Abstract
The budding yeast S. cerevisiae divides asymmetrically and is an excellent model system for asymmetric cell division. As for other asymmetrically dividing cells, proper spindle positioning along the mother-daughter polarity axis is crucial for balanced chromosome segregation. Thus, a surveillance mechanism named Spindle Position Checkpoint (SPOC) inhibits mitotic exit and cytokinesis until the mitotic spindle is properly oriented, thereby preventing the generation of cells with aberrant ploidies. The small GTPase Tem1 is required to trigger a Hippo-like protein kinase cascade, named Mitotic Exit Network (MEN), that is essential for mitotic exit and cytokinesis but also contributes to correct spindle alignment in metaphase. Importantly, Tem1 is the target of the SPOC, which relies on the activity of the GTPase-activating complex (GAP) Bub2-Bfa1 to keep Tem1 in the GDP-bound inactive form. Tem1 forms a hetero-trimeric complex with Bub2-Bfa1 at spindle poles (SPBs) that accumulates asymmetrically on the bud-directed spindle pole during mitosis when the spindle is properly positioned. In contrast, the complex remains symmetrically localized on both poles of misaligned spindles. We have recently shown that Tem1 residence at SPBs depends on its nucleotide state and, importantly, asymmetry of the Bub2-Bfa1-Tem1 complex does not promote mitotic exit but rather controls spindle positioning.
Keywords: asymmetric cell division, Kar9, mitotic exit network, spindle positioning, spindle position checkpoint, spindle pole bodies, Tem1
Spindle Positioning in Budding Yeast
In asymmetrically dividing cells, proper spindle positioning is crucial to ensure the unequal fate of daughter cells. In stem cells, derangement of the mechanisms controlling asymmetric cell division, including spindle positioning, affects the developmental fate of daughter cells and can promote tumorigenesis.(reviewed in ref.1).
The budding yeast Saccharomyces cerevisiae divides asymmetrically and has long been used as model system to study the mechanisms underlying asymmetric cell division. Because accurate spindle positioning is critical for asymmetric cell division, 2 redundant pathways are responsible for correct spindle positioning in S. cerevisiae: a pathway dependent on the microtubule-binding protein Kar9 and another pathway requiring the minus end-directed motor dynein. The Kar9 pathway is supposed to act mainly in metaphase and to mediate the sliding of astral microtubule ends along actin cables.2 Conversely, the dynein-dependent pathway is thought to act predominantly in anaphase.3 However, its ability to compensate for the lack of the Kar9 pathway indicates that is likely already active in metaphase.4 Consistent with the notion that the 2 spindle positioning mechanisms are largely redundant, kar9 or dynein single mutants display only mild spindle mispositioning, while double mutants are lethal.5 One critical feature of Kar9 is its asymmetric localization to the astral microtubules emanating only from the bud-oriented SPB.2,6-8 The asymmetry of Kar9 ensures that only one pole of the mitotic spindle is oriented toward the bud. Interestingly, also dynein is localized asymmetrically at spindle poles, with a strong bias for the bud-directed SPB.9 Asymmetry of Kar9 and dynein, however, seems to be controlled by different mechanisms.7,9,10
The Tem1 GTPase
The budding yeast TEM1 gene was identified through a genetic screen in 1994 as encoding for a novel GTP-binding protein.11 The deletion of TEM1 was lethal, and the tem1 temperature-sensitive mutants arrested at telophase, indicating that TEM1 is required to exit from mitosis. Later on, Tem1 was shown to be indeed a Rab-like GTPase.12 The counterpart of Tem1 in S. pombe, named Spg1, is also a GTPase that is dispensable for mitotic exit and solely required for cytokinesis.13 In spite of the essential roles played by these GTPases in yeasts, no mammalian counterpart has been identified so far.
