Abstract
Xeroderma pigmentosum group G (XPG), one of key components of nucleotide excision repair pathway (NER), is involved in excision repair of UV-induced DNA damage. Single nucleotide polymorphisms (SNPs) in the XPG gene have been reported to associate with the clinical outcome of various cancer patients. We aimed to assess the impact of four potentially functional SNPs (rs2094258 C>T, rs2296147 T>C, rs751402 G>A, and rs873601 G>A) in the XPG gene on prognosis in colorectal cancer (CRC) patients. A total of 1901 patients diagnosed with pathologically confirmed CRC were genotyped for four XPG polymorphisms. Cox proportional hazards model analysis controlled for several confounding factors was conducted to compute hazard ratios (HRs) and 95% confidence intervals (CIs). Of the four included SNPs, only rs2296147 was shown to significantly affect progression-free survival (PFS) in CRC. Patients carrying rs2296147 CT/TT genotype had a significantly shorter median 10 years PFS than those carrying CC genotype (88.5 months vs. 118.1 months), and an increased progression risk were observed with rs2296147 (HR = 1.324, 95% CI = 1.046–1.667). Moreover, none of the four SNPs were associated with overall survival. In conclusion, our study showed that XPG rs2296147 CT/TT variants conferred significant survival disadvantage in CRC patients in term of PFS. XPG rs2296147 polymorphism could be predictive of unfavorable prognosis of CRC patients.
Keywords: colorectal cancer, xeroderma pigmentosum group G, single nucleotide polymorphism, progression-free survival, overall survival
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer-related death in the world (http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx). Incidence of CRC dramatically varies from region to region. It ranks fifth in the commonly diagnosed malignancies in China, with 253, 427 new cases diagnosed and 139, 416 cancer deaths in 2012 (http://globocan.iarc.fr/Pages/fact_sheets_population.aspx).
It ranks the third in the commonly diagnosed malignancies in males and the second in females, with 1.4 million new cases diagnosed and 693, 900 cancer deaths in 2012 [1]. Therefore, it remains a major public health problem in China. Physicians make treatment plan depending on the clinical stage, performance status, and molecular characteristic of the tumor. Generally, surgery is used to treat the early stage of CRC, while the combination of 5-fluorouracil (5-FU), irinotecan, and oxaliplatin (FOLFOXIRI) is administrated to late-stage patients as the standard first-line chemotherapy to improve the prognosis [2]. The prognosis of colorectal cancer has been gradually improved over the past decades, with a 5-year relative survival of 65% and less than 50% in high- and low-income countries, respectively [3].
CRC is a complex disease, and both environmental and genetic factors contribute to oncogenesis. Diet (e.g., red meat) [4], smoking [5], drinking [6] and obesity [7] are well-known risk factors for CRC, although the underlying mechanisms remain clarified. Apart from those environmental factors, numerous evidence suggests that DNA repair systems also play an important role in modifying the risk of CRC [8–10]. For example, SNPs in XPD have an effect on the prognosis of CRC patients who were treated with oxaliplatin and 5-fluorouracil. Comparing to XPD 751Lys/Lys genotype, patients carrying Lys/Gln genotypes had more prone to chemotherapy failure and patients carrying ≥ 1 Gln had shorter median disease progression [8]. DNA repair systems include nucleotide excision repair (NER), base excision repair (BER), mis-match repair (MMR) and double-strand break repair (DSBR) pathways [11]. Of them, NER is responsible for repairing ultraviolet light (UV)-DNA damage, bulky DNA adducts (thymine dimers and 6, 4-photoproducts). NER functions properly by orchestrating different functional proteins involved in this pathway for the recognition of DNA lesion, incision, repair, and ligation [12]. Xeroderma pigmentosum group G (XPG) is one of the critical proteins in the NER pathway, encoded by excision repair cross-complementation group 5 (ERCC5) [12]. Accumulating studies have shown that the functional single nucleotide polymorphisms (SNPs) in the XPG gene may modify DNA repair capacity, and consequently increase the instability of genome. Additionally, such SNPs may also influence the ability to repair DNA damage caused by chemotherapeutic drugs, which enhances chemotherapeutic sensitivity and improve prognosis of CRC patients [13–15]. XPG is located on chromosome 13q33, containing 15 exons. Its protein product, an 1186-amino acid protein, is a structure-specific endonuclease responsible for the 3′ incision of DNA damage during NER, which has preference for the binding site of the single strand and the double strand of the degeneration bubble [16].
