Nonsyntenic genes are overrepresented among differential, nonadditive, and allelic expression patterns in root tissues of maize hybrids during the early developmental manifestation of heterosis.
Abstract
Distantly related maize (Zea mays) inbred lines display an exceptional degree of genomic diversity. F1 progeny of such inbred lines are often more vigorous than their parents, a phenomenon known as heterosis. In this study, we investigated how the genetic divergence of the maize inbred lines B73 and Mo17 and their F1 hybrid progeny is reflected in differential, nonadditive, and allelic expression patterns in primary root tissues. In pairwise comparisons of the four genotypes, the number of differentially expressed genes between the two parental inbred lines significantly exceeded those of parent versus hybrid comparisons in all four tissues under analysis. No differentially expressed genes were detected between reciprocal hybrids, which share the same nuclear genome. Moreover, hundreds of nonadditive and allelic expression ratios that were different from the expression ratios of the parents were observed in the reciprocal hybrids. The overlap of both nonadditive and allelic expression patterns in the reciprocal hybrids significantly exceeded the expected values. For all studied types of expression - differential, nonadditive, and allelic - substantial tissue-specific plasticity was observed. Significantly, nonsyntenic genes that evolved after the last whole genome duplication of a maize progenitor from genes with synteny to sorghum (Sorghum bicolor) were highly overrepresented among differential, nonadditive, and allelic expression patterns compared with the fraction of these genes among all expressed genes. This observation underscores the role of nonsyntenic genes in shaping the transcriptomic landscape of maize hybrids during the early developmental manifestation of heterosis in root tissues of maize hybrids.
Maize (Zea mays) is one of the most prolific crops with a global production of 1.02 billion tons in 2014. In the past 50 years, global maize production has increased almost 5-fold (http://faostat3.fao.org/). Between 50% to 60% of this increase has been attributed to genetic improvements in hybrid breeding (Duvick, 2005).
Heterosis, or hybrid vigor, describes the superior performance of heterozygous F1 hybrid progeny relative to the average of their homozygous parental inbred lines (midparent heterosis) or the better performing parent (best parent heterosis; Falconer and Mackay, 1996). The phenomenon of heterosis was first scientifically reported by Charles Darwin after he compared the performance of self-pollinated maize progeny to cross-pollinated maize plants (Darwin, 1876). In the early twentieth century, heterosis was rediscovered by Edward M. East (East, 1908) and George H. Shull (Shull, 1908). The first commercial hybrid maize was introduced in the 1930s (Duvick, 2005). Since then, heterosis has been widely exploited in plant breeding and agriculture. Approximately 95% of U.S. maize and 55% of the rice (Oryza sativa) acreage in China are planted with hybrid seeds (Ding et al., 2014).
The phenotypic effect of heterosis is most evident in adult plants and can be monitored for instance as increased plant height, biomass, yield, fertility, and improved resistance to abiotic or biotic stress (Falconer and Mackay, 1996). Nevertheless, it has been demonstrated that heterosis can already be observed a few days after germination for seedling root traits such as lateral root density or primary root length (Hoecker et al., 2006). Several genetic hypotheses have been proposed that make the combination of a remarkable number of genes responsible for heterosis. These genetic hypotheses attribute heterosis to the complementation of superior dominant alleles (dominance hypothesis), allelic interactions at one or multiple loci (overdominance hypothesis), or epistatic interactions between nonallelic genes (for review, see Birchler et al., 2006; Hochholdinger and Hoecker, 2007).
On the level of gene expression, the prevalence of expression patterns in hybrids different from the average of their parental values, i.e. nonadditive expression, widely varied between different species, tissues, and genotypes (Guo et al., 2006; Stupar and Springer, 2006; Swanson-Wagner et al., 2006; Uzarowska et al., 2007; Stupar et al., 2008; Hoecker et al., 2008b; Paschold et al., 2012). Nonadditive gene expression is the result of allelic interactions that modify regulatory networks that lead to gene activity patterns different from average parental values (Stupar and Springer, 2006). Furthermore, unexpected allelic expression patterns were observed in hybrids that deviated from the allelic ratios of their parents (Guo et al., 2004; Stupar and Springer, 2006; Springer and Stupar, 2007; Paschold et al., 2012; Song et al., 2013). Although nonadditive expression patterns and unexpected allelic ratios have been observed in hybrids of different species, none of these expression types have yet been associated with the evolutionary origin of these genes.
The evolutionary history of flowering plants included several rounds of hybridization and polyploidization events followed by fractionation and rediploidization (for review, see Doyle et al., 2008). Within the lineage leading to maize, a last whole genome duplication occurred between ∼4.8 and ∼11.9 million years ago by the hybridization of two progenitors, which led to a tetraploid genome (Swigonová et al., 2004). Comparisons of the duplicated regions of the genome of modern maize with orthologous regions of the unduplicated genomes of rice (Oryza sativa) and sorghum (Sorghum bicolor) indicated that the maize genome had lost many of its duplicated genes (Haberer et al., 2005). Based on the synteny to sorghum, the genome of maize can be subdivided into two subgenomes, which contain pairs of duplicated genes shared by both subgenomes but also single-copy genes present in only one of the two subgenomes (Schnable et al., 2011). In total, 19,365 genes, representing 49% of the 39,656 maize genes of the filtered gene set (FGSv2), were assigned to these two subgenomes (Schnable et al., 2011). Beside these syntenic maize genes, the genome of modern maize is complemented by a set of nonsyntenic genes that lack syntenic orthologs in other grass species (Schnable et al., 2011). These 20,291 (51%) maize genes (FGSv2) likely evolved by single gene duplication after the last whole genome duplication event (Woodhouse et al., 2010).
Along the longitudinal axis, maize roots are structurally divided into a subterminal meristematic zone at the terminal end, followed by the elongation and differentiation zones (Hochholdinger, 2009). In the meristematic zone, new root cells are formed by cell division and are dislocated proximally into the elongation zone, where they elongate. Finally, cells are translocated into the differentiation zone. Cells in the differentiation zone display diverse functions. In transverse orientation, the stele with the pericycle as its outermost cell layer contains xylem and primary phloem elements functioning in the transport of water, nutrients, and photosynthates. The surrounding cortical parenchyma consists of the endodermis, multiple layers of cortex tissue, and the epidermis that connects the root to the rhizosphere. Hence, roots represent gradients of development, with very young undifferentiated cells at the distal end near the root cap and fully differentiated cells toward the proximal end of the root (Hochholdinger, 2009).
A previous study of our research group revealed dynamic root tissue-specific patterns of single-parent expression (SPE), an extreme instance of expression complementation in which genes are expressed in both reciprocal hybrids but only in one parental inbred line (Paschold et al., 2014). In this study, this dataset was surveyed to determine tissue-specific nonadditive, differential, and allelic gene expression patterns and the relation of these expression patterns to the phylogenetic origin of these genes. We demonstrated that nonsyntenic genes were highly overrepresented among nonadditive and allelic expression patterns in hybrids, suggesting a possible role of these genes in the early developmental manifestation of heterosis in maize roots.