During the past 2 decades, several studies shed light on the role of Tem1 as a molecular switch for activation of the Mitotic Exit Network (MEN), an essential kinase cascade that promotes mitotic exit and cytokinesis and is organized similarly to the Septation Initiation Network (SIN) in fission yeast and the Hippo pathway in metazoans (reviewed in ref.14). The MEN effector of Tem1 is the Ste20-like kinase Cdc15,15-18 which in turn promotes the activation of the downstream Mob1-Dbf2 kinase complex (LATS-NDR in the Hippo pathway).19,20 that ultimately leads to the release and activation of Cdc14,21-23 the main CDK-counteracting phosphatase that triggers mitotic exit through dephosphorylation of mitotic CDK substrates.24 One notable feature of the MEN is that most of its components, including the Tem1 GTPase, reside at spindle pole bodies (SPBs), i.e. the yeast microtubule-organizing centers (Fig. 1A-B). Thus, not only Tem1 is necessary for cell cycle progression, but is also ideally localized to get positional feedback from the spindle poles. Not surprisingly, Tem1 is the target of the Spindle Position Checkpoint (SPOC), a surveillance mechanism that blocks mitotic exit when the spindle is not properly oriented, in order to prevent the formation of aneuploid and polyploid cells after cytokinesis (reviewed in. ref.25,26, see below).
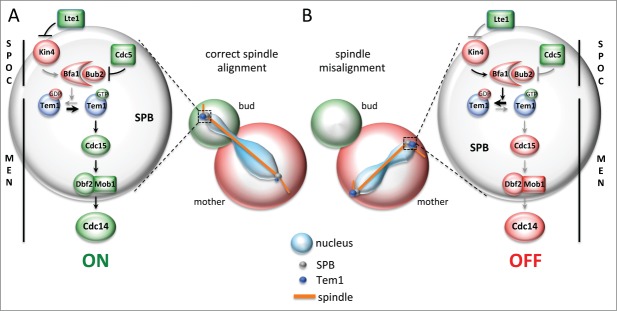
Figure 1.
Asymmetry of the Tem1 GTPase at SPBs is broken upon spindle misalignment. (A) When cells align properly their mitotic spindles along the mother-to-bud polarity axis the Tem1 GTPase localizes preferentially to the bud-directed SPB and triggers the Mitotic Exit Network (MEN) kinase cascade (Cdc15, Dbf2-Mob1) that leads to the activation of Cdc14 phosphatase and mitotic exit. Two separated compartments for inhibition and activation of the MEN are defined in the mother cell and in the bud by Kin4 and Lte1, respectively, and depicted in red and green. (B) Upon spindle misalignment Tem1 localizes symmetrically on both SPBs and the Spindle Position Checkpoint (SPOC) inhibits Tem1 through the GTPase-Activating Protein (GAP) complex Bub2-Bfa1, thereby restraining the MEN until the spindle repositions correctly. The GAP is in turn kept active by the kinase Kin4, which counteracts the inhibitory phosphorylation of the GAP by the Polo kinase Cdc5.
