Abstract
Aneuploidy, an unbalanced karyotype in which one or more chromosomes are present in excess or reduced copy number, causes an array of known phenotypes including proteotoxicity, genomic instability, and slowed proliferation. However, the molecular consequences of aneuploidy are poorly understood and an unbiased investigation into aneuploid cell biology is lacking. We performed high-throughput screens for genes the deletion of which has a synthetic fitness cost in aneuploid Saccharomyces cerevisiae cells containing single extra chromosomes. This analysis identified genes that, when deleted, decrease the fitness of specific disomic strains as well as those that impair the proliferation of a broad range of aneuploidies. In one case, a chromosome-specific synthetic growth defect could be explained fully by the specific duplication of a single gene on the aneuploid chromosome, highlighting the ability of individual dosage imbalances to cause chromosome-specific phenotypes in aneuploid cells. Deletion of other genes, particularly those involved in protein transport, however, confers synthetic sickness on a broad array of aneuploid strains. Indeed, aneuploid cells, regardless of karyotype, exhibit protein secretion and cell-wall integrity defects. Thus, we were able to use this screen to identify novel cellular consequences of aneuploidy, dependent on both specific chromosome imbalances and caused by many different aneuploid karyotypes. Interestingly, the vast majority of cancer cells are highly aneuploid, so this approach could be of further use in identifying both karyotype-specific and nonspecific stresses exhibited by cancer cells as potential targets for the development of novel cancer therapeutics.
Keywords: aneuploidy, synthetic lethality, dosage imbalance, protein transport
ANEUPLOIDY, defined as an imbalanced karyotype in which the copy number of one or more chromosomes deviates from base ploidy, has myriad phenotypic consequences on both the cellular and organismal levels. In humans, aneuploidy is the leading cause of spontaneous abortions, and aneuploid organisms display severe developmental defects exemplified by the growth delays and mental retardation characteristic of trisomy 21, or Down syndrome. Paradoxically, the vast majority of cancer cells are also aneuploid; recent estimates indicate that >90% of solid tumors harbor at least one aneuploid chromosome (Weaver and Cleveland 2006; Nagaoka et al. 2012; Chen et al. 2015).
Across both cellular and organismal aneuploid model systems, the expression of genes present on imbalanced chromosomes causes a set of fitness defects including slowed proliferation—in particular, delays in the G1 phase of the cell cycle (Torres et al. 2007; Williams et al. 2008; Thorburn et al. 2013). Aneuploidy also induces a characteristic stress-associated transcriptional program called the “environmental stress response,” causes multiple forms of genomic instability, and broadly disrupts protein homeostasis (Torres et al. 2007; Sheltzer et al. 2011; Stingele et al. 2012; Oromendia et al. 2012; Dephoure et al. 2014). In principle, these phenotypes shared among many different aneuploidies could be due to copy-number imbalances of specific genes the misexpression of which has particular cellular consequences or due to the aggregate effect of imbalances in the levels of many genes. Recent work suggests that the proliferation defects of aneuploid yeast cells cannot be explained by changes in the copy number of specific dosage-sensitive genes (Bonney et al. 2015). In contrast, specific drug sensitivities of aneuploid yeast strains in some cases are attributable to gene-specific effects (Chen et al. 2012, 2015). Although some effects of genomic imbalance have been characterized, an unbiased investigation into the molecular consequences of aneuploidy is lacking.
The development of a high-throughput synthetic lethal screening technology, synthetic genetic array (SGA) analysis, has enabled the unbiased, genome-wide interrogation of novel aspects of yeast biology (Tong 2001; Baryshnikova et al. 2010; Costanzo et al. 2010). This method utilizes the Saccharomyces cerevisiae deletion collection as a basis for screens to identify genes the deletion of which causes synthetic lethality or synthetic sickness when combined with a genetic manipulation of interest (Giaever et al. 2002). In this work, we used stable haploid yeast strains that carry an additional copy of single yeast chromosomes, henceforth known as disomes, as query strains in these screens to further investigate the biology of aneuploid cells.
Using this technique, we identified a number of candidate genes the deletion of which negatively impacts the fitness of aneuploid cells. We identified a subset of these candidate gene deletions that either affect the fitness of specific disomes or impair proliferation in a large number of different disomic yeast strains. We then used this analysis to identify pathways the function of which is compromised in disomic yeast cells. Notably, we have discovered previously unknown phenotypes of aneuploid cells, namely defects in the secretory pathway and in the integrity of the cell wall. Importantly, the utility of this method to uncover commonalities of aneuploid cells as well as chromosome-specific phenotypes could ultimately be utilized to selectively target aneuploid cells in the context of cancer therapy.
