual flowers of cucumber are formed by selective arrestment of carpel or stamen development (Malepszy and Niemirowicz-Szczytt, 1991; Hao et al., 2003; Bai et al., 2004). Three ethylene synthesis enzymes, i.e. F (CsACS1G), M (CsACS2), and A (CsACS11), have been shown to regulate the development of unisexual flowers in cucurbits (Galun, 1962; Kamachi et al., 1997; Trebitsh et al., 1997; Mibus and Tatlioglu, 2004; Li et al., 2009; Boualem et al., 2015).
Flower opening is essential for pollination and thus successful sexual reproduction. Although intensive studies have been performed on flowering and five pathways (photoperiod, vernalization, autonomous, gibberellin, and aging pathways) have been identified to be involved in the regulation of flowering in model plants (Komeda, 2004), little is known about the mechanisms of flower opening. Depending on species, flower opening may be either nocturnal or diurnal, and either single or repetitive. Flower opening requires differential elongation growth or osmotic oscillations, followed by influx of water into cells (van Doorn and van Meeteren, 2003; Yamada et al., 2009b). Prior to opening, osmotic solute levels are rapidly increased by breaking down polysaccharides (starch or fructan) into monosaccharides, combining with sugar uptake from the apoplast (Hammond, 1982; Yamane et al., 1991; van Doorn and van Meeteren, 2003; Yamada et al., 2009a). Increased osmotic pressure contributes to the reduction of petal water potential, which facilitates water influx into the cells, and thus results in cell enlargement (Shinozaki et al., 2014). Cell wall extensibility is believed to be a limiting factor for petal expansion (Yamada et al., 2009b). The expressions of genes related to cell wall function such as those that encode xyloglucan endotransglucosylase/hydrolase and expansins are positively correlated with the petal growth during flower opening (Harada et al., 2011; Dai et al., 2012; Shinozaki et al., 2014). Hormones, including ethylene, gibberellins, and auxin, regulate petal growth and flower opening. Ethylene promotes flower opening in carnation, orchids, petunia, and some rare rose cultivars, but inhibits flower opening in other rose cultivars (Tang and Woodson, 1996; Jones and Woodson, 1997; Bui and O’Neill, 1998; Macnish et al., 2010). Ethylene inhibits rose opening via promoting the expression of DELLA genes, whose products repress cell expansion by binding to the promoters of cell wall synthesis genes (Luo et al., 2013). Exogenous gibberellin application promotes flower opening in plants such as Ipomoea nil (Raab and Koning, 1987), Gaillardia grandiflora (Koning, 1984), and Limonium sinuatum (Steinitz and Cohen, 1982). Gibberellic acid (GA) promotes cell elongation by opposing the function of DELLA proteins, and consistently GA-deficient Arabidopsis mutant plants display reduced petal growth (Cheng et al., 2004). Auxin has been reported to inhibit flower opening in Ipomoea (Kaihara and Takimoto, 1983). Mutation in the auxin signaling gene AUXIN RESPONSE FACTOR8 resulted in significantly larger petals with increased cell number and cell expansion (Varaud et al., 2011). Loss-of-function of genes implicated in auxin effects such as PETAL LOSS and miRNA319a resulted in reduced petal growth in Arabidopsis (Nag et al., 2009; Koyama et al., 2010; Varaud et al., 2011; Sauret-Güeto et al., 2013). In addition, light, temperature, and circadian clock are also involved in the regulation of flower opening (McClung and Gutiérrez, 2010; Ruelland and Zachowski, 2010; van Doorn and Kamdee, 2014). Nevertheless, the molecular mechanism of timing control of flower opening remains largely unknown, mainly due to lack of mutants or lines that display appreciable changes in the timing of flower opening.
In this study, we introduce a unique cucumber line ‘6547’ that produces super ovaries with 4-d to 5-d delay of corolla opening when nutrient supply is abundant. A super ovary refers to an ovary that is larger than normal at anthesis. It was first defined by Rudich et al. (1977) based on the phenotype of a parthenocarpic monoecious cucumber line (Nitsch, 1952; Nitsch et al., 1952). The main difference in the formation of normal and super ovaries in line ‘6547’ is the delayed corolla growth and opening of the super ovary. The resulted commercially mature cucumber fruits from the two types of ovaries are indistinguishable in length, but the fruits from super ovaries are straight and have flowers remaining on the tip, which are regarded as more attractive and preferred in the fresh market. To understand the underlying mechanisms for corolla opening in cucumber, we conducted detailed comparisons of the physiological, cytological, nutritional, and transcriptomic features of the super and normal ovaries across four stages of corolla development, and found that cell division and cell expansion, nutrient availability, photosynthesis, protein degradation, signaling kinases, and hormones were implicated in the timing control of corolla opening, and that cytokinin (CK) and nutritional cues acted as the predominant factors orchestrating the progression of female flower opening in cucumber.
RESULTS
Resource-Dependent Genesis and Morphological and Cytological Characterization of Super Ovaries
The ‘6457’ line belongs to the Southern China type of cucumbers, producing light-green fruits covered by sparse spines and warts (Fig. 1). It was first identified in 2002 to have a unique feature of spontaneously producing super ovaries with delayed corolla opening. It was then stabilized through selfing for over 10 generations. Under standard growth condition, the ratio of super ovaries produced by the ‘6457’ line was 29.1%. To dissect the developmental mechanism of the super ovary in cucumber, the ‘6457’ line was subjected to two distinct cultivation conditions, the super ovary induction treatment and the normal ovary induction treatment. Under the super ovary induction treatment, in which fertilizer and water were oversupplied (130% of standard) and only one fruit per plant was kept to eliminate interfruit competition, 87.4% of plants (n = 322) produced super ovaries. Under the normal ovary induction treatment, in which fertilizer and water were under-supplied (50% of standard) and two fruits per plant were kept, 100% of plants (n = 300) produced normal ovaries. Super and normal ovaries produced under the above two growth conditions were used for further characterization hereafter.
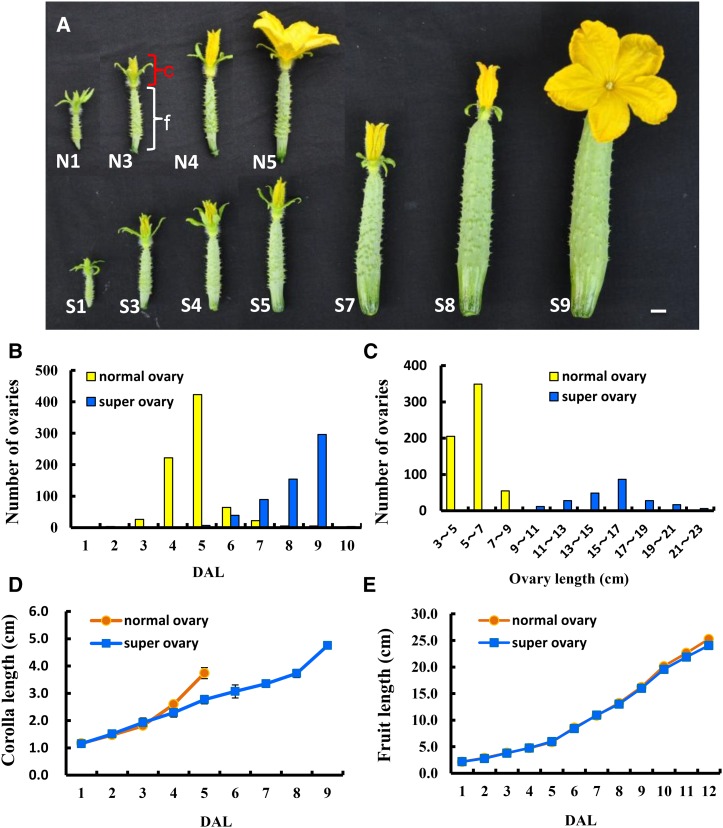
Figure 1.
Morphological characterization of the normal ovary and super ovary in cucumber. A, Progression of corolla opening in the normal ovary (N; top) and super ovary (S; bottom). The numbers refer to DAL. B, Quantification of the days to blooming in the normal ovary and super ovary. C, The fruit at anthesis was on average much longer in the super ovary than the normal ovary. D and E, Development rates of corolla length (D) and fruit length (E) in the normal ovary and super ovary. Bar = 1 cm. c, Corolla (red square bracket); f, fruit (white square bracket).
The normal ovaries mostly bloomed at 5 d after labeling (DAL; labeling was done when an ovary became visible), while most super ovaries bloomed at 9 DAL (Fig. 1, A and B). The average fruit length at anthesis from the normal and super ovary producing plants was 5.8 cm and 14.4 cm, respectively (Fig. 1C). Accordingly, the corolla at anthesis was much larger in the super ovary, although corollas from both ovaries accelerated their growth 1 d before anthesis (Fig. 1D). However, fruits resulted from super and normal ovaries displayed no significant difference in their growth rates, and the fruits at the commercially mature stage showed no appreciable difference in length (Fig. 1E). As for seed production, although both normal and super ovaries had a high percentage of seeded fruit if hand pollination was carried out at DAL 5 (when the normal ovary was at anthesis), the average number of seeds per fruit was significantly reduced in the super ovary (45.8) as compared with that in the normal ovary (166.2; Supplemental Table S1). No seeds were produced if pollination was performed at DAL 9 (when the super ovary was at anthesis; Supplemental Table S1). Therefore, super ovaries have two major advantages over normal ovaries in cucumber production. First, super ovaries produce straight fruits with higher market values due to delayed corolla opening, inability to be fertilized at anthesis, and no seed production in the open field. Normal ovaries sequentially undergo fertilization and seed development, thus frequently giving rise to fruits with a big-belly shape in the open field. Second, fruits from the super ovaries have flowers remaining on the tip and appear freshly harvested, which is a favorable feature for customers in the Asian vegetable market.
As seen from the above characterization, one fundamental difference between super and normal ovaries was the temporally differential growth and opening of corolla. Based on the color and shape, corolla development can be divided into four stages: green bud, green-yellow bud, yellow bud, and flowering. In the normal ovary, 1 DAL (N1), 3 DAL (N3), 4 DAL (N4), and 5 DAL (N5) indicated the green bud, green-yellow bud, yellow bud, and flowering stages, respectively, whereas in the super ovary, these four stages typically corresponded to 1 DAL (S1), 3 to 5 DAL (S3, S4, and S5), 7 to 8 DAL (S7 and S8), and 9 DAL (S9), respectively (Fig. 2A). Thus, the corolla progression between stages was much delayed in the super ovary, especially during the green-yellow bud and yellow bud stages.
