Abstract
Many questions about human genetic history can be addressed by examining the patterns of shared genetic variation between sets of populations. A useful methodological framework for this purpose is F-statistics that measure shared genetic drift between sets of two, three, and four populations and can be used to test simple and complex hypotheses about admixture between populations. This article provides context from phylogenetic and population genetic theory. I review how F-statistics can be interpreted as branch lengths or paths and derive new interpretations, using coalescent theory. I further show that the admixture tests can be interpreted as testing general properties of phylogenies, allowing extension of some ideas applications to arbitrary phylogenetic trees. The new results are used to investigate the behavior of the statistics under different models of population structure and show how population substructure complicates inference. The results lead to simplified estimators in many cases, and I recommend to replace F3 with the average number of pairwise differences for estimating population divergence.
Keywords: admixture, gene flow, phylogenetics, population genetics, phylogenetic network
FOR humans, whole-genome genotype data are now available for individuals from hundreds of populations (Lazaridis et al. 2014; Yunusbayev et al. 2015), opening up the possibility to ask more detailed and complex questions about our history (Pickrell and Reich 2014; Schraiber and Akey 2015) and stimulating the development of new tools for the analysis of the joint history of these populations (Reich et al. 2009; Patterson et al. 2012; Pickrell and Pritchard 2012; Lipson et al. 2013; Ralph and Coop 2013; Hellenthal et al. A simple and intuitive approach that has quickly gained in popularity are the F-statistics, introduced by Reich et al. (2009) and summarized in Patterson et al. (2012). In that framework, inference is based on “shared genetic drift” between sets of populations, under the premise that shared drift implies a shared evolutionary history. Tools based on this framework have quickly become widely used in the study of human genetic history, both for ancient and for modern DNA (Green et al. 2010; Reich et al. 2012; Lazaridis et al. 2014; Allentoft et al. 2015; Haak et al. 2015).
Some care is required with terminology, as the F-statistics sensu Reich et al. (2009) are distinct, but closely related to Wright’s fixation indexes (Wright 1931; Reich et al. 2009), which are also often referred to as F-statistics. Furthermore, it is necessary to distinguish between statistics (quantities calculated from data) and the underlying parameters (which are part of the model) (Weir and Cockerham 1984).
In this article, I mostly discuss model parameters, and I therefore refer to them as drift indexes. The term F-statistics is used when referring to the general framework introduced by Reich et al. (2009), and Wright’s statistics are referred to as FST or f.
Most applications of the F-statistic framework can be phrased in terms of the following six questions:
Treeness test: Are populations related in a tree-like fashion (Reich et al. 2009)?
Admixture test: Is a particular population descended from multiple ancestral populations (Reich et al. 2009)?
Admixture proportions: What are the contributions from different populations to a focal population (Green et al. 2010; Haak et al. 2015)?
Number of founders: How many founder populations are there for a certain region (Reich et al. 2012; Lazaridis et al. 2014)?
Complex demography: How can mixtures and splits of population explain demography (Patterson et al. 2012; Lipson et al. 2013)?
Closest relative: What is the closest relative to a contemporary or ancient population (Raghavan et al. 2014)?
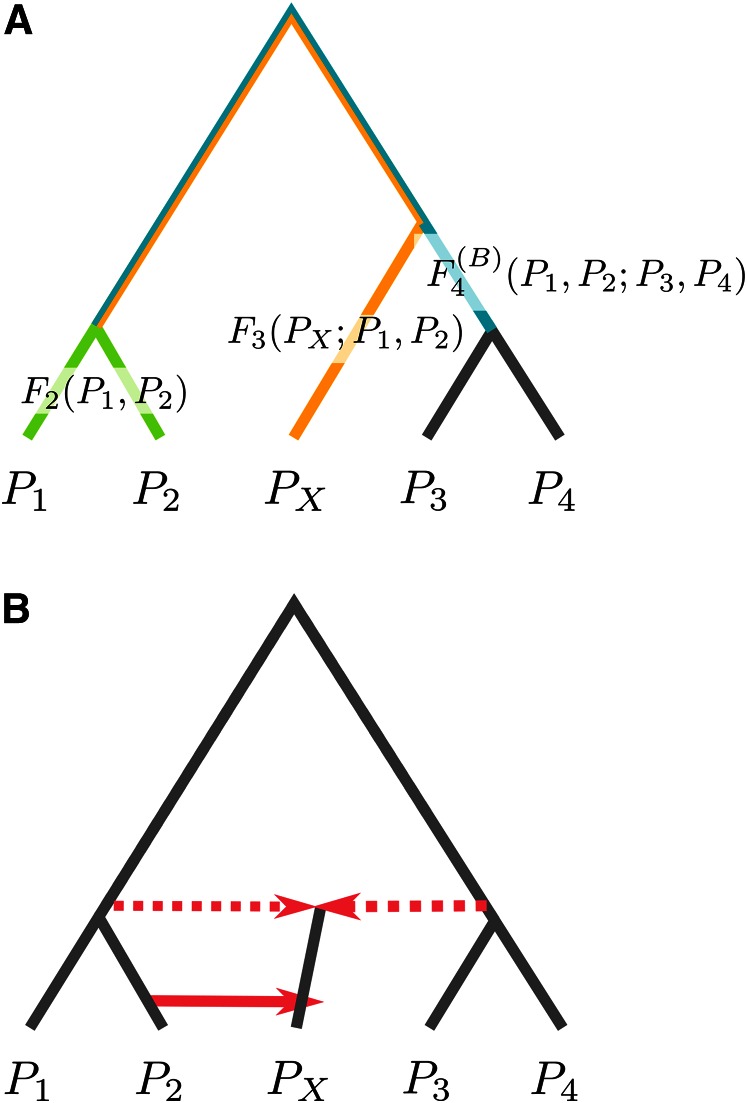
The demographic models under which these questions are addressed, and that motivated the drift indexes, are called population phylogenies and admixture graphs. The population phylogeny (or population tree) is a model where populations are related in a tree-like fashion (Figure 1A), and it frequently serves as the null model for admixture tests. The branch lengths in the population phylogeny correspond to how much genetic drift occurred, so that a branch that is subtended by two different populations can be interpreted as the “shared” genetic drift between these populations. The alternative model is an admixture graph (Figure 1B), which extends the population phylogeny by allowing edges that represent population mergers or a significant exchange of migrants.
Figure 1.
(A) A population phylogeny with branches corresponding to F2 (green), F3 (yellow), and (blue). (B) An admixture graph extends a population phylogeny by allowing gene flow (red, solid line) and admixture events (red, dotted line).
Under a population phylogeny, the three F-statistics proposed by Reich et al. (2009), labeled F2, F3, and F4, have interpretations as branch lengths (Figure 1A) between two, three, and four taxa, respectively. Assume populations are labeled as P1, P2, …. Then
F2(P1, P2) corresponds to the path on the phylogeny from P1 to P2.
F3 (PX; P1, P2) represents the length of the external branch from PX to the (unique) internal vertex connecting all three populations. Thus, the first parameter of F3 has a unique role, whereas the other two can be switched arbitrarily.
(P1, P2; P3, P4) represents the internal branch from the internal vertex connecting P1 and P2 to the vertex connecting P3 and P4 (Figure 1A, blue).
If the arguments are permuted, some F-statistics will have no corresponding internal branch. In particular, it can be shown that in a population phylogeny, one F4 index will be zero, implying that the corresponding internal branch is missing. This is the property that is used in the admixture test. For clarity, I add the superscript if I need to emphasize the interpretation of F4 as a branch length and to emphasize the interpretation as a test statistic. For details, see the F4 subsection in Methods and Results.
In an admixture graph, there is no longer a single branch length corresponding to each F-statistic, and interpretations are more complex. However, F-statistics can still be thought of as the proportion of genetic drift shared between populations (Reich et al. 2009). The basic idea exploited in addressing all six questions outlined above is that under a tree model, branch lengths, and thus the drift indexes, must satisfy some constraints (Buneman 1971; Semple and Steel 2003; Reich et al. 2009). The two most relevant constraints are that (i) in a tree, all branches have positive lengths (tested using the F3-admixture test) and (ii) in a tree with four leaves, there is at most one internal branch (tested using the F4-admixture test).
