GAL4-UAS enhancer trap populations flag a range of cell-type-specific promoters in vivo.
Abstract
Cell-type-specific gene expression is essential to distinguish between the numerous cell types of multicellular organism. Therefore, cell-type-specific gene expression is tightly regulated and for most genes RNA transcription is the central point of control. Thus, transcriptional reporters are broadly used markers for cell identity. In Arabidopsis (Arabidopsis thaliana), a recognized standard for cell identities is a collection of GAL4/UAS enhancer trap lines. Yet, while greatly used, very few of them have been molecularly characterized. Here, we have selected a set of 21 frequently used GAL4/UAS enhancer trap lines for detailed characterization of expression pattern and genomic insertion position. We studied their embryonic and postembryonic expression domains and grouped them into three groups (early embryo development, late embryo development, and embryonic root apical meristem lines) based on their dominant expression. We show that some of the analyzed lines are expressed in a domain often broader than the one that is reported. Additionally, we present an overview of the location of the T-DNA inserts of all lines, with one exception. Finally, we demonstrate how the obtained information can be used for generating novel cell-type-specific marker lines and for genotyping enhancer trap lines. The knowledge could therefore support the extensive use of these valuable lines.
Differences among the numerous cell types of multicellular organism are instructed by unique cell-type-specific gene expression. Understanding how cell identities are genetically controlled is therefore a major challenge in developmental biology. While epigenetic, transcriptional, and posttranscriptional mechanisms all contribute to cell-type-specific gene expression, gene activity primarily depends on being actively transcribed. Hence, transcriptional reporters, such as gene promoter-reporter fusions or enhancer trap lines, are widely used markers for cell identity in model organisms (De Rybel et al., 2013; Levesque et al., 2006; Fendrych et al., 2014). Marker genes are thus essential tools for determining identity in for example mutant cells (Sabatini et al., 2003; Wolters et al., 2011; Mylona et al., 2002). The need for robust identity markers is especially urgent in plants, where cell identity is very flexible and can easily be reprogrammed during regeneration (Sugimoto et al., 2010; Sena et al., 2009), somatic embryo induction (de Vries et al., 1988), or even under influence of environmental signals (e.g. hydropatterning or stress; Ikeda-Iwai et al., 2003). In addition, cell-type-specific markers are often used to drive ectopic gene expression as part of studying gene function (Weijers et al., 2006; Waki et al., 2013).
Many markers have been generated in the model plant Arabidopsis (Arabidopsis thaliana), but in plant developmental biology, a collection of GAL4/UAS enhancer trap lines has become an accepted standard for cell identities. These so-called “Haseloff” lines (Haseloff, 1999), as well as later derivations of the same principle (Ckurshumova et al., 2009; Gardner et al., 2009), thus provide an essential resource in Arabidopsis biology. The GAL4/UAS system is a two-component gene expression system widely used for targeted gene misexpression. It was first developed for use in Drosophila melanogaster (Brand and Perrimon, 1993) and later also successfully optimized for Arabidopsis (Haseloff, 1999) and other model organisms (Scheer and Campos-Ortega, 1999; Kawakami et al., 2004; Ornitz et al., 1991; Hartley et al., 2002). The two-component system requires two lines: one that contains GAL4-VP16, the DNA-binding domain of the yeast GAL4 transcriptional activator fused to the potent Herpes simplex VP16 transcriptional activation domain (Sadowski et al., 1988), which can be driven by a (characterized) cell-type-specific promoter. Another line carries a selected target gene placed under control of a GAL4-dependent promoter (Upstream Activation Sequence [UAS]) that is silent in the absence of GAL4. Genetic crossing between these two lines will specifically activate the target gene in particular tissue or cell types.
The “Haseloff” GAL4/UAS collection (Haseloff, 1999) consists of around 250 plant lines. In these lines, a T-DNA carrying GAL4-VP16 and modified UAS-driven GFP-gene (mGFP5ER) is randomly inserted into the Arabidopsis genome. A minimal CaMV 35S promoter is placed upstream of GAL4-VP16, and expression depends on insertion near an endogenous enhancer element. Thus, GFP expression in these enhancer trap lines reports the activity of genomic enhancer sequences in the proximity of minimal promoter elements (Haseloff, 1999). The set of 250 lines was selected based on GFP expression in the root. From this set, a number of lines, expressed in defined domains of the root, have risen to prominence as very widely used tools in root biology (Sabatini et al., 1999; Levesque et al., 2006; De Rybel et al., 2013; Xu et al., 2013). In addition, as the lines cell-specifically express GAL4-VP16, introduction of a second UAS-driven gene will target expression of that gene in the GFP-marked domain. This strategy has also been widely used for local misexpression (Weijers et al., 2006; Kang et al., 2013; Sabatini et al., 2003).
Yet, while the “Haseloff” lines are extensively used in the Arabidopsis community, very few of them have been molecularly characterized (Cary et al., 2002; Laplaze et al., 2005; Gardner et al., 2009). Thus, it is not known where the insertions are located and what gene expression these lines actually report or if perhaps the insertion disrupts the gene it is inserted in. A specific caveat is that the lines were generated in an ecotype (C24) that is no longer commonly used, and intercrossing with other ecotypes such as Columbia causes strong phenotypic variation, including extreme delay of flowering in the F1 generation. Knowledge of insertion sites would allow migration of the useful expression driver into other ecotypes. Finally, knowing insertion sites in these lines would facilitate introgression in mutant backgrounds as plants could be easily tested for homozygosity or heterozygosity of the marker using PCR.
Here, we selected a set of the 21 most commonly used GAL4/UAS-GFP enhancer trap lines for a detailed characterization of expression and transgene insertion. Given that most of the cell identities in the root are specified during embryogenesis, we extended the description of expression to the embryo. In addition, we explored other seedling organs for GFP expression. We report the genomic insertion sites of these enhancer trap lines and show that this information can be used to infer expression of neighboring genes, as well as to generate novel cell-type-specific markers, as well as to genotype enhancer trap lines. We expect that this resource will aid the extensive use of these important identity markers in the community.
RESULTS AND DISCUSSION
Twenty-one GAL4/UAS-GFP enhancer trap lines from the “Haseloff” collection (Haseloff, 1999) were selected based on their widespread use in the Arabidopsis community (Mylona et al., 2002; Webb et al., 2002; Weijers et al., 2006; Møller et al., 2009; Tsugeki et al., 2010; Rademacher et al., 2012; Wenzel et al., 2012; De Rybel et al., 2013; Waki et al., 2013) and on their documented expression pattern (Haseloff 1999; http://data.plantsci.cam.ac.uk/Haseloff/). For each line, we first confirmed and extended the expression analysis and determined the genomic coordinates of the T-DNA insertion. As a proof of concept, we show how this information can be applied to generate cell-type-specific reporter lines and for genotyping of enhancer trap lines.
Expression Patterns of GAL4/UAS Enhancer Trap Lines
We preselected GAL4/UAS lines for root and embryo expression that are among the most used/popular ones from the “Haseloff” collection (Mylona et al., 2002; Webb et al., 2002; Weijers et al., 2006; Møller et al., 2009; Tsugeki et al., 2010; Wenzel et al., 2012; De Rybel et al., 2013; Waki et al., 2013). Although the expression patterns of some of the selected enhancer trap lines were previously described with respect to embryo development and/or root development (e.g. J0571, Q0990, and J0121; Laplaze et al., 2005; Wenzel et al., 2012), the expression of all 21 lines was systematically characterized in detail at different stages of development and in different parts of the plant (including embryo, root, leaf, shoot apical meristem [SAM], and cotyledons; Table I).