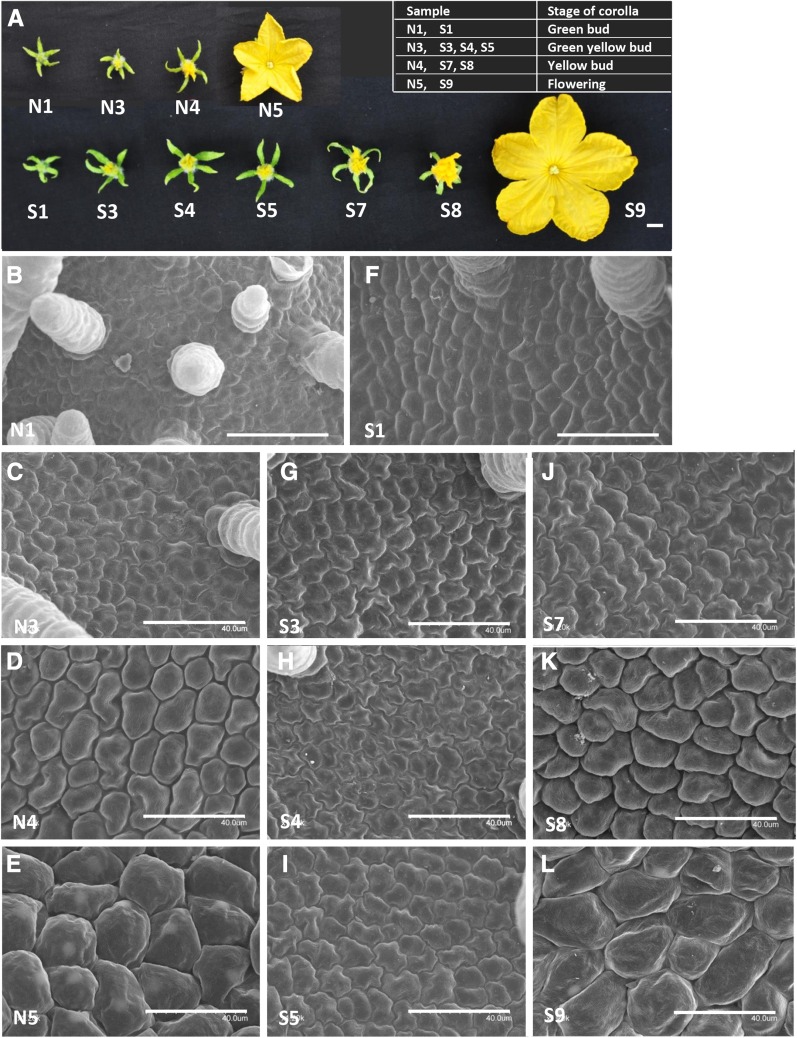
Figure 2.
Cytological characterizations of petals in the normal ovary and super ovary in cucumber. A, The four typical stages of corolla development in the normal ovary (top) and super ovary (bottom) at which samples were collected for transcriptome analyses: green bud, green-yellow bud, yellow bud, and flowering. B to L, Scanning electron micrographs of the adaxial side of petals in the normal ovary (B–E) and super ovary (F–L). Note that the cell size was progressively increased from green bud to flowering stage in both normal ovary and super ovary, but cell division and cell expansion persisted for a longer period of time in the super ovary, especially during the green-yellow bud (G–I) and yellow bud stages (J and K). Bars represented 1 cm in A and 40 μm in B to L.
To explore cytological differences in the corolla development between normal and super ovaries, scanning electron microscopy was used to examine the adaxial side of petals at each stage. In the normal ovary, there was no notable change in the cell size from green bud to green-yellow bud (Fig. 2, B and C), but there was dramatic cell expansion from yellow bud to flowering (Fig. 2, D and E). Considering that the corolla length was progressively increasing (Fig. 1D), these data suggested that the cells were mostly in cell division during the green-yellow bud stage and were in cell expansion since the yellow bud stage. In the super ovary, on the other hand, although the cell sizes in the initial stage (S1, green bud) and the final stage (S9, flowering) were the same as those in the normal ovary, cell size remained generally unchanged from S1 to S5, followed by gradual cell expansion from S5 to S9 (Fig. 2, F–L); this suggests that both cell division and cell expansion persisted for a longer period of time in the corolla of the super ovary, resulting in enlarged size of corolla (Fig. 1D).
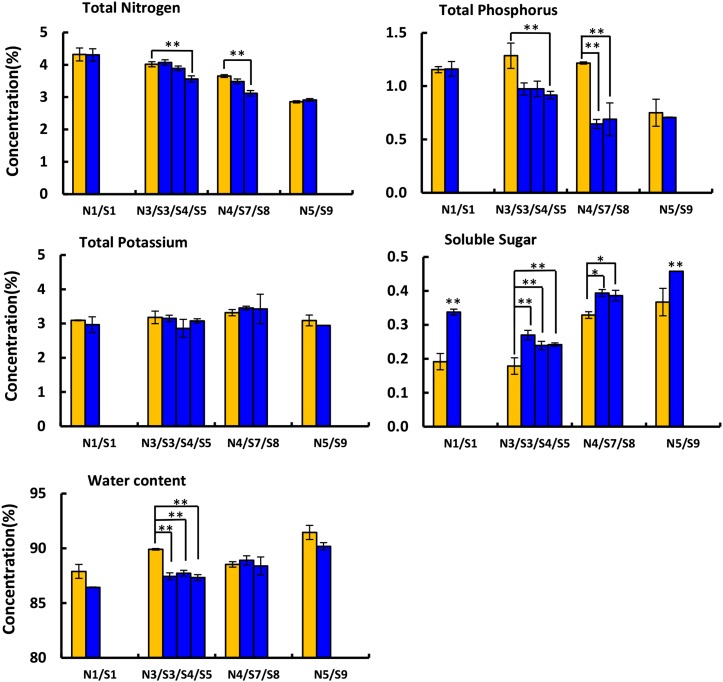
Since the genesis of the super ovary is closely linked to nutrient supplies, the contents of total nitrogen (N), phosphorus (P), potassium (K), sugars, and water in the corolla of normal and super ovaries were analyzed across all four stages (Fig. 3). The concentrations of N and P decreased with corolla opening, but the reduction was significantly greater in the super ovary during the green-yellow bud (N3; S3, S4, and S5) and yellow bud (N4; S7 and S8) stages (Fig. 3). The concentration of K showed no difference between normal and super ovaries. Soluble sugars, on the other hand, were significantly increased in the super ovary during all four stages (Fig. 3). In both normal and super ovaries, water content increased with the development of corolla (Fig. 3), which is consistent with previous findings that water potential was higher in petals than leaves at anthesis (Trolinder et al., 1993). Although super ovaries were supplied with abundant water (130% standard), the increase of water content was smaller than normal ovaries during the green-yellow bud stage (Fig. 3), implying that the hastened flower opening in normal ovaries may be a stress adaption in cucumber (Fig. 1A).
Figure 3.
Nutritional measurements in the corollas of the normal ovary and super ovary. The levels of total N, total P, total K, soluble sugars, and water content in the corollas of the normal ovary (orange) and super ovary (blue) at four developmental stages: green bud (N1 and S1), green-yellow bud (N3, S3, S4, and S5), yellow bud (N4, S7, and S8), and flowering (N5 and S9). At least three replicates were performed and bars represent standard deviations. Asterisks and double asterisks represent significant difference as compared with that in the normal ovary at P < 0.05 and P < 0.01, respectively (Duncan’s multiple range test).
Distinct Transcriptomic Signatures during the Progression of Corolla Opening in Normal and Super Ovaries
To identify genes and gene networks regulating corolla opening, we performed time-course RNA-seq analysis of the corollas in female flowers of super and normal ovaries, including samples from the green bud (N1; S1), green-yellow bud (N3; S3, S4, and S5), yellow bud (N4; S7 and S8), and flowering stages (N5; S9; Fig. 2A), and three biological replicates for each time point. High-throughput RNA sequencing generated 40.15 to 57.04 million pair-ended reads for each sample, with a total of 1.548 billion paired-end reads (193-Gb sequences) for all 33 libraries (Supplemental Table S2). Clean reads (32.52–47.75 million) were mapped to the cucumber genome using the software TopHat (Huang et al., 2009; Trapnell et al., 2009). The expression level of each gene was analyzed with the Python package “HT-seq” (Anders et al., 2015) and 23,247 genes with at least 1 transcript per million (TPM) among three replicates were considered expressed.
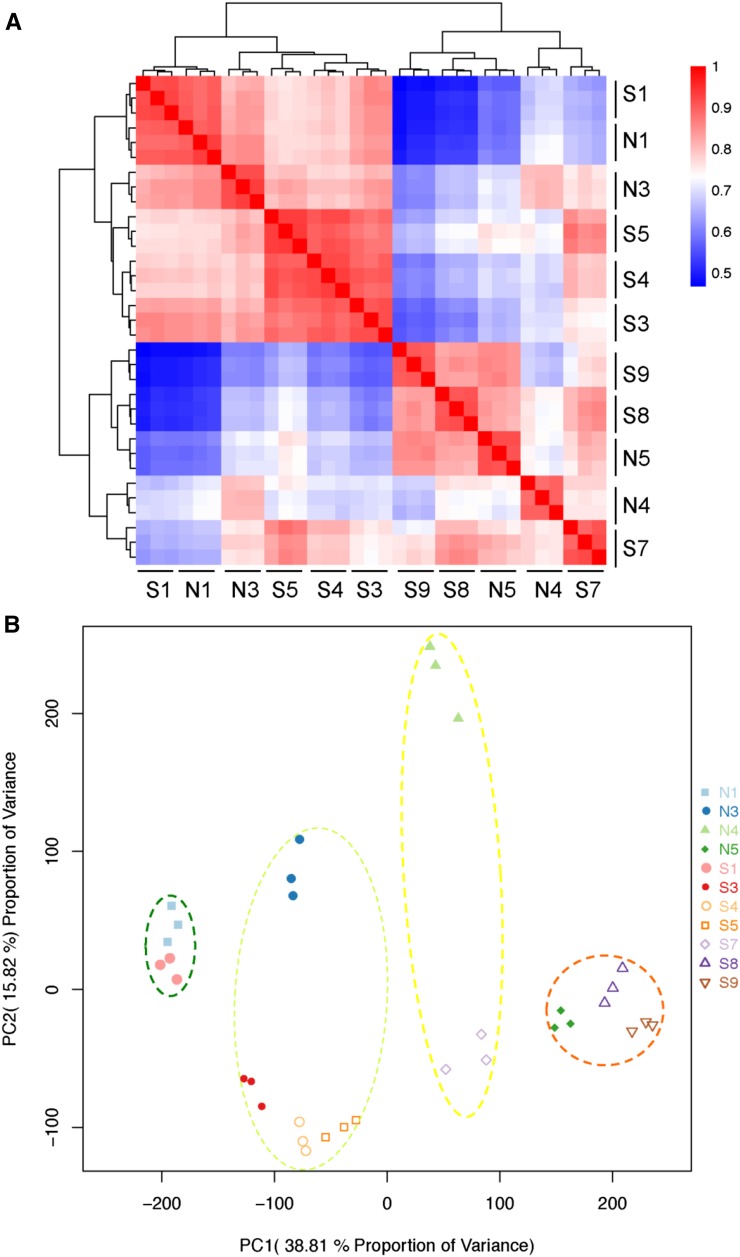
The correlation clustering (Fig. 4A) and principal component analysis (PCA; Fig. 4B) of the 33 RNA-seq datasets showed four distinct groups corresponding to the sequential stages of corolla development: green bud (N1; S1), green-yellow bud (N3; S3, S4, and S5), yellow bud (N4; S7), and flowering (N5; S9). Although S8 is phenotypically the yellow bud stage, PCA analysis showed that it had an expression pattern resembling that of the flowering stage, which may be because gene expression generally progressed prior to phenotypic transformation. The wider area occupied by groups 2 (green-yellow circle) and 3 (yellow circle) than groups 1 (green circle) and 4 (orange circle) in the PCA analysis indicated that normal and super ovaries had larger differences in gene expression profiles during the green-yellow bud and yellow bud stages (Fig. 4B), which underlay the delayed corolla opening in the super ovary.