The goal of this article is to give a broad overview on the theory, ideas, and applications of F-statistics. Our starting point is a brief review on how genetic drift is quantified in general and how it is measured using F2. I then propose an alternative definition of F2 that allows us to simplify applications and study them under a wide range of population structure models. I then review some basic properties of distance-based phylogenetic trees, show how the admixture tests are interpreted in this context, and evaluate their behavior. Many of the results that are highlighted here are implicit in classical (Wahlund 1928; Wright 1931; Cavalli-Sforza and Edwards 1967; Felsenstein 1973, 1981; Cavalli-Sforza and Piazza 1975; Slatkin 1991; Excoffier et al. 1992) and more recent work (Patterson et al. 2012; Pickrell and Pritchard 2012; Lipson et al. 2013), but often not explicitly stated or given in a different context.
Methods and Results
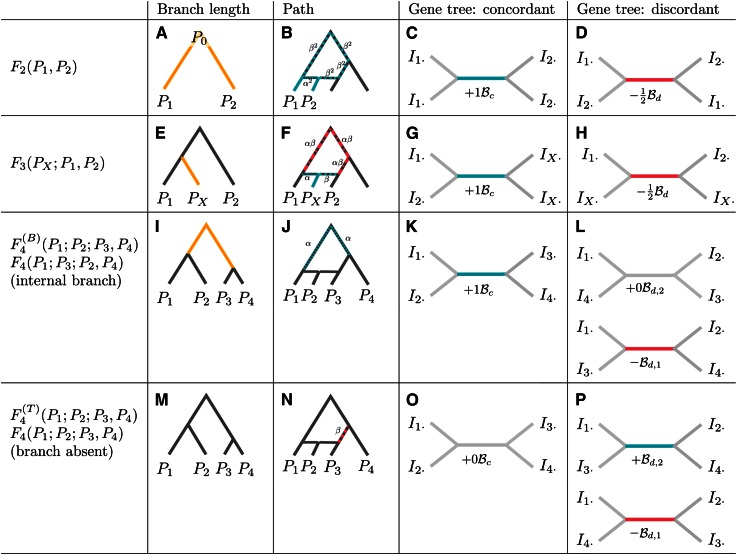
The next sections discuss the F-statistics, introducing different interpretations and giving derivations for some useful expressions. A graphical summary of the three interpretations of the statistics is given in Figure 2, and the main formulas are summarized in Table 1.
Figure 2.
Interpretation of F-statistics. F-statistics can be interpreted as branch lengths in a population phylogeny (A, E, I, and M), as the overlap of paths in an admixture graph (B, F, J, and N, see also Figure S1), and in terms of the internal branches of gene genealogies (see Figure 4, Figure S2, and Figure S3). For gene trees consistent with the population tree, the internal branch contributes positively (C, G, and K), and for discordant branches, internal branches contribute negatively (D and H) or zero (L). F4 has two possible interpretations; depending on how the arguments are permuted relative to the tree topology, it may reflect either the length of the internal branch [I–L, ] or a test statistic that is zero under a population phylogeny [M–P, ]. For the admixture test, the two possible gene trees contribute to the statistic with different sign, highlighting the similarity to the D-statistic (Green et al. 2010) and its expectation of zero in a symmetric model.
Table 1. Summary of equations.
| Drift Measure | F2 (P1, P2) | F3 (PX; P1, P2) | F4(P1, P2, P3, P4) |
|---|---|---|---|
| Definition | |||
| F2 | — | ||
| Coalescence times | |||
| Variance | |||
| Branch length |
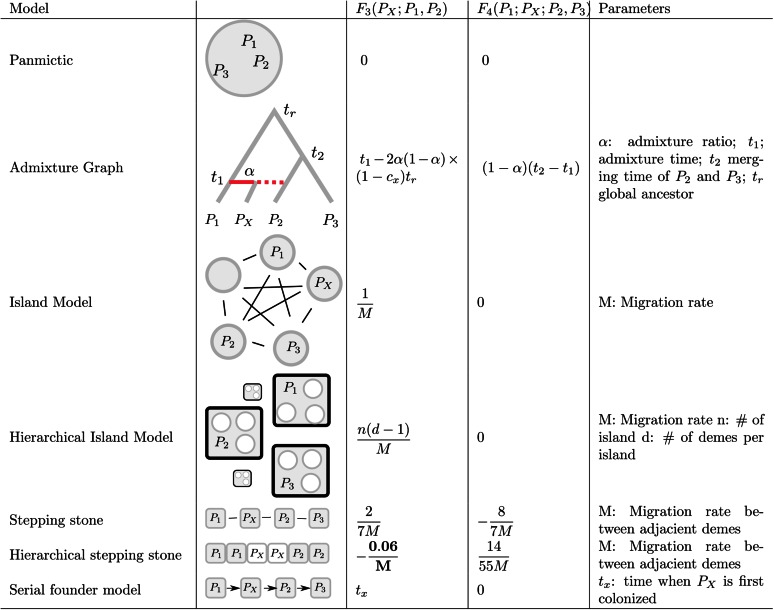
A constant of proportionality is omitted for coalescence times and branch lengths. Derivations for F2 are given in the main text, and F3 and F4 are a simple result of combining Equation 16 with Equations 20b and 24b. and correspond to the average length of the internal branch in a gene genealogy concordant and discordant with the population assignment, respectively (see Gene tree branch lengths section).
Throughout this article, populations are labeled as P1, P2, … , Pi, … . Often, PX will denote an admixed population. The allele frequency pi is defined as the proportion of individuals in Pi that carry a particular allele at a biallelic locus, and throughout this article I assume that all individuals are haploid. However, all results hold if instead of haploid individuals, an arbitrary allele of a diploid individual is used. I focus on genetic drift only and ignore the effects of mutation, selection, and other evolutionary forces.
Measuring genetic drift—F2
The purpose of F2 is simply to measure how much genetic drift occurred between two populations, i.e., to measure genetic dissimilarity. For populations P1 and P2, F2 is defined as
| (1) |
(Reich et al. 2009). The expectation is with respect to the evolutionary process, but in practice F2 is estimated from hundreds of thousands of loci across the genome (Patterson et al. 2012), which are assumed to be nonindependent replicates of the evolutionary history of the populations.
Why is F2 a useful measure of genetic drift? As it is infeasible to observe changes in allele frequency directly, the effect of drift is assessed indirectly, through its impact on genetic diversity. Most commonly, genetic drift is quantified in terms of (i) the variance in allele frequency, (ii) heterozygosity, (iii) probability of identity by descent, (iv) correlation (or covariance) between individuals, and (v) the probability of coalescence (two lineages having a common ancestor). In the next sections I show how F2 relates to these quantities in the cases of a single population changing through time and a pair of populations that are partially isolated.
Single population:
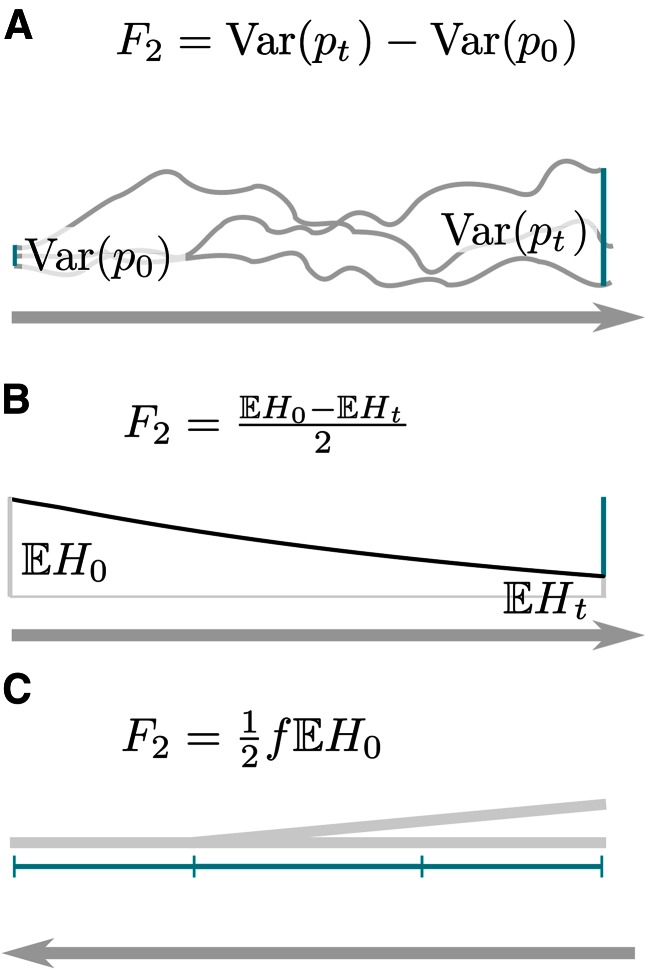
I assume a single population, measured at two time points (t0 and t), and label the two samples P0 and Pt. Then F2 (P0, Pt) can be interpreted in terms of the variances of allele frequencies:
| (2) |
Here, I used to obtain lines three and five. It is worth noting that this result holds for any model of genetic drift where the expected allele frequency is the current allele frequency and increments are independent. For example, this interpretation of F2 holds also if genetic drift is modeled as a Brownian motion (Cavalli-Sforza and Edwards 1967).
