A weak mutation in an enzyme that attaches a GPI anchor to proteins permits analysis of post-embryonic functions of GPI-anchored proteins.
Abstract
GPI-anchored proteins (GPI-APs) are essential for plant growth and development; knockout mutations in enzymes responsible for anchor biosynthesis or attachment are gametophyte or embryo lethal. In a genetic screen targeted to identify genes regulating stomata formation, we discovered a missense mutation in the Arabidopsis (Arabidopsis thaliana) homolog of GPI8/PIG-K, a Cys protease that transfers an assembled GPI anchor to proteins. The Arabidopsis genome has a single copy of AtGPI8, and the atgpi8-1 mutation reduces the efficiency of this enzyme, leading to reduced accumulation of GPI-anchored proteins. While the atgpi8-1 mutation strongly disrupts plant growth, it is not lethal. Phenotypic analysis of atgpi8-1 mutants suggests that GPI-APs are important for root and shoot growth, stomata formation, apical dominance, transition to flowering, and male gametophyte viability. In addition, atgpi8-1 mutants accumulate higher levels of callose and have reduced plasmodesmata permeability. Genetic interactions of atgpi8-1 with mutations in ERECTA family (ERf) genes suggest the existence of a GPI-AP in a branch of the ERf signaling pathway that regulates stomata formation. Activation of the ERf signal transduction cascade by constitutively active YODA rescues stomata clustering in atgpi8-1, indicating that a GPI-AP functions upstream of the MAP kinase cascade. TOO MANY MOUTHS (TMM) is a receptor-like protein that is able to form heterodimers with ERfs. Our analysis demonstrates that tmm-1 is epistatic to atgpi8-1, indicating that either TMM is a GPI-AP or there is another GPI-AP regulating stomata development whose function is dependent upon TMM.
A protein can be integrated into a membrane through a polypeptide sequence or via a lipid anchor. While the modification of proteins with lipids is universal, the structure of the lipid anchors and the mechanism of their attachment differ in prokaryotes and eukaryotes. Several different types of lipids attach eukaryotic proteins to the inner leaflet of the plasma membrane. The attachment of eukaryotic proteins to the outer leaflet occurs only via GPI anchors. The biosynthesis of GPI glycolipid is a multistep process that relies on more than 20 proteins (Maeda and Kinoshita, 2011). It begins on the cytoplasmic side of the endoplasmic reticulum (ER) and continues to the luminal side where the anchor is attached to the C terminus of a protein. The GPI is transferred en bloc by the GPI transamidase (GPI-T) complex consisting of five proteins (Kinoshita, 2014). From the ER, GPI-anchored proteins (GPI-APs) are transported to the Golgi where the anchor undergoes remodeling. The backbone structure of a GPI anchor is highly conserved, but it can be remodeled in a variety of ways (Fujita and Kinoshita, 2012).
Proteins destined to acquire a GPI anchor have two signal peptides. On the N terminus there is an ER targeting signal, and on the C terminus there is a signal directing the attachment of the GPI anchor. The latter is usually 17 to 31 amino acids long; it has no consensus sequence but has four specific regions: an unstructured, approximately 10 amino acid-long linker region; small amino acids at the site of GPI anchor addition (the ω site) and the ω+2 site; a hydrophilic spacer region of 5 to 10 amino acids; and a hydrophobic tail of 15 to 20 amino acids. Efficient cleavage and addition of a GPI anchor depends on the marginal hydrophobicity of the C-terminal region when proprotein possibly transitions from a transmembrane form to a soluble form within the ER (Galian et al., 2012). Gpi8p/PIG-K and GPAA1/GAA1 are the catalytic subunits of GPI-T (Kinoshita, 2014). Gpi8p/PIG-K is a Cys protease that hydrolyzes a peptide bond between residues ω and ω+1. GPAA1/GAA1 catalyzes the next step, which is the formation of a bond between the ω site amino acid and the terminal ethanolamine phosphate of GPI (Eisenhaber et al., 2014). Failure to cleave the GPI attachment signal from precursor proteins prevents their O-glycosylation and leads to the retention of corresponding proteins in the ER, where they are subsequently degraded (Field et al., 1994).
The intracellular transport and localization of GPI-APs differs from that of proteins with transmembrane domains (Muñiz and Zurzolo, 2014). Both types of proteins are transported from the ER to the Golgi by COPII vesicles, but transmembrane proteins are directly recruited on the cytoplasmic side by Sec23/24 proteins while GPI-APs have to be recognized by p24 transmembrane cargo receptors (Muniz and Riezman, 2015). In mammalian epithelial cells, GPI anchors promote polar transport of GPI-APs to the apical membrane (Lisanti et al., 1989; Brown and Rose, 1992). Multiple lines of evidence suggest that GPI-AP association with lipid rafts, sphingolipid and cholesterol-rich nano-scale domains in the membrane, is critical for their trafficking and function (Maeda and Kinoshita, 2011).
According to the UniProt database, a eukaryotic organism can contain anywhere from 60 to 160 different GPI-APs, including receptors, adhesion molecules, proteoglycans, and various enzymes. This is an approximate number, since computational predictions are imprecise and proteomic approaches can underestimate the number of GPI-APs due to some having a low abundance. GPI-APs are essential for the viability of yeast and protozoa (Leidich et al., 1994; Nagamune et al., 2000). The loss of GPI anchoring is embryonic lethal in mammals and plants (Nozaki et al., 1999; Lalanne et al., 2004; Gillmor et al., 2005). In plants, GPI-APs are involved in various metabolic and developmental processes, including cellulose biosynthesis, callose metabolism, morphogenesis, and different aspects of reproduction (Schindelman et al., 2001; Lalanne et al., 2004; Gillmor et al., 2005; Levy et al., 2007; Capron et al., 2008; Simpson et al., 2009; Harpaz-Saad et al., 2011; Li et al., 2013; Cheung et al., 2014). Here, for the first time to our knowledge, we investigate the role of GPI-APs in stomata formation.
Stomata complexes are formed by a series of cell divisions and cell fate transitions. The pathway is initiated by asymmetric division in a subset of protodermal cells called meristemoid mother cells. A meristemoid mother cells gives rise to a smaller triangular meristemoid cell and a larger stomatal lineage ground cell. In Arabidosis (Arabidopsis thaliana), meristemoids typically undergo one to three rounds of asymmetric division, after which they differentiate into an oval guard mother cell, which then divides symmetrically to generate a pair of guard cells. In the wild type, two stomata do not form directly adjacent to each other; they are separated by at least one pavement cell (Sachs, 1991). Stomata formation is regulated by both positive and negative regulators. The negative regulators are Leu-rich repeat receptor-like kinases from the ERECTA family (ERfs; Shpak et al., 2005). In Arabidopsis, this family consists of three genes: ERECTA, ERL1, and ERL2. The activity of these receptors is regulated by a family of secreted Cys-rich peptides from the EPF/EPFL family (Shimada et al., 2011). The agonist and antagonist ligands compete for binding to the ERf receptor complexes fine-tuning stomata patterning in an organ specific manner (Lee et al., 2015). TOO MANY MOUTHS (TMM) is a receptor-like protein also involved in regulation of stomata development (Nadeau and Sack, 2002). It can form heterodimers with ERECTA and ERL1 in vivo and has been shown to directly bind EPF2 and STOMAGEN but not EPF1 in vitro (Lee et al., 2012, 2015). While TMM was the first gene linked with regulation of stomata development (Yang and Sack, 1995), its precise molecular function is still unclear. The phenotype of tmm mutants is organ specific (Geisler et al., 1998). In cotyledons and on the abaxial side of sepals and rosette leaves, this mutation increases the number of stomata and leads to their clustering. In stems, there are no stomata, and meristemoids differentiate into pavement cells instead of guard mother cells. Based on these mutant phenotypes, TMM seems to both inhibit and promote stomata development in an organ-specific manner. Recently, it was hypothesized that these seemingly opposing functions of TMM in leaves and stems might be related to the different availability of ligands in those organs rather than differences in TMM function (Abrash et al., 2011; Shpak, 2013).
To further our understanding of the mechanisms regulating stomata development, we have analyzed a novel mutant exhibiting increased stomata clustering. This mutant was found in an enhancer genetic screen that used Arabidopsis erl1 erl2 mutations as a background. Positional cloning determined that the mutation had occurred in a gene homologous to yeast GPI8, a gene encoding one of the catalytic subunits of an enzymatic complex responsible for the attachment of GPI anchors to selected proteins. We named the mutant atgpi8-1 and examined it for its potential to illuminate the importance of GPI anchoring for plant growth and stomata formation. Our analyses suggest that one or more GPI-anchored protein(s) functions in the ERf/TMM signaling pathway.