Figure 4.
Global analyses of RNA-seq data from different corolla samples. A and B, Correlations (A) and the PCA (B) of gene expressions in corolla samples from the normal ovary and super ovary.
A total of 14,282 unique differentially expressed genes (DEGs) were identified through the R package edgeR with a false discovery rate (FDR) less than 0.05 and a fold change greater than 2 (Robinson et al., 2010; Supplemental Table S3). To verify DEGs identified by RNA-seq, 10 DEGs representing the green bud, yellow bud, and flowering stages (Supplemental Fig. S1) were chosen for quantitative real-time RT-PCR (qRT-PCR) analysis using independently collected samples. The qRT-PCR data showed the same expression patterns as those in the RNA-seq analysis (Supplemental Fig. S1), and the Pearson’s correlation was 0.89 between qRT-PCR and RNA-seq data, indicating high reliability of the RNA-seq results.
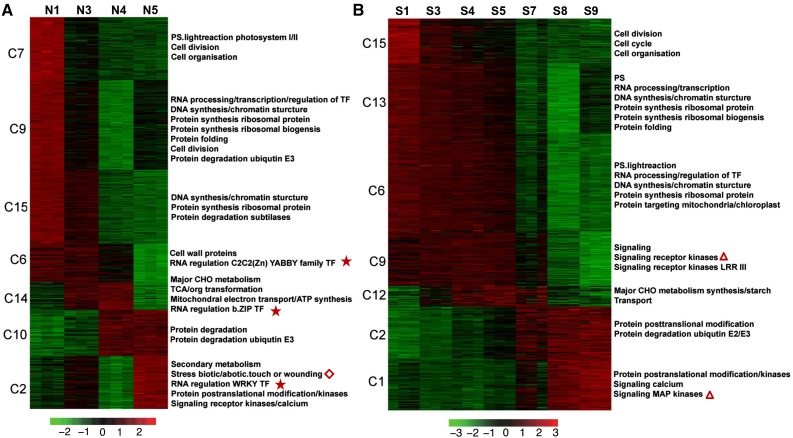
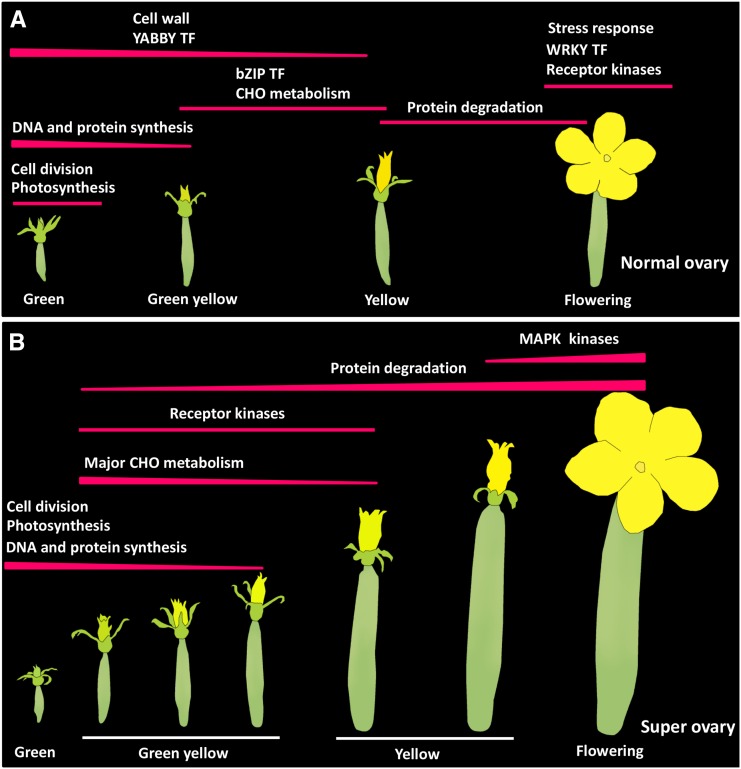
Using the K-means clustering method, 11,875 DEGs expressed in the normal ovary and 12,728 DEGs expressed in the super ovary were classified into 15 coexpression modules, respectively (Fig. 5, A and B, Supplemental Table S4). In the normal ovary, sequential gene expression featured the four developmental stages of corolla opening. Cell division-related genes were specifically expressed in the green bud stage (N1). Genes encoding transcription factors (TFs) and genes related to DNA and protein synthesis were highly expressed in the green bud stage and weakly expressed in the green-yellow bud stage (N3). Genes encoding cell wall proteins and YABBY family TFs were highly expressed in the first two stages, and weakly expressed in the yellow bud stage (N4). Major CHO metabolism and bZIP TF genes were specifically up-regulated in N3 and N4, while genes related to protein degradation were specifically expressed in the N4 and flowering stages (N5). Stress-related genes including genes encoding WRKY TFs and signaling kinases were highly up-regulated in the N5 (Fig. 5A). Gene expression patterns in the super ovary were generally similar to those in the normal ovary when S3, S4, and S5 were considered as the green-yellow bud stage, and S7 and S8 as the yellow bud stage. However, there were three key distinctions in the super ovary. (1) Genes encoding receptor kinases were expressed much earlier. Signaling kinase genes were significantly overrepresented in cluster 9 (highly expressed in the green bud stage and green-yellow stages) and cluster 1 (highly expressed in the yellow bud and flowering stages; triangles in Fig. 5B). (2) Genes encoding TFs such as YABBY, bZIP, and WRKY family members were not enriched in any of the 15 clusters (stars in Fig. 5A). (3) Stress-related genes were not overrepresented in the super ovary (diamond in Fig. 5A).
Figure 5.
Gene expression pattern and functional category over the time course of corolla development in the normal ovary and super ovary. A and B, Heat maps showing the expression patterns of genes in seven selected K-means clusters in the normal ovary (A) and super ovary (B). For each gene, the value shown was the Z-score value of normalized TPM. The red color indicated higher expression while green indicated lower expression. Overrepresented (FDR < 0.05) functional categories were indicated on the right.
Next, transcriptomic data were subject to three types of pairwise comparisons (Supplemental Fig. S2). Type 1 DEGs were genes with differential expression at two adjacent time points (Supplemental Fig. S2A). In the normal ovary, 1929 DEGs were up-regulated and 1937 DEGs were down-regulated from N1 to N3, and the number of DEGs was progressively increased along with the progression of corolla opening. In the super ovary, the numbers of type 1 DEGs were generally reduced as compared with those in the normal ovary, implying reduced molecular changes between time points, consistent with the morphological and cytological data of retarded transitions between stages in the super ovary (Figs. 1 and 2). Specifically, 1243, 531, and 501 genes were differentially expressed in S3/S1, S4/S3, and S5/S4, respectively, followed by a sharp increase in the number of DEGs (2628–3570 DEGs) that occurred from S5 to S9 (Supplemental Fig. S2A). Type 2 DEGs were identified by comparisons of the transcriptome at a later time point with that at N1 or S1 (Supplemental Fig. S2B). In both normal and super ovaries, the numbers of type 2 DEGs increased along with corolla development from the green bud stage to blooming, with more dramatic increase in the super ovary (Supplemental Fig. S2B). Type 3 DEGs were derived from comparisons of transcriptomes between super and normal ovaries at the same time point or the same developmental stage (Supplemental Fig. S2C). Compared with S1/N1 (green/green) and S3/N3 (green-yellow/green-yellow), S4/N4 (green-yellow/yellow) and S5/N5 (green-yellow/flowering) had significantly more DEGs, suggesting that the developmental stage is a more important factor than the time interval in patterning gene expression during corolla opening. Within the same developmental stage, significantly more DEGs were identified in the green-yellow bud stage (S3/N3, S4/N3, and S5/N3) and yellow bud stage (S7/N4 and S8/N4), as compared with the number of DEGs during the green bud stage or flowering stage (Supplemental Fig. S2C), further supporting that green-yellow and yellow bud stages may lay the molecular basis for delayed corolla opening in the super ovary.
Transcriptomic Differentiation of the Normal and Super Ovaries during Corolla Opening
To dissect the molecular differentiation during corolla opening between the normal and super ovaries, the type 3 DEGs were further functionally classified using the MapMan program (Fig. 6). Genes related to cell wall were highly overrepresented in the genes that were up-regulated in the super ovary during green-yellow (S4/N3 and S5/N3), yellow bud (S7/N4 and S8/N4), and flowering stages (S9/N5; diamond in Fig. 6). The cell wall is essential for cell division and cell expansion (Quatrano and Shaw, 1997; Sampathkumar et al., 2014), and up-regulated cell wall-related genes in the super ovary were correlated with the elongated period of cell division and cell expansion as revealed by the cytological data (Fig. 2). In addition, genes related to hormone metabolism were up-regulated in the normal ovary during the green-yellow bud stage (S3/N3, S4/N3, and S5/N3), but were up-regulated in the super ovary during the yellow bud stage (S7/N4 and S8/N4; triangle in Fig. 6), suggesting that activation of hormone-related genes were delayed in the super ovary during corolla opening. In addition, signaling genes was significantly enriched in the up-regulated genes in the super ovary (star in Fig. 6).
Figure 6.
Enrichment of MapMan functional categories in the DEGs between the normal ovary and super ovary. The heat map showed the enrichment in each category of DEGs. The significance of enrichment (FDR) was indicated by the color ladder.