An elegant way to introduce the use of F2 in terms of expected heterozygosities Ht (Figure 3B) and identity by descent (Figure 3C) is the duality
| (3) |
This equation is due to Tavaré (1984), who also provided the following intuition: Given nt individuals are sampled at time t, let E denote the event that all individuals carry allele x, conditional on allele x having frequency p0 at time t0. There are two components to this: First, the frequency will change between t0 and t, and then all nt sampled individuals need to carry x.
Figure 3.
Measures of genetic drift in a single population. Shown are interpretations of F2 in terms of (A) the increase in allele frequency variance; (B) the decrease in heterozygosity; and (C) f, which can be interpreted as probability of coalescence of two lineages or the probability that they are identical by descent.
In a diffusion framework,
| (4) |
On the other hand, one may argue using the coalescent: For E to occur, all nt samples need to carry the x allele. At time t0, they had n0 ancestral lineages, who all carry x with probability p0. Therefore,
| (5) |
Equating (4) and (5) yields Equation 3.
In the present case, the only relevant cases are nt = 1,2, since
where f is the probability that two lineage sampled at time t coalesce before time t0.
This yields an expression for F2 by conditioning on the allele frequency p0,
where H0 = 2p0 (1 – p0) is the heterozygosity. Integrating over yields
| (6) |
and it can be seen that F2 increases as a function of f (Figure 3C). This equation can also be interpreted in terms of probabilities of identity by descent: f is the probability that two individuals are identical by descent in Pt given their ancestors were not identical by descent in P0 (Wright 1931), and is the probability two individuals are not identical in P0.
Furthermore, (equation 3.4 in Wakeley 2009) and therefore
| (7) |
which shows that F2 measures the decay of heterozygosity (Figure 3A). A similar argument was used by Lipson et al. (2013) to estimate ancestral heterozygosities and to linearize F2.
These equations can be rearranged to make the connection between other measures of genetic drift and F2 more explicit:
| (8a) |
| (8b) |
| (8c) |
Pairs of populations:
Equations 8b and 8c describing the decay of heterozygosity are–of course–well known by population geneticists, having been established by Wright (1931). In structured populations, very similar relationships exist when the number of heterozygotes expected from the overall allele frequency, is compared with the number of heterozygotes present due to differences in allele frequencies between populations Hexp (Wahlund 1928; Wright 1931).
In fact, already Wahlund showed by considering the genotypes of all possible matings in two subpopulations (table 3 in Wahlund 1928) that for a population made of two subpopulations with equal proportions, the proportion of heterozygotes is reduced by
from which it follows that
| (9) |
Furthermore, but and therefore
| (10) |
Finally, the original definition of F2 was as the numerator of FST (Reich et al. 2009), but FST can be written as from which follows
| (11) |
Covariance interpretation:
To see how F2 can be interpreted as a covariance, define Xi and Xj as indicator variables that two individuals from the same population sample have the A allele, which has frequency p1 in one and p2 in the other population. If individuals are equally likely to be sampled from either population,
Justification for F2:
The preceding arguments show how the usage of F2 for both single and structured populations can be justified by the similar effects of F2 on different measures of genetic drift. However, what is the benefit of using F2 instead of the established inbreeding coefficient f and fixation index FST? Recall that Wright motivated f and FST as correlation coefficients between alleles (Wright 1921, 1931). Correlation coefficients have the advantage that they are easy to interpret, as, e.g., FST = 0 implies panmixia and FST = 1 implies complete divergence between subpopulations. In contrast, F2 depends on allele frequencies and is highest for intermediate-frequency alleles. However, F2 has an interpretation as a covariance, making it simpler and mathematically more convenient to work with. In particular, variances and covariances are frequently partitioned into components due to different effects, using techniques such as analysis of variance and analysis of covariance (e.g., Excoffier et al. 1992).
F2 as branch length:
Reich et al. (2009) and Patterson et al. (2012) proposed to partition “drift” (as previously established, measured by covariance, allele frequency variance, or decrease in heterozygosity) between different populations into contribution on the different branches of a population phylogeny. This model has been studied by Cavalli-Sforza and Edwards (1967) and Felsenstein (1973) in the context of a Brownian motion process. In this model, drift on independent branches is assumed to be independent, meaning that the variances can simply be added. This is what is referred to as the additivity property of F2 (Patterson et al. 2012).
To illustrate the additivity property, consider two populations P1 and P2 that split recently from a common ancestral population P0 (Figure 2A). In this case, p1 and p2 are assumed to be independent conditional on p0, and therefore Then, using (2) and (10),
Alternative proofs of this statement and more detailed reasoning behind the additivity assumption can be found in Cavalli-Sforza and Edwards (1967), Felsenstein (1973), Reich et al. (2009), and Patterson et al. (2012).
Lineages are not independent in an admixture graph, and so this approach cannot be used. Reich et al. (2009) approached this by conditioning on the possible population trees that are consistent with an admixture scenario. In particular, they proposed a framework of counting the possible paths through the graph (Reich et al. 2009; Patterson et al. 2012). An example of this representation for F2 in a simple admixture graph is given in Supplemental Material, Figure S1, with the result summarized in Figure 2B. Detailed motivation behind this visualization approach is given in Appendix 2 of Patterson et al. (2012). In brief, the reasoning is as follows: Recall that and interpret the two terms in parentheses as two paths between P1 and P2, and F2 as the overlap of these two paths. In a population phylogeny, there is only one possible path, and the two paths are always the same; therefore F2 is the sum of the length of all the branches connecting the two populations. However, if there is admixture, as in Figure 2B, both paths choose independently which admixture edge they follow. With probability α they will go left, and with probability β = 1 − α they go right. Thus, F2 can be interpreted by enumerating all possible choices for the two paths, resulting in three possible combinations of paths on the trees (Figure S1), and the branches included will differ, depending on which path is chosen, so that the final F2 is made of an average of the path overlap in the topologies, weighted by the probabilities of the topologies.
However, one drawback of this approach is that it scales quadratically with the number of admixture events, making calculations cumbersome when the number of admixture events is large. More importantly, this approach is restricted to panmictic subpopulations and cannot be used when the population model cannot be represented as a weighted average of trees.
Gene tree Interpretation:
For this reason, I propose to redefine F2, using coalescent theory (Wakeley 2009). Instead of allele frequencies on a fixed admixture graph, coalescent theory tracks the ancestors of a sample of individuals, tracing their history back to their most recent common ancestor. The resulting tree is called a gene tree (or coalescent tree). Gene trees vary between loci and will often have a different topology from that of the population phylogeny, but they are nevertheless highly informative about a population’s history. Moreover, expected coalescence times and expected branch lengths are easily calculated under a wide array of neutral demographic models.
In a seminal article, Slatkin (1991) showed how FST can be interpreted in terms of the expected coalescence times of gene trees,
where and are the expected coalescence times of two lineages sampled in two different populations and the same population, respectively.
Unsurprisingly, given the close relationship between F2 and FST, an analogous expression exists for F2 (P1, P2): The derivation starts by considering F2 for two samples of size 1. I then express F2 for arbitrary sample sizes in terms of individual-level F2 and obtain a sample-size independent expression by letting the sample size n go to infinity.
In this framework, I assume that mutation is rare such that there is at most one mutation at any locus. In a sample of size 2, let be an indicator random variable that individual i has a particular allele. For two individuals, F2 (I1, I2) = 1 implies I1 = I2, whereas F2 (I1, I2) = 0 implies I1 ≠ I2. Thus, F2(I1, I2) is another indicator random variable with the parameter equal to the probability that a mutation happened on the tree branch between I1 and I2.
