An extracellular proteolytic cascade involving two matrix metalloproteinases and the subtilisin-like proteinase P69B attenuates epidermal cell death as a prerequisite for tomato plant development.
Abstract
In contrast to mammalian matrix metalloproteinases (MMPs) that play important roles in the remodeling of the extracellular matrix in animals, the proteases responsible for dynamic modifications of the plant cell wall are largely unknown. A possible involvement of MMPs was addressed by cloning and functional characterization of Sl2-MMP and Sl3-MMP from tomato (Solanum lycopersicum). The two tomato MMPs were found to resemble mammalian homologs with respect to gelatinolytic activity, substrate preference for hydrophobic amino acids on both sides of the scissile bond, and catalytic properties. In transgenic tomato seedlings silenced for Sl2/3-MMP expression, necrotic lesions were observed at the base of the hypocotyl. Cell death initiated in the epidermis and proceeded to include outer cortical cell layers. In later developmental stages, necrosis spread, covering the entire stem and extending into the leaves of MMP-silenced plants. The subtilisin-like protease P69B was identified as a substrate of Sl2- and Sl3-MMP. P69B was shown to colocalize with Sl-MMPs in the apoplast of the tomato hypocotyl, it exhibited increased stability in transgenic plants silenced for Sl-MMP activity, and it was cleaved and inactivated by Sl-MMPs in vitro. The induction of cell death in Sl2/3-MMP-silenced plants depended on P69B, indicating that Sl2- and Sl3-MMP act upstream of P69B in an extracellular proteolytic cascade that contributes to the regulation of cell death in tomato.
A characteristic feature of plant cells is their cell wall, which determines the shape of the cell and the biomechanical properties of plant organs. It is composed largely of polysaccharides that form a structurally complex network consisting of cellulose microfibrils embedded in a matrix of hemicelluloses and pectins (Cosgrove, 2005). This polysaccharide frame accounts for about 90% of the cell wall dry weight and is reinforced by structural proteins, including Gly-, Pro-, and Hyp-rich (glyco)proteins (Albersheim et al., 2011). Above and beyond its structural role, the cell wall also is functionally important, as it is critically involved in growth and differentiation. It is reorganized and modified during cellular growth and organ development and ultimately may be disassembled during processes such as organ abscission and fruit ripening. The cell wall also is the interface for plant interactions with their biotic environment, as it is in the aqueous environment of this extracellular compartment, where plants first encounter their pathogens and symbionts (Hématy et al., 2009; Doehlemann and Hemetsberger, 2013; Bellincampi et al., 2014; Rich et al., 2014). For pathogens and symbionts alike, the cell wall can serve as a barrier, as a reservoir of signaling molecules, or as a source of nutrients. Cell wall modifications during biotic interactions range from reinforcement by callose-rich cell wall appositions at the attempted site of pathogen penetration to massive breakdown of the extracellular wall matrix (Hückelhoven, 2007; Underwood, 2012). Thus, although superficially rigid, the plant cell wall is not static but a highly dynamic structure. Extracellular enzymes are responsible for maintaining the physical and biological functions of the cell wall as well as cell wall dynamics. Indeed, hundreds of cell wall proteins have been identified, and many of them are enzymes potentially involved in cell wall remodeling, including enzymes acting on carbohydrates, peroxidases, and proteases (Chivasa et al., 2002; Bayer et al., 2006; Jamet et al., 2006; Albenne et al., 2013).
Similar to the cell wall in plants, the extracellular matrix in vertebrates is extensively remodeled in the course of tissue growth, organ development, and disease. The principal actors in these processes are enzymes belonging to the family of matrix metalloproteinases (MMPs; Nagase and Woessner, 1999; Lemaître and D’Armiento, 2006). An amphibian collagenase was discovered as the first MMP and is responsible for tissue lysis in the tail of a tadpole undergoing metamorphosis (Gross and Lapiere, 1962). Twenty-four MMPs have since been identified in vertebrates (23 in humans), the primary function of which has long been thought to be the degradation of the extracellular matrix as part of normal development and pathogenesis (Visse and Nagase, 2003; Lemaître and D’Armiento, 2006). More recently, however, MMPs also have been implicated in processes other than extracellular matrix turnover, including the regulation of other proteins by limited proteolysis, the mobilization of growth factors and cytokines, and the shedding of membrane-anchored proteins into circulation (Kessenbrock et al., 2010; Rodríguez et al., 2010).
Considering the prominent role of MMPs during tissue growth and remodeling in vertebrates, we addressed the question of whether MMPs may be similarly important for plant growth and development. MMP homologs are present in plant genomes, but our knowledge of MMP function is still rudimentary (Flinn, 2008; Marino and Funk, 2012). Sequence comparison with vertebrate MMPs revealed a domain architecture that is most similar to matrilysins (human MMP-7 and MMP-26), with which plant MMPs share a signal peptide for secretion, a prodomain containing a conserved Cys switch motif (PRCGXPD) responsible for the latency of the proenzyme, and the catalytic domain characterized by a conserved zinc-binding motif (HEXGHXXGXXH) at the active site. The hinge region and C-terminal hemopexin-like domain, typically present in all vertebrate MMPs but matrylisins, is not found in the plant enzymes (Flinn, 2008).
Plant MMPs have been described in several species, including soybean (Glycine max; Graham et al., 1991; McGeehan et al., 1992; Pak et al., 1997; Liu et al., 2001), cucumber (Cucumis sativus; Delorme et al., 2000), pine (Pinus spp.; Ratnaparkhe et al., 2009), Nicotiana benthamiana (Schiermeyer et al., 2009; Mandal et al., 2010), tomato (Solanum lycopersicum; Li et al., 2015), and Arabidopsis (Arabidopsis thaliana; Maidment et al., 1999; Golldack et al., 2002). Some of these enzymes have been characterized with respect to activity and specificity for small peptide substrates in vitro, but their physiological substrates and functions in vivo remain elusive (Flinn, 2008; Marino and Funk, 2012; Marino et al., 2014). Largely based on expression patterns, plant MMPs have been implicated in a variety of physiological processes, including seed development and germination (Ratnaparkhe et al., 2009), programmed cell death (Delorme et al., 2000), and pathogen defense (Liu et al., 2001; Frick and Schaller, 2002; Schiermeyer et al., 2009). In a few studies, the role of plant MMPs was addressed more directly by loss-of-function analysis. For At2-MMP, a pleiotropic role in Arabidopsis development was suggested by the late-flowering, early-senescence, and growth-inhibited phenotype of the corresponding transfer DNA insertion mutant (Golldack et al., 2002). Gene silencing by RNA interference (RNAi) revealed a role for Mt1-MMP during the symbiotic interaction of nitrogen-fixing rhizobia with Medicago truncatula (Combier et al., 2007). Li et al. (2015) used virus-induced gene silencing (VIGS) to down-regulate each of the five tomato MMP genes individually and found that one of them, Sl3-MMP, contributes to pathogen resistance. In neither case, however, was the physiological substrate identified or the specific function revealed.
We addressed these questions for two MMPs from tomato, Sl2-MMP and Sl3-MMP, which were found to act as negative regulators of epidermal cell death. P69B, an extracellular protease of the subtilase family, was identified as a major substrate of these two tomato MMPs. MMP-mediated regulation of cell death depended on P69B, indicating that tomato MMPs act upstream of P69B in an extracellular proteolytic cascade for cell death control.
RESULTS
Cloning of Sl2-MMP and Sl3-MMP
Based on ESTs that were identified in a database search at the Solanaceae Genomics Network (http://solgenomics.net/), we cloned two tomato MMPs (accession nos. HE819181 and HE819182). These clones later turned out to be identical to Sl2-MMP and Sl3-MMP that were identified by Li et al. (2015) as two out of five MMP genes in the most recent release of the tomato genome sequence (ITAG2.40). The Sl2- and Sl3-MMP genes encode proteins of 363 and 367 residues sharing 75% amino acid sequence identity (Supplemental Fig. S1). The typical preproprotein structure of MMPs was observed for both sequences, including predicted (http://www.cbs.dtu.dk/services/SignalP/) signal peptides of 22 (Sl2-MMP) and 21 (Sl3-MMP) amino acids, a prodomain with the conserved autoinhibitory Cys switch, and a catalytic domain comprising the HEXGHXXGXXH zinc-binding motif that is characteristic for the MMP family (Supplemental Fig. S1). A glycosylphosphatidylinositol (GPI) anchor for plasma membrane anchoring was predicted for both proteins (http://mendel.imp.ac.at/sat/gpi/gpi_server.html). A potential ω-site for GPI-anchor attachment (Eisenhaber et al., 2003; Schultz et al., 2004) was observed close to their C termini as well as the hydrophobic, membrane-spanning region, which attaches the immature proteins to the membrane until it is replaced by the GPI anchor during passage through the secretory pathway (Supplemental Fig. S1).