Table I. Overview of the GAL4/UAS-GFP expression patterns and GAL4 insertion positions.
n.d., Not detected; RAM, root apical meristem.
| Driver Line/ARBC Seed Stock No. | Expression Pattern |
Insertion Position (from the ATG) | Flanking Gene(s) | Chromosomal Position | Strand | Short Gene Description | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Embryo | Root | Other | |||||||
| Group 1: Early embryo development lines | |||||||||
| Q0990/N9217 | Provascular cells | Vasculature | n.d. | −15,383 bp | AT5G43810 | 17,596,556 | Forward | Encodes AGO10, a member of the EIF2C (elongation initiation factor 2c)/Argonaute class of proteins | |
| −823 bp | AT5G43800 | Forward | Transposable element gene | ||||||
| −3,074 bp | AT5G43790 | Complement | Pentatricopeptide repeat superfamily protein | ||||||
| Q2500/N9135 | Ground tissue, mesophyll precursors, protoderm of cotyledons | Ground tissue, pericycle, and QC | Cotyledon junction and guard mother cells | +1,105 bp | AT4G02590 | 1,139,201 | Complement | Unfertilized embryo sac 12 (UNE12), bHLH transcription factor | |
| −4,939 bp | AT4G02600 | Forward | MLO1; a member of a large family of seven-transmembrane domain proteins specific to plants, homologs of the barley mildew resistance locus o (MLO) protein | ||||||
| Group 2: Late embryo development lines | |||||||||
| J0571/N9094 | Ground tissue, basal protoderm cells | Ground tissue, some QC cells | Guard and pavement cells | −732 bp | AT4G39900 | 18,511,021 | Complement | Unknown protein | Miyashima et al. (2011) |
| AT4G39910 | Complement | Encodes a nuclear ubiquitin-specific protease | |||||||
| J2731 | Vascular tissue | Cells above QC | Guard and pavement cells | −2,418 bp | AT1G21040 | 7,365,752 | Complement | Transposable element gene | Møller et al. (2009) |
| −1,106 bp | AT1G21050 | Forward | Unknown protein | ||||||
| M0148/N9303 | Cells surrounding SAM | Pericycle cells | Cells surrounding SAM | −2,369 bp | AT1G78300 | 29,459,514 | Forward | G-box binding factor GF14 omega encoding a 14-3-3 protein | |
| −604 bp | AT1G78290 | Complement | Encodes a member of SNF1-related protein kinase (SnRK2) family whose activity is activated by ionic (salt) and nonionic (mannitol) osmotic stress and dehydration | ||||||
| M0164/N9307 | Vascular strands in embryonic cotyledon primordia | Pericycle | Shoot vasculature | −4,452 bp | AT1G09520 | 3,076,482 | Complement | Domains: zinc-finger, PHD-type, conserved site | |
| −733 bp | AT1G09530 | Forward | (PIF3) Transcription factor interacting with photoreceptors phyA and phyB | ||||||
| −1,763 bp | AT4G00440 | 193,413 | Forward | TON1 RECRUITING MOTIF15 (TRM15) | |||||
| −5,881 bp | AT4G00430 | Complement | A member of the plasma membrane intrinsic protein subfamily PIP1 | ||||||
| M0167/N9308 | Intersection of the two cotyledon primordia | n.d. | Cotyledon junction | −4,452 bp | AT1G09520 | 3,076,482 | Complement | Domains: zinc-finger, PHD-type, conserved site | |
| −733 bp | AT1G09530 | Forward | (PIF3) Transcription factor interacting with photoreceptors phyA and phyB | ||||||
| M0223/N9336 | Intersection of the two cotyledon primordia | n.d. | Cotyledon junction | −4,461 bp | AT3G15150 | 5,108,543 | Complement | Encodes a SUMO E3 ligase that regulates endocycle onset and meristem maintenance | Cary et al. (2002) |
| −264 bp/−1,368 bp | AT3G15160/AT3G15170 | Forward | Unknown protein/CUC1, encodes a transcription factor involved in SAM formation and auxin-mediated lateral root formation | ||||||
| Group 3: Embryonic RAM lines | |||||||||
| J1092/N9147 | Future RAM and LRC cells | RAM, LRC | n. d. | +81 bp | AT2G28400 | 12,148,920 | Complement | Unknown protein | |
| −1,459 bp | AT2G28401 | Forward | Unknown protein | ||||||
| J3281/N9128 | Future RAM | Young vascular tissue; columella cells | n.d. | −6,847 bp | AT5G62230 | 24,989,586 | Forward | Encodes a receptor-like kinase that, together with ER and ERL2 governs the initial decision of protodermal cells to either divide proliferatively to produce pavement cells or divide asymmetrically to generate stomatal complexes | |
| +424 bp | AT5G62220 | Complement | Encodes a Golgi apparatus-localized galactosyltransferase involved in galactosyl-substitution of xyloglucan at position 2 | ||||||
| J3411/N9131 | Future RAM and LRC cells | RAM, LRC | n. d. | −5,801 bp | AT2G36360 | 15,253,324 | Complement | Gal oxidase/kelch repeat superfamily protein | |
| AT2G36370/80 | Forward | Ubiquitin-protein ligases/pleiotropic drug resistance 6 (PDR6) | |||||||
| M0028/N9274 | Future RAM | Columella cells | n.d. | −1,463 bp | AT4G21690 | 11,525,766 | Forward | Gibberellin 3-oxidase 3 (GA3OX3) | |
| −6,189 bp | AT4G21680 | Complement | Encodes a nitrate transporter (NRT1.8); functions in nitrate removal from the xylem sap | ||||||
| Q0680/N9209 | Future RAM | Columella cells | n.d. | −1,749 bp | AT2G45200 | 18,641,389 | Complement | Encodes a member of the GOS1 (Golgi SNARE) gene family | |
| −494 bp | AT2G45210 | Forward | SAUR36,SAUR-like auxin-responsive protein family | ||||||
| Q1630/N9227 | Future RAM | Columella cells | n.d. | −1,749 bp | AT2G45200 | 18,641,389 | Complement | Encodes a member of the GOS1 (Golgi SNARE) gene family | |
| −494 bp | AT2G45210 | Forward | SAUR36,SAUR-like auxin-responsive protein family | ||||||
| Other lines | |||||||||
| J0121/N9090 | Seed Coat | Xylem pole pericycle cells | Young leaf primordia | −3,701 bp | AT5G01750 | 286,333 | Forward | Unknown protein | Laplaze et al. (2005) |
| −1,253 bp | AT5G01747 | Forward | Encodes a microRNA that targets several genes containing NAC domains including NAC1 | ||||||
| −5,051 bp | AT5G01740 | Forward | Nuclear transport factor 2 (NTF2) family protein | ||||||
| Q0171/N9207 | n. d. | Root cap | n.d. | −9,611 bp | AT1G04690 | 1,304,051 | Forward | Potassium channel β-subunit 1 (KAB1) | |
| AT1G04680 | Complement | Pectin lyase-like superfamily protein | |||||||
| RM1000 | n. d. | Leaf margins | n.d. | −649 bp | AT2G42010 | 17,538,639 | Complement | Phospholipase D (PLDβ) | |
| AT2G42030 | Complement | RING/U-box superfamily protein | |||||||
| No expression lines | |||||||||
| J2501/N9121 | n. d. | n.d. | n.d. | −4,314 bp | AT1G17744 | 6,106,109 | Complement | Unknown protein | |
| AT1G17745/50 | Forward | Encodes a 3-phosphoglycerate dehydrogenase/encodes PEPR2, a plasma membrane Leu-rich repeat receptor kinase functioning as a receptor for the Pep1 and Pep2 peptides | |||||||
| J2661/N9187 | n. d. | n.d. | n.d. | −129 bp | AT2G01010 | 3,577 | Forward | 18S rRNA | |
| AT2G01008 | Forward | Unknown protein | |||||||
| −129 bp | AT3G41768 | 14,197,548 | Forward | 18S rRNA | |||||
| AT3G41761 | Forward | Other RNA | |||||||
| M0136/N9302 | n. d. | n.d. | n.d. | −3,605 bp | AT1G32140 | 11,568,438 | Complement | F-box family protein | |
| AT1G32150/60 | Forward | Encodes a G group bZIP(68) transcription factor family member that can bind cis-elements with an ACGT core, such as G-box, Hex, C-box, and As-1/unknown protein | |||||||
Based on confocal microscopy observations, 15 out of 21 GAL4/UAS enhancer trap lines showed GFP expression during embryo development (Table I), while three (J0121, Q0171, and RM1000) were expressed only later during postembryonic development (Table I; Supplemental Fig. 1, F–J). In the remaining three lines (J2501, J2661, and M0136), no GFP signal could be detected. In previous studies, the expression pattern of J2661 and J2501 was described to be restricted to the pericycle (Levesque et al., 2006; Petersson et al., 2009). A plausible explanation for the lack of GFP expression in our analysis might be a gene silencing event or differences in growth conditions between labs.