RESULTS
Transcriptome Relationships and Tissue-Specific Expression of Four Primary Root Tissues of the Maize Inbred Lines B73 and Mo17 and Their Reciprocal Hybrids
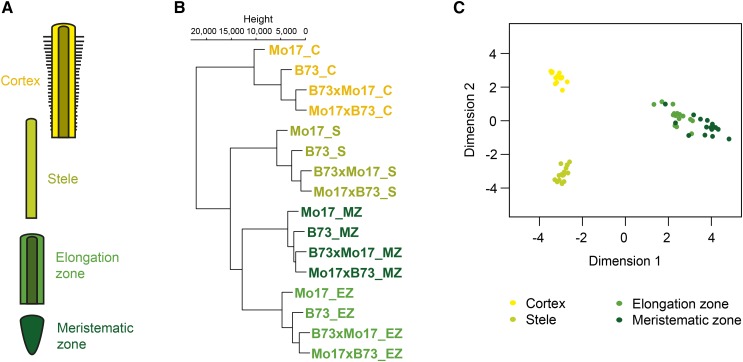
Relationships of RNA-sequencing (RNA-seq) samples of the meristematic zone, the elongation zone, and the cortex and stele of the differentiation zone of maize primary roots (Fig. 1A) of the inbred lines B73 and Mo17 and their reciprocal F1 hybrids B73xMo17 and Mo17xB73 were explored in a hierarchical cluster analysis (Fig. 1B) and in a multidimensional scaling plot (Fig. 1C). Each of the sixteen genotype/tissue combinations was represented by four biological replicates. The replicated samples of each root tissue marked with the same color code formed a cluster irrespective of their genotype. This indicates that the four distinct tissues of a single genotype display a higher degree of transcriptomic dissimilarity than the same tissues of different genotypes. Furthermore, among the four tissues, the meristematic and the elongation zone displayed the highest similarity (Fig. 1, B and C). On the genotype level, the transcriptomes of the two reciprocal hybrids that contain an identical nuclear genome were closely related in all four tissues, while the transcriptomes of the parental inbred lines were more distantly related (Fig. 1B).
Figure 1.
Overview of RNA-seq sample relationship. A, Color-coded schematic of the four mechanically separated maize primary root tissues: yellow, cortex (C); light green, stele (S); green, elongation zone (EZ); dark green, meristematic zone (MZ). B, Hierarchical cluster analysis of normalized read counts among all four replicates of each RNA-seq sample. C, Multidimensional scaling plot of all 64 RNA-seq samples.
Determination of Differentially Expressed Genes between Different Tissues
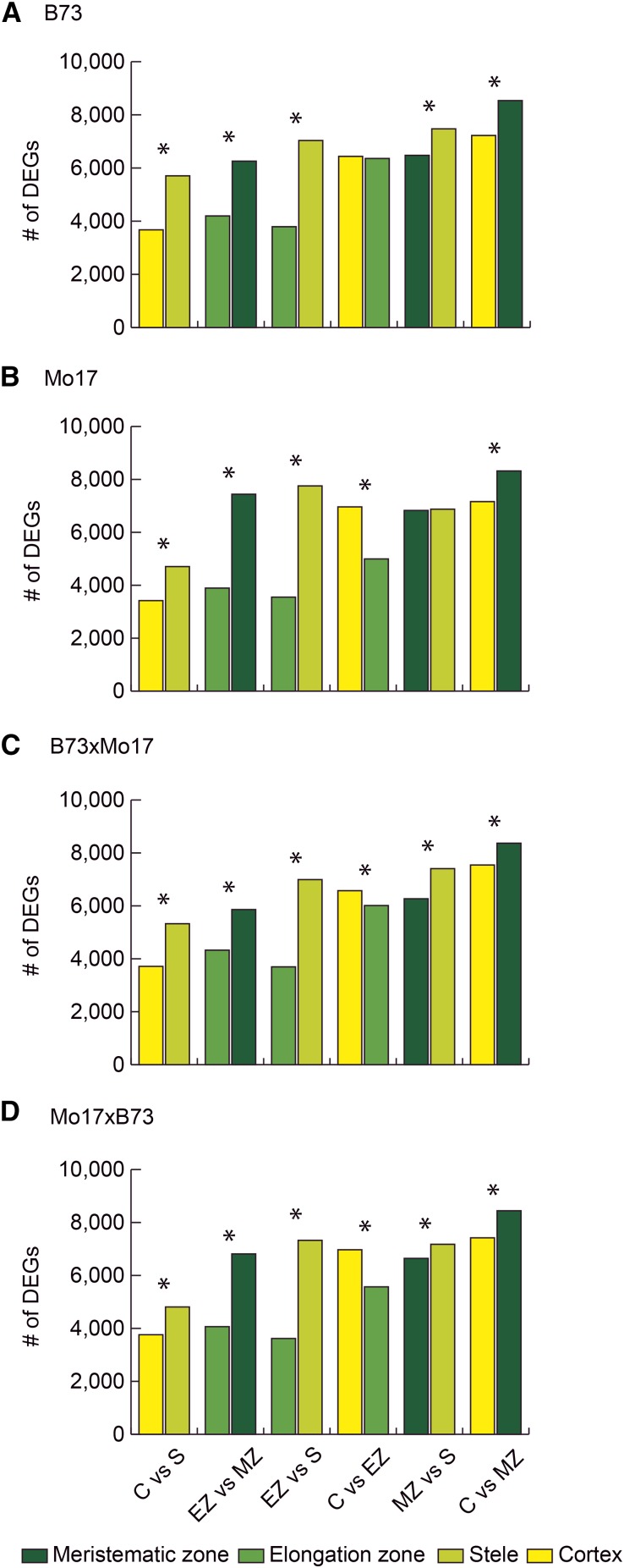
To determine the number of differentially expressed genes between the four different primary root tissues, six pairwise comparisons were performed per genotype (Fig. 2). For each comparison two bars are displayed in Figure 2 indicating the number of genes preferentially expressed in the color-coded tissue. In all four genotypes, the relative number of differentially expressed genes in the six comparisons was similar. The comparison between cortex and stele, which are both complementary tissues of the differentiation zone and between the elongation and the meristematic zone, revealed the smallest numbers of differentially expressed genes (Fig. 2; Supplemental Data File S1). In contrast, the highest numbers of differentially expressed genes were observed in the comparisons of the meristematic zone with cortex and with stele. These tissues are located at opposite poles of the root, and the high number of differentially expressed genes underscores the dissimilarity between these primary root tissues. In all three pairwise comparisons of the stele with other tissues, a higher number of genes was preferentially expressed in the stele compared with the other tissues (Fig. 2). In pairwise comparisons of the remaining three tissues, a higher number of genes was predominantly expressed in the meristematic zone compared with cortex or elongation zone, while in the comparison of cortex versus elongation zone a higher number of differentially expressed genes was preferentially expressed in the cortex. These tendencies were observed for all comparisons and were statistically significant for 22 of 24 comparisons (Fig. 2). Cross-comparison of the differentially expressed genes between the different root tissues among the four maize genotypes B73, Mo17, and their reciprocal hybrids showed that between 44% (cortex versus stele) and 68% (cortex versus meristematic zone) of differentially expressed genes were conserved among the four genotypes (Supplemental Fig. S1). Among the genes that were differentially expressed in only one genotype, the parental inbred lines displayed between 2 to 5 times more such genes than the reciprocal hybrids (Supplemental Fig. S1).
