Abstract
Several Artemisia species are used as herbal medicines including the dried aerial parts of Artemisia capillaris, which are used as Artemisiae Capillaris Herba (known as “Injinho” in Korean medicinal terminology and “Yin Chen Hao” in Chinese). In this study, we developed tools for distinguishing between A. capillaris and 11 other Artemisia species that grow and/or are cultured in China, Japan, and Korea. Based on partial nucleotide sequences in the internal transcribed spacer (ITS) that differ between the species, we designed primers to amplify a DNA marker for A. capillaris. In addition, to detect other Artemisia species that are contaminants of A. capillaris, we designed primers to amplify DNA markers of A. japonica, A. annua, A. apiacea, and A. anomala. Moreover, based on random amplified polymorphic DNA analysis, we confirmed that primers developed in a previous study could be used to identify Artemisia species that are sources of Artemisiae Argyi Folium and Artemisiae Iwayomogii Herba. By using these primers, we found that multiplex polymerase chain reaction (PCR) was a reliable tool to distinguish between A. capillaris and other Artemisia species and to identify other Artemisia species as contaminants of A. capillaris in a single PCR.
1. Introduction
The genus Artemisia belongs to the Asteraceae family and is composed of 500 species that are mainly found in Asia, Europe, and North America [1, 2]. Over 350 species in the genus Artemisia are grown in Asia, including China, Korea, and Japan [1]. Several Artemisia species have long been used for the treatment of disease in modern and traditional medicine [2, 3]. For example, the dried aerial parts of A. capillaris are used as Artemisiae Capillaris Herba (“Injinho” in Korean medicinal terminology and “Yin Chen Hao” in Chinese) [4], which controls fever [2], protects the liver [5], and inhibits inflammatory responses [6]. However, the dried leaves of A. capillaris are often mistaken for those of A. japonica. Moreover, young A. capillaris leaves that are harvested in early spring are similar to those of A. argyi and A. princeps [7], which are sources of Artemisiae Argyi Folium (“Aeyup” in Korean and “Ai Ye” in Chinese) that is used for the treatment of pain, vomiting, and bleeding in the uterus [8].
Because of the morphological similarities among the dried and/or sliced shoots and leaves of Artemisia species, some are traded as other species in traditional herbal medicine markets [5, 7]. To resolve this problem, various molecular biology techniques that are based on plant genetic information, such as gene nucleotide sequences (rbcL, matK, or a combination of both), have been used for plant identification and authentication, including medicinal plants [3]. Other gene sequences have been used to discriminate specific medicinal plants from an adulterant or substitute, for example, the trnL-F intergenic spacer for Coptis spp., matK for Rheum spp., and psbA-trnH for Phyllanthus spp. [9–11]. Internal transcribed spacer (ITS) sequences are effective discriminatory tools, and the ITS2 region in particular can be used as a universal DNA barcode for identifying plants and animals [12], including medicinal plants in the family Fabaceae [13] and genus Artemisia [14].
Random amplified polymorphic DNA- (RAPD-) based DNA markers have been used previously for authenticating medicinal plants [3]. In a previous study conducted on six Artemisia species that mainly grow and/or are cultured in Korea (A. princeps, A. argyi, A. capillaris, A. iwayomogi, A. japonica, and A. keiskeana), we discriminated both A. princeps and A. argyi from other Artemisia species using a sequence-characterized amplified region (SCAR) marker, which was based on RAPD results [7]. Using the same method, we identified A. iwayomogi, which is a source of Artemisiae Iwayomogii Herba (“Haninjin” in Korean medicinal terminology) that has been prescribed as a substitute for Artemisiae Capillaris Herba in Korea [15]. However, we were unable to discriminate A. capillaris from A. japonica using the RAPD-based method [5, 7].
In this study, we discriminated A. capillaris from other Artemisia species, particularly A. japonica, by exploiting sequence differences in a specific region of the ITS. We used 12 Artemisia species, six from our previous study and 6 additional Artemisia species, which grow and/or are cultivated in China, Korea, and Japan: A. asiatica, A. montana, and A. lavandulaefolia, which are sources of Artemisiae Argyi Folium in China and Korea [4, 16, 17]; A. annua and A. apiacea, which are sources of Artemisiae Annuae Herba (“Chung-ho” in Korean medical terminology and “Qing Hao” in Chinese) [4, 18] used for the treatment of malaria [19]; and A. anomala, which is a source of Artemisiae Anomalae Herba (“Yugino” in Korean medicinal terminology and “Liu ji nu” in Chinese) [4, 17] used for the treatment of fever and inflammation [20]. A. anomala was included to increase the reliability of the discrimination of A. capillaris. In addition, we tested the effectiveness of multiplex polymerase chain reaction (PCR) to detect contamination of A. capillaris products with those from other Artemisia species. We used primers based on ITS sequences to discriminate among the Artemisia species and two RAPD-based primer sets to discriminate between Artemisia species that are sources of Artemisiae Argyi Folium and Artemisiae Iwayomogii Herba [5, 7].
2. Materials and Methods
2.1. Plant Materials
The fleshy aerial parts, including the leaves, of Artemisia species that grow and/or are cultivated in China, Korea, and Japan were collected (Table 1). The samples were dried at room temperature, frozen, and stored at −80°C. The authenticity of the samples was verified by the Korea Institute of Oriental Medicine (KIOM) and the Department of Herbology, Wonkwang University. The voucher samples were deposited in the KIOM and the Department of Herbology.
Table 1.
Artemisia plants used to determine the internal transcribed spacer (ITS) sequence.