In the following, we describe in detail the expression patterns observed for the remaining 18 lines, categorized by their dominant expression domain. All expression patterns are summarized in Table I.
Early Embryo Development
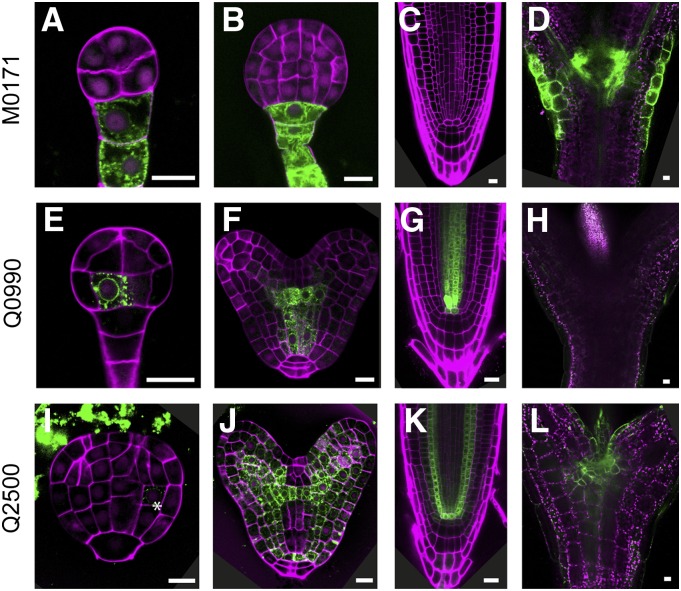
A first group, consisting of three lines (M0171, Q0990, and Q2500), showed cell-type-specific expression during early embryo development (Fig. 1). The GFP expression of M0171 was observed as early as the octant cell stage in all suspensor cells, and only later on, when the suspensor is no longer present, the expression was switched on in the cotyledon junction (Fig. 1, A–D; Supplemental Fig. 1A). Subsequently, during postembryonic development, M0171 expression remained cotyledon junction specific (Fig. 1D). Expression of the Q0990 line commenced at dermatogen stage in the inner basal cells (Fig. 1E), which are the precursors of ground and vascular tissues (Peris et al., 2010; ten Hove et al., 2015). The expression of this line remains vascular tissue specific later during embryogenesis as well as postembryonically (Fig. 1, F–H). This line has been extensively used as vascular marker (De Rybel et al., 2013; Levesque et al., 2006; Donner et al., 2009) and in misexpression studies (Wenzel et al., 2012; Weijers et al., 2006).
Figure 1.
GFP expression in early embryogenesis lines. GFP fluorescence in preglobular or globular stage embryos (A, E, and I), late globular or heart stage embryos (B, F, and J), root tips (C, G, and K), and shoot apex (D, H, and L) of M0171 (A–D), Q0990 (E–H), and Q2500 (I–L) lines. Magenta counterstaining in A, B, E, F, I, and J is Renaissance fluorescence, propidium iodide in C, G, and K, and chlorophyll autofluorescence in D, H, and L. Bars = 10 μm.
In Q2500 embryos, very weak expression was first detected during late globular stage in the ground tissue precursors (Fig. 1I). Additional expression foci were found with the progression of embryogenesis (at heart stage) in the mesophyll precursors and protoderm of future cotyledons. Expression extended through the ground tissue of the hypocotyl and root and also included the pericycle and quiescent center (QC; Fig. 1J). Postembryonically, the GFP expression of Q2500 is found in the root ground tissue, pericycle, QC (Fig. 1K), and cotyledon junction as predicted by its expression during embryogenesis, but also in the leaf marking the guard mother cells (Fig. 1L; Supplemental Fig. 1C). In contrast to the other two lines in this group, Q2500 has broader expression domain found in multiple tissue types.
Late Embryo Development
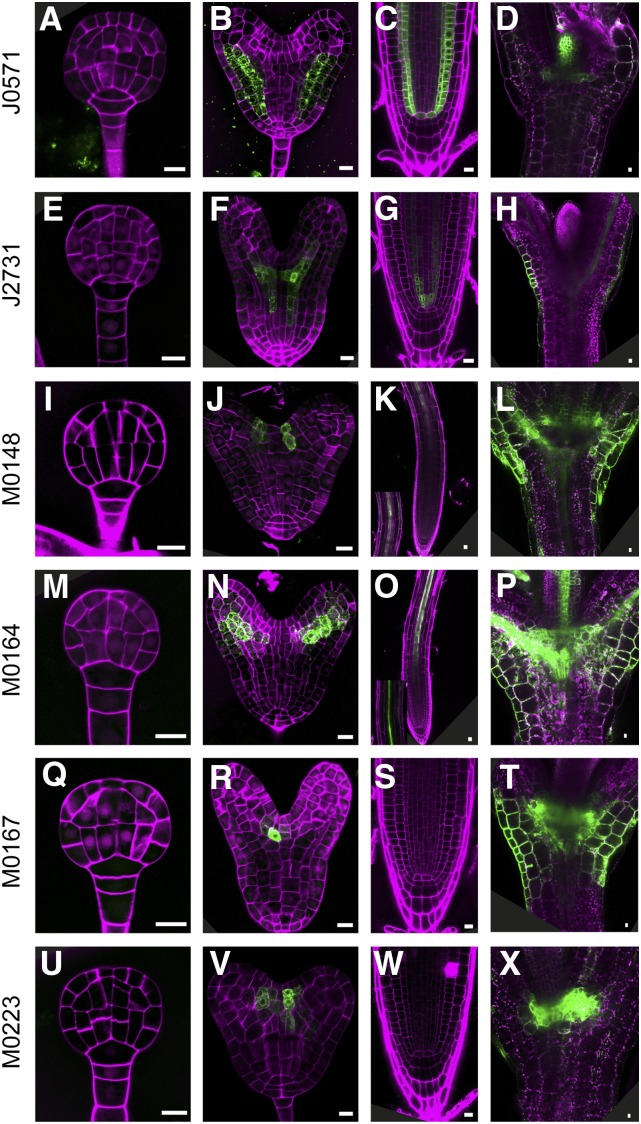
The second group consists of six lines (J0571, J2731, M0148, M0164, M0167, and M0223) whose expression marks specific embryonic tissues during late embryo development. The first line in this group is J0571, which is widely used as ground tissue specific marker (Mylona et al., 2002; Tsugeki et al., 2010; Xu et al., 2013; Zhang et al., 2015) and for targeted gene expression studies (Haseloff, 1999; Wenzel et al., 2012; Waki et al., 2013). Indeed, starting from heart stage, GFP expression in J0571 was observed in the ground tissue, but weak expression could be also seen in the basal protoderm cells (Fig. 2, A and B). In addition to the ground tissue expression in the postembryonic root, GFP could detected in some, but not all, QC cells, and this line showed strong GFP expression in the shoot meristem margins, young leaf primordia and in the leaf epidermis (guard and pavement cells; (Fig. 2, C and D; Supplemental Fig. 1D). The expression pattern in line J2731 is very remarkable. In developing embryos, its expression was first observed in vascular tissue at early heart stage, but at a distance of one to two cells from the QC (Fig. 2, E and F). This pattern is exactly opposite to that of recently reported “stem cell” zone markers, which are expressed in the cells closest to the lens-shaped cell, but not in the cells above it (Wendrich et al., 2015b). Strikingly, J2731 roots reveal a GFP signal in the cells immediately above the QC (Fig. 2G), which suggests that expression shifts toward more juvenile vascular cells as development progresses. Very strong expression was also detected in the guard mother cells and in the pavement cells, similar to line Q2500 (Supplemental Fig. 1, C and E).