Subcellular Localization of Sl2- and Sl3-MMP
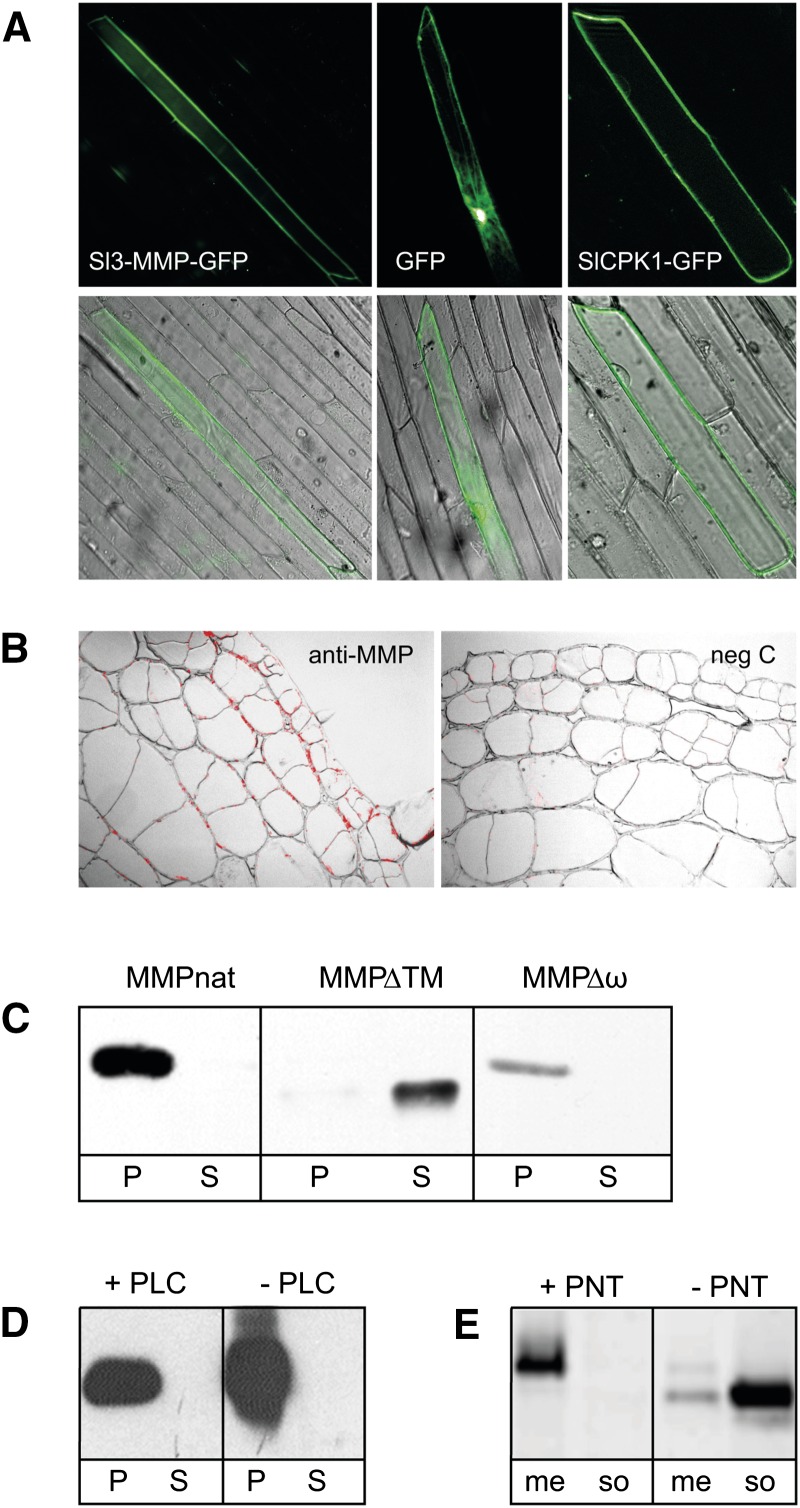
Li et al. (2015) reported plasma membrane localization of Sl3-MMP and attributed it to the presence of a C-terminal transmembrane domain rather than a GPI anchor. Therefore, we readdressed the subcellular localization of Sl3-MMP and particularly the mode of membrane anchoring. A GFP fusion construct was generated, in which the catalytic domain of Sl3-MMP was replaced by GFP, while the predicted N- and C-terminal targeting sequences were retained. The fusion construct was transiently expressed in onion (Allium cepa) epidermal cells, and its subcellular localization was analyzed by fluorescence microscopy (Fig. 1A). GFP fluorescence was detected at the cell periphery. Comparison with a bona fide plasma membrane protein and with GFP as a cytoplasmic control suggested plasma membrane localization of the Sl3-MMP-GFP fusion protein (Fig. 1A). In a similar experiment, Li et al. (2015) observed plasma membrane localization of an Sl3-MMP-GFP fusion before and after plasmolysis, thus excluding the possibility that Sl3-MMP is secreted into the apoplast. Consistent with this finding, a polyclonal antiserum directed against Sl2- and Sl3-MMP specifically labeled the circumference of epidermal and cortical cells in tissue sections of tomato hypocotyls (Fig. 1B).
Figure 1.
Subcellular localization of Sl2- and Sl3-MMP. A, Plasma membrane targeting of Sl3-MMP-GFP. The catalytic domain of Sl3-MMP was replaced by GFP, and the fusion construct was transiently expressed in onion epidermal cells (left). Subcellular localization was analyzed by fluorescence microscopy and compared with a cytoplasmic control (GFP without any additional targeting signals; center) and a bona fide plasma membrane protein (SlCPK1; right; Rutschmann et al., 2002). Top and bottom rows show the signal in the green fluorescence channel (excitation wavelength [λex], 470/20 nm; emission wavelength [λem], 525/50 nm) alone and in overlay with the corresponding differential interference contrast image, respectively. B, Immunolocalization of Sl2/3-MMPs. Sl-MMPs were detected histochemically in cross sections (5 µm) of tomato hypocotyls using the IgG fraction of a polyclonal anti-Sl3-MMP serum and Alexa Fluor 568-labeled goat anti-rabbit IgG as the primary and secondary antibodies, respectively (left). The IgG fraction from which Sl3-MMP antibodies had specifically been depleted was used in the negative control (right). The red fluorescence signal (λex, 550/25 nm; λem, 605/70 nm) is shown in an overlay with the corresponding bright-field images (100×). C, Sl-MMP targeting in transgenic cell cultures. The cell pellet (P) and culture supernatant (S) from cell cultures expressing full-length Sl3-MMP (MMPnat), a truncated protein lacking the C-terminal membrane-spanning region (MMPΔTM), and a site-directed mutant lacking the ω site (MMPΔω) were separated by centrifugation and analyzed on western blots using a polyclonal anti-Sl3-MMP serum. Western blots were developed by enhanced chemiluminescence. D, PI-PLC insensitivity of Sl3-MMP. PI-PLC-treated (+PLC) and untreated (−PLC) MMPnat-expressing cell cultures were centrifuged to obtain the cell pellet (P) and culture supernatant (S), which were analyzed on western blots for the presence/absence of Sl3-MMP. E, Shedding of Sl-MMPs. Protein was extracted from the MMPnat-expressing cell culture in the presence (+PNT) or absence (−PNT) of 1,10-phenanthroline, then extracts were separated by ultracentrifugation into membrane (me) and soluble (so) fractions and analyzed on western blots for the presence of Sl3-MMP.
To further confirm membrane localization and to obtain evidence for GPI anchoring, we generated transgenic tomato suspension cell cultures stably expressing full-length Sl3-MMP (MMPnat), MMPΔω, in which the ω-site for GPI anchor attachment had been destroyed by site-directed mutagenesis, and MMPΔTM, in which the C-terminal hydrophobic region had been deleted. After centrifugation, MMPnat and MMPΔω were found in the cell sediment, while MMPΔTM was located in the culture medium (Fig. 1C). Similarly, in leaf extracts from transgenic plants expressing the same MMP constructs, MMPnat and MMPΔω were associated with membranes, while MMPΔTM was contained in the soluble fraction (Supplemental Fig. S2). These data indicate that MMPΔTM is targeted to the secretory pathway and secreted, while MMPnat and MMPΔω remain membrane associated.
Sensitivity to bacterial phosphoinositol-specific phospholipase C (PI-PLC) is frequently used as a diagnostic tool for GPI anchoring (Varma and Hendrickson, 2010). Treatment with PI-PLC did not release MMPnat from membranes, thus failing to provide direct evidence for GPI anchoring (Fig. 1D). This negative result, however, did not preclude GPI anchoring, since PI-PLC insensitivity, as a result of specific modifications of the inositol moiety, has been reported previously for GPI-anchored proteins in plants and other organisms (Morita et al., 1996; Ferguson, 1999; Coppinger et al., 2004). Interestingly, despite its insensitivity to bacterial PI-PLC, Sl3-MMP was released from the membrane by an endogenous shedding activity, thus providing indirect evidence for GPI anchoring. Shedding was inhibited by 1,10-phenanthroline, a chelator of divalent metal ions (Fig. 1E). A similar metal ion-dependent shedding activity releasing GPI-anchored arabinogalactan proteins was observed in membrane vesicles from rose (Rosa spp.) cells and was attributed to an endogenous phospholipase activity (Svetek et al., 1999). Cleavage of the GPI anchor by endogenous (glycosyl)phosphatidylinositol-specific phospholipases C or D, which are known to depend on divalent cations in other systems, is thus likely to be responsible for the shedding of both Sl3-MMP and arabinogalactan proteins (Bütikofer and Brodbeck, 1993; Li et al., 1994; Williams and Katan, 1996; Oxley and Bacic, 1999).
Catalytic Properties of Sl2- and Sl3-MMP
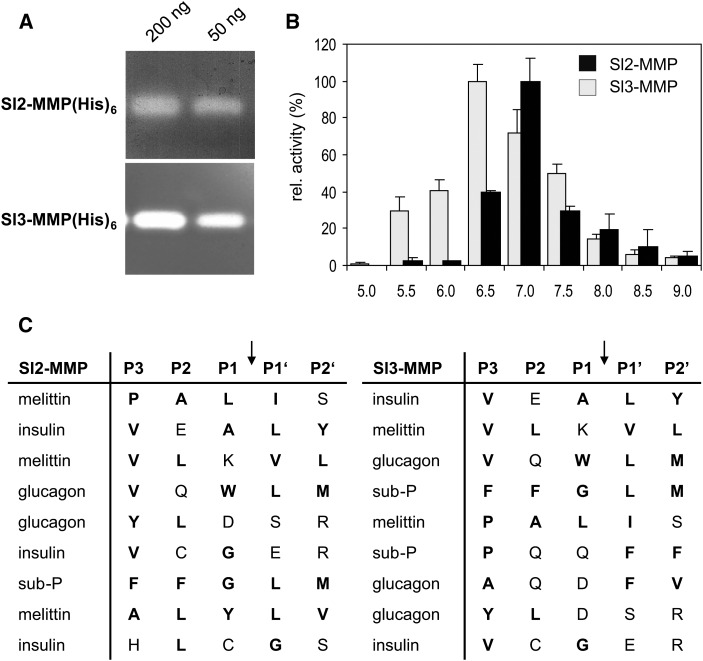
The two MMPs were obtained as recombinant His-tagged proteins after heterologous expression in Escherichia coli (Supplemental Fig. S3). Both recombinant proteins were found to be catalytically active and exhibited gelatinolytic activity (Fig. 2). To address the substrate preference of Sl2- and Sl3-MMP(His)6, we offered a variety of synthetic peptides as substrates and analyzed the cleavage products by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF)-mass spectrometry (MS). Of the six peptides tested, four were cleaved by both MMPs (oxidized insulin B chain, mellitin, glucagon, and substance P; no cleavage was observed for systemin and neurotensin). The alignment and ranking of the observed cleavage sites (Fig. 2C) revealed a similar preference of the two MMPs for hydrophobic amino acids on both sides of the scissile bond, particularly in the P1′ and P3 positions (i.e. the residues immediately downstream and three amino acids upstream of the cleavage site; Schechter and Berger, 1967). Pro also was frequent in P3 (Fig. 2C). The observed specificity resembles that of Arabidopsis MMP1 to MMP4 (Marino et al., 2014) and of mammalian MMPs, which require a large hydrophobic residue (typically Leu or Ile) in P1′ (Visse and Nagase, 2003; Schilling and Overall, 2008). We thus chose a fluorogenic peptide substrate comprising the scissile bond of collagen (Leu in P1′ and Pro in P3; Neumann et al., 2004) to analyze the steady-state kinetic properties of tomato MMPs. The activity of Sl2-MMP(His)6 and Sl3-MMP(His)6 exhibited maxima at pH 7 and 6.5, respectively (Fig. 2B), and was found to be stimulated by Ca2+ up to a concentration of 0.5 mm. Apparent Km values of 19.9 and 19.5 µm and catalytic efficiencies (turnover number [kcat]/Km) of 2.4 and 1 × 104 m−1 s−1 were determined for Sl2-MMP(His)6 and Sl3-MMP(His)6, respectively. Thus, the biochemical properties of Sl-MMP1(His)6 and Sl-MMP2(His)6 are very similar to each other, and they also resemble mammalian MMPs with respect to gelatinolytic activity, stimulation by calcium, substrate preference, and catalytic constants (Stracke et al., 2000; Neumann et al., 2004; Schilling and Overall, 2008). As the only difference between the activities of the two MMPs, Sl2-MMP exhibited a slightly more alkaline pH optimum, suggesting that the contribution of Sl2-MMP to total MMP activity may increase when the extracellular milieu becomes more alkaline (e.g. under stress conditions).
