Abstract
Dosage compensation adjusts the expression levels of genes on one or both targeted sex chromosomes in heterogametic species. This process results in the normalized transcriptional output of important and essential gene families encoded on multiple chromosomes. The mechanisms of dosage compensation have been studied in many model organisms, including Drosophila melanogaster (fly), Caenorhabditis elegans (worm), and Mus musculus (mouse). Although the mechanisms of dosage compensations differ among these species, all of these processes rely on the initial discrimination of the X chromosome from autosomes. Recently, a new paradigm for how the X chromosome is targeted for regulation was identified in Drosophila. This mechanism involves a newly identified zinc finger protein, CLAMP. Here, we review important factors involved in dosage compensation across species with special focus on the fly. Understanding how the newly identified CLAMP protein is involved in X targeting in the fly could provide key insights into how the X chromosome is initially identified across species.
Keywords: dosage compensation, Drosophila, zinc finger protein, transcription factor
Introduction
Concurrent with the evolution of sex chromosomes, an imbalance in gene copy number developed between heterogametic and homogametic sexes (Charlesworth 1978). Therefore, species-specific mechanisms co-evolved to equalize this potentially lethal gene dose imbalance (Livernois et al. 2012). These mechanisms, collectively called dosage compensation, adjust the expression levels of genes on one or both targeted sex chromosomes. This process results in the normalized transcriptional output of important and essential gene families encoded on multiple chromosomes (Disteche 2012).
The mechanisms of dosage compensation have been studied in many model organisms including, but not limited to, Drosophila melanogaster (fly), Caenorhabditis elegans (worm), and Mus musculus (mouse). Although the mechanisms of dosage compensations differ among these species, all of these processes rely on the initial discrimination of the X chromosome from autosomes.
Targeting of the X chromosome is followed by additional changes in gene expression, which are highly species specific. However, recently, a new paradigm for how the X chromosome is targeted for regulation was identified in Drosophila (Larschan et al. 2012; Soruco et al. 2013). This mechanism involves a newly identified zinc finger protein, CLAMP, which could provide key new insights into how the X chromosome is initially identified across species. Here, we review important factors involved in dosage compensation across species with special focus on the fly to understand how the newly identified CLAMP protein could integrate into current models of X identification.
Similarities among dosage compensation mechanisms across species
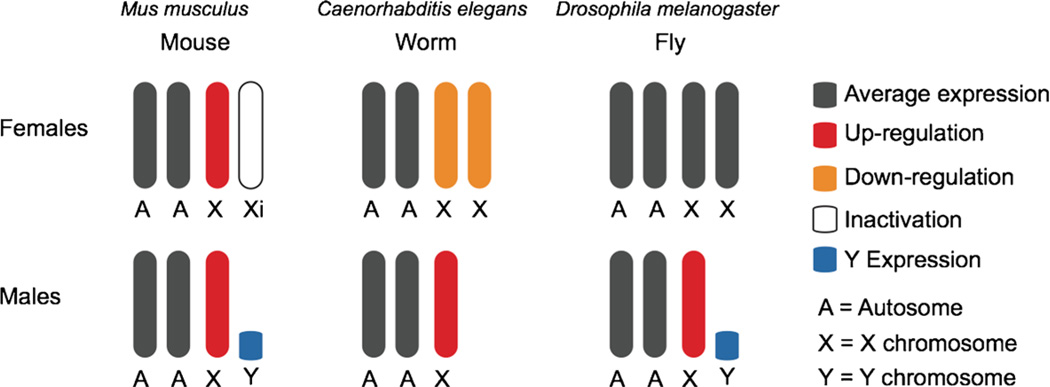
In the mouse, in which males are XY and females are XX, one X chromosome is upregulated in both males and females to equalize gene dosage between X-linked and autosomal genes (Fig. 1). Additionally, one female X chromosome is randomly inactivated, thereby equalizing gene dosage between males and females. The upregulation of X-linked genes is caused by the histone 4 lysine 16 acetyl (H4K16ac) chromatin mark found at promoters of active genes on the active X chromosome (Deng et al. 2013). This histone modification has been correlated with an increase in RNA polymerase II (RNA Pol II) at active genes and increases in X-linked transcription (Deng et al. 2013).
Fig. 1.
Dosage compensation patterns across species. Increased transcription of the X chromosome is required for dosage compensation in Mus musculus (mouse), Caenorhabditis elegans (worm), and Drosophila melanogaster (fly). In the mouse, one X chromosome is upregulated in both sexes. A second step of regulation takes place in which one X chromosome is randomly inactivated in mouse females. In the worm, the X chromosome is upregulated in males, and both X chromosomes are downregulated by half in hermaphrodite females. In the fly, only in males the X chromosome is upregulated
The process of X inactivation is regulated by a number of noncoding RNAs. The Xist noncoding RNA coats the length of the silenced X chromosome in a multistep process as follows (Jonkers et al. 2008; Jeon et al. 2012). First, initial transcription of Xist is activated by an additional noncoding RNA, Jpx (Tian et al. 2010). Next, spreading of Xist first targets gene-rich regions followed by gene poor regions (Simon et al. 2013). Xist RNA is tethered to the X chromosome by the zinc finger protein ying yang 1 (YY1) and the CCCTC-binding factor (CTCF) (Chao et al. 2002; Donohoe et al. 2007; Jeon and Lee 2011). Third, targeting of the Polycomb Repressive 2 (PCR2) complex, which deposits the histone 3 lysine 27 tri-methylation (H3K27me3) modification, leads to the chromatin condensation and silencing of X-linked genes (Plath et al. 2003; Simon et al. 2013). The active X escapes silencing through transcription of the noncoding RNA Tsix, which is antisense to the Xist transcript and acts as a repressor of Xist transcription (Jeon et al. 2012).