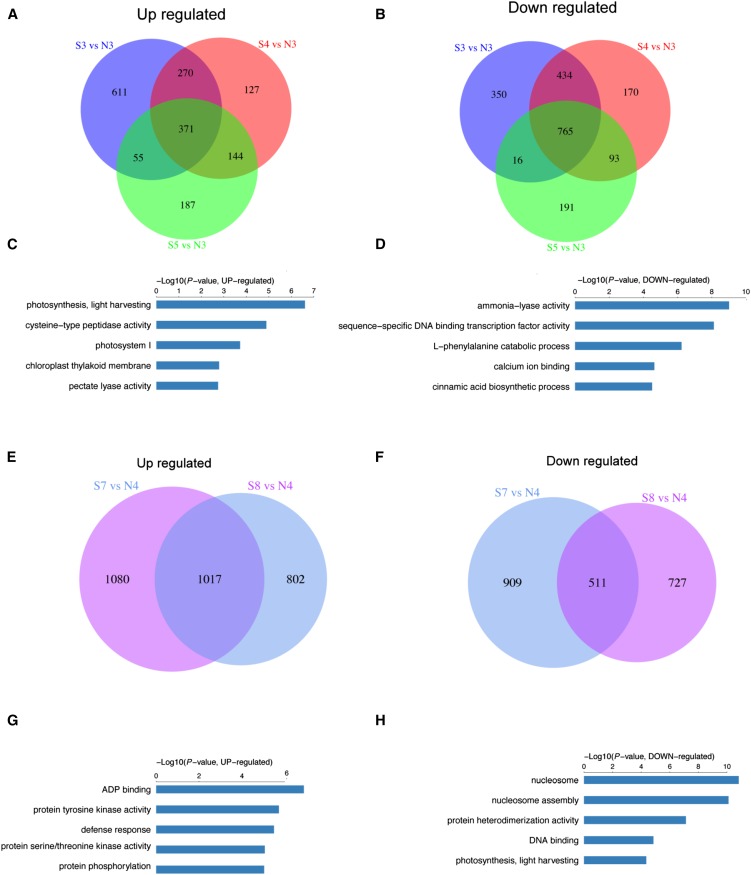
Because the phenotypic and transcriptomic data suggested that the differentiation of the super ovary from the normal ovary occurred in the green-yellow bud and yellow bud stages (Figs. 1 and 2, Supplemental Fig. S2C), we next investigated the type 3 DEGs during these two stages. During green-yellow stage, 371 genes were commonly up-regulated while 765 genes were commonly down-regulated in the corollas of the super ovary (Fig. 7, A and B). GO enrichment analysis showed that “photosynthesis, light harvesting” and “cysteine-type peptidase activity” were the most significantly enriched GO terms in the up-regulated DEGs in the super ovary (Fig. 7C), suggesting that photosynthesis and protein degradation were elevated in the corolla of the super ovary at green-yellow stage. “Ammonia-lyase activity” and “sequence-specific DNA binding transcription factor activity” were the top two enriched GO terms in the down-regulated DEGs (Fig. 7D). Specially, 46 TFs were commonly down-regulated while only six TFs were commonly up-regulated in the super ovary (Fig. 7, C and D), suggesting that most TFs were repressed in the super ovary during green-yellow bud stage. For example, the normalized expression level of a putative basic helix-loop-helix TF (Csa3G002860) was 1.9 in N3, but only 0.2, 0.3, and 0.9 in S3, S4, and S5, respectively; and the normalized expression level of a putative RAD-like1 TF (Csa3G857030) was 3.8 in N3, but only 1.5, 1.3, and 1.5 in S3, S4, and S5, respectively.
Figure 7.
GO term enrichment of the common DEGs between the normal ovary and super ovary at green-yellow and yellow stages. A and B, Venn diagrams of the overlapping DEGs that were up-regulated (A) or down-regulated (B) at the green-yellow bud stage (S3/N3, S4/N3, and S5/N3). C and D, Bar plot showing the top 5 GO terms of commonly up-regulated (C) and down-regulated (D) DEGs corresponding to A and B, respectively. E and F, Venn diagrams of the overlapping DEGs that were up-regulated (E) or down-regulated (F) at the yellow bud stage (S7/N4, S8/N4). G and H, Bar plot showing the top 5 GO terms of commonly up-regulated (G) or down-regulated (H) DEGs corresponding to E and F, respectively.
During the yellow bud stage, 1017 type 3 DEGs were commonly up-regulated and 511 type 3 DEGs were commonly down-regulated in the corolla of the super ovary (Fig. 7, E and F). GO enrichment analysis showed that “ADP binding” and “protein tyrosine kinase activity” were the most significantly enriched GO terms in the up-regulated DEGs in the super ovary (Fig. 7G), indicating that signaling kinase genes were activated in the super ovary during the yellow bud stage. For example, many MAP kinase family genes such as CsMAPKKK5-2 (Csa2g60650) were significantly down-regulated at N4, while no such repression was observed in the super ovary at S7 and S8 (Supplemental Fig. S3). In addition, most MAP kinase family genes were activated from yellow bud to flowering stage in the normal ovary, and such activation was elevated in the super ovary, especially during yellow bud stage (Supplemental Fig. S3). For example, the expression of putative CsMAPKKK21-2 (Csa2G278170) increased from 0.43 at N3 to 2.73 at N4 in the normal ovary, while it dramatically changed from 0.43 at green-yellow stage (S5) to 10.0 at yellow stage (S8) in the super ovary (Supplemental Fig. S3; Supplemental Table S5). In the commonly down-regulated DEGs in the super ovary, genes related to nucleosome assembly were highly overrepresented (Fig. 7H).
Of particular interest was the temporal compartmentation of enriched hormone metabolism genes (Fig. 6) in the corolla between normal and super ovaries, especially those related to CK and gibberellin’s metabolism (Supplemental Table S6). As shown in Figure 8, genes encoding UGTs and CK oxidases (CKXs) in the CK pathway were the top two groups of genes that were differentially expressed in the corollas of super and normal ovaries (Fig. 8). UGTs function in CK conjugation by converting active CKs into inactive stable storage forms (Schmülling et al., 2003). Many UGTs were specifically expressed in the early stages of corolla development. For example, Csa7G063950 (UGT85A2) and Csa5G196580 (UGT85A2) were highly expressed in the green bud stage and weakly expressed in the green-yellow bud stage in both normal and super ovaries (Supplemental Table S5). In contrast, some other UGT genes such as Csa7G059150 (UGT85A2) and Csa7G063980 (UGT85A2) were specifically up-regulated in the later stages of corolla development only in the normal ovary (Fig. 8A; Supplemental Table S6). Importantly, genes encoding CKXs such as CKX3 and CKX6 that function in CK degradation (Bilyeu et al., 2001) were significantly up-regulated during the flowering stage in the normal ovary, but such up-regulation was greatly reduced in the super ovary. These data suggested that CK conjugation and degradation were reduced in the super ovary during the later stages of corolla opening.
Figure 8.
Expressions of key CK and gibberellin pathway genes across developmental stages in the normal and super ovaries. A and B, Heat-map expression profiles of the CK- (A) and gibberellin-related (B) genes at different stages of corolla development. The colors in the squares represent the Z-score values of normalized expression levels.
Putative GA biosynthesis genes GA5, GA2, CYP88A3, GA1, and KAO2 were specifically expressed in the green bud stage in the normal ovary, but were expressed in both green bud and green-yellow bud stages (S3, S4, and S5) in the super ovary (Fig. 8B). Particularly, Csa3G903540 (KAO2) was expressed 5.4-fold higher in the super ovary compared with the normal ovary during green bud stage. On the other hand, GA2ox8, GA2ox1, and GA2ox, which are involved in GA metabolism, were highly expressed in the flowering stage, but their expression levels were reduced in the super ovary (Olszewski et al., 2002; Sun, 2008). Genes encoding the homologs of GA receptors GID1B and DELLA proteins (RGA1 and RGL2) that function in GA signaling (Olszewski et al., 2002; Sun, 2008) were mainly expressed in the flowering stage in both normal and super ovaries (Fig. 8B).
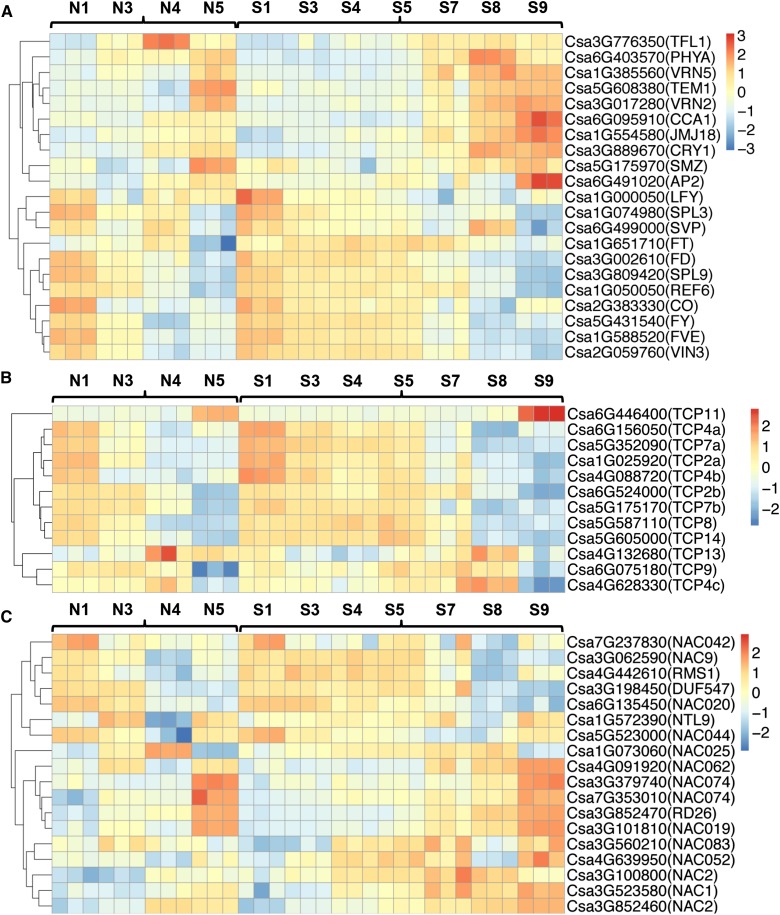
Previous studies showed that five pathways, including photoperiod, vernalization, autonomous, gibberellin, and aging pathways, were involved in regulation of the timing of flowering in model plants (Komeda, 2004). To investigate whether flowering regulators participate in flower opening in cucumber, homologs of flowering genes were identified from all DEGs and the corresponding heat-map expression profiles were compared between normal and super ovaries across all stages (Fig. 9A). Negative regulators of flowering such as TERMINAL FLOWER1 and SCHLAFMUTZE were down-regulated and positive regulators such as CIRCADIAN CLOCK ASSOCIATED1, JUMONJI DOMAIN-CONTAINING PROTEIN18, CRYPTOCHROME1, and APETALA2 were significantly up-regulated in the super ovary at the same developmental stages (Fig. 9A), suggesting that flowering regulators may contribute to the delayed corolla opening in super ovary in the opposite way than they do for flowering control. In addition, TCP and NAC TFs have been shown to regulate organ size in flower petals (Pei et al., 2013; van Doorn and Kamdee, 2014). Here we found that genes encoding five putative TCP TFs (TCP11, TCP4a, TCP7a, TCP2, and TCP4b) displayed higher expression in the super ovary (Fig. 9B), with Csa6G446400 (a putative TCP11) being specifically expressed in the flowering stage (63.2 TPM in the normal ovary and 210.4 TPM in the super ovary; Supplemental Table S7). In Arabidopsis, TCP 11 and TCP7 belong to class I subfamily, whereas TCP4 and TCP2 belong to class II subfamily, and all the four genes positively regulate organ growth through cell proliferation (Viola et al., 2011; Aguilar-Martínez and Sinha, 2013; Li, 2015). Similarly, genes encoding multiple NAC TFs such as NAC062, NAC052, NAC2, and NAC1 were up-regulated in the super ovary (Fig. 9C), including the homolog of RhNAC2 (Csa3G852460) that regulates cell expansion through binding to the promoter of RhEXPA4 in rose petals and Arabidopsis (Dai et al., 2012). These data suggested that the enlarged petal size in the super ovary may be caused by increased cell proliferation and cell expansion mediating by TCP and NAC TFs.