Figure 2.
Catalytic properties of Sl2- and Sl3-MMP. A, Gelatinolytic activity of Sl-MMPs. Zymograms of recombinant His6-tagged Sl2- and Sl3-MMP (50 and 200 ng) on gelatin-containing polyacrylamide gels are shown. B, pH optima of Sl2- and Sl3-MMP. The activity of the recombinant His6-tagged proteins was analyzed using a fluorigenic peptide substrate in a continuous assay under steady-state conditions, and the results are shown in percentage of the maximum (7.4 pmol min−1 at pH 7 for Sl2-MMP [black bars] and 7.3 pmol min−1 at pH 6.5 for Sl2-MMP [gray bars]). Data represent means of three independent experiments ± sd. C, Substrate preferences of Sl2- and Sl3-MMP. The recombinant His6-tagged proteins were incubated with synthetic peptide substrates, and the cleavage products were analyzed by MALDI-TOF-MS. The cleaved amino acid sequences were aligned with respect to the cleavage site (arrows) and ranked according to the rate of cleavage. Hydrophobic and nonpolar amino acid residues are highlighted in boldface. The full-length peptide sequences are given in Supplemental Methods S1.
Silencing of Sl2- and Sl3-MMP by RNAi
Considering the highly similar biochemical characteristics and their partly overlapping expression patterns in roots, hypocotyls, stems, leaves, and flowers of tomato plants (Supplemental Fig. S4), Sl2- and Sl3-MMP are likely to serve overlapping and possibly redundant functions. Therefore, in order to reveal any loss-of-function phenotype that may not be apparent when the two MMPs are silenced individually (Li et al., 2015), we used RNAi as an approach to achieve posttranscriptional silencing of both MMPs simultaneously (Wesley et al., 2001). A hairpin (HP) construct comprising 490 bp from the 5′ end of the Sl3-MMP complementary DNA (cDNA) with an overall sequence identity of 79% with Sl2-MMP was cloned in the sense and antisense orientations under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter and stably introduced into the tomato genome by Agrobacterium tumefaciens-mediated transformation. Three independent transformants were chosen for further analysis (HP1, HP2, and HP4). Expression levels were analyzed in the homozygous HP lines in Figure 3, confirming the silencing of Sl2- and Sl3-MMP in tomato hypocotyls and strong down-regulation in leaves. The Sl2-MMP transcript was barely detectable in leaf tissue, while residual transcript of Sl3-MMP was still present in leaves but not in hypocotyls of the transgenic lines (Fig. 3A). At the protein level, Sl2- and Sl3-MMP were undetectable in leaves of silenced tomato plants (Fig. 3B).
Figure 3.
Silencing of Sl2/3-MMP expression by RNAi. A, Semiquantitative reverse transcription (RT)-PCR analysis of Sl2-MMP and Sl3-MMP expression in hypocotyls and leaves of 24-d-old tomato wild-type seedlings (WT) and three independent transgenic lines harboring an HP construct for posttranscriptional gene silencing (HP1, HP2, and HP4). After RT, cDNAs of Sl2- and Sl3-MMP and the actin control were detected by 35 and 30 cycles of PCR, respectively. Tomato genomic DNA was used as a control (C). B, Phenotypes and Sl-MMP protein levels of RNAi plants. At top, a 2-week-old tomato plant silenced for Sl-MMP expression is shown on the left, next to a wild-type seedling on the right. Ten plants of the segregating F1 progeny of a primary RNAi transformant were analyzed for the presence of the silencing construct by PCR amplification of the nptII antibiotic resistance marker (middle) and Sl2/3-MMP expression by western-blot analysis (bottom). The seedling phenotype (necrotic hypocotyl) was found to cosegregate with the silencing construct and the loss of Sl2/3-MMP protein expression.
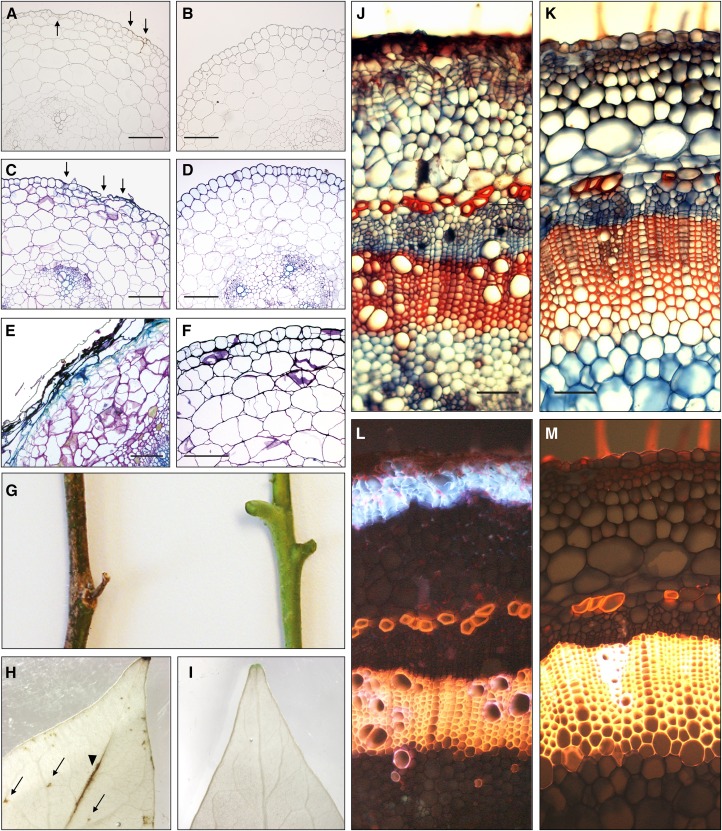
HP lines did not show any difference in germination or early seedling establishment until necrotic lesions became apparent at the base of the hypocotyl (Fig. 3B). In the segregating progeny of the primary transformants, this phenotype cosegregated with both the presence of the silencing construct and the loss of Sl2/3-MMP expression (Fig. 3B). Closer inspection of the Sl2/3-MMP loss-of-function phenotype in cross sections of tomato hypocotyls revealed that cell death initiated in the epidermis and then progressed to include outer cortical cell layers (Fig. 4, A–F). These are the tissues where Sl-MMPs were detected immunohistochemically (Fig. 1B), thus linking the cell death phenotype to the loss of MMP function and suggesting a role for these proteases in cell death control. The remaining stem cortex appeared disorganized in HP lines compared with wild-type plants, with cells being irregular in size and shape (Fig. 4E). During later developmental stages, necrosis spread. In fully grown tomato plants, it covered the entire stem and also extended to the leaves (Fig. 4, G–M), where lesions were first visible at the epidermis over the leaf veins, and were later detected as distinct spots within the leaf blade (Fig. 4H). Despite extensive spontaneous cell death, HP lines survived to flower, bear fruit, and set seed. The recovery of MMP-silenced plants involved a massive wound-healing response resulting in the formation of a secondary dermal tissue (wound periderm) to replace the lost epidermis. Periderm formation was initiated in the stem cortex below the regions of cell death, resulting in columnar files of cork cells (Fig. 4J) with highly fluorescent cell walls indicating the accumulation of phenolic material and suberin incorporation (Fig. 4L; Supplemental Fig. S5). The data show that MMPs are required for the normal development of tomato plants, particularly of the stem epidermis and cortex, and they further suggest that Sl2/3-MMP expression in the wild type suppresses the onset of spontaneous cell death.
Figure 4.
Sl2/3-MMP loss-of-function phenotypes. A to F, Hypocotyl cross sections (5 µm; 0.5 cm above the crown) of Sl2/3-MMP-silenced plants (A, C, and E) and wild-type plants (B, D, and F). The onset of cell death is indicated by arrows in unstained (A versus B) and Toluidine Blue-stained (C versus D) sections of 2-week-old seedlings. Deterioration of the epidermis, loss of cellular organization in the stem cortex, and accumulation of phenolic material as visualized by Toluidine Blue staining are apparent in 4-week-old seedlings (E versus F). Bars = 200 µm. G to I, Sl2/3-MMP loss-of-function phenotypes in fully grown tomato plants. G, Stem segments of 2-month-old HP and wild-type plants. H and I, Cell death in leaves. Destaining in boiling ethanol reveals necrotic lesions on top of the main vein (arrowhead) and within the leaf blade (arrows) in leaflets of silenced plants (H) but not in the wild type (I). J to M Formation of a secondary dermal tissue (wound periderm) in stems of fully developed tomato plants silenced for Sl2/3-MMP expression (J and L) compared with wild-type controls (K and M). The periderm is visible as distinct files of cells at the periphery of the stem section (hand cut, stained with safranin/Alcian Blue; J). In the same section, blue autofluorescence (excitation at 365 nm) indicates the accumulation of phenolic compounds in cell walls of suberized peridermal cork cells (L). Bars = 100 µm.
Extracellular Substrates of Sl2- and Sl3-MMPs
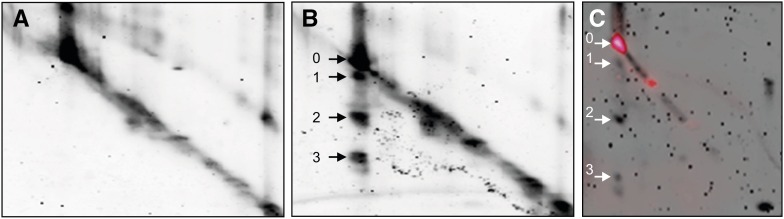
To narrow down the specific function in cell death control, we aimed to identify the substrate(s) of Sl2/3-MMP. Reasoning that potential substrates are likely to be more abundant in plants lacking the cognate protease, apoplastic proteins were isolated from HP lines and tested as substrates of Sl2/3-MMP in a two-dimensional electrophoretic mobility shift assay (2D-EMSA). Apoplastic proteins were separated in a single lane by SDS-PAGE, and the lane was excised and in-gel digested with recombinant Sl2- or Sl3-MMP(His)6. The gel strip was then subjected to SDS-PAGE in the second dimension, and cleaved proteins were identified by their altered electrophoretic mobility (Fig. 5, A and B; Supplemental Fig. S6). Major degradation products (arrows 1–3 in Fig. 5B; arrows 2.1–2.3 in Supplemental Fig. S6) were excised from the gel, digested with trypsin, and subjected to MS analysis. Peptide fingerprints identified the subtilisin-like tomato protease (subtilase) P69B as the major degradation product in each of the three spots (Supplemental Figs. S6 and S7). Fragments of α-l-arabinofuranosidase/β-d-xylosidase and α-mannosidase were detected as less abundant degradation products of Sl3-MMP in spots 2 and 3 (Fig. 5B).