RESULTS
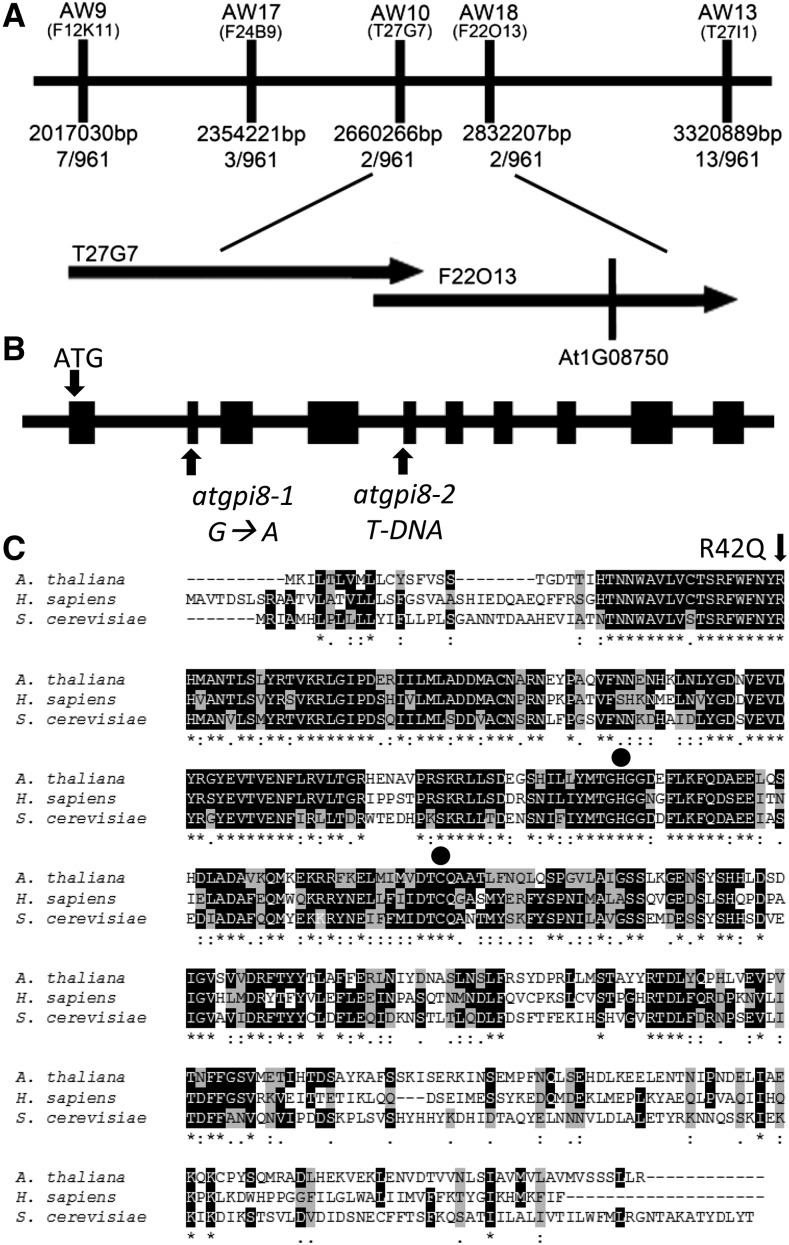
Positional Cloning of atgpi8-1
A search was performed in a population of ethyl methanesulfonic acid-mutagenized M2 erl1 erl2 seedlings for mutants containing stomata clusters. The screen led to the identification of the 2094 mutant. Through map-based cloning the mutation was determined to be located on the long arm of chromosome 1 between bp 2660266 and 2832207 (Fig. 1A). A sequence analysis of genes in this region uncovered a mutation in At1g08750; a G to A substitution at base pair 125 that resulted in replacement of Arg42 to Gln42 (Fig. 1B). Homology analysis of At1g08750 revealed that the protein has high amino acid sequence similarity to Saccharomyces cerevisiae GPI8 (77%) and Homo sapiens PIG-K (69%; Fig. 1C). GPI8 and PIG-K encode one of the catalytic subunits of the GPI-T. Genome data suggest that the overall mechanism of GPI anchoring is conserved between plants, yeast, and animals, since homologs for most essential genes are present (Eisenhaber et al., 2001, 2003a). Since Arabidopsis contains only one gene similar to GPI8/PIG-K, At1g08750 is most likely a catalytic subunit of plant GPI-T; we therefore named the gene AtGPI8 and the mutation atgpi8-1. Although knockout of GPI8 is lethal in S. cerevisiae, a mutation in the first conserved His produces a partially functional GPI8 (Benghezal et al., 1996; Meyer et al., 2000). The Arg42 to Gln42 mutation is one amino acid ahead of this residue. The atgpi8-1 mutation does not affect expression of the gene at the transcriptional level (Supplemental Fig. S1).
Figure 1.
Positional cloning of AtGPI8-1. A, Fine mapping of AtGPI8. The atgpi8-1 mutation was mapped to the upper arm of chromosome 1 between molecular markers AW9 and AW13. The number of recombinants obtained is indicated. Markers are positioned to scale. The corresponding BAC clones and the location of the AtGPI8 locus (At1g08750) are indicated. B, The structure of the AtGPI8 gene and the position of mutations. Boxes indicate exons and thick lines introns. G to A substitution in atgpi8-1 results in Arg42→Gln42. Location of the T-DNA insertion for atgpi8-2 is shown. C, Alignment of Arabidopsis AtGPI8, S. cerevisiae GPI8, and human PIG-K predicted protein sequences. Identical residues are colored black, similar residues are colored gray. Residues labeled with an asterisk are conserved, those labeled with a colon have conservation between groups of strongly similar properties, and those labeled with a period have conservation between groups of weakly similar properties. The conserved amino acids in predicted active sites are marked by circles. The position of the missense mutation in atgpi8-1 is marked with an arrow.
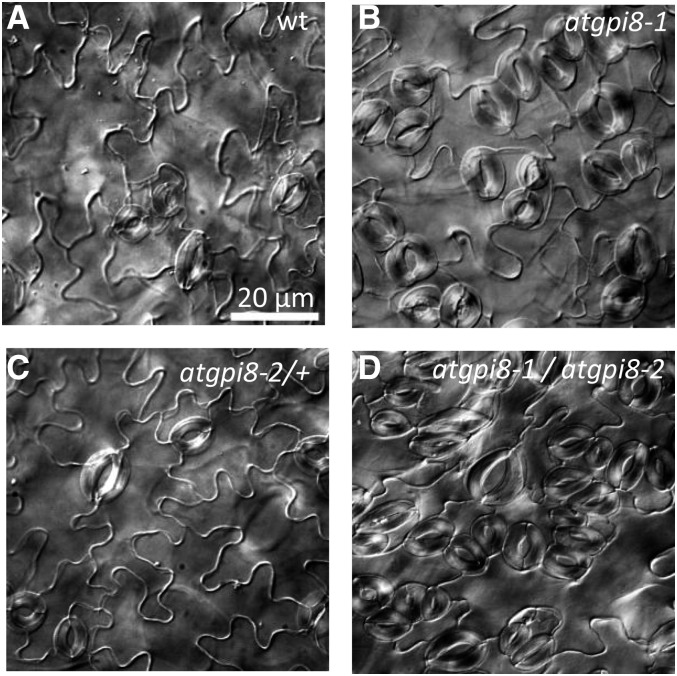
To confirm that the observed phenotype is due to the mutation in AtGPI8, we performed allelic analysis using an available T-DNA insertion line, CS853564 or atgpi8-2. The atgpi8-2 mutant is distributed as a heterozygous line, and it does not segregate out homozygous plants. Genotyping of 64 atgpi8-2/+ plant offspring identified 36% of atgpi8-2/+ and 66% of the wild type. For an allelism test, we crossed atgpi8-1 with atgpi8-2/+ and genotyped the F1 progeny. The identified atgpi8-1/atgpi8-2 seedlings displayed strong stomata clustering in cotyledons (Fig. 2). In addition, atgpi8-1/atgpi8-2 plants were severely dwarfed, never flowered, and did not survive into maturity (Supplemental Fig. S2). To test if other phenotypes were due to the AtGPI8 mutation, we expressed AtGPI8-EGFP under its endogenous promoter in atgpi8-1 mutants, which led to rescue of the majority of the observed phenotypes (Supplemental Fig. S3). These results confirmed that the positional cloning identified the mutation responsible for the phenotypes observed in the 2094 mutant. Unfortunately, the AtGPI8-EGFP construct is not useful for monitoring expression of AtGPI8, since no fluorescence was detected.
Figure 2.
The atgpi8-1 mutation leads to formation of stomata clusters and is allelic with atgpi8-2. Cleared differential interference contrast images of abaxial epidermis of mature cotyledons of wild type (A), atgpi8-1 (B), atgpi8-2/+(C), and atgpi8-1/atgpi8-2 (D). All images are under the same magnification.