It has been reported that the SNPs in the XPG gene play a vital role in the outcomes of various cancers, including gastric carcinoma [17, 18], non-small cell lung cancer (NSCLC) [19], breast cancer [20]. However, the association between CRC and XPG polymorphisms remain controversial [21]. Therefore, the aim of our study was to assess the association of four potentially functional SNPs of XPG (rs2094258 C>T, rs2296147 T>C, rs751402 G>A, and rs873601 G>A) with over-all survival (OS) and progression-free survival (PFS) of CRC in 1901 Chinese CRC patients.
RESULTS
Patient characteristics and clinicopathological cutcomes
The demographic and clinicopathological characteristics of 1901 patients were shown in Table 1. Patients aged between 13 to 91 years, with a median age of 57.05 years. Body mass index (BMI) ranged from 13.19–41.36 (median: 22.38) and 60.5% of the patients were males. Most of patients did not have unhealthy lifestyle, such as smoking and drinking, and most of the patients with later Dukes stages. Patients with rectum and colon cancer accounted for 54.2% and 45.8% of CRC, respectively. After surgery and chemotherapy, 96.8% patients showed good response and about 1/3 patients later underwent recurrence or metastasis.
Table 1. Demographic and clinical data of patients.
| Variables | Number | Range/Percentage (%) |
|---|---|---|
| Age (years) 58.00 | 916 (> 58.00) | 13–91 (range) |
| BMI (weight/hight2) 22.23 | 840 (> 22.23) | 13.19–41.36 (range) |
| Patients with PFS | 1516 | 79.7 |
| Patients without PFS | 385 | 20.3 |
| Sex | ||
| Male | 1150 | 60.5 |
| Female | 751 | 39.5 |
| Smoking status | ||
| Never | 1381 | 73.2 |
| Ever | 505 | 26.8 |
| Drinking status | ||
| Never | 1599 | 85.0 |
| Ever | 282 | 15.0 |
| Dukes stage | ||
| A | 207 | 10.9 |
| B | 629 | 33.2 |
| C | 608 | 32.1 |
| D | 449 | 23.7 |
| Tumor site | ||
| Rectum | 977 | 54.2 |
| Colon | 824 | 45.8 |
| Therapeutic response | ||
| Better | 1716 | 96.8 |
| Worse | 57 | 3.2 |
| Recurrence/Metastasis status | ||
| No | 1197 | 63.0 |
| Yes | 704 | 37.0 |
| Therapeutic methods | ||
| Surgery | 1001 | 52.7 |
| Chemotherapy | 708 | 37.2 |
| Radiotherapy | 59 | 3.1 |
BMI, body mass index.