Regulation of the Tem1 GTPase
As long as the spindle is not properly positioned at the bud neck and oriented along the mother-bud polarity axis, the SPOC prevents Tem1 activation (reviewed in ref.25,26), thereby preventing premature exit from mitosis or aberrant mitosis. The key actor in this control is the two-component GTPase-activating protein (GAP) Bub2-Bfa1 that inactivates Tem1 by stimulating GTP hydrolysis.12,27 (Fig. 1B). The GAP activity of the Bub2-Bfa1 complex resides on Bub2, which carries a TBC domain (Tre-2, Bub2 and Cdc16; reviewed in ref.28), whereas Bfa1 mediates Bub2 interaction with Tem1 and acts as guanine-nucleotide dissociation inhibitor (GDI), stabilizing Tem1 in its GDP- or GTP-bound state.12,27,29 Interestingly, Bfa1 can inhibit Tem1 and mitotic exit also in a Bub2-independent manner, likely by competing with Cdc15 for Tem1 interaction30 and possibly by locking Tem1 in the GDP-inactive state.29
Although activation of most GTPases requires nucleotide exchange factors (GEFs) that catalyze the release of GDP and promote its replacement by GTP, the identity of the GEF(s) for Tem1, if any, remains elusive. The ability of recombinant Tem1 to efficiently load guanine nucleotides in vitro.12,27 raises the possibility that Tem1 does not need a GEF. The putative GEF Lte1, which activates Tem1 in vivo and has long been thought to catalyze Tem1 binding to GTP, does not seem in fact to bear GEF activity for Tem1 in vitro.29
The activity of the GAP complex Bub2-Bfa1 is itself finely regulated by the partition of the cell in 2 distinct regulatory compartments: the mother- and the bud-compartment, which respectively inhibit and activate Tem1 and the downstream MEN cascade.31 (Fig. 1A-B). These regulatory compartments are defined by the activity of 2 asymmetrically distributed proteins, i.e. Kin4 and Lte1, that localize specifically in the mother cell and in the bud, respectively.32-36 In the mother compartment, the Kin4 protein kinase activates the Bub2-Bfa1 GAP complex by direct phosphorylation of Bfa1, thereby preventing its inhibitory phosphorylation by the Polo kinase Cdc5.33,35,37 Moreover, it creates a docking site on Bfa1 for the 14-3-3 protein Bmh1, which in turn weakens the association of Bfa1 with the SPB and accelerates the turnover of the GAP complex at SPBs.38 In the bud compartment, Lte1 prevents Kin4 spreading into the mother cell, thus allowing the Cdc5 kinase to inactivate the GAP through Bfa1 phosphorylation.39-41 Therefore, when both SPBs remain in the mother cell following spindle mispositioning (Fig. 1B), Tem1 and the MEN are kept inactive, while translocation of one SPB into the bud promotes Tem1 activation and allows mitotic exit.42 SPB-localized Tem1 thus acts as a sensor that relays the positional information from the spindle and translates it into a biochemical signal to license or prevent mitotic exit.
Regulation of Tem1 Localization
Besides on its nucleotide state, Tem1 activity also depends on its localization. Indeed, Tem1 loading onto the SPBs is an essential step to trigger the mitotic exit cascade.43,44 A tem1 mutant that fails to localize at SPBs is deficient in mitotic exit,15 while artificially tethering Tem1 to SPBs is sufficient to drive out of mitosis cells experiencing spindle misalignment.44 The downstream MEN kinases are also recruited to SPBs once Tem1 gets activated. Localization at spindle poles of MEN components, including the inhibitory GAP Bub2-Bfa1 and the kinase Kin4, is critical for MEN activity and regulation and is mainly accomplished at the cytoplasmic face of the SPBs through association to the SPB scaffold Nud1.15,35,43,44 Consistently, nud1 temperature-sensitive mutants fail to exit mitosis and arrest in telophase.43 Several observations indicate that Tem1 activation is necessary but not sufficient to promote mitotic exit during the unperturbed cell cycle. Indeed, covalent tethering of Tem1 to SPBs promotes its activation and increases Cdc15 levels at spindle poles, but does not promote premature mitotic exit.44 Similarly, fusion of Cdc15 to the SPBs leads to premature activation of the Dbf2 kinase, but not premature mitotic exit.17 Inhibition of Cdc15 and Dbf2-Mob1 by cyclinB-CDKs is likely responsible for the inability of active Tem1 to trigger mitotic exit in metaphase and provides a safety mechanism to ensure the correct order of cell cycle events.45 Indeed, cyclinB degradation at the onset of anaphase is a requisite that must be satisfied for MEN to get fully active.