| Number | Medicinal name | Name of the plant species | Place of collection | Voucher number |
|
| ||||
| 1 | Artemisiae Argyi Folium | A. asiatica | Bonghwa, Korea | WKUARE04 |
| 2 | WKUARE24 | |||
| 3 | WKUARE25 | |||
| 4 | A. montana | Kyoto, Japan | WKUARE76 | |
| 5 | WKUARE77 | |||
| 6 | WKUARE78 | |||
| 7 | Jeonju, Korea | WKUARE40 | ||
| 8 | WKUARE41 | |||
| 9 | A. lavandulaefolia | Sichuan, China | WKUARE66 | |
| 10 | WKUARE57 | |||
| 11 | WKUARE58 | |||
| 12 | WKUARE67 | |||
| 13 | A. argyi | Suwon, Korea | WKUARE05 | |
| 14 | WKUARE06 | |||
| 15 | WKUARE30 | |||
| 16 | WKUARE31 | |||
| 17 | Guangxi, China | WKUARE27 | ||
| 18 | Sichuan, China | WKUARE59 | ||
| 19 | A. princeps | Jeonju, Korea | WKUARE43 | |
| 20 | WKUARE44 | |||
| 21 | WKUARE45 | |||
| 22 | Uiseong, Korea | WKUARE01 | ||
| 23 | Ganghwa, Korea | WKUARE55 | ||
| 24 | WKUARE56 | |||
|
| ||||
| 25 | Artemisiae Capillaris Herba | A. capillaris | Suwon, Korea | WKUARE33 |
| 26 | WKUARE34 | |||
| 27 | Jeonju, Korea | WKUARE35 | ||
| 28 | WKUARE46 | |||
| 29 | WKUARE47 | |||
| 30 | Nishi, Japan | WKUARE79 | ||
| 31 | WKUARE80 | |||
| 32 | Sichuan, China | WKUARE52 | ||
|
| ||||
| 33 | Artemisiae Iwayomogii Herba | A. iwayomogi | Suwon, Korea | WKUARE37 |
| 34 | Jinan, Korea | WKUARE10 | ||
| 35 | Pohang, Korea | WKUARE68 | ||
| 36 | WKUARE69 | |||
| 37 | Jeonju, Korea | WKUARE11 | ||
| 38 | WKUARE48 | |||
| 39 | WKUARE49 | |||
|
| ||||
| 40 | Artemisiae Annuae Herba | A. annua | Namwon, Korea | WKUARE20 |
| 41 | Yeongcheon, Korea | WKUARE21 | ||
| 42 | Sichuan, China | WKUARE60 | ||
| 43 | WKUARE61 | |||
| 44 | WKUARE62 | |||
| 45 | A. apiacea | Sichuan, China | WKUARE63 | |
| 46 | WKUARE53 | |||
| 47 | WKUARE54 | |||
| 48 | Nishi, Japan | WKUARE81 | ||
| 49 | WKUARE82 | |||
|
| ||||
| 50 | Artemisiae Anomalae Herba | A. anomala | Sichuan, China | WKUARE64 |
| 51 | WKUARE65 | |||
| 52 | Pohang, Korea | WKUARE73 | ||
| 53 | WKUARE74 | |||
| 54 | WKUARE75 | |||
|
| ||||
| 55 | Artemisiae Japonicae Herba | A. japonica | Suwon, Korea | WKUARE39 |
| 56 | Jeonju, Korea | WKUARE50 | ||
| 57 | WKUARE51 | |||
| 58 | Namryung, China | WKUARE17 | ||
| 59 | Nishi, Japan | WKUARE83 | ||
| 60 | WKUARE84 | |||
|
| ||||
| 61 | Artemisia Keiskeanae Herba | A. keiskeana | Suwon, Korea | WKUARE16 |
| 62 | Uiseong, Korea | WKUARE15 | ||
| 63 | Pohang, Korea | WKUARE70 | ||
| 64 | WKUARE71 | |||
| 65 | WKUARE72 | |||
2.2. Preparation of Genomic DNA
Genomic DNA from each sample was extracted in accordance with the instruction manual for the NucleoSpin® Plant II (Macherey-Nagel, Duren, Germany). To improve DNA quality, phenolic compounds and polysaccharides were removed using 10% cetyltrimethylammonium bromide and 0.7 M NaCl. After the purity and amount of the prepared genomic DNA were determined using a NanoDrop™ DN-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA), the DNA was diluted to 10 ng/μL and stored.
2.3. PCR Amplification
2.3.1. Amplification of ITS
A PCR for the amplification of the ITS, including the 5.8S rRNA coding region, was conducted using a T-personal cycler (Biometra, Goettingen, Germany) according to the protocol by White et al. [21]. In brief, 1.2 pmol of ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) primers, 1 U Taq polymerase (ABgene, Epsom, UK), and 20 ng of genomic DNA extracted from each sample were used for the PCR amplification. During the 35-cycle PCR process, predenaturation was conducted for 5 min at 95°C and denaturation for 30 s at 95°C. The annealing process was conducted for 30 s at 52°C and the extension process for 1 min at 72°C. A final reaction step was conducted for 7 min at 72°C. The amplified products were separated on 1.2% agarose gel and revealed by staining with ethidium bromide (Sigma-Aldrich, St. Louis, MO, USA). The amplified PCR products were analyzed using MyImage (Seoulin Biotechnology, Seoul, Korea) and purified using a LaboPass™ Gel Kit (Cosmo Genetech, Seoul, Korea).
2.3.2. Amplification of DNA and SCAR Markers
In brief, 1.2 pmol of primers, 1 U Taq polymerase (ABgene), and 50 ng of genomic DNA extracted from each Artemisia species were used for the PCR amplification. During the 23-cycle PCR process, predenaturation was conducted for 5 min at 95°C and denaturation for 30 s at 95°C. In general, the annealing process was conducted for 30 s at 53.5°C for the amplification of the DNA markers. However, to amplify the DNA markers for A. capillaris, A. japonica, A. apiacea, A. annua, and A. anomala, this process was conducted for 15–30 s at 54–58°C. The extension process was conducted for 20 s (except for A. apiacea, which had 30 s) at 72°C, and a final reaction step was conducted for 5 min at 72°C. To amplify an internal standard for the evaluation of the PCR conduct, a primer set (AYF/AYR) was used to amplify a 94 bp sequence. The amplified products were separated on 1.2% agarose gel and revealed by staining with ethidium bromide (Sigma-Aldrich). The amplified PCR products were then analyzed using MyImage (Seoulin Biotechnology).
2.3.3. Multiplex PCR
For the multiplex PCR amplification, 0.07 pmol of the primers Fb and R7; 0.14 pmol of the primers AYF and AYR; 0.7 pmol of the primer Aam F3; 1.7 pmol of the primers AC F4, ACJ R3, and Aap R2; 3.4 pmol of the primers 2F1, 2F3, AJ F1, AC R3, Aap F1, AA F3, and Aa R4; 1x PrimeSTAR® Max DNA Polymerase (Takara Bio Inc., Kusatsu, Japan); and 20 ng of genomic DNA extracted from each Artemisia species were used. During the 30-cycle PCR process, predenaturation was conducted for 10 min at 95°C and denaturation for 10 s at 95°C. The annealing process was conducted for 5 s at 56.5°C and the extension process for 10 s at 72°C. A final reaction step was conducted for 7 min at 72°C. The amplified products were separated on 2% agarose gel and revealed by staining with ethidium bromide (Sigma-Aldrich). In order to amplify an internal standard for the evaluation of the PCR, the AYF/AYR primer set was used to amplify a 94 bp sequence. The amplified PCR products were then analyzed using MyImage (Seoulin Biotechnology).
2.4. Nucleotide Sequencing of the PCR Products
The nucleotide sequences of the PCR products were directly determined using the primers ITS1 and ITS4 by Macrogen (Seoul, Korea). In other cases, the PCR products resolved by agarose electrophoresis were cloned using a pGEM®-T Easy Vector System I (Promega, Madison, WI, USA). The nucleotide sequences of the subcloned PCR products were determined by Macrogen.