Association between XPG SNPs and prognosis of CRC
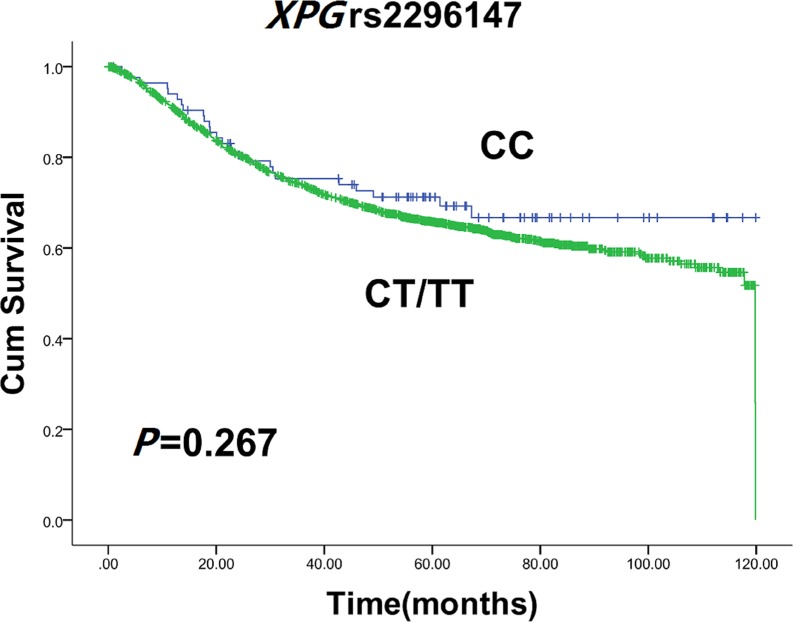
Genotype frequency distributions of four XPG SNPs were summarized in Table 2. The SNPs were analyzed for association with PFS and OS in patients with CRC. A significant association was found between XPG rs2296147 variant genotypes and PFS in CRC. XPG rs2296147 T>C polymorphism led to a decrease in median 10-year PFS time of 88.5 months for carriers of rs2296147 CT/TT genotype, when compared with 118.1 months for patients with CC genotype (log-rank test, P = 0.020) (Figure 1). Univariate Cox proportional hazards model analysis indicated that age, BMI, dukes stage, recurrence/metastasis status, and therapeutic response could influence 10 years OS or PFS (Table 3). After adjusting for those potential confounding factors, multivariate Cox proportional hazards model showed that patients carrying rs2296147 CT/TT genotype had an the hazard ratios (HR) of 1.324 (95% CI = 1.046–1.667) for developing progression in comparison non-carriers. It suggested that XPG rs2296147 CT/TT genotype might be an independent predictor of poor prognosis in CRC (Table 4). No association with PFS was observed for other SNPs (rs2094258, rs751402 and rs873601). Moreover, we failed to find any significant association for the XPG rs2296147 with 10-year OS (Figure 2), as well as the other three polymorphisms.
Table 2. Kaplan-Meier method and Cox proportional hazards model analysis of associations between the genotypes of XPG and CRC prognosis.
| Variants | 10 years OS | 10 years PFS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| event | Median (months) | Log-rank P | HR (95% CI) | P a | event | Median (months) | Log-rank P | HR (95%CI) | P b | |
| rs2094258 | ||||||||||
| TT | 83 | 106.067 | 1.000 | 76 | 87.667 | 1.000 | ||||
| CT | 254 | 21.267 | 0.357 | 0.973 (0.747, 1.268) | 0.842 | 208 | 98.633 | 0.169 | 0.85 (0.655, 1.108) | 0.232 |
| CC | 265 | 119.800 | 0.945 | 0.942 (0.724, 1.223) | 0.649 | 227 | 83.967 | 0.728 | 1.019 (0.785, 1.323) | 0.889 |
| CT/CC | 519 | 119.800 | 0.600 | 0.978 (0.865, 1.106) | 0.722 | 435 | 92.033 | 0.363 | 0.964 (0.854, 1.090) | 0.561 |
| rs2296147 | ||||||||||
| CC | 25 | 20.067 | 1.000 | 18 | 118.067 | 1.000 | ||||
| CT | 200 | 119.800 | 0.392 | 1.381 (0.837, 2.276) | 0.206 | 178 | 84.867 | 0.024 | 1.780 (1.093, 2.898) | 0.021 |
| TT | 377 | 119.867 | 0.258 | 1.492 (0.915, 2.433) | 0.108 | 315 | 89.567 | 0.025 | 1.740 (1.080, 2.803) | 0.023 |
| CT/TT | 577 | 119.800 | 0.267 | 1.205(0.946,1.535) | 0.131 | 493 | 88.500 | 0.020 | 1.324 (1.046, 1.667) | 0.020 |
| rs751402 | ||||||||||
| AA | 83 | 119.867 | 1.000 | 76 | 83.867 | 1.000 | ||||
| AG | 263 | 119.800 | 0.489 | 0.953 (0.733, 1.240) | 0.721 | 216 | 89.133 | 0.384 | 0.851 (0.655, 1.107) | 0.851 |
| GG | 256 | 117.800 | 0.567 | 0.893 (0.688, 1.161) | 0.399 | 219 | 92.033 | 0.521 | 0.830 (0.639, 1.080) | 0.830 |
| AG/GG | 519 | 119.800 | 0.478 | 0.960 (0.850, 1.085) | 0.515 | 435 | 91.233 | 0.414 | 0.917 (0.811, 1.036) | 0.165 |
| rs873601 | ||||||||||
| AA | 155 | 117.800 | 1.000 | 144 | 85.300 | 1.000 | ||||
| AG | 286 | 119.800 | 0.347 | 1.028 (0,831, 1.272) | 0.799 | 227 | 92.900 | 0.119 | 0.856 (0.694, 1.056) | 0.856 |
| GG | 161 | 119.867 | 0.822 | 0.994 (0.780, 1.267) | 0.964 | 140 | 93.267 | 0.502 | 0.969 (0.766, 1.226) | 0.969 |
| AG/GG | 447 | 119.800 | 0.574 | 1.008 (0.912, 1.113) | 0.877 | 367 | 93.267 | 0.162 | 0.946 (0.859, 1.043) | 0.265 |
CRC, colorectal cancer; HR, hazard ratio; CI, confidence interval; OS, overall survival; PFS, progression-free survival.