Now, instead of a single individual I1, consider a sample of n1 individuals: The sample allele frequency is And the sample F2 is
The first three terms can be grouped into n1 terms of the form F2 (I1,i, I2), and the last two terms can be grouped into terms of the form F2 (I1,i, I1,j), one for each possible pair of samples in P1.
Therefore,
| (12) |
where the second sum is over all pairs in P1. This equation is equivalent to equation 22 in Felsenstein (1973).
As I can switch the labels and obtain the same expression for a second population Taking the average over all I2,j yields
| (13) |
Thus, I can write F2 between the two populations as the average number of differences between individuals from different populations, minus some terms including differences within each sample.
Equation 13 is quite general, making no assumptions on where samples are placed on a tree. In a coalescence framework, it is useful to make the assumptions that all individuals from the same population have the same branch length distribution; i.e., for all pairs of samples (x1, x2) and (y1, y2) from populations Pi and Pj. Second, I assume that all samples correspond to the leaves of the tree, so that I can estimate branch lengths in terms of the time to a common ancestor Tij. Finally, I assume that mutations occur at a constant rate of on each branch. Taken together, these assumptions imply that for all individuals from populations Pi, Pj. this simplifies (13) to
| (14) |
which, for the cases of n = 1, 2, was also derived by Petkova et al. (2014). In some applications, F2 might be calculated only for segregating sites in a large sample. As the expected number of segregating sites is (with Ttot denoting the total tree length), taking the limit where is meaningful (Slatkin 1991; Petkova et al. 2014):
| (15) |
In either of these equations, or θ acts as a constant of proportionality that is the same for all statistics calculated from the same data. Since interest is focused on the relative magnitude of F2 or whether a sum of F2 values is different from zero, this constant has no impact on inference.
Furthermore, a population-level quantity is obtained by taking the limit when the numbers of individuals n1 and n2 go to infinity:
| (16) |
Unlike FST, the mutation parameter θ does not cancel. However, for most applications, the absolute magnitude of F2 is of little interest, since only the sign of the statistics is used for most tests. In other applications F-statistics with presumably the same θ (Reich et al. 2009) are compared. In these cases, θ can be regarded as a constant of proportionality and will not change the theoretical properties of the F-statistics. It will, however, influence statistical properties, as a larger θ implies more mutations and hence more data.
Estimator for F2:
An estimator for F2 can be derived using the average number of pairwise differences πij as an estimator for θTij (Tajima 1983). Thus, a natural estimator for F2 is
| (17) |
Strikingly, the estimator in Equation 17 is equivalent to that given by Reich et al. (2009) in terms of the sample allele frequency and sample size ni:
The last line is the same as equation 10 in the appendix of Reich et al. (2009).
However, while the estimators are identical, the underlying modeling assumptions are different: The original definition considered only loci that were segregating in an ancestral population; loci not segregating there were discarded. Since ancestral populations are usually unsampled, this is often replaced by ascertainment in an outgroup (Patterson et al. 2012; Lipson et al. 2013). In contrast, Equation 17 assumes that all markers are used, which is the more natural interpretation for sequence data.
Gene tree branch lengths:
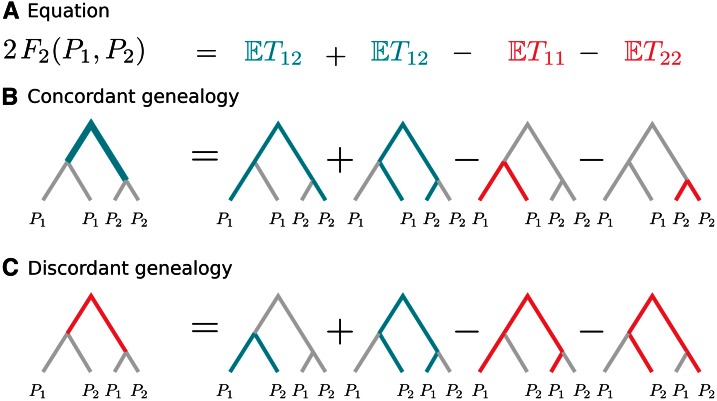
An important feature of Equation 16 is that it depends only on the expected coalescence times between pairs of lineages. Thus, the behavior of F2 can be fully characterized by considering a sample of size 4, with two random individuals taken from each population. This is all that is needed to study the joint distribution of T12, T11, and T22 and hence F2. By linearity of expectation, larger samples can be accommodated by summing the expectations over all possible quartets.
For a sample of size 4 with two pairs, there are only two possible unrooted tree topologies: one, where the lineages from the same population are more closely related to each other [called concordant topology, ] and one where lineages from different populations coalesce first [which I refer to as discordant topology, ]. The superscripts refers to the topologies being for F2, and I discard them in cases where no ambiguity arises.
Conditioning on the topology yields
Figure 4 contains graphical representations for (Figure 4B) and (Figure 4C), respectively.
Figure 4.
(A–C) Schematic explanation of how F2 behaves conditioned on a gene tree. (A) Equation with terms corresponding to the branches in the tree below. Blue terms and branches correspond to positive contributions, whereas red branches and terms are subtracted. Labels represent individuals randomly sampled from that population. External branches cancel out, so only the internal branches have nonzero contribution to F2. In the concordant genealogy (B), the contribution is positive (with weight 2), and in the discordant genealogy (C), it is negative (with weight 1). The mutation rate as constant of proportionality is omitted.
In this representation, T12 corresponds to a path from a random individual from P1 to a random individual from P2, and T11 represents the path between the two samples from P1.
For the internal branch is always included in T12, but never in T11 or T22. External branches, on the other hand, are included with 50% probability in T12 on any path through the tree. T11 and T22, on the other hand, consist only of external branches, and the lengths of the external branches cancel.
On the other hand, for the internal branch is always included in T11 and T22, but only half the time in T12. Thus, they contribute negatively to F2, but only with half the magnitude of As for each T contains exactly two external branches, cancelling the external branches from T12.
An interesting way to represent F2 is therefore in terms of the internal branches over all possible gene genealogies. Denote the unconditional average length of the internal branch of as and the average length of the internal branch in as Then, F2 can be written in terms of these branch lengths as
| (18) |
resulting in the representation given in Figure 2, C and D.
As a brief sanity check, consider the case of a population without structure. In this case, the branch length is independent of the topology and is twice as likely as and hence from which it follows that F2 will be zero, as expected in a randomly mating population
This argument can be transformed from branch lengths to observed mutations by recalling that mutations occur on a branch at a rate proportional to its length. F2 is increased by doubletons that support the assignment of populations (i.e., the two lineages from the same population have the same allele), but reduced by doubletons shared by individuals from different populations. All other mutations have a contribution of zero.
Testing treeness
Many applications consider tens or even hundreds of populations simultaneously (Patterson et al. 2012; Pickrell and Pritchard 2012; Haak et al. 2015; Yunusbayev et al. 2015), with the goal to infer where and between which populations admixture occurred. Using F-statistics, the approach is to interpret as a measure of dissimilarity between P1 and P2, as a large F2 value implies that populations are highly diverged. Thus, the strategy is to calculate all pairwise F2 indexes between populations, combine them into a dissimilarity matrix, and ask whether that matrix is consistent with a tree.
One way to approach this question is by using phylogenetic theory: Many classical algorithms have been proposed that use a measure of dissimilarity to generate a tree (Fitch et al. 1967; Saitou and Nei 1987; Semple and Steel 2003; Felsenstein 2004) and what properties a general dissimilarity matrix needs to have to be consistent with a tree (Buneman 1971; Cavalli-Sforza and Piazza 1975), in which case the matrix is also called a tree metric (Semple and Steel 2003). Thus, testing for admixture can be thought of as testing treeness.
For a dissimilarity matrix to be consistent with a tree, there are two central properties it needs to satisfy: First, the length of all branches has to be positive. This is strictly not necessary for phylogenetic trees, and some algorithms may return negative branch lengths (e.g. Saitou and Nei 1987); however, since in our case branches have an interpretation of genetic drift, negative genetic drift is biologically nonsensical, and therefore negative branches should be interpreted as a violation of the modeling assumptions and hence of treeness.
The second property of a tree metric important in the present context is a bit more involved: A dissimilarity matrix (written in terms of F2) is consistent with a tree if for any four populations Pi, Pj, Pk, and Pl,
| (19) |
that is, if the sums of pairs of distances are compared, two of these sums will be the same, and no smaller than the third one. This theorem, due to Buneman (1971, 1974), is called the four-point condition or sometimes, more modestly, the “fundamental theorem of phylogenetics.” A proof can be found in Semple and Steel (2003, Chap. 7).