Figure 2.
Differential expression between four different maize primary root tissues. Differentially expressed genes (DEGs) were identified on the basis of six pairwise comparisons between the four analyzed root tissues: cortex (C, yellow), stele (S, light green), elongation zone (EZ, green), and meristematic zone (MZ, dark green) among the four maize genotypes B73 (A), Mo17 (B), and their reciprocal hybrids B73xMo17 (C) and Mo17xB73 (D). The number of differentially expressed genes while controlling the FDR at 5% and with a cutoff of |log2(FC)| > 1 are shown. For each pairwise comparison, two color-coded bars illustrate the tissue preference of the differentially expressed genes. Significant differences (α ≤ 5%) between the two bars per comparison are indicated by an asterisk.
Determination of Differentially Expressed Genes between Different Genotypes
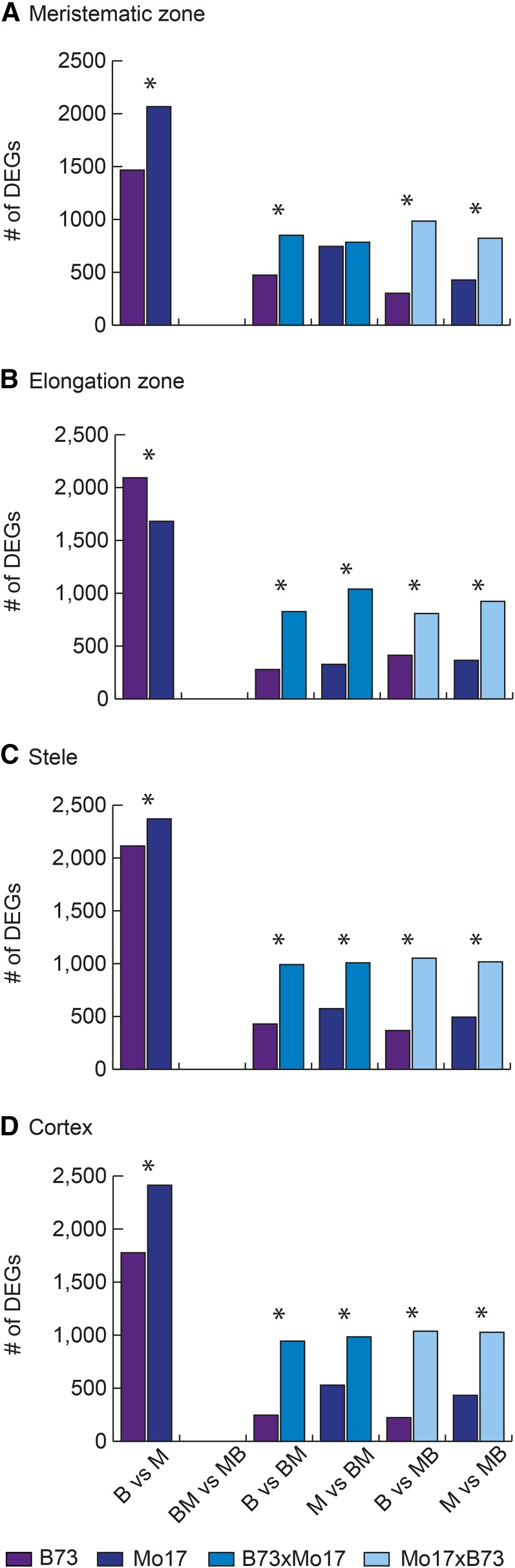
Differential gene expression between the four different genotypes was determined for each primary root tissue (Fig. 3). For all four tissues, the pairwise comparison between the two parental inbred lines B73 and Mo17 revealed the highest number of differentially expressed genes. In contrast, no differential gene expression was observed between the two reciprocal hybrids for any of the four tissues. For all four possible parent-hybrid comparisons, in each tissue between 1,104 (B73 versus B73xMo17 in elongation zone) and 1,582 (Mo17 versus B73xMo17 in stele) differentially expressed genes were estimated by controlling the false discovery rate (FDR) at 5% and an absolute log2 fold change (FC) > 1 (Fig. 3; Supplemental Data File S1). Hence, the number of differentially expressed genes in parent-hybrid comparisons was substantially below parent-parent comparisons but higher than in hybrid-hybrid comparisons. These numbers reflect the genetic relationship of the analyzed genotypes. While the reciprocal hybrids, which contain an identical nuclear genome, did not display any differentially expressed genes, the genetically distinct parents displayed a high degree of differential gene expression. Intermediate numbers of differentially expressed genes were observed in comparisons of hybrids with one of the parental inbred lines. Hybrids and parental inbred lines share 50% of their nuclear genomes. In all inbred versus hybrid comparisons, a significantly higher number of differentially expressed genes was preferentially expressed in the hybrids. This conserved trend was statistically significant for 15 of 16 comparisons (Fig. 3). Irrespective of the compared genotypes, the overlap of differentially expressed genes showed that fewer genes (between 7% and 9%) were differentially expressed in all four primary root tissues, and a substantially higher number of genes (between 28% and 47%) was exclusively expressed in one tissue (Supplemental Fig. S2).
Figure 3.
Differential expression between four different maize genotypes. Differentially expressed genes (DEGs) were identified on the basis of six pairwise comparisons between the four analyzed maize genotypes: B73 (B, purple), Mo17 (M, dark blue), B73xMo17 (BM, blue), and Mo17xB73 (MB, light blue) for each of the four primary root tissues: meristematic zone (A), elongation zone (B), stele (C), and cortex (D). The number of differentially expressed genes while controlling the FDR at 5% and with a cutoff of |log2(FC)| > 1 are displayed. For each pairwise comparison, two color-coded bars illustrate the genotype preference of the differentially expressed genes. Significant differences (α ≤ 5%) between the two bars per comparison are indicated by an asterisk.