Similar to the mouse, dosage compensation in the worm involves both activation and repression (Fig. 1). The X chromosome is upregulated in hermaphroditic females (XX) and males (X0) in a process not well understood but thought to be mediated by H4K16ac (Deng et al. 2011). Then, in hermaphrodites, the expression of both X chromosomes is downregulated by half. The Dosage Compensation Complex (DCC), which shares components with the condensin complex, reduces expression of genes on both X chromosomes by 50 % (Meyer 2010; Wood et al. 2010). One possible mechanism for this would be via chromatin condensation.
Once again, binding of the DCC to the X chromosome takes place in a multistep process. First, sequence-specific targeting of the DCC to recruitment element on X (rex) sites occurs on both X chromosomes. Second, in a step that remains poorly understood, the DCC then spreads to dependent on X (dox) sites within gene promoters resulting in X-specific targeting at active genes (Jans et al. 2009; Pferdehirt et al. 2011). Third, reduced expression of X-linked genes is likely due to a decrease in RNA Pol II recruitment to the transcription start site of genes on the hermaphrodite X chromosomes (Kruesi et al. 2013).
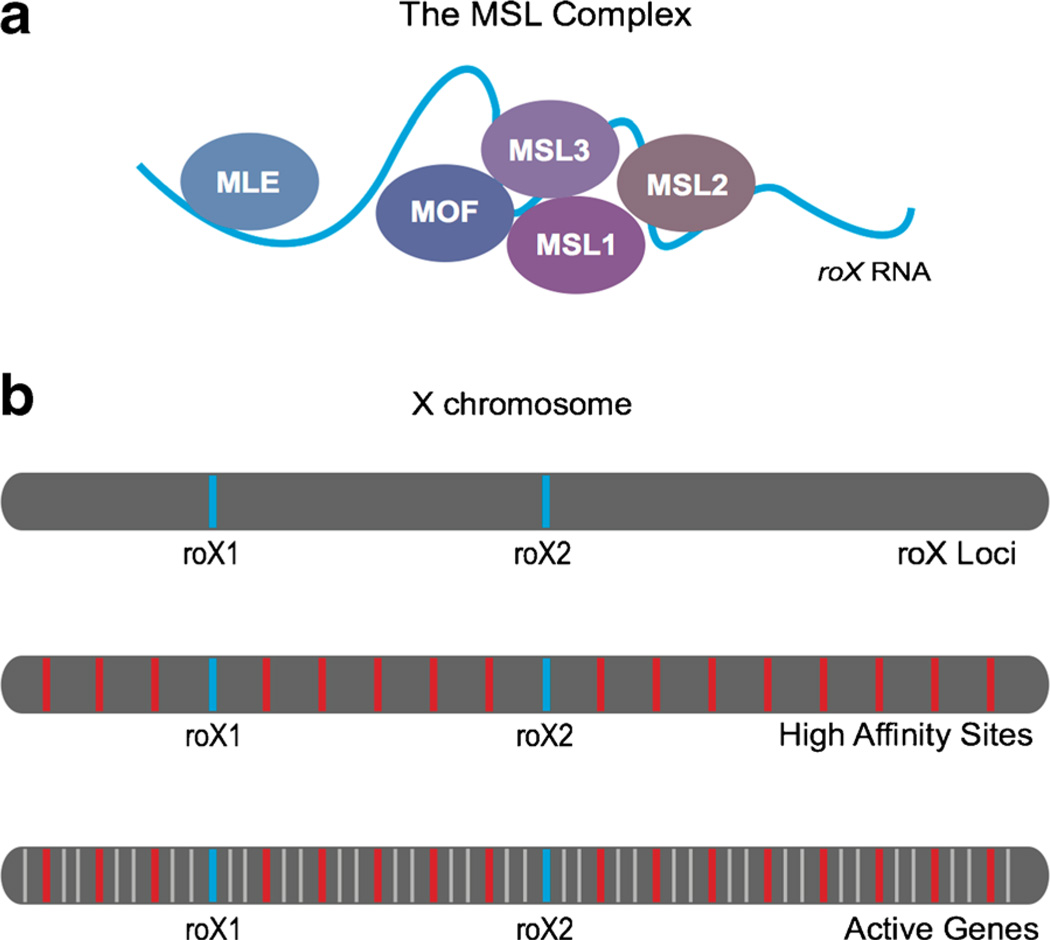
In the fly, dosage compensation of the X chromosome involves only a single type of gene regulation, the conserved upregulation of the single male X chromosome. In the fly, the single X chromosome in males (XY) is upregulated approximately 2-fold, while no regulation takes place in females (XX) (Larschan et al. 2011) (Fig. 1). The Male Specific Lethal (MSL) complex is the master regulator of this process and is only present in males (Fig. 2a). Similar to the multistep processes in the worm and the mouse, the MSL complex is also thought to target the X in a multistep process involving both nucleation and spreading. First, it is thought that the MSL complex targets the RNA on X (roX) loci, which are two sites on the X chromosome that encode the noncoding RNA components of the complex (Kelley et al. 1999; Bai et al. 2004) (Fig. 2b). Subsequent spreading of the MSL complex from the roX loci takes place through the targeting of a series high affinity sites. These sites are approximately 1.5 kb in length and are able to recruit very high levels of MSL complex (Alekseyenko et al. 2008; Sural et al. 2008). Additionally, the high affinity sites contain smaller 21-bp MSL Recognition Element (MRE) sequence motifs necessary for MSL complex recruitment. The MRE motif is found throughout the genome and is approximately 2-fold enriched on the X (Alekseyenko et al. 2008). Due to the thousands of autosomal MRE motifs, X-linked MREs alone are not sufficient to promote X specificity (Alekseyenko et al. 2008). Second, the MSL complex then targets active genes in a sequence independent step involving recognition of the histone 3 lysine 36 tri-methylation (H3K36me3) histone modification that marks the bodies of active genes (Larschan et al. 2007; Sural et al. 2008) (Fig. 2b). Third, the MSL complex deposits the H4K16ac mark upon targeting. Similar to the mouse, this mark has been linked to an open chromatin state and a 1.4-fold increase in RNA Pol II into gene bodies (Gelbart et al. 2009; Larschan et al. 2011).
