Abstract
In most metazoans, embryonic development is orchestrated by a precise series of cellular behaviors. Understanding how such events are regulated to achieve a stereotypical temporal progression is a fundamental problem in developmental biology. In this review, we argue that studying the regulation of the cell cycle in early embryonic development will reveal novel principles of how embryos accurately measure time. We will discuss the strategies that have emerged from studying early development of Drosophila embryos. By comparing the development of flies to that of other metazoans, we will highlight both conserved and alternative mechanisms to generate precision during embryonic development.
Keywords: Cell Cycle, timing, embryonic development, quantitative biology, signaling, transcription
Regulation of the cell cycle in early embryos: the need for speed and precision
When an egg develops into an organism, cells undergo an extraordinary population expansion and they obtain a spatial and temporal identity that will then determine their fate. This process poses a high risk for error amplification resulting from stochastic cellular decisions and unusually fast time scales [1]. Nonetheless, embryonic development is a precise process. It is thought that there must be complex developmental programs with correcting mechanisms in place to avoid the transmission of errors [2]. The Drosophila embryo provides a good model to study these problems since development is controlled in a highly stereotypical and reproducible pattern. Moreover, tools and methodologies for Drosophila are extensive and allow for careful interrogation of the dynamics of regulatory pathways quantitatively. In this review, we will propose that studying the cell cycle of early Drosophila embryos provides a unique system to dissect the molecular mechanisms ensuring precise temporal control of development.
A widespread phenomenon in the development of living organisms is the remodeling of the cell cycle to allow for remarkably fast cell cycles prior to gastrulation [3]. This pattern is particularly conserved in organisms that lay eggs, which develop externally, such as insects, amphibians and fish. The need for such exceptionally rapid cell cycle programs is most likely linked to the fact that eggs that develop externally subsist entirely on the maternal nutrients contained within the egg [4]. To make sure that embryos have sufficient nutrients, mothers lay very large eggs. However, such big size poses severe challenges for the regulation of embryonic development, since it is probably difficult for a single diploid nucleus to transcribe genes so efficiently to keep up with the vast demand of a very large cytoplasm. Therefore, embryos remain transcriptionally silent while they undergo several rounds of extremely rapid cleavage divisions [3]. The end result of this phase is an embryo with thousands of cells, which are now ready to take on developmental programs and gastrulation through transcriptional regulation.
The developmental strategy outlined above highlights the need for speed and synchrony in the regulation of the cleavage divisions, as the proper execution of the developmental programs that drive morphogenesis requires that these programs be initiated at very similar time across large spatial scales (Fig. 1A). How such features are achieved remain largely uncharacterized, although recent studies have started to shed light on this important problem. Before discussing these insights, we need to quickly review the early steps of embryonic development as well as our molecular understanding of cell cycle regulation.
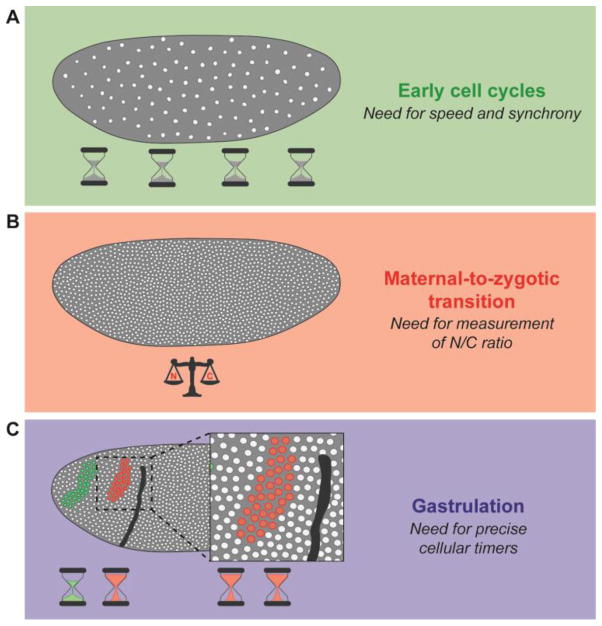
Fig. 1. Challenges in leading a precisely timed dynamic program in Drosophila embryos.
(A) Cell cycles in early syncytial embryos are fast and synchronous which is primarily achieved by preloading the embryo with maternal gene products, omitting gap phases and progressing rapidly through S-phase. (B) During the maternal-to-zygotic transition, embryos execute a series of regulatory changes in response to the N/C ratio. (C) Cell cycle regulation during gastrulation is primarily controlled by precise cellular timers. Nuclei located in the same mitotic domain divide synchronously, whereas each mitotic domain enters mitosis at different times. Green circles depict nuclei in mitotic domain 1 and red circles depict those in mitotic domain 5.
Cell cycle dynamics during early Drosophila development
The Drosophila egg is an oval-shaped cell about 500μm long and 150μm in diameter. Despite its large size, development of the Drosophila embryo follows a precisely timed dynamic program [5]. After fertilization, the egg goes through 13 rapid and synchronous divisions, which take place in a syncytium (i.e. a common cytoplasm not divided by membranes) [6]. These early cell cycles are exceptionally fast: nuclei undergo 13 mitotic divisions in 2–2.5 hours, whereas an average tissue culture cell takes 8–24 hours to go through one cell cycle [7]. These unusual speeds are achieved by omitting gap phases, having very short S-phases, and depending on maternally-loaded gene products to direct development. Nuclei in the early embryo therefore alternate between S-phase and mitosis during the early cycles. Gap phases (G1 and G2) canonically serve as pauses in the cell cycle during which cells grow or exit the cell cycle in the presence of unfavorable growth conditions or inhibitory signals from other cells [8]. However, the nuclei in the syncytial fruit fly embryo have all of the nutrients needed for development and embryos do not grow in size. Therefore, gap phases are dispensable in these initial cycles, which allows for a faster cycling times.
Cell cycle regulation and the mitotic switch
The embryonic cell cycle is driven by a regulatory network of proteins centered on the cyclin-dependent kinase (Cdk1) and the anaphase promoting complex (APC) [9]. Even though concentration of Cdk1 is constant, Cdk1 activity oscillates as nuclei go through several rounds of cell cycles due to oscillations in the levels of its regulatory subunits. These oscillations result in the phosphorylation of downstream components of the cell cycle machinery, which then lead to the initiation of cell cycle events [10].