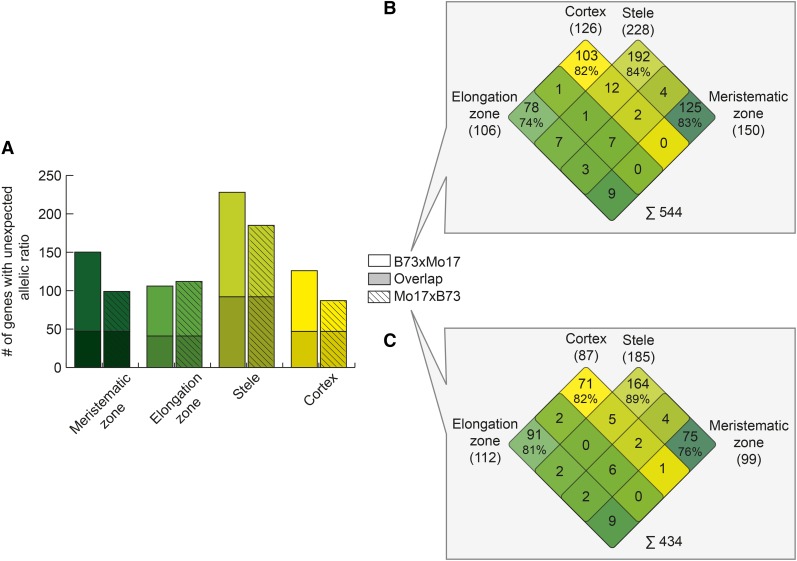
Genes Exhibiting Unexpected Allelic Expression Ratios in Hybrids Are Tissue-Specific
For each tissue, the allelic ratio was determined in the hybrids and compared with the allelic expression ratio of the two parental inbred lines. The number of genes for which these ratios were significantly different (unexpected allelic ratios) is summarized in Figure 4A. In total, between 87 (Mo17xB73 in cortex) and 228 (B73xMo17 in stele) genes displayed unexpected allelic ratios (Supplemental Table S1, column Observed). Overall, between 39% (41/106 in elongation zone) and 54% (47/87 in cortex) of unexpected allelic ratios were conserved between the reciprocal hybrids (Fig. 3A, dark shaded bars), which was significantly higher than expected (Supplemental Table S1, column Expected) as calculated by a χ2 test. For each hybrid, the genes displaying unexpected allelic expression ratios were compared between the four primary root tissues. For B73xMo17, this comparison indicated that 74% to 84% of the genes with unexpected allelic ratios were exclusively expressed in one primary root tissue and only 1.3% of these genes were common in all four root tissues (Fig. 4B). Similarly, for Mo17xB73, between 76% and 89% of the genes with differential allelic expression were tissue-specific and only 1.4% of these genes were expressed in all four tissues (Fig. 4C). In total, 739 unique genes displaying unexpected allelic expression ratios were identified in this study (Supplemental Data File S1).
Figure 4.
Genes with unexpected allelic expression in the hybrids. A, Number of genes showing unexpected allelic ratios in both hybrids (plain: B73xMo17; shaded: Mo17xB73) for each primary root tissue. The overlapping genes between the two hybrids within each tissue are highlighted by the darker color. B and C, Overlap of genes with unexpected allelic ratio between the four different primary root tissues in B73xMo17 (B) and in Mo17xB73 (C). The numbers in parentheses indicate the total number of genes with unexpected allelic ratio in the corresponding root tissue.
Nonadditive Gene Expression Patterns Are Conserved between Reciprocal Hybrids in All Four Primary Root Tissues
For each tissue, gene expression levels in the reciprocal hybrids relative to their parental inbred lines were characterized based on nine gene expression classes, which were derived from the classes previously defined by Hoecker et al. (2008a). Subsequently, the subset of genes showing expression levels that deviate significantly from the midparent value was determined in these nine classes. The number of genes in each expression class was estimated by combining three pairwise t tests that compared the expression level of the hybrid with each parent and between both parents (Supplemental Table S2). According to this classification scheme most of the expressed genes in the hybrids revealed no significant difference in their expression relative to their parents (class 4). Genes that show expression levels between the parental values or exhibited low- or high-parent expression were abundant and were assigned to expression classes 1, 2, and 3, respectively. Genes that exhibited either above high-parent (36 genes in B73xMo17; 17 genes in Mo17xB73) or below low-parent (147 genes in B73xMo17; 36 genes in Mo17xB73) expression in the hybrids were assigned to the expression classes 5 to 8. Genes with ambiguous gene expression levels in the hybrids were assigned to class 9. In total, 1,056 and 824 nonadditively expressed genes were exclusively identified in the hybrids B73xMo17 and Mo17xB73, respectively. Among those, 439 nonadditive genes overlapped between the reciprocal hybrids, which substantially exceeded the expected number of 35 overlapping genes as demonstrated by a χ2 test (Supplemental Table S2).
Each expression class contained a subset of genes that exhibited nonadditive gene expression, i.e. gene expression in these hybrids was significantly different from the mean of expression of the two parental inbred lines. The highest number of nonadditively expressed genes was observed in gene expression class 1 (between 205 in cortex and 432 in meristematic zone) and class 3 (between 201 in meristematic zone and 339 in elongation zone; Supplemental Table S2). Within each class, the overlap of the nonadditively expressed genes between the two hybrids was significantly higher than expected. Lower numbers of nonadditively expressed genes were observed in gene expression class 2 and the classes to which genes with extreme expression patterns were assigned to (classes 5–8). For each tissue, the observed number of common genes in both hybrids significantly exceeded the number of genes expected purely by chance. Consistent with the definition of the gene expression pattern of class 4, no nonadditively expressed genes were observed in this class.
Among the genes with unexpected allelic ratios, between 26 (Mo17xB73, elongation zone) and 58 (B73xMo17, meristematic zone) genes showed nonadditive gene expression patterns (Supplemental Table S1). The comparison of the nonadditively expressed genes with unexpected allelic ratios between the two reciprocal hybrids B73xMo17 and Mo17xB73 within each primary root tissue revealed that a significantly higher number of these genes is common in both hybrids than expected.