Informally, this statement can be understood by noting that on a tree, two of the pairs of distances will include the internal branch, whereas the third one will not and therefore be shorter. Thus, the four-point condition can be colloquially rephrased as “any four-taxa tree has at most one internal branch.”
Why are these properties useful? It turns out that the admixture tests based on F-statistics can be interpreted as tests of these properties: The F3 test can be interpreted as a test for the positivity of a branch and the F4 as a test of the four-point condition. Thus, the working of the two test statistics can be interpreted in terms of fundamental properties of phylogenetic trees, with the immediate consequence that they may be applied as treeness tests for arbitrary dissimilarity matrices.
An early test of treeness, based on a likelihood ratio, was proposed by Cavalli-Sforza and Piazza (1975): They compared the likelihood of the observed F2 matrix to that induced by the best-fitting tree (assuming Brownian motion), rejecting the null hypothesis if the tree likelihood is much lower than that of the empirical matrix. In practice, however, finding the best-fitting tree is a challenging problem, especially for large trees (Felsenstein 2004), and so the likelihood test proved to be difficult to apply. From that perspective, the F3 and F4 tests provide a convenient alternative: Since treeness implies that all subsets of taxa are also trees, the ingenious idea of Reich et al. (2009) was that rejection of treeness for subtrees of sizes 3 (for F3) and 4 (for F4) is sufficient to reject treeness for the entire tree. Furthermore, tests on these subsets also pinpoint the populations involved in the non-tree-like history.
F3: Three population statistic
In the previous section, I showed how F2 can be interpreted as a branch length, as an overlap of paths, or in terms of gene trees (Figure 2). Furthermore, I gave expressions in terms of coalescence times, allele frequency variances, and internal branch lengths of gene trees. In this section, I give analogous results for F3.
Reich et al. (2009) defined F3 as
| (20a) |
with the goal to test whether PX is admixed. Recalling the path interpretation detailed in Patterson et al. (2012), F3 can be interpreted as the shared portion of the paths from PX to P1 with the path from PX to P1. In a population phylogeny (Figure 2E) this corresponds to the branch between PX and the internal node. Equivalently, F3 can also be written in terms of F2 (Reich et al. 2009):
| (20b) |
If F2 in Equation 20b is generalized to an arbitrary tree metric, Equation 20b is known as the Gromov product in phylogenetics (Semple and Steel 2003). The Gromov product is a commonly used operation in classical phylogenetic algorithms to calculate the length of the portion of a branch shared between P1 and P2 (Fitch et al. 1967; Felsenstein 1973; Saitou and Nei 1987), consistent with the notion that F3 is the length of an external branch in a population phylogeny.
In an admixture graph, there is no longer a single external branch; instead all possible trees have to be considered, and F3 is the (weighted) average of paths through the admixture graph (Figure 2F).
Combining Equations 16 and 20b gives an expression of F3 in terms of expected coalescence times:
| (20c) |
Similarly, an expression in terms of variances is obtained by combining Equation 2 with Equation 20b,
| (20d) |
which was also noted by Pickrell and Pritchard (2012).
Outgroup F3 statistics:
A simple application of the interpretation of F3 as a shared branch length are the “outgroup” F3 statistics proposed by Raghavan et al. (2014). For an unknown population PU, they wanted to find the most closely related population from a panel of k extant populations They did this by calculating F3 (PO; PU, Pi), where PO is an outgroup population that was assumed widely diverged from PU and all populations in the panel. This measures the shared drift (or shared branch) of PU with the populations from the panel, and high F3 values imply close relatedness.
However, using Equation 20c, the outgroup F3 statistic can be written as
| (21) |
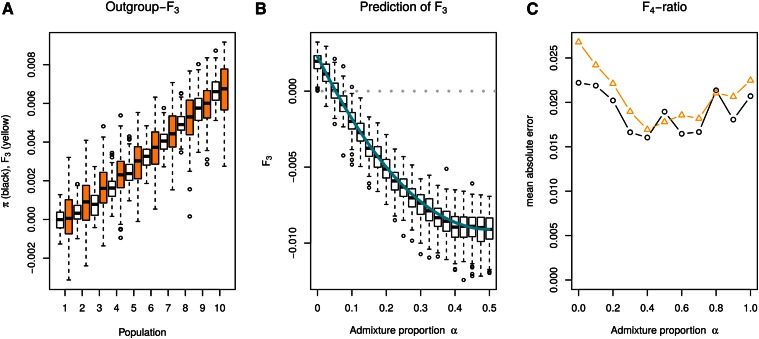
Of these four terms, and do not depend on Pi. Furthermore, if PO is truly an outgroup, then all should be the same, as pairs of individuals from the panel population and the outgroup can coalesce only once they are in the joint ancestral population. Therefore, only the term is expected to vary between different panel populations, suggesting that using the number of pairwise differences, is largely equivalent to using F3 (PO; PU, Pi). I confirm this in Figure 5A by calculating outgroup F3 and for a set of increasingly divergent populations, with each population having its own size, sample size, and sequencing error probability. Linear regression confirms the visual picture that has a higher correlation with divergence time (R2 = 0.90) than F3 (R2 = 0.73). Hence, the number of pairwise differences may be a better metric for population divergence than F3.
Figure 5.
Simulation results. (A) Outgroup F3 statistics (yellow) and (white) for a panel of populations with linearly increasing divergence time. Both statistics are scaled to have the same range, with the first divergence between the most closely related populations set to zero. F3 is inverted, so that it increases with distance. (B) Simulated (boxplots) and predicted (blue) F3 statistics under a simple admixture model. (C) Comparison of F4 ratio (yellow triangles, Equation 29) and ratio of differences (black circles, Equation 31).
F3 admixture test:
However, F3 is motivated and primarily used as an admixture test (Reich et al. 2009). In this context, the null hypothesis is that F3 is nonnegative; i.e., the null hypothesis is that the data are generated from a phylogenetic tree that has positive edge lengths. If this is not the case, the null hypothesis is rejected in favor of the more complex admixture graph. From Figure 2F it may be seen that drift on the path on the internal branches (red) contributes negatively to F3. If these branches are long enough compared to the branch after the admixture event (blue), then F3 will be negative. For the simplest scenario where PX is admixed between P1 and P2, Reich et al. (2009) provided a condition when this is the case (equation 20 in supplement 2 of Reich et al. 2009). However, since this condition involves F-statistics with internal, unobserved populations, it cannot be used in practical applications. A more useful condition is obtained using Equation 20c.
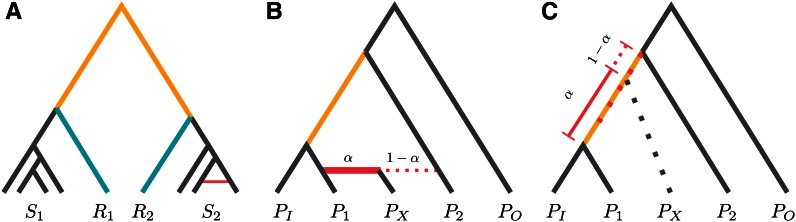
In the simplest admixture model, an ancestral population splits into P1 and P2 at time tr. At time t1, the populations mix to form PX, such that with probability α, individuals in PX descend from individuals from P1, and with probability (1 − α), they descend from P2 (see Figure 7 for an illustration). In this case, F3 (PX; P1, P2) is negative if
| (22) |
where cx is the probability two individuals sampled in PX have a common ancestor before t1. For a randomly mating population with changing size N(t),
Thus, the power of F3 to detect admixture is large (1) if the admixture proportion α is close to 50%; (2) if the ratio between the times of the original split and the time of secondary contact is large; and (3) if the probability of coalescence before the admixture event in PX is small, i.e., the size of PX is large.
Figure 7.
Expectations for F3 and F4 under select models. The constant factor is omitted.
A more general condition for negativity of F3 is obtained by considering the internal branches of the possible gene tree topologies, analogously to that given for F2 in the Gene tree branch lengths section. Since Equation 20c includes only two individuals from PX are needed and one each from P1 and P2 to study the joint distribution of all terms in (20c). The minimal case therefore contains again just four samples (Figure S2).