Fig. 2.
The MSL complex identifies the X chromosome in multiple steps. a The male specific lethal (MSL) complex is comprised of five proteins and one structural, noncoding RNA. b The MSL complex targets the X chromosome in multiple steps. First, it is thought that the roX loci (blue) are targeted during incorporation of a roX RNA into the complex. Second, a series of high affinity sites (red), containing MSL recognition element (MRE) motifs, are targeted. Finally, spreading from high affinity sites to the bodies of active genes (gray) results in an approximate two-fold increase in X-linked genes
Drosophila as a model for X identification
In all three major model organisms in which dosage compensation has been studied, it is clear that initial X chromosome identification is required for all X-linked gene regulation (Lucchesi et al. 2005; Payer and Lee 2008) (Fig. 1). Additionally, the correlation of the H4K16ac mark with an increase in gene expression could indicate a conserved step that is also linked to X identification and therefore retained during evolution from the fly to the mouse (Gelbart et al. 2009; Deng et al. 2013). However, the mechanism by which H4K16ac is specifically targeted to the X chromosome remains poorly understood in the mouse and worm. Because Drosophila dosage compensation involves depositing of the H4K16ac mark by the MSL complex and not additional silencing steps, it is a powerful model with which to study the initial X-identification step of dosage compensation.
Although the MSL complex is able to identify the MRE sequence elements in vivo, none of the MSL proteins have significant specificity for the MRE sequence in vitro (Fauth et al. 2010). Therefore, the identification of a factor that physically links the MSL complex to the MRE elements was key to deciphering the early stages of X targeting. Furthermore, the candidate protein necessary for targeting MRE sequences would have to employ a mechanism that stabilizes interaction with the MSL complex on the X chromosome and not on autosomes. Therefore, the mechanism by which the MSL complex targets MRE sequences within high affinity sites on the X chromosome and not autosomes has remained elusive.
Recently, a previously unstudied protein was found to be the regulator of Drosophila dosage compensation (Larschan et al. 2012; Soruco et al. 2013). The zinc finger protein chromatin linked adapter for MSL proteins (CLAMP) specifically identifies MRE sequences. Similar to zinc finger proteins functioning in mouse dosage compensation, the CLAMP protein links the MSL complex and noncoding RNAs to the X chromosome. Notably, CLAMP is enriched at the important roX loci, the MSL high affinity sites, and on the X overall (Larschan et al. 2012; Soruco et al. 2013). This is the first time a factor has linked the MSL complex to the X chromosome DNA and the roX loci, which are both required for MSL complex targeting. To understand this new interaction more clearly, the function of CLAMP in recruiting the MSL complex components is discussed in more detail below.
Protein components of the MSL complex
The MSL complex is the ribonucleoprotein activator complex responsible for increasing transcript levels on the X chromosome. It is comprised of one of two redundant, long noncoding roX RNAs and at least five proteins: Male Specific Lethal 1 (MSL1),Male Specific Lethal 2 (MSL2), Male Specific Lethal 3 (MSL3), Maleless (MLE) and Males Absent on the First (MOF) (Fig. 2a) (Fukunaga et al. 1975; Belote and Lucchesi 1980; Gorman et al. 1995; Hilfiker et al. 1997; Ilik et al. 2013). All of the protein components of the MSL complex were identified in genetic screens for mutations that cause male-specific lethality (Fukunaga et al. 1975; Belote and Lucchesi 1980; Gorman et al. 1995; Hilfiker et al. 1997). However, all of the components except for MSL2 are not male specific as they are also found in females (Kelley et al. 1997).
MSL1 and MSL2 are the core structural components and nucleate complex assembly (Copps et al. 1998). MSL1 plays a scaffolding role that organizes the other components: MSL2, MSL3, MLE, and MOF (Copps et al. 1998; Scott et al. 2000; Li et al. 2005; Kadlec et al. 2011). MSL1 can also form a homodimer with itself suggesting that the MSL complex may function as a heteroduplex potentially being comprised of two complete complex structures (Hallacli et al. 2012).
MSL2 is the limiting component of the complex and is translationally repressed in females by the master regulator of sex-determination, Sex Lethal (Sxl) (Kelley et al. 1997; Beckmann et al. 2005). MSL2 is an ubiquitin ligase, which ubiquitylates H2B lysine 34 when associated with MSL1 (Wu et al. 2011). Additionally, MSL2 can ubiquitylate other MSL components when overexpressed potentially controlling protein turnover and final stability of the assembled complex (Villa et al. 2012).
MSL3 has a chromo-domain region with specificity for the H3K36me3 histone modification associated with actively transcribed genes (Larschan et al. 2007; Kim et al. 2010).MOF is the major histone acetyl-transferase (HAT) in Drosophila specific for H4K16ac. Similar to the mouse, this histone mark in the fly is thought to mediate the increased transcriptional activity that dosage compensates the male X chromosome (Smith et al. 2000, 2001). MOF is also a member of the Nonspecific Lethal (NSL) Complex, found at the promoters of many constitutively active genes on both the X and autosomes (Lam et al. 2012).