Given its essential role in cell cycle progression, Cdk1 has many regulators of its kinase activity. To be activated, Cdk1 first requires binding of regulatory proteins called cyclins. Once Cdk1 is bound by a cyclin partner, it can be phosphorylated at Thr161 by Cdk1-activating kinase (CAK) which is required for enzymatic activity [11]. Surprisingly, CAK activity is not regulated by any known cell-cycle control pathway and it is maintained at high levels throughout the cell cycle. Therefore, the activating phosphorylation of Cdk1 is not rate limiting. Even though the activating phosphorylation of Cdk1 is not regulated, two inhibitory phosphorylations in Cdk1 are highly regulated and play an important role in the dynamics of Cdk1 activity [8]. One of them is found at a conserved tyrosine residue (Tyr15) and the other is found in animal cells at a threonine residue (Thr14). Tyr15 and Thr14 are located near the ATP-binding pocket and most likely block Cdk1 activity by interfering with the orientation of ATP phosphates [8].
The kinases that are responsible for adding these inhibitory phosphorylations are Wee1 and Myt1. Cdc25 phosphatases (String and Twine in Drosophila) are in charge of removing the inhibitory phosphorylations. Hence, there are four Cdk1 isoforms and the active form of Cdk1 is phosphorylated on Thr161 but not on Tyr15/Thr14. Finally, Cdk1 can be indirectly regulated by the regulators of Cdc25 and Wee1. For example, Chk1 kinase (Grapes in Drosophila) can indirectly inhibit Cdk1 by activating the Cdk1 inhibitor, Wee1, and inhibiting the Cdk1 activator, Cdc25 [7, 8].
The described cell cycle control system generates robust, switch-like and adaptable changes in Cdk activity which lead to all-or-none transitions of cell cycle events. This is because the Cdk1 regulatory network includes feedback loops and other regulatory interactions that lead to irreversible activation and inactivation of cyclin-Cdk1 complexes [12]. Wee1 and Cdc25 provide the basis for the rapid activation of the mitotic switch. Both enzymes are regulated by active cyclin-Cdk1 complexes: Wee1 is inhibited and Cdc25 is activated [13, 14]. Thus, active Cdk1 activates its activator and inhibits its inhibitor, generating a positive feedback loop and a double negative (positive) feedback loop, respectively. These feedbacks have the important property of generating a bistable system, which rapidly transitions from a low state of Cdk1 activity to a high state [15–18]. Bistability also provides hysteresis, i.e. the activity of Cdk1 is dependent of its history, a property which helps with the irreversible nature of entry into mitosis [16, 17].
Mitotic exit is driven by a negative feedback loop. Active cyclin-Cdk1 complexes activate the APC, which results in the polyubiquitination and degradation of cyclin [19–21], resetting Cdk1 complexes to their inactive, interphase state. The Cdk1-APC system behaves as a time-delayed negative feedback, a property which plays an important role in regulating the oscillatory activity of Cdk1 [22]. Other feedback mechanisms have been described that play a role in ensuring the proper abrupt regulation of anaphase [23, 24].
Proteins phosphorylated by Cdk1 during mitosis must be dephosphorylated to reset the cycle to the next interphase. In metazoans, PP2A phosphatases play a crucial role in these dephosphorylation events. Importantly, Cdk1 has an active role in downregulating the activity of PP2A through a negative feedback mechanism, mediated by the activity of the Greatwall kinase and the endosulfine inhibitor [25, 26]. This feedback mechanism seems to significantly contribute to changing the phosphorylation-dephosphorylation balance of Cdk1 substrates during the embryonic cycles [26].
Regulation of the early cell cycles
The early nuclear cycles of the Drosophila embryo demonstrate several specialized mechanisms by which very rapid cell cycles can be implemented [7, 27]. First, all the required cell cycle components are loaded in the embryo at extremely high levels maternally. Specifically, the high Cdk1 activity is able to drive DNA replication of the several nuclei present in the embryo with extreme speed [7, 27]. Experiments analyzing the activity of Cdk1 have suggested that these early nuclear cycles could proceed in the presence of very little oscillations in Cdk1 activity [27]. However, cyclin degradation is still required for mitotic exit events during syncytial cycles [28]. How can cell cycle events be triggered in the absence of oscillations in Cdk1 activity, but still require cyclin degradation? One possibility is that Cdk1 activity oscillates only locally in regions surrounding nuclei and spindles to regulate mitosis, so that analysis of its activity by biochemical methods (which report total activity in the embryo) would not show oscillations. Drosophila embryos also undergo cortical contractions during the early cycles which span the entire surface of the embryo, even when nuclei are inside and far away [29]. These cortical contractions are linked to Cdk1 activity [29]. How could Cdk1 activity regulate these oscillatory events at the cortex if its activity was only oscillating close to nuclei which are 100–200 microns away? Based on these facts, we propose that even small amounts of cyclin degradation can trigger mechanisms that result in effective oscillatory activities. A possible model is that a decrease in the activity of Cdk1 causes an increase in the activity of the opposing phosphatase PP2A. Such small changes could be amplified through feedback mechanisms to generate a state of high PP2A/low Cdk1 activity. We speculate that an alternation between high Cdk1/low PP2A and low Cdk1/high PP2A states due to an oscillation based on signaling mechanisms might be a strategy for fast regulation of the cell cycles, which avoids the longer timescales possibly associated with resynthesizing cyclins fully.
Synchrony is the other striking feature of the temporal regulation of the early nuclear cycles of Drosophila embryos [5]. These nuclear cycles are synchronized to ensure that morphogenesis is properly executed at later stages. The mechanisms of synchronization remain poorly understood. Diffusion across the common cytoplasm is too slow to synchronize the cell cycle, as it would typically take proteins hundreds of minutes to diffuse across the embryo. On the other hand, chemical waves have the ability to travel much faster than diffusion [30]. In fact, classical experiments have shown that mitotic events happen in a wave-like fashion in the embryo [5]. Two alternative mechanisms have been proposed. Chang and Ferrell have analyzed the mitotic pattern in vitro (using Xenopus egg extracts) and have proposed that coupling diffusion and bistability in the Cdk1 regulatory network can generate waves of Cdk1 activity [31]. They observed waves of mitotic entry and exit and demonstrated that they could be altered by inhibition of Wee1, which they interpreted as evidence for the existence of Cdk1 waves. However, analysis of the physical properties of the mitotic waves during Drosophila development has indicated that the process might be more complex [32]. Mitotic waves undergo a progressive and significant slowdown during development [32]. In principle, such slowdown is difficult to reconcile with a stereotypical chemical mechanism, such as the bistable Cdk1 system or waves of calcium signaling. Therefore, the authors proposed that mitotic waves might be the result of the coupling between an excitable mechanical system and the biochemical machinery regulating the cell cycle [32]. Overall, the nature of the mitotic waves in Drosophila embryos and the molecular and physical mechanisms of their regulation remain to be elucidated.