Figure 2.
GFP expression in late embryogenesis lines. GFP fluorescence in globular stage embryos (A, E, I, M, Q, and U), heart stage embryos (B, F, J, N, R, and V), root tips (C, G, K, O, S, and W) and shoot apex (D, H, L, P, T, and X) of J0571 (A–D), J2731 (E–H), M0148 (I–L), M0164 (M–P), M0167 (Q–T), and M0223 (U-X) lines. Magenta counterstaining in (A, B, E, F, I, J, M, N, Q, R, U, and V) is Renaissance fluorescence, propidium iodide in C, G, K, O, S, and W, and chlorophyll autofluorescence in D, H, L, P, T, and X. Bars = 10 μm.
During embryogenesis, expression of M0148 was found in two narrow files of cells surrounding the SAM, while postembryonically, the marker was expressed in pericycle cells in the differentiation zone of the root (Fig. 2, I–K) and in a broad domain surrounding the SAM (Fig. 2L). M0164 marks the vascular strands in the embryonic cotyledon primordia (Fig. 2N). Like M0148, GFP signal of M0164 was detected in the root pericycle (Fig. 2O). Furthermore, strong expression was found in shoot vascular tissues (Fig. 2P). Finally, lines M0167 and M0223 have similar expression in the intersection of the two cotyledon primordia (Fig. 2, Q–X).
Embryonic RAM Lines
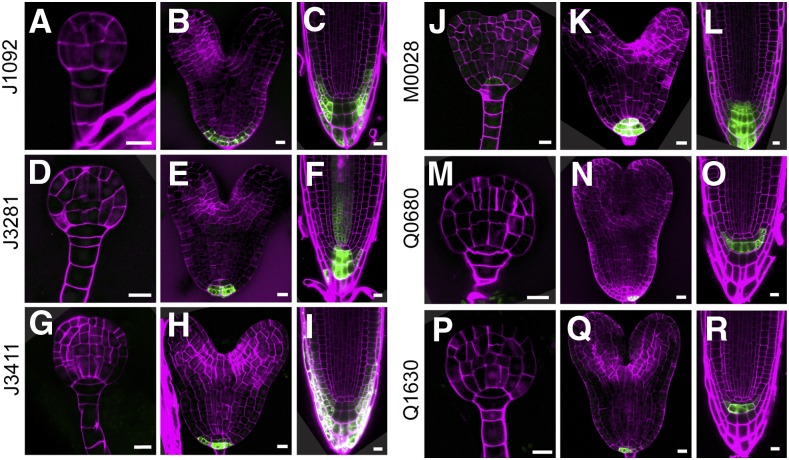
Six lines (J1092, J3281, J3411, M0028, Q0680, and Q1630) are expressed mainly or specifically in the embryonic root meristem. All lines in this group show similar expression patterns observed in the future RAM cells and in the case of J1092 and J3411 extended to the lateral root cap (LRC) precursors (Fig. 3, C and I). The marked cell types originate from the uppermost suspensor cell, specified as hypophysis at early globular stage of embryonic development. An asymmetric division of the hypophysis then generates a lens-shaped cell from which the QC will form and a basal cell, which is the precursor of the root cap (Scheres et al., 1994; ten Hove et al., 2015). The expression of these lines during postembryonic root development fully recapitulates their embryonic expression domain (Fig. 3) and could not be found in other parts of the plant body, except J3281, whose expression in the postembryonic root extends to include the young vascular tissue (Fig. 3F). Hence, the lines in this group are excellent candidates for RAM/columella markers.
Figure 3.
GFP expression in embryonic root meristem lines. GFP fluorescence in globular stage embryos (A, D, G, J, M, and P), heart stage embryos (B, E, H, K, N, and Q), and root tips (C, F, I, L, O, and R) of J1092 (A–C), J3281 (D–F), J3411 (G–I), M0028 (J–L), Q0680 (M–O), and Q1630 (P–R) lines. Magenta counterstaining in A, B, D, E, G, H, J, K, M, N, P, and Q is Renaissance fluorescence and propidium iodide in C, F, I, L, O, and R. Bars = 10 μm.
Even though lines J0121, Q0171, and RM1000 did not fall in any of the above-described groups, they possess interesting expression patterns. J0121 is one of the most frequently used xylem pole pericycle marker lines (Laplaze et al., 2005; Parizot et al., 2008; Sugimoto et al., 2010), but we could also detect a very distinct GFP signal in the seed coat and young leaf primordia (Supplemental Fig. 1, H and F). The remaining two lines, Q0171 and RM1000, show very specific expression in the postembryonic root cap and in the leaf margins, respectively (Supplemental Fig. 1, I and J).
Identification of Insert Location
To molecularly characterize the GAL4/UAS enhancer trap lines, as well as to identify novel cell-type-specific promoters during embryo development, we determined the genomic coordinates of the T-DNA insertion. To amplify the genomic DNA flanking the T-DNA inserts, we performed thermal asymmetric interlaced PCR (TAIL-PCR; Liu et al., 1995) with degenerate primers and a set of nested primers in the T-DNA left border. The TAIL-PCR products were subsequently sequenced.
The insert location in lines J0571, J0121, J2731, M0167, and M0223 were previously reported (Miyashima et al., 2011; Laplaze et al., 2005; Møller et al., 2009; Cary et al., 2002), yet, as the description of some were not very explicit, we included these lines as well. In the following, we will focus on notable cases only. However, a detailed overview of all insert locations and the flanking genes is presented in Table I.
We were able to map the T-DNA insert positions in all lines (Table I; Supplemental Fig. 2), with one notable exception. Despite repeated attempts, we did not manage to amplify a T-DNA flank from the M0171 line. Since the line clearly expresses GFP, has been used to transactivate other genes (Rademacher et al., 2012), and is kanamycin resistant, the T-DNA must be intact. Therefore, the inability to amplify flanking regions may be related to the genomic location (e.g. high GC content or repeats). We also included the three lines (J2501, J2661, and M0136), which did not show any GFP expression, but identified insertion sites for each, suggesting that the lack GFP expression is not due to absence of an insertion.