Furthermore, P1 and P2 are exchangeable, and thus there are again just two distinct gene genealogies, a concordant one where the two lineages from PX are most closely related and a discordant genealogy where the lineages from PX merge first with the other two lineages. A similar argument to that for F2 shows (presented in Figure S2) that F3 can be written as a function of just the internal branches in the topologies,
| (23) |
where and are the lengths of the internal branches in and respectively, and similar to F2, concordant branches have twice the weight of discordant ones. Again, the case of all individuals coming from a single populations serves as a sanity check: In this case is twice as likely as and all branches are expected to have the same length, resulting in F3 being zero. However, for F3 to be negative, note that needs to be more than two times longer than Since mutations are proportional to and F3 can be interpreted as a test whether mutations that agree with the population tree are more than twice as common as mutations that disagree with it.
I performed a small simulation study to test the accuracy of Equation 22. Parameters were chosen such that F3 has a negative expectation for α > 0.05, and I find that the predicted F3 fitted very well with the simulations (Figure 5B).
F4: Four population study
The second admixture statistic, F4, is defined as
| (24a) |
(Reich et al. 2009). Similarly to F3, F4 can be written as a linear combination of F2,
| (24b) |
which leads to
| (24c) |
As four populations are involved, there are 4! = 24 possible ways of arranging the populations in Equation 24a. However, there are four possible permutations of arguments that will lead to identical values, leaving only six unique F4 values for any four populations. Furthermore, these six values come in pairs that have the same absolute value and a different sign [i.e., ], leaving only three unique absolute values, which correspond to the three possible tree topologies. Of these three, one F4 can be written as the sum of the other two, leaving just two independent possibilities:
As for F3, Equation 24b can be generalized by replacing F2 with an arbitrary tree metric. In this case, Equation 24b is known as a tree split (Buneman 1971), as it measures the length of the overlap of the branch lengths between the two pairs. As there are two independent F4 indexes for a fixed tree, there are two different interpretations for the F4 indexes. Consider the tree from Figure 1A: can be interpreted as the overlap between the paths from P1 to P2 and from P3 to P4. However, these paths do not overlap in Figure 1A, and therefore F4 = 0. This is how F4 is used as a test statistic. On the other hand, measures the overlap between the paths from P1 to P3 and from P2 to P4, which is the internal branch in Figure 1A, and will be positive.
It is cumbersome that the interpretation of F4 depends on the ordering of its arguments. To make the intention clear, instead of switching the arguments around for the two interpretations, I introduce the superscripts (T) (for test) and (B) (for branch length):
| (25a) |
| (25b) |
Four-point condition and F4:
Tree splits, and hence F4, are closely related to the four-point condition (Buneman 1971, 1974), which, informally, states that a (sub)tree with four populations will have at most one internal branch. Thus, if data are consistent with a tree, will be the length of that branch, and will be zero. Figure 2, I–L, corresponds to the internal branch and Figure 2, M–P, to the “zero” branch.
Thus, in the context of testing for admixture, testing that F4 is zero is equivalent to checking whether there is in fact only a single internal branch. If that is not the case, the population phylogeny is rejected. This statement can be generalized to arbitrary tree metrics: The four-point condition (Buneman 1971) can be written as
| (26) |
for any permutations of the samples. This implies that two of the sums need to be the same and larger than the third one. The claim is that if the four-point condition holds, at least one of the F4 values will be zero, and the others will have the same absolute value.
Without loss of generality, assume that
Simply plugging this into the three possible F4 equations yields
where
It is worth noting that the converse is false. If
the four-point condition is violated, but is still zero, and the other two F4 values have the same magnitude.
Gene trees:
Evaluating F4 in terms of gene trees and their internal branches, there are three different gene tree topologies that have to be considered, whose interpretation depends on whether the branch length or test-statistic interpretation is considered.
For the branch length [], the gene tree corresponding to the population tree has a positive contribution to F4, and the other two possible trees have a zero and negative contribution, respectively (Figure S3). Since the gene tree corresponding to the population tree is expected to be most frequent, F4 will be positive and can be written as
| (27) |
This equation is slightly different from those for F2 and F3, where the coefficient for the discordant genealogy was half that for the concordant genealogy. Note, however, that F4 includes only one of the two discordant genealogies. Under a tree, both discordant genealogies are equally likely (Durand et al. 2011), and thus the expectation of F4 will be the same.
In contrast, for the admixture test statistic [], the contribution of the concordant genealogy will be zero, and the discordant genealogies will contribute with coefficients −1 and +1, respectively and thus the expectation of F4 as a test statistic
| (28) |
is zero under the null hypothesis. Furthermore, the statistic is closely related to the ABBA-BABA or D-statistic also used to test for admixture (Green et al. 2010; Durand et al. 2011), which includes a normalization term and conditions on alleles being derived. In our notation the expectation of D is
and thus, and D are different test statistics for the same null hypothesis.
Rank test:
Two major applications of F4 use its interpretation as a branch length. First, the rank of a matrix of all F4 statistics is used to obtain a lower bound on the number of admixture events required to explain data (Reich et al. 2012). The principal idea of this approach is that the number of internal branches in a genealogy is bounded to be at most n − 3 in an unrooted tree. Since each F4 is a sum of the length of tree branches, all F4 indexes should be sums of n − 3 branches or n − 3 independent components. This implies that the rank of the matrix (see, e.g., section 4 in McCullagh 2009) is at most n − 3, if the data are consistent with a tree. However, admixture events may increase the rank of the matrix, as they add additional internal branches (Reich et al. 2012). Therefore, if the rank of the matrix is r, the number of admixture events is at least r − n + 3.
One issue is that the full F4 matrix has size and may thus become rather large. Furthermore, in many cases only admixture events in a certain part of the phylogeny are of interest. To estimate the minimum number of admixture events on a particular branch of the phylogeny, Reich et al. (2012) proposed to find two sets of test populations S1 and S2 and two reference populations for each set R1 and R2 that are presumed unadmixed (see Figure 6A). Assuming a phylogeny, all (S1, R1; S2, R2) will measure the length of the same branch, and all (S1, R1; S2, R2) should be zero. Since each admixture event introduces at most one additional branch, the rank of the resulting matrix will increase by at most one, and the rank of either the matrix of all or the matrix of all may reveal the number of branches of that form.
Figure 6.
Applications of F4. (A) Visualization of rank test to estimate the number of admixture events. F4 (S1, R1, S2, R2) measures a branch absent from the phylogeny and should be zero for all populations from S1 and S2. (B) Model underlying admixture ratio estimate (Green et al. 2010). PX splits, and the mean coalescence time of PX with PI gives the admixture proportion. (C) If the model is violated, α measures where on the internal branch in the underlying genealogy PX (on average) merges.
Admixture proportion:
The second application is by comparing branches between closely related populations to obtain an estimate of mixture proportion or how much two focal populations correspond to an admixed population (Green et al. 2010):
| (29) |
Here, PX is the population whose admixture proportion is estimated; P1 and P2 are the potential contributors, where I assume that they contribute with proportions α and 1 − α, respectively; and PO, PI are reference populations with no direct contribution to PX (see Figure 6B). PI has to be more closely related to one of P1 or P2 than the other, and PO is an outgroup.
The canonical way (Patterson et al. 2012) to interpret this ratio is as follows: The denominator is the branch length from the common ancestor population from PI and P1 to the common ancestor of PI with P2 (Figure 6C, yellow line). The numerator has a similar interpretation as an internal branch (Figure 6C, red dotted line). In an admixture scenario (Figure 6B), this is not unique and is replaced by a linear combination of lineages merging at the common ancestor of PI and P1 (with probability α) and lineages merging at the common ancestor of PI with P2 (with probability 1 − α).
Thus, a more general interpretation is that α measures how much closer the common ancestor of PX and PI is to the common ancestor of PI and P1 and the common ancestor of PI and P2, indicated by the red dotted line in Figure 6C. This quantity is defined also when the assumptions underlying the admixture test are violated and, if the assumptions are not carefully checked, might lead to misinterpretations of the data. In particular, α is well defined in cases where no admixture occurred or in cases where either one of P1 and P2 did not experience any admixture.
Furthermore, it is evident from Figure 6 that if all populations are sampled at the same time, and therefore
| (30) |
Thus,
| (31) |
is another estimator for α that can be used even if no outgroup is available. I compare Equations 29 and 31 for varying admixture proportions in Figure 5C, using the mean absolute error in the admixture proportion. Both estimators perform very well, but (31) performs slightly better in cases where the admixture proportion is low. However, in most cases this minor improvement possibly does not negate the drawback that Equation 31 is applicable only when populations are sampled at the same time.