Nonadditively, Allelic, and Differentially Expressed Genes Are Overrepresented among Nonsyntenic Genes
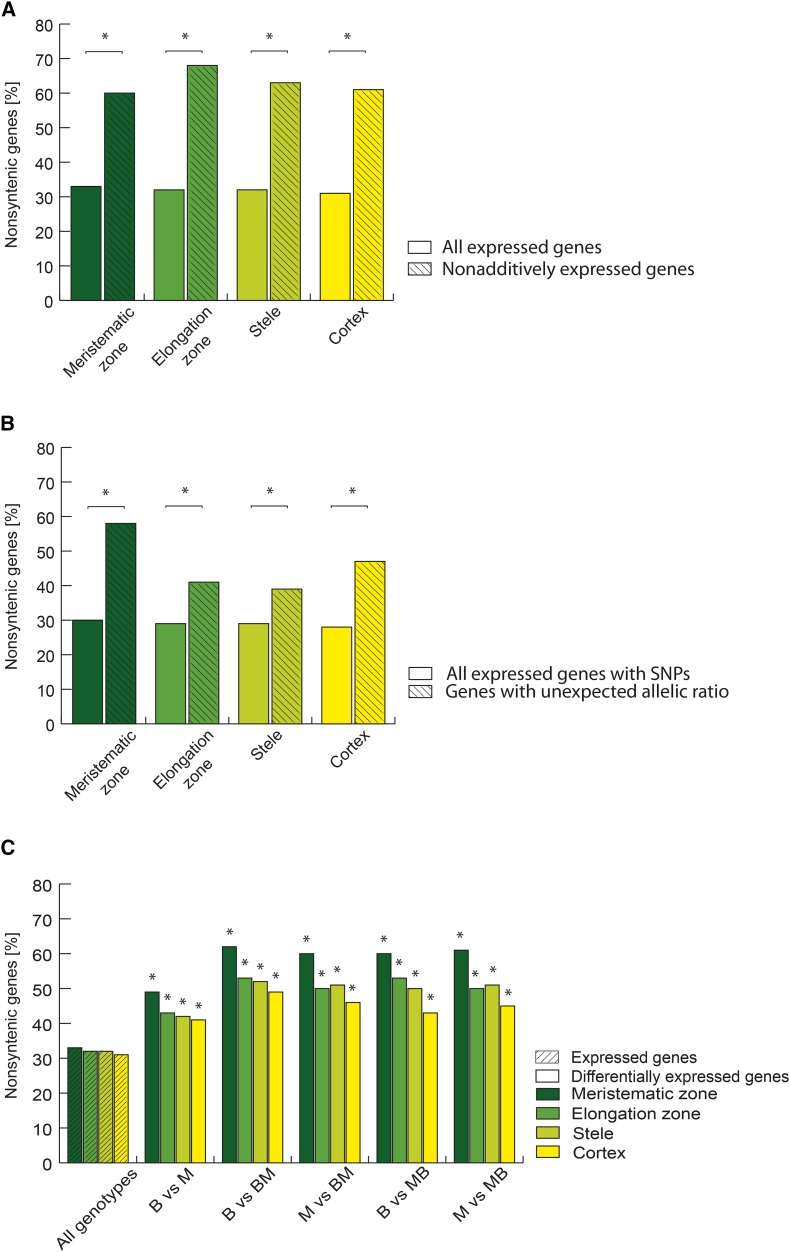
To study their evolutionary origin, differentially and nonadditively expressed genes, and genes exhibiting unexpected allelic ratios, were compared with the set of nonsyntenic genes that constitute 51% of all genes in the filtered gene set (FGSv2) of the maize genome (Schnable et al., 2011). Among the genes expressed in each of the four tissues, only between 31% (6,653/21,276 in cortex) and 33% (6,860/20,899 in meristematic zone) were nonsyntenic (Fig. 5A; Supplemental Data File S1). Hence, nonsyntenic genes were substantially underrepresented among all expressed genes relative to their fraction of all genes present in FGSv2. Remarkably, in the elongation zone, 68% (501/740) of the nonadditively expressed genes were nonsyntenic and thus significantly overrepresented compared with their prevalence among all expressed genes (Fig. 5A). Similar values were observed for nonadditively expressed genes in the remaining three primary root tissues investigated: cortex 61% (407/666), meristematic zone 60% (667/1,115), and stele 63% (435/686). Similarly, nonsyntenic genes were significantly overrepresented among the genes with unexpected allelic expression ratios relative to all expressed genes with single nucleotide polymorphisms (SNPs) in each of the four root tissues (Fig. 5B). Among the expressed genes with SNPs, only between 28% (3,596/12,963 in cortex) and 30% (3,809/12,903 in meristematic zone) were nonsyntenic. In comparison, 47% (78/166) of genes with unexpected allelic ratios were nonsyntenic in cortex, 41% (73/177) in the elongation zone, 58% (118/202) in the meristematic zone, and 39% (124/321) in the stele (Fig. 5B).
Figure 5.
Distribution of nonsyntenic genes among different classes of genes. A, Proportion of nonsyntenic genes among nonadditively expressed genes (shaded) in the hybrids relative to all expressed genes (not shaded) in each of the four primary root tissues. B, Proportion of nonsyntenic genes among genes showing unexpected allelic ratios (shaded) in the hybrids relative to all expressed genes containing an SNP (not shaded) in each of the four primary root tissues. C, Proportion of nonsyntenic genes among differentially expressed genes [FDR ≤ 5%; |log2(FC)| > 1] (not shaded) between the four different genotypes B73 (B), Mo17 (M), B73xMo17 (BM), and Mo17xB73 (MB) in cortex, stele, elongation, and meristematic zone relative to all expressed genes in all genotypes (shaded). Significant differences (α ≤ 5%) compared with all expressed genes are indicated with an asterisk.
Furthermore, nonsyntenic genes were significantly overrepresented among genes differentially expressed between the four genotypes compared with all expressed genes (Fig. 5C; Supplemental Data File S1). Their fraction ranged from 41% (1,704/4,190 of B73 versus Mo17 in cortex) to 62% (818/1,323 of B73 versus B73xMo17 in meristematic zone; Fig. 5C). However, among all expressed genes in the different tissues, only between 30% (cortex) and 33% (meristematic zone) were nonsyntenic.
Functional Classification of Nonsyntenic Nonadditively Expressed Genes
To identify overrepresented biological and molecular functions among the nonsyntenic genes with nonadditive and allelic expression patterns, a Gene Ontology (GO) analysis was performed. Only for the nonsyntenic nonadditively expressed genes of the cortex and the elongation zone, significantly enriched GO terms were identified (Supplemental Table S3). Most prominently, the biological processes “death” and “cell death” with the subcategories “programmed cell death” and “apoptosis” were overrepresented in the cortex samples of the reciprocal hybrids. Moreover, in B73xMo17, the molecular processes “iron ion binding” and “tetrapyrrole binding” with its subcategory “heme binding” were significantly enriched. Furthermore, in the reciprocal hybrid Mo17xB73, the molecular process “monooxygenase activity” was overrepresented. Finally, in the elongation zone, the molecular process “aspartic-type peptidase activity” and its subcategory “aspartic-type endopeptidase activity” were overrepresented among the nonsyntenic nonadditively expressed genes in Mo17xB73.
DISCUSSION
In this study, differential, nonadditive, and allelic gene expression patterns of the maize inbred lines B73 and Mo17 and their reciprocal hybrid progeny were analyzed in four maize primary root tissues (Fig. 1A), based on a RNA-seq dataset from our laboratory (Paschold et al., 2014).
Hierarchical clustering and multidimensional scaling plots of all samples revealed high correlation of gene expression in the adjacent meristematic and elongation zones and only low correlation of these zones with cortex and stele tissue of the differentiation zone (Fig. 1, B and C). This correlation reflects the developmental gradient of roots along the longitudinal axis with the youngest undifferentiated cells in the meristematic zone of the root tip (Ishikawa and Evans, 1995). Moreover, the low correlation of the cortex and stele can be explained by the functional differences of the disparate cell types present in these tissues (Saleem et al., 2010). As illustrated by these correlation analyses, overall gene expression is more divergent between different tissues of a genotype than between different genotypes within a tissue. This observation is supported by similar distributions of differentially expressed genes identified between the four root tissues in the four genotypes (Fig. 2). Less differentially expressed genes were estimated between adjacent tissues, whereas high numbers of differentially expressed genes were observed in comparisons of the differentiation zone with the meristematic or elongation zone. On the proteome level, a similar distribution of differentially accumulated proteins between the same four primary root tissues of B73 seedlings at the same developmental stage was observed (Marcon et al., 2015).