Cell cycle lengthening at the maternal-to-zygotic transition
Reliably after 13 synchronous mitoses, there is a long pause in the cell cycle, which coincides with a great increase in zygotic transcription at the maternal-to-zygotic transition (MZT). Understanding how the MZT is timed has proved an ideal system to understand the temporal regulation of developmental transitions. The cell cycles preceding the MZT get gradually longer (from 9 minutes at cycle 10 to 18 minutes at cycle 13) due to the activation of the DNA replication checkpoint [7, 33, 34]. The cell cycle slowdown is explained by a lengthening of S-phase, as mitosis has an invariant duration through all the syncytial cycles. Modulation of Cdk1 activity is required for the increased duration of S-phase [35, 36]. Following this gradual lengthening, a much more dramatic one is observed at cycle 14 when S-phase duration increases from 13 to 50 minutes and a G2 phase is introduced [36, 37]. Moreover, the onset of cycle 14 coincides with the MZT, which is characterized by a great increase in the transcriptional activity of the embryo [38]. Importantly, transcriptional activation is not a complete on-off switch and transcription can be detected in cycles preceding the MZT [39]. Transcription might in fact play a role in inducing the DNA replication stresses that cause the activation of the checkpoint [40].
Essentially, all the important remodeling events of the cell cycle at the MZT are triggered by post-translational inactivation of Cdk1 [27]. While cyclins are rapidly resynthesized at cycle 14, Cdk1 is kept inactive by inhibitory phosphorylation [7, 27]. This inactivation is the result of a change in the balance between phosphorylation-dephosphorylation by Wee1 and Cdc25 phosphatases. Farrell et al. showed that expression of cdc25 mRNA in early cycle 14 triggered early replication of satellite sequences that resulted in decreased S-phase duration [35]. Lowering Cdk1 activity in cycle 13 lengthened the pre-MBT S-phase and prematurely triggered the start of late replication. This led to the conclusion that Cdc25 and a decrease in Cdk1 activity between cycle 13 and 14 lengthen cycle 14 through the slowdown of DNA replication and by incorporation of a G2 phase. Moreover, the activity of Wee1 is required for the lengthening of cycle 11 to cycle 13 and wee1 mutants are unable to execute the MZT properly and cause embryonic lethality [3, 41, 42]. Collectively, these experiments support the idea that the remodeling of Cdk1 activity and the cell cycle shows the features of an ultrasensitive switch. During cycle 10 to cycle 13, changes in the balance between inhibition and activation of Cdk1 have small gradual effects on cell cycle duration. This gradual increase in cell cycle duration places the system close to a threshold so that a rapid switch-like transition can be triggered at the MZT.
How is Cdc25 regulated such that it becomes limiting in cycle 14, which then allows Cdk1 to become inhibited through phosphorylation? The egg is deposited with two forms of cdc25 that encode two different proteins called String and Twine. Immunoblots show that String starts to decrease slowly during the blastoderm cycles and is undetectable at cycle 13 [27]. Twine, on the other hand, is relatively stable before the MBT, but during early cycle 14 it is abruptly destabilized and destroyed [43, 44]. Even though there is significant destruction of maternal transcripts during cycle 14, RNAi knockdown of string and twine does not cause cell cycle arrest, suggesting that downregulating the mRNA is not sufficient to explain the cell cycle changes [44]. In fact, combining a method to measure the lifetime of proteins (tagged with the photoswitchable fluorescent protein Dronpa) in living embryos and mathematical modeling, it was shown that the levels of String and Twine are mainly regulated by changes in their lifetime [43].
Wee1, String and Twine are differential players in the switch-like mechanism that causes cell cycle remodeling. String levels decrease gradually beginning around cycle 9 [45]. This gradual degradation results in complete clearance of String protein by cycle 13 [43–45]. Coincidently with this decrease in String levels, Wee1 activity increases due to activation by the DNA replication checkpoint effector kinase Chk1 [41, 42]. Twine levels on the other hand are stable until the beginning of cycle 14 when transcription of tribbles and other unidentified zygotic genes trigger its rapid degradation [43, 44]. Such rapid degradation of Twine is absolutely required for arresting the cell cycle, as expression of a more stable Twine mutant results in an extra mitosis prior to the MZT [43]. Other genetic manipulations involving Twine support its contribution to cell cycle control during the MZT. Embryos from flies that had two additional copies of twine would occasionally execute the MZT one cycle late [45, 46]. Furthermore, mothers that were germline deficient for string and heterozygous for twine would lay eggs that would frequently cellularize one cycle early [45]. Embryos with 4 copies of twine and deficient for fruhstart, a cyclin-dependent kinase inhibitor (CKI) that is transcribed in early cycle 14, almost always display an extra pre-MBT division [46].
Comparison of String and Twine degradation in haploid and diploid embryos indicates that their degradation is clearly regulated by the nuclear-to-cytoplasmic ratio (N/C ratio) [43, 44]. Moreover, String but not Twine degradation seem to be sensitive to the DNA replication checkpoint [43]. Based on these observations, we propose the following model for the regulation of the MZT through the N/C ratio. The gradual increase of the N/C ratio during development triggers the gradual activation of the DNA replication checkpoint, which in turn increases the activity of Wee1 and probably represses the activity of String by contributing to its degradation or inactivation. This gradual cell cycle remodeling facilitates the activation of gene expression, so that a rapid switch-like increase in zygotic transcription is observed at the onset of cycle 14. Among the several zygotic genes activated, there are few whose activity is required to target Twine for degradation and cause the significant remodeling of the cell cycle observed at the MZT [44]. The identification of these genes and the elucidation of their transcriptional regulation, as well as the mechanisms by which they target Twine for degradation, will provide fundamental insights on the timing of the MZT. Moreover, we argue that quantitative experiments of cell cycle dynamics and of the dynamics of Cdc25 and Cdk1 activities will reveal fundamental principles of regulation of cell cycle remodeling.
The role of the N/C ratio has been of great interest in the field ever since early experiments showed that manipulating this ratio caused a change in the time of MZT events in Xenopus egg extracts [47]. This suggested that some events require a particular amount of DNA to take place. Manipulating the N/C ratio in Drosophila also suggested that the N/C ratio plays a role in the slowing of the cell cycle (Fig. 1B) [48]. Embryos with half the amount of DNA of a normal embryo require an extra cycle to trigger the slowdown of the cell cycle. Another study generated embryos with varying amounts of DNA and was able to show that there is a threshold of around 70% of the DNA that would normally be present at cycle 14 that determines timing of the MZT. Embryos with less than 75% of the normal amount of DNA go through an extra cell cycle before the MZT, whereas embryos with more than 134% often undergo one fewer cycle before the MZT [49].