2.5. Alignment of the DNA Sequences and Construction of a Dendrogram
The DNA sequences were manually edited and aligned by ClustalW multiple sequence alignment in BioEdit v7.0.9 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). A dendrogram was constructed using the neighbor-joining method [22] in the MEGA6 program [23] with 1000 bootstrap iterations. Evolutionary distances were computed using the maximum composite likelihood method [24] in MEGA6.
3. Results
3.1. Determination and Analysis of ITS Sequences
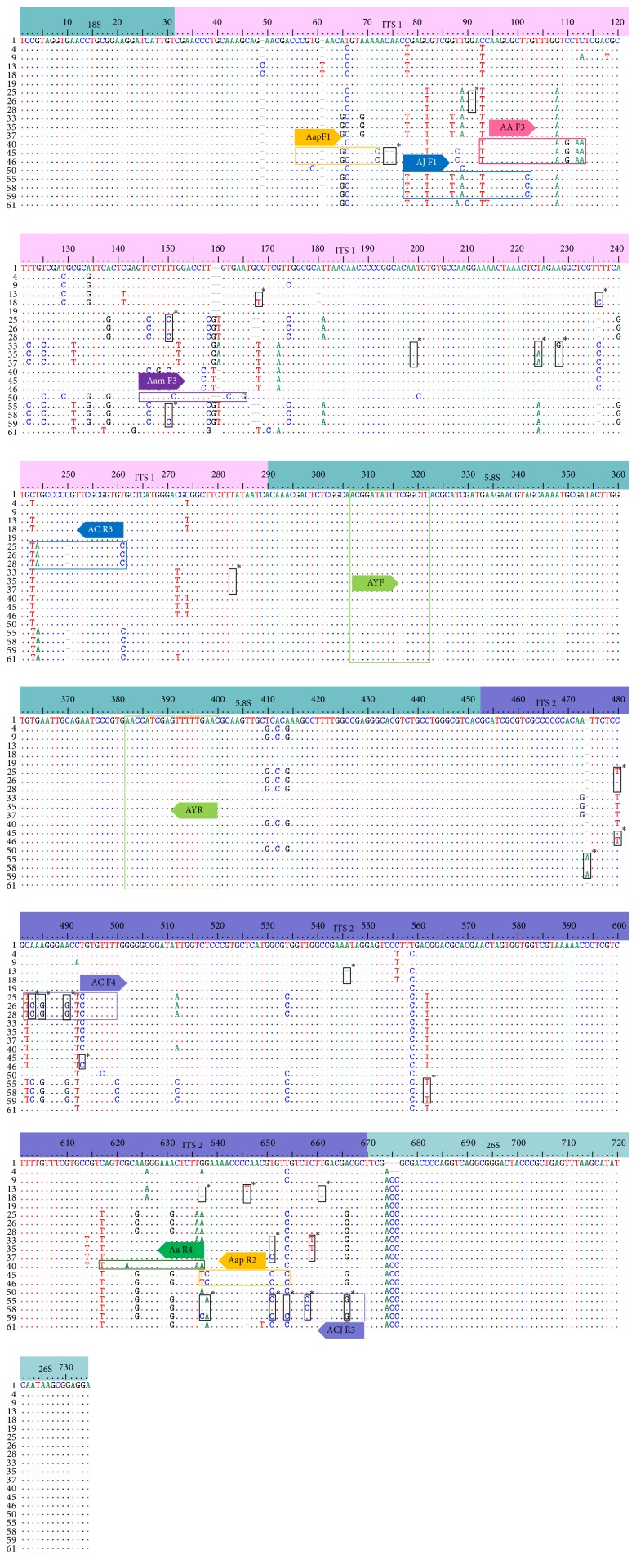
The 726–731 bp nucleotide sequences of the ITS, including the 5.8S region, were determined in 65 samples of 12 Artemisia species (Table 1). Parts of the ITS sequences of each Artemisia species are presented in Figure 1 and were deposited in GenBank (accession numbers KT965653–KT965672). As shown in Figure 1, in the intraspecific samples of five Artemisia species (A. argyi, A. capillaris, A. iwayomogi, A. apiacea, and A. japonica), 4–9 bp differences in the ITS1 and ITS2 sequences were detected. In the case of A. japonica (sample numbers 55, 58, and 59), there were 8 bp differences in the ITS2 region and a 1 bp difference in the ITS1 region. These differences resulted mainly from substitutions (mostly base transitions) and a deletion. In A. apiacea (sample numbers 45 and 46), two base deletions in ITS1 and two substitutions in ITS2 were detected in sample number 45.
Figure 1.
Multiple alignments of nucleotide sequences of the internal transcribed spacer (ITS) among Artemisia species. The dots indicate consensus nucleotides and the dashes represent gaps. Numbers represent sample numbers (see Table 1). Bold arrows indicate the primers used to amplify DNA markers of the Artemisia species, and colored boxes represent nucleotide sequences as well as the positions of the ITS in the primers. Black boxes with an asterisk indicate variations in the nucleotides within species.
To determine whether each Artemisia species could be identified by interspecific ITS sequence differences, we constructed a dendrogram based on the ITS sequences. As outgroups, we used GenBank sequences of Aster yomena (accession number HQ154048.1) and Chrysanthemum coronarium (accession number EF577292.1) in the family Asteraceae, in which Artemisia is included (Figure 2). As shown in Figure 2, each Artemisia species was classified into a separate group on the dendrogram. All of the A. japonica samples that exhibited excessive intraspecific ITS sequence variation were sorted into a group. Fortunately, the A. capillaris samples were separate from the A. japonica samples on the dendrogram. In addition, both A. annua and A. apiacea, which are sources of Artemisia Annuae Herba, were classified into the same cluster on the dendrogram (Figure 2). Interestingly, A. argyi, A. princeps, A. montana, A. lavandulaefolia, and A. asiatica, which are sources of Artemisiae Argyi Folium, were classified into only one cluster.
Figure 2.