To determine whether the atgpi8-1 mutation alters the metabolism of GPI-APs, we outcrossed the GPI-anchored protein GFP-SKU5 into the mutant (Sedbrook et al., 2002). We observed a decreased accumulation of GFP-SKU5 in the membrane protein fraction compared with the transmembrane receptor-like protein BAK1 (Fig. 3). When a GPI anchor cannot be transferred to the corresponding protein, it is retained in the ER and degraded (Field et al., 1994). For example, in pnt mutants deficient in biosynthesis of GPI anchors, the GFP-SKU5 protein and several other GPI-APs could not be detected (Gillmor et al., 2005). Therefore, the reduced accumulation of GPI-APs in atgpi8-1 suggests that the mutation leads to a partial loss of enzyme function.
Figure 3.
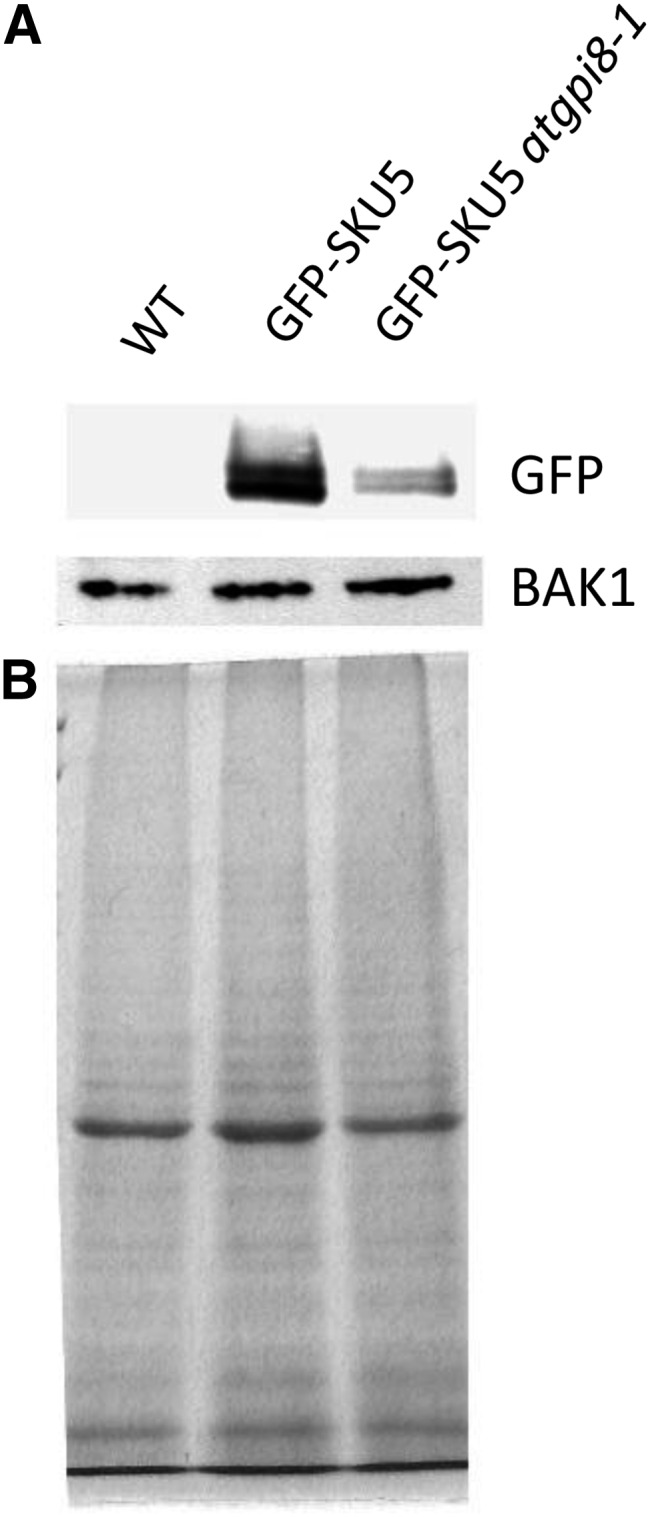
Expression of GFP-SKU5 is decreased in atgpi8-1. A, Protein gel detection of GFP-SKU5 and BAK1 in the membrane fractions of 8-d-old seedlings. B, A Coomassie-stained gel shows the total proteins in the membrane fraction.
The atgpi8 Mutations Affect Many Developmental Processes
Studies of mutations that disrupt the GPI anchoring process suggest that it is essential during the early stages of organism development, since mutants rarely survive past embryogenesis (Leidich et al., 1994; Nozaki et al., 1999; Lalanne et al., 2004; Gillmor et al., 2005). The identification of the nonlethal atgpi8-1 mutation affords us a unique opportunity to explore the importance of GPI anchoring during later stages of plant development. In atgpi8-1, the early postgermination growth of aboveground organs is very minimally affected, with 15-d-old cotyledons and first rosette leaves being of similar size in the mutant and the wild type (Fig. 4, A–C). However, root growth and hypocotyl elongation are significantly reduced in mutant seedlings of the same age (Fig. 4, A, H, and I). Leaves formed later in development are smaller in atgpi8-1, and at day 30 the wild-type and atgpi8-1 plants noticeably differ in size (Fig. 4D). The atgpi8-1 mutation also leads to reduced internode and pedicel elongation that results in formation of more compact inflorescence apices (Fig. 4, F, G, L, and N). The final height of atgpi8-1 plants is only moderately reduced since the number of internodes is increased (Fig. 4, K and O). An increased number of cauline branches in atgpi8-1 suggests that GPI anchoring is important for axillary shoot formation (Fig. 4M). The transition to flowering in atgpi8-1 is delayed, with the mutant plants bolting at 39.5 ± 3.3 d (±sd here and below) versus at 21.1 ± 1.1 d in the wild type (Fig. 4, E and J). All these phenotypes are rescued by expression of AtGPI8-EGFP in atgpi8-1 (Supplemental Fig. S3). However, the root elongation defects are only partially rescued, which could be due to additional mutations retained after three backcrosses or because the selected promoter region did not contain all of the necessary elements for the correct expression of the gene.
Figure 4.
Growth phenotypes of atgpi8-1 plants. A to C, During seedling development the atgpi8-1 mutation leads to reduced growth of roots and petioles (A) but not blades of cotyledons (B) or first two leaves (C). Images are of 15-d-old seedlings. D and E, Size differences of 30-d-old (D) and 60-d-old (E) wild-type and atgpi8-1 plants. F and G, Inflorescences of wild-type and atgpi8-1. H to L, Morphometric analysis of wild-type (gray bars) and atgpi8-1 (white bars) seedlings and mature plants. H, Hypocotyl length of 7-d-old etiolated seedlings (n = 15). I, Root length of 7-d-old seedlings (n = 10). J, Days until flowering (n = 20). K, Plant height (n = 18). L, Length of pedicels (n = 40). M, Number of cauline branches (n = 20). N, Number of siliques on the main stem (n = 40). H to N, Values are mean ± sd. O, Distance between pedicels (n = 40). Values are mean ± se. All values in H to O are statistically significant with P < 0.0001 based on Student’s t test.
Since the progeny of atgpi8-2/+ plants contained only heterozygotes and no homozygotes, we investigated gametophyte viability using reciprocal crosses between the wild type and atgpi8-2/+. The cross with atgpi8-2/+ as a male produced F1 progeny that were all wild type (20 plants genotyped), suggesting that this mutation leads to male gametophyte lethality. The cross with atgpi8-2/+ as a female produced F1 that was 43% atgpi8-2/+ and 57% wild type (37 plants genotyped), suggesting that the atgpi8-2 mutation does not have a strong impact on female gametophyte viability. The T-DNA insertion of atgpi8-2 includes a BASTA resistance gene. The progeny of atgpi8-2/+ contained 54.5% BASTA sensitive seedlings that were not statistically different from the 50% expected from plants with nonfunctional male gametophyte and viable female gametophyte (ntotal = 371; χ2 = 2.9, P = .087). As only 16.1% of atgpi8-1/+ progeny were mutants, which is significantly less than the expected 25% (ntotal = 87; χ2 = 3.89, P = .0485), male gametophyte viability is likely reduced in atgpi8-1 as well.
Our analysis of atgpi8 mutants implies that GPI-APs play important roles in multiple developmental processes, including lateral organ growth, axillary shoot formation, and transition to flowering. In addition, GPI-APs are essential for male gametophyte viability but do not have a strong effect on female gametophytes.