Pairwise comparison of gene expression levels revealed a high number of differentially expressed genes between the two parental inbred lines B73 and Mo17 but no differentially expressed genes between the genetically identical reciprocal hybrids of a specific tissue (Fig. 3). Phylogenetic analyses demonstrated that the inbred lines B73 and Mo17 are distantly related (Jiao et al., 2012; Lorenz and Hoegemeyer, 2013). Similarly, mapping of Mo17 genomic sequences to the B73 reference genome revealed that only 63% of the sequences were identical in both genotypes (Wei and Wang, 2013). This finding supports the notion that transcriptome diversity in the roots of young maize hybrids is conditioned by genomic differences of the parental inbred lines. A similar distribution of the differentially expressed genes between the two maize inbred lines and their reciprocal hybrids was previously reported for aboveground tissues of 11-d-old maize seedlings (Stupar et al., 2008) and for whole primary roots of the same developmental stage as described here (Paschold et al., 2012). As suggested by the dominance hypothesis, the combination of two diverse genomes in a hybrid might lead to their integration or the complementation of deleterious alleles and form a superior plant (Jones, 1917).
Remarkably, when comparing gene expression levels between hybrids and one parent in the four primary root tissues, in all instances more differentially expressed genes were preferentially expressed in hybrids than in the parental inbred line (Fig. 3). This result is consistent with the previous findings obtained in maize top ear shoots (Qin et al., 2013), in which approximately two-thirds of the differentially expressed genes were up-regulated in the F1 hybrid relative to the parental inbred lines. Similar results were observed in early maize ear inflorescences (Ding et al., 2014).
Unexpected allelic gene expression describes the deviation of the allelic expression ratios in the hybrids from the expression ratios of their parental inbred lines (Hochholdinger and Hoecker, 2007). In whole maize primary roots, 840 genes were reported for which the expression ratio of the two parental alleles in the reciprocal hybrids significantly deviated from the expected allelic ratio of the inbred lines (Paschold et al., 2012). In this study, a total of 739 genes were identified that show unexpected allelic expression ratios (Fig. 4). Unexpected allelic expression patterns might be predetermined by the epigenetic status of the parental genomes and the activation of transposable elements or uniparental expression of noncoding RNAs (for review, see Groszmann et al., 2013). In this study, between 15% (stele) and 35% (meristematic zone) of genes with unexpected allelic expression ratios in hybrids revealed also nonadditive expression patterns, i.e. expression levels in hybrids that deviated significantly from the midparent value. This observation supports the notion that altered allelic activity in hybrids could contribute to nonadditive gene expression (Paschold et al., 2012). In this study, approximately 8% of all expressed genes were nonadditively expressed, which is similar to results previously observed in whole primary maize roots of the same developmental stage (Paschold et al., 2012). Another remarkable observation was the high degree of conservation of both the nonadditively expressed genes and those showing unexpected allelic expression ratios between the two reciprocal hybrids (Fig. 4; Supplemental Tables S1 and S2). This underscores the importance of nuclear genome composition in reciprocal hybrids irrespective of the origin of the two parental alleles. Such a high degree of expression conservation between reciprocal hybrids was already observed in whole primary maize roots (Paschold et al., 2012).
The genome of an ancient maize progenitor underwent several duplications, including that of a paleopolyploid ancestor (Paterson et al., 2004) and an additional whole genome duplication ∼5 to 12 million years ago (Blanc and Wolfe, 2004a; Swigonová et al., 2004). This resulted in the divergence of modern maize from its close and unduplicated relative sorghum (Paterson et al., 2009). Comparative analyses of the maize and sorghum genomes identified syntenic paralogs between these species and nonsyntenic maize genes that developed after this genome duplication event (Woodhouse et al., 2010; Schnable et al., 2011). In a comparative genomic study in Arabidopsis (Arabidopsis thaliana), it was shown that disease resistance genes were significantly overrepresented among genes found in nonsyntenic regions (Freeling et al., 2008). This indicates that nonsyntenic genes are instrumental to cope with the selective fluctuating environment, which requires constant adaptations to new conditions. In contrast, syntenic genes may encode functions that are under continuous selection. We determined if the evolutionary origin of genes correlates with the intraspecific transcriptome diversity between parental inbred lines B73 and Mo17 and their reciprocal hybrids. To this end, all genes displaying differential, nonadditive, or allelic expression patterns were surveyed concerning their synteny with sorghum, hence their evolutionary origin. Syntenic genes were shaped by more than 5 million years of natural selection (Blanc and Wolfe, 2004b). Therefore, they are likely instrumental for maize development and thus highly conserved between different maize inbred lines. In general, nonsyntenic genes were expressed significantly less frequently in this study compared with their relative genomic proportion in the maize FGSv2. Nevertheless, nonsyntenic genes were significantly overrepresented among the nonadditively expressed genes and genes with unexpected allelic expression ratios in the hybrids (Fig. 5, A and B). In addition, nonsyntenic genes were significantly overrepresented among differentially expressed genes (Fig. 5C). This finding suggests that genotype-specific gene expression patterns in the inbred lines B73 and Mo17 and their hybrid progeny are determined to a disproportionally high level by evolutionarily younger genes. Although the maize tetraploidy with subsequent genome fractionation occurred already between 5 and 12 million years ago, there is evidence that biased gene loss and expression continues today (Woodhouse et al., 2010; Schnable et al., 2011). This ongoing change in genome structure might at least in part explain the remarkable genetic diversity found among different maize lines and the overrepresentation of nonsyntenic differentially expressed genes in this study. A similar observation was made in the four maize primary root tissues, where nonsyntenic genes were significantly overrepresented among genes displaying SPE patterns (Paschold et al., 2014). SPE is an extreme instance of expression complementation in which genes are expressed in both reciprocal hybrids but only in one of the two parental inbred lines (Paschold et al., 2012). Hence, this pattern represents a special instance of the dominance model of complementation on the gene expression level.
Among the nonsyntenic nonadditively expressed genes, the GO terms death and cell death were significantly enriched in the cortex in the reciprocal hybrids. Those genes are involved in several developmental processes in roots, e.g. the development of root cortical aerenchyma. Root cortical aerenchyma is formed by enlarged gas spaces in the root cortex that are the result of cell death or the separation of cells (Evans, 2003). It has been demonstrated that maize genotypes with higher root cortical aerenchyma formation have growth advantages over genotypes with lower root cortical aerenchyma formation under certain conditions (Postma and Lynch, 2011). To date no systematic comparisons of related inbred hybrid combinations are available with respect to root cortical aerenchyma formation. It will therefore be interesting to see in future experiments if the overrepresentation of cell death and apoptosis genes in the cortex is a general feature of maize hybrids and if this observation is related to an increase in root cortical aerenchyma in hybrids.
CONCLUSION
In summary, nonsyntenic genes were significantly overrepresented among nonadditive, differential, and unexpected allelic expression patterns relative to their prevalence among all expressed genes. These observations underscore the role of these genes in shaping the transcriptomic landscape of the reciprocal maize hybrids B73xMo17 and Mo17xB73, which might be associated with the developmental manifestation of heterosis.
MATERIALS AND METHODS
RNA-Seq and Allelic Read Calling
The RNA-seq dataset analyzed in this study was generated in our research group and described in detail by Paschold et al. (2014). In brief, primary roots of a length of 2 to 4 cm of the inbred lines B73, Mo17, and their reciprocal hybrids B73xMo17 and Mo17xB73 were separated into meristematic zone, elongation zone, cortex, and stele. The 64 cDNA libraries were loaded on two flow cells according to a split plot design (supplemental table S1 in Paschold et al., 2014). Subsequently, RNA-seq (100 bp, single read) of high-quality RNA (RNA integrity number ≥ 7.2; Schroeder et al., 2006) was performed with a GAIIx genome analyzer equipped with on-instrument sequencing control software SCS Version 2.8 and real-time analysis RTA1.8.7 in four biological replicates per tissue and genotype combination. Base calling and run statistics were performed with the genome analyzer data analysis pipeline OLB Version 1.8.0.