P values were calculated after adjustment for BMI, dukes stage, recurrence/metastasis status and therapeutic response.
P values were calculated after adjustment for age and dukes stage.
Figure 1. Kaplan-Meier estimates of 10 years PFS with XPG rs2296147 CT/TT and CC genotypes in CRC patients.
Table 3. Cox proportional hazards model analysis of associations between demographic and clinical data of patients and CRC prognosis.
| Variables | P | HR | 95% CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| 10 years OS | ||||
| BMI | 0.001 | 0.956 | 0.932 | 0.982 |
| Therapeutic response | 0.000 | 2.835 | 2.509 | 3.203 |
| Dukes stage | 0.000 | 3.613 | 3.226 | 4.046 |
| Recurrence/Metastasis status | 0.000 | 4.710 | 3.993 | 5.556 |
| 10 years PFS | ||||
| Age | 0.002 | 0.990 | 0.984 | 0.996 |
| Dukes stage | 0.000 | 2.016 | 1.817 | 2.238 |
CRC, colorectal cancer; HR, hazard ratio; CI, confidence interval; OS, overall survival; BMI, body mass index; PFS, progression-free survival.
Table 4. Kruskal-Wallis analysis of associations between the demographic and clinical data of patients and genotypes of XPG.
| Variables | rs2094258 | rs2296147 | rs751402 | rs873601 | ||||
|---|---|---|---|---|---|---|---|---|
| Pa | Pb | Pa | Pb | P1 | P2 | P1 | P2 | |
| Sex | 0.693 | 0.399 | 0.721 | 0.231 | 0.799 | 0.313 | 0.172 | 0.122 |
| Age | 0.107 | 0.026 | 0.666 | 0.544 | 0.121 | 0.054 | 0.063 | 0.106 |
| Smoking status | 0.509 | 0.537 | 0.576 | 0.539 | 0.410 | 0.898 | 0.929 | 0.800 |
| Drinking status | 0.299 | 0.510 | 0.170 | 0.271 | 0.282 | 0.713 | 0.671 | 0.789 |
| BMI | 0.682 | 0.764 | 0.612 | 0.812 | 0.523 | 0.615 | 0.830 | 0.919 |
| Therapeutic response | 0.538 | 0.865 | 0.230 | 0.181 | 0.232 | 0.550 | 0.824 | 0.963 |
| Tumor site | 0.569 | 0.241 | 0.708 | 0.408 | 0.751 | 0.818 | 0.796 | 0.447 |
| Dukes stage | 0.346 | 0.309 | 0.957 | 0.834 | 0.699 | 0.442 | 0.637 | 0.590 |
BMI, body mass index.
P values were calculated for 10 years OS.
P values were calculated for 10 years PFS.
Figure 2. Kaplan-Meier estimates of 10 years OS with XPG rs2296147 CT/TT and CC genotypes in CRC patients.
DISCUSSION
In the present study, we found that XPG rs2296147 CT/TT genotypes were correlated with poor 10-year PFS in CRC when compared with CC genotype. In other words, XPG rs2296147 CC genotype was associated with favorable prognosis regarding 10- year PFS, suggesting that XPG rs2296147 CC homozygous variation is a protective factor for the 10 years PFS in CRC. However, incidence of this homozygous variation is relatively low. There was no significant association between the rest of polymorphisms and clinical outcome of CRC patients.