Figure 9.
Expressions of flowering-related genes and TCP and NAC TF-encoding genes across developmental stages in the normal and super ovaries. A to C, Heat-map expression profiles of flowering-related genes (A), TCP TF genes (B), and NAC TF genes (C) at different stages of corolla development in cucumber. The colors in the squares represent the Z-score values of normalized expression levels.
Decisive Roles of Hormone Modulation in the Genesis of the Super Ovary
Phytohormones play essential regulatory roles in most flower developmental processes (de Jong et al., 2009; Bartrina et al., 2011; Kumar et al., 2014). Our transcriptomic data showed that genes related to the metabolism of hormones, especially CKs and gibberellins, were differentially expressed in the corollas of two types of ovaries (Figs. 6 and 8). Therefore, we measured the levels of CKs (3-indole propionic acid, IPA; trans-zeatin riboside, ZR; dihydrogen zeatin riboside, DHZR), gibberellins (GA3, GA4), auxin (3-indole acetic acid [IAA]), abscisic acid (ABA), jasmonic acid (JA), and brassinosteroid (BR) in the corollas and fruits of normal and super ovaries at the green-yellow bud (N3; S3–S5), yellow bud (N4 and S7), and flowering (N5 and S9) stages. As shown in Figure 10, the concentrations of IPA and ZR were significantly reduced in the corolla of normal ovary during the flowering stage (N5), but remained unchanged in the super ovary. GA3 and GA4 concentrations increased during the green-yellow bud and yellow bud stages in the super ovary. The level of ABA decreased in the yellow bud stage, and that of JA dramatically increased at the flowering stage in the corolla of the super ovary. No significant difference was observed for the IAA and BR levels between the corollas of normal and super ovaries at all three stages examined (Fig. 10). On the other hand, the concentration of CKs was similar in the fruit, and GA4 increased during the green-yellow bud stage, whereas IAA was significantly decreased in the fruit of the super ovary in both green-yellow bud and flowering stages (Supplemental Fig. S4A). During the green-yellow bud stage, levels of ABA and BR were reduced in the fruit of the super ovary (Supplemental Fig. S4A). As such, the concentrations of CKs, gibberellins, and JA were higher, while that of ABA in the corolla of the super ovary. In the fruit, the level of gibberellins was increased, while the levels of IAA, ABA, and BR were reduced in the super ovary compared with those in the normal ovary.
Figure 10.
Hormone measurements in the corollas of normal ovary and super ovary in cucumber. The concentrations of IPA, ZR, DHZR, gibberellins (GA3, GA4), auxin (IAA), ABA, JA, and BR in the corollas of normal ovary (orange) and super ovary (blue) at three stages: green-yellow bud (N3, S3, and S5), yellow bud (N4 and S7), and flowering (N5 and S9). Three biological replicates were performed, and the bars represent sd. Double asterisks represent significant difference as compared with the concentration in the normal ovary at P < 0.01(unpaired t test).
To further investigate whether CK is a dominant regulator of the super ovary genesis, different concentrations of thidiazuron (TDZ) were applied to the normal ovary during the green bud stage. As shown in Table I, TDZ treatments significantly induced the super ovary production as compared with the water control, and the 2.5 mg L−1 TDZ treatment displayed the best effect. The frequency of the super ovary increased from 12.5% to 75% upon the TDZ treatment, and corolla opening was delayed from 1 to approximately 2 d, to 4 to approximately 5 d. However, unlike in the super ovary, TDZ treatment dramatically accelerated the growth rate of fruit (Fig. 1E; Supplemental Fig. S4B).
Table I. Effects of exogenous CK treatment on the super ovary production.
| TDZ | Percentage of Super Ovaries | Ovary Length at Anthesis | Delayed Days for Corolla Opening |
|---|---|---|---|
| mg/L | cm | ||
| Control | 0 | 3.9 | 0 |
| 1.25 | 12.5 | 6.8* | 1 to approximately 2 |
| 2.5 | 75.0 | 11.2** | 4 to approximately 5 |
| 5 | 25.0 | 7.5** | 2 to approximately 3 |
| 10 | 37.5 | 7.9** | 2 to approximately 3 |
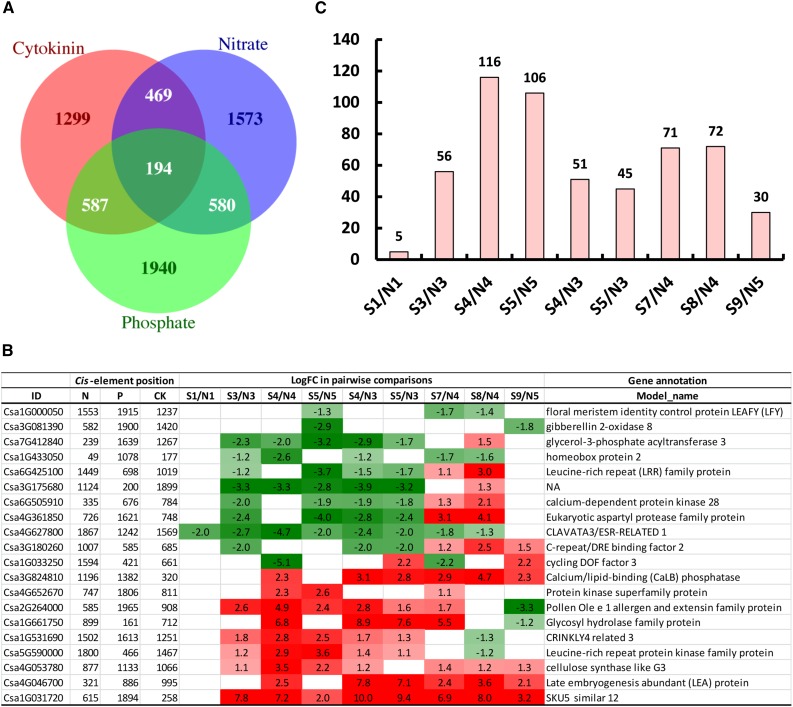
To connect CK functioning and nutritional regulation during the super ovary genesis, we analyzed the cis-regulatory elements for CK, N, and P in the 2-kb promoter regions of 11,127 type 3 DEGs (Konishi and Yanagisawa, 2010; Ramireddy et al., 2013; Schünmann et al., 2004; Fig. 11A), and found 2549 genes with CK cis-element 5′-AAGAT(T/C)TT-3′, 2816 genes with nitrate response motif 5′-(T/A)7A(G/C)TCA-3′, and 3301 genes with P-related cis-element 5′-GNATATNC-3′. A total of 194 genes had all three kinds of cis-elements, including genes involved in signaling kinases/phosphatases, TFs, cell wall, and proteases (Fig. 11B; Supplemental Table S8). In particular, the putative CLAVATA3 (Csa4G627800) that functions in meristem maintenance and contains the cis-elements for N, P, and CK at 1867, 1242, and 1569 bp of promoter, respectively, was significantly down-regulated in the super ovary. The putative SKU5 similar 12 gene (Csa1G031720) that mediates cell-wall expansion and contains the cis-elements for N, P, and CK at 615, 1894, and 258 bp of promoter, respectively, was dramatically up-regulated in the super ovary (Supplemental Table S8; Fletcher et al., 1999; Sedbrook et al., 2002). The distribution of the 194 DEGs with all three cis-elements across the four developmental stages correlated with the number of DEGs in each pairwise comparison. S4/N4 (green-yellow/yellow) and S5/N5 (green-yellow/flowering) had the most DEGs because different stages were compared. Within the same developmental stages, the yellow bud stage (S7/N4 and S8/N4) had the highest number of DEGs, followed by the green-yellow bud stage (S3/N3, S4/N4, and S5/N3) and the flowering stage (S9/N5), and the green bud stage (S1/N1) had the least number of DEGs with cis-elements for N, P, and CK (Fig. 11C).
Figure 11.
cis-Elements for CK, N, and P response in the promoters of type 3 DEGs. A, Venn diagrams of the overlapping DEGs with all three cis-elements for CK, N, and P response. B, Detailed information of 20 selected DEGs with all three cis-elements across stages. C, Distribution of the 194 overlapping DEGs with all three cis-elements in the pairwise comparisons between the normal ovary and super ovary.
DISCUSSION
Corolla Opening Is Featured by Four Distinct Stages along with Sequential Gene Expression in Cucumber
Corolla opening is an evolutionary breakthrough for progeny proliferation of higher flowering plants (van Doorn and Kamdee, 2014). In agricultural crops such as cucumber, the timing of corolla opening is not only a very interesting biological phenomenon, but also an economically important issue that requires extensive investigation. Based on color transition in the corolla, flower opening is divided into four stages in cucumber, including green bud, green-yellow bud, yellow bud, and flowering (Fig. 1A). For most cucumber varieties, this process takes 4 d to 5 d and it is insensitive to nutrient conditions. Cucumber inbred line ‘6457’ produces super ovaries with 4 d to 5 d delay of corolla opening under abundant nutrient condition, and produces normal ovaries within the normal timeframe of flower opening under limited nutrient supply mostly for seed production. Cytological data and transcriptomic signatures indicate that N1 and S1 belong to the green bud stage; N3 and S3, S4, and S5 are in the green-yellow bud stage; N4 and S7, S8 are in the yellow bud stage; and N5 and S9 belong to the flowering stage (Figs. 2, 4; Supplemental Fig. S2). Further, we found that the expressions of some key genes were sequentially activated across stages of corolla development in the normal ovary (Fig. 12). Cell division- and photosynthesis-related genes were specifically activated at the green bud stage; DNA and protein synthesis genes were highly expressed in the green bud stage and then descending until the green-yellow stage. Expressions of cell wall-related genes and YABBY genes were decreased progressively from green to yellow bud stage. Major CHO metabolism and bZIP TF genes were specifically activated in the green-yellow and yellow bud stages. Protein degradation-related genes were specifically activated during the yellow and flowering stages, whereas genes related to stress response, WRKY, and signaling kinases were specifically activated during the flowering stage (Figs. 12A and 5A). In the super ovary, such sequential gene activation modules were generally maintained, except for a few fundamental differences. Firstly, activation of cell division and photosynthesis genes lasted for a longer period of time until the green-yellow bud stage, which is likely the reason for the enlarged corolla size and increased sugar content in the super ovary (Figs. 12B, 2, and 3). Secondly, expression of most TF genes was delayed or repressed during the green-yellow bud stage in the super ovary (Fig. 7D), and the expression of YABBY, bZIP, and WRKY TF family genes was specifically enhanced in the normal ovary (Figs. 12B and 5B). The activities of some TCP and NAC TF genes were increased in the super ovary, which may have contributed to the enlarged petal size through their function in cell proliferation and cell expansion (Fig. 9, B and C). Thirdly, activation of signaling kinases was promoted in the super ovary (Figs. 12B, 7, and 5B, Supplemental Fig. S3). Fourthly, protein degradation was up-regulated during the green-yellow bud stage in the super ovary, which explains the decreased total N and P content (Figs. 12B, 3, and 7). Fifth, and lastly, stress-related genes were specifically up-regulated in the normal ovary during the flowering stage, but not in the super ovary (Figs. 12B and 5B). As such, this study not only provides the most comprehensive gene expression modules across stages of corolla development in both normal and super ovaries, but also offers valuable resources that can be used for comparative studies of flower opening in other plant species.