MLE is weakly associated with the MSL complex through interactions with the roX RNAs (Copps et al. 1998).MLE contains two RNA binding domains, which recognize specific secondary structures of the roX RNAs necessary for incorporation of the roX RNAs into the complex (Izzo et al. 2008; Maenner et al. 2013). In addition to functioning in the MSL complex, MLE affects splicing and RNA editing genome wide (Reenan et al. 2000).
A more loosely associated factor is the serine threonine kinase JIL-1 (Jin et al. 2000; Li et al. 2005). JIL-1 has specificity for histone 3 serine 10 (H3S10) and maintains euchromatin boundaries to prevent heterochromatin formation (Wang et al. 2011). It typically colocalizes with H3K36me3 and H4K16ac marks associated with MSL complex targeting (Regnard et al. 2011). However, the specific role of JIL-1 in Drosophila dosage compensation remains unknown.
RNA components of the MSL complex
As with dosage compensation in the mouse, Drosophila noncoding RNAs play an important role in dosage compensation. The noncoding RNAs, roX1 and roX2, are essential for complex targeting and function (Franke and Baker 1999; Deng and Meller 2006; Li et al. 2008). Although the roX RNAs are very different in sequence composition and length (approximately 3.7 and 0.6 kb, respectively), they are functionally redundant (Meller and Rattner 2002; Ilik et al. 2013). Both contain conserved minimal tandem repeat stem loop features necessary for MLE-mediated incorporation into the complex (Meller et al. 2000; Ilik et al. 2013). However, only one RNA transcript, either roX1 or roX2, is incorporated at a time into a functioning MSL complex (Ilik et al. 2013). This suggests that spreading outwards from individual roX loci is likely to occur.
The RNAs are highly unstable unless incorporated into the complex and differ greatly in their transcriptional control. The roX1 locus is transcribed early during development from an MSL-independent promoter, generating a shorter RNA transcript (Meller et al. 2000; Lim and Kelley 2012). This likely generates the initial functional MSL complex early in development. Later, MSL-mediated transcription of a long roX1 transcript and/or the roX2 transcript is then likely responsible for global compensation (Lim and Kelley 2012). Importantly, the roX genes are proximal to MRE sequences that recruit MSL complex and act as enhancers to increase roX gene transcription (Park et al. 2003; Bai et al. 2004). In females, roX1 transcription is repressed by the nucleosome remodeling factor (NURF301) repressor complex in females (Bai et al. 2007). The repression of roX1 RNA transcription, along with the destabilization of MSL2 by Sxl in females, functions to prevent MSL complex formation in females (Kelley et al. 1997; Beckmann et al. 2005; Bai et al. 2007).
The location of the roX loci on the X chromosome is thought to serve as an important element involved in the initial stages of X targeting. However, in mutant flies lacking both roX loci on the X chromosome, but containing a roX locus inserted on an autosome, the MSL complex can move in trans to identify the X chromosome (Kelley et al. 1999; Park et al. 2002; Oh et al. 2003). This indicates that the location of the roX loci on the X is not sufficient to initiate X targeting. Additionally, changing the levels of roX RNA transcribed from an autosomal transgene alters the levels of MSL complex spreading in cis with a maximal spreading of 5 Mb (Kelley et al. 2008; Park et al. 2010). Importantly, spreading across the entire autosome does not occur, likely because factors that are associated with high affinity sites are not as dense on autosomes compared with the X chromosome. Spreading of the MSL complex along the X chromosome is an active area of investigation.
It has been hypothesized that the roX loci are the first sites of MSL targeting because roX RNAs are necessary for complex integrity (Deng and Meller 2006). However, this has yet to be demonstrated in vivo. Therefore, there must be additional elements identifying the X chromosome, which allows the MSL complex to spread along the length of the entire X chromosome.
High affinity sites and MRE DNA elements
In the absence of the peripheral MSL complex components (MSL3, MOF, and MLE), the core MSL complex components, MSL1 and MSL2, can only identify a subset of the wild-type sites on the X chromosome (Lyman et al. 1997; Gu et al. 1998). This finding leads to a multistep model for MSL complex targeting the X chromosome, which is facilitated through the tethering of MSL complex components to the X chromosome. By mapping the localization of MSL complex core components in the absence of MSL3, high-resolution characterization of MSL targeting led to the identification of 150 evenly spaced high affinity sites, called chromatin entry sites (CES) (Alekseyenko et al. 2008). The CES retain very high levels of MSL complex compared to surrounding sequence and are retained in msl3 mutant embryos.
Importantly, from these high affinity sites, a 21-bp sequence was found to be required for MSL complex recruitment and is approximately 2-fold enriched on the X chromosome and 4-fold enriched when focusing the analysis to active genes (Alekseyenko et al. 2008). This 21-bp DNA sequence, termed the MSL Recognition Element (MRE), has a highly conserved 8-bpGA-repeat core. Mutational analysis of both the 8-bp core sequence and the flanking sequence of the MRE motif showed that the entire MRE is necessary for MSL recruitment (Alekseyenko et al. 2008). However, there are approximately 13,000 MRE sequences found throughout the genome, and they are only about 2-fold enriched on the X chromosome. Therefore, in the context of the entire genome where only 3.5% of MREs map to high affinity sites, MRE sequences alone are not sufficient for accurate MSL complex targeting (Alekseyenko et al. 2012).
In an alternative approach, low formaldehyde crosslinking conditions and individual RNAi treatments against MLE, MOF, and MSL3 in male S2 cells, identified 130 high affinity sites (Straub et al. 2008). The high affinity sites from this study are located in noncoding gene regions and have almost complete overlap with the original 150 high affinity sites (Alekseyenko et al. 2008). In summary, these diverse approaches identified the same group of high affinity sites, which are likely to be key initial sites of X identification.