Figure 5.
2D-EMSA for the identification of substrates of Sl2- and Sl3-MMP. Apoplastic proteins were isolated from leaves of 5-week-old transgenic tomato plants silenced for Sl2/3-MMP expression and separated by SDS-PAGE. The lanes were excised, infiltrated with recombinant Sl3-MMP(His)6 in reaction buffer (B) or buffer alone (A), incubated overnight, and subjected to SDS-PAGE in the second dimension. Fluorescence (TOPLAB Ruby) staining revealed undigested proteins in the diagonal and proteins cleaved by Sl3-MMP(His)6 with increased electrophoretic mobility below (spots 1–3). Equivalent results were obtained after in-gel digestion with Sl2-MMP(His)6 (Supplemental Fig. S6). Sl2/3-MMP cleavage products (spots 1–3) and the corresponding undigested protein (spot 0) were excised and subjected to peptide fingerprint analysis by tryptic digestion and MALDI-TOF-MS (Supplemental Fig. S7). A, Buffer-infiltrated control. B, Digestion of apoplastic proteins by Sl3-MMP(His)6. C, Zymography reveals proteolytic activity for spot 0 but not for spots 1 to 3.
In order to identify the MMP cleavage site(s) in P69B, the N termini of two abundant P69B fragments (spots 2 and 3 in Fig. 5B) were determined by Edman degradation. Cleavage occurred at Phe-482, between the His and Ser residues of the catalytic triad (Supplemental Fig. S7B). Cleavage at this position thus is expected to inactivate P69B, which was confirmed by zymography (Fig. 5C). The activity of intact P69B (and two other apoplastic proteases) was detected on the diagonal line formed by uncleaved proteins, but not for the P69B fragments (spots 1–3 in Fig. 5C). The observation that residual uncleaved P69B was catalytically active (spot 0 in Fig. 5C) indicates that the protein retained its native conformation after SDS-PAGE. Therefore, Sl2-MMP(His)6 and Sl3-MMP(His)6 appear to act upon native P69B rather than the denatured polypeptide chain. To further support this notion, we transiently expressed (His)6-tagged P69B in N. benthamiana leaves. Recombinant P69B(His)6 was purified under native conditions and found to be a substrate of Sl3-MMP(His)6 in vitro (Supplemental Fig. S8). In the following, we addressed the question of whether P69B also may be a substrate of tomato MMPs in vivo. If P69B were a physiological substrate, it would be expected to accumulate in cell walls of plants silenced for Sl2/3-MMP expression.
Comparative Proteomics of Wild-Type and MMP-Silenced Hypocotyls
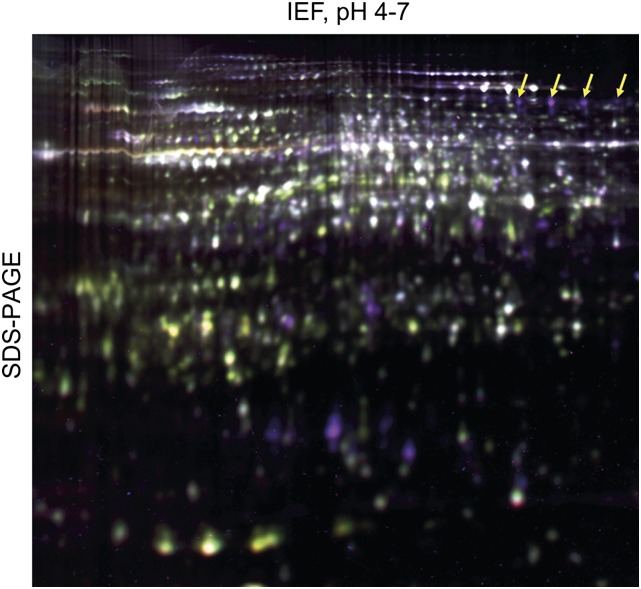
To compare relative protein abundance in tomato wild-type plants and HP lines, we used two-dimensional (2D)-difference gel electrophoresis (DIGE) in a proteomics approach. Protein extracts from hypocotyls of wild-type plants and HP lines (four experiments involving three independent HP lines) were differentially labeled with Cy3 and Cy5 fluorescent dyes, spiked with a Cy2-labeled internal standard, and separated by 2D electrophoresis (a representative gel is shown in Fig. 6). A total of 1,132 protein spots were detected on all four gels, 124 of which showed greater than 2-fold differential expression (significant at P < 0.01 and a false discovery rate < 5%; Supplemental Fig. S9). Fifty-six of these spots were identified by MS. They represented 38 different proteins, including 20 that were more abundant in HP lines compared with the wild type (Table I) and 18 less abundant proteins (Supplemental Table S1). P69B was identified in seven of the protein spots that were significantly stronger in HP lines than in the wild type (the four most abundant ones are indicated in Fig. 6). Multiple isoforms, as observed for P69B and other differentially expressed proteins, are likely to result from posttranslational modification. Tomato subtilases including P69B are in fact known to be modified posttranslationally by N-linked glycosylation at multiple sites (Bykova et al., 2006; Cedzich et al., 2009).
Figure 6.
2D-DIGE comparison of hypocotyl proteomes from wild-type plants and HP lines. Protein extracts (50 µg) of hypocotyls from 4-week old plants silenced for Sl2/3-MMP expression and wild-type tomato were differentially labeled with Cy3 and Cy5. After the addition of Cy2-labeled internal standard (an aliquot of pooled samples of all extracts in the DIGE experiment), proteins were separated by isoelectric focusing (IEF; pH 4–7) in the first dimension and SDS-PAGE in the second. Fluorescence signals were scanned in all three channels (Typhoon Trio Imager), and an overlay of the three images is shown. Up- and down-regulated proteins appear as purple and green spots, respectively; proteins of equal abundance are shown in white. Arrows indicate the position of the four major spots identified as P69B by MS.
Table I. Proteins showing higher abundance in HP compared with wild-type plants.
After preparative 2D electrophoresis, differentially expressed proteins were identified by electrospray ionization-MS. Spot identifiers correspond to those indicated in Supplemental Figure S9. Expression ratio is given as the ratio (HP to wild type) of protein abundance as determined by 2D-DIGE analysis (Fig. 6). For each spot, the identity of the protein(s) it contained is indicated by the locus identifier(s) in the tomato genome (http://solgenomics.net/organism/Solanum_lycopersicum/genome) and the corresponding annotation. Protein identifications were accepted only if they could be established at greater than 99% probability and contained at least two peptides, each identified at greater than 95% probability (for details, see Supplemental Methods S1). If multiple spots were identified as isoforms of the same protein (spot count > 1), the given spot identifier corresponds to the spot with the largest difference in expression. Proteins that were less abundant in HP lines compared with wild-type plants are shown in Supplemental Table S1.
| Spot Identifier | Spot Count | Ratio | Locus | Solanaceae Genomics Network Annotation |
|---|---|---|---|---|
| 6006 | 7 | 57.1 | Solyc08g079870 | Subtilisin-like protease (P69B) |
| 5656 | 1 | 28.5 | Solyc08g008210 | V-type proton ATPase subunit E |
| Solyc06g081980 | Pyridoxyl biosynthesis lyase pdx6 | |||
| 5826 | 2 | 12.7 | Solyc09g090980 | Major allergen Mal d 1 (PR-10) |
| 5671 | 1 | 9.6 | Solyc10g055810 | Endochitinase (PR-3) |
| 5827 | 1 | 7.3 | Solyc01g079820 | Redoxin domain protein |
| 5816 | 1 | 6.1 | Solyc01g100030 | Deoxyuridine 5′-triphosphate nucleotide hydrolase (dUTPase) |
| 5644 | 2 | 5.2 | Solyc07g051850 | Aspartic proteinase (cyprosin-like) |
| 5909 | 1 | 4.3 | Solyc00g174340 | Pathogenesis-related protein 1b (PR-1b) |
| 5642 | 1 | 3.6 | Solyc08g080140 | dTDP-4-dehydrorhamnose reductase |
| 6015 | 1 | 3.4 | Solyc11g069270 | β-Galactosidase |
| 5731 | 1 | 3.2 | Solyc01g088480 | Adenylate kinase |
| 5587 | 1 | 3.2 | Solyc05g052600 | Fru-1,6-bisphosphatase class 1 |
| 6025 | 2 | 3.1 | Solyc06g073190 | Fructokinase |
| Solyc09g082060 | Cys synthase | |||
| 6183 | 5 | 3.0 | Solyc07g052350 | 3-Isopropylmalate dehydratase large subunit (aconitate hydratase) |
| 5273 | 1 | 2.8 | Solyc05g050970 | Transketolase1 |
| 5271 | 1 | 2.5 | Solyc08g079170 | Stress-induced protein, sti1-like protein |
| 5696 | 1 | 2.3 | Solyc03g025850 | Remorin1 |
| 6040 | 1 | 2.1 | Solyc05g054760 | Dehydroascorbate reductase |
| 5593 | 1 | 2.1 | Solyc05g014470 | Glycerinaldehyde-3-phosphate dehydrogenase |
| 6144 | 1 | 2.0 | Solyc03g123830 | Phosphoglycerate dehydrogenase |
| Solyc04g045340 | Phosphoglucomutase |
Other proteins that differed in abundance between hypocotyls of HP lines and the wild type included many that are involved in defense responses and in sugar metabolism (Table I). Up-regulation of these proteins is likely to be a consequence of cell death in the epidermis and stem cortical tissues and the subsequent formation of a wound periderm (Ginzberg, 2008). Defense proteins are abundant in the proteomes of oak (Quercus spp.) and potato (Solanum tuberosum) periderm, where they contribute to the protection provided by the periderm against pathogen invasion (Barel and Ginzberg, 2008; Chaves et al., 2009; Ricardo et al., 2011). Consistently, several pathogenesis-related (PR) defense proteins showed enhanced levels in HP lines compared with the wild type, including PR-1b (4.3-fold), PR-3 (9.6-fold), and PR-10 (12.7-fold; Table I). The accumulation of proteins involved in carbohydrate metabolism (trans-ketolase, fructokinase, Fru-1,6-bisphosphatase, glycerinaldehyde 3-phosphate dehydrogenase, phosphoglycerate dehydrogenase, phosphoglucomutase, and aconitase) likely reflects changes in energy supply compensating for the loss of photosynthetically active tissue as a result of cell death in the stem cortex (Fig. 4, J and L; Supplemental Fig. S5). Consistent with the reduced number of chloroplasts in the stem cortex (Supplemental Fig. S5), photosynthetic proteins (chlorophyll a/b-binding protein and Rubisco large and small subunit) were frequent among those found to be depleted in HP lines (Supplemental Table S1).