It is worth mentioning that not all MZT-associated events are affected by the N/C ratio. In haploid Drosophila embryos, cellularization begins in cycle 14 as it normally would but it is forced to abort and restart due to the early mitosis [39, 48]. Further confirmation was obtained by a recent study that arrested embryos in cycle 12 or 13 using RNAi against cyclins and noted that cellularization starts at the normal time of what would have been cycle 14 [50]. This finding suggests that the downregulation of Cdk1 activity can overcome the need for a cycle 14 N/C ratio for some MZT events. Additionally, this conclusion also implies other timers involved in the regulation of MZT events are independent of the N/C ratio. Could N/C ratio-dependent transcription time the cell cycle transition at the MZT? A study made use of high-throughput methods to compare expression of several hundred genes in normal versus haploid embryos. They found that some genes are N/C-dependent and others are time-dependent [49]. Given that transcription is required for cell cycle slowing at the MZT, one attractive model is that N/C ratio-dependent transcription influences the onset of Twine destruction. Therefore, a small number of N/C ratio-dependent transcripts could be the trigger for the dramatic slowdown of cycle 14, but these remain to be elucidated.
Timing of mitosis at the onset of gastrulation
Gastrulation, like the rest of Drosophila embryogenesis, is unusually fast. True, the mitotic cycle at the onset of gastrulation is about 8 times longer than the early cycles (~75 vs. ~10 minutes). But this cycle is still ten times faster than any other larval or adult cell cycle [51]. More impressive is the coordination of cell proliferation with the ongoing morphogenic processes. During gastrulation, massive cytoskeletal rearrangements occur throughout the embryo at the same time as cells enter the first post-MZT mitosis [52]. Interference between dividing cells and tissue morphogenesis would be devastating. The embryo’s solution to this problem is precise temporal coordination of cell division and morphogenetic events.
At the onset of Drosophila gastrulation, twenty-five groups of cells across the surface of the blastoderm enter mitosis in a reproducible spatial and temporal pattern [53]. While the timing of mitosis between domains is asynchronous, deviation in timing of each dividing cell within a single domain is only about 2 minutes [54]. Mitotic domain 1, for instance, is comprised of about sixty cells. When the time is right, these cells enter mitosis together with minimal error. This kind of precision is unprecedented and represents a great model for stereotypical temporal patterning. We will argue that this kind of temporal precision requires precise cellular timers (Fig. 1C).
Before considering the timing of mitosis 14, we will first briefly review ideas for developmental precision. Biological processes are riddled with noise [55]. This is principally the result of variations in gene transcription and other stochastic processes. Noise in gene expression has essentially two sources, which are described as intrinsic and extrinsic noise [56]. Intrinsic noise describes the fluctuations which are generated by events specific to the gene of interest. For example, thermodynamic fluctuations involved in transcription factor binding, Pol II docking, and so on, produce noise in transcriptional output. Extrinsic noise, on the other hand, describes the noise that is independent of the behavior of indivual genes, but has to do with global cellular properties. For example, a gene could be expressed at very different levels in different cells, depending on the cell status, transcriptional capacity, etc. Recent measurements of mRNA in Drosophila suggest that intrinsic noise in transcription is about 45% [57]. How is noise reconciled with developmental precision?
Experiments addressing the mechanisms of developmental precision and the filtering of noise in biological processes have essentially revealed two strategies [1, 58, 59]. A first strategy for developmental precision involves regulating every transition very accurately. A second strategy suggests that noise in cellular behaviors might be later filtered by correcting mechanisms. The first approach requires that both the developmental inputs and the pathways responding to them are regulated precisely. A notable example of such precise regulation is the Bicoid/Hunchback relationship at cell cycle 14 in Drosophila embryogenesis [58]. The maternally deposited bicoid mRNA is positioned at the anterior pole of the embryo, and Bicoid protein diffuses to form a concentration gradient along the anteroposterior axis [60, 61]. Each cell in the embryo “measures” the concentration of Bicoid and “decides” whether to transcribe hunchback. Impressively, Hunchback expression is confined sharply to half of the embryo. Two important features are important for ensuring this precision. Firstly, the hunchback promoter/enhancer is capable of sensing small differences in Bicoid concentration through an ultrasensitive response (Hill coefficient of 5). And secondly, the Bicoid gradient is extremely reproducible among embryos; fluctuations in Bicoid concentration are as low as ~10%. Even so, the precision with which the concentration of Bicoid can be estimated by the hunchback enhancer is limited by the intrinsic physical limits imposed on counting random molecular encounters [58, 62, 63]. Using physical estimates, it was established that some other mechanism must play a role in the establishment of the observed precision of hunchback expression [58]. Analyzing the correlations between Bicoid and Hunchback levels, Gregor et al. proposed that a spatial averaging mechanism across few nuclei is sufficient for the establishment of an accurate Hunchback expression [58]. Similarly, a role for temporal and spatial integration might extend to the precise regulation of all the gap genes, which participate in the establishment of the Drosophila body plan [57]. These experiments are notable, since they argue that the simple mechanisms of spatiotemporal integration (the noise levels of any stable mRNA or protein will be reduced over time) can determine the precision of development. However, other developmental transitions are often very rapid and it is not yet clear whether different mechanisms of noise suppression exist for such rapid transitions.
An alternative approach for generating precision in development involves the deployment of correcting mechanisms. Neural tube patterning in zebrafish is an excellent example of this method. During neural tube development, sharply defined cell progenitor domains form along the ventral-dorsal axis [64]. A concentration gradient of the signaling protein Sonic Hedgehog (Shh) specifies cell location in space and time [64]. However, neural tube formation is a highly dynamic process, and cell movement of progenitor cells can disrupt any inherent precision in the morphogen gradient. The canonical “French flag model” [65] for morphogen gradient guided organization can only be part of the story. Xiong et al. (2013) propose that a cell sorting mechanism based on cell-cell adhesion corrects for the noisy Shh signaling [66]. Cells use the Shh concentration as a first approximation for specifying location, but monitor an alternative criterion to adjust for any prior mistakes in positioning.
Cells do not have the luxury to correct the all-or-none decision to enter mitosis after it has been made. Mitosis 14 only happens once and cells get it right every time. So what ensures this precision? First of all, these cells are peculiar in that they regulate entry into mitosis at the G2/M transition, rather than with the standard accumulation of cyclins [37, 67]. Although the necessary cyclins (A, B, and B3) are present, cyclin-Cdk1 complexes are held inactivated by Wee1 kinases [7, 27]. And, as discussed above, all maternally deposited Cdc25 molecules are degraded at the mid-zygotic transition. Hence, cells are left delicately positioned in G2 waiting in anticipation for developmental inputs that inform them to transcribe the limiting reagent, cdc25/string [37, 67]. String protein dephosphorylates Cdk1, and activated Cdk1 triggers entry into mitosis. The timing of mitosis 14 thus depends on two important regulatory steps. First, string transcription is turned on at the right time in response to developmental inputs. And second, cells measure the accumulation of String accurately despite variation in gene transcription. It was recently shown that these two aspects of regulation are independent [54] and therefore precision must be ensured at both steps, as there is no correction mechanism.