XPG gene is mapped to chromosome 13q33, encoding an 1186 amino acid structure-specific endonuclease. During the process of NER, XPG is involved in the incision on 5′ side of damage and the maintenance of stability of TFIIH. Up to now, although an increasing number of studies focus on the common genetic variations in the NER pathway, studies on the association of XPG SNPs and the clinical outcome of CRC patients were limited. To the best of our knowledge, our study was the first large-scale case study to investigate the relationship between XPG polymorphisms of and prognosis of CRC. The XPG polymorphisms have been reported to affect the platinum-based chemotherapy sensitivity and prognosis of various cancers, including gastric cancer [18] and NSCLC [22]. Zhou et al. [19] found XPG rs2296147 and rs2094258 polymorphisms were associated with PFS and OS in NSCLC. Zhang et al. [23] also reported that down-regulation of XPG activity caused by rs2296147 polymorphism was correlated with increased OS. There are a number of studies evaluating the influence of the XPG SNPs on the risk and therapeutic response of CRC [15, 24–26]; however few have explored the association of XPG SNPs with the prognosis of CRC patients. CRC is one of the most common cancer worldwide, with approximately 55% of the cases occurring in the developed regions. So far, very few CRC-related studies involve Chinese populations [27, 28]. The current study might be the largest one that investigated the association of interest solely in Chinese by far. The association of polymorphisms in the NER pathway with CRC remains inconclusive. Moreno et al. [14] carried out a case-control study to assess gene-environment interactions by genotyping 28 SNPs in the 15 DNA repair genes among 377 CRC patients and 329 controls. Their results highlighted the important influence of SNPs in the DNA repair genes on the response to chemotherapy and prognosis of CRC patients. Meanwhile, Du et al. [25] found that XPG Asp1104His polymorphism was associated with a significantly increased risk of CRC, especially in Asian populations. However, Mort et al. [29] investigated polymorphisms in the NER genes (XPD, XPF, XPG, ERCC1) and failed to prove the important role of studied SNPs in protection against CRC. Zhu et al. [30] performed a meta-analysis to explore the relationship between the ERCC5/XPG Asp1104His polymorphism and cancer risk under the recessive genetic model, and found null association between the polymorphism and the risk of CRC. In contrast, our large-scale study provided evidence of the robust association between the XPG SNPs and the prognosis of CRC patients.
The XPG rs2296147 polymorphism is located in the 5′ untranslated regions (UTR), which was predicted to influence activity of transcription factor binding sites (TFBS) [31]. Cartharius et al. [32] found that XPG rs2296147 is a putative P53 transcription factor-binding site. Blomquist et al. [33] found that XPG rs2296147 is associated with altered allele-specific expression of XPG transcript in normal human bronchial epithelium, so the rs2296147 polymorphism may be important to XPG expression. Our study is the first one to validate the association between XPG rs2296147 polymorphism and risk of CRC, previous studies focus on how this site act on the clinical outcome of platinum-based chemotherapy in NSCLC patients [19, 22, 23, 34, 35], susceptibility of prostate cancer patients [36, 37], gastric cancer [38, 39], and breast cancer [20], but the number was limited. Our study is the first one to clarify the relationship between rs2296147 polymorphism and CRC survival.
In conclusion, our results indicated that XPG rs2296147 CT/TT was correlated with the prognosis of CRC patients. XPG rs2296147 polymorphism could be used as an independent predictive marker for the prognosis in CRC. Our study identified prognostic value of XPG SNPs. It might also serve as a molecular biomarker in individualized treatmen of colorectal cancer in future.
MATERIALS AND METHODS
Subject
A total of 1901 patients with histologically confirmed colorectal carcinoma were recruited from Sun Yat-Sen University Cancer Center between January 2000 to May 2010. We recruited patients without restrictions on age, sex, ethnicity, or clinical stage. We gained demographic and clinical data from medical record review, including age, sex, smoking status, drinking status, tumor site, therapeutic response, recurrence/metastasis status, and Dukes stage. Individuals who smoked cigarettes less than 1 package were defined as never smokers, while the others were ever smokers. An individual who drank alcohol less than 50 ml was defined as a never drinker, while the others were ever drinkers. All the patients were followed-up every year by telephone and deaths were recorded and confirmed by local Public Security Bureau until April 2015. All patients with informed consent donated their blood sample for the study.