Tem1, together with its GAP Bub2-Bfa1, is present on both SPBs in metaphase, when it accumulates preferentially on the SPB that is most proximal to the bud.27,46-48 (Fig. 2A). Its localization becomes even more asymmetric in anaphase, when it keeps concentrating on the bud-directed SPB (Fig. 1A). Conversely, if the spindle fails to elongate properly along the cell polarity axis, the Bub2-Bfa1-Tem1 complex is symmetrically retained on both SPBs and shows high turnover, exchanging with its cytoplasmic pool.42,46,48 (Fig. 1B). Surprisingly, chimeric proteins obtained by fusing Bfa1 or Bub2 to both SPBs cause unscheduled mitotic exit upon spindle misalignment,46,49 similar to constitutive recruitment of Tem1 to SPBs. These data led to the idea that the Bub2-Bfa1 complex, while acting as inhibitory GAP on Tem1, also stabilizes Tem1 at SPBs where it becomes active.44,49 Since the Bub2-Bfa1 complex is completely dispensable for mitotic exit, in its absence Tem1 is likely recruited to SPBs through another receptor, that has been proposed to be the SPB scaffold Nud1.44 Consistently, in the absence of Bub2-Bfa1 Tem1 levels at SPBs are drastically reduced throughout most of the cell cycle, to then sharply rise in telophase.48 The molecular basis for SPB-driven Tem1 activation remains elusive. A recent proposal envisions that an as-yet unidentified GEF for Tem1 resides at SPBs.50
Figure 2.
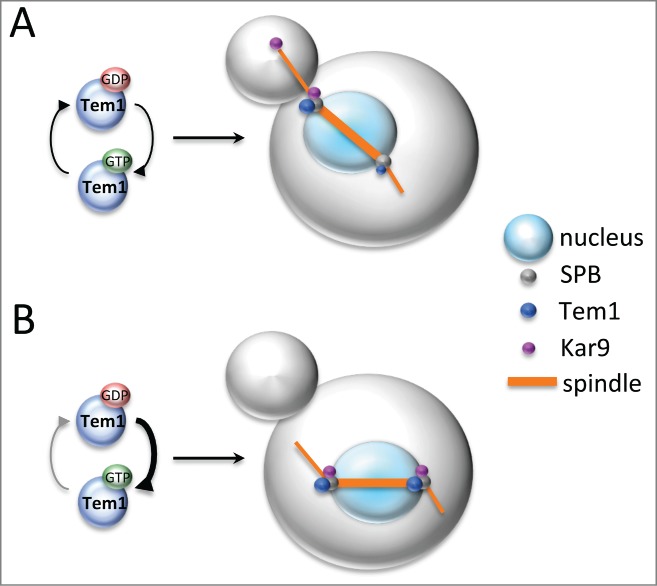
Tem1 asymmetry contributes to asymmetric distribution of Kar9 and proper spindle positioning. (A) Partial asymmetry of Tem1 at SPBs is already established in metaphase and contributes to the asymmetry of Kar9 on the bud-directed SPB and the tip of the astral microtubules that nucleate from it. Kar9 asymmetry is in turn necessary for proper spindle positioning in metaphase. (B) Constitutive activation or SPB tethering of Tem1 breaks its asymmetry at SPBs leading to a more symmetric distribution of Kar9 and spindle position defects. See text for further details.