An area of recent development is how these estimates can be extended to more populations. A simple approach is to assume a fixed series of admixture events, in which case admixture proportions for each event can be extracted from a series of F4 ratios (Lazaridis et al. 2014, SI 13). A more sophisticated approach estimates mixture weights using the rank of the F4 matrix, as discussed in the Rank test section (Haak et al. 2015, SI 10). Then, it is possible to estimate mixture proportions, using a model similar to that introduced in the program structure (Pritchard et al. 2000), by obtaining a low-rank approximation for the F4 matrix.
Population structure models
Here, I use Equation 16 together with Equations 20b and 24b to derive expectations for F3 and F4 under some simple models.
Panmixia:
In a randomly mating population (with arbitrary population size changes), P1 and P2 are taken from the same pool of individuals and therefore
Island models:
A (finite) island model has D subpopulations of size 1 each. Migration occurs at rate M between subpopulations. It can be shown (Strobeck 1987) that satisfies
| (32) |
with solution This results in the equation in Figure 7. The derivation of coalescence times for the hierarchical island models is marginally more complicated, but similar. It is given in Slatkin and Voelm (1991).
Admixture models:
These are the models for which the F-statistics were originally developed. Many details, applications, and the origin of the path representation are found in Patterson et al. (2012). For simplicity, I look at the simplest possible tree with four populations, where PX is admixed from P1 and P2 with contributions α and respectively. I assume that all populations have the same size and that this size is 1. Then,
| (33) |
Here, cx is the probability that the two lineages from PX coalesce before the admixture event.
Thus, F3 is negative if
| (34) |
which is more likely if α is large, the admixture is recent, and the overall coalescent is far in the past.
For F4, omitting the within-population coalescence time of 1,
Stepping-stone models:
For the stepping-stone models, I have to solve the recursions of the Markov chains describing the location of all lineages in a sample of size 2. For the standard stepping-stone model, I assumed there were four demes, all of which exchange migrants at rate M. This results in a Markov chain with the following five states: (i) lineages in same deme, (ii) lineages in demes 1 and 2, (iii) lineages in demes 1 and 3, (iv) lineages in demes 1 and 4, and (v) lineages in demes 2 and 3. Note that the symmetry of this system allows collapsing some states. The transition matrix for this system is
| (35) |
Once lineages are in the same deme, the system terminates as the time to coalescence time is independent of the deme in isotropic migration models (Strobeck 1987) and cancels from the F-statistics.
Therefore, the vector v of the expected time until two lineages are in the same deme is found using standard Markov chain theory by solving v = (I − T)−1)1, where T is the transition matrix involving only the transitive states in the Markov chain (all but the first state), and 1 is a vector of 1’s.
Finding the expected coalescence time involves solving a system of five equations. The terms involved in calculating the F-statistics (Table 1) are the entries in v corresponding to these states.
The hierarchical case is similar, except there are six demes and 10 equations. Representing states as lineages being in demes (same), (1, 2), (1, 3), (1, 4), (1, 5), (1, 6), (2, 3), (2, 4), (2, 5), (3, 4),
As in the nonhierarchical case, solving this system yields all pairwise coalescence times. Then, all I have to do is average the coalescence times over all possibilities; e.g.,
| (36) |
For F4, I assume that demes 1 and 2 are in P1, demes 3 and 4 are in PX, and demes 5 and 6 correspond to P2 and P3, respectively.
Range expansion model:
I use a serial founder model with no migration (Peter and Slatkin 2015), where I assume that the expansion is recent enough such that the effect of migration after the expansion finished can be ignored. Under that model, I assume that samples P1 and P2 are taken from demes D1 and D2, with D1 closer to the origin of the expansion and populations with high identification numbers even farther away from the expansion origin. Then where is the time required for a lineage sampled farther away in the expansion to end up in D1. (Note that t1 depends only on the deme that is closer to the origin.) Thus, for three demes,
and
More interesting is
A hierarchical stepping-stone model, where demes are combined into populations, is the only case I studied (besides the admixture graph) where F3 can be negative. This effect indicates that admixture and population structure models may be the two sides of the same coin: Admixture is a (temporary) reduction in gene flow between individuals from the same population. Finally, for a simple serial founder model without migration, I find that F3 measures the time between subsequent founder events.
Simulations
Simulations were performed using ms (Hudson 2002). Specific commands used are
ms 466 100 -t 100 -r 10 100000 -I 12 22 6 61 49 57 33 43 34 40 84 13 24 -en 0 2 7.2 -en 0 3 .2 -en 0 4 .4 -en 0 5 .2 -en 0 6 4.4 -en 0 7 3.2 -en 0 8 4.8 -en 0 9 0.2 -en 0 10 3.2 -en 0 11 0.2 -en 0 12 0.7 -ej 0.01 2 1 -ej 0.02 3 1 -ej 0.04 4 1 -ej 0.06 5 1 -ej 0.08 6 1 -ej 0.10 7 1 -ej 0.12 8 1 -ej 0.14 9 1 -ej 0.16 10 1 -ej 0.18 11 1 -ej 0.3 12 1
for the outgroup F3 statistic (Figure 5A). Sample sizes and population sizes were picked randomly, but kept the same over all 100 replicates. Additionally, I randomly assigned each population an error rate uniformly between 0 and 0.05. Errors were introduced by adding additional singletons and flipping alleles at that rate.
For Figure 5B, the command was
ms 301 100 -t 10 -I 4 100 100 100 1 -es 0.001 2 $ALPHA -ej 0.03 2 1 -ej 0.03 5 3 -ej 0.3 3 1 -ej 0.31 4 1
with the admixture proportion $ALPHA set to increments of 0.025 from 0 to 0.5, with 200 data sets generated per $ALPHA.
Finally, data for Figure 5C were simulated using
ms 501 100 -t 50 -r 50 10000 -I 6 100 100 100 100 100 1 -es 0.001 3 $ALPHA -ej 0.03 3 2 -ej 0.03 7 4 -ej 0.1 2 1 -ej 0.2 4 1 -ej 0.3 5 1 -ej 0.31 6 1
Here, the admixture proportion $ALPHA was varied in increments of 0.1 from 0 to 1, again with 200 data sets generated per $ALPHA.
F3 and F4 statistics were calculated using the implementation from Pickrell and Pritchard (2012).
Estimation and testing
In this article, I focused almost exclusively on the theoretical properties of the F-statistics, glancing over the statistical problems of how they are estimated. Many procedures are implemented in the software package ADMIXTOOLS and described in Patterson et al. (2012). Alternatively, the software package treemix (Pickrell and Pritchard 2012) contains lightwight alternatives for calculating F3 and F4 statistics. Both use a block-jackknife approach to estimate standard errors, taking linkage between markers into account.
Discussion
There are three main ways to interpret F-statistics: In the simplest case, they represent branches in a population phylogeny. In the case of an admixture graph, the idea of shared drift in terms of paths is most convenient. Finally, the expressions in terms of coalescence times and the lengths of the internal branches of gene genealogies are useful for more complex scenarios. This last interpretation makes the connection to the ABBA-BABA statistic explicit and allows the investigation of the behavior of the F-statistics under arbitrary demographic models.
If drift indexes exist for two, three, and four populations, should there be corresponding quantities for five or more populations (e.g., Pease and Hahn 2015)? Two of the interpretations speak against this possibility: First, a population phylogeny can be fully characterized by internal and external branches, and it is not clear how a five-population statistic could be written as a meaningful branch length. Second, all F-statistics can be written in terms of four-individual trees, but this is not possible for five samples. This seems to suggest that there may not exist a five-population statistic as general as the three F-statistics I discussed here, but they will still be useful for questions pertaining to a specific demographic model.
A well-known drawback of F3 is that it may have a positive expectation under some admixture scenarios (Patterson et al. 2012). Here, I showed that F3 is positive if and only if the branch supporting the population tree is longer than the two branches discordant with the population tree. Note that this is (possibly) distinct from the probabilities of tree topologies, although the average branch length of the internal branch in a topology and the probability of that topology are frequently strongly correlated. Thus, negative F3 values indicate that individuals from the admixed population are likely to coalesce with individuals from the two other populations, before they coalesce with other individuals from their own population!