High-quality reads of all genotypes were aligned to the B73 reference genome using the short-read nucleotide alignment program GSNAP (http://research-pub.gene.com/gmap/ version 2012-01-11; Schnable et al., 2009), which was indexed by applying GMAP_BUILD (k-mer size of 15 and step size 3). For subsequent analyses only uniquely mapping reads with a maximum of two mismatches out of 36 bp and with ideally 5 bp tails for every 76 bp were used. Furthermore, stacked reads that share the same start and end coordinate, sequencing direction, and sequence were discarded from the set of uniquely mapping reads. The remaining set of reads was aligned to the filtered gene set (http://ftp.maizesequence.org/release-5b/filtered-set/ release 5b.60) of the B73 reference genome derived from the Maize Genome Sequencing Project using a Perl script. Mapping Mo17 reads to the B73 genome introduces a slight bias because Mo17 reads that contain polymorphisms beyond the parameters defined above are at disadvantage of being aligned to the B73 genome sequence. However, in this dataset this bias is very small. A total of 88.1% of B73 reads mapped to the B73 genomic sequence, while only a slightly smaller fraction of Mo17 reads (87.4% of reads) mapped to the B73 genome sequence (see supplemental table S2 in Paschold et al., 2014).The set of allelic reads was determined with the help of a Perl script by extracting the uniquely mapping reads to any of the 4,034,683 B73-Mo17 SNP positions, which were previously identified between B73 and Mo17 by the 123SNP software (http://schnablelab.plantgenomics.iastate.edu/software). Only uniquely mapping reads were considered, as this mapping procedure did not allow for quantifying sequence reads that map to repeats throughout the genome. Therefore, transcript isoforms of a given gene could not be distinguished.
Analysis of Differential Gene Expression and Allelic Expression
The following analyses were applied to genes that were declared expressed if represented by a minimum of five mapped reads in all four replicates of at least one sample. The raw sequencing reads were normalized by sequencing depth and were log2-transformed to meet the assumptions of linear models. Furthermore, the mean-variance relationship within the count data were estimated and precision weights for each observation were computed (Law et al., 2014). Prior to further data analyses, sample relations were analyzed based on multidimensional scaling using the plotMDS function of the Bioconductor package limma (Smyth, 2005) in R (R version 3.1.1 2014-07-10, limma_3.20.9). The distance between each pair of samples was estimated as the root-mean-square deviation for the top 500 genes with the largest standard deviations between samples. Additionally, a hierarchical cluster analysis was performed using the Euclidean distance of the average values of the four tissues and genotypes. To analyze the differences in expression between the primary root tissues and between the four genotypes, two different linear mixed models were fitted based on the complete dataset within the Bioconductor package limma (Smyth, 2005) in R. According to the experimental design used for sequencing, the model for the analysis of the differentially expressed genes between the different tissues included fixed effects for tissue, genotype, the interaction of both treatment factors, and for block. In addition, normally distributed random effects for main- and subplot were included. Both the main- and subplot represent in a split plot design a randomization unit and need therefore be represented by a random error term each. For the estimation of the differences in expression between the four genotypes within a specific primary root tissue, the statistical model was in such a way modified that it corresponds to a complete block design. Therefore, the model included a fixed effect for genotype and block and a normally distributed random error term. After model fit, an empirical Bayes approach was applied to shrink the sample variances toward a common value (Smyth, 2004). Hypothesis tests were performed using the contrasts.fit function of the Bioconductor package limma (Smyth, 2005). The resulting p values of the performed pairwise t tests were used to determine the total number of differentially expressed genes for each comparison by controlling the FDR ≤ 5% to adjust for multiple testing (Benjamini and Hochberg, 1995). Significant differences in the number of up- and down-regulated genes for each of the tissue and genotype pairwise comparisons were determined based on a McNemar test (McNemar, 1947) at a significance level of alpha ≤ 5%.
To estimate the differences in allelic gene expression between the parental inbred lines and the two reciprocal hybrids within a specific primary root tissue, a linear mixed model was fitted in R. The model is composed of a treatment model explaining the genotypic and allelic effects and of an interaction term of both factors. Additionally, a fixed effect for block and a normally distributed random error term were included in the analysis. Using the allelic read counts, the hypothesis test that the ratio of the expression of the parental alleles in both inbred lines equals to the ratio of the expression of the parental alleles in a specific hybrid was performed by applying the contrasts.fit function in limma (Smyth, 2005). The hypothesis test was performed for both reciprocal hybrids separately. The resulting p values were FDR-adjusted (Benjamini and Hochberg, 1995) and used to determine the number of genes showing unexpected allelic ratios.
Analysis and Classification of Gene Expression Patterns in the Hybrids
Gene expression levels in the hybrids were classified according to nine gene expression classes defined previously (Paschold et al., 2012). The expression classes describe the level of gene expression in a hybrid relative to the gene expression level of both parents. The contrasts.fit function of the limma package estimates coefficients and standard errors for a given set of contrasts (Smyth, 2005). The estimated coefficients that resulted from each hybrid-parent comparison within a specific root tissue directly supply the estimated expression level for each gene separately. The pairwise comparisons between two genotypes resulting in an adjusted p ≤ 0.05 were used for the classification of the gene expression in the hybrid. The parental inbred line with the larger estimated expression level within a given gene was termed high parent (HP), whereas the other parent was labeled as low parent (LP).
In a second step, gene expression levels of the hybrid were compared with midparent values (MPV) to determine the number of nonadditively expressed genes. For each hybrid, a contrast was fitted within the linear mixed model framework of limma to compare the estimated log-expression value of the hybrid to the mean of the log-expression values of both parents. The determination of nonadditively expressed genes was performed for each of the nine gene expression classes separately.
Determination of Syntenic and Nonsyntenic Maize Genes and Singular Enrichment Analysis
The lists comprising genes with differential, nonadditive, and allelic expression patterns were compared with the genes assigned to the maize 1 and maize 2 subgenomes (Schnable et al., 2011; http://www.skraelingmountain.com/datasets.php).
The Web-based agriGO platform (http://bioinfo.cau.edu.cn/agriGO/analysis.php) was used to assign GO functional categories to nonsyntenic genes with nonadditive and allelic expression patterns. Singular enrichment analysis computed overrepresented categories in these two gene sets by comparing them with GO terms in the set of all expressed genes using Fisher’s exact test (Du et al., 2010).
Accession Numbers
Raw sequencing data are stored at the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP029742.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overlap of differentially expressed genes of all tissue comparisons.
Supplemental Figure S2. Overlap of differentially expressed genes of all genotype comparisons.
Supplemental Table S1. Genes showing unexpected allelic expression patterns in each tissue.