Mechanisms for spreading of the MSL complex and increasing transcription of X-linked genes
Once the roX loci and high affinity sites are targeted through MRE-mediated activity, the MSL complex then spreads in part through the function of the MSL3 chromo-domain interaction with the H3K36me3 chromatin mark to approximately 1,000 active X-linked genes (Larschan et al. 2007; Sural et al. 2008). Spreading of the MSL complex from high affinity sites is sequence independent because an active autosomal gene that does not contain any MRE sequence can be inserted on the X chromosome and is targeted by MSL complex (Gorchakov et al. 2009).
The H4K16ac histone mark deposited on the X chromosome by the MSL complex physically decompacts chromatin by abrogated internucleosome contacts (Shogren-Knaak et al. 2006). It is likely that increased H4K16ac over gene bodies promotes more rapid release of paused RNA polymerase II into gene bodies (Larschan et al. 2011). However, the exact mechanism by which transcription is upregulated approximately 2-fold remains unknown and is a critical question for future study.
An alternative mechanism has been proposed in which the core components of the MSL complex counteract high levels of transcriptional activation that would otherwise be mediated by H4K16ac (Prestel et al. 2010; Sun et al. 2013). However, a mechanism for how this repression could function has not been identified.
The CLAMP protein: a newly discovered regulator of Drosophila dosage compensation
Despite many recent advances in the understanding of Drosophila dosage compensation, the mechanism that is responsible for X-targeting has remained elusive for the following reasons: First, none of the MSL complex components are able to recognize the MRE sequence specifically. Second, while the roX RNAs play an important role in the initial targeting, the roX loci are not sufficient for this process because the MSL complex containing the RNAs can travel in trans from the autosomes to the X chromosome. Third, though the MRE sequences are enriched on the X, they are also found on autosomes. Consequently, the precise mechanism by which the MSL complex is able to initially distinguish the X from the autosomes is not known.
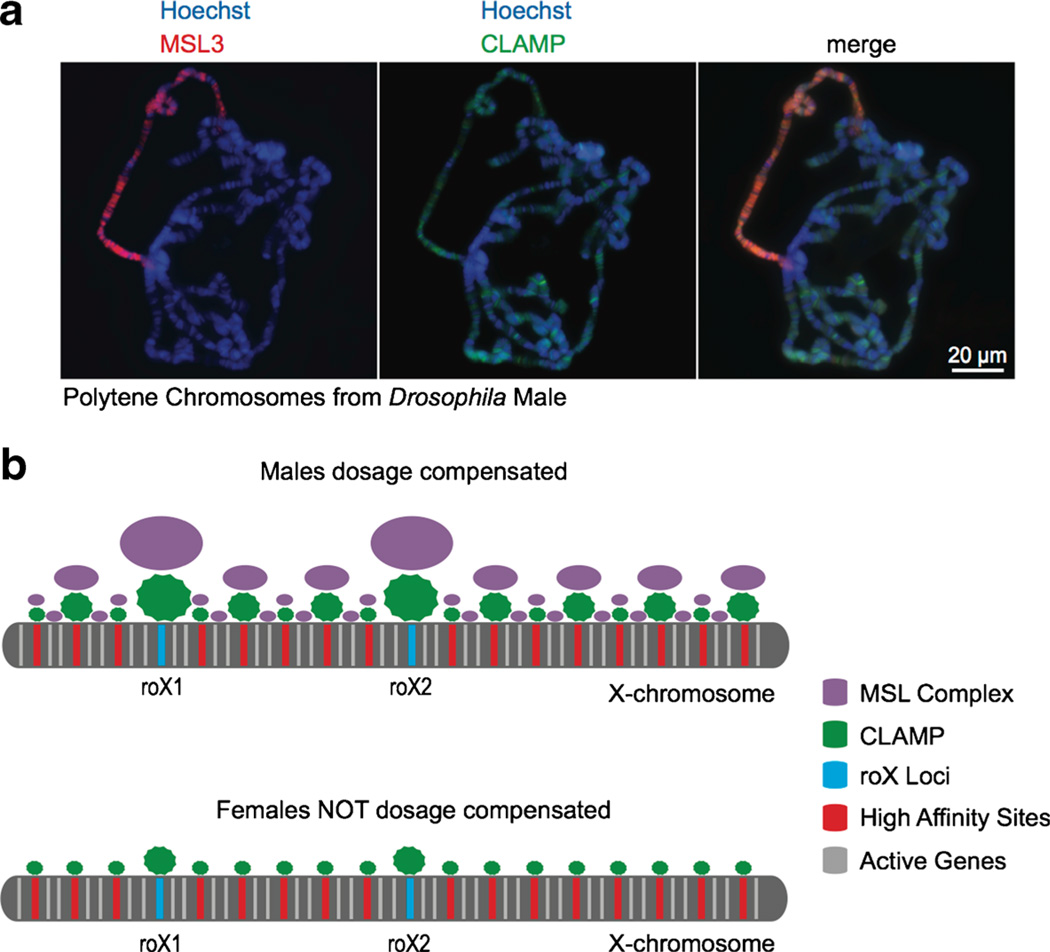
Recently, a new cell-based screening strategy was used that allowed for the identification of factors that have both male-specific roles in dosage compensation as well as roles in transcription in both males and females (Larschan et al. 2012). Analogous to the condensin complexes in C. elegans, it was hypothesized that non-sex-specific factors may have an additional role in dosage compensation. Such factors with non-sex-specific roles would have been missed in previous screens, which only identified male-specific lethal factors. In this way, the zinc finger protein CLAMP was identified, which provides the first physical link between the MSL complex and the male X chromosome (Fig. 3a). The CLAMP protein is able to directly recognize MRE sequences throughout the genome and is found enriched on the X compared to autosomes, specifically at the high affinity sites (Fig. 3b). Furthermore, CLAMP is bound to MRE sequences at the roX loci and increases roX2 transcription levels (Soruco et al. 2013).