The atgpi8-1 Mutant Has Decreased Plasmodesmata Conductivity
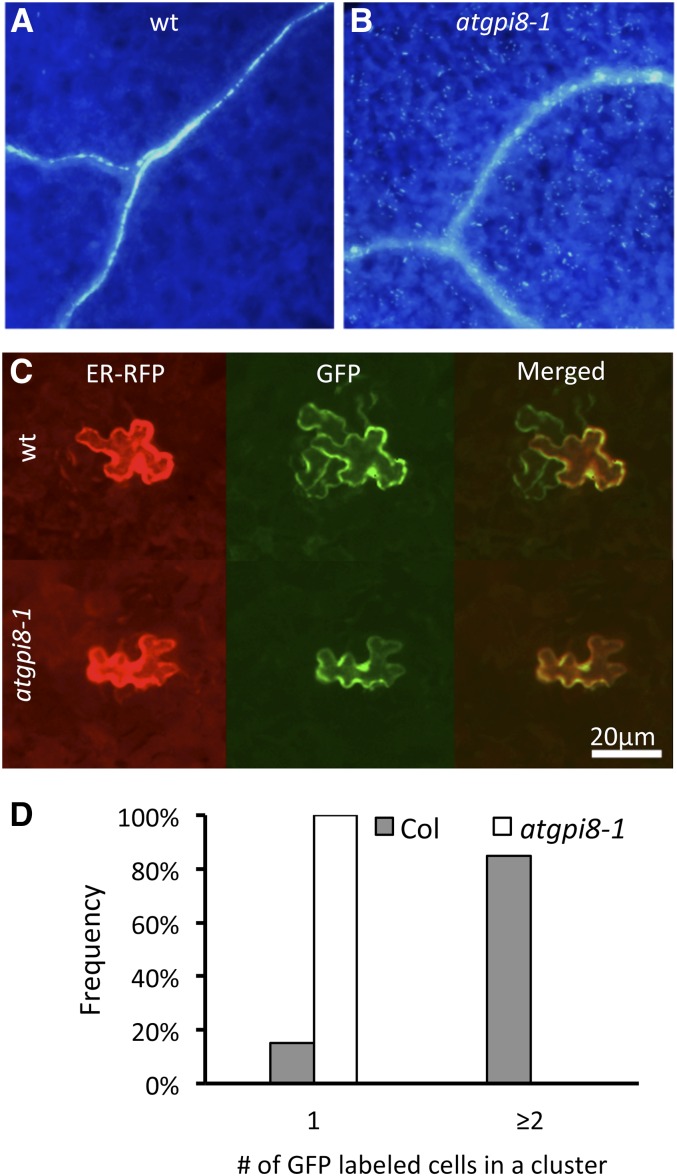
Atgpi8-1 was isolated as a mutant by its substantial stomata clustering (Fig. 2B). One of the potential causes of stomata cluster formation is an increase in plasmodesmata conductivity as in chorus and kobito1-3, two mutants with stomata clustering and multiple other developmental defects (Guseman et al., 2010; Kong et al., 2012). The accumulation of callose at the neck regions of plasmodesmata has a strong impact on conductivity; a decrease in callose deposition leads to plasmodesma opening (Iglesias and Meins, 2000; Levy et al., 2007; Guseman et al., 2010). Aniline blue staining detected increased callose accumulation in atgpi8-1, which was particularly evident in the vasculature and thick inner walls of guard cells (Fig. 5, A and B). To determine if the atgpi8-1 mutation also has an impact on the proper size exclusion limit of plasmodesmata, we performed a cell to cell mobility assay. Two plasmids, one carrying a gene encoding GFP and the other a gene encoding ER-localized RFP, both under control of the 35S cauliflower mosaic virus (CaMV) promoter, were cobombarded into the abaxial epidermis of 7-d-old seedlings. During bombardment, the particle gun usually transforms individual epidermal cells that can be confirmed by analysis of RFP expression, as this protein cannot move to neighboring cells due to its ER retention. If plasmodesmata are open, GFP can be detected in the surrounding cells due to diffusion. In wild-type seedlings, we observed that in 85% of transformation events, GFP was able to move to the neighboring cells while no GFP movement was observed in atgpi8-1 seedlings (Fig. 5, C and D). The average cluster size of cells expressing GFP was 4.0 ± 2.1 (±sd) for the wild type and 1.0 ± 0 for atgpi8-1. These data strongly suggest that plasmodesmata conductivity in atgpi8-1 is significantly decreased, and therefore formation of stomata clusters cannot be caused by changes in the plasmodesmata structure.
Figure 5.
Increased callose accumulation and decreased plasmodesmata conductivity of atgpi8-1. A and B, The accumulation of callose in 17-d-old cotyledons of atgpi8-1 (B) is increased compared to wild type (A) as determined by aniline blue staining. C and D, Analysis of GFP movement in the epidermis of 7-d-old seedlings suggests decreased plasmodesmata conductivity in atgpi8-1. C, Representative images of the abaxial side of the epidermis expressing cobombarded ER-localized RFP (left), GFP (center), and both merged (right). D, Distribution analysis of the number of cells in clusters expressing GFP from a single transformation event based on RFP expression; n = 40, P < 0.0001.
Synergistic Interactions of ERfs and AtGPI8 during Stomata Formation
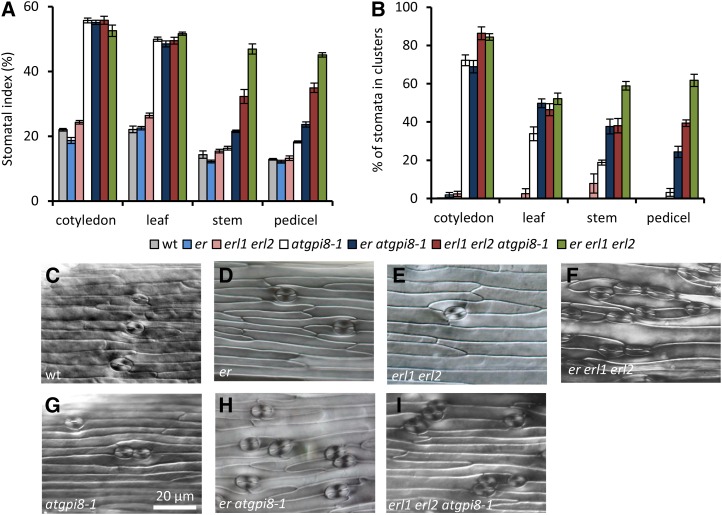
Since GPI anchoring has not previously been associated with stomata formation, we were especially interested to investigate the impact of the atgpi8-1 mutation on epidermis differentiation. Analysis of the epidermis in cotyledons, rosette leaves, stems, and pedicels demonstrated an increase in both the stomatal index (SI) and stomata clustering in atgpi8-1 versus the wild type (Fig. 6). The change in epidermis development is especially dramatic on the abaxial side of cotyledons and leaves, with SI being increased 2.5 times in atgpi8-1 cotyledons and 2.3 times in atgpi8-1 leaves. While <1% of stomata are in clusters in the wild type, 72.2 ± 6.3% and 33.9 ± 8.9% of stomata are in clusters on the abaxial side of atgpi8-1 cotyledons and rosette leaves, respectively. These data suggest the existence of a GPI-anchored protein that works to inhibit stomata formation.
Figure 6.
Genetic interactions of ERECTA family genes and AtGPI8 during stomata development. A and B, Analysis of stomata formation in different organs of wild type, er, erl1 erl2, atgpi8-1, er atgpi8-1, erl1 erl2 atgpi8-1, and er erl1 erl2. SI (A) and percent of stomata in clusters (B) were measured in the abaxial epidermis of mature cotyledons and first rosette leaf as well as in the epidermis of stems and pedicels. Values are means ± sd, n = 6. C to I, Representative images of stem epidermis in wild type, atgpi8-1, er, erl1 erl2, er atgpi8-1, er atgpi8-1, and er erl1 erl2. All images are under the same magnification.
ERfs are plasma membrane-localized receptors that are known to inhibit stomata formation (Shpak et al., 2005). To investigate whether a potential GPI-AP functions in the ER signaling pathway, we analyzed genetic interactions between mutants of ERf genes and atgpi8-1. In cotyledons and rosette leaves, stomata formation is not altered by addition of the er or erl1 erl2 mutations to atgpi8-1, possibly because the stomatal phenotype of atgpi8-1 is already very strong in those organs (Fig. 6, A and B). Thus, in cotyledons and rosette leaves, the SI and the percent of stomata in clusters are identical in atgpi8-1 and er erl1 erl2 (Fig. 6, A and B). However, in stems and pedicels where the atgpi8-1 phenotype is not so strong, we observed synergistic interactions between atgpi8-1 and er and between atgpi8-1 and erl1 erl2 (Fig. 6). For example, in stems the atgpi8-1, er, and erl1 erl2 mutations do not increase SI on their own, but the combination of er or erl1 erl2 with atgpi8-1 increases SI to 21.5 ± 0.9% and 32.3 ± 5.3%, respectively, from 14.3 ± 2.8% (±sd here and below) in the wild type (Fig. 6A). In pedicels of atgpi8-1, only 3.2 ± 4.9% of stomata are in clusters and no stomata clusters were detected in wild type, er, or erl1 erl2 pedicels. However, pedicels of atgpi8-1 er and atgp8-1 erl1 erl2 contained 24.3 ± 7.1% and 39.4 ± 9.2% of stomata in clusters, respectively. At the same time, in stems and pedicels the synergistic interaction between erl1 erl2 and atgpi8-1 does not increase the SI or the percent of stomata in clusters to the same level as in er erl1 erl2 (Fig. 6, A and B).