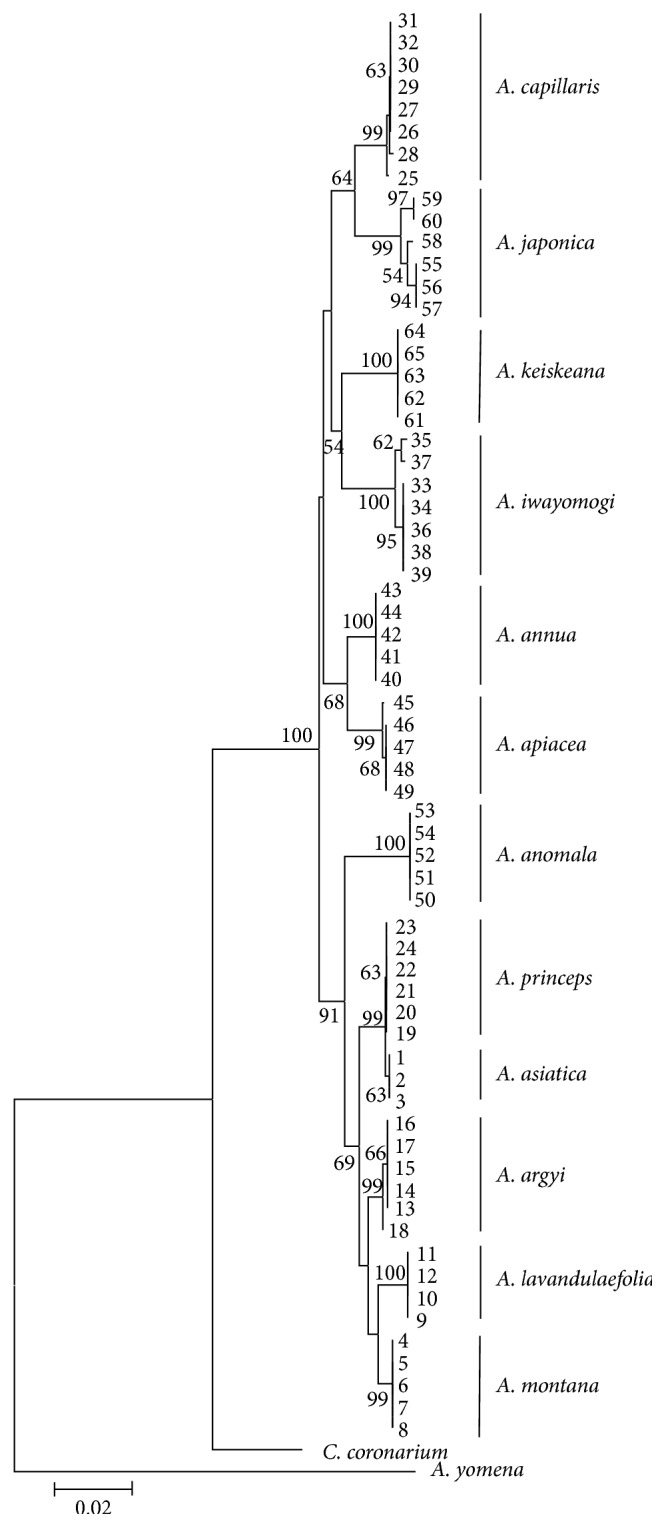
Dendrogram based on the internal transcribed spacer (ITS) sequences presented in Figure 1. ITS sequences of Aster yomena (accession number HQ154048.1) and Chrysanthemum coronarium (accession number EF577292.1) in GenBank were used as outgroups. The unit of evolutionary distance was the number of base substitutions per site; bootstrap values of over 50% are indicated on the branches of the dendrogram.
3.2. Discrimination of A. capillaris from Other Artemisia Species by Differences in ITS Sequences
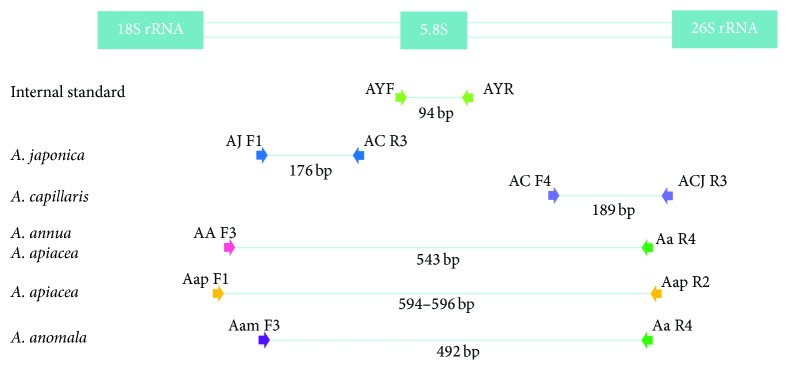
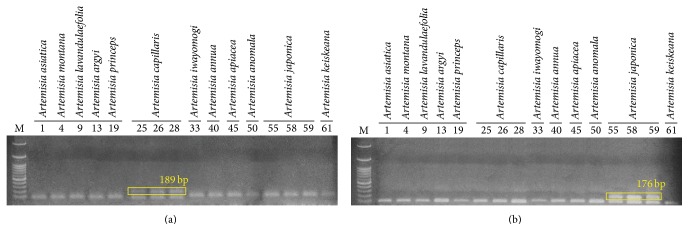
Based on the results shown in Figure 2, we could discriminate A. capillaris from other Artemisia species, at least from the 11 Artemisia species used in this study, by differences in the ITS sequences. It was difficult to discriminate A. capillaris from A. japonica, which was close to A. capillaris on the dendrogram and exhibited significant variation in its ITS sequence. Most of the variation in the ITS sequences among the intraspecific A. japonica samples was found in the ITS2 region (Figure 1); therefore, we excluded the ITS2 region when designing primers to amplify specific DNA markers for A. japonica. As shown in Figures 1 and 3, we designed the primer set AC F4/ACJ R3 in order to amplify a 189 bp PCR product in the ITS1 region that only appeared in A. capillaris samples (Figures 3 and 4(a)). Subsequently, we designed the AJ F1/AC R3 primer set in order to amplify a 176 bp PCR product in ITS2 that only appeared in A. japonica samples (Figures 3 and 4(b)).
Figure 3.
Relative positions of the primers designed to amplify DNA markers of Artemisia species on the internal transcribed spacer and the expected size of the polymerase chain reaction products.
Figure 4.
Polymerase chain reaction products of the primer sets AC F4/ACJ R3 (a) and AJ F1/AC R3 (b) from 12 Artemisia species. Lane numbers are listed in Table 1. M: 100 bp ladder.
Based on these results, we suggest that two primer sets (AJ F1/AC R3 and AC F4/ACJ R3) could be used to discriminate A. capillaris not only from A. japonica but also from other Artemisia species.