Figure 12.
Diagram of gene expression modules during different stages of corolla development in cucumber. A and B, Sequential activations of gene expression in the normal ovary (A) and super ovary (B) at green bud, green-yellow bud, yellow bud, and flowering stages of corolla development in cucumber.
Hormone Modulation Was Sufficient for the Genesis of the Super Ovary
Phytohormones such as ethylene, gibberellins, and auxin have been documented to mediate flower opening (van Doorn and Kamdee, 2014). Ethylene promoted or hastened flower opening depending on different species or even different cultivars in rose (Reid et al., 1989; Jones and Woodson, 1997). GA promotes cell elongation through repressing DELLA proteins, and thus GA-deficient Arabidopsis mutant plants exhibited reduced petal growth (Cheng et al., 2004). Consistently, here we found that gibberellins were increased in the super ovary during the early stages of corolla development, which corresponds to the enlarged corolla size (Figs. 1 and 10). Time-course RNA-seq analysis showed that the increased GA levels were due to the up-regulation of GA biosynthesis genes in the super ovary (Fig. 8B). Although previous studies showed that exogenous gibberellin application promoted flower opening in several species (Steinitz and Cohen, 1982; Koning, 1984; Raab and Koning, 1987), the role of endogenous GA in cucumber flower opening needs further characterization. Auxin was shown to regulate cell proliferation and cell expansion, and mutation of genes implicated in auxin effects such as AUXIN RESPONSE FACTOR8 and PETAL LOSS resulted in a change of petal size (Varaud et al., 2011; Sauret-Güeto et al., 2013). Further, flower opening can be strongly promoted by the IAA (auxin indoleacetic acid; van Doorn et al., 2013). Although no significant difference was observed for auxin level in the corollas of normal and super ovaries, IAA concentration was dramatically reduced in the fruit of the super ovary (Supplemental Fig. S4). Considering that only female flower blooming was delayed in the line ‘6457’, while male flower opening was unaffected, the reduced IAA level in the fruit may have contributed to the delayed corolla opening in the super ovary. In addition, we found that genes encoding UGTs and CKXs were significantly down-regulated in the super ovary (Fig. 8A). Consistently, reduction of CK content was smaller in the super ovary during the later stages of corolla development (Fig. 10), suggesting that the higher level of CKs observed in the super ovary is due to reduced CK conjugation and degradation. Further, exogenous CK TDZ treatment significantly delayed flower opening and accelerated fruit growth, thus partially mimicking the genesis of the super ovary (Table I; Supplemental Fig. S4B; and the CK levels in the fruit (Supplemental Fig. S4A). Therefore, CKs appear to be the primary regulator for flower opening in cucumber, whereas GA and auxin may be coordinately involved in the size control of corolla and fruit.
Nutrient Supplies Played an Essential Role in Regulating Corolla Opening in Cucumber
The corolla development of most cucumber varieties takes 4 d to 5 d from green bud to flowering stages, and this process is insensitive to nutrient conditions. The unique cucumber inbred line ‘6457’ produces super ovaries or normal ovaries under abundant or insufficient nutrient conditions, respectively, suggesting that nutrient availability plays an essential role in corolla opening of this cucumber line. N and P are the major fertilizer components used in this study, and function as critical macronutrients participating in various physiological and biological processes in plants (Prinsi et al., 2009). We found that both N and P levels were decreased with the progression of corolla development, but the reduction was significantly greater in the super ovary during the green-yellow bud (S3, S4, and S5) and yellow bud (S7 and S8) stages (Fig. 3), which may be attributed to the much greater size of corolla in the super ovary under these stages (Fig. 1). Considering that N and P are essential for constitution of cellular components such as proteins and nucleic acids (Rouached et al., 2010; Chiou and Lin, 2011; Simons et al., 2014; Krapp, 2015), reduction of N and P levels in the super ovary is consistent with the enhanced protein degradation mediated by cysteine-type peptidase activity during the green-yellow bud stage (Fig. 7C), and up-regulated signaling kinase activity during the yellow bud stage (Fig. 7G). N availability is closely related to N assimilates and soluble sugars in plants (Walch-Liu et al., 2000). In the super ovary, soluble sugars were significantly increased during all four stages (Fig. 3), which is consistent with abundant N supply and prolonged photosynthesis in the super ovary (Figs. 5B and 7C). N nutritional status directly affects the CK level as well. N supplementation promotes CK accumulation in the bare root and xylem sap of maize, while low N causes a significant decrease in the CK concentration in wheat (Garnica et al., 2010; Takei et al., 2001). On the contrary, CKs regulate carbohydrate mobilization and N signaling (Ashikari et al., 2005; Sakakibara, 2006). Our data showed that the concentration of CKs was higher in the super ovary (Fig. 10), and that exogenous CK treatment mimicked the super ovary phenotype (Table I). Moreover, 194 DEGs contains all three cis-regulatory elements for CK, N, and P. Thus, it is intriguing to speculate that nutrient supplies and CKs function coordinately, or nutrient availability acts upstream of CKs, to regulate the genesis of the super ovary.
Cytological and transcriptomic data showed that the primary difference between normal and super ovaries is in the middle stages: i.e. the green-yellow and yellow bud stages (Fig. 2; Supplemental Fig. S2). Compared with the super ovary, in the normal ovary the increment of water content was greater during the green-yellow bud stage (Fig. 3); the stress hormone ABA (Leng et al., 2014) was higher in both green-yellow and yellow bud stages (Fig. 10); and genes related to stress response and WRKY TFs were specifically activated during the flowering stage (Fig. 5A), indicating that the hastened flower opening in the normal ovary was a response to nutritional stress. The WRKY family is an enormous family that has 74 members in Arabidopsis and more than 90 members in rice (Oryza sativa; Ülker and Somssich, 2004). Previous studies indicate that WRKY TFs play important roles in stress responses (Ren et al., 2010; Qin et al., 2013; Yan et al., 2014). For example, rice WRKY13 regulates the cross talk between drought and disease-resistance signaling pathways by binding to different DNA sequence motifs (Xiao et al., 2013). Arabidopsis WRKY33 responds to pathogen infection through functions downstream of MPK3/MPK6 (Mao et al., 2011). WRKY TFs have also been shown to regulate flowering time in Arabidopsis (Robatzek and Somssich, 2002; Miao et al., 2004; Ülker and Somssich, 2004). Overexpression of MlWRKY12, the homolog of AtWRKY12 in Miscanthus lutarioriparius, led to early flowering phenotype and significantly increased the expression of flowering-related genes such as CONSTANS, FLOWERING LOCUS T, LEAFY, APETALA1, CAULIFLOWER, and FRUITFULL in Arabidopsis (Yu et al., 2013). However, flowering-promoting genes including LEAFY, CONSTANS, and APETALA2 were down-regulated while flowering-repressing genes such as TERMINAL FLOWER1 and SCHLAFMUTZE were up-regulated in the normal ovary (Fig. 9A), suggesting that the major function of WRKY up-regulation in the normal ovary may be nutritional stress response instead of promoting flowering.
In addition, receptor-like kinase-related genes were significantly up-regulated in the super ovary during the green-yellow and yellow bud stages (Figs. 5B and 7G). Similarly, most MAP kinase family genes were more significantly activated from the yellow bud to flowering stage in the super ovary (Supplemental Fig. S3). Receptor-like kinases regulate the signaling pathways involved in multiple developmental processes (Walker, 1994). For example, the protein phosphorylation/dephosphorylation mediated by receptor-like kinases plays an important role in the initiation of carnation flower opening (Harada et al., 2010; Shinozaki et al., 2014). Leu-rich repeat receptor-like kinases PAN1 and PAN2 have been shown to promote polarization of subsidiary mother-cell divisions during stomatal development in maize (Cartwright et al., 2009; Zhang et al., 2012). Therefore, the enlarged organ size and delayed flower opening in the super ovary may be partially caused by the function of signaling kinases. Given that the line ‘6457’ is sensitive to nutritional availability, it is intriguing to speculate that the key genes responsible for the unique feature of ‘6457’ may be epigenetically regulated, by DNA or histone modifications such as phosphorylation and methylation. Future studies using map-based cloning or bulked segregant analysis-sequencing (Michelmore et al., 1991) to clone the key gene(s) in ‘6457’ would be promising to dissect the precise genetic regulation of the timing of flower opening in cucumber.
MATERIALS AND METHODS
Plant Materials and Treatments
The cucumber (Cucumis sativus) inbred line ‘6457’ was grown in the greenhouse of Hebei Normal University of Science and Technology (Hebei), and subjected to two distinct cultivation treatments. The first treatment was super ovary induction treatment, in which massive fertilizer and water (130% of standard) was applied and only one fruit was kept per plant to eliminate interfruit competition. The second treatment was normal ovary induction treatment, in which insufficient fertilizer and water (50% of standard) was applied and two fruits were kept per plant. Pest control was performed according to standard practices. The lengths of the labeled ovary and corolla were measured every day, and an ovary with length greater than 9.0 cm at anthesis was considered a super ovary.
Scanning Electron Microscopy
Corollas at 1, 3, 4, and 5 DAL from the normal ovary treatment and corollas at 1, 3, 4, 5, 7, 8, and 9 DAL from the super ovary treatment were cut and fixed with 2.5% glutaraldehyde at 4°C for 24 h, washed with PBS (pH 7.2) three times, and post-fixed in 1% (v/v) OsO4. The samples were then dehydrated three times through an ethanol series (30, 50, 70, 80, 90, and 100%), critical-point-dried using a desiccator (HCP-2; Hitachi), with a bulb sputter-coated with gold palladium (EiKO IB-3). Images were taken with a model number S-4700 scanning electron microscope (Hitachi) using a 2 kV accelerating voltage.
RNA Preparation for RNA-Seq
Corolla samples at 1, 3, 4, and 5 DAL from the normal ovary treatment, and at 1, 3, 4, 5, 7, 8, and 9 DAL from the super ovary treatment, were collected for RNA-seq analysis. Samples from five corollas were pooled as one biological replicate and three biological replicates were performed for each time point. Total RNAs were extracted using the RNA Extraction Kit (Huayueyang). RNA was checked on 1% agarose gels to detect possible degradation and contamination, and was then examined by a Nano Photometer Spectrophotometer (Implen USA) for RNA purity. A Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies) was used to measure the RNA concentration, and an RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 System (Agilent Technologies) was used to evaluate RNA integrity. Only RNA samples that passed the quality tests were chosen for RNA-seq analysis.
RNA-Seq Library Construction and Sequencing
RNA-seq library construction was performed using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) following manufacturer’s instructions and four index codes were added to attribute sequences to different samples (Wang et al., 2009). RNA-seq libraries were sequenced on an Illumina HiSEquation 2000 platform (NEB) to generate 125-bp paired-end reads.