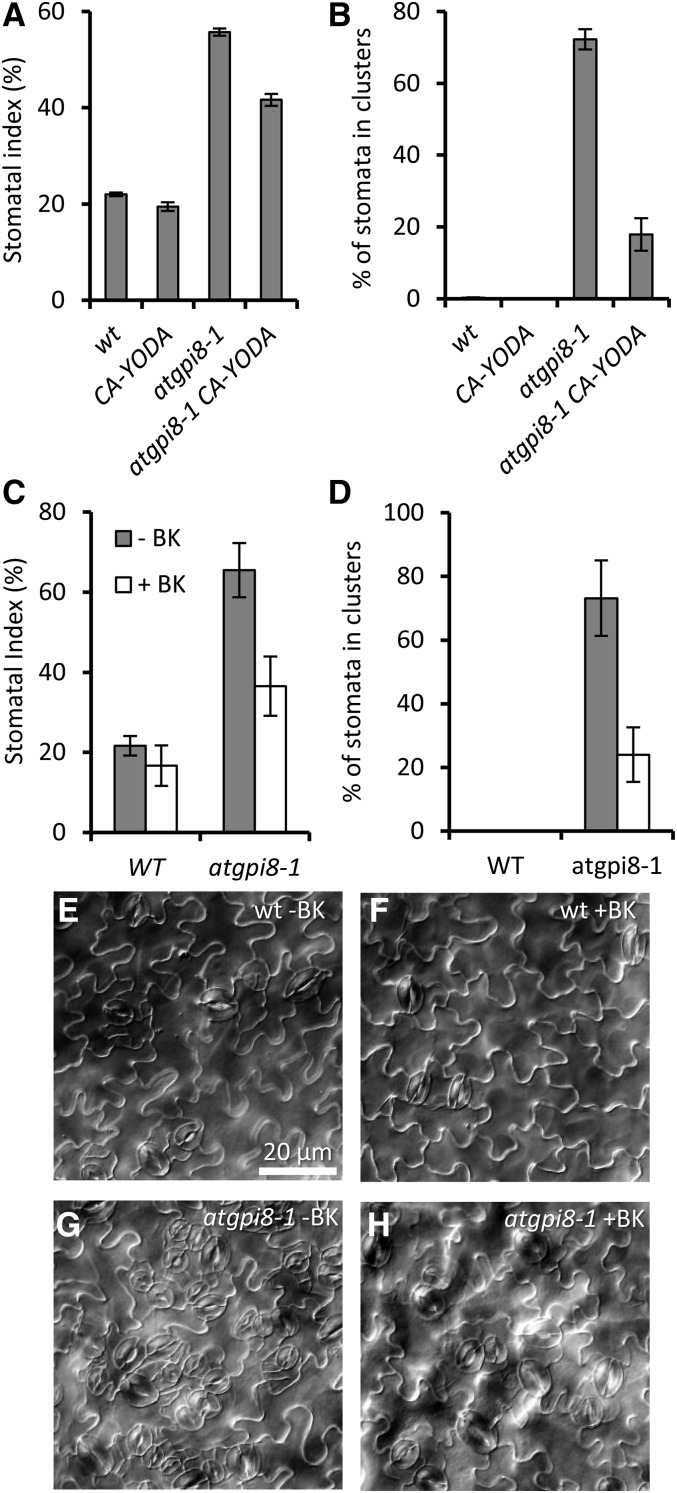
To further investigate the connection between GPI anchoring and the ER signaling pathway, we analyzed genetic interactions of AtGPI8 with YODA (YDA), a MAPKK kinase that functions downstream of TMM and the ERfs (Bergmann et al., 2004; Meng et al., 2012). The N terminus of YDA contains a negative regulatory domain, and its deletion produces a constitutively active YDA (CA-YDA) that inhibits stomata development and promotes stem and pedicel elongation when expressed in the wild type (Bergmann et al., 2004). We transformed the CA-YDA construct into atgpi8/+ and then analyzed stomata development in F2 progeny of a transgenic line selected based on increased length of pedicels and stems. The F2 progeny segregated CA-YDA in wild-type and atgpi8-1/+ backgrounds as well as CA-YDA atgpi8-1 plants. While in this particular line CA-YDA did not statistically significantly change development of stomata on its own (P < .04), it was able to partially rescue atgpi8-1 plants, decreasing both SI and stomata clustering in the mutant (Fig. 7, A and B). Thus, the GPI-anchored protein that functions in stomata formation is likely to be upstream of YDA. It has been previously reported that a GSK3 kinase regulates stomata development downstream of TMM and the ERf and upstream of YDA (Kim et al., 2012). To further validate the existence of a GPI-anchored protein in the ER signaling pathway, we examined whether bikinin, a GSK3 kinase inhibitor, could rescue the stomata phenotype of atgpi8-1. Similar to its effect on stomata development in tmm and er erl1 erl2, bikinin decreased the stomata index and stomata clustering in atgpi8-1 seedlings (Fig. 7, C–H). Growth in the presence of bikinin decreased SI on the abaxial side of atgpi8-1 cotyledons from 73 ± 12% to 33 ± 8% and stomata clustering from 70 ± 14.8% to 16 ± 12%. Thus, a GPI-anchored protein is likely to function upstream of a GSK3 kinase. Taken together, these data strongly suggest that a GPI-AP regulating formation of stomata functions in the ERf signaling pathway upstream of the MAP kinase cascade.
Figure 7.
CA-YDA and bikinin partially rescue the epidermal phenotype of atgpi8-1. Expression of CA-YDA decreases SI (A) and reduces percent of stomata in clusters (B) in abaxial epidermis of atgpi8-1 mature cotyledons. Values are mean ± sd for six seedlings. The effect of 30 μm bikinin on SI (C) and the percent of stomata in clusters (D) in abaxial epidermis of wild-type and atgpi8-1 cotyledons. Values are mean ± sd for six seedlings. E to H, Representative images of abaxial epidermis of wild-type and atgpi8-1 cotyledons grown on the media with (+BK) and without (−BK) bikinin.
The tmm-1 Mutation Is Epistatic to the atgpi8-1 Mutation
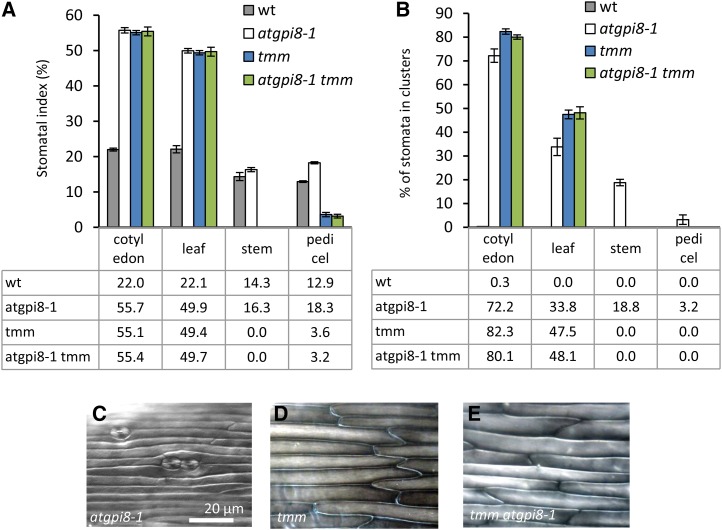
The receptor-like protein TMM forms heterodimers with ERfs (Lee et al., 2012). In contrast to ERfs that always inhibit stomata development, TMM inhibits stomata development in cotyledons and leaves but promotes it in stems and pedicels (Geisler et al., 1998). To understand the genetic interactions between TMM and GPI-AP involved in stomata formation, we outcrossed atgpi8-1 into the tmm-1 mutant and analyzed SI and stomata clustering. Cotyledons and leaves of tmm-1 and atgpi8-1 have very similar phenotypes: increased SI and massive stomata clustering (Fig. 8, A and B). The only difference observed was slightly weaker clustering of stomata in atgpi8-1 (Fig. 8, A and B). No additive effects of the tmm-1 and atgpi8-1 mutations were observed in leaves and cotyledons, and the phenotype of atgpi8-1 tmm-1 was identical to the phenotype of tmm-1 (Fig. 8, A and B). In stems and pedicels the phenotypes of tmm-1 and atgpi8-1 were quite different. There were no stomata formed in stems of tmm-1, while in atgpi8-1 stomata developed and formed clusters (Fig. 8, C and D). The pedicels of tmm-1 formed a greatly reduced number of stomata and no stomata clusters were observed (Fig. 8, A and B). In contrast, the pedicels of atgpi8-1 had an increased SI and some stomata clustering (Fig. 8, A and B). The phenotype of atgpi8-1 tmm-1 epidermis in stems and pedicels was identical to tmm-1 (Fig. 8, A, B, and E), suggesting that tmm-1 is epistatic to atgpi8-1.
