ABSTRACT
Multiciliated cells (MCCs), which are present in specialized vertebrate tissues such as mucociliary epithelia, project hundreds of motile cilia from their apical membrane. Coordinated ciliary beating in MCCs contributes to fluid propulsion in several biological processes. In a previous work, we demonstrated that microRNAs of the miR-34/449 family act as new conserved regulators of MCC differentiation by specifically repressing cell cycle genes and the Notch pathway. Recently, we have shown that miR-34/449 also modulate small GTPase pathways to promote, in a later stage of differentiation, the assembly of the apical actin network, a prerequisite for proper anchoring of centrioles-derived neo-synthesized basal bodies. We characterized several miR-34/449 targets related to small GTPase pathways including R-Ras, which represents a key and conserved regulator during MCC differentiation. Direct RRAS repression by miR-34/449 is necessary for apical actin meshwork assembly, notably by allowing the apical relocalization of the actin binding protein Filamin-A near basal bodies. Our studies establish miR-34/449 as central players that orchestrate several steps of MCC differentiation program by regulating distinct signaling pathways.
KEYWORDS: actin network, airway epithelium, microRNAs, miR-34/449, miR-34, miR-449, Motile cilia, multiciliated cells, small GTPase, Xenopus larval skin
To date, the small GTPase superfamily encompasses 5 major subfamilies: Ras, Rho, Rab, Arf/Arl, and Ran, on the basis of sequence and function similarities.1 Small GTPases are monomeric G proteins acting as GDP/GTP-regulated molecular switches. They represent versatile spatio-temporal regulators which activity is controlled by GTP binding and hydrolysis. Their activity is finely modulated by numerous molecular actors to mediate crosstalk between key signaling pathways implicated in several biological functions, such as cell proliferation, differentiation, cell shape, membrane- and cytoskeleton-related cellular processes.1,2 A growing body of work has highlighted the roles of various small GTPases in cilia formation and function, as well as in ciliopathies.3-9
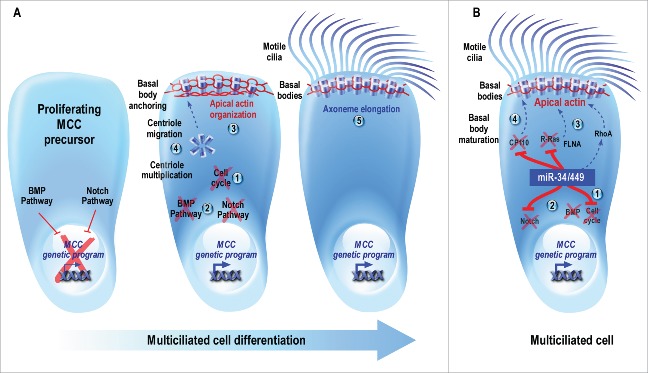
Cilia are microtubule-based membrane projections found in most eukaryotic cells, which play important roles in locomotion, fluid transport, and sensory perception.10 The primary cilium is a single non-motile cilium that functions as a cell antenna to perceive chemical or mechanical cues. In contrast, the motile cilium forms a micrometer-scale whip-like organelle found either as a single cilium per cell (nodal cilium or flagellum on sperm for instance) or up to hundreds at the surface of specialized cells, named multiciliated cells (MCCs). In all vertebrates, MCCs line the luminal surface of some tissues such as the airways, the cerebral ventricles, the oviducts and the efferent ducts of testis.10-13 More species-specific instances of MCCs are represented by the embryonic epidermis of amphibians, the olfactory placodes of fish and the pronephros of both amphibians and fish.14-18 The coordinated beating of motile cilia allows the evacuation from airways of inhaled particles trapped in mucus, the circulation of the cerebrospinal fluid or the progression of the embryo along the genital tract.13 Any dysfunction or reduction of the number of motile cilia can cause or worsen the symptoms of many diseases such as ciliopathies (e.g primary ciliary dyskinesia) or chronic respiratory diseases (e.g cystic fibrosis, asthma or chronic obstructive pulmonary disease).19-21 The formation of multiple motile cilia (a process called multiciliogenesis) occurs during embryonic development or regeneration of some specialized epithelia. It involves multiple events including: (1) cell cycle exit, (2) acquisition of the MCC identity under the control of both the BMP (bone morphogenetic protein) and Notch signaling pathways, (3) reorganization of the apical actin network, (4) massive multiplication of centrioles, which then migrate and anchor in the apical actin meshwork to become basal bodies, and finally, (5) each basal body at the base of each cilium act as microtubule organizing center from which an axoneme elongates (Fig. 1A).8,10,22-24
Figure 1.