Identification of Insert Location in the Early Embryo Development Lines
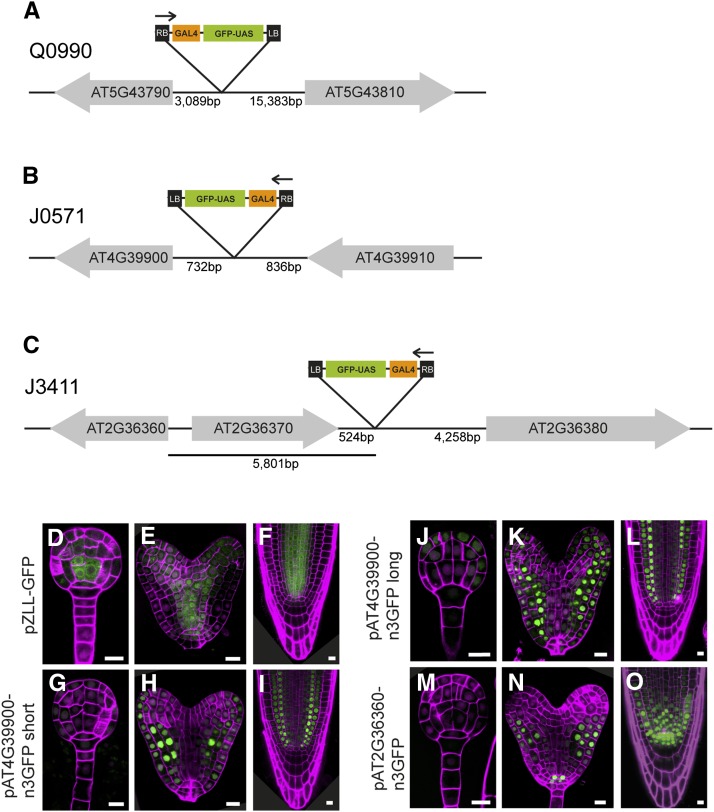
Intriguingly, in the vascular-specific Q0990 line, the insertion is mapped to a large intergenic region 15.4 kb upstream of the ATG of AT5G43810 and 3 kb upstream of the ATG of AT5G43790 (Fig. 4A). The T-DNA is in the same coding direction as AT5G43810, which encodes ARGONAUTE10 (AGO10; known also as ZWILLE [ZLL]; Moussian et al., 1998), a member of the AGO/ELONGATION INITIATION FACTOR 2C class of proteins. The other gene, AT5G43790, is in the opposite coding direction to the T-DNA insertion and encodes putative pentatricopeptide repeat superfamily protein. In addition, a hypothetical transposable element (AT5G43800) is located between the Q0990 insertion and the ZLL gene. In theory, the insertion should report on the activity of local enhancer elements that normally drive expression of one or both of the adjacent genes. According to microarray data (Brady et al., 2007; Belmonte et al., 2013), expression of both genes is found in the vasculature in the root, but only ZLL is expressed during embryogenesis (Moussian et al., 1998; Tucker et al., 2008). We generated a construct containing the promoter region of ZLL fused to GFP and tested its expression pattern in wild-type plants. As shown in Figure 4, the pattern of expression of pZLL-GFP is identical to the one of Q0990 (Fig. 1, E–H), which suggests that a regulatory element(s) in the ZLL promoter (downstream of the insertion) is responsible for the GFP expression pattern in Q0990.
Figure 4.
Identification of T-DNA insertion sites and validation of neighboring gene expression. A to C, Schematics illustrating the insertion site of the enhancer trap T-DNA in Q0990 (A), J0571 (B), and J3411 (C) lines. Neighboring genes are indicated as gray arrows, starting at the gene’s ATG and pointing toward the gene’s stop codon. The schematics are not drawn to scale and distances (in base pairs) are indicated. The orientation of the T-DNA insertions is indicated by an arrow over the T-DNA map (starting from the right border). D to O, Expression of transcriptional fusions of gene promoters to nuclear lamina-localized GFP (D–F) or nuclear 3xGFP (G–O) in globular stage embryos (D, G, J, and M), heart stage embryos (E, H, K, and N), and root tips (F, I, L, and O). D to F, pZLL-GFP (related to Q0990). G to L, short version (G–I) or long version (J–L) of the At4G39900 promoter fused to n3GFP (related to J0571). M to O, pAt2G36360-n3GFP (related to J3411). Magenta counterstaining in D, E, G, H, J, K, M, and N) is Renaissance fluorescence and propidium iodide in F, I, L, and O. Bars = 10 μm.
Q2500 enhancer trap line is inserted on chromosome IV in the third exon of UNFERTILIZED EMBRYO SAC12 (UNE12; AT4G02590), 1.1 kb downstream of its ATG and coding in the same direction (Table I; Supplemental Fig. 2). UNE12 is a bHLH transcription factor involved in the double fertilization event forming the zygote and the endosperm (Pagnussat et al., 2005). The other gene flanking the T-DNA, in the opposite direction, is AT4G02600. The latter encodes MLO1, a member of a large plant-specific family of seven-transmembrane domain proteins. As shown by microarray experiments (Brady et al., 2007; Moussian et al., 1998), both UNE12 and MLO1 are expressed in the ground tissue, including pericycle, but only UNE12 is expressed during embryo development with a peak at heart stage and might thus be responsible for the specific expression pattern in Q2500. Interestingly, the T-DNA insertion in this line (third exon of UNE12) does not induce the developmental defects that were reported for loss of function alleles (Pagnussat et al., 2005).
Identification of Insert Location in the Late Embryo Development Lines
The T-DNA insert in line J0751 is, as reported previously by Miyashima et al. (2011), in the intergenic region between AT4G39900 and AT4G39910, which encode an unknown protein and a nuclear ubiquitin-specific protease (UBP3), respectively. The insert is located in the promoter region of AT4G39900, 732 bp upstream of the start codon and oriented in the same direction (Fig. 4B), which makes it more likely to have ground tissue-specific expression. To test if this is true, we made reporter lines using 3- and 1.7-kb fragments upstream of the start codon of AT4G39900 and fused both to sensitive nuclear-localized 3xGFP (n3GFP). Both short and long promoter fusions showed slightly broader expression patterns compared to J0571 (Fig. 4, G–L). During embryogenesis, weak expression of pAT4G39900-n3GFP was first detected in globular-stage embryos in the protoderm and in the ground tissue precursor cells (Fig. 4J). Later on, during heart stage, the expression is seen in the same domains with a peak in the ground tissue (Fig. 4, H and K). In the postembryonic root, GFP expression in the epidermis was not observed, but in addition to the ground tissue, a very weak expression could be detected in the vasculature, yet the occasional QC expression in J0571 was not observed in these lines (Fig. 4, I and L). Fusion of a shorter genomic DNA fragment, adjacent to the insertion point, may help to limit expression to the ground tissue. In addition, thorough examination of the AT4G39900 promoter using deletions should help to elucidate this point.