Figure 8.
TMM is epistatic to AtGPI8. A and B, Analysis of stomata formation in different organs of wild type, atgpi8-1, tmm, and atgpi8-1 tmm. SI (A) and percent of stomata in clusters (B) were measured in the abaxial epidermis of mature cotyledons and first rosette leaf as well as in the epidermis of stems and pedicels. Values are means ± sd, n = 6. Values in the table represent the mean. C to E, Representative images of stem epidermis in atgpi8-1, tmm, and tmm atgpi8-1. All images are under the same magnification.
DISCUSSION
The GPI-anchor biosynthesis pathway has been studied intensively in yeast and in humans, but less is known about this pathway in plants. In Arabidopsis, homologous genes exist for almost all the known components of the GPI anchoring pathway, but currently the only genes that have been studied are SETH1, SETH2, PEANUT1 (PNT1), and APTG1 (Lalanne et al., 2004; Gillmor et al., 2005; Dai et al., 2014). These genes encode enzymes that catalyze specific steps in the biosynthesis of the anchor. In this paper, we describe a mutation in a protein that is required for attachment of the anchor to the designated proteins. The yeast and mammal GPI transamidase complexes consist of five subunits, two of which have enzymatic activity (Kinoshita, 2014). Arabidopsis has homologs of all five subunits, with all but one subunit encoded by only one gene: At1g08750 is a homolog of Gpi8/PIG-K, At5g19130 of Gaa1p/GAA1, At3g07180 of Gpi17p/PIG-S, and At3g07140 of Gpi16p/PIG-T. Arabidopsis has two genes homologous to Gab1p/ PIG-U: At1G12730 and At3g27325. Of the five GPI-T subunits GPI8p/PIG-K/AtGPI8 is the most conserved. Amino acid sequence similarity of AtGPI8 is 77% with S. cerevisiae Gpi8p and 69% with H. sapiens PIG-K. The similarity of other Arabidopsis GPI-T subunits with their human and yeast homologs is <50%. In a genetic screen, we identified a point mutation in At1g08750 that results in replacement of Arg42 to Gln42. Gpi8p/PIG-K/AtGPI8 is a Cys protease with a Cys/His catalytic dyad that hydrolyzes a peptide bond in proteins containing a signal for GPI anchoring. Mutation of either Cys or His in the active site of Gpi8p abolishes activity of the enzyme (Meyer et al., 2000). The mutation of another His (in yeast His-54) leads to partial loss of Gpi8p function (Meyer et al., 2000). The amino acid substitution in atpgi8-1 is directly adjacent to His-43 that is homologous to yeast His-54. Accumulation of the GPI-AP SKU5 is reduced in the atgpi8-1 mutant, suggesting a partial loss of function due to the Arg-42 mutation. Thus, our results corroborate the importance of this region for Gpi8p/PIG-K/AtGPI8 subunit function.
Studies of mutants with damaged biosynthesis and attachment of the GPI anchor reveal functions of GPI-APs during plant growth and development. Previous analysis of mutations in PNT1, SETH1, SETH2, and APTG1 demonstrated that GPI-APs are essential for male fertility. In seth1 and seth2, pollen grains develop normally but pollen tube germination and growth are drastically reduced, and pnt1 is characterized by reduced pollen transmission (Lalanne et al., 2004; Gillmor et al., 2005). In the aptg1 mutant, pollen tubes are able to elongate but suffer a reduced ability to find micropyles (Dai et al., 2014). Unsurprisingly, the atgpi8-2 knockout mutation cannot be transmitted through the male gametophyte.
GPI-APs play important roles during embryogenesis. Aptg1 embryos are arrested at a globular stage and pnt1 embryos undergo delayed morphogenesis (Gillmor et al., 2005; Dai et al., 2014). The pnt1 mutants are seedling lethal and can proliferate only as callus, indicating that GPI-APs are required for coordinated multicellular growth (Gillmor et al., 2005). As a consequence, the role of GPI-APs in the later stages of plant development has been investigated only on the level of individual proteins. This study of the atgpi8-1 mutant allowed us to explore the role of GPI-APs on a more global scale at the later developmental stages. First, we noticed that root growth is sensitive to interference with GPI-AP biosynthesis. Since GPI-APs promote the accumulation of crystalline cellulose and decrease the accumulation of xyloglucans, pectins, and callose in the cell wall, they are expected to have a strong impact on cell elongation (Gillmor et al., 2005). Likewise, specific GPI-APs such as COBRA, SOS5, and SKU5 have been linked with cell elongation, including expansion of cells in roots (Schindelman et al., 2001; Sedbrook et al., 2002; Shi et al., 2003; Roudier et al., 2005; Xu et al., 2008). Our analysis of atgpi8-1 suggests that GPI-APs promote plasmodesmata permeability and are essential for reduced callose accumulation. This is not surprising, as β-1,3-glucanases, enzymes that degrade callose, are GPI-APs (Elortza et al., 2003; Elortza et al., 2006), and mutation of AtBG_ppap, a plasmodesmata-localized β-1,3-glucan synthase, increases callose accumulation and reduces plasmodesmata conductivity (Levy et al., 2007).
We speculate that increased callose accumulation in atgpi8-1 might disrupt proper root development due to impaired cell-to-cell communication as previously observed in gain-of-function mutants of the callose synthase CALS3 (Vatén et al., 2011). Our other findings concern the importance of GPI-APs for apical dominance and transition to flowering. Whether those developmental processes are regulated by specific GPI-APs or through changes in callose deposition is a question for the future. The callose lining in sieve plate pores is essential for normal phloem transport and as a result for the movement of FLOWERING LOCUS T (Barratt et al., 2011; Rinne et al., 2011).
We also find that GPI-APs are important for stomata formation. Analysis of genetic interactions uncovered a synergy between AtGPI8 and the ERf genes implying the existence of a GPI-AP in the ERf signaling pathway. The ability of constitutively active YODA to rescue stomata clustering in atgpi8-1 corroborates this hypothesis and suggests that a GPI-AP functions upstream of the MAP kinase cascade. TMM is a receptor-like protein that can potentially be a GPI-AP. TMM was previously computationally identified as a putative GPI-AP by two independent methods (Borner et al., 2002; Eisenhaber et al., 2003b). However, analysis of plants expressing TMM fused with GFP at the C terminus that are expected to prevent addition of the GPI anchor implied that TMM can function as a transmembrane protein (Nadeau and Sack, 2002). Analysis of genetic interactions demonstrated that tmm-1 is epistatic to atgpi8-1, indicating that either TMM is a GPI-AP or there is another GPI-AP whose function in stomata formation is entirely dependent on TMM. Future analysis of TMM attachment to the plasma membrane should distinguish between these two possibilities. In mammalian cells, the majority of GPI-APs are polar localized and are associated with lipid rafts (Lisanti et al., 1989; Brown and Rose, 1992; Maeda and Kinoshita, 2011). The presence of a GPI-AP in the ERf signaling pathway suggests the possibility that the ERf receptors directly interact with that protein and the receptor complexes might be targeted to lipid rafts. Lipid rafts have been proposed to act as signaling platforms where receptor complexes are stabilized (van Zanten et al., 2009). Alternatively, a GPI-AP might facilitate transport of ERfs to the plasma membrane or promote polar localization of ERf complexes in epidermal cells. The GPI-APs LORELEI and LRE-like GPI-AP1 function as chaperones of the receptor-like kinase FERONIA delivering it to the plasma membrane (Li et al., 2015). A GPI-linked lipid transfer protein has recently been shown to be dynamically targeted to specific regions of epidermal cells (Ambrose et al., 2013). Finally, a GPI-AP might contribute to specificity of ligand binding. In mammalian cells, the GPI-APs CRIPTO and GFRα function as coreceptors of corresponding transmembrane receptor kinases and define the specificity of ligand binding (Yeo and Whitman 2001; Klein et al., 1997). Thus, the existence of a GPI-AP in the ERf signaling pathway poses new questions about the mechanism of ERf receptor function.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col) was used as the wild type. The mutant atgpi8-1 was obtained from an ethyl methanesulfonic acid-mutagenized (0.3% for 14 h) screen in an erl1-2 erl2-1 population (Shpak et al., 2004). Individual M2 seed lines were grown on modified Murashige and Skoog media plates supplemented with 1× Gamborg B5 vitamins and 1% w/v Suc and screened for stomata patterning defects in cotyledons. Atgpi8-2 (CS853564) and tmm-1 (CS6140) were obtained from the Arabidopsis Biological Resource Center. The er-105 and er-105 erl1-2 erl2-1 mutants have been described previously (Torii et al., 1996; Shpak et al., 2004). Plants were grown on a soil mixture of a 1:1 ratio of Promix PGX (Premier Horticulture) and Vermiculite (Pametto Vermiculite) and were supplemented with Miracle-Gro (Scotts) and approximately 3.5 mg/cm3 of Osmocote 15-9-12 (Scotts). All plants were grown at 20°C under long-day conditions (18 h light/6 h dark).