(A) Schematic description of multiciliogenesis. Proliferating MCC precursors must undergo (1) cell cycle exit followed by (2) the inhibition of BMP and Notch pathways, 2 early events required for the entry into MCC differentiation. (3) In maturing MCCs, the apical actin cytoskeleton is reorganized into a dense cortical meshwork. (4) In addition, a massive multiplication of centrioles occurs, followed by their migration toward the apical membrane, where they mature and anchor to the actin meshwork to form ciliary organizing centers known as basal bodies. (5) Finally, MCC maturation is achieved through the elongation of axonemes from basal bodies to form multiple motile cilia. These motile cilia subsequently orient and beat in a coordinated manner to generate robust directional fluid flow at the surface of the epithelium. (B) Model illustrating the conserved roles of miR-34/449 during vertebrate MCC differentiation. The specific expression of miR-34/449 in immature MCCs allows (1) the exit from the cell cycle and (2) the entry into differentiation through the direct repression of the Notch pathway. As a result, the MCC genetic program is triggered. As miR-34/449 expression is itself repressed by the activation of the Notch signaling, these miRNAs accumulate in maturing MCCs, thus maintaining a double negative feedback loop. Then, miR-34/449 can regulate subsequent steps such as (3) the reorganization of the apical actin network, by directly repressing R-Ras, modulating the RhoA activity, and allowing the apical relocalization of FLNA, and (4) basal body maturation by directly repressing CP110. Plain lines indicate direct interactions; dotted lines identify pathways that may or may not be direct.
Several key regulators of multiciliogenesis have been identified so far, such as the transcription factors FoxJ1 (Forkhead box protein J1), MYB, some members of the RFX family (regulatory factor X),25,26 the geminin-related nuclear protein Multicilin,27 Grainyhead-like 2 (Grhl2)28 or cyclin O (CCNO).29 It is also well known that an early inhibition of the Notch pathway drives vertebrate MCC differentiation.22,23,30 In a recent work, we showed that inhibition of BMP signaling is an additional early event required to trigger MCC differentiation in the Xenopus embryonic epidermis and in regenerating human airway primary cultures.24 Using these 2 models, we also established a link between multiciliogenesis, the Notch pathway and the miR-34/449 superfamily of microRNAs (miRNAs or miR).8,22,30 MiRNAs belong to a class of small single-stranded and non-coding regulatory RNAs that control many biological processes by limiting the stability or the translation of their target mRNAs.31 Based on their strong conservation in vertebrates, their high sequence homology and their common targets, miR-34 and miR-449 can be classified into a single miR-34/449 superfamily. Members of the miR-34 family have been detected in MCCs from invertebrates to vertebrates, while the miR-449 family appears restricted to vertebrate MCCs.8,14,22,32-35 Our previous data established miR-34/449 miRNAs as new key regulators of vertebrate multiciliogenesis that directly repress the cell cycle and the Notch pathway, 2 early events required for MCC differentiation.22 Our findings have been confirmed in 2 studies showing that miR-34/449-knockout mice display multiciliogenesis defects, causing altered mucociliary clearance, respiratory distress and post-natal lethality.32,33 Our own work also revealed a double negative feedback loop between Notch and miR-449: miR-449 represses Notch activity, which in return enhances miR-449 expression.8,22 This feedback loop allows the accumulation of miR-34/449 molecules in maturing MCCs, providing an opportunity for them to repress additional targets at later stages of multiciliogenesis. Consistent with our hypothesis, Song and colleagues proposed that the centriolar protein CP110 represents a late target of miR-34/449 that should be suppressed to allow basal body maturation.32
Another important prerequisite for basal body docking during multiciliogenesis is the apical actin meshwork reorganization, a process controlled by several factors including FoxJ1,9,36,37 Multicilin,27 the ERK7 mitogen-activated protein kinase38 and the small GTPase RhoA (Ras homolog family member A).9 Focal adhesion proteins are also required for the interaction between basal bodies and the apical actin network during multiciliogenesis.39 GTPases of the RHO family such as RhoA interact with different effectors to control actin cytoskeleton formation in a wide range of cellular processes.40 For instance, Rho GTPase signaling could interact with the planar cell polarity pathway to regulate the assembly of apical actin filaments, as well as the docking and planar polarization of the basal bodies in MCCs.41 The action of small GTPases on actin cytoskeletal