We have recently shown a close link between Tem1 activity and the establishment of its asymmetry at spindle poles.49 The hyperactive TEM1-Q79L mutant allele, where a leucine replaces the catalytic glutamine in the G domain, is completely refractory to stimulation of GTP hydrolysis by Bub2-Bfa1 both in vitro and in vivo, without affecting its interaction with the GAP. The mutant protein shows a markedly increased symmetry at anaphase spindle poles and also impairs the asymmetry of Bub2 and Bfa1.49 Likewise, mutations abolishing Bub2-Bfa1 GAP activity perturb the asymmetry of the Tem1-Bub2-Bfa1 complex, indicating that GTP hydrolysis promotes asymmetry of both Tem1 and its GAP at SPBs.27,49
Tem1 in the Control of Spindle Positioning
Although the most prominent MEN function is closely coupled with the final stages of mitosis, recent observations showed that MEN components also have important functions earlier on during the cell cycle. These include a role in orientation of the mitotic spindle in metaphase and in establishing Kar9 asymmetry.51 Indeed, MEN downregulation by inactivation of Nud1, Tem1, Cdc15 or Dbf2, prevents the Kar9 protein to properly accumulate on the aster microtubules of the bud-directed SPB in early mitosis and allows it to bind also the second spindle pole. This leads to spindle misorientation due to the ability of both asters to pull along actin cable in the bud. At the heart of this mechanism there is the Dbf2-dependent phosphorylation of Kar9 that stabilizes its asymmetric binding to the bud-directed SPB. Consistently, the downstream Cdc14 phosphatase is not involved in Kar9 distribution.51 Localization of Tem1 and its downstream kinases at SPBs is likely critical for the control of Kar9 asymmetry. Indeed, the lack of the Bub2-Bfa1 complex, which prevents Tem1 recruitment to SPBs in early mitosis, compromises Kar9 asymmetry and spindle orientation in a similar way to MEN inactivation.49
Our recent data indicate that Tem1 asymmetry plays a critical role in directing Kar9 asymmetry. Indeed, chimeric proteins that stabilize Tem1 at both SPBs or expression of the constitutively active Tem1-Q79L GTPase that remains more symmetric at SPBs both impair Kar9 asymmetry and spindle positioning.49 (Fig. 2B). Thus, at a first glance inactivation or hyperactivation of Tem1 similarly impact on Kar9 asymmetry. One notable difference, however, is that while Tem1 inactivation causes only spindle orientation defects (where orientation is defined by the angle between the metaphase spindle and the bud neck),51 Tem1 hyperactivation causes in addition spindle mispositioning (where position is defined by the distance between the proximal SPB and the bud neck).49 The molecular bases of this difference remain to be established. One possibility to be explored further is that the absolute amounts and/or turnover of Kar9 at SPBs differ in the 2 conditions.
Conclusions
One crucial question that remains unsolved is how Tem1 activity toward mitotic exit is restrained until telophase while being already operational in metaphase toward spindle positioning. One possible scenario is that only the fraction of Tem1 at SPBs gets activated in metaphase, while a more global activation of Tem1 in the cytoplasm is required for mitotic exit.52 A differential requirement of different pools of Tem1 in processes that must occur in different cell cycle phases would intrinsically ensure the proper coordination between spindle positioning and mitotic exit.
Another important issue that remains to be addressed is what triggers Tem1 activation at SPBs. As mentioned above, it is possible that an unidentified GEF resides at SPBs. Alternatively, Tem1 might not need a GEF; rather, at SPBs it could be simply refractory to inhibition by its GAP. It is worth noticing that the Polo kinase Cdc5, which downregulates the GAP activity of the Bub2-Bfa1 complex,41,53 is found at both SPBs during metaphase54 and is therefore likely part of this regulatory mechanism. Moreover, Tem1 itself is subject to post-translational modifications, such as phosphorylation and ubiquitylation,55 that could contribute to its regulation.
The control of Tem1 and the MEN is a beautiful example of the strategies that eukaryotic cells have evolved to couple spindle positioning and cell cycle progression. Surveillance mechanisms analogous to the SPOC have been identified in asymmetrically dividing stem cells,56 highlighting their importance in ensuring the right fate of a cell lineage and preserving the proper ploidy of cells.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize with all the colleagues whose work could not be cited due to space limitations.
Funding
Work in S. Piatti's lab is supported by the Ligue Nationale contre le Cancer, the Fondation ARC pour la Recherche sur le Cancer (grant PJA20141201926) and the Fondation pour la Recherche Médicale (grant DEQ20150331740).