3.3. Discrimination of Artemisia Species That Are Sources of Artemisiae Annuae Herba and Artemisiae Anomalae Herba by Differences in ITS Sequences
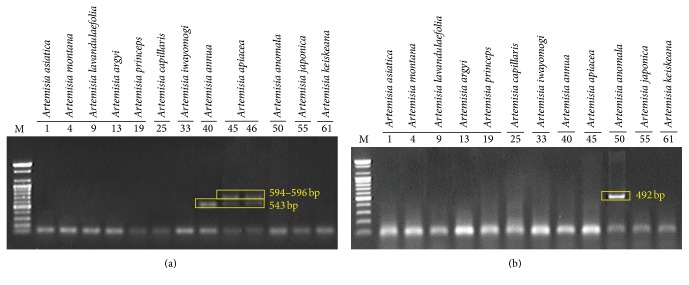
We developed DNA markers in order to detect contamination of A. capillaris by other Artemisia species. As shown in Figure 2, A. annua and A. apiacea, which are sources of Artemisiae Annuae Herba, were close together on the dendrogram in a similar manner as A. capillaris and A. japonica. Therefore, we attempted to find region(s) in ITS1 and ITS2 to discriminate both A. annua and A. apiacea from other Artemisia species. As shown in Figures 1 and 3, we designed the AA F3/Aa R4 primer set in order to amplify a 543 bp PCR product in both A. annua and A. apiacea simultaneously as a common DNA marker. Subsequently, we designed primers to amplify a specific DNA marker to discriminate A. annua from A. apiacea. Based on the differences found in the ITS1 and ITS2 sequences, we designed the Aap F1/Aap R2 primer set in order to amplify a 594 (in sample number 45, which had a 2 bp deletion) or 596 bp (in sample number 46) PCR product that only appeared in A. apiacea samples (Figures 1 and 3). Based on amplifications of the one or two PCR products expected on the gel (Figure 5(a)), we confirmed that the AA F3/Aa R4 and Aap F1/Aap R2 primer sets could discriminate not only A. annua from A. apiacea but also these two species from other Artemisia species. In the case of A. anomala, we designed an Aam F3/Aa R4 primer set in order to amplify a 492 bp PCR product in A. anomala samples (Figures 1 and 3) and confirmed that the expected 492 bp single band of the PCR product only appeared in A. anomala samples (Figure 5(b)).
Figure 5.
Polymerase chain reaction products of the primer sets AA F3/Aa R4 and Aap F1/Aap R2 (a) and Aam F3/Aa R4 (b) from 12 Artemisia species. Lane numbers are listed in Table 1. M: 100 bp ladder.
3.4. Detection of Contamination by Other Artemisia Species Using Multiplex PCR
As shown in Figures 1 and 2, differences in the ITS sequences could discriminate five Artemisia species—A. asiatica, A. montana, A. lavandulaefolia, A. argyi, and A. princeps—that are sources of Artemisiae Argyi Folium and A. iwayomogi that is a source of Artemisiae Iwayomogii Herba from the six other Artemisia species. However, designing primers in order to amplify DNA markers for these species based on differences in the ITS sequences was difficult. Therefore, we tested the usability of the Fb/R7 and 2F1/2F3 primer sets in order to amplify SCAR markers that were developed in previous studies with six Artemisia species [5, 7]. We confirmed that the Fb/R7 primer set amplified a 254 bp SCAR marker in samples of not only A. princeps and A. argyi but also A. asiatica, A. lavandulaefolia, and A. montana (data not shown). Furthermore, we confirmed that the 2F1/2F3 primer set amplified a 364 or 365 bp SCAR marker only in A. iwayomogi, and that this marker was not amplified in any other species, including A. asiatica, A. montana, A. lavandulaefolia, A. annua, A. apiacea, or A. anomala (data not shown). Therefore, these two RAPD-based primer sets could detect contamination by these Artemisia species in addition to the six other Artemisia species. Using the multiplex PCR method, we tested the reliability of these two primer sets and those developed based on the ITS sequences to discriminate A. capillaris from other Artemisia species and to detect contamination by other Artemisia species. For the multiplex PCR process, we randomly selected one sample from each Artemisia species listed in Table 1. As shown in Figure 6, these primer sets functioned reliably, not only to discriminate A. capillaris from other Artemisia species, but also to simultaneously detect contamination by other Artemisia species in a single PCR process.
Figure 6.
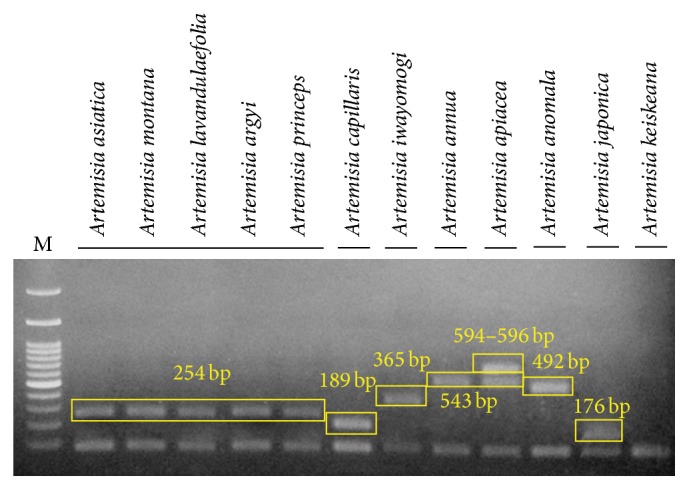
Multiplex polymerase chain reaction products using the primers shown in Figures 1 and 3 from 12 randomly selected Artemisia species. Genomic DNA from a randomly chosen sample of each Artemisia species was used for PCR amplification. M: 100 bp ladder.
Finally, by mixing genomic DNA isolated from different Artemisia species at varying content ratios, we tested the reliability of this PCR method to detect contamination of other Artemisia species, such as A. japonica, A. princeps, and A. iwayomogi, which are mostly found in Korea and are easily misused. As shown in Figure 7, the multiplex PCR detected two Artemisia species that had been mixed at ratios of 9 : 1 and 19 : 1. Furthermore, the multiplex PCR detected a mixture of three Artemisia species (A. capillaris with A. japonica and A. princeps or A. capillaris with A. japonica and A. iwayomogi) at ratios of 8 : 1 : 1 and 18 : 1 : 1 (Figure 7). Therefore, we suggest that the multiplex PCR method is an accurate tool to discriminate A. capillaris from other Artemisia species and could be used to determine whether A. capillaris samples have been mixed with other Artemisia species.
Figure 7.