dynamics is regulated by a complex network of interactions with additional GTPases, such as the Ras family member R-Ras,42 and other regulatory factors including guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs) and GDP-dissociation inhibitors (GDIs)43,44 as well as miRNAs.45 In that context, we have demonstrated that miR-34/449 control apical actin meshwork formation during multiciliogenesis by acting on small GTPase pathways.8 On the one hand, miR-34/449 silencing blocks the formation of both apical actin network and motile cilia in human airway epithelium and frog epidermis, although RhoA activity remains detectable. On the other hand, the expression of miR-34/449 in human respiratory cells increased the formation of actin stress fibers, focal adhesion and RhoA activity, in a Notch pathway independent manner (Fig. 1B). We therefore hypothesized that apical actin network reorganization during multiciliogenesis is controlled by additional miR-34/449 targets, independent of the Notch pathway and possibly acting downstream or in parallel to RhoA.8 Accordingly, we identified and validated several additional miR-34/449 transcript targets related to small GTPase pathways including ARHGAP1, ARHGDIB, and RRAS which encodes ARHGAP1, RhoGDI2 and R-Ras, respectively. In human airway epithelium, expression of ARHGAP1, ARHGDIB, and RRAS was restricted to basal cells and was specifically down-regulated at both transcript and protein levels during MCC differentiation in response to miR-34/449 expression. Silencing of ARHGAP1 or RRAS using siRNAs (small interfering RNA) at an early stage of differentiation affected both apical actin remodeling and multiciliogenesis. RhoA activity was increased by RRAS silencing but was unaffected by ARHGAP1 or ARHGDIB silencing. Thus, the induction of RhoA activity by miR-34/449 may at least in part involve the silencing of RRAS, notwithstanding possible contributions by additional regulators. Incidentally, these observations suggested that ARHGAP1 and R-Ras are probably necessary at early stages of multiciliogenesis. The absence of effect of ARHGAP1 and ARHGDIB silencing on RhoA activity is probably in line with the existence of redundant or compensatory mechanisms controlling RhoA activity in the context of multiciliogenesis. During Xenopus MCC differentiation, rras was expressed in inner layer epidermal cells that were negative for MCC markers and in an anti-correlated manner with miR-34/449. Furthermore, the pattern of expression of both arhgap1 and arhgdib was not consistent with a link to multiciliogenesis in Xenopus. Taken together, these observations indicated that R-Ras could be a bona fide target, conserved from amphibian to human and contributing to the formation of the apical actin network during multiciliogenesis. In both species, the specific blockade of miR-34/449 binding to the RRAS transcripts using protector oligonucleotides suppressed the apical actin network reorganization and multiciliogenesis, whereas RhoA activation at the apical surface was still detectable, thus mimicking the effects obtained by miR-34/449 inhibition. Importantly, RRAS silencing was able to restore apical actin network formation and multiciliogenesis in Xenopus embryos protected against the action of miR-34/449.
R-Ras has been reported to interact with Filamin A (FLNA), an actin-binding protein involved in actin cytoskeleton remodeling, with the Rho signaling pathway, and could regulate basal body positioning through its interaction with Meckelin.46-50 MiR-34/449 expression in maturing MCCs caused the apical relocalization of FLNA in the vicinity of basal bodies. This step may be important for stabilizing the apical actin network, which is an essential prerequisite for basal body anchoring. Collectively, our findings indicate that miR-34/449 control vertebrate MCC differentiation by successively i) inducing cell cycle arrest, ii) directly repressing the Notch pathway, iii) promoting the apical actin network reorganization through R-Ras silencing and RhoA pathway modulation (Fig. 1B).
Our works, together with others, illustrate the intricate role played by the miR-34/449 superfamily in coordinating a complex differentiation program and open up new perspectives for the study of ciliopathies etiology. The elucidation of mechanisms governing multiciliogenesis may allow the emergence of more specific therapeutic strategies for treating diseases associated with ciliary disorders.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Our works are supported by CNRS, the French Government (National Research Agency, ANR) through the “Investments for the Future” LABEX SIGNALIFE (ANR-11-LABX-0028-01) and FRANCE GENOMIQUE (ANR-10-INBS-09-03 and ANR-10-INBS-09-02), with supports from the Canceropôle PACA, and by grants from ANR (MITHRA, ANR-12-EMMA-0015-01), Vaincre la Mucoviscidose, and FRM (DEQ20130326464).