References
- 1.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet 2007; 8:462-72; PMID:17510666; http://dx.doi.org/ 10.1038/nrg2103 [DOI] [PubMed] [Google Scholar]
- 2.Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol 2000; 10:1497-506; PMID:11114516; http://dx.doi.org/ 10.1016/S0960-9822(00)00837-X [DOI] [PubMed] [Google Scholar]
- 3.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 1995; 130:687-700; PMID:7622568; http://dx.doi.org/ 10.1083/jcb.130.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell 2000; 11:3949-61; PMID:11071919; http://dx.doi.org/ 10.1091/mbc.11.11.3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol 1998; 140:377-90; PMID:9442113; http://dx.doi.org/ 10.1083/jcb.140.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cepeda-Garcia C, Delgehyr N, Juanes Ortiz MA, ten Hoopen R, Zhiteneva A, Segal M. Actin-mediated delivery of astral microtubules instructs Kar9p asymmetric loading to the bud-ward spindle pole. Mol Biol Cell 2010; 21:2685-95; PMID:20534809; http://dx.doi.org/ 10.1091/mbc.E10-03-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 2003; 112:561-74; PMID:12600318; http://dx.doi.org/ 10.1016/S0092-8674(03)00119-3 [DOI] [PubMed] [Google Scholar]
- 8.Miller RK, Matheos D, Rose MD. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol 1999; 144:963-75; PMID:10085294; http://dx.doi.org/ 10.1083/jcb.144.5.963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell 2006; 10:425-39; PMID:16580990; http://dx.doi.org/ 10.1016/j.devcel.2006.02.018 [DOI] [PubMed] [Google Scholar]
- 10.Maekawa H, Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev 2004; 18:1709-24; PMID:15256500; http://dx.doi.org/ 10.1101/gad.298704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirayama M, Matsui Y, Toh EA. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol 1994; 14:7476-82; PMID:7935462; http://dx.doi.org/ 10.1128/MCB.14.11.7476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geymonat M, Spanos A, Smith SJ, Wheatley E, Rittinger K, Johnston LH, Sedgwick SG. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J Biol Chem 2002; 277:28439-45; PMID:12048186; http://dx.doi.org/ 10.1074/jbc.M202540200 [DOI] [PubMed] [Google Scholar]
- 13.Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev 1997; 11:1519-34; PMID:9203579; http://dx.doi.org/ 10.1101/gad.11.12.1519 [DOI] [PubMed] [Google Scholar]
- 14.Hergovich A, Hemmings BA. Hippo signalling in the G2/M cell cycle phase: lessons learned from the yeast MEN and SIN pathways. Semin Cell Dev Biol 2012; 23:794-802; PMID:22525225; http://dx.doi.org/ 10.1016/j.semcdb.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asakawa K, Yoshida S, Otake F, Toh EA. A Novel Functional Domain of Cdc15 Kinase Is Required for Its Interaction With Tem1 GTPase in Saccharomyces cerevisiae. Genetics 2001; 157:1437-50; PMID:11290702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr Biol 2001; 11:784-8; PMID:11378390; http://dx.doi.org/ 10.1016/S0960-9822(01)00228-7 [DOI] [PubMed] [Google Scholar]
- 17.Rock JM, Amon A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev 2011; 25:1943-54; PMID:21937712; http://dx.doi.org/ 10.1101/gad.17257711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visintin R, Amon A. Regulation of the mitotic exit protein kinases cdc15 and dbf2. Mol Biol Cell 2001; 12:2961-74; PMID:11598184; http://dx.doi.org/ 10.1091/mbc.12.10.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mah AS, Jang J, Deshaies RJ. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc Natl Acad Sci U S A 2001; 98:7325-30; PMID:11404483; http://dx.doi.org/ 10.1073/pnas.141098998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rock JM, Lim D, Stach L, Ogrodowicz RW, Keck JM, Jones MH, Wong CC, Yates JR 3rd, Winey M, Smerdon SJ, et al.. Activation of the Yeast Hippo Pathway by Phosphorylation-Dependent Assembly of Signaling Complexes. Science 2013; 340:871-5; PMID:23579499; http://dx.doi.org/ 10.1126/science.1235822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J Cell Biol 2009; 184(4):527-39; PMID:19221193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 1999; 97:233-44; PMID:10219244; http://dx.doi.org/ 10.1016/S0092-8674(00)80733-3 [DOI] [PubMed] [Google Scholar]