M0164 is the only line in our selection that harbors two insertions, one on chromosome I and another on chromosome IV. On chromosome I, the T-DNA insert is oriented in the same direction and 4.5 kb upstream of AT1G09520 and 733 bp upstream of the start codon of AT1G09530, but in reverse orientation. AT1G09520 encodes an unknown protein, which possesses zinc-finger and PHD-type domains, whereas AT1G09530 encodes PHYTOCHROME INTERACTING FACTOR3 (PIF3), which is bHLH transcription factor that interacts with photoreceptors phyA and phyB (Ni et al., 1998; Table I; Supplemental Fig. 2). On chromosome IV, M0164 enhancer trap T-DNA was inserted between AT4G00440 and AT4G00430, encoding TON RECRUITING MOTIF15 (TRM15) and PLASMA MEMBRANE INTRINSIC PROTEIN 1;4 (PIP1;4), respectively. The GAL4 T-DNA insertion was about 1.8 kb upstream of and with the same orientation as TRM15. Based on the microarray-predicted expression patterns (Brady et al., 2007) and including the fact that M0164 has two insertions on different chromosomes, it is very challenging to predict which gene(s) underlies the specific expression pattern of M0164. Interestingly, we mapped M0167 enhancer trap T-DNA insertion on chromosome I to exactly the same position as one of the insertions identified in M0164 (Table I; Supplemental Fig. 2). Thus, because M0164 has a broader expression pattern than M0167 (Fig. 2, M–P and Q–T), it is likely that M0167 is a derivative of M0164.
Identification of Insert Location in the Embryonic RAM Lines
In line J3281, the T-DNA insertion was positioned 424 bp downstream of the start codon and in reverse orientation of AT5G62220 and 6.8 kb upstream of AT5G62230 (in the same orientation; Table I; Supplemental Fig. 2). The latter encodes ERECTA-LIKE1 (ERL1; Shpak et al., 2004), an ERECTA (ER) family Leu-rich repeat-receptor-like kinase, which together with ER and ERL2 specifies aerial organ size by promoting cell proliferation (Shpak et al., 2004). In addition, ERL1 is involved in specification of stomatal stem cell fate and differentiation of guard cells (Shpak et al., 2005). As reported previously, the expression of ERL1 marks the actively proliferating organs like shoot meristem, leaf primordia, and young developing flowers (Shpak et al., 2004). According to microarray data, expression of ERL1 can be also found in the root vascular tissue (Brady et al., 2007) consistent with the observed pattern of expression in J3281.
J3411 enhancer trap T-DNA is inserted in an intergenic region between AT2G36370 and AT2G36380 and is coding in the opposite direction to the two flanking genes (Table I; Figure 4C). AT2G36370 and AT2G36380 encode ubiquitin-protein ligase and ATP-BINDING CASETTE G34 (also known as PLEIOTROPIC DRUG RESISTENCE6), respectively. Both flanking genes have broad expression domains as shown by microarray experiments (Birnbaum et al., 2003), and it is therefore less likely that one of them is responsible for the specific GAL4-GFP expression pattern in J3411. However, the T-DNA was inserted upstream and it was in the same orientation as AT2G36360 (Fig. 4C). The latter encodes Gal oxidase/kelch repeat superfamily protein with unknown function. To determine if the J3411 expression pattern is due to regulatory DNA sequence in the AT2G36360 promoter, a 3.9-kb fragment upstream of the AT2G36360 ATG was fused to n3GFP and was subsequently introduced in wild-type Col-0 plants. pAT2G36360-n3GFP was detected in both embryos and roots. In embryos, the GFP signal was first detected at early heart stage, in the future QC and columella cells, and expanded to include the ground tissue and the protoderm later on (Fig. 4, M and N). In the postembryonic root, the expression was broader including the vasculature and columella initials, but it was absent from the distal columella cells (Fig. 4O). Thus, the specific GAL4 expression of J3411 is not replicated by the expression of pAT2G36360-n3GFP. A possible scenario is that the expression pattern observed in line J3411 may be due to regulatory elements located in the genomic DNA instantly upstream of the T-DNA insert. A similar case has been previously reported, where the expression of AT5G65590, a Dof zinc-finger transcription factor, which is flanking the insert in the guard cell specific line E1728, did not match the expression of E1728. Alternatively, fusion of a DNA fragment neighboring the T-DNA insert in E1728 and the GAL4 TATA box to uidA reporter gene showed strong GUS activity in guard cells (Gardner et al., 2009).
In line M0028, the T-DNA insert was found to be 1.46 kb upstream from the ATG of GIBBERELLIN3-OXIDASE3 (GA3OX3), in the same coding direction (AT4G21690) and 6.2 kb upstream from the ATG of NITRATE TRANSPORTER 1.8 (NRT1.8; also known as NPF7.2), being in the opposite direction of the T-DNA (AT4G21680; Supplemental Fig. 2). The expression of GA3OX3 has been studied in detail and it was found in heart and torpedo stage embryos, near the junction between the embryo axis and cotyledons (Hu et al., 2008). Although, the T-DNA insert in M0028 is in a very close proximity of GA3OX3 and in the same orientation, GA3OX3 has very different expression pattern. Further analysis on the intergenic region between GA3OX3 and NPF7.2 will provide more information about the elements responsible for the observed columella-specific M0028 expression pattern.
The last two lines in this group, Q0680 and Q1630, report the same expression pattern (Fig. 3, M–O and P–R). Moreover, the TAIL-PCR mapping results indicated that insert position of the enhancer traps in both lines is the same, located between AT2G45200 and AT2G45210, which code for GOLGI SNARE12 (GOS12) and SMALL AUXIN UPREGULATED 36 (SAUR36), respectively. The GAL4 gene was found 1.75 kb upstream of the start codon of AT2G45200 (GOS12), in the same coding orientation, and only 494 bp upstream of the start codon of AT2G45210 (SAUR36), but in the opposite orientation (Table I; Supplemental Fig. 2). GOS12 is predicted to be expressed in the root tip with a peak in the columella initials, while SAUR36 is expected to be more specific to the vasculature (Brady et al., 2007). It is difficult to speculate which gene is more likely to report the cell-specific GAL4 expression of Q0680/Q1630 based on the predicted expression patterns, since both genes are generally expressed. In this case, the T-DNA insert is closer to SAUR36, although it is in the opposite orientation. In line M0223, the T-DNA was also found to be in the opposite orientation, upstream of CUC1 (AT3G15170; see also Table I; Supplemental Fig. 2), and Cary et al. (2002) showed that this is the gene reporting the expression of M0223.
Ambiguous Insertion Sites
In line J2501, the T-DNA insertion is located on chromosome I between a gene encoding 3-phosphoglycerate dehydrogenase (AT1G17745) and a plasma membrane Leu-rich repeat receptor kinase (PERP2; AT1G17750). In this case, the T-DNA is oriented in the opposite direction to the two flanking genes (Table I; Supplemental Fig. 2). Both genes are expressed in the root stele (including also the ground tissue), as shown by microarray experiments (Birnbaum et al., 2003), and it is not clear which might be responsible for the GAL4 expression pattern in J2501. The next gene, which is 4.3 kb downstream and in the same orientation, is an unknown gene, AT1G17744. In J2661, the T-DNA was located in a duplicated region present in both chromosomes II and III. In chromosome II, the insert is found in the very end of the chromosome between the first and the second gene, namely, AT2G01008 (unknown gene) and AT2G01010 (18S rRNA), and the T-DNA was in the same direction as both genes. Since the T-DNA is only 129 bp upstream of the second gene, it is possible that this gene is preferentially of exclusively expressed in the pericycle. The situation in chromosome III is similar with the T-DNA inserted between AT3G41768 and AT3G41761, encoding 18S rRNA and other RNA, respectively. The same as in chromosome II, the T-DNA is 129 bp upstream of the 18S rRNA. The T-DNA insert in M0136 is positioned in the opposite direction, between a transcription factor bZIP68 (AT1G32150) and an unknown protein (AT1G32160), but it is in the same direction and 3.6 kb upstream of an F-box family protein (AT1G32140; Table I; Supplemental Fig. 2).