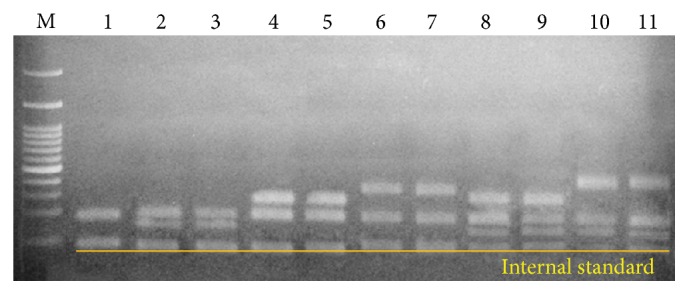
Multiplex polymerase chain reaction products by using two or three combined primer sets from mixed genomic DNA isolated from two or three Artemisia species at different content ratios. Primer set AC F4/ACJ R3 amplified a 189 bp DNA marker to detect A. capillaris; AJ F1/AC R3 amplified a 176 bp DNA marker to detect A. japonica; Fb/R7 amplified a 254 bp DNA marker to detect A. princeps; 2F1/2F3 amplified a 365 bp DNA marker to detect A. iwayomogi. Lane 1: A. capillaris; Lane 2: A. capillaris and A. japonica (9 : 1); Lane 3: A. capillaris and A. japonica (19 : 1); Lane 4: A. capillaris and A. princeps (9 : 1); Lane 5: A. capillaris and A. princeps (19 : 1); Lane 6: A. capillaris and A. iwayomogi (9 : 1); Lane 7: A. capillaris and A. iwayomogi (19 : 1); Lane 8: A. capillaris, A. japonica, and A. princes (8 : 1 : 1); Lane 9: A. capillaris, A. japonica, and A. princeps (18 : 1 : 1); Lane 10: A. capillaris, A. japonica, and A. iwayomogi (8 : 1 : 1); Lane 11: A. capillaris, A. japonica, and A. iwayomogi (18 : 1 : 1); M: 100 bp ladder, yellow underline: 96 bp internal standard amplified by AYF/AYR primer set.
4. Discussion
Medicinal plants have long been used to treat disease in traditional and modern medicine [1]. However, because of the substitution and adulteration of medicinal plants with closely related species, the value of the original drug decreases and in some cases can make it lethal when substituted or contaminated with toxic adulterant plant(s) [3]. Therefore, the authentication of medicinal plants is crucial. As mentioned previously, A. capillaris, which is a source of Artemisiae Capillaris Herba, should be discriminated from not only A. japonica, A. argyi, and A. princeps but also other Artemisia species that grow and/or are cultivated in Korea and China and could contaminate the products of A. capillaris. Artemisia species, including A. capillaris, are a valuable source of new drugs and essential oils, and their unique chemical compositions and pharmacological activity are species-specific [1, 2, 5]. In this context, we developed a method to discriminate A. capillaris from other Artemisia species and to detect contamination among Artemisia species.
The DNA barcode is a powerful tool for identifying and discriminating between species of animal, plant, and fungus. The sequence at the 5′ end of cytochrome c oxidase subunit 1 (CO1) in the mitochondrial genome is used for animal taxonomic classification [25, 26]; however, plants cannot currently be identified by the sequence of a single locus [3]. Therefore, the Consortium for the Barcode of Life (CBOL) Plant Working Group proposed a combination of sequences of matK in the nuclear genome and rbcL in the chloroplast genome to identify plants [3]. However, the discriminatory power of the combined matK and rblL loci is low, particularly when discriminating between closely related species, such as 36 species in the genus Dendrobium [27]. Instead, by using a single matK sequence, medicinal plants in the subfamily Rauvolfioideae and genus Rheum have been successfully discriminated from each other [28, 29]. Despite the relatively low level of variation found in rbcL sequences in 48 plant genera including Amaranthus, Angelica, and Ilex, their combination with trnH-psbA intergenic spacer sequences increased the identification and discrimination success rate from 79% to 88% [30]. Therefore, to identify or discriminate between specific medicinal plants and closely related species, other single loci, besides CO1, or a combination of loci, besides matK and rbcL, have been used. For example, various Dendrobium Sw. species have been discriminated between them using a single sequence of the trnH-psbA intergenic spacer [31]. In addition, the trnL-trnF intergenic spacer sequence clearly discriminated Cardiocrinum giganteum from C. giganteum var. yunnanense and C. cordatum [32].
Of the various DNA barcode loci used, the ITS is one of the most useful. Multiple copies of the ITS are tandemly located at one or more chromosomal loci, and there are hundreds or thousands of ITS repeats in the nuclear genome. Furthermore, the ITS, including ITS1, 5.8S rRNA, and ITS2, is relatively small and ranges from 400 bp to under 1000 bp long [33]. Because of the presence of high copy numbers of the ITS and its small size, the ITS is easily amplified by PCR [34]. The level of variation among interspecific ITS sequences is high, so they can be used for the identification of plants at the specific, generic, and even family levels [35]. In contrast, levels of variation within intraspecific ITS sequences are often very low [34]. Concerted evolution should homogenize the sequences of ITS repeats that exist in a species by high-frequency unequal crossing over or gene conversion [36, 37]. The ITS2 sequence in particular has been used to identify medicinal plants that belong to the genera Swartzia and Artemisia in the family Fabaceae [13, 14]. In addition, ITS2 sequences, combined with rbcL sequences, have been used for detecting the contamination and substitution of products from 42 medicinal plants, including Achillea racemose and Urtica dioica, in Canada and the USA [38].
As shown in Figures 1 and 2, discriminating A. capillaris from A. japonica and 10 other Artemisia species was based upon differences in ITS sequences among the Artemisia species. Using the RAPD method with nonspecific primers, we were unable to discriminate A. capillaris from A. japonica in a previous study [5, 7]. Here, we were able to discriminate A. capillaris from A. japonica because of differences in nucleotide sequences, particularly in the ITS2 region (Figure 1). Basing discrimination on differences in ITS sequences was conducted cautiously, because of the considerable sequence variation found in the ITS sequences, particularly among intraspecific A. japonica samples. We also observed this variation in the A. japonica ITS sequences deposited in GenBank (accession numbers AM398882, AY548200, GU724289, JF326554, JX051713, and KC493078). Therefore, we confirmed the discriminatory power of the ITS sequences by using the A. capillaris and A. japonica ITS sequences deposited in GenBank. The deposited A. japonica sequences, together with the A. japonica sequences determined in this study (sample numbers 55–60), were clearly discriminated from both the deposited (accession numbers AY548201 and KC493083) and determined (sample numbers 25–32) A. capillaris sequences (data not shown).
Therefore, differences in ITS sequences can be used to discriminate among Artemisia species, despite the large variations observed in the ITS sequences of specific Artemisia species. Lee et al. [39] compared ITS sequences among Artemisia species that grow naturally in Korea, including two varieties and one subspecies of A. japonica. They estimated the pairwise divergence value as 0.004 between the varieties and subspecies based on the Kiura-2 parameter. Because we could not find any sequence information in their article or GenBank, we were unable to determine how many nucleotide variations exist between the varieties and subspecies of A. japonica. However, based on the results of their study, we suggest that the intraspecific ITS sequence variation detected in A. japonica could result from the different varieties and/or subspecies of A. japonica used for the determination of the ITS sequences.