SNP selected and genotyping
Genomic DNA was obtained from the buffy coat fraction of each blood sample using a Qiagen Blood DNA Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions. The candidate XPG SNPs were selected as reported previously [40]. Finally, four SNPs (rs2296147, rs2094258, rs751402, rs873601) in the XPG gene were chosen and included in the analysis. All these four selected SNPs were genotyped by using TaqMan real-time PCR as described previously [40]. For quality control, approximately 10% of the samples were randomly selected and repeatedly genotyped, and the results confirmed 100% concordance.
Statistical methods
OS was defined as the time from the date of pathologically confirmed to the date of death or last clinical follow-up. PFS was calculated from the date of the pathologically confirmed to the progression of the disease, death without progression, or last clinical follow-up. Numerical variable in this study were expressed as mean and percentage. Survival distributions were estimated by using the Kaplan-Meier method and difference in the survival was tested using the log-rank test. To estimate the association of the four XPG SNPs with PFS and OS in CRC, the HR and 95% CI were calculated by univariate Cox proportional hazards model. Multivariate Cox model were performed to compute adjusted HR and 95% CI, after adjusting for potential risk factors. Homozygous variant genotype severed as a reference group. All tests were two-sided and P < 0.05 was considered to be significant. All statistics were conducted by SPSS 19.0 software.
ACKNOWLEDGMENTS AND FUNDING
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81502046), Special Financial Grant from the China Postdoctoral Science Foundation (Grant No. 2014T70836), the Natural Science Foundation of Guangdong Province (Grant No. 2015A030310324), and the National Science Fund for Distinguished Young Scholars (Grant No. 81325018).
Footnotes
Authors' contributions
All authors contributed significantly to this work. F.W., S.-D. Z., H.-M. X., X.-Z. L., W.-Q. X., and X.-Z. L. performed the research study and collected the data; F. W., R.-X. H. and T.-M. W. analyzed the data; J. H. and W.-H. J. designed the research study; F.W., J.-H. Z., J. H. and W.-H. J. wrote the paper, and F.W. and B.-H. X. prepared Figure 1 and Tables 1–3. All authors reviewed the manuscript. In addition, all authors approved the final draft.
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Stintzing S. Management of colorectal cancer. F1000Prime Rep. 2014;6:108. doi: 10.12703/P6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.Azeem S, Gillani SW, Siddiqui A, Jandrajupalli SB, Poh V, Syed Sulaiman SA. Diet and Colorectal Cancer Risk in Asia—a Systematic Review. Asian Pac J Cancer Prev. 2015;16:5389–5396. doi: 10.7314/apjcp.2015.16.13.5389. [DOI] [PubMed] [Google Scholar]
- 5.Hannan LM, Jacobs EJ, Thun MJ. The association between cigarette smoking and risk of colorectal cancer in a large prospective cohort from the United States. Cancer Epidemiol Biomarkers Prev. 2009;18:3362–3367. doi: 10.1158/1055-9965.EPI-09-0661. [DOI] [PubMed] [Google Scholar]
- 6.Cho S, Shin A, Park SK, Shin HR, Chang SH, Yoo KY. Alcohol Drinking, Cigarette Smoking and Risk of Colorectal Cancer in the Korean Multi-center Cancer Cohort. J Cancer Prev. 2015;20:147–152. doi: 10.15430/JCP.2015.20.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Yang Y, Wang F, Zhang P, Shi C, Zou Y, Qin H. Obesity and risk of colorectal cancer: a systematic review of prospective studies. PLoS One. 2013;8:e53916. doi: 10.1371/journal.pone.0053916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong Y, Liu JW, Gao YJ, Zhou T, Chen YM. Relationship between DNA repair gene XPD751 single-nucleotide polymorphisms and prognosis of colorectal cancer. Genet Mol Res. 2015;14:5390–5398. doi: 10.4238/2015.May.22.8. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Deng X, Wang Z, Wang Q, Liu Y. Genetic polymorphisms of DNA repair genes XRCC1 and XRCC3 and risk of colorectal cancer in Chinese population. Asian Pac J Cancer Prev. 2012;13:665–669. doi: 10.7314/apjcp.2012.13.2.665. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002;511:145–178. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 12.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 13.Scharer OD. XPG: its products and biological roles. Adv Exp Med Biol. 2008;637:83–92. doi: 10.1007/978-0-387-09599-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, Gonzalez S, Guino E, Capella G, Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 15.Monzo M, Moreno I, Navarro A, Ibeas R, Artells R, Gel B, Martinez F, Moreno J, Hernandez R, Navarro-Vigo M. Single nucleotide polymorphisms in nucleotide excision repair genes XPA, XPD, XPG and ERCC1 in advanced colorectal cancer patients treated with first-line oxaliplatin/fluoropyrimidine. Oncology. 2007;72:364–370. doi: 10.1159/000113534. [DOI] [PubMed] [Google Scholar]
- 16.Emmert S, Schneider TD, Khan SG, Kraemer KH. The human XPG gene: gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:1443–1452. doi: 10.1093/nar/29.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng N, Liu JW, Sun LP, Xu Q, Duan ZP, Dong NN, Yuan Y. Expression of XPG protein in the development, progression and prognosis of gastric cancer. PloS one. 2014;9:e108704. doi: 10.1371/journal.pone.0108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue MH, Li GY, Wu XJ, Zhang CX, Zhang CF, Zhu KX. Genetic variability of genes in NER pathway influences the treatment outcome of gastric cancer. Int J Clin Exp Pathol. 2015;8:5563–5569. [PMC free article] [PubMed] [Google Scholar]
- 19.Zou HZ, Zhao YQ. XPG polymorphisms are associated with prognosis of advanced non-small cell lung cancer treated with platinum-based doublet chemotherapy. Genet Mol Res. 2015;14:500–506. doi: 10.4238/2015.January.26.3. [DOI] [PubMed] [Google Scholar]
- 20.Na N, Dun E, Ren L, Li G. Association between ERCC5 gene polymorphisms and breast cancer risk. Int J Clin Exp Pathol. 2015;8:3192–3197. [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Deng J, Xiong Y, Wang S, Xiong W. Genetic association between ERCC5 rs17655 polymorphism and lung cancer risk: evidence based on a meta-analysis. Tumour Biol. 2014;35:5613–5618. doi: 10.1007/s13277-014-1742-2. [DOI] [PubMed] [Google Scholar]
- 22.Yuli Y, Zhe S, Xia W, Siqing L, Zhenxuan W, Yu-Hua Z, Bing S, Jun-Wei C. XPG is a novel biomarker of clinical outcome in advanced non-small-cell lung cancer. Pak J Med Sci. 2013;29:762–767. doi: 10.12669/pjms.293.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T, Sun J, Lv M, Zhang L, Wang X, Ren JC, Wang B. XPG is predictive gene of clinical outcome in advanced non-small-cell lung cancer with platinum drug therapy. Asian Pac J Cancer Prev. 2013;14:701–705. doi: 10.7314/apjcp.2013.14.2.701. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Wu HZ, Zhang YN, Kang H, Sun MJ, Wang EH, Yang XL, Lian MQ, Yu ZJ, Zhao L, Olopade OI, Wei MJ. DNA repair genes XPC, XPG polymorphisms: relation to the risk of colorectal carcinoma and therapeutic outcome with Oxaliplatin-based adjuvant chemotherapy. Mol Carcinog. 2012;51(Suppl 1):E83–93. doi: 10.1002/mc.21862. [DOI] [PubMed] [Google Scholar]
- 25.Du H, Zhang X, Du M, Guo N, Chen Z, Shu Y, Zhang Z, Wang M, Zhu L. Association study between XPG Asp1104His polymorphism and colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:6700. doi: 10.1038/srep06700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun K, Gong A, Liang P. Predictive impact of genetic polymorphisms in DNA repair genes on susceptibility and therapeutic outcomes to colorectal cancer patients. Tumour Biol. 2015;36:1549–1559. doi: 10.1007/s13277-014-2721-3. [DOI] [PubMed] [Google Scholar]