Bioinformatics Analysis of RNA-Seq Data
The methods for RNA-seq analysis were as previously described by Zhao et al. (2016). In short, the raw reads were preprocessed to remove low-quality regions using the SolexaQA tool (version 2.2; http://solexaqa.sourceforge.net/) and adapter sequences were removed using the Cutadapt tool (version 1.3; http://cutadapt.readthedocs.io/en/stable/guide.html; Martin, 2011). Clean reads were mapped to the cucumber genome sequence (http://cucumber.genomics.org.cn, ver. 2i) using the software TopHat (Trapnell et al., 2009; Huang et al., 2009). Read counts of annotated genes were obtained by the Python software HTSeq-count (Anders et al., 2015). The genes with an expression level of at least 1 TPM in at least three samples were retained for further analysis. The R package edgeR (Bioconductor, https://www.bioconductor.org/), which uses counts per gene in different samples from HTSeq-count as input and performs data normalization using the trimmed mean of M-values method, was used to identify DEGs (Robinson et al., 2010). The genes with at least 2-fold change in expression and a FDR of less than 0.05 were considered to be differentially expressed. Gene expressions were first normalized to 1 TPM according to the total number of mapped clean reads in each of 33 libraries. The log2 values of normalized expressions were used to build an expression matrix and the Prcomp function in R (R-Core-Team, 2014) was used for principle component analysis.
MapMan Software Analysis of DEGs
To obtain detailed functional categorization of DEGs, we used the on-line web tool Mercator (MapMan, http://mapman.gabipd.org/web/guest/mapman; Lohse et al., 2014) to obtain the cucumber protein annotation mapping file for MapMan with default parameters and then used the Java software MapMan to assign functional bins for each gene (Thimm et al., 2004). Fisher’s exact test was used to examine whether a functional category was significantly overrepresented in our selected genes against the set of all genes with MapMan annotations, and the Benjamini Hochberg method was used to adjust P-values for multiple testing. The K-means method was used to cluster all the DEGs identified through all pairwise comparisons and the R-package, the software “pheatmap” (Kolde, 2011), was used to draw the heat map of each cluster. For each cluster, overrepresented functional categories were determined using the software MapMan.
Gene Ontology Term Enrichment Analysis
We combined blast2GO (https://www.blast2go.com/; Conesa et al., 2005) and interProScan (http://www.ebi.ac.uk/interpro/search/sequence-search; Jones et al., 2014) search results to assign gene ontology (GO) terms to cucumber genes. The GO term enrichment analysis was conducted for up- and down-regulated DEGs, respectively, using the R package TopGO (Bioconductor, Alexa et al., 2006). Adrian Alexi’s improved weighted scoring algorithm and Fisher’s test were used to determine the significance of GO term enrichment. Significantly enriched GO terms were identified as those with a p-value less than 0.05. The software VennDiagram (http://creately.com/Draw-Venn-Diagrams-Online; Chen and Boutros, 2011) was used to indicate the overlaps of different DEGs.
Nutritional Measurements in Corollas
Major nutrients (N, P, and K), soluble sugars, and water content were measured with independently generated corolla samples at the same developmental stages as those in RNA-seq. Total N was analyzed using the Semimicro-Kjeldahl method (Bremer, 1965; Hesse, 1971). Total P was measured as previously described by Bray and Kurtz (1945). The flame emission photometry method was used for total K assay, and water content was measured following the standard protocol. Soluble sugars were determined as described by Beck et al. (2014).
Endogenous Hormone Measurement
To examine the levels of auxin, CK, gibberellin, JA, BRs, and ABA in the super ovary and normal ovary, the ovary and corolla were separately harvested and immediately frozen in liquid N until further use. Sample extraction and hormone measurements were performed using enzyme-linked immunosorbent assays as previously described by Maldiney et al. (1986), Abdala et al. (1996), Cui et al. (2005), and Swaczynova et al. (2007). Standard auxin (IAA), IPA, ZR, DHZR, gibberellins (GA3, GA4), ABA, JA, and BR (Sangon Biotech) were used for calibration. The results were presented as mean ± se of three biological replicates and three technical replicates.
Exogenous CK Treatment
Line ‘6457’ under the normal ovary induction system was used for CK treatment. Plants with uniform growth were selected, and the female flowers (including both corolla and ovaries) at the green bud stage (2 to approximately 3 cm long) were dipped with different concentrations of TDZ solution: 1.25, 2.5, 5, and 10 mg/L. Water was used as negative control. Eight female flowers from different plants were used for each treatment. Fruit length and flowering time were recorded every day since treatment.
Identification of cis-Elements for CK, N, and P
The 2-kb DNA sequences upstream of the transcription start site of the type 3 DEGs were used for cis-element identification. A custom Perl script was used to scan for cis-elements 5′-AAGAT(T/C)TT-3′ for CK, 5′-(T/A)7A(G/C)TCA-3′ for N, and 5′-GNATATNC-3′ for P. The overlap of DEG response to CK, N, and P was obtained using the software VennDiagram within the R package (Chen and Boutros, 2011).
Quantitative Real-Time RT-PCR
qRT-PCR analyses were performed with independently generated corolla samples at the same developmental stages as those in RNA-seq. Primers for qRT-PCR were designed using the Primer 5 software and synthesized by Sangon Biotech. cDNAs were reverse-transcribed from total RNA using the PrimeScript RT reagent Kit (Takara), and qRT-PCR analyses were performed on an ABI PRISM 7500 Real-Time PCR System (Applied Biosystems). The cucumber UBI gene was used as an internal control to normalize the expression data (Wan et al., 2010). Each qRT-PCR experiment was repeated three times. The relative expression of each gene was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001) and the sd was calculated among three biological replicates. The gene-specific primers were listed in Supplemental Table S9.
Accession Numbers
Sequencing data were deposited to the Gene Expression Omnibus database at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE76358.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. qRT-PCR analysis of selected genes in corollas at different stages in the normal and super ovaries.
Supplemental Figure S2. Bar plot showing the number of DEGs between different samples.
Supplemental Figure S3. Expression of MAP kinase family genes across developmental stages in the normal ovary and super ovary.
Supplemental Figure S4. Hormone measurements and CK effect on the fruit growth rate in cucumber.
Supplemental Table S1. Quantifications of seeded fruits and seeds per fruit in the normal and super ovaries depending on the timing of hand pollination.
Supplemental Table S2. Summary of transcriptome sequencing data.
Supplemental Table S3. DEGs from the pairwise comparisons in Figure S2.
Supplemental Table S4. K-means clustering of the DEGs corresponding to Figure 5.
Supplemental Table S5. Expression levels of the MAP kinase family genes in Supplemental Figure S3.
Supplemental Table S6. Expression levels of the CK- and GA-related genes corresponding to those in Figure 8.
Supplemental Table S7. Expression levels of the flowering-related genes, TCP, and NAC family genes corresponding to those in Figure 9.
Supplemental Table S8. Detailed information of the 194 overlapping DEGs with CK, N, and P cis-elements in the promoters corresponding to Figure 11.
Supplemental Table S9. Primers used for qRT-PCR assays in this study.
Supplementary Material
Acknowledgments
We thank the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences for providing the cucumber genome and annotation data, and members of the Zhang, Yan, and Liu Labs for discussions and technical help.
Glossary
- DAL, DALs
day or days after labeling
- DEG, DEGs
differentially expressed gene or genes
- FDR
false discovery rate
- GO
gene ontology
- PCA
principal component analysis
- TF, TFs
transcription factor, transcription factors
- TPM
transcript per million
- TDZ
thidiazuron
Footnotes
This work was supported by grants from the Hebei Vegetable Innovation Projects of Modern Agricultural Industry Technology System to L.Y., National Basic Research of China 973 program under grant number 2012CB113900 and Chinese Universities Scientific Fund under grant number 2015NX004 to X.Z., the 100-Talent Program of the Chinese Academy of Sciences to R.L., and Natural Science Foundation of Hebei under grant number C2015407051 to C.S.
Articles can be viewed without a subscription.
References
- Abdala G, Castro G, Guinazu MM, Tizio R, Miersch O (1996) Occurrence of jasmonic acid in organs of Solanum tuberosum L. and its effect on tuberization. Plant Growth Regul 19: 139–143 [Google Scholar]
- Aguilar-Martínez JA, Sinha N (2013) Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 4: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Bai SL, Peng YB, Cui JX, Gu HT, Xu LY, Li YQ, Xu ZH, Bai SN (2004) Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220: 230–240 [DOI] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck TK, Jensen S, Bjoern GK, Kidmose U (2014) The masking effect of sucrose on perception of bitter compounds in brassica vegetables. J Sens Stud 29: 190–200 [Google Scholar]
- Bilyeu KD, Cole JL, Laskey JG, Riekhof WR, Esparza TJ, Kramer MD, Morris RO (2001) Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol 125: 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Camps C, Lemhemdi A, Morin H, Sari MA, Fraenkel-Zagouri R, Kovalski I, Dogimont C, Perl-Treves R, et al. (2015) A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350: 688–691 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Bray RH, Kurtz LT (1945) Determination of total, organic, and available forms of phosphorus in soils. Soil Sci 59: 39–45 [Google Scholar]
- Bremer JM. (1965) Total nitrogen. Methods of soil analysis (Part 2). Chemical and microbiological properties. Agron Monogr 9: 1149–1178 [Google Scholar]
- Bui AQ, O’Neill SD (1998) Three 1-aminocyclopropane-1-carboxylate synthase genes regulated by primary and secondary pollination signals in orchid flowers. Plant Physiol 116: 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright HN, Humphries JA, Smith LG (2009) PAN1: a receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science 323: 649–651 [DOI] [PubMed] [Google Scholar]
- Chen H, Boutros PC (2011) VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Cui D, Neill SJ, Tang Z, Cai W (2005) Gibberellin-regulated XET is differentially induced by auxin in rice leaf sheath bases during gravitropic bending. J Exp Bot 56: 1327–1334 [DOI] [PubMed] [Google Scholar]
- Dai F, Zhang C, Jiang X, Kang M, Yin X, Lü P, Zhang X, Zheng Y, Gao J (2012) RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol 160: 2064–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson JA, Frommer WB (2013) Plant science. Jack of all trades, master of flowering. Science 339: 659–660 [DOI] [PubMed] [Google Scholar]
- de Jong M, Mariani C, Vriezen WH (2009) The role of auxin and gibberellin in tomato fruit set. J Exp Bot 60: 1523–1532 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Calderon-Urrea A (1993) Sex determination in flowering plants. Plant Cell 5: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Galun E. (1962) Study of the inheritance of sex expression in the cucumber. The interaction of major genes with modifying genetic and non-genetic factors. Genetica 32: 134–163 [Google Scholar]
- Garnica M, Houdusse F, Zamarreño AM, Garcia-Mina JM (2010) The signal effect of nitrate supply enhances active forms of cytokinins and indole acetic content and reduces abscisic acid in wheat plants grown with ammonium. J Plant Physiol 167: 1264–1272 [DOI] [PubMed] [Google Scholar]
- Hammond J. (1982) Changes in amylase activity during rose bud opening. Sci Hortic (Amsterdam) 16: 283–289 [Google Scholar]