The first level of regulation is a question of how the activity of an enhancer is regulated precisely in time and space. The string enhancer contains cis-regulatory elements that respond to numerous transcriptional activators and repressors, and is regulated in a combinatorial manner [68]. Presumably, integration of several developmental inputs guarantees the reliability in spatial patterning of the mitotic domains. But what sets the timing of mitosis 14? There are essentially two available models. One hypothesis is that genes encoding spatial information also encode temporal information [51]. After all, the spatial distribution of transcription factors in the embryo changes with time, beginning with the initial conditions of maternally deposited content like bicoid. This is an attractive hypothesis because it eliminates the need for a separate time keeping molecule. If it does turn out that each of the twenty-five mitotic domains are timed by patterning genes encoding spatial regulation, then each mitotic domain is principally on its own clock, and this clock is linked to the rate of accumulation/degradation of its regulators. An alternative model posits the existence of a single universal clock. In such a scenario, the spatial patterning genes might prime the string enhancer in each mitotic domain. The rapid accumulation of, say, some further transcriptional activator would then act as the final trigger of string transcription. The different sets of spatial patterning genes would then, presumably, determine the threshold for the time keeping molecule. Different thresholds would account for the distinctive times at which each mitotic domain enters mitosis. Teasing apart these (and unforeseen) models will improve our understanding of timing in development.
Regardless, temporal regulation of string transcription relies on its enhancer sensing the correct concentration of the appropriate transcription factors at the right time. Integration of several developmental inputs can help increase the reliability of spatial precision. But this is very much a function of how long the enhancer has to sample its environment. Accuracy is easy to achieve given enough time; making an accurate and fast decision is more challenging. The enhancer cannot be too sensitive, lest it risk the chance of firing prematurely. Nor can the enhancer be too stubborn. There is considerable interest in understanding the architecture of developmental enhancers [69] as well as the tradeoffs in optimization between weak versus strong transcription factor binding sites [70]. The core questions generally concern spatial precision and transcriptional output. Questions addressing precise temporal dynamics of promoters/enhancers in development are relatively unexplored [71]. Recently, there are two noteable examples. Lagha et al. found that Pol II pausing at promoters helps ensure the rapid and synchrounous transcription of snail to coordinate the invagination of the mesoderm in Drosophila gastrulation [71]. Additionally, Ferraro et al. found evidence for “transcriptional memory” as a means to ensure synchronous gene transcription: cells that transcribe a developmental gene in one cell cycle, can be primed to transcribe that gene in the next cycle [72]. But Pol II pausing does not obviate the need for transcriptional regulators to kick Pol II into action, and string, for instance, is not expressed in the prior cell cycle. The question remains how an ~500bp piece of DNA can quickly read its environment. What determines the structure of an “optimal” enhancer for making quick decisions? Theoretically, this is reminiscent of the “exploit-explore” problem, in which a sensor optimizes the time spent collecting information before making a choice [73]. Certain minimal string enhancer elements can be used to recapitulate the spatial and temporal patterning of string in the mitotic domains [68]. The regulation of string transcription is a promising model system for posing this kind of question.
The second aspect regulating the timing of mitosis 14 involves the ability of cells to process String accumulation accurately. Even if the “optimized” enhancer turns on at the right time, there is invariably going to be noise in the output of this process. As mentioned above, the variability in transcription of any given enhancer in Drosophila appears to be about 45% [57]. These measurements were made on spatial patterning genes in the early Drosophila syncytium. In that context, the authors suggest that error in protein expression is minimized through mRNA stability and diffusion of mRNA transcripts. However, transcription of string happens post-cellularization, and therefore spatial averaging of this kind is not possible. Moreover, the time interval between transcriptional activation of string and mitosis is very short. The embryo solved this problem by acquiring “memory”, i.e., the ability to integrate (Fig. 2). Cells behave as short-term integrators whereby they measure the concentration of String over periods of about 2 minutes before making the decision to enter mitosis [54].
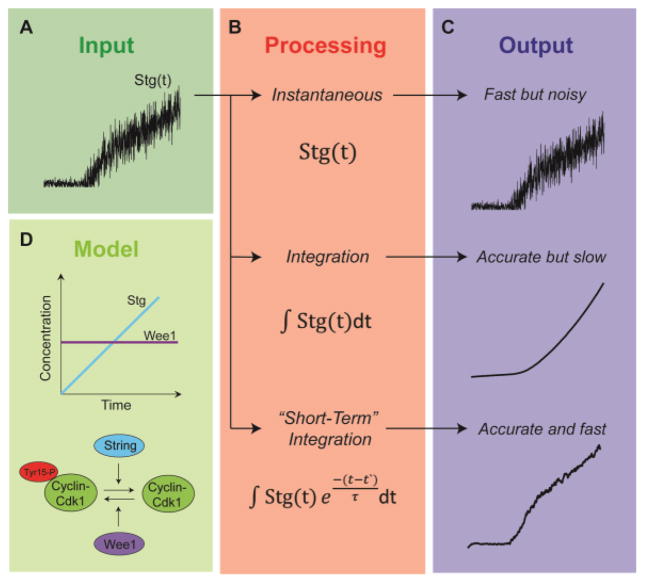
Fig. 2. Cellular information processing.
(A) The expression of String protein signals cells to enter mitosis 14 at the onset of Drosophila gastrulation. Here is a graphical illustration of the noise present in String concentration over time. (B) Cells can interpret or “process” the concentration of String (as an input) in myriad ways. We provide a schematic of just three simple tactics of information processing. Cells can respond to a particular threshold concentration of String at a particular time, or cells can use an integration-type method, whereby the concentration of String is integrated over the whole history or a particular time interval. (C) The output generated by each processing strategy is shown. The ouput generated by the instantaneous method is subject to any noise present in the input. The integration method is able to smoothen noise, but in doing so, generates a sluggish response, which loses the abrupt change in String concentration. The short-term integration method is superior for making a decision which is both quick and accurate, because it filters noise while maintaining the abrupt change in String concentration. (D) A diagram of the covalent modification cycle between Wee1, String, and Cdk1 is provided. A linear influx of String overcomes the constant concentration of Wee1. Under certain paramters this is enough to trigger an ultrasensitive response.
Interestingly, the canonical feedback between Cdk1 and its regulators does not play a role in the switch-like entry into mitosis at gastrulation. Mutants for wee1 and string that disable feedback do not have an effect on the timing or entry into mitosis 14 [54]. One potential explanation for this might be that the time frame is too fast for feedback to make a difference. So where does mitosis 14 get its ultrasensitivity? The all-or-none response of mitosis 14 is a property of far from equilibrium covalent modification cycles. During interphase, the concentration of Wee1 and inactive Cdk1 are both constant and the system is effectively at equilibrium. The behavior of this system changes abruptly with the rapid influx of String [74]. Theoretical arguments indicate that the rapid linear accumulation of an activator enzyme like String is enough to induce a sigmoidal response. Furthermore, this type of mechanism is highly adaptable. The integration time before entering mitosis can be “tuned” by changing either the rate of String accumulation, or the basal level of Wee1 [74].