Genotyping Enhancer Trap Lines
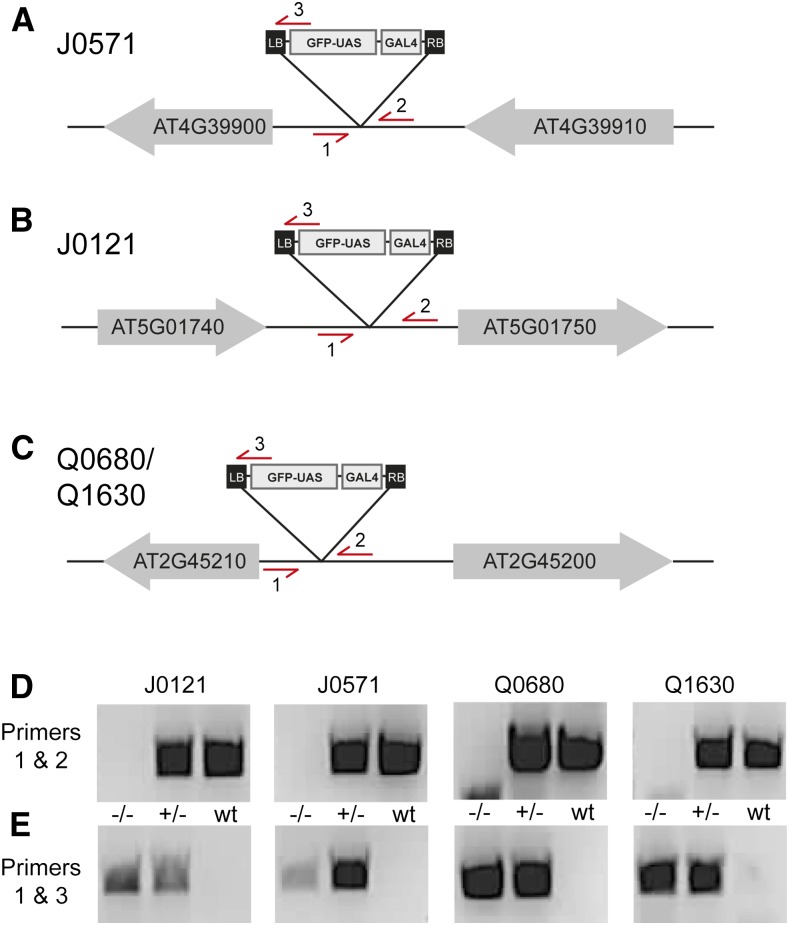
We have characterized the expression patterns and the insert locations of 21 GAL4 enhancer trap lines. This information can now be used for several purposes, one of which being the development of primer pairs that can identify presence and absence of the insertion, which is useful for PCR genotyping during introgression. As a proof of principle, we designed genotyping primers for four lines. We chose two of the most used GAL4 driver lines, J0121 and J0571, and Q0680 and Q1630, where we can verify the suggestion that these two lines harbor the same enhancer trap insertion.
For all four lines, two pairs of primers were generated. One pair binds to the genomic sequence flanking the left and the right border of the T-DNA insert and it should amplify DNA only in absence of insertion (Fig. 5, A–C). The other pair of primers uses a primer in the left border of the T-DNA (the same as the third specific primer used for the TAIL-PCRs) and a primer in the genomic sequence in the proximity of the left T-DNA border. Contrary to the first primer pair, this one should amplify DNA only in presence of insertion. We have tested the primers on wild-type plants as well as on homo- and heterozygous plants and the results were as expected (Fig. 5, D and E).
Figure 5.
Genotyping GAL4/UAS transgenes. A to C, Schematics illustrating the insertion site of the enhancer trap T-DNA in J0571 (A), J0121 (B), and Q0680/Q1630 (C) lines. Neighboring genes are indicated as gray arrows, starting at the gene’s ATG and pointing toward the gene’s stop codon. Primers used for genotyping are indicated as red arrows. In each case, the transgene should be amplified using primers 1 and 3, while the wild type is amplified with primers 1 and 2. D and E, PCR amplification of the wild-type fragment (D; primers 1 and 2) or the insert-specific fragment (E; primers 1 and 3) in genomic DNA from plants homozygous (−/−), heterozygous (+/−), or wild-type (wt) for the J0517, J0121, Q0680, and Q1630 lines. Note that fragments of the appropriate size are amplified according to the expected genotype of each line.
Concluding Remarks
We have selected 21 widely used GAL4/UAS enhancer trap lines for detailed characterization of expression pattern and genomic insertion site. First, we systematically documented their expression patterns and grouped them into three groups based on their dominant expression domains. Our microscopic observations demonstrate that the expression patterns of many of the selected GAL4 enhancer trap lines are often expressed in a domain that extends beyond the one that is often reported. This finding should be taken as a cautionary note in two ways. First, the expression may not be taken as evidence for a cell type in its strictest sense, but rather as a regional marker in the local context. Second, when using these GAL4/UAS drivers to target local misexpression, one should be aware that the target gene is in fact misexpressed in a broader domain, which might cause more pleiotropic effects that could otherwise be interpreted as non-cell-autonomous effects of misexpression. Furthermore, we present an overview of the genomic positions of the T-DNA insertion of all lines, with one exception: M0171. This identified two duplications: Line M0167 is likely a derivative of M0164, and lines Q0680 and Q1630 share exactly the same expression pattern as well as the same insert position. In addition, the detailed insertion maps should help to rationalize the cause of local gene expression and as such will provide a useful tool in studying the genes close to the insertion sites. In addition, the expression patterns can now in principle be migrated to other ecotype backgrounds or used to generate simpler promoter-reporter fusions based on the genes close to the insertion site.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The GAL4-GFP enhancer trap lines used here (Table I) are part of a collection of Arabidopsis (Arabidopsis thaliana) lines generated in C24 ecotype (Haseloff, 1999) and are available from the Nottingham Arabidopsis Stock Center. The new transgenic lines generated in this study are Columbia (Col-0) ecotype.
All Arabidopsis seeds were surface-sterilized and dried seeds were subsequently grown on half-strength Murashige and Skoog plates with or without antibiotics selection at 22°C in standard long-day (16:8 h light:dark) growth conditions. After 2 weeks of growth, the seedlings were transferred to soil and further grown under the same conditions.
Microscopy
Confocal microscopy was performed as according to Llavata-Peris et al. (2013) with minor modifications. Briefly, for imaging of embryos, ovules were isolated and mounted in a 4% paraformaldehyde/5% glycerol/1× PBS solution including 1.5% SCRI Renaissance Stain 2200 (R2200; Renaissance Chemicals) for counterstaining of embryos. After applying the coverslip, the embryos were squeezed out of the ovules, and R2200 and GFP fluorescence were visualized by excitation at 405 and 488 nm and detection between 430 to 470 and 500 to 535 nm, respectively. For imaging of roots, 5-d-old-seedlings were incubated in 10 μg/mL propidium iodide solution for 1 to 2 min, and GFP and propidium iodide were visualized by excitation at 488 nm and detection between 500 to 535 nm and 630 to 700 nm, respectively. All confocal imaging was performed on a Leica SP5 II system equipped with hybrid detectors.
Mapping of the T-DNA Insertion Sites
Genomic DNA was isolated from all GAL4-GFP enhancer trap line seedlings using CTAB (cetyltrimethylammonium bromide) extraction buffer (1% CTAB, 100 mm Tris-HCl, pH 8.0, 20 mM EDTA, pH 8.0, and 1.5 m NaCl, water) and was afterward precipitated with isopropanol and washed with 70% ethanol. TAIL-PCR was performed as described previously (Liu et al., 1995) with minor modifications using the specific left border primers listed in Supplemental Table S1. Some PCR products were cloned into pGEM-T vector (Promega), and the PCR products were then sequenced for identification of the T-DNA insertion sites. Database searches were done using the BLAST through the NCBI website (www.ncbi.nlm.nih.gov).