For practical purposes, it is useful to know how the admixture tests perform under demographic models different from population phylogenies and admixture graphs and in which cases the assumptions made for the tests are problematic. In other words, under which demographic models is population structure distinguishable from a tree? Equation 16 enables the derivation of expectations for F3 and F4 under a wide variety of models of population structure (Figure 7). The simplest case is that of a single panmictic population. In that case, all F-statistics have an expectation of zero, consistent with the assumption that no structure and therefore no population phylogeny exists. Under island models, F4 is also zero, and F3 is inversely proportional to the migration rate. Results are similar under a hierarchical island model, except that the number of demes has a small effect. This corresponds to a population phylogeny that is star-like and has no internal branches, which is explained by the strong symmetry of the island model. Thus, looking at different F3 and F4 statistics may be a simple heuristic to see whether data are broadly consistent with an island model; if F3 values vary a lot between populations, or if F4 is substantially different from zero, an island model might be a poor choice. When looking at a finite stepping-stone model, F3 and F4 are both nonzero, highlighting that F4 (and the ABBA-BABA D-statistic) is susceptible to migration between any pair of populations. Thus, for applications, F4 should be used as an admixture test only if there is good evidence that gene flow between some pairs of the populations was severely restricted.
Overall, when F3 is applicable, it is remarkably robust to population structure, requiring rather strong substructure to yield false positives. Thus, it is a very striking finding that in many applications to humans, negative F3 values are commonly found (Patterson et al. 2012), indicating that for most human populations, the majority of markers support a discordant gene tree, which suggests that population structure and admixture are widespread and that population phylogenies are poorly suited to describe human evolution.
Ancient population structure was proposed as possible confounder for the D-statistic and F4 statistic (Green et al. 2010). Here, I show that nonsymmetric population structure such as in stepping-stone models can lead to nonzero F4 values, showing that both ancestral and persisting population structure may result in false positives when assumptions are violated.
Furthermore, I showed that F2 can be seen as a special case of a tree metric and that using F-statistics is equivalent to using phylogenetic theory to test hypotheses about simple phylogenetic networks (Huson et al. 2010). From this perspective, it is worth raising again the issue pointed out by Felsenstein (1973) of how and when allele-frequency data should be transformed for within-species phylogenetic inference. While F2 has become a de facto standard, different transformations of allele frequencies might be useful in some cases, as both F3 and F4 can be interpreted as tests for treeness for arbitrary tree metrics.
This relationship provides ample opportunities for interaction between these currently diverged fields: Theory (Huson and Bryant 2006; Huson et al. 2010) and algorithms for finding phylogenetic networks such as Neighbor-Net (Bryant and Moulton 2004) may provide a useful alternative to tools specifically developed for allele frequencies and F-statistics (Patterson et al. 2012; Pickrell and Pritchard 2012; Lipson et al. 2013), particularly in complex cases. On the other hand, the tests and different interpretations described here may be useful to test for treeness in other phylogenetic applications, and the complex history of humans may provide motivation to further develop the theory of phylogenetic networks and stress its usefulness for within-species demographic analyses.
Acknowledgments
I thank Heejung Shim, Rasmus Nielsen, John Novembre, and all members of the Novembre laboratory for helpful comments and discussions. I am further grateful for comments from Nick Patterson and an anonymous reviewer. B.M.P. is supported by a Swiss National Science Foundation early postdoctoral mobility fellowship. Additional funding for this work was provided by National Institutes of Health grant R01 HG007089 to John Novembre.
Footnotes
Communicating editor: S. Ramachandran
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183913/-/DC1.
Literature Cited
- Allentoft M. E., Sikora M., Sjögren K.-G., Rasmussen S., Rasmussen M., et al. , 2015. Population genomics of Bronze Age Eurasia. Nature 522: 167–172. [DOI] [PubMed] [Google Scholar]
- Bryant D., Moulton V., 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21: 255–265. [DOI] [PubMed] [Google Scholar]
- Buneman, P., 1971 The recovery of trees from measures of dissimilarity, Mathematics in the Archaeological and Historical Sciences. [Google Scholar]
- Buneman P., 1974. A note on the metric properties of trees. J. Comb. Theory Ser. B 17: 48–50. [Google Scholar]
- Cavalli-Sforza, L. L. and A. W. F. Edwards, 1967 Phylogenetic analysis: models and estimation procedures. Evolution 21: 550–570. [DOI] [PubMed]
- Cavalli-Sforza L. L., Piazza A., 1975. Analysis of evolution: evolutionary rates, independence and treeness. Theor. Popul. Biol. 8: 127–165. [DOI] [PubMed] [Google Scholar]
- Durand E., Patterson N., Reich D., Slatkin M., 2011. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Smouse P. E., Quattro J. M., 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1973. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 25: 471–492. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1981. Evolutionary trees from gene frequencies and quantitative characters: finding maximum likelihood estimates. Evolution 35: 1229–1242. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 2004. Inferring Phylogenies. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Fitch W. M., Margoliash E., 1967. Construction of phylogenetic trees. Science 155: 279–284. [DOI] [PubMed] [Google Scholar]
- Green, R., J. Krause, A. Briggs, T. Maricic, U. Stenzel et al., 2010 A draft sequence of the Neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed]
- Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., et al. , 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenthal G., Busby G. B. J., Band G., Wilson J. F., Capelli C., et al. , 2014. A genetic atlas of human admixture history. Science 343: 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Huson D. H., Bryant D., 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Huson, D. H., R. Rupp, and C. Scornavacca, 2010 Phylogenetic Networks: Concepts, Algorithms and Applications. Cambridge University Press, Cambridge/London/New York. [Google Scholar]
- Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., et al. , 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M., Loh P.-R., Levin A., Reich D., Patterson N., et al. , 2013. Efficient moment-based inference of admixture parameters and sources of gene flow. Mol. Biol. Evol. 30: 1788–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P., 2009. Marginal likelihood for distance matrices. Stat. Sin. 19: 631. [Google Scholar]
- Patterson, N. J., P. Moorjani, Y. Luo, S. Mallick, N. Rohland et al., 2012 Ancient admixture in human history. Genetics 192: 1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease J. B., Hahn M. W., 2015. Detection and polarization of introgression in a five-taxon phylogeny. Syst. Biol. 64: 651–662. [DOI] [PubMed] [Google Scholar]
- Peter B. M., Slatkin M., 2015. The effective founder effect in a spatially expanding population. Evolution 69: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova, D., J. Novembre, and M. Stephens, 2014 Visualizing spatial population structure with estimated effective migration surfaces. Nat. Genet. 48: 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Pritchard J. K., 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8: e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Reich D., 2014. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 30: 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M., Skoglund P., Graf K. E., Metspalu M., Albrechtsen A., et al. , 2014. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph P., Coop G., 2013. The geography of recent genetic ancestry across Europe. PLoS Biol. 11: e1001555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., 2009. Reconstructing Indian population history. Nature 461: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Patterson N., Campbell D., Tandon A., Mazieres S., et al. , 2012. Reconstructing Native American population history. Nature 488: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schraiber J. G., Akey J. M., 2015. Methods and models for unravelling human evolutionary history. Nat. Rev. Genet. 16: 727–740. [DOI] [PubMed] [Google Scholar]
- Semple, C., and M. A. Steel, 2003 Phylogenetics. Oxford University Press, London/New York/Oxford. [Google Scholar]
- Slatkin M., 1991. Inbreeding coefficients and coalescence times. Genet. Res. 58: 167–175. [DOI] [PubMed] [Google Scholar]
- Slatkin M., Voelm L., 1991. FST in a hierarchical island model. Genetics 127: 627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck C., 1987. Average number of nucleotide differences in a sample from a single subpopulation: a test for population subdivision. Genetics 117: 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S., 1984. Line-of-descent and genealogical processes, and their applications in population genetics models. Theor. Popul. Biol. 26: 119–164. [DOI] [PubMed] [Google Scholar]
- Wahlund S., 1928. Zusammensetzung von populationen und korrelationserscheinungen vom standpunkt der vererbungslehre aus betrachtet. Hereditas 11: 65–106. [Google Scholar]
- Wakeley J., 2009. Coalescent Theory: An Introduction. Roberts & Co. Greenwood Village, CO. [Google Scholar]
- Weir, B. S., and C. C. Cockerham, 1984 Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed]
- Wright S., 1921. Systems of mating. Genetics 6: 111–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunusbayev B., Metspalu M., Metspalu E., Valeev A., Litvinov S., et al. , 2015. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 11: e1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]