References
- [1].Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci 2005; 118:843-6; PMID:15731001; http://dx.doi.org/ 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- [2].Burridge K, Wennerberg K. Rho and Rac take center stage. Cell 2004; 116:167-79; PMID:14744429; http://dx.doi.org/ 10.1016/S0092-8674(04)00003-0 [DOI] [PubMed] [Google Scholar]
- [3].Deretic D. Crosstalk of Arf and Rab GTPases en route to cilia. Small GTPases 2013; 4:70-7; PMID:23567335; http://dx.doi.org/ 10.4161/sgtp.24396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang Q, Hu J, Ling K. Molecular views of Arf-like small GTPases in cilia and ciliopathies. Exp Cell Res 2013; 319:2316-22; PMID:23548655; http://dx.doi.org/ 10.1016/j.yexcr.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Epting D, Slanchev K, Boehlke C, Hoff S, Loges NT, Yasunaga T, Indorf L, Nestel S, Lienkamp SS, Omran H, et al.. The Rac1 regulator ELMO controls basal body migration and docking in multiciliated cells through interaction with Ezrin. Development 2015; 142:1553; PMID:25852201; http://dx.doi.org/ 10.1242/dev.124214 [DOI] [PubMed] [Google Scholar]
- [6].Li Y, Hu J. Small GTPases and cilia. Protein Cell 2011; 2:13-25; PMID:21337006; http://dx.doi.org/ 10.1007/s13238-011-1004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li Y, Ling K, Hu J. The emerging role of Arf/Arl small GTPases in cilia and ciliopathies. J Cell Biochem 2012; 113:2201-7; PMID:22389062; http://dx.doi.org/ 10.1002/jcb.24116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chevalier B, Adamiok A, Mercey O, Revinski DR, Zaragosi LE, Pasini A, Kodjabachian L, Barbry P, Marcet B. miR-34/449 control apical actin network formation during multiciliogenesis through small GTPase pathways. Nat Commun 2015; 6:8386; PMID:26381333; http://dx.doi.org/ 10.1038/ncomms9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pan J, You Y, Huang T, Brody SL. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J Cell Sci 2007; 120:1868-76; PMID:17488776; http://dx.doi.org/ 10.1242/jcs.005306 [DOI] [PubMed] [Google Scholar]
- [10].Brooks ER, Wallingford JB. Multiciliated cells. Curr Biol 2014; 24:R973-82; PMID:25291643; http://dx.doi.org/ 10.1016/j.cub.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Takeda S, Narita K. Structure and function of vertebrate cilia, towards a new taxonomy. Differentiation 2012; 83:S4-11; PMID:22118931; http://dx.doi.org/ 10.1016/j.diff.2011.11.002 [DOI] [PubMed] [Google Scholar]
- [12].Kirchhoff C, Osterhoff C, Samalecos A. HE6/GPR64 adhesion receptor co-localizes with apical and subapical F-actin scaffold in male excurrent duct epithelia. Reproduction 2008; 136:235-45; PMID:18469038; http://dx.doi.org/ 10.1530/REP-08-0078 [DOI] [PubMed] [Google Scholar]
- [13].Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 2007; 8:880-93; PMID:17955020; http://dx.doi.org/ 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- [14].Wang L, Fu C, Fan H, Du T, Dong M, Chen Y, Jin Y, Zhou Y, Deng M, Gu A, et al.. miR-34b regulates multiciliogenesis during organ formation in zebrafish. Development 2013; 140:2755-64; PMID:23698347; http://dx.doi.org/ 10.1242/dev.092825 [DOI] [PubMed] [Google Scholar]
- [15].Hansen A, Zeiske E. The peripheral olfactory organ of the zebrafish, Danio rerio: an ultrastructural study. Chem Senses 1998; 23:39-48; PMID:9530968; http://dx.doi.org/ 10.1093/chemse/23.1.39 [DOI] [PubMed] [Google Scholar]
- [16].Dubaissi E, Papalopulu N. Embryonic frog epidermis: a model for the study of cell-cell interactions in the development of mucociliary disease. Dis Model Mech 2011; 4:179-92; PMID:21183475; http://dx.doi.org/ 10.1242/dmm.006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Malicki J, Avanesov A, Li J, Yuan S, Sun Z. Analysis of cilia structure and function in zebrafish. Methods Cell Biol 2011; 101:39-74; PMID:21550439; http://dx.doi.org/ 10.1016/B978-0-12-387036-0.00003-7 [DOI] [PubMed] [Google Scholar]