P69B was the protein that showed the strongest up-regulation in MMP-silenced plants. The most prominent isoform of P69B was 57-fold more abundant in HP lines compared with wild-type plants (Table I). Increased abundance of P69B in hypocotyls of MMP-silenced compared with wild-type plants also was confirmed by immunoblot analysis using an antiserum directed against a P69B peptide (Supplemental Fig. S10) P69B transcript levels, on the other hand, did not differ between wild-type and MMP-silenced plants (Supplemental Fig. S11), indicating that increased stability of the protein rather than up-regulation at the transcriptional level accounts for the accumulation of P69B. Increased stability of the P69B protein in HP lines is fully consistent with P69B being a substrate of Sl2- and Sl3-MMP not only in vitro but also in vivo. α-l-Arabinofuranosidase/β-d-xylosidase and α-mannosidase, which were identified as minor cleavage products of Sl3-MMP in vitro (Fig. 5), were not among the proteins that accumulated in MMP-silenced plants (Table I) and thus are unlikely to be MMP substrates in vivo.
Accumulation of P69B in Sl2/3-MMP-Silenced Plants Facilitates Cell Death Induction
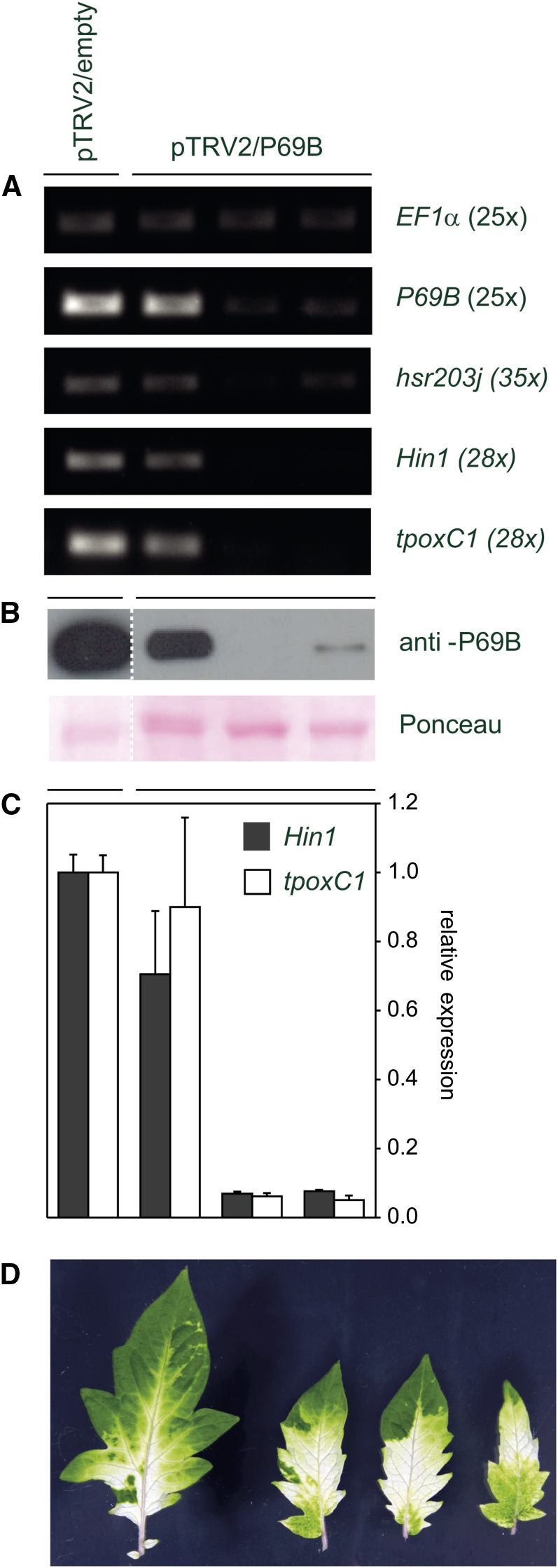
In order to address the question of whether there is a causal link between the increased stability of P69B and the induction of cell death, we used VIGS to suppress the expression of P69B in Sl2/3-MMP-deficient plants and monitored the expression of three well-established cell death markers, Hin1, hsr203J, and tpoxC1 (Pontier et al., 1994, 1999; Gopalan et al., 1996; Hiraga et al., 2000; Rivas et al., 2004). Cell death markers were expressed in Sl2/3-MMP-deficient control plants that were mock infiltrated with the empty silencing vector. Infiltration of the P69B silencing construct led to a considerable reduction of P69B transcript (Fig. 7A) and protein levels (Fig. 7B) compared with mock-infiltrated controls. Residual protein levels were to be expected because VIGS is never complete, as indicated by the variegated phenotype that is commonly observed in controls silenced for phytoene desaturase (Fig. 7D; Liu et al., 2002a, 2002b). Importantly, the reduction in P69B transcript and protein levels resulted in a concomitant reduction in the expression of cell death markers (Fig. 7, A and C). These data indicate that the expression of cell death markers and, by inference, the cell death phenotype of Sl2/3-MMP-deficient HP plants depend on P69B, and they support the conclusion that the accumulation of P69B in Sl2/3-MMP-silenced plants is responsible at least in part for the onset of cell death.
Figure 7.
Expression of cell death markers in Sl2/3-MMP-silenced plants depends on P69B. Sl2/3-MMP RNAi plants were infiltrated with A. tumefaciens carrying a VIGS construct for silencing of P69B (pTRV2/P69B) or an empty vector control (pTRV2/empty). A, Expression of cell death markers correlates with P69B expression. The expression of P69B and the cell death markers hsr203j, Hin1, and tpoxC1 was analyzed by RT-PCR in three pTRV2/P69B-infiltrated plants and the empty vector control. B, Western-blot analysis of the VIGS plants shown in A. The expression of P69B was analyzed in pTRV2/P69B-infiltrated and control plants using a polyclonal antiserum specific for P69B. The membrane was stained with Ponceau Red as a control of protein loading and transfer. C, Expression of cell death markers depends on P69B. The expression of Hin1 and tpoxC1 was analyzed by quantitative PCR in three pTRV2/P69B-infiltrated plants and is given as fractions of the empty vector control. Values represent means of two replicates ± sd. D, Silencing of phytoene desaturase. Control plants were infiltrated with pTRV2/PDS. Incomplete silencing of phytoene desaturase is visualized by the variegated phenotype of individual leaflets of pTRV2/PDS-infiltrated plants.
DISCUSSION
The earliest consequences of Sl2- and Sl3-MMP loss of function we observed were isolated events of cell death in the epidermis of tomato hypocotyls silenced for MMP expression by RNAi (Fig. 4, A and C). Cell death then spread to include the entire epidermis and cells of the outer stem cortex (Fig. 4E), precisely the tissues in which immunolocalization detected Sl2/3-MMPs in wild-type plants (Fig. 1B). Subsequent phenotypic changes included the formation of a secondary dermal tissue, with cork cells in columnar rows (Fig. 4J), autofluorescent cell walls likely due to the incorporation of aromatic suberin polymers (Fig. 4L; Supplemental Fig. S5), and the accumulation of defense proteins (Table I). These are hallmarks of the wound periderm, formation of which is typically initiated to replace the epidermis when the latter is damaged (Barel and Ginzberg, 2008; Ginzberg, 2008; Chaves et al., 2009). Thus, wound periderm formation may be viewed as a consequence of the cell death phenotype of Sl2/3-MMP-silenced plants and, therefore, is unlikely to be directly linked to a reduction in Sl2/3-MMP activity. Epidermal cell death, on the other hand, is viewed as a direct result of Sl2/3-MMP silencing and implies Sl2- and Sl3-MMP activity in cell death control.
The function of Sl2- and Sl3-MMP was addressed recently by others using gene-specific VIGS to down-regulate the expression of each of the two genes individually (Li et al., 2015). The down-regulation of Sl3-MMP impaired resistance against fungal (Botrytis cinerea) and bacterial (Pseudomonas syringae pv tomato) pathogens, indicating that there is no redundancy between Sl2- and Sl3-MMP with respect to their role in plant defense. On the other hand, spontaneous cell death was not reported for either Sl2- or Sl3-MMP-silenced plants (Li et al., 2015), suggesting that the two MMPs act redundantly with respect to cell death control. Overlapping and partially redundant functions for Sl2- and Sl3-MMP are consistent with our biochemical data, including similar steady-state enzyme kinetics, similar substrate specificity, and cleavage of P69B by both proteases.
Proteases that were shown previously to play a role in the control of plant cell death include enzymes from two of the major catalytic classes, namely saspases (Coffeen and Wolpert, 2004) and phytaspases (Chichkova et al., 2010) among the Ser peptidases and metacaspases (Coll et al., 2010) and vacuolar processing enzyme (Hatsugai et al., 2004; Rojo et al., 2004) as representatives of the Cys peptidases. The data presented here for Sl2- and Sl3-MMP now implicate metalloproteinases in the regulation of plant cell death as well. To narrow down their specific functions, we first characterized Sl2- and Sl3-MMP at the biochemical level in vitro. The enzymatic properties of the two tomato MMPs resembled those of mammalian homologs, particularly with respect to their specificity for hydrophobic amino acids on the carboxy side of the hydrolyzed bond (i.e. the P1′ position). The P1′ residue of the substrate is accommodated by the S1′ substrate-binding pocket of the enzyme, which is particularly important for substrate selectivity in mammalian MMPs (Overall and Kleifeld, 2006; Tallant et al., 2010). Structural modeling indicated that this also is the case in Arabidopsis (Marino et al., 2014). The S1′ pocket of the five Arabidopsis MMPs was modeled as a deep hydrophobic channel, matching the substrate specificity observed for these enzymes, which also is characterized by the preference for large hydrophobic residues in P1′ (Marino et al., 2014). The second most important position with respect to cleavage site selection appears to be P3, which is occupied by Pro or small hydrophobic residues (Val and Ala) in four of the five Arabidopsis MMPs (Marino et al., 2014) and also in the two tomato MMPs (Fig. 2C).