Map-Based Cloning of atgpi8-1
A mapping population was created by crossing atgpi8-1 erl1 erl2 to the Landsberg erecta ecotype. A bulk segregant analysis using DNA from 35 F2 atgpi8-1-like seedlings revealed a linkage to the long arm of chromosome 1 between the SSLP markers NGA 280 and NGA 392. Fine mapping within this region using 961 F2 mutant plants localized the mutation to be between SSLP markers AW10 and AW18 in a segment of 172 kb that included two BACs (T27G7 and F22O13). Sequencing performed in this region identified a single G-A substitution at position 559 bp in the At1g08750 gene. The SSLP markers for map-based cloning were designed from the information provided by the Monsanto Arabidopsis Polymorphism and Ler Sequence Collection (http://www.Arabidopsis.org/Cereon/index.jsp) as well as the Arabidopsis Mapping Platform (http://amp.genomics.org.cn/). For primer sequences and amplified fragment sizes in both Col and Landsberg erecta, see Supplemental Table S1.
Sequence Alignment
Full-length amino acid sequences of AtGPI8 homologs from Saccharomyces cerevisiae (GPI8; accession NP_010618) and Homo Sapiens (PIG-K; accession CAI21820) were retrieved from the NCBI database and aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Crosses and Genotyping
To obtain the atgpi8-1 mutant, atgpi8-1 erl1 erl2 plants were crossed with Col. The atgpi8-1 plants were identified both by the short root phenotype and by genotyping. The primers AtGPI8 F1 (5′-GACTGGAGTTCCATCGTGG-3′) and AtGPI8 R1N (5′-CGTGACTCTGGAGTTCTTCTGC-3′) were used to amplify a 1.5-kb fragment containing a BsrFI restriction site in the wild type but not in atgpi8-1. In the wild type, restriction digestion of this fragment results in two 750-bp fragments. Genotyping of erl1-2 and erl2-1 was performed as described by Shpak et al. (2004).
For the allelism test, the atgpi8-1 mutant was crossed with atgpi8-2/+. The atgpi8-1/atgpi8-2 plants were identified in the F1 generation. The presence of the atgpi8-1 mutation was confirmed by genotyping as described above. The presence of atgpi8-2 was determined through PCR using AtGPI8 1780 (5′-GGTTGATACTTGCCAAGCTG-3′) and AtGPI8 2171.rc (5′ -GCTTCAGATTGGTGTATCTG-3′) primers and the left border primer p745 (5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′). The size of the PCR product is 400 bp in the wild type and 200 bp in atgpi8-2.
To construct a plasmid carrying cAtGPI8-EGFP (pESH 504), the cDNA sequence of AtGPI8 was cloned behind the 2.1-kb AtGPI8 promoter sequence and in frame with EGFP into a pZP222 vector. A 35S terminator was placed after the stop codon of EGFP. The pESH504 plasmid was introduced into an Agrobacterium tumefaciens strain GV3101/pMP90 by electroporation and into atgpi8-1 mutants by the floral dip method. While atgpi8-1 plants produced very few T1 seeds, we still were able to identify multiple transgenic lines after selection on gentamycin. The atgpi8-1 genotype of the selected T1 lines was confirmed as described above. In a separate experiment, atgpi8-1/+ plants were transformed with a plasmid containing CA-YDA (Lukowitz et al., 2004). Transgenic plants carrying CA-YDA were selected on soil using 0.02% Finale (Bayer CropScience) with 0.005% of Silwet l-77 (Lehle Seeds) and then genotyped for atgpi8-1.
To generate er atgpi8-1, atgpi8-1 was crossed with er-105. The presence of er-105 was determined phenotypically in the F2 generation as er-105 plants are shorter, have clustered inflorescence and short, blunt siliques. To generate tmm atgpi8-1, atgpi8-1 was crossed to tmm-1. Plants homozygous for tmm-1 were selected based on the absence of stomata in the stems. The presence of the atgpi8-1 mutation in both of these crosses was confirmed by genotyping. Atgpi8-1 expressing GFP-SKU5 was generated by crossing the mutant with the transgenic plants (Sedbrook et al., 2002) and selecting for the atgpi8-1 short root phenotype and GFP fluorescence in the F2 generation.
Analysis of Plant Development and Growth
The SI and stomata clustering were measured in cotyledons and leaves of 17-d-old seedlings and in stems and pedicels of mature plants using Differential Interference Contrast (DIC) microscopy. To analyze the effects of bikinin on stomata development, seedlings were grown on Murashige and Skoog plates supplemented with 30 µM bikinin for 7 days. For DIC microscopy, plant tissues were fixed overnight in ethanol:acetic acid (9:1) and cleared in a chloral hydrate solution (chloral hydrate:water:glycerol 8:1:1) for approximately 24 h. Structure of epidermis was observed using a Nikon Eclipse 80i microscope with DIC optics, and pictures were obtained with a 12-megapixel cooled color DXM-1200c (Nikon) camera. The number of stomata were counted using NSI-Elements BR 2.30.
Plant height, the number of siliques and cauline branches, and internode and pedicel length were measured at full maturity at 60 d for wild type and at 90 d for atgpi8-1. The number of cauline branches was determined by counting the number of lateral branches on the main stem. To analyze callose accumulation, 7-d-old seedlings were fixed overnight in a solution of ethanol:acetic acid (9:1), rinsed in 90% ethanol, incubated for 30 min in 0.09M sodium phosphate buffer (pH 9), and finally submerged for 1 h in 0.01% aniline blue dissolved in the indicated buffer. A Nikon Eclipse 80i epifluorescence microscope with a 12-megapixel cooled color camera and a UV-2A filter (Nikon) was used to observe the seedlings immediately after incubation.
Transient Transformation of Seedlings and Cell-to-Cell Mobility Assay
The abaxial epidermis of 7-day-old Arabidopsis seedlings was transformed with the vectors pAVA 321 (CaMV 35S::mGFPS65T; von Arnim et al., 1998) and pAN456 (CaMV 35S::RFP with ER retention signal; Nelson et al., 2007). During transformation, 1.1-μm tungsten M-17 microcarriers (Bio-Rad) were fired at 400 psi using a PSD-1000/He particle bombardment system (Bio-Rad). The fluorescence was observed 18 h postbombardment using a Zeiss Axio Observer.Z1 microscope, and images were obtained with 1.3-megapixel cooled black and white ORCA-AG (Hamamatsu) camera. The presence of RFP designated transformed cells. Protein mobility was established by analyzing GFP fluorescence.
Protein Gel-Blot Analysis
Eight-day-old GFP-SKU5, GFP-SKU5 atgpi8-1, and wild-type seedlings were ground in liquid nitrogen to a fine powder, and then 3 volumes of extraction buffer (100 mm Tris-HCl at pH 8.8, 150 mm NaCl, 1 mm EDTA, 20% glycerol, 20 mm NaF, 1 mm PMSF, 1:1000 Complete protease inhibitor cocktail [Sigma]) were added to the ground material. The extracts were sonicated and centrifuged at 10,000 g for 10 min at 4°C to remove insoluble debris. The supernatant was ultra-centrifuged at 100,000 g for 1 h at 4°C to obtain the crude membrane fraction as a pellet. Membrane pellets were suspended in 100 mM potassium phosphate with pH 7.4. The protein gel-blot analysis was performed as described previously with minor modifications (Shpak et al., 2003). Proteins were separated by 9% or 10% SDS-PAGE. The primary anti-GFP polyclonal antibody (Life Technology) was used at a dilution of 1:5,000 following by the secondary HRP Conjugated Goat Anti-Rabbit IgG antibody (Pierce) at a dilution of 1:45,000. Primary anti-BAK1 polyclonal antibody (Agrisera) was used at a dilution of 1:5,000 following by the secondary HRP Conjugated Goat Anti-Rabbit IgG antibody (Agrisera) at a dilution of 1:5,000. The detection of GFP-SKU5 and BAK1 was performed with SuperSignal West Pico Rabbit IgG detection kit (Pierce).
Reverse Transcription-PCR
Total RNA was isolated from 12-d-old Arabidopsis seedlings using a Spectrum Plant Total RNA Kit (Sigma). First-strand cDNA was synthesized from 785 ng of RNA with a ProtoScript M-MuLV Taq RT-PCR Kit (New England Biolabs) according to the manufacturer’s instructions. PCR was performed with the first-strand synthesized cDNA at 95°C for 2 mins, varying cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s, followed by a final 72°C for 5 min. The primers 5′-GGTTGATACTTGCCAAGCTG-3′ and 5′-TCATCGTAGTAAAGATGATGAGACCATTAC-3′ were used to amplify AtGPI8 and the primers 5′-GCCATCCAAGCTGTTCTCTC-3′ and 5′-GCTCGTAGTCAACAGCAACAA-3′ were used to amplify ACTIN-2 (At3g18780) as a control. PCR products were separated on 1% agarose gel and visualized with ethidium bromide staining.