Covalent modification cycles behave as low-pass filters [75], i.e. systems which are able to filter noise of frequencies higher than a characteristic one (the inverse of the integration time). Cells might solve the problem of making decisions rapidly and accurately using covalent modification cycles in order to integrate over the appropriate timescale [54] or to approximate the statistical estimates that guarantee the optimal tradeoff of speed and accuracy [73].
It would be interesting to investigate if similar biochemical mechanisms play a role in the precise transcriptional regulation of string. How might such mechanisms operate? An intriguing idea is that the transcription factors that regulate string induce a covalent modification (e.g. methylation) of histones. Since enzymes reversing these modifications must be present in the embryo, the string enhancer would be regulated by a covalent modification cycle. As the levels of a transcriptional activator increase the rate of modification would increase, but the enhancer would respond to the levels integrated over the timescale intrinsic to the rates of the covalent cycle, similarly to what was observed for the regulation of mitosis by String.
Comparison with other organisms
The rapid cell cycle stage as well as the progressive slowing appear to be a widespread occurrence linked to the maternal-to-zygotic transition in a number of species, suggesting that it may be an integral component of the early cycles. The fact that it is so conserved also suggests that this might be an evolutionary trait that was adopted as a basic strategy for organisms that lay eggs. We will briefly review what is known about early cell cycle control in Xenopus laevis and zebrafish.
At a glance, Xenopus early cell cycles are very similar to those of Drosophila. The Xenopus embryo goes through 11 rapid cell cycles, which last approximately 30 minutes each, followed by a 12th cycle that is slightly longer, which is defined as the mid-blastula transition (MBT, considered equivalent to the MZT in other organisms). Cycles 13 through 15 get progressively longer, lasting 50, 99 and 253 minutes, respectively [76]. The early gastrula transition (EGT) occurs during the extended cycle 15 during which gastrulation begins. The Xenopus MBT is dependent on the N/C ratio, but is not dependent on zygotic transcription [47, 77], although recent experiments show that perturbations that advance the onset of zygotic gene expression result in early cell cycle lengthening [78]. Cell cycle regulation transitions from a rapid alternation of S and M phases to a cycle with gap phases (G1 and G2) at cycle 12. A major player of such transition is the degradation of cyclin E, which remarkably is not controlled by the N/C ratio but responds to a timing mechanism [79]. Chk1 also becomes activated during the Xenopus MBT (cycle 11–12) and is required for cell cycle lengthening (Shimuta et al., 2002). O’Farrell et al. suggested that alignment of embryonic development by gastrulation would give the best understanding of cell cycle remodeling events when comparing Drosophila and mice [3]. What would happen if we took the same approach when comparing Drosophila and Xenopus and aligned cycle 14 in Drosophila with cycle 15 in Xenopus?
From cycle 10 to cycle 13 the Drosophila embryo undergoes a two-fold change in cell cycle duration, which is not very dissimilar to the 3-fold change observed between cycle 11 and cycle 14 in a Xenopus embryo. Similarly, in both organisms this lengthening is not affected by treating embryos with alpha-amanitin [45, 47] an inhibitor of transcriptional elongation whose effects on the MBT must be however carefully evaluated [40]. Finally, in both organisms cell cycle lengthening would coincide with the earliest time of activation of the DNA replication checkpoint. These observations would lead one to speculate that in both Drosophila and Xenopus the first effect of increasing N/C ratio is to trigger an activation of the DNA replication checkpoint and a lengthening of the cell cycle. Such gradual lengthening prepares the embryo for a more abrupt remodeling of the cell cycle prior to the onset of gastrulation, when transcriptional mechanisms take over the regulation of the cell cycle to ensure proper coupling with morphogenesis. However, there are caveats in pushing this model too far. Most notably, gap phases are only introduced in cycle 14 in Drosophila, while they become important at cycle 12 in Xenopus. We propose that further quantitative analysis of cell cycle regulation during early development of Drosophila and Xenopus is required to fully elucidate the common principles of cell remodeling through development.
Two interesting models have recently been proposed for how the N/C ratio might trigger the MBT in Xenopus embryos. Collart et al. provided evidence that the titration of factors required for DNA replication contribute to cell cycle remodeling and might represent the mechanism by which embryos sense the N/C ratio [80]. Amodeo et al. found evidence that titration of histone proteins plays a role in coupling the N/C ratio to activation of zygotic gene expression and cell cycle remodeling [78]. These two models and the observation that activation of zygotic gene expression contributes to the activation of the DNA replication checkpoint in Drosophila [40] suggest that titration of factors required for DNA replication or repressors of zygotic gene expression are two parallel mechanisms by which the DNA replication checkpoint is activated at the Xenopus MBT.
The zebrafish embryo goes through 9 rapid cell cycles (15 minutes each), slows a little in cycles 10 through 11, and undergoes a dramatic slowdown in cycle 12. Cells divide synchronously until cycle 11, where they start to show the first asynchronies [81]. The N/C ratio can also time the MBT slowing as in Drosophila and Xenopus [82]. However, the dramatic slowdown does not depend on transcription. As in Drosophila, the slowing of the cell cycle arises from a longer S-phase at the MBT, even though it does not require transcription. Both Cdc25 and Cdk1 are significantly downregulated at the MBT and overexpression of Cdc25a or inhibitory-phosphorylation resistant Cdk1 causes continuous rapid divisions [83]. However, upregulating Cdk1 activity does not prevent S-phase lengthening, suggesting that even though Cdc25 and Cdk1 may be responsible for the introduction of a G2 phase, the mechanism to increase S-phase duration is different than Drosophila’s and is not dependent on Cdk1.
Like Drosophila, zebrafish display stereotypical mitotic domains. Rather than twenty-five domains, zebrafish have only three, and unlike Drosophila, the mitotic domains in zebrafish do not correlate with the embryonic cell fate map [84]. Bouldin et al. used FUCCI (fluorescence ubiquitination-mediated cell cycle indicator) lines to track proliferating cells during gastrulation, somitogenesis, and tail-bud formation. The authors found that following the rapid cell divisions at gastrulation, posterior progenitor cells pause in S/G2, and that this pause correlates with the downregulation of the zebrafish mitotic phosphatase cdc25a [85]. Ectopic expression of cdc25a using a heat-shock promoter induces cell division in the posterior progenitor cells leading the authors to hypothesize that cdc25a exclusion mediates lengthening of the G2 phase during somitogenesis. This is reminiscent of cell cycle regulation in Drosophila gastrulation, which uses G2/M cell cycle regulation to obtain high temporal precision. The same reasoning may also apply in zebrafish. Cell division of the posterior progenitor cells is shut down in the absence of cdc25a expression. Upon entry into the somites, cells re-express cdc25a and this triggers mitosis. The G2/M transition may be a general method for carefully timing and compartmentalizing cell division. Local developmental inputs present in the somites (like the mitotic domains) can quickly activate cdc25a transcription. Bouldin et al. speculate that the timing of this cell cycle is made precise so that it can coordinate with the well-known segmentation clock. We propose that studying the timing of cell cycle during somitogenesis provides an excellent system for studying precise developmental timers at a later stage of embryogenesis.