Fig. 3.
The role of the CLAMP protein in X-targeting. a The MSL complex specifically targets the X chromosome in males (left), while the CLAMP zinc finger protein is targeted throughout the genome (middle). b CLAMP is enriched at high affinity sites on the X chromosome, including the roX loci, in the absence of MSL complex in females (bottom). A cooperative interaction leads to increased levels of both factors when co-localized on the X chromosome (top)
CLAMP directly interacts with MRE DNA sequences, and it is enriched on the X chromosome even in the absence of the MSL complex (Fig. 3b). Therefore, enrichment of CLAMP on the X chromosome in the absence of the MSL complex could mark the high affinity sites as “seed sites” prior to MSL targeting (Fig. 3b). Additionally, the CLAMP protein and MSL complex have a cooperative relationship, which results in increased occupancy of both factors when colocalized. Therefore, once the initial X identification occurs, the cooperative interactions between CLAMP and MSL complex are likely to further increase the specificity of X identification (Soruco et al. 2013) (Fig. 3b).
Unique X chromosome characteristics
There are several additional features that distinguish the X chromosome from autosomes independent of MSL complex. The X chromosome has an increased GC content in the 10-kb regions surrounding high affinity sites and a decrease in GC content within 1 kb of functional MREs (Alekseyenko et al. 2012). However, GC content is not sufficient to predict functional MREs. Importantly, using the modENCODE histone modifications ChIP-seq library as a training data set, H3K36me3 and JIL-1 association were predictive marks for high affinity sites on the X chromosome independent of MSL complex. Additionally, significant nucleosome depletion at high affinity sites was also noted even in the absence of MSL complex (Oh et al. 2004; Alekseyenko et al. 2012). However, future work is needed to determine the relative contributions of these elements during the targeting process.
Nuclear domain
Future experiments investigating the formation of a three-dimensional nuclear domain could reveal a domain that allows the X to be distinguished from autosomes prior to MSL complex recruitment. It is also possible that the CLAMP protein, through its glutamine-rich domain could mediate the formation of a subnuclear domain. Other glutamine-rich domains nucleate aggregates and trap coactivators, suggesting that they may mediate nuclear domain formation (Wilkins and Lis 1999; Kato et al. 2012). Additionally, a nuclear domain could maintain high concentrations of important transcriptional machinery within close proximity to the X chromosome, facilitating increased transcription levels after MSL complex is recruited. Understanding the mechanism of X targeting during dosage compensation is likely to provide new insights into essential cellular mechanisms that allow transcription factors to find their targets and precisely regulate the genome.
Evolutionary considerations
With the discovery of the CLAMP protein as an important regulator of Drosophila dosage compensation, experiments further defining the role of CLAMP during the early stages of X identification are eagerly anticipated. Recently, an increased accumulation of GA repeats on the X chromosome was noted when comparing D. melanogaster to Drosophila miranda, which contains a newly evolved X chromosome (Alekseyenko et al. 2013). Therefore, evolution of X-specific GA repeats could be the precise mechanism that has allowed for an increase in CLAMP targeting to the X chromosome.
To gain insight into the evolution of dosage compensation, it is interesting to consider the non-sex-specific roles of the previously unstudied CLAMP protein. It is possible that the MSL complex co-opted the more ancient CLAMP protein because it recognized GA-rich sequences that are enriched on the X chromosome (Ellison and Bachtrog 2013). Positive selection may have resulted in increased GA repeats on the X chromosome due to their interaction with the highly conserved CLAMP protein, or vice versa. Clearly, a physical interaction between CLAMP and MSL complex would be required for this. This would likely be mediated through the glutamine-rich domain of CLAMP because glutamine-rich domains are known to be involved in protein–protein interactions (Wang et al. 2013; Gonzalez Nelson et al. 2014).
However, there are other transcription factors that bind GA-repeat sequences. For example, GAGA factor (GAF), which binds GA-rich sequences genome wide, is notably decreased at high affinity sites in Drosophila male cells in contrast to CLAMP that is found enriched (Alekseyenko et al. 2012; Soruco et al. 2013). Therefore, it is possible that there is an X-specific interaction between CLAMP and the MSL complex that is not favored between GAF and MSL complex. This interaction could be mediated through the glutamine-rich domain of CLAMP, the characteristics of the surrounding chromatin, or both.
There are GA-rich motifs at the 5′ ends of many genes, and therefore, CLAMP is found at the transcription start site of many transcribed genes on all chromosomes. On the X chromosome, CLAMP is additionally enriched the 3′ end of genes where MSL complex is also typically localized (Alekseyenko et al. 2006; Soruco et al. 2013). This spatial separation could also isolate the two distinct functions of the CLAMP protein: the male-specific dosage compensation function and the non-sex-specific function. This idea is supported by a recent study using new sonication techniques, which revealed different MSL subcomplexes or core components acting over varying gene regions (Straub et al. 2013)
Summary
There are numerous protein factors, RNA and DNA elements, that have evolved to ensure precise identification of the X chromosome during dosage compensation. Non-sex-specific factors are necessary for dosage compensation in both the fly and worm. Similarly, noncoding RNAs guide dosage compensation components in the fly and the mouse. Shared across all three species are DNA motifs necessary in directing initial targeting factors and the H4K16ac histone modification associated with X upregulation. The discovery of the CLAMP protein in Drosophila has emphasized zinc finger proteins in initial X-targeting in the mouse and the fly. Importantly, it has greatly increased our overall understanding of X chromosome targeting in the fly. This advance could provide a long-awaited link to additional proteins, motifs, noncoding RNAs, and chromatin states that are essential in dosage compensation across species.