While biochemical characterization and cleavage site identification are important first steps toward the identification of potential substrates and the physiological role of a protease, predicting substrates from consensus cleavage sites is not yet a reliable approach, as it neglects the potential contribution of exosites to substrate recognition and the requirement of physical colocalization of a protease with its substrate in the same cellular compartment (Overall and Blobel, 2007). Therefore, we used an EMSA as a more direct approach to identify extracellular tomato proteins that are cleaved by recombinant MMPs in vitro, revealing subtilase P69B, α-l-arabinofuranosidase/β-d-xylosidase, and α-mannosidase as potential substrates. Of these three proteins, only P69B exhibited increased stability in MMP-silenced plants and, thus, is supported as a physiologically relevant substrate of Sl2- and Sl3-MMP in vivo.
We further showed that the induction of cell death markers in Sl2/3-MMP-deficient plants is attenuated upon silencing of P69B, indicating that Sl-MMPs act upstream of P69B in the context of cell death control. This observation suggests that the accumulation of P69B in Sl2/3-MMP-silenced plants is at least in part responsible for the onset of cell death, implying a cell death-promoting, proapoptotic activity for P69B. Such an activity would be consistent with the apparent involvement of P69B in plant defense against biotrophic pathogens. P69B is one of two closely related tomato subtilases that are induced in tomato plants in response to biotrophic pathogens (citrus exocortis viroid, Phytophthora infestans, and P. syringae) and regulated by the defense hormone salicylic acid (Fischer et al., 1989; Tornero et al., 1996, 1997; Jordá et al., 2000). Consistent with its relevance for defense, P69B is targeted by P. infestans virulence factors. During the infection process, the pathogen secretes the Kazal-like proteinase inhibitors Epi1 and Epi10 that bind and inactivate P69B (Tian et al., 2004, 2005). Inhibition of any cell death-promoting activity of P69B would obviously benefit the pathogen during its biotrophic phase of growth. In fact, a cell death-promoting activity has been shown for subtilases in other plant species, including saspases in oat (Coffeen and Wolpert, 2004) and phytaspases in N. benthamiana and rice (Oryza sativa; Chichkova et al., 2010; Vartapetian et al., 2011), but not for P69B in tomato. We also did not observe cell death when P69B was transiently overexpressed in tomato (Supplemental Fig. S12). This negative result indicates that P69B activity alone is not sufficient to trigger cell death. Additional factors or stimuli appear to be involved, likely including downstream targets of P69B activity. The downstream targets of cell death-promoting subtilases, including saspases and phytaspases, are still unknown. The identification of the substrates of these proteases will provide further insight into the mechanisms of cell death control and remains an important goal for future research.
MATERIALS AND METHODS
Cloning and Expression of Sl2-MMP and Sl3-MMP
The cDNA of Sl3-MMP (accession no. HE819181) was obtained as EST cLEC-15-M21 from the Clemson University Genomics Institute. Database searches at the Solanaceae Genomics Network (http://solgenomics.net) identified three overlapping ESTs (cLHT-22-F10 from Solanum habrochaites and cLEC-52-A32 and cLEW-23-A21 from tomato [Solanum lycopersicum]) comprising the open reading frame of a second MMP. Primers were derived from the EST contig (for sequences of all primers used in this study, see Supplemental Methods S1) and used to PCR amplify Sl2-MMP from tomato (‘UC82B’) genomic DNA (accession no. HE819182). For the generation of expression constructs, the catalytic domains lacking their C-terminal membrane anchor were amplified for Sl2-MMP (residues 146–342) and Sl3-MMP (residues 151–346) and cloned into pET21d(+) (Novagen/Merck, VWR International) in frame with a C-terminal (His)6 tag. The constructs were introduced into Escherichia coli BL21-CodonPlus (DE3)-RIL (Agilent Technologies). Protein expression was induced by 1 mm isopropylthio-β-galactoside. After 4 h at 30°C, cells were harvested and lysed using BugBuster (Novagen/Merck) according to the manufacturer’s instructions. The recombinant proteins were purified by metal chelate affinity chromatography on nickel-nitrilotriacetic acid agarose (Qiagen) and dialyzed against 50 mm MES-HCl, pH5.5.
Characterization of Sl-MMP Activity
The hydrolysis of synthetic peptide substrates was analyzed by MALDI-TOF-MS as described (Huet et al., 2008; Cedzich et al., 2009). Zymography was performed on 10% polyacrylamide gels copolymerized with 0.5% gelatin. Protein samples were separated under nonreducing conditions using the Laemmli buffer system (Laemmli, 1970). After electrophoresis, the gel was washed twice for 15 min in 2% Triton X-100, incubated overnight in 50 mm Tris-HCl, pH 7, 1 mm ZnCl2, and 2% Triton X-100, and subsequently stained with Coomassie Brilliant Blue R250.
For steady-state kinetic analyses, Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 (M-2350; Bachem) was used as a peptide substrate. Standard assays contained 7 µm substrate and 500 µm CaCl2 in 1 mL of 100 mm Na2HPO4/NaH2PO4, pH 7, for Sl2-MMP or 100 mm sodium acetate buffered at pH 6.5 for Sl3-MMP. The reaction was initiated by the addition of enzyme (0.1 and 0.5 µg of recombinant Sl2- and Sl3-MMP, respectively), and cleavage of the internally quenched peptide substrate was monitored in a Cary Eclipse (Agilent Technologies) fluorescence spectrophotometer (λex, 325 nm; λem, 400 nm). To derive kinetic constants, substrate concentrations were varied between 1 and 20 µm, and data from three independent experiments were fitted to the Michaelis-Menten equation using a nonlinear least-squares method in the enzyme kinetics module of Sigmaplot software (Systat Software). pH optima were determined in 100 mm Tris-HCl (pH 9–8), 100 mm Na2HPO4/NaH2PO4 (pH 7.5 and 7), or 100 mm sodium acetate (pH 6.5–5). Ca2+ dependence was analyzed at optimum reaction conditions with the addition of CaCl2 at concentrations ranging from 0 to 1 mm.
Expression of Sl3-MMP Variants in Transgenic Cell Cultures and Plants
The open reading frames for MMPnat, MMPΔTM, and MMPΔω were amplified by PCR from the Sl3-MMP cDNA and cloned into the multiple cloning site of pART7 (Gleave, 1992) under the control of the CaMV 35S promoter and OCS terminator (Supplemental Methods S1). The entire expression cassette was cut out with NotI and transferred into pART27 (Gleave, 1992). Transgenic tomato (Solanum peruvianum) cell lines were generated by particle bombardment as described (Cedzich et al., 2009; Pickel et al., 2010). Transgenic tomato plants were generated by Agrobacterium tumefaciens-mediated transformation as described (Bosch et al., 2014).
Subcellular Localization of Sl-MMPs
The subcellular localization of Sl3-MMP was analyzed by transient expression of an EGFP fusion protein as described previously (Strassner et al., 2002). Briefly, EGFP was equipped with the N- and C-terminal targeting sequences of Sl3-MMP by replacing the central part of the Sl3-MMP cDNA (nucleotides 108–905) with the EGFP open reading frame (Clontech Laboratories). The fusion construct was introduced into onion (Allium cepa) epidermal cells by particle bombardment. EGFP fluorescence was analyzed after 2 d using a Zeiss AxioImager equipped with ApoTome and filter set 38 HE (λex, bandpass (BP) 470/40; beam splitter (Farbteiler, FT) 495; λem, BP 525/50 nm; Carl Zeiss MicroImaging). Control proteins were used for plasma membrane and cytoplasmic localization as described (Rutschmann et al., 2002). Targeting of Sl-MMPs was further analyzed by fractionation of cell cultures followed by western blot (for details, see Supplemental Methods S1). A polyclonal antiserum raised against recombinant Sl3-MMP(His)6 (Eurogentec) and goat anti-rabbit IgG coupled to horseradish peroxidase (Calbiochem/Merck) were used as primary and secondary antibodies, respectively. SuperSignal West Dura Extended Duration Substrate (Pierce/Thermo Fisher Scientific) was used for enhanced chemiluminescence detection according to the manufacturer’s instruction.
(Immuno)histochemistry and Microscopy
Tomato hypocotyl segments were fixed in 5% formaldehyde, dehydrated in a graded ethanol series, and embedded in Technovit 7100 (Heraeus Kulzer). Five-micrometer sections were mounted on SuperFrost Plus microscope slides (Menzel). For bright-field microscopy on Zeiss Axioskop 2 plus (Carl Zeiss MicroImaging), Toluidine Blue (0.03 g per 100 mL of water) was used to stain phenols, and Alcian Blue and safranin were used to stain cellulosic and lignified cell walls, respectively (Kukkola et al., 2003). Autofluorescence of suberized cell walls was detected with filter set 02 (λex, colored glass (G) 365; FT 395; λem, long bandpass (LP) 420; Carl Zeiss MicroImaging). For immunocytochemistry, sections were incubated in 1% SDS (1 h), washed in phosphate-buffered saline (PBS)/Tween (0.01% Tween 20 in PBS; 3× 5 min), and blocked with 5% bovine serum albumin in PBS/Tween (1 h). The IgG fraction (25 µg mL−1 PBS/Tween) of the polyclonal Sl3-MMP(His)6 antiserum was used for immunodetection. For negative controls, the same IgG fraction was used after specific depletion of antibodies directed against Sl3-MMP by negative chromatography on HiTrap N-hydroxysuccinimide-activated HP Sepharose (GE Healthcare) coupled to recombinant Sl3-MMP1(His)6. Alexa Fluor 568-labeled goat anti-rabbit IgG (1:1,000; Life Technologies) was used as a secondary antibody. SlowFade Gold antifade reagent (Life Technologies) was added prior to microscopy on AxioImager Z1 equipped with ApoTome and filter set 43 HE (λex, BP 550/25; beam splitter, FT 570; λem, BP 605/70; Carl Zeiss MicroImaging).
Silencing of Sl-MMPs by RNAi
A total of 490 bp of the Sl3-MMP cDNA (nucleotides 13–503) with an overall sequence identity of 79% with Sl2-MMP was amplified by PCR, and the product was cloned in the sense and antisense orientations into pHANNIBAL (Wesley et al., 2001). The entire expression cassette comprising the CaMV 35S promoter, Sl3-MMP-HP, and the OCS terminator was cut out with NotI and transferred into pART27 (Gleave, 1992). Transgenic tomato plants were generated by A. tumefaciens-mediated transformation as described (Bosch et al., 2014). Stable integration of the transgene and the independence of transformation events were confirmed by Southern-blot analysis. Silencing of Sl2- and Sl3-MMP in transgenic HP lines was confirmed by western-blot analysis (as described above) and RT-PCR.