Supplemental Table S2. Gene expression classification in the reciprocal hybrids within each tissue.
Supplemental Table S3. Functional categorization of nonsyntenic nonadditively expressed genes in the reciprocal hybrids.
Supplemental Data File S1. Gene lists for all analyzed expression patterns in the four tissues of the primary maize roots.
Supplementary Material
Acknowledgments
We thank Hans-Peter Piepho (Institute for Crop Sciences, Biostatistics Unit, University of Hohenheim, Stuttgart, Germany) for advice in statistical questions.
Glossary
- SPE
single-parent expression
- FDR
false discovery rate
- FC
fold change
- SNP
single nucleotide polymorphism
- GO
Gene Ontology
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (Grant HO2249-9/3 to F.H.).
Articles can be viewed without a subscription.
References
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57: 289–300 [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S (2006) Unraveling the genetic basis of hybrid vigor. Proc Natl Acad Sci USA 103: 12957–12958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004a) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004b) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. (1876) The effects of cross- and self-fertilization in the vegetable kingdom, Ed 1. John Murray [Google Scholar]
- Ding H, Qin C, Luo X, Li L, Chen Z, Liu H, Gao J, Lin H, Shen Y, Zhao M, et al. (2014) Heterosis in early maize ear inflorescence development: a genome-wide transcription analysis for two maize inbred lines and their hybrid. Int J Mol Sci 15: 13892–13915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN. (2005) Genetic progress in yield of United States maize (Zea mays L.). Maydica 50: 193–202 [Google Scholar]
- East EM. (1908) Inbreeding in corn. Conn Agric Exp Sta Rpt 1907: 419–428 [Google Scholar]
- Evans DE. (2003) Aerenchyma formation. New Phytol 161: 35–49 [Google Scholar]
- Falconer DS, Mackay TFC (1996) Introduction to Quantitative Genetics, Ed 4. Longman, Harlow, UK [Google Scholar]
- Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D (2008) Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res 18: 1924–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES (2013) The role of epigenetics in hybrid vigour. Trends Genet 29: 684–690 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Yang X, Crasta O, Zinselmeier C, Smith OS, Bowen B (2006) Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet 113: 831–845 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS (2004) Allelic variation of gene expression in maize hybrids. Plant Cell 16: 1707–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, Fuks G, Butler E, Wing RA, Rounsley S, Birren B, et al. (2005) Structure and architecture of the maize genome. Plant Physiol 139: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F. (2009) The maize root system: morphology, anatomy and genetics. In Bennetzen J, Hake S, eds, Handbook of Maize. Springer, New York, pp 145–160 [Google Scholar]
- Hochholdinger F, Hoecker N (2007) Towards the molecular basis of heterosis. Trends Plant Sci 12: 427–432 [DOI] [PubMed] [Google Scholar]
- Hoecker N, Keller B, Muthreich N, Chollet D, Descombes P, Piepho HP, Hochholdinger F (2008a) Comparison of maize (Zea mays L.) F1-hybrid and parental inbred line primary root transcriptomes suggests organ-specific patterns of nonadditive gene expression and conserved expression trends. Genetics 179: 1275–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker N, Keller B, Piepho HP, Hochholdinger F (2006) Manifestation of heterosis during early maize (Zea mays L.) root development. Theor Appl Genet 112: 421–429 [DOI] [PubMed] [Google Scholar]
- Hoecker N, Lamkemeyer T, Sarholz B, Paschold A, Fladerer C, Madlung J, Wurster K, Stahl M, Piepho HP, Nordheim A, et al. (2008b) Analysis of nonadditive protein accumulation in young primary roots of a maize (Zea mays L.) F1-hybrid compared to its parental inbred lines. Proteomics 8: 3882–3894 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML (1995) Specialized zones of development in roots. Plant Physiol 109: 725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, et al. (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44: 812–815 [DOI] [PubMed] [Google Scholar]
- Jones DF. (1917) Dominance of linked factors as a means of accounting for heterosis. Genetics 2: 466–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK (2014) voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Hoegemeyer T (2013) The phylogenetic relationships of US maize germplasm. Nat Genet 45: 844–845 [DOI] [PubMed] [Google Scholar]
- Marcon C, Malik WA, Walley JW, Shen Z, Paschold A, Smith LG, Piepho HP, Briggs SP, Hochholdinger F (2015) A high-resolution tissue-specific proteome and phosphoproteome atlas of maize primary roots reveals functional gradients along the root axes. Plant Physiol 168: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNemar Q. (1947) Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 12: 153–157 [DOI] [PubMed] [Google Scholar]
- Paschold A, Jia Y, Marcon C, Lund S, Larson NB, Yeh CT, Ossowski S, Lanz C, Nettleton D, Schnable PS, et al. (2012) Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Res 22: 2445–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Larson NB, Marcon C, Schnable JC, Yeh CT, Lanz C, Nettleton D, Piepho HP, Schnable PS, Hochholdinger F (2014) Nonsyntenic genes drive highly dynamic complementation of gene expression in maize hybrids. Plant Cell 26: 3939–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma JA, Lynch JP (2011) Root cortical aerenchyma enhances the growth of maize on soils with suboptimal availability of nitrogen, phosphorus, and potassium. Plant Physiol 156: 1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Scheuring CF, Wei G, Zhi H, Zhang M, Huang JJ, Zhou X, Galbraith DW, Zhang HB (2013) Identification and characterization of a repertoire of genes differentially expressed in developing top ear shoots between a superior hybrid and its parental inbreds in Zea mays L. Mol Genet Genomics 288: 691–705 [DOI] [PubMed] [Google Scholar]
- Saleem M, Lamkemeyer T, Schützenmeister A, Madlung J, Sakai H, Piepho HP, Nordheim A, Hochholdinger F (2010) Specification of cortical parenchyma and stele of maize primary roots by asymmetric levels of auxin, cytokinin, and cytokinin-regulated proteins. Plant Physiol 152: 4–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull G. (1908) The composition of a field of maize. J Hered 4: 296–301 [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: e3. [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2005) Limma: linear models for microarray data. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds, Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Song G, Guo Z, Liu Z, Cheng Q, Qu X, Chen R, Jiang D, Liu C, Wang W, Sun Y, et al. (2013) Global RNA sequencing reveals that genotype-dependent allele-specific expression contributes to differential expression in rice F1 hybrids. BMC Plant Biol 13: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM (2007) Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Gardiner JM, Oldre AG, Haun WJ, Chandler VL, Springer NM (2008) Gene expression analyses in maize inbreds and hybrids with varying levels of heterosis. BMC Plant Biol 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Springer NM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173: 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103: 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14: 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzarowska A, Keller B, Piepho HP, Schwarz G, Ingvardsen C, Wenzel G, Lübberstedt T (2007) Comparative expression profiling in meristems of inbred-hybrid triplets of maize based on morphological investigations of heterosis for plant height. Plant Mol Biol 63: 21–34 [DOI] [PubMed] [Google Scholar]
- Wei X, Wang X (2013) A computational workflow to identify allele-specific expression and epigenetic modification in maize. Genomics Proteomics Bioinformatics 11: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, Freeling M (2010) Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.