- 23.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus [see comments]. Nature 1999; 398:818-23; PMID:10235265; http://dx.doi.org/ 10.1038/19775 [DOI] [PubMed] [Google Scholar]
- 24.Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk- dependent phosphorylation. Mol Cell 1998; 2:709-18; PMID:9885559; http://dx.doi.org/ 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- 25.Caydasi AK, Ibrahim B, Pereira G. Monitoring spindle orientation: Spindle position checkpoint in charge. Cell Div 2010; 5:28; PMID:21143992; http://dx.doi.org/ 10.1186/1747-1028-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraschini R, Venturetti M, Chiroli E, Piatti S. The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans 2008; 36:416-20; PMID:18481971; http://dx.doi.org/ 10.1042/BST0360416 [DOI] [PubMed] [Google Scholar]
- 27.Fraschini R, D'Ambrosio C, Venturetti M, Lucchini G, Piatti S. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J Cell Biol 2006; 172:335-46; PMID:16449187; http://dx.doi.org/ 10.1083/jcb.200507162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuwald AF. A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem Sci 1997; 22:243-4; PMID:9255064; http://dx.doi.org/ 10.1016/S0968-0004(97)01073-6 [DOI] [PubMed] [Google Scholar]
- 29.Geymonat M, Spanos A, de Bettignies G, Sedgwick SG. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J Cell Biol 2009; 187:497-511; PMID:19948498; http://dx.doi.org/ 10.1083/jcb.200905114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ro HS, Song S, Lee KS. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2002; 99:5436-41; PMID:11959999; http://dx.doi.org/ 10.1073/pnas.062059999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan LY, Amon A. Spindle position is coordinated with cell-cycle progression through establishment of mitotic exit-activating and -inhibitory zones. Mol Cell 2010; 39:444-54; PMID:20705245; http://dx.doi.org/ 10.1016/j.molcel.2010.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiroli E, Fraschini R, Beretta A, Tonelli M, Lucchini G, Piatti S. Budding yeast PAK kinases regulate mitotic exit by two different mechanisms. J Cell Biol 2003; 160:857-74; PMID:12642613; http://dx.doi.org/ 10.1083/jcb.200209097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, Lai L, Shokat KM, Amon A. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell 2005; 19:223-34; PMID:16039591; http://dx.doi.org/ 10.1016/j.molcel.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 34.Hofken T, Schiebel E. A role for cell polarity proteins in mitotic exit. Embo J 2002; 21:4851-62; PMID:12234925; http://dx.doi.org/ 10.1093/emboj/cdf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira G, Schiebel E. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol Cell 2005; 19:209-21; PMID:16039590; http://dx.doi.org/ 10.1016/j.molcel.2005.05.030 [DOI] [PubMed] [Google Scholar]
- 36.Seshan A, Bardin AJ, Amon A. Control of lte1 localization by cell polarity determinants and cdc14. Curr Biol 2002; 12:2098-110; PMID:12498684; http://dx.doi.org/ 10.1016/S0960-9822(02)01388-X [DOI] [PubMed] [Google Scholar]