For the discrimination of A. capillaris from A. japonica, which were closer to each other than to any other Artemisia species on the dendrogram (Figure 2), the primer sets AC F4/ACJ R3 (that amplified a 189 bp DNA marker in A. capillaris) and AJ F1/AC R3 (that amplified a 176 bp DNA marker in A. japonica) were designed (Figures 1 and 3). Despite the fact that there was not a remarkable difference in the sizes of the DNA markers for A. capillaris and A. japonica, they were clearly separated on 2% agarose gel after 40 min of gel running (Figure 6).
The method of amplifying double DNA markers of specific species was used to discriminate A. annua from A. apiacea, which were close to each other on the dendrogram (Figure 2). A 543 bp DNA marker was only amplified in A. annua using the AA F3/Aa R4 primer set, and both the 543 and 594 bp (or 596 bp, depending on the presence of a base deletion) DNA markers were amplified in A. apiacea using the AA F3/Aa R4 and Aap F1/Aap R2 primer sets, respectively (Figures 5(a) and 5(b)). For the discrimination of the five Artemisia species that are sources of Artemisiae Argyi Folium, we first determined whether the nonspecific UBC primer 329 (5′-GCGAACCTCC-3′), which amplified a unique 850 bp PCR product only in A. argyi and A. princeps in a previous study [7], could amplify the same PCR product in three additional Artemisia species (A. asiatica, A. montana, and A. lavandulaefolia). Using samples from the 12 species, we confirmed that the same PCR products were amplified in these three Artemisia species (data not shown). We then confirmed that the Fb/R7 primer set, which was designed to amplify a 254 bp SCAR marker based on the sequence of an 850 bp PCR product [7], amplified the same-sized DNA marker in the three additional Artemisia species (data not shown). For the discrimination of A. iwayomogi, we tested whether the nonspecific UBC primer 391 (5′-GCGAACCTCG-3′), which amplified four kinds of PCR products that ranged in size from 707 to 719 bp in A. iwayomogi in a previous study [5], could amplify the same PCR products in six additional Artemisia species (A. asiatica, A. montana, A. lavandulaefolia, A. apiacea, A. annua, and A. anomala). We confirmed that the UBC primer 391 amplified PCR products only in A. iwayomogi. In addition, we confirmed that the 2F1/2F3 primer set, which was designed to amplify a 365 bp SCAR marker based on the sequences of four PCR products [5], amplified the same-sized DNA marker only in A. iwayomogi (data not shown). Based on these results, we were convinced that the Fb/R7 and 2F1/2F3 primer sets could discriminate A. capillaris not only from the five Artemisia species that are sources of Artemisiae Argyi Folium but also from A. iwayomogi.
Using primer sets based on the ITS sequences and RAPD results to discriminate among the Artemisia species, we evaluated the multiplex PCR method to discriminate A. capillaris and to detect contamination of A. capillaris by randomly selecting each sample of Artemisia species (Figure 6) and mixed samples of A. capillaris with A. japonica, A. princeps, and A. iwayomogi (Figure 7). Therefore, we suggest that the multiplex PCR method is an accurate tool to discriminate A. capillaris from other Artemisia species and could be used to determine whether A. capillaris samples have been mixed with samples from other Artemisia species, at least those tested in this study.
5. Conclusion
To differentiate among A. capillaris plants that produce Artemisiae Capillaris Herba and 11 other Artemisia species, 726–731 bp ITS nucleotide sequences in 65 samples were determined and analyzed. Based on differences found in partial ITS nucleotide sequences between the species, we designed the primer sets AC F4/ACJ R3 to amplify a 189 bp PCR product and AJ F1/AC R3 to amplify a 176 bp PCR product in A. capillaris and A. japonica, respectively. To detect traces of other Artemisia species in A. capillaris, we designed the primer set AA F3/Aa R4 to amplify a 543 bp product in A. annua, the primer set Aap F1/Aap R2 to amplify a 594–596 bp product in A. apiacea, and the primer set Aam F3/Aa R4 to amplify a 492 bp product in A. anomala. In addition, we confirmed that the primer sets Fb/R7 and 2F1/2F3, which had been developed in a previous study based on RAPD, could be used to amplify 254 bp products in A. princeps, A. argyi, A. asiatica, A. lavandulaefolia, and A. montana, which are sources of Artemisiae Argyi Folium, and to amplify 364 or 365 bp products in A. iwayomogi. Therefore, we demonstrate that the discrimination of A. capillaris from and the detection of contamination by other Artemisia species can be reliably performed by multiplex PCR using these primers.
Acknowledgments
This study was part of Konkuk University's research support program for its faculty on sabbatical leave in 2013.
Disclosure
Eui Jeong Doh's present address is Center for Metabolic Function Regulation, Wonkwang University, Iksan 570-749, Republic of Korea.
Competing Interests
The authors declare that there are no competing interests regarding the publication of this paper.
Authors' Contributions
Eui Jeong Doh and Seung-Ho Paek contributed equally to this work.