Arabidopsis Genome Initiative
Arabidopsis Genome Initiative numbers for the genes discussed here are as follows: AtGPI8 (At1g08750), ER (At2g26330), ERL1 (At5g62230), ERL2 (At5g07180), TMM (At1g80080), YODA (At1g63700), and SKU5 (At4g12420).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Semi-quantitative RT-PCR analysis of AtGPI8 and ACTIN transcripts in 12-day-old seedlings.
Supplemental Figure S2. At 30 days growth and development of atgpi8-1/atgpi8-2 plants is drastically impeded compared to wild type.
Supplemental Figure S3. The expression of AtGPI8-EGFP under the endogenous promoter rescued the majority of phenotypes observed in atgpi8-1 mutants.
Supplemental Table S1. Primers used to amplify SSLPs during positional cloning of atgpi8-1.
Supplementary Material
Acknowledgments
We thank Albrecht von Arnim, Andreas Nebenführ, Wolfgang Lukowitz, and John Sedbrook for sharing with us the plasmid constructs (pAVA 321, pAN456, CA-YDA) and seeds of transgenic plants expressing GFP-SKU5. We thank Anastasia Aksenova for helping with the analysis of the bikinin effect on stomata formation and Richard Maradiaga for help with the genotyping of mutants.
Glossary
Footnotes
This work was supported by the National Science Foundation (IOS-0843340 to E.D.S.).
Articles can be viewed without a subscription.
References
- Abrash EB, Davies KA, Bergmann DC (2011) Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, DeBono A, Wasteneys G (2013) Cell geometry guides the dynamic targeting of apoplastic GPI-linked lipid transfer protein to cell wall elements and cell borders in Arabidopsis thaliana. PLoS One 8: e81215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt DH, Kölling K, Graf A, Pike M, Calder G, Findlay K, Zeeman SC, Smith AM (2011) Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol 155: 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A (1996) Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J 15: 6575–6583 [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Borner GH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Rose JK (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68: 533–544 [DOI] [PubMed] [Google Scholar]
- Capron A, Gourgues M, Neiva LS, Faure JE, Berger F, Pagnussat G, Krishnan A, Alvarez-Mejia C, Vielle-Calzada JP, Lee YR, et al. (2008) Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20: 3038–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Li C, Zou YJ, Wu HM (2014) Glycosylphosphatidylinositol anchoring: control through modification. Plant Physiol 166: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XR, Gao XQ, Chen GH, Tang LL, Wang H, Zhang XS (2014) ABNORMAL POLLEN TUBE GUIDANCE1, an endoplasmic reticulum-localized mannosyltransferase homolog of GLYCOSYLPHOSPHATIDYLINOSITOL10 in yeast and PHOSPHATIDYLINOSITOL GLYCAN ANCHOR BIOSYNTHESIS B in human, is required for arabidopsis pollen tube micropylar guidance and embryo development. Plant Physiol 165: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F (2001) Post-translational GPI lipid anchor modification of proteins in kingdoms of life: analysis of protein sequence data from complete genomes. Protein Eng 14: 17–25 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Eisenhaber S, Kwang TY, Grüber G, Eisenhaber F (2014) Transamidase subunit GAA1/GPAA1 is a M28 family metallo-peptide-synthetase that catalyzes the peptide bond formation between the substrate protein’s omega-site and the GPI lipid anchor’s phosphoethanolamine. Cell Cycle 13: 1912–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Maurer-Stroh S, Novatchkova M, Schneider G, Eisenhaber F (2003a) Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. BioEssays 25: 367–385 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, Schultz CJ, Borner GH, Dupree P, Eisenhaber F (2003b) Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elortza F, Mohammed S, Bunkenborg J, Foster LJ, Nühse TS, Brodbeck U, Peck SC, Jensen ON (2006) Modification-specific proteomics of plasma membrane proteins: identification and characterization of glycosylphosphatidylinositol-anchored proteins released upon phospholipase D treatment. J Proteome Res 5: 935–943 [DOI] [PubMed] [Google Scholar]
- Elortza F, Nühse TS, Foster LJ, Stensballe A, Peck SC, Jensen ON (2003) Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol Cell Proteomics 2: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Field MC, Moran P, Li W, Keller GA, Caras IW (1994) Retention and degradation of proteins containing an uncleaved glycosylphosphatidylinositol signal. J Biol Chem 269: 10830–10837 [PubMed] [Google Scholar]
- Fujita M, Kinoshita T (2012) GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim Biophys Acta 1821: 1050–1058 [DOI] [PubMed] [Google Scholar]
- Galian C, Björkholm P, Bulleid N, von Heijne G (2012) Efficient glycosylphosphatidylinositol (GPI) modification of membrane proteins requires a C-terminal anchoring signal of marginal hydrophobicity. J Biol Chem 287: 16399–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Yang M, Sack FD (1998) Divergent regulation of stomatal initiation and patterning in organ and suborgan regions of the Arabidopsis mutants too many mouths and four lips. Planta 205: 522–530 [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Lukowitz W, Brininstool G, Sedbrook JC, Hamann T, Poindexter P, Somerville C (2005) Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman JM, Lee JS, Bogenschutz NL, Peterson KM, Virata RE, Xie B, Kanaoka MM, Hong Z, Torii KU (2010) Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741 [DOI] [PubMed] [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Iglesias VA, Meins F Jr (2000) Movement of plant viruses is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J 21: 157–166 [DOI] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T. (2014) Enzymatic mechanism of GPI anchor attachment clarified. Cell Cycle 13: 1838–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RD, Sherman D, Ho WH, Stone D, Bennett GL, Moffat B, Vandlen R, Simmons L, Gu Q, Hongo JA, et al. (1997) A GPI-linked protein that interacts with Ret to form a candidate neurturin receptor. Nature 387: 717–721 [DOI] [PubMed] [Google Scholar]
- Kong D, Karve R, Willet A, Chen MK, Oden J, Shpak ED (2012) Regulation of plasmodesmatal permeability and stomatal patterning by the glycosyltransferase-like protein KOBITO1. Plant Physiol 159: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GHH, Lilley KS, Dupree P, Grossniklaus U, Twell D (2004) SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YCL, Putarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidich SD, Drapp DA, Orlean P (1994) A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J Biol Chem 269: 10193–10196 [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL (2007) A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ge FR, Xu M, Zhao XY, Huang GQ, Zhou LZ, Wang JG, Kombrink A, McCormick S, Zhang XS, et al. (2013) Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J 74: 486–497 [DOI] [PubMed] [Google Scholar]
- Lisanti MP, Caras IW, Davitz MA, Rodriguez-Boulan E (1989) A glycophospholipid membrane anchor acts as an apical targeting signal in polarized epithelial cells. J Cell Biol 109: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Kinoshita T (2011) Structural remodeling, trafficking and functions of glycosylphosphatidylinositol-anchored proteins. Prog Lipid Res 50: 411–424 [DOI] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S (2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Benghezal M, Imhof I, Conzelmann A (2000) Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry 39: 3461–3471 [DOI] [PubMed] [Google Scholar]
- Muniz M, Riezman C (2016) Trafficking of glycosylphosphatidyl inositol anchored proteins from the endoplasmic reticulum to the cell surface. J Lipid Res 57: 352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz M, Zurzolo C (2014) Sorting of GPI-anchored proteins from yeast to mammals: common pathways at different sites? J Cell Sci 127: 2793–2801 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nagamune K, Nozaki T, Maeda Y, Ohishi K, Fukuma T, Hara T, Schwarz RT, Sutterlin C, Brun R, Riezman H, et al. (2000) Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc Natl Acad Sci USA 97: 10336–10341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J (1999) Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab Invest 79: 293–299 [PubMed] [Google Scholar]
- Rinne PL, Welling A, Vahala J, Ripel L, Ruonala R, Kangasjärvi J, van der Schoot C (2011) Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-β-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23: 130–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. (1991) Stomata as an Example of Meristmoid Development. Pattern Formation in Plant Tissues. Cambridge University Press, Cambridge, UK, pp 101–117 [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Sugano SS, Hara-Nishimura I (2011) Positive and negative peptide signals control stomatal density. Cell Mol Life Sci 68: 2081–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED. (2013) Diverse roles of ERECTA family genes in plant development. J Integr Plant Biol 55: 1238–1250 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Lakeman MB, Torii KU (2003) Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ (2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten TS, Cambi A, Koopman M, Joosten B, Figdor CG, Garcia-Parajo MF (2009) Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci USA 106: 18557–18562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ (2008) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Sack FD (1995) The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C, Whitman M (2001) Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell 7: 949–957 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.