Cloning and Plant Transformation
For generating transcriptional fusions, up to 4-kb fragments upstream of the ATG were amplified from genomic DNA using Phusion Flash PCR Master Mix (Thermo Scientific) and cloned into vector pPLV104 using ligation-independent cloning (Wendrich et al., 2015a) and primers listed in Supplemental Table S1. All inserts were completely sequenced and transformed into Col-0 wild-type Arabidopsis plants by simplified floral dipping (De Rybel et al., 2011).
Genotyping of the GAL4-GFP Enhancer Trap Lines
Genomic DNA from homozygous and heterozygous GAL4-GFP plants and C24 wild-type plants was isolated as mentioned above using CTAB extraction buffer. Four microliters of the isolated DNA was subsequently used for PCR amplification with homemade purified recombinant Taq DNA polymerase. The program was as follows: 35 cycles (95°C, 30 s; 65°C, 30 s; 72°C, 30 s). The primers used for genotyping are listed in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Additional GFP expression patterns.
Supplemental Figure S2. Schematic representation of the GAL4 insertion positions.
Supplemental Table S1. Oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank Jim Haseloff for generating the extremely useful collection of GAL4/UAS lines and sharing these with the Arabidopsis community. We acknowledge the Nottingham Arabidopsis Stock Center for distributing seeds, Fons de Vogel and Akshita Chordia for assistance in microscopy and insert mapping, and our colleagues Bert De Rybel, Kuan-Ju Lu, and Joakim Palovaara for comments on the manuscript.
Glossary
- SAM
shoot apical meristem
- QC
quiescent center
- LRC
lateral root cap
- TAIL-PCR
thermal asymmetric interlaced PCR
- CTAB
cetyltrimethylammonium bromide
Footnotes
Articles can be viewed without a subscription.
This work was supported by grants from the Netherlands Organization for Scientific Research (ALW-NSFC Plant Development Collaborative Grant 846.11.001 to D.W. and ALW-VENI Grant 863.12.010 to C.A.t.H.) and the European Research Council (Starting Grant CELLPATTERN; contract number 281573 to D.W.).
References
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Cary AJ, Che P, Howell SH (2002) Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J 32: 867–877 [DOI] [PubMed] [Google Scholar]
- Ckurshumova W, Koizumi K, Chatfield SP, Sanchez-Buelna SU, Gangaeva AE, McKenzie R, Berleth T (2009) Tissue-specific GAL4 expression patterns as a resource enabling targeted gene expression, cell type-specific transcript profiling and gene function characterization in the Arabidopsis vascular system. Plant Cell Physiol 50: 141–150 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D (2013) A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 24: 426–437 [DOI] [PubMed] [Google Scholar]
- De Rybel B, van den Berg W, Lokerse A, Liao CY, van Mourik H, Möller B, Peris CL, Weijers D (2011) A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol 156: 1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SC, Booij H, Meyerink P, Huisman G, Wilde HD, Thomas TL, van Kammen A (1988) Acquisition of embryogenic potential in carrot cell-suspension cultures. Planta 176: 196–204 [DOI] [PubMed] [Google Scholar]
- Donner TJ, Sherr I, Scarpella E (2009) Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, Nowack MK (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 24: 931–940 [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Baker AJ, Assie JM, Poethig RS, Haseloff JP, Webb AA (2009) GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J Exp Bot 60: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley KO, Nutt SL, Amaya E (2002) Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc Natl Acad Sci USA 99: 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J. (1999) GFP variants for multispectral imaging of living cells. Methods Cell Biol 58: 139–151 [DOI] [PubMed] [Google Scholar]
- Hu J, Mitchum MG, Barnaby N, Ayele BT, Ogawa M, Nam E, Lai W-C, Hanada A, Alonso JM, Ecker JR, et al. (2008) Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20: 320–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Umehara M, Satoh S, Kamada H (2003) Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J 34: 107–114 [DOI] [PubMed] [Google Scholar]
- Kang YH, Song S-K, Schiefelbein J, Lee MM (2013) Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol 163: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M (2004) A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell 7: 133–144 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martinière A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J (2005) GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56: 2433–2442 [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, Scheres B, Benfey PN (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Llavata-Peris C, Lokerse A, Möller B, De Rybel B, Weijers D (2013) Imaging of phenotypes, gene expression, and protein localization during embryonic root formation in Arabidopsis. Methods Mol Biol 959: 137–148 [DOI] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313 [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jürgens G, Laux T (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J 17: 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Linstead P, Martienssen R, Dolan L (2002) SCHIZORIZA controls an asymmetric cell division and restricts epidermal identity in the Arabidopsis root. Development 129: 4327–4334 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Moreadith RW, Leder P (1991) Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci USA 88: 698–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L-F, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J, et al. (2008) Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol 146: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris CI, Rademacher EH, Weijers D (2010) Green beginnings - pattern formation in the early plant embryo. Curr Top Dev Biol 91: 1–27 [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Freire Rios A, Borst JW, Lukowitz W, Jürgens G, Weijers D (2012) Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell 22: 211–222 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA (1999) Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev 80: 153–158 [DOI] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P (1994) Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487 [Google Scholar]
- Sena G, Wang X, Liu H-Y, Hofhuis H, Birnbaum KD (2009) Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Lu KJ, Weijers D (2015) Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142: 420–430 [DOI] [PubMed] [Google Scholar]
- Tsugeki R, Ditengou FA, Palme K, Okada K (2010) NO VEIN facilitates auxin-mediated development in Arabidopsis. Plant Signal Behav 5: 1249–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Hinze A, Tucker EJ, Takada S, Jürgens G, Laux T (2008) Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development 135: 2839–2843 [DOI] [PubMed] [Google Scholar]
- Waki T, Miyashima S, Nakanishi M, Ikeda Y, Hashimoto T, Nakajima K (2013) A GAL4-based targeted activation tagging system in Arabidopsis thaliana. Plant J 73: 357–367 [DOI] [PubMed] [Google Scholar]
- Webb M, Jouannic S, Foreman J, Linstead P, Dolan L (2002) Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3--a katanin-p60 protein. Development 129: 123–131 [DOI] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G (2006) Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10: 265–270 [DOI] [PubMed] [Google Scholar]
- Wendrich JR, Liao CY, van den Berg WA, De Rybel B, Weijers D (2015a) Ligation-independent cloning for plant research. Methods Mol Biol 1284: 421–431 [DOI] [PubMed] [Google Scholar]
- Wendrich JR, Möller BK, Uddin B, Radoeva T, Lokerse AS, De Rybel B, Weijers D (2015b) A set of domain-specific markers in the Arabidopsis embryo. Plant Reprod 28: 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel CL, Marrison J, Mattsson J, Haseloff J, Bougourd SM (2012) Ectopic divisions in vascular and ground tissues of Arabidopsis thaliana result in distinct leaf venation defects. J Exp Bot 63: 5351–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H, Anders N, Geldner N, Gavidia R, Jürgens G (2011) Coordination of apical and basal embryo development revealed by tissue-specific GNOM functions. Development 138: 117–126 [DOI] [PubMed] [Google Scholar]
- Xu P, Yuan D, Liu M, Li C, Liu Y, Zhang S, Yao N, Yang C (2013) AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol 161: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jiao Y, Liu Z, Zhu Y-X (2015) ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat Commun 6: 6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.