- 37.Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol 2007; 179:423-36; PMID:17967947; http://dx.doi.org/ 10.1083/jcb.200705197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caydasi AK, Micoogullari Y, Kurtulmus B, Palani S, Pereira G. The 14-3-3 protein Bmh1 functions in the spindle position checkpoint by breaking Bfa1 asymmetry at yeast centrosomes. Mol Biol Cell 2014; 25:2143-51; PMID:24850890; http://dx.doi.org/ 10.1091/mbc.E14-04-0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertazzi DT, Kurtulmus B, Pereira G. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J Cell Biol 2011; 193:1033-48; PMID:21670215; http://dx.doi.org/ 10.1083/jcb.201101056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falk JE, Chan LY, Amon A. Lte1 promotes mitotic exit by controlling the localization of the spindle position checkpoint kinase Kin4. Proc Natl Acad Sci U S A 2011; 108:12584-90; PMID:21709215; http://dx.doi.org/ 10.1073/pnas.1107784108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 2001; 107:655-65; PMID:11733064; http://dx.doi.org/ 10.1016/S0092-8674(01)00580-3 [DOI] [PubMed] [Google Scholar]
- 42.Molk JN, Schuyler SC, Liu JY, Evans JG, Salmon ED, Pellman D, Bloom K. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol Biol Cell 2004; 15:1519-32; PMID:14718561; http://dx.doi.org/ 10.1091/mbc.E03-09-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis [In Process Citation]. Embo J 2000; 19:6475-88; PMID:11101520; http://dx.doi.org/ 10.1093/emboj/19.23.6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valerio-Santiago M, Monje-Casas F. Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J Cell Biol 2011; 192:599-614; PMID:21321099; http://dx.doi.org/ 10.1083/jcb.201007044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konig C, Maekawa H, Schiebel E. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J Cell Biol 2010; 188:351-68; PMID:20123997; http://dx.doi.org/ 10.1083/jcb.200911128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caydasi AK, Pereira G. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev Cell 2009; 16:146-56; PMID:19154725; http://dx.doi.org/ 10.1016/j.devcel.2008.10.013 [DOI] [PubMed] [Google Scholar]
- 47.Monje-Casas F, Amon A. Cell polarity determinants establish asymmetry in MEN signaling. Dev Cell 2009; 16:132-45; PMID:19154724; http://dx.doi.org/ 10.1016/j.devcel.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell 2000; 6:1-10; PMID:10949022; http://dx.doi.org/ 10.1016/S1097-2765(05)00017-1 [DOI] [PubMed] [Google Scholar]
- 49.Scarfone I, Venturetti M, Hotz M, Lengefeld J, Barral Y, Piatti S. Asymmetry of the budding yeast Tem1 GTPase at spindle poles is required for spindle positioning but not for mitotic exit. PLoS Genet 2015; 11:e1004938; PMID:25658911; http://dx.doi.org/ 10.1371/journal.pgen.1004938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hotz M, Barral Y. The Mitotic Exit Network: new turns on old pathways. Trends Cell Biol 2014; 24:145-52; PMID:24594661; http://dx.doi.org/ 10.1016/j.tcb.2013.09.010 [DOI] [PubMed] [Google Scholar]
- 51.Hotz M, Leisner C, Chen D, Manatschal C, Wegleiter T, Ouellet J, Lindstrom D, Gottschling DE, Vogel J, Barral Y. Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell 2012; 148:958-72; PMID:22385961; http://dx.doi.org/ 10.1016/j.cell.2012.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caydasi AK, Lohel M, Grunert G, Dittrich P, Pereira G, Ibrahim B. A dynamical model of the spindle position checkpoint. Mol Syst Biol 2012; 8:582; PMID: 22580890; http://dx.doi.org/ 10.1038/msb.2012.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG. In Vitro Regulation of Budding Yeast Bfa1/Bub2 GAP Activity by Cdc5. J Biol Chem 2003; 278:14591-4; PMID:12637549; http://dx.doi.org/ 10.1074/jbc.C300059200 [DOI] [PubMed] [Google Scholar]
- 54.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. Embo J 1998; 17:1336-49; PMID:9482731; http://dx.doi.org/ 10.1093/emboj/17.5.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 2013; 10:676-82; PMID:23749301; http://dx.doi.org/ 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature 2008; 456(7222):599-604 [DOI] [PMC free article] [PubMed] [Google Scholar]