Gel-Shift Assay for Substrate Identification
Apoplastic proteins were extracted from 9 g of leaf material of 5-week-old HP plants by vacuum (100 mbar) infiltration in 300 mL of 50 mm MES-HCl, pH 5, and centrifugation (500g for 10 min). The apoplastic wash fluid was cleared by centrifugation (14,000g for 5 min), and proteins were precipitated with 5 volumes of acetone at −20°C. The precipitate was resuspended in one-tenth volume of 100 mm MES-HCl, pH 6.5, 1 mm phenylmethylsulfonyl fluoride, 100 µm leupeptin, and 10 µm E-64, and 20 µL was separated in a single lane by 12% SDS-PAGE. The lane was excised from the gel, washed successively in water (10 min) and acetonitrile (10 min), and subsequently infiltrated with 100 mm MES-HCl, pH 6.5, 500 µm CaCl2, and 10 µg mL−1 Sl2-MMP(His)6, Sl3-MMP(His)6, or buffer alone for controls. After overnight incubation at 37°C, the gel strip was loaded onto a second gel and separated by SDS-PAGE in the second dimension under identical conditions. After electrophoresis, gels were incubated for 1 h in 10% ethanol/7% glacial acetic acid, stained with TOPLAB Ruby (1:10,000; Supplemental Methods S1), and fixed overnight in 40% ethanol/10% glacial acetic acid. Fluorescence was recorded using a Typhoon Trio Imager (GE Healthcare) at 488 nm, and protein spots were excised for MS analysis (Supplemental Methods S1). For the detection of protease activity, two-dimensional SDS-PAGE was done on 12% gels containing 0.5 µg of gelatin conjugated to 5-carboxyfluorescein (FAM-gelatin; λex, 490 nm; λem, 520 nm; Interchim). After electrophoresis, gels were developed for zymography as described above and scanned to record the fluorescence resulting from the cleavage of FAM-gelatin (λex, 488 nm; Typhoon Trio Imager; GE Healthcare). Gels were stained subsequently with TOPLAB Ruby and scanned to detect total protein.
2D-DIGE Analysis
2D-DIGE was performed to compare the hypocotyl proteome of 4-week-old wild-type tomato and HP lines involving four comparisons (including dye swap) using three independent HP lines (2× HP-1, HP-4, and HP-5). All 2D-DIGE reagents were from GE Healthcare and were used according to the manufacturer’s instructions. Protein extracts were prepared in 10% (w/v) TCA and 0.07% (v/v) β-mercaptoethanol in acetone. The precipitate was washed in acetone containing 0.07% (v/v) β-mercaptoethanol and lyophilized. Protein was solubilized in 2 m thiourea, 7 m urea, 2% (w/v) dithiothreitol (DTT), and 4% (w/v) CHAPS, and its concentration was determined using the 2D Quant Kit (GE Healthcare) with bovine serum albumin as a standard. Each protein sample (50 µg) adjusted to pH 8.5 was labeled with one of three CyDye DIGE minimal dyes (Cy3 and Cy5 for samples, and Cy2 for the internal standard made up of equal parts of all samples in the experiment). Samples to be compared and internal standard were mixed with an equal volume of sample buffer, applied to 24-cm rehydrated immobilized pH gradient strips, pH 4 to 7, by cup loading, and subjected to isoelectric focusing (63.5 kV × h, 75 µA) in an ETTAN IPGphor 3 instrument (GE Healthcare). Immobilized pH gradient strips were equilibrated (15 min) in 75 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, and 1% DTT and then incubated in the same buffer with 2.5% iodoacetamide instead of DTT. The strips were subsequently placed on top of 10% polyacrylamide gels and electrophoresed overnight at 1.5 kW and 10°C in an Ettan DALTsix electrophoresis system (GE Healthcare). Gels were scanned at different wavelengths using a Typhoon Trio Imager (GE Healthcare) and evaluated using the Delta 2D (version 3.6) software package (DeCodon). One of the gels was selected as a master gel, to which all other gels were aligned using vectors that were determined by the software and edited manually. Spot detection was performed automatically with manual editing on the aligned gel picture. Delta 2D and MeV were used for quantitative spot analysis and statistical evaluation (Saeed et al., 2003, 2006). A total of 124 of 1,132 spots showed a greater than 2-fold change in abundance at P < 0.01 (unpaired Student’s t test) and a false discovery rate < 5%. False discovery rate at 2-fold or greater change was determined using Significance Analysis of Microarrays (two-class unpaired Student’s t test, all permutations, positive constant (s0) = 10) according to Tusher et al. (2001). For protein identification, a preparative 2D gel was run under identical conditions and stained with TOPLAB Ruby as described above. Protein spots of interest were cut out and subjected to MS analysis (Supplemental Methods S1).
VIGS of P69B in Sl2/3-MMP-Deficient HP Lines
The bipartite tobacco rattle virus system was used for VIGS, with the two virus components on two transfer DNA vectors, pTRV1 and pTRV2, used for agroinfiltration (Ratcliff et al., 2001; Liu et al., 2002a). Gene-specific cDNA fragments of P69B and phytoene desaturase as a control were cloned into pTRV2 to generate the silencing constructs pTRV2/P69B and pTRV2/PDS, respectively. The constructs were infiltrated into tomato seedlings; systemic spread and expression of the silencing constructs was confirmed by PCR. Five weeks after agroinfiltration, the expression of P69B and the cell death markers Hin1, hsr203j, and tpoxC1 (Rivas et al., 2004) was analyzed by RT-PCR and western-blot analysis. Further details are given in Supplemental Methods S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HE819182 and HE819181 for Sl2-MMP and Sl3-MMP, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Primary structures of Sl2- and Sl3-MMP.
Supplemental Figure S2. Membrane association of Sl-MMP variants.
Supplemental Figure S3. Expression of Sl2-MMP(His)6 and Sl3-MMP(His)6 in E. coli.
Supplemental Figure S4. RT-PCR analysis of Sl2- and Sl3-MMP expression in tomato tissues.
Supplemental Figure S5. Sl2/3-MMP loss-of-function phenotype in stems of fully grown tomato plants.
Supplemental Figure S6. Identification of tomato subtilase P69B as a substrate of Sl2-MMP.
Supplemental Figure S7. Identification of tomato subtilase P69B as a substrate of Sl3-MMP.
Supplemental Figure S8. Degradation of recombinant P69B(His)6 by Sl3-MMP(His)6 in vitro.
Supplemental Figure S9. Identification of proteins differentially expressed between HP and wild-type plants.
Supplemental Figure S10. Western-blot analysis of P69B abundance in wild-type versus MMP-silenced plants.
Supplemental Figure S11. RT-PCR analysis of P69B expression in wild-type versus MMP-silenced plants.
Supplemental Figure S12. Transient expression of P69B in tomato leaves.
Supplemental Table S1. Proteins of lower abundance in HP lines compared with wild-type plants.
Supplemental Methods S1. Extended Materials and Methods
Acknowledgments
We thank Iris Klaiber, Brigitte Rösingh, Renate Frei, Jutta Babo, and Denisa Heindel for excellent technical assistance, the University of Hohenheim Service Unit Mass Spectrometry for help with proteomic analyses, Stephen H. Howell (Iowa State University) for introducing us to the 2D-DIGE technique, the Clemson University Genomics Institute for EST clones, and Commonwealth Scientific and Industrial Research Organization Plant Industry for the pHANNIBAL/pKANNIBAL vector system.
Glossary
- RNAi
RNA interference
- VIGS
virus-induced gene silencing
- GPI
glycosylphosphatidylinositol
- MALDI-TOF
matrix-assisted laser-desorption ionization time of flight
- HP
hairpin
- cDNA
complementary DNA
- CaMV
cauliflower mosaic virus
- EMSA
electrophoretic mobility shift assay
- MS
mass spectrometry
- 2D
two-dimensional
- DIGE
difference gel electrophoresis
- PBS
phosphate-buffered saline
- RT
reverse transcription
- DTT
dithiothreitol
Footnotes
This work was supported by the State of Baden-Württemberg, within the framework of a collaborative program between the University of Hohenheim and the Hebrew University of Jerusalem, and by a travel grant from the Arabidopsis Functional Genomics Network of the German Research Foundation (to D.Z.).
Articles can be viewed without a subscription.