References
- 1.Abad M. J., Bedoya L. M., Apaza L., Bermejo P. The Artemisia L. genus: a review of bioactive essential oils. Molecules. 2012;17(3):2542–2566. doi: 10.3390/molecules17032542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joshi R. K. Artemisia capillaris: medicinal uses and future source for commercial uses from western himalaya of Uttrakhand. Asian Journal of Research in Pharmaceutical Science. 2013;3(3):137–140. [Google Scholar]
- 3.Techen N., Parveen I., Pan Z., Khan I. A. DNA barcoding of medicinal plant material for identification. Current Opinion in Biotechnology. 2014;25:103–110. doi: 10.1016/j.copbio.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Korea Food and Drug Administration. The Korean Herbal Pharmacopoeia. Seoul, Republic of Korea: KFDA Press; 2011. [Google Scholar]
- 5.Lee M. Y., Doh E. J., Kim E. S., Kim Y. W., Ko B. S., Oh S.-E. Application of the multiplex PCR method for discrimination of Artemisia iwayomogi from other Artemisia herbs. Biological and Pharmaceutical Bulletin. 2008;31(4):685–690. doi: 10.1248/bpb.31.685. [DOI] [PubMed] [Google Scholar]
- 6.Hong S. H., Seo S. H., Lee J. H., Choi B. T. The aqueous extract from Artemisia capillaris Thunb. inhibits lipopolysaccharide-induced inflammatory response through preventing NF-κB activation in human hepatoma cell line and rat liver. International Journal of Molecular Medicine. 2004;13(5):717–720. [PubMed] [Google Scholar]
- 7.Lee M. Y., Doh E. J., Park C. H., et al. Development of SCAR marker for discrimination of Artemisia princeps and A. argyi from other Artemisia herbs. Biological and Pharmaceutical Bulletin. 2006;29(4):629–633. doi: 10.1248/bpb.29.629. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Q.-C., Kiyohara H., Yamada H. Anti-complementary neutral polysaccharides from leaves of Artemisia princeps . Phytochemistry. 1993;35(1):73–77. doi: 10.1016/s0031-9422(00)90512-0. [DOI] [PubMed] [Google Scholar]
- 9.Doh E. J., Lee M. Y., Ko B. S., Oh S.-E. Differentiating Coptis chinensis from Coptis japonica and other Coptis species used in Coptidis Rhizoma based on partial trnL-F intergenic spacer sequences. Genes & Genomics. 2014;36(3):345–354. doi: 10.1007/s13258-014-0172-2. [DOI] [Google Scholar]
- 10.Xu G., Wang X., Liu C., et al. Authentication of official Da-huang by sequencing and multiplex allele-specific PCR of a short maturase K gene. Genome. 2013;56(2):109–113. doi: 10.1139/gen-2012-0182. [DOI] [PubMed] [Google Scholar]
- 11.Srirama R., Gurumurthy B. R., Senthilkumar U., Ravikanth G., Shaanker R. U., Shivanna M. B. Are mini DNA-barcodes sufficiently informative to resolve species identities? An in silico analysis using Phyllanthus . Journal of Genetics. 2014;93(3):823–829. doi: 10.1007/s12041-014-0432-6. [DOI] [PubMed] [Google Scholar]
- 12.Yao H., Song J., Liu C., et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013102.e13102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao T., Yao H., Song J., et al. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. Journal of Ethnopharmacology. 2010;130(1):116–121. doi: 10.1016/j.jep.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Liu M.-Z., Song J.-Y., Luo K., Lin Y.-L., Liu P., Yao H. Identification of nine common medicinal plants from Artemisia L. by DNA barcoding sequences. Chinese Traditional and Herbal Drugs. 2012;43(7):1393–1397. [Google Scholar]
- 15.Kim Y. H., Jeong M. Y., Lee N. K., et al. Antimicrobial effect on the periodontal pathogens and anti-inflammatory effect of Artemisiae Iwayomogii Herba. The Korea Journal of Herbology. 2008;23(2):1–8. [Google Scholar]
- 16.Pharmacopoeia Committee of the DPRK. Pharmacopoeia of Democratic People's Republic of Korea. 7th. Pyeongyang, Democratic People's Republic of Korea: Medicine and Science Press; 2011. [Google Scholar]
- 17.Zhonghua Bencao. Zhonghua Bencao. Shanghai, China: Shanghai Science and Technology Press; 1999. [Google Scholar]
- 18.Chinese Pharmacopoeia Committee. Chinese Pharmacopoeia Commission. Beijing, China: China Medical Science and Technology Press; 2010. [Google Scholar]
- 19.Willcox M. L., Bodeker G., Bourdy G., et al. Artemisia annua as a traditional herbal antimalarial. In: Willcox M. L., Bodeker G., Rasoanaivo P., editors. Traditional Medicinal Plants and Malaria. Boca Raton, Fla, USA: CRC Press; 2004. pp. 43–59. [Google Scholar]
- 20.Tan X., Wang Y.-L., Yang X.-L., Zhang D.-D. Ethyl acetate extract of Artemisia anomala S. moore displays potent anti-inflammatory effect. Evidence-Based Complementary and Alternative Medicine. 2014;2014:10. doi: 10.1155/2014/681352.681352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White T. J., Bruns T., Lee S. J., Taylor J. W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 1990;18:315–322. [Google Scholar]
- 22.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hebert P. D., Cywinska A., Ball S. L. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebert P. D. N., Ratnasingham S., de Waard J. R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B: Biological Sciences. 2003;270(supplement 1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh H. K., Parveen I., Raghuvanshi S., Babbar S. B. The loci recommended as universal barcodes for plants on the basis of floristic studies may not work with congeneric species as exemplified by DNA barcoding of Dendrobium species. BMC Research Notes. 2012;5, article 42 doi: 10.1186/1756-0500-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahadani P., Sharma G. D., Ghosh S. K. Identification of ethnomedicinal plants (Rauvolfioideae: Apocynaceae) through DNA barcoding from northeast India. Pharmacognosy Magazine. 2013;9(35):255–263. doi: 10.4103/0973-1296.113284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D.-Y., Fushimi H., Cai S.-Q., Komatsu K. Molecular analysis of Rheum species used as Rhei Rhizoma based on the chloroplast matK gene sequence and its application for identification. Biological and Pharmaceutical Bulletin. 2004;27(3):375–383. doi: 10.1248/bpb.27.375. [DOI] [PubMed] [Google Scholar]
- 30.Kress W. J., Erickson D. L. A two-locus global DNA barcode for land plants: the coding rbcL gene complemnets non-coding trnH-psbA spacer region. PLoS ONE. 2007;2(6, article e508) doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao H., Song J.-Y., Ma X.-Y., et al. Identification of Dendrobium species by a candidate DNA barcode sequence: the chloroplast psbA-trnH intergenic region. Planta Medica. 2009;75(6):667–669. doi: 10.1055/s-0029-1185385. [DOI] [PubMed] [Google Scholar]
- 32.Li M., Ling K. H., Lam H., et al. Cardiocrinum seeds as a replacement for Aristolochia fruits in treating cough. Journal of Ethnopharmacology. 2010;130:429–432. doi: 10.1016/j.jep.2010.04.040. [DOI] [PubMed] [Google Scholar]
- 33.Li M., Cao H., But P. P.-H., Shaw P.-C. Identification of herbal medicinal materials using DNA barcodes. Journal of Systematics and Evolution. 2011;49(3):271–283. doi: 10.1111/j.1759-6831.2011.00132.x. [DOI] [Google Scholar]
- 34.Álvarez I., Wendel J. F. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29(3):417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 35.Baldwin B. G., Sanderson M. J., Porter J. M., Wojciechowski M. F., Campbell C. S., Donoghue M. J. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82(2):247–277. doi: 10.2307/2399880. [DOI] [Google Scholar]
- 36.Ainouche M. L., Bayer R. J. On the origins of the tetraploid Bromus species (section Bromus, Poaceae): insights from internal transcribed spacer sequences of nuclear ribosomal DNA. Genome. 1997;40(5):730–743. doi: 10.1139/g97-796. [DOI] [PubMed] [Google Scholar]
- 37.Ganley A. R. D., Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Research. 2007;17(2):184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newmaster S. G., Grguric M., Shanmughanandhan D., Ramalingam S., Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Medicine. 2013;11(1, article 222) doi: 10.1186/1741-7015-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Lee J. H., Park C. B., Park C. G., Moon S. G. A phylogenetic analysis of Korean Artemisia L. based on ITS sequences. Korean Journal of Plant Resources. 2010;23(4):293–302. [Google Scholar]