- [18].Wessely O, Tran U. Xenopus pronephros development–past, present, and future. Pediatr Nephrol 2011; 26:1545-51; PMID:21499947; http://dx.doi.org/ 10.1007/s00467-011-1881-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Livraghi A, Randell SH. Cystic fibrosis and other respiratory diseases of impaired mucus clearance. Toxicol Pathol 2007; 35:116-29; PMID:17325980; http://dx.doi.org/ 10.1080/01926230601060025 [DOI] [PubMed] [Google Scholar]
- [20].Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, Olbrich H, Dougherty GW, Raidt J, Werner C, Amirav I, et al.. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Commun 2014; 5:4418; PMID:25048963; http://dx.doi.org/ 10.1038/ncomms5418 [DOI] [PubMed] [Google Scholar]
- [21].Wallmeier J, Al-Mutairi DA, Chen CT, Loges NT, Pennekamp P, Menchen T, Ma L, Shamseldin HE, Olbrich H, Dougherty GW, et al.. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Genet 2014; 46:646-51; PMID:24747639; http://dx.doi.org/ 10.1038/ng.2961 [DOI] [PubMed] [Google Scholar]
- [22].Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi LE, Cibois M, Robbe-Sermesant K, Jolly T, Cardinaud B, Moreilhon C, et al.. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol 2011; 13:693-9; PMID:21602795; http://dx.doi.org/ 10.1038/ncb2358 [DOI] [PubMed] [Google Scholar]
- [23].Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 1999; 126:4715-28; PMID:10518489. [DOI] [PubMed] [Google Scholar]
- [24].Cibois M, Luxardi G, Chevalier B, Thome V, Mercey O, Zaragosi LE, Barbry P, Pasini A, Marcet B, Kodjabachian L. BMP signalling controls the construction of vertebrate mucociliary epithelia. Development 2015; 142:2352-63; PMID:26092849; http://dx.doi.org/ 10.1242/dev.118679 [DOI] [PubMed] [Google Scholar]
- [25].Jerber J, Thomas J, Durand B. [Transcriptional control of ciliogenesis in animal development]. Biol Aujourdhui 2012; 206:205-18; PMID:23171843; http://dx.doi.org/ 10.1051/jbio/2012023 [DOI] [PubMed] [Google Scholar]
- [26].Tan FE, Vladar EK, Ma L, Fuentealba LC, Hoh R, Espinoza FH, Axelrod JD, Alvarez-Buylla A, Stearns T, Kintner C, et al.. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development 2013; 140:4277-86; PMID:24048590; http://dx.doi.org/ 10.1242/dev.094102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stubbs JL, Vladar EK, Axelrod JD, Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 2012; 14:140-7; PMID:22231168; http://dx.doi.org/ 10.1038/ncb2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gao X, Bali AS, Randell SH, Hogan BL. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J Cell Biol 2015; 211(3):669-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Funk MC, Bera AN, Menchen T, Kuales G, Thriene K, Lienkamp SS, Dengjel J, Omran H, Frank M, Arnold SJ. Cyclin O (Ccno) functions during deuterosome-mediated centriole amplification of multiciliated cells. EMBO J 2015; 34:1078-89; PMID:25712475; http://dx.doi.org/ 10.15252/embj.201490805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marcet B, Chevalier B, Coraux C, Kodjabachian L, Barbry P. MicroRNA-based silencing of Delta/Notch signaling promotes multiple cilia formation. Cell Cycle 2011; 10:2858-64; PMID:21857154; http://dx.doi.org/ 10.4161/cc.10.17.17011 [DOI] [PubMed] [Google Scholar]
- [31].Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132:9-14; PMID:18191211; http://dx.doi.org/ 10.1016/j.cell.2007.12.024 [DOI] [PubMed] [Google Scholar]