- 27.Negandhi AA, Hyde A, Dicks E, Pollett W, Younghusband BH, Parfrey P, Green RC, Savas S. MTHFR Glu429Ala and ERCC5 His46His polymorphisms are associated with prognosis in colorectal cancer patients: analysis of two independent cohorts from Newfoundland. PLoS One. 2013;8:e61469. doi: 10.1371/journal.pone.0061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paszkowska-Szczur K, Scott RJ, Gorski B, Cybulski C, Kurzawski G, Dymerska D, Gupta S, van de Wetering T, Masojc B, Kashyap A, Gapska P, Gromowski T, Kladny J, et al. Polymorphisms in nucleotide excision repair genes and susceptibility to colorectal cancer in the Polish population. Mol Biol Rep. 2015;42:755–764. doi: 10.1007/s11033-014-3824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mort R, Mo L, McEwan C, Melton DW. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. Br J Cancer. 2003;89:333–337. doi: 10.1038/sj.bjc.6601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu ML, Wang M, Cao ZG, He J, Shi TY, Xia KQ, Qiu LX, Wei QY. Association between the ERCC5 Asp1104His polymorphism and cancer risk: a meta-analysis. PLoS One. 2012;7:e36293. doi: 10.1371/journal.pone.0036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohrenweiser H. Survey of polymorphic sequence variation in the immediate 5′ region of human DNA repair genes. Mutat Res. 2007;616:221–226. doi: 10.1016/j.mrfmmm.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 33.Blomquist TM, Crawford EL, Willey JC. Cis-acting genetic variation at an E2F1/YY1 response site and putative p53 site is associated with altered allele-specific expression of ERCC5 (XPG) transcript in normal human bronchial epithelium. Carcinogenesis. 2010;31:1242–1250. doi: 10.1093/carcin/bgq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He C, Duan Z, Li P, Xu Q, Yuan Y. Role of ERCC5 promoter polymorphisms in response to platinum-based chemotherapy in patients with advanced non-small-cell lung cancer. Anticancer Drugs. 2013;24:300–305. doi: 10.1097/CAD.0b013e32835bd6ce. [DOI] [PubMed] [Google Scholar]
- 35.Hu W, Pan J, Zhao P, Yang G, Yang S. Genetic polymorphisms in XPG could predict clinical outcome of platinum-based chemotherapy for advanced non-small cell lung cancer. Tumour Biol. 2014;35:5561–5567. doi: 10.1007/s13277-014-1732-4. [DOI] [PubMed] [Google Scholar]
- 36.Yang B, Chen WH, Wen XF, Liu H, Liu F. Role of DNA repair-related gene polymorphisms in susceptibility to risk of prostate cancer. Asian Pac J Cancer Prev. 2013;14:5839–5842. doi: 10.7314/apjcp.2013.14.10.5839. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XJ, Liu P, Zhu F. Polymorphisms of DNA repair-related genes with susceptibility and prognosis of prostate cancer. Genet Mol Res. 2014;13:4419–4424. doi: 10.4238/2014.January.24.20. [DOI] [PubMed] [Google Scholar]
- 38.Duan Z, He C, Gong Y, Li P, Xu Q, Sun LP, Wang Z, Xing C, Yuan Y. Promoter polymorphisms in DNA repair gene ERCC5 and susceptibility to gastric cancer in Chinese. Gene. 2012;511:274–279. doi: 10.1016/j.gene.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 39.Yang WG, Zhang SF, Chen JW, Li L, Wang WP, Zhang XF. SNPs of excision repair cross complementing group 5 and gastric cancer risk in Chinese populations. Asian Pac J Cancer Prev. 2012;13:6269–6272. doi: 10.7314/apjcp.2012.13.12.6269. [DOI] [PubMed] [Google Scholar]
- 40.He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP, Wang YN, Sun MH, Zhou XY, Yang YJ, Wang JC, Jin L, Wei QY, et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235–1244. doi: 10.1007/s00439-012-1152-8. [DOI] [PubMed] [Google Scholar]