References
- Albenne C, Canut H, Jamet E (2013) Plant cell wall proteomics: the leadership of Arabidopsis thaliana. Front Plant Sci 4: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albersheim P, Darvill A, Roberts K, Sedoroff R, Staehelin A (2011) Plant Cell Walls: From Chemistry to Biology. Garland Science, New York [Google Scholar]
- Barel G, Ginzberg I (2008) Potato skin proteome is enriched with plant defence components. J Exp Bot 59: 3347–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ (2006) Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6: 301–311 [DOI] [PubMed] [Google Scholar]
- Bellincampi D, Cervone F, Lionetti V (2014) Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front Plant Sci 5: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Wright LP, Gershenzon J, Wasternack C, Hause B, Schaller A, Stintzi A (2014) Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol 166: 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bütikofer P, Brodbeck U (1993) Partial purification and characterization of a (glycosyl) inositol phospholipid-specific phospholipase C from peanut. J Biol Chem 268: 17794–17802 [PubMed] [Google Scholar]
- Bykova NV, Rampitsch C, Krokhin O, Standing KG, Ens W (2006) Determination and characterization of site-specific N-glycosylation using MALDI-Qq-TOF tandem mass spectrometry: case study with a plant protease. Anal Chem 78: 1093–1103 [DOI] [PubMed] [Google Scholar]
- Cedzich A, Huttenlocher F, Kuhn BM, Pfannstiel J, Gabler L, Stintzi A, Schaller A (2009) The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3). J Biol Chem 284: 14068–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pinheiro C, Paiva JAP, Planchon S, Sergeant K, Renaut J, Graça JA, Costa G, Coelho AV, Ricardo CP (2009) Proteomic evaluation of wound-healing processes in potato (Solanum tuberosum L.) tuber tissue. Proteomics 9: 4154–4175 [DOI] [PubMed] [Google Scholar]
- Chichkova NV, Shaw J, Galiullina RA, Drury GE, Tuzhikov AI, Kim SH, Kalkum M, Hong TB, Gorshkova EN, Torrance L, et al. (2010) Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J 29: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Robertson D, Yu XL, Knox JP, Bolwell P, Slabas AR (2002) Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis 23: 1754–1765 [DOI] [PubMed] [Google Scholar]
- Coffeen WC, Wolpert TJ (2004) Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16: 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P (2010) Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Combier JP, Vernié T, de Billy F, El Yahyaoui F, Mathis R, Gamas P (2007) The MtMMPL1 early nodulin is a novel member of the matrix metalloendoproteinase family with a role in Medicago truncatula infection by Sinorhizobium meliloti. Plant Physiol 144: 703–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ (2004) Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J 40: 225–237 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Delorme VG, McCabe PF, Kim DJ, Leaver CJ (2000) A matrix metalloproteinase gene is expressed at the boundary of senescence and programmed cell death in cucumber. Plant Physiol 123: 917–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G, Hemetsberger C (2013) Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol 198: 1001–1016 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, Schultz CJ, Borner GHH, Dupree P, Eisenhaber F (2003) Glycosylphosphatidylinositol lipid anchoring of plant proteins: sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MAJ. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci 112: 2799–2809 [DOI] [PubMed] [Google Scholar]
- Fischer W, Christ U, Baumgartner M, Erismann KH, Mösinger E (1989) Pathogenesis-related proteins from tomato. II. Biochemical and immunological characterization. Physiol Mol Plant Pathol 35: 67–83 [Google Scholar]
- Flinn BS. (2008) Plant extracellular matrix metalloproteinases. Funct Plant Biol 35: 1183–1193 [DOI] [PubMed] [Google Scholar]
- Frick UB, Schaller A (2002) cDNA microarray analysis of fusicoccin-induced changes in gene expression in tomato plants. Planta 216: 83–94 [DOI] [PubMed] [Google Scholar]
- Ginzberg I. (2008) Wound-periderm formation. In Schaller A, ed, Induced Plant Resistance to Herbivory. Springer, Heidelberg, Germany, pp 131–146 [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Golldack D, Popova OV, Dietz KJ (2002) Mutation of the matrix metalloproteinase At2-MMP inhibits growth and causes late flowering and early senescence in Arabidopsis. J Biol Chem 277: 5541–5547 [DOI] [PubMed] [Google Scholar]
- Gopalan S, Wei W, He SY (1996) hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J 10: 591–600 [DOI] [PubMed] [Google Scholar]
- Graham JS, Xiong J, Gillikin JW (1991) Purification and developmental analysis of a metalloendoproteinase from the leaves of Glycine max. Plant Physiol 97: 786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Lapiere CM (1962) Collagenolytic activity in amphibian tissues: a tissue culture assay. Proc Natl Acad Sci USA 48: 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- Hématy K, Cherk C, Somerville S (2009) Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol 12: 406–413 [DOI] [PubMed] [Google Scholar]
- Hiraga S, Ito H, Yamakawa H, Ohtsubo N, Seo S, Mitsuhara I, Matsui H, Honma M, Ohashi Y (2000) An HR-induced tobacco peroxidase gene is responsive to spermine, but not to salicylate, methyl jasmonate, and ethephon. Mol Plant Microbe Interact 13: 210–216 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. (2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Huet Y, Strassner J, Schaller A (2008) Cloning, expression and characterization of insulin-degrading enzyme from tomato (Solanum lycopersicum). Biol Chem 389: 91–98 [DOI] [PubMed] [Google Scholar]
- Jamet E, Canut H, Boudart G, Pont-Lezica RF (2006) Cell wall proteins: a new insight through proteomics. Trends Plant Sci 11: 33–39 [DOI] [PubMed] [Google Scholar]
- Jordá L, Conejero V, Vera P (2000) Characterization of P69E and P69F, two differentially regulated genes encoding new members of the subtilisin-like proteinase family from tomato plants. Plant Physiol 122: 67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z (2010) Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141: 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkola EM, Koutaniemi S, Gustafsson M, Karhunen P, Ruel K, Lundell TK, Saranpää P, Brunow G, Teeri TH, Fagerstedt KV (2003) Localization of dibenzodioxocin substructures in lignifying Norway spruce xylem by transmission electron microscopy-immunogold labeling. Planta 217: 229–237 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lemaître V, D’Armiento J (2006) Matrix metalloproteinases in development and disease. Birth Defects Res C Embryo Today 78: 1–10 [DOI] [PubMed] [Google Scholar]
- Li D, Zhang H, Song Q, Wang L, Liu S, Hong Y, Huang L, Song F (2015) Tomato Sl3-MMP, a member of the matrix metalloproteinase family, is required for disease resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000. BMC Plant Biol 15: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Hollfelder K, Huang KS, Low MG (1994) Structural features of GPI-specific phospholipase D revealed by proteolytic fragmentation and Ca2+ binding studies. J Biol Chem 269: 28963–28971 [PubMed] [Google Scholar]
- Liu Y, Dammann C, Bhattacharyya MK (2001) The matrix metalloproteinase gene GmMMP2 is activated in response to pathogenic infections in soybean. Plant Physiol 127: 1788–1797 [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002a) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Maidment JM, Moore D, Murphy GP, Murphy G, Clark IM (1999) Matrix metalloproteinase homologues from Arabidopsis thaliana: expression and activity. J Biol Chem 274: 34706–34710 [DOI] [PubMed] [Google Scholar]
- Mandal MK, Fischer R, Schillberg S, Schiermeyer A (2010) Biochemical properties of the matrix metalloproteinase NtMMP1 from Nicotiana tabacum cv. BY-2 suspension cells. Planta 232: 899–910 [DOI] [PubMed] [Google Scholar]
- Marino G, Funk C (2012) Matrix metalloproteinases in plants: a brief overview. Physiol Plant 145: 196–202 [DOI] [PubMed] [Google Scholar]
- Marino G, Huesgen PF, Eckhard U, Overall CM, Schröder WP, Funk C (2014) Family-wide characterization of matrix metalloproteinases from Arabidopsis thaliana reveals their distinct proteolytic activity and cleavage site specificity. Biochem J 457: 335–346 [DOI] [PubMed] [Google Scholar]
- McGeehan G, Burkhart W, Anderegg R, Becherer JD, Gillikin JW, Graham JS (1992) Sequencing and characterization of the soybean leaf metalloproteinase: structural and functional similarity to the matrix metalloproteinase family. Plant Physiol 99: 1179–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita N, Nakazato H, Okuyama H, Kim Y, Thompson GA Jr (1996) Evidence for a glycosylinositolphospholipid-anchored alkaline phosphatase in the aquatic plant Spirodela oligorrhiza. Biochim Biophys Acta 1290: 53–62 [DOI] [PubMed] [Google Scholar]
- Nagase H, Woessner JF Jr (1999) Matrix metalloproteinases. J Biol Chem 274: 21491–21494 [DOI] [PubMed] [Google Scholar]
- Neumann U, Kubota H, Frei K, Ganu V, Leppert D (2004) Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal Biochem 328: 166–173 [DOI] [PubMed] [Google Scholar]
- Overall CM, Blobel CP (2007) In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol 8: 245–257 [DOI] [PubMed] [Google Scholar]
- Overall CM, Kleifeld O (2006) Towards third generation matrix metalloproteinase inhibitors for cancer therapy. Br J Cancer 94: 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley D, Bacic A (1999) Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc Natl Acad Sci USA 96: 14246–14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JH, Liu CY, Huangpu J, Graham JS (1997) Construction and characterization of the soybean leaf metalloproteinase cDNA. FEBS Lett 404: 283–288 [DOI] [PubMed] [Google Scholar]
- Pickel B, Constantin MA, Pfannstiel J, Conrad J, Beifuss U, Schaller A (2010) An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew Chem Int Ed Engl 49: 202–204 [DOI] [PubMed] [Google Scholar]
- Pontier D, Gan S, Amasino RM, Roby D, Lam E (1999) Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol Biol 39: 1243–1255 [DOI] [PubMed] [Google Scholar]
- Pontier D, Godiard L, Marco Y, Roby D (1994) hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J 5: 507–521 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Ratnaparkhe SM, Egertsdotter EM, Flinn BS (2009) Identification and characterization of a matrix metalloproteinase (Pta1-MMP) expressed during loblolly pine (Pinus taeda) seed development, germination completion, and early seedling establishment. Planta 230: 339–354 [DOI] [PubMed] [Google Scholar]
- Ricardo CPP, Martins I, Francisco R, Sergeant K, Pinheiro C, Campos A, Renaut J, Fevereiro P (2011) Proteins associated with cork formation in Quercus suber L. stem tissues. J Proteomics 74: 1266–1278 [DOI] [PubMed] [Google Scholar]
- Rich MK, Schorderet M, Reinhardt D (2014) The role of the cell wall compartment in mutualistic symbioses of plants. Front Plant Sci 5: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas S, Rougon-Cardoso A, Smoker M, Schauser L, Yoshioka H, Jones JD (2004) CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J 23: 2156–2165 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rodríguez D, Morrison CJ, Overall CM (2010) Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim Biophys Acta 1803: 39–54 [DOI] [PubMed] [Google Scholar]
- Rojo E, Martín R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sánchez-Serrano JJ, Baker B, et al. (2004) VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Rutschmann F, Stalder U, Piotrowski M, Oecking C, Schaller A (2002) LeCPK1, a calcium-dependent protein kinase from tomato: plasma membrane targeting and biochemical characterization. Plant Physiol 129: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A (1967) On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 27: 157–162 [DOI] [PubMed] [Google Scholar]
- Schiermeyer A, Hartenstein H, Mandal MK, Otte B, Wahner V, Schillberg S (2009) A membrane-bound matrix-metalloproteinase from Nicotiana tabacum cv. BY-2 is induced by bacterial pathogens. BMC Plant Biol 9: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling O, Overall CM (2008) Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol 26: 685–694 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Ferguson KL, Lahnstein J, Bacic A (2004) Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana: endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. J Biol Chem 279: 45503–45511 [DOI] [PubMed] [Google Scholar]
- Stracke JO, Hutton M, Stewart M, Pendás AM, Smith B, López-Otin C, Murphy G, Knäuper V (2000) Biochemical characterization of the catalytic domain of human matrix metalloproteinase 19: evidence for a role as a potent basement membrane degrading enzyme. J Biol Chem 275: 14809–14816 [DOI] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601 [DOI] [PubMed] [Google Scholar]
- Svetek J, Yadav MP, Nothnagel EA (1999) Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J Biol Chem 274: 14724–14733 [DOI] [PubMed] [Google Scholar]
- Tallant C, Marrero A, Gomis-Rüth FX (2010) Matrix metalloproteinases: fold and function of their catalytic domains. Biochim Biophys Acta 1803: 20–28 [DOI] [PubMed] [Google Scholar]
- Tian M, Benedetti B, Kamoun S (2005) A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol 138: 1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S (2004) A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem 279: 26370–26377 [DOI] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P (1996) Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA 93: 6332–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P (1997) Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem 272: 14412–14419 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W. (2012) The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma Y, Hendrickson T (2010) Methods to study GPI anchoring of proteins. ChemBioChem 11: 623–636 [DOI] [PubMed] [Google Scholar]
- Vartapetian AB, Tuzhikov AI, Chichkova NV, Taliansky M, Wolpert TJ (2011) A plant alternative to animal caspases: subtilisin-like proteases. Cell Death Differ 18: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visse R, Nagase H (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res 92: 827–839 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Williams RL, Katan M (1996) Structural views of phosphoinositide-specific phospholipase C: signalling the way ahead. Structure 4: 1387–1394 [DOI] [PubMed] [Google Scholar]