Acknowledgments
We are grateful to Dr. Leila Rieder for insights and critical review of the manuscript.
Abbreviations
- H4K16ac
Histone 4 lysine 16 acetylation
- RNA Pol II
RNA polymerase II
- Xist
X inactive specific transcript
- Enox or Jpx
Expressed neighbor of Xist
- YY1
Ying yang 1
- CTCF
CCCTC-binding factor
- PCR2
Polycomb repressive 2
- H3K27me3
Histone 3 lysine 27 tri-methylation
- Tsix
Antisense to Xist
- DCC
Dosage compensation complex
- rex
Recruitment element on X
- dox
Dependent on X
- MSL
Male specific lethal complex
- roX
RNA on the X
- MRE
MSL recognition elements
- H3K36me3
Histone 3 lysine 36 tri-methylation
- CLAMP
Chromatin-linked adapter for MSL proteins
- MSL1
Male specific lethal 1
- MSL2
Male specific lethal 2
- MSL3
Male specific lethal 3
- MLE
Maleless
- MOF
Males absent on the first
- Sxl
Sex lethal
- H2B
Histone 2B
- HAT
Histone acetyl-transferase
- NSL
Nonspecific lethal
- JIL-1
Serine protein kinase
- H3S10
Histone 3 Serine 10
- NURF301
Nucleosome remodeling factor
- CES
Chromatin entry sites
- modENCODE
Model organism encyclopedia of DNA elements
- ChIP-seq
Chromatin immunoprecipitaiton followed with next gen sequencing
- GAF
GAGA factor
Footnotes
Conflict of interest The author declares that they have no conflict of interest.
References
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Ho JW, Peng S, Gelbart M, Tolstorukov MY, Plachetka A, Kharchenko PV, Jung YL, Gorchakov AA, Larschan E, et al. Sequence-specific targeting of dosage compensation in Drosophila favors an active chromatin context. PLoS Genet. 2012;8:e1002646. doi: 10.1371/journal.pgen.1002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Ellison CE, Gorchakov AA, Zhou Q, Kaiser VB, Toda N, Walton Z, Peng S, Park PJ, Bachtrog D, et al. Conservation and de novo acquisition of dosage compensation on newly evolved sex chromosomes in Drosophila. Genes Dev. 2013;27:853–858. doi: 10.1101/gad.215426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Alekseyenko AA, Kuroda MI. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 2004;23:2853–2861. doi: 10.1038/sj.emboj.7600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Larschan E, Kwon SY, Badenhorst P, Kuroda MI. Regional control of chromatin organization by noncoding roX RNAs and the NURF remodeling complex in Drosophila melanogaster. Genetics. 2007;176:1491–1499. doi: 10.1534/genetics.107.071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann K, Grskovic M, Gebauer F, Hentze MW. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell. 2005;122:529–540. doi: 10.1016/j.cell.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Belote JM, Lucchesi JC. Male-specific lethal mutations of Drosophila melanogaster. Genetics. 1980;96:165–186. doi: 10.1093/genetics/96.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci U S A. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copps K, Richman R, Lyman LM, Chang KA, Rampersad-Ammons J, Kuroda MI. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J. 1998;17:5409–5417. doi: 10.1093/emboj/17.18.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Meller VH. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics. 2006;174:1859–1866. doi: 10.1534/genetics.106.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Ma W, Nguyen DK, Hiatt JB, Noble WS, Shendure J, Disteche CM. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev Cell. 2013;25:55–68. doi: 10.1016/j.devcel.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Ellison CE, Bachtrog D. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science. 2013;342:846–850. doi: 10.1126/science.1239552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauth T, Muller-Planitz F, Konig C, Straub T, Becker PB. The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome. Nucleic Acids Res. 2010;38:3209–3221. doi: 10.1093/nar/gkq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4:117–122. doi: 10.1016/s1097-2765(00)80193-8. [DOI] [PubMed] [Google Scholar]
- Fukunaga A, Tanaka A, Oishi K. Maleless, a recessive autosomal mutant of Drosophila melanogaster that specifically kills male zygotes. Genetics. 1975;81:135–141. doi: 10.1093/genetics/81.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol. 2009;16:825–832. doi: 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Nelson AC, Paul KR, Petri M, Flores N, Rogge RA, Cascarina SM, Ross ED. Increasing prion propensity by hydrophobic insertion. PLoS One. 2014;9:e89286. doi: 10.1371/journal.pone.0089286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda MI. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev. 2009;23:2266–2271. doi: 10.1101/gad.1840409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M, Franke A, Baker BS. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development. 1995;121:463–475. doi: 10.1242/dev.121.2.463. [DOI] [PubMed] [Google Scholar]
- Gu W, Szauter P, Lucchesi JC. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev Genet. 1998;22:56–64. doi: 10.1002/(SICI)1520-6408(1998)22:1<56::AID-DVG6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hallacli E, Lipp M, Georgiev P, Spielman C, Cusack S, Akhtar A, Kadlec J. Msl1-mediated dimerization of the dosage compensation complex is essential for male X-chromosome regulation in Drosophila. Mol Cell. 2012;48:587–600. doi: 10.1016/j.molcel.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo A, Regnard C, Morales V, Kremmer E, Becker PB. Structure-function analysis of the RNA helicase maleless. Nucleic Acids Res. 2008;36:950–962. doi: 10.1093/nar/gkm1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 2009;23:602–618. doi: 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr Opin Genet Dev. 2012;22:62–71. doi: 10.1016/j.gde.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang Y, Johansen J, Johansen KM. JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J Cell Biol. 2000;149:1005–1010. doi: 10.1083/jcb.149.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Monkhorst K, Rentmeester E, Grootegoed JA, Grosveld F, Gribnau J. Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle. Mol Cell Biol. 2008;28:5583–5594. doi: 10.1128/MCB.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Hallacli E, Lipp M, Holz H, Sanchez-Weatherby J, Cusack S, Akhtar A. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol. 2011;18:142–149. doi: 10.1038/nsmb.1960. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Wang J, Bell L, Kuroda MI. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Lee OK, Shim YK. Transcription rate of noncoding roX1 RNA controls local spreading of the Drosophila MSL chromatin remodeling complex. Mech Dev. 2008;125:1009–1019. doi: 10.1016/j.mod.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Blus BJ, Chandra V, Huang P, Rastinejad F, Khorasanizadeh S. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol. 2010;17:1027–1029. doi: 10.1038/nsmb.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KC, Muhlpfordt F, Vaquerizas JM, Raja SJ, Holz H, Luscombe NM, Manke T, Akhtar A. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012;8:e1002736. doi: 10.1371/journal.pgen.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Soruco MM, Lee OK, Peng S, Bishop E, Chery J, Goebel K, Feng J, Park PJ, Kuroda MI. Identification of chromatin-associated regulators of MSL complex targeting in Drosophila dosage compensation. PLoS Genet. 2012;8:e1002830. doi: 10.1371/journal.pgen.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Parry DA, Scott MJ. The amino-terminal region of Drosophila MSL1 contains basic, glycine-rich, and leucine zipper-like motifs that promote X chromosome binding, self-association, and MSL2 binding, respectively. Mol Cell Biol. 2005;25:8913–8924. doi: 10.1128/MCB.25.20.8913-8924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Schiemann AH, Scott MJ. Incorporation of the noncoding roX RNAs alters the chromatin-binding specificity of the Drosophila MSL1/MSL2 complex. Mol Cell Biol. 2008;28:1252–1264. doi: 10.1128/MCB.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CK, Kelley RL. Autoregulation of the Drosophila noncoding roX1 RNA Gene. PLoS Genet. 2012;8:e1002564. doi: 10.1371/journal.pgen.1002564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livernois AM, Graves JA, Waters PD. The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity. 2012;108:50–58. doi: 10.1038/hdy.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–1753. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Muller M, Frohlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol Cell. 2013;51:174–184. doi: 10.1016/j.molcel.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Park Y, Kuroda MI. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 2003;17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Bone JR, Kuroda MI. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol. 2004;14:481–487. doi: 10.1016/j.cub.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- Park Y, Mengus G, Bai X, Kageyama Y, Meller VH, Becker PB, Kuroda MI. Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol Cell. 2003;11:977–986. doi: 10.1016/s1097-2765(03)00147-3. [DOI] [PubMed] [Google Scholar]
- Park SW, Oh H, Lin YR, Park Y. MSL cis-spreading from roX gene up-regulates the neighboring genes. Biochem Biophys Res Commun. 2010;399:227–231. doi: 10.1016/j.bbrc.2010.07.059. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 2011;25:499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, dela Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Prestel M, Feller C, Straub T, Mitlohner H, Becker PB. The activation potential of MOF is constrained for dosage compensation. Mol Cell. 2010;38:815–826. doi: 10.1016/j.molcel.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Reenan RA, Hanrahan CJ, Ganetzky B. The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Regnard C, Straub T, Mitterweger A, Dahlsveen IK, Fabian V, Becker PB. Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet. 2011;7:e1001327. doi: 10.1371/journal.pgen.1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MJ, Pan LL, Cleland SB, Knox AL, Heinrich J. MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. EMBO J. 2000;19:144–155. doi: 10.1093/emboj/19.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- Soruco MM, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 2013;27:1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Zabel A, Gilfillan GD, Feller C, Becker PB. Different chromatin interfaces of the Drosophila dosage compensation complex revealed by high-shear ChIP-seq. Genome Res. 2013;23:473–485. doi: 10.1101/gr.146407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Fernandez HR, Donohue RC, Li J, Cheng J, Birchler JA. Male-specific lethal complex in Drosophila counteracts histone acetylation and does not mediate dosage compensation. Proc Natl Acad Sci U S A. 2013;110:E808–E817. doi: 10.1073/pnas.1222542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R, Forne I, Muller M, Imhof A, Straub T, Becker PB. MSL2 combines sensor and effector functions in homeostatic control of the Drosophila dosage compensation machinery. Mol Cell. 2012;48:647–654. doi: 10.1016/j.molcel.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Wang C, Cai W, Li Y, Deng H, Bao X, Girton J, Johansen J, Johansen KM. The epigenetic H3S10 phosphorylation mark is required for counteracting heterochromatic spreading and gene silencing in Drosophila melanogaster. J Cell Sci. 2011;124:4309–4317. doi: 10.1242/jcs.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CI, Alekseyenko AA, LeRoy G, Elia AE, Gorchakov AA, Britton LM, Elledge SJ, Kharchenko PV, Garcia BA, Kuroda MI. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol. 2013;20:202–209. doi: 10.1038/nsmb.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins RC, Lis JT. DNA distortion and multimerization: novel functions of the glutamine-rich domain of GAGA factor. J Mol Biol. 1999;285:515–525. doi: 10.1006/jmbi.1998.2356. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell. 2011;43:132–144. doi: 10.1016/j.molcel.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]