- [32].Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, et al.. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 2014; 510:115-20; PMID:24899310; http://dx.doi.org/ 10.1038/nature13413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A 2014; 111:E2851-7; PMID:24982181; http://dx.doi.org/ 10.1073/pnas.1407777111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Comazzetto S, Di Giacomo M, Rasmussen KD, Much C, Azzi C, Perlas E, Morgan M, O'Carroll D. Oligoasthenoteratozoospermia and infertility in mice deficient for miR-34b/c and miR-449 loci. PLoS Genet 2014; 10:e1004597; PMID:25329700; http://dx.doi.org/ 10.1371/journal.pgen.1004597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D. Ancient animal microRNAs and the evolution of tissue identity. Nature 2010; 463:1084-8; PMID:20118916; http://dx.doi.org/ 10.1038/nature08744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gomperts BN, Gong-Cooper X, Hackett BP. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J Cell Sci 2004; 117:1329-37; PMID:14996907; http://dx.doi.org/ 10.1242/jcs.00978 [DOI] [PubMed] [Google Scholar]
- [37].Huang T, You Y, Spoor MS, Richer EJ, Kudva VV, Paige RC, Seiler MP, Liebler JM, Zabner J, Plopper CG, et al.. Foxj1 is required for apical localization of ezrin in airway epithelial cells. J Cell Sci 2003; 116:4935-45; PMID:14625387; http://dx.doi.org/ 10.1242/jcs.00830 [DOI] [PubMed] [Google Scholar]
- [38].Miyatake K, Kusakabe M, Takahashi C, Nishida E. ERK7 regulates ciliogenesis by phosphorylating the actin regulator CapZIP in cooperation with Dishevelled. Nat Commun 2015; 6:6666; PMID:25823377; http://dx.doi.org/ 10.1038/ncomms7666 [DOI] [PubMed] [Google Scholar]
- [39].Antoniades I, Stylianou P, Skourides PA. Making the connection: ciliary adhesion complexes anchor basal bodies to the actin cytoskeleton. Dev Cell 2014; 28:70-80; PMID:24434137; http://dx.doi.org/ 10.1016/j.devcel.2013.12.003 [DOI] [PubMed] [Google Scholar]
- [40].Guilluy C, Garcia-Mata R, Burridge K. Rho protein crosstalk: another social network? Trends Cell Biol 2011; 21:718-26; PMID:21924908; http://dx.doi.org/ 10.1016/j.tcb.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet 2008; 40:871-9; PMID:18552847; http://dx.doi.org/ 10.1038/ng.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jeong HW, Nam JO, Kim IS. The COOH-terminal end of R-Ras alters the motility and morphology of breast epithelial cells through Rho/Rho-kinase. Cancer Res 2005; 65:507-15; PMID:15695393. [PubMed] [Google Scholar]
- [43].Boulter E, Garcia-Mata R. RhoGDI: A rheostat for the Rho switch. Small Gtpases 2010; 1:65-8; PMID:21686121; http://dx.doi.org/ 10.4161/sgtp.1.1.12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol 2011; 12:493-504; PMID:21779026; http://dx.doi.org/ 10.1038/nrm3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu M, Bi F, Zhou X, Zheng Y. Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol 2012; 22:365-73; PMID:22572609; http://dx.doi.org/ 10.1016/j.tcb.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Adams M, Simms RJ, Abdelhamed Z, Dawe HR, Szymanska K, Logan CV, Wheway G, Pitt E, Gull K, Knowles MA, et al.. A meckelin-filamin A interaction mediates ciliogenesis. Hum Mol Genet 2012; 21:1272-86; PMID:22121117; http://dx.doi.org/ 10.1093/hmg/ddr557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gawecka JE, Griffiths GS, Ek-Rylander B, Ramos JW, Matter ML. R-Ras regulates migration through an interaction with filamin A in melanoma cells. PLoS One 2010; 5:e11269; PMID:20585650; http://dx.doi.org/ 10.1371/journal.pone.0011269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Griffiths GS, Grundl M, Allen JS 3rd, Matter ML. R-Ras interacts with filamin a to maintain endothelial barrier function. J Cell Physiol 2011; 226:2287-96; PMID:21660952; http://dx.doi.org/ 10.1002/jcp.22565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol 2000; 2:888-92; PMID:11146652; http://dx.doi.org/ 10.1038/35046533 [DOI] [PubMed] [Google Scholar]
- [50].Shang X, Cancelas JA, Li L, Guo F, Liu W, Johnson JF, Ficker A, Daria D, Geiger H, Ratner N, et al.. R-Ras and Rac GTPase cross-talk regulates hematopoietic progenitor cell migration, homing, and mobilization. J Biol Chem 2011; 286:24068-78; PMID:21572048; http://dx.doi.org/ 10.1074/jbc.M111.226951 [DOI] [PMC free article] [PubMed] [Google Scholar]