Conclusions
Understanding how cellular behaviors are regulated in a temporally accurate manner during development remains a fundamental open question. In this review, we have argued that studying the cell cycle will reveal mechanisms of temporal regulation during embryonic development. The cell cycles of early embryos undergo significant remodeling and offer the opportunity to dissect several different strategies of regulation. For example, the early cell cycles of Drosophila embryos are extremely rapid and synchronous across a large syncytium which is transcriptionally silent. On the other hand, during gastrulation the cell cycle is primarily regulated transcriptionally by developmental inputs. Analyzing the molecular mechanisms that ensure the precision of cell cycle timing during development has the potential to reveal novel principles in areas ranging from cell signaling to transcriptional regulatory networks.
Acknowledgments
We apologize to the authors in the field, whose work was not cited for sake of brevity. We acknowledge discussion with Amir Momen-Roknabadi (Princeton University) and Massimo Vergassola (UCSD) on several aspects of temporal regulation of embryonic development. Work in our laboratory on the timing mechanisms of cell cycle during development is partially funded by an NIH grant (R00-HD074670). V.E.D. is supported by a Faculty for the Future Fellowship from the Schlumberger Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balazsi Gb, Van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144:910–25. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nature Reviews Genetics. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- 3.O’Farrell PH, Stumpff J, Su TT. Embryonic Cleavage Cycles: How Is a Mouse Like a Fly? Current Biology. 2004;14:35–45. doi: 10.1016/j.cub.2003.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Farrell PH. Growing an embryo from a single cell: A hurdle in animal life. Cold Spring Harbor Perspectives in Biology. 2015;7 doi: 10.1101/cshperspect.a019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. Journal of Cell Science. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- 6.Rabinowitz M. Studies on the cytology and early embryology of the egg of Drosophila melanogaster. J Morphol. 1941;69 [Google Scholar]
- 7.Farrell JA, O’Farrell PH. From Egg to Gastrula: How the Cell Cycle Is Remodeled During the Drosophila Mid-Blastula Transition. Annual Review of Genetics. 2014;48:269–94. doi: 10.1146/annurev-genet-111212-133531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan DO. The cell cycle: principles of control. Sunderland, MA; London;: Published by New Science Press in association with Oxford University Press; Distributed inside North America by Sinauer Associates, Publishers; 2007. [Google Scholar]
- 9.Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annual Review of Cell and Developmental Biology. 1997;13:261–91. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 10.Murray aW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–80. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 11.Harper JW, Elledge SJ. The role of Cdk7 in CAK function, a retro-retrospective. Genes and Development. 1998;12:285–9. doi: 10.1101/gad.12.3.285. [DOI] [PubMed] [Google Scholar]
- 12.Lindqvist A, Rodríguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. Journal of Cell Biology. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGowan CH, Russell P. Cell cycle regulation of human WEE1. The EMBO journal. 1995;14:2166–75. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2--cyclin B and its involvement in the self-amplification of MPF at mitosis. The EMBO journal. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novák B, Tyson JJ. Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. Journal of Cell Science. 1993;106:1153–68. doi: 10.1242/jcs.106.4.1153. [DOI] [PubMed] [Google Scholar]
- 16.Pomerening JR, Sontag ED, Ferrell JE. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nature cell biology. 2003;5:346–51. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 17.Sha WW, Moore JJ, Chen KK, Lassaletta ADAD, Yi C-SCS, Tyson JJJJ, et al. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:975–80. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pomerening JR, Sun YK, Ferrell JE. Systems-level dissection of the cell-cycle oscillator: Bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–78. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 19.King RW, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20s complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–88. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 20.King RW, Deshaies RJ, Peters J-M, Kirschner MW. How Proteolysis Drives the Cell Cycle. Science. 1996;274:1652–9. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 21.Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen LH, Luca FC, et al. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. Journal of Biological Chemistry. 1994;269:4940–6. [PubMed] [Google Scholar]
- 22.Yang Q, Ferrell JE. The Cdk1–APC/C cell cycle oscillator circuit functions as a time-delayed, ultrasensitive switch. Nat Cell Biol. 2013;15:519–25. doi: 10.1038/ncb2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–7. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tischer T, Hörmanseder E, Mayer TU. The APC/C Inhibitor XErp1/Emi2 Is Essential for Xenopus Early Embryonic Divisions. Science. 2012;338:520–4. doi: 10.1126/science.1228394. [DOI] [PubMed] [Google Scholar]
- 25.Mochida S, Maslen SL, Skehel M, Hunt T. Greatwall Phosphorylates an Inhibitor of Protein Phosphatase 2A That Is Essential for Mitosis. Science. 2010;202508 doi: 10.1126/science.1195689. [DOI] [PubMed] [Google Scholar]
- 26.Glover DM. The overlooked greatwall: a new perspective on mitotic control. Open Biology. 2012;2 doi: 10.1098/rsob.120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar BA, Sprenger F, Duronio RJ, Leopold P, O’Farrell PH. Distinct molecular mechanisms regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes and Development. 1994;8:440–52. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O’Farrell PH. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes & Development. 1998;12:1495–503. doi: 10.1101/gad.12.10.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Royou A, Sullivan W, Karess R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. The Journal of Cell Biology. 2002;158:127–37. doi: 10.1083/jcb.200203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keener JTaJ. Singular Perturbation Theory of Spiral Waves in Excitable Media. Physica D. 1988;32 [Google Scholar]
- 31.Chang JB, Ferrell JE. Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature. 2013;500:603–7. doi: 10.1038/nature12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Idema T, Dubuis JO, Kang L, Manning ML, Nelson PC, Lubensky TC, et al. The syncytial Drosophila embryo as a mechanically excitable medium. PloS one. 2013;8:e77216. doi: 10.1371/journal.pone.0077216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sibon OC, Stevenson Va, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–7. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- 34.Fogarty P, Campbell SD, Abu-Shumays R, Phalle BdS, Yu KR, Uy GL, et al. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Current Biology. 7:418–26. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 35.Farrell JA, Shermoen AW, Yuan K, O’Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes and Development. 2012;26:714–25. doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shermoen AW, McCleland ML, O’Farrell PH. Developmental control of late replication and s phase length. Current Biology. 2010;20:2067–77. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar BA, O’Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–80. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tadros W, Westwood JT, Lipshitz HD. The Mother-to-Child Transition. Developmental Cell. 2007;12:847–9. doi: 10.1016/j.devcel.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early drosophila development. Cell. 1986;44:871–7. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- 40.Blythe SA, Wieschaus EF. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell. 2015;160:1169–81. doi: 10.1016/j.cell.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price D, Rabinovitch S, O’Farrell PH, Campbell SD. Drosophila wee1 has an essential role in the nuclear divisions of early embryogenesis. Genetics. 2000;155:159–66. doi: 10.1093/genetics/155.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stumpff J, Duncan T, Homola E, Cambell SD, Su TT. Drosophila Wee1 Kinase Regulates Cdk1 and Mitotic Entry during Embryogenesis. Current Biology. 2004;14:2143–8. doi: 10.1016/j.cub.2004.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Talia S, She R, Blythe SA, Lu X, Zhang QF, Wieschaus EF. Posttranslational control of Cdc25 degradation terminates drosophila’s early cell-cycle program. Current Biology. 2013;23:127–32. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrell JA, O’Farrell PH. Mechanism and regulation of Cdc25/twine protein destruction in embryonic cell-cycle remodeling. Current Biology. 2013;23:118–26. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes and Development. 1996;10:1966–77. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- 46.Großhans J, Müller HAJ, Wieschaus E. Control of cleavage cycles in Drosophila embryos by frühstart. Developmental Cell. 2003;5:285–94. doi: 10.1016/s1534-5807(03)00208-9. [DOI] [PubMed] [Google Scholar]
- 47.Newport J, Kirschner M. A major developmental transition in early xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–86. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 48.Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–72. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- 49.Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development (Cambridge, England) 2009;136:2101–10. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCleland ML, O’Farrell PH. RNAi of Mitotic Cyclins in Drosophila Uncouples the Nuclear and Centrosome Cycle. Current Biology. 2008;18:245–54. doi: 10.1016/j.cub.2008.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Follette PJ, O’Farrell PH. Connecting cell behavior to patterning: Lessons from the cell cycle. Cell. 1997;88:309–14. doi: 10.1016/s0092-8674(00)81869-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar BA, Lehner CF. Developmental Control of Cell Cycle Regulators: A Fly’s Perspective. Science. 1996;274:1646–52. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- 53.Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development (Cambridge, England) 1989;107:1–22. [PubMed] [Google Scholar]
- 54.Di Talia S, Wieschaus EF. Short-Term Integration of Cdc25 Dynamics Controls Mitotic Entry during Drosophila Gastrulation. Developmental Cell. 2012;22:763–74. doi: 10.1016/j.devcel.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raj A, van Oudenaarden A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell. 2008;135:216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science (80-) 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 57.Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154:789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the Limits to Positional Information. Cell. 2007;130:153–64. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrödinger E. What is life? The physical aspect of the living cell: Cambridge [Eng.] Vol. 1945 The University press; 1945. [Google Scholar]
- 60.Driever W, Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989:138–43. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 61.Struhl G, Struhl K, Macdonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–73. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- 62.Berg HC, Purcell EM. Physics of chemoreception. Biophysical Journal. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bialek W, Setayeshgar S. Physical limits to biochemical signaling. Proceedings of the National Academy of Sciences. 2005;102:10040–5. doi: 10.1073/pnas.0504321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briscoe J, Small S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015;142:3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolpert L. Positional information and the spatial pattern of cellular differentiation. Journal of theoretical biology. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 66.Xiong F, Tentner AR, Huang P, Gelas A, Mosaliganti KR, Souhait L, et al. Specified neural progenitors sort to form sharp domains after noisy Shh signaling. Cell. 2013;153:550–61. doi: 10.1016/j.cell.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.OFP H, Lakich D, Lehner CF, Edgar BA. Directing Cell Division During Development Science. 1989;246:635–40. doi: 10.1126/science.2683080. [DOI] [PubMed] [Google Scholar]
- 68.Lehman Da, Patterson B, Johnston La, Balzer T, Britton JS, Saint R, et al. Cis-regulatory elements of the mitotic regulator, string/Cdc25. Development (Cambridge, England) 1999;126:1793–803. doi: 10.1242/dev.126.9.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife. 2015;4:1–14. doi: 10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine M. Suboptimization of developmental enhancers. Science (New York, NY) 2015;350:325–8. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lagha M, Bothma Jacques P, Esposito E, Ng S, Stefanik L, Tsui C, et al. Paused Pol II Coordinates Tissue Morphogenesis in the Drosophila Embryo. Cell. 2013;153:976–87. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferraro T, Esposito E, Mancini L, Ng S, Lucas T, Coppey M, et al. Transcriptional Memory in the Drosophila Embryo. Current Biology. 2015:1–7. doi: 10.1016/j.cub.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siggia ED, Vergassola M. Decisions on the fly in cellular sensory systems. Proceedings of the National Academy of Sciences. 2013:3704–12. doi: 10.1073/pnas.1314081110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Di Talia S, Wieschaus EF. Simple biochemical pathways far from steady state can provide switchlike and integrated responses. Biophysical Journal. 2014;107:L1–L4. doi: 10.1016/j.bpj.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Detwiler PB, Ramanathan S, Sengupta A, Shraiman BI. Engineering Aspects of Enzymatic Signal Transduction: Photoreceptors in the Retina. Biophysical Journal. 79:2801–17. doi: 10.1016/S0006-3495(00)76519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howe JA, Howell M, Hunt T, Newport JW. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes and Development. 1995;9:1164–76. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- 77.Newport J, Kirschner M. A major developmental transition in early xenopus embryos: II. control of the onset of transcription. Cell. 1982;30:687–96. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 78.Amodeo AA, Jukam D, Straight AF, Skotheim JM. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1086–95. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Howe Ja, Newport JW. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:2060–4. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science (New York, NY) 2013;341:893–6. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–56. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 82.Zhang M, Kothari P, Mullins M, Lampson MA. Regulation of zygotic genome activation and DNA damage checkpoint acquisition at the mid-blastula transition. Cell Cycle. 2014;13:3828–38. doi: 10.4161/15384101.2014.967066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dalle Nogare DE, Pauerstein PT, Lane ME. G2 acquisition by transcription-independent mechanism at the zebrafish midblastula transition. Developmental Biology. 2009;326:131–42. doi: 10.1016/j.ydbio.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 84.Kane Da, Warga RM, Kimmel CB. Mitotic domains in the early embryo of the zebrafish. Nature. 1992;360:735–7. doi: 10.1038/360735a0. [DOI] [PubMed] [Google Scholar]
- 85.Bouldin CM, Snelson CD, Farr GH, Kimelman D. Restricted expression of cdc25a in the tailbud is essential for formation of the zebrafish posterior body. Genes and Development. 2014;28:384–95. doi: 10.1101/gad.233577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]