- Hao YJ, Wang DH, Peng YB, Bai SL, Xu LY, Li YQ, Xu ZH, Bai SN (2003) DNA damage in the early primordial anther is closely correlated with stamen arrest in the female flower of cucumber (Cucumis sativus L.). Planta 217: 888–895 [DOI] [PubMed] [Google Scholar]
- Harada T, Torii Y, Morita S, Masumura T, Satoh S (2010) Differential expression of genes identified by suppression subtractive hybridization in petals of opening carnation flowers. J Exp Bot 61: 2345–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Torii Y, Morita S, Onodera R, Hara Y, Yokoyama R, Nishitani K, Satoh S (2011) Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening. J Exp Bot 62: 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse PR. (1971) A Textbook of Soil Chemical Analysis. William Clowes, London, UK [Google Scholar]
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Jones ML, Woodson WR (1997) Pollination-induced ethylene in carnation (role of stylar ethylene in corolla senescence). Plant Physiol 115: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaihara S, Takimoto A (1983) Effect of plant growth regulators on flower opening of pharbitis nil. Plant Cell Physiol 24: 309–316 [Google Scholar]
- Kamachi S, Sekimoto H, Kondo N, Sakai S (1997) Cloning of a cDNA for a 1-aminocyclopropane-1-carboxylate synthase that is expressed during development of female flowers at the apices of Cucumis sativus L. Plant Cell Physiol 38: 1197–1206 [DOI] [PubMed] [Google Scholar]
- Kolde R. (2011) pheatmap: Pretty Heatmaps. http://cran.r-project.org/package=pheatmap [Google Scholar]
- Komeda Y. (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55: 521–535 [DOI] [PubMed] [Google Scholar]
- Koning RE. (1984) The roles of plant hormones in the growth of the corolla of Gaillardia grandiflora (Asteraceae) ray flowers. Am J Bot 71: 1–8 [Google Scholar]
- Konishi M, Yanagisawa S (2010) Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J 63: 269–282 [DOI] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A. (2015) Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Curr Opin Plant Biol 25: 115–122 [DOI] [PubMed] [Google Scholar]
- Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65: 4561–4575 [DOI] [PubMed] [Google Scholar]
- Leng P, Yuan B, Guo Y (2014) The role of abscisic acid in fruit ripening and responses to abiotic stress. J Exp Bot 65: 4577–4588 [DOI] [PubMed] [Google Scholar]
- Li S. (2015) The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal Behav 10: e1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang S, Liu S, Pan J, Zhang Z, Tao Q, Shi Q, Jia Z, Zhang W, Chen H, et al. (2009) Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 182: 1381–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B (2014) Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ 37: 1250–1258 [DOI] [PubMed] [Google Scholar]
- Luo J, Ma N, Pei H, Chen J, Li J, Gao J (2013) A DELLA gene, RhGAI1, is a direct target of EIN3 and mediates ethylene-regulated rose petal cell expansion via repressing the expression of RhCesA2. J Exp Bot 64: 5075–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnish AJ, Leonard RT, Borda AM, Nell TA (2010) Genotypic variation in the postharvest performance and ethylene sensitivity of cut rose flowers. HortScience 45: 790–796 [Google Scholar]
- Maldiney R, Leroux B, Sabbagh I, Sotta B, Sossountzov L, Miginiac E (1986) A biotin-avidin-based enzyme-immunoassay to quantify three phytohormone: auxin, abscisic-acid and zeatin-riboside. J Immunol Methods 90: 151–158 [Google Scholar]
- Malepszy S, Niemirowicz-Szczytt K (1991) Sex determination in cucumber (Cucumis sativus L.) as a model system for molecular biology. Plant Sci 80: 39–47 [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- McClung CR, Gutiérrez RA (2010) Network news: prime time for systems biology of the plant circadian clock. Curr Opin Genet Dev 20: 588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Mibus H, Tatlioglu T (2004) Molecular characterization and isolation of the F/f gene for femaleness in cucumber (Cucumis sativus L.). Theor Appl Genet 109: 1669–1676 [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch JP. (1952) Plant hormones in the development of fruits. Q Rev Biol 27: 33–57 [DOI] [PubMed] [Google Scholar]
- Nitsch JP, Kurtz EB, Liverman JL, Went FW (1952) The development of sex expression in cucurbit flowers. Am J Bot 39: 32–43 [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl): S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Ma N, Tian J, Luo J, Chen J, Li J, Zheng Y, Chen X, Fei Z, Gao J (2013) An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol 163: 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsi B, Negri AS, Pesaresi P, Cocucci M, Espen L (2009) Evaluation of protein pattern changes in roots and leaves of Zea mays plants in response to nitrate availability by two-dimensional gel electrophoresis analysis. BMC Plant Biol 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Tian Y, Han L, Yang X (2013) Constitutive expression of a salinity-induced wheat WRKY transcription factor enhances salinity and ionic stress tolerance in transgenic Arabidopsis thaliana. Biochem Biophys Res Commun 441: 476–481 [PubMed] [Google Scholar]
- Quatrano RS, Shaw SL (1997) Role of the cell wall in the determination of cell polarity and the plane of cell division in Fucus embryos. Trends Plant Sci 2: 15–21 [Google Scholar]
- Raab MM, Koning RE (1987) Interacting roles of gibberellin and ethylene in corolla expansion of ipomoea nil (Convolvulaceae). Am J Bot 74: 921–927 [DOI] [PubMed] [Google Scholar]
- Ramireddy E, Brenner WG, Pfeifer A, Heyl A, Schmülling T (2013) In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol 54: 1079–1092 [DOI] [PubMed] [Google Scholar]
- R-Core-Team (2014) R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Reid MS, Dodge LL, Mor Y, Evans RY (1989) Effects of ethylene on rose opening. Acta Hortic 261: 220–251 [Google Scholar]
- Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE (1995) Dioecy and its correlates in the flowering plants. Am J Bot 82: 596–606 [Google Scholar]
- Robatzek S, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3: 288–299 [DOI] [PubMed] [Google Scholar]
- Rudich J, Baker LR, Sell HM (1977) Parthenocarpy in Cucumis-sativus L as affected by genetic parthenocarpy, thermo-photoperiod, and femaleness. J Am Soc Hortic Sci 102: 225–228 [Google Scholar]
- Ruelland E, Zachowski A (2010) How plants sense temperature. Environ Exp Bot 69: 225–232 [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sampathkumar A, Yan A, Krupinski P, Meyerowitz EM (2014) Physical forces regulate plant development and morphogenesis. Curr Biol 24: R475–R483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E (2013) JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol 11: e1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T, Werner T, Riefler M, Krupková E, Bartrina y Manns I (2003) Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116: 241–252 [DOI] [PubMed] [Google Scholar]
- Schünmann PH, Richardson AE, Vickers CE, Delhaize E (2004) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136: 4205–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y, Tanaka R, Ono H, Ogiwara I, Kanekatsu M, van Doorn WG, Yamada T (2014) Length of the dark period affects flower opening and the expression of circadian-clock associated genes as well as xyloglucan endotransglucosylase/hydrolase genes in petals of morning glory (Ipomoea nil). Plant Cell Rep 33: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Simons M, Saha R, Guillard L, Clément G, Armengaud P, Cañas R, Maranas CD, Lea PJ, Hirel B (2014) Nitrogen-use efficiency in maize (Zea mays L.): from ‘omics’ studies to metabolic modelling. J Exp Bot 65: 5657–5671 [DOI] [PubMed] [Google Scholar]
- Steinitz B, Cohen A (1982) Gibberellic acid promotes flower bud opening on detached flower stalks of statice (Limonium sinuatum L.). HortScience 17: 903–904 [Google Scholar]
- Sun TP. (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6: e0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaczynova J, Novak O, Hauserova E, Fuksova K, Sisa M, Kohout L, Strnad M (2007) New techniques for the estimation of naturally occurring brassinosteroids. J Plant Growth Regul 26: 1–14 [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T (2001) Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol 42: 85–93 [DOI] [PubMed] [Google Scholar]
- Tang X, Woodson WR (1996) Temporal and spatial expression of 1-aminocyclopropane-1-carboxylate oxidase mRNA following pollination of immature and mature petunia flowers. Plant Physiol 112: 503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Staub JE, O’Neill SD (1997) Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiol 113: 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolinder NL, Mcmichael BL, Upchurch DR (1993) Water relations of cotton flower petals and fruit. Plant Cell Environ 16: 755–760 [Google Scholar]
- Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Dole I, Celikel FG, Harkema H (2013) Opening of Iris flowers is regulated by endogenous auxins. J Plant Physiol 170: 161–164 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Kamdee C (2014) Flower opening and closure: an update. J Exp Bot 65: 5749–5757 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, van Meeteren U (2003) Flower opening and closure: a review. J Exp Bot 54: 1801–1812 [DOI] [PubMed] [Google Scholar]
- Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M (2011) AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23: 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH (2011) The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435: 143–155 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Neumann G, Bangerth F, Engels C (2000) Rapid effects of nitrogen form on leaf morphogenesis in tobacco. J Exp Bot 51: 227–237 [DOI] [PubMed] [Google Scholar]
- Walker JC. (1994) Structure and function of the receptor-like protein kinases of higher plants. Plant Mol Biol 26: 1599–1609 [DOI] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399: 257–261 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Cheng H, Li X, Xiao J, Xu C, Wang S (2013) Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol 163: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Norikoshi R, Suzuki K, Imanishi H, Ichimura K (2009a) Determination of subcellular concentrations of soluble carbohydrates in rose petals during opening by nonaqueous fractionation method combined with infiltration-centrifugation method. Planta 230: 1115–1127 [DOI] [PubMed] [Google Scholar]
- Yamada K, Takahashi R, Fujitani C, Mishima K, Yoshida M, Joyce DC, Yamaki S (2009b) Cell wall extensibility and effect of cell-wall-loosening proteins during rose flower opening. J Jpn Soc Hortic Sci 78: 242–251 [Google Scholar]
- Yamane K, Kawabata S, Sakiyama R (1991) Changes in water relations, carbohydrate contents and acid invertase activity associated with perianth elongation during anthesis of cut gladiolus flowers. J Jpn Soc Hortic Sci 60: 421–428 [Google Scholar]
- Yampolsky H, Yampolsky C (1922) Distribution of sex forms in the phanerogamic flora. Bibl Genet 3: 1–62 [Google Scholar]
- Yan H, Jia H, Chen X, Hao L, An H, Guo X (2014) The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol 55: 2060–2076 [DOI] [PubMed] [Google Scholar]
- Yu Y, Hu R, Wang H, Cao Y, He G, Fu C, Zhou G (2013) MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci 212: 1–9 [DOI] [PubMed] [Google Scholar]
- Zhang X, Facette M, Humphries JA, Shen Z, Park Y, Sutimantanapi D, Sylvester AW, Briggs SP, Smith LG (2012) Identification of PAN2 by quantitative proteomics as a leucine-rich repeat-receptor-like kinase acting upstream of PAN1 to polarize cell division in maize. Plant Cell 24: 4577–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Li Y, Ding L, Yan S, Liu M, Jiang L, Zhao W, Wang Q, Yan L, Liu R, Zhang X (2016) Phloem transcriptome signatures underpin the physiological differentiation of the pedicel, stalk and fruit of cucumber (Cucumis sativus L.). Plant Cell Physiol 57: 19–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.