Materials and Methods
Yeast strains and plasmids
Disomes used in this study are derivatives of those published in Torres et al. (2007) or were generated using the same method. Disomes used for screening were crossed to the Boone lab starting strain for SGA technology, Y7092, with the genotype can1delta::STE2pr-Sp_his5 lyp1delta his3delta1 leu2delta0 ura3delta0 met15delta0. De novo gene deletions were generated using published methods (Longtine et al. 1998) in a wild-type W303 yeast strain. Disomes carrying candidate gene deletions were constructed by crosses. Karyotypes of key disomic strains were verified by comparative genomic hybridization as described (Torres et al. 2007) and analyzed with Java TreeView. All strains are listed in Supporting Information, Table S1.
Synthetic lethal screens
The SGA screens were performed robotically in triplicate, as previously described (Tong and Boone 2012) using MATα disomes as query strains. Data were analyzed using SGAtools, a normalization and scoring methodology developed for small-scale screens (Wagih et al. 2013). Validations were performed by crossing deletions generated de novo into disomic strains lacking the screen-specific markers, followed by tetrad dissection and fitness measurements of the resultant disomic deletion mutants.
Doubling time analysis
Cells were grown overnight at room temperature in yeast extract/peptone medium containing 2% glucose (YPD) and diluted to OD600 = 0.1 the next morning. The growth rate of these cultures at 25° was measured in triplicate using a BioTek plate reader to take measurements every 15 min for 24 hr. Data were accumulated using Gen5 BioTek software. The period of exponential growth was used to calculate doubling time using GraphPad Prism software. Data shown are the average of two to four biological replicates performed on different days.
Assessment of disomic drug sensitivities
Overnight cultures in YPD were diluted to OD600 = 1.0, and 1.9 μl of 10-fold serial dilutions were spotted on YPD plates that contained 250 μg/ml Calcofluor white, 10 μg/ml Congo red, 50 μg/ml Congo red, 16 μg/ml fluconazole, 32 μg/ml fluconazole, or 200 μg/ml Brefeldin A. Plates were incubated at 30° for 2 days before images were taken.
Lucifer yellow uptake experiments
Uptake of Lucifer yellow (LY) was assayed essentially as described (Duncan et al. 2001). Briefly, 1 ml of cells at OD600 = 0.1–0.2 was resuspended in 100 μl YPD containing 4 mg/ml Lucifer Yellow carbohydrazide (Sigma-Aldrich). Cells were incubated at room temperature for 1 hr before 1 ml ice-cold 50 mM potassium phosphate buffer containing 10 mM NaN and 10 mM NaF was added. Cells were washed three times and resuspended in 20 μl of the same buffer before imaging using a Zeiss Axioplan 2 microscope with a Hamamatsu OCRA-ER digital camera. Image analysis was performed using Volocity software.
Zymolyase sensitivity assay
Sensitivity to zymolyase was assayed as described (Castrejon et al. 2006). Briefly, doubling times were measured as described above with the addition of 10, 25, or 50 μg/ml of zymolyase (20T, MP Biomedicals). The ratio of doubling times with zymolyase to those without was calculated and plotted relative to wild type.
Detection of immature Ccw14p protein intermediates
Cells were grown to exponential phase at room temperature (with the exception of ipl1-321, which was grown initially at 25° and shifted to 30° for 3 hr immediately before cells were collected). Cell pellets were suspended in 2 ml 5% trichloroacetic acid, and after incubation at 4° for 10 min, pellets were washed with acetone. Dried pellets were then suspended in 100 μl breakage buffer (50 mM Tris–HCl at pH 7.5, 1 mM EDTA, 2.75 mM DTT, Roche complete protease inhibitor), glass beads were added, and cells were broken by beating for 5 min on a Biospec minibead beater; 3× SDS sample buffer was added and samples were boiled for 5 min and centrifuged briefly. Lysates were loaded onto 10% SDS-polyacrylamide gels for electrophoresis and transferred onto nitrocellulose membranes before probing with rabbit Ccw14 antiserum (1:1000) or mouse anti-Pgk1 (1:10,000, A-6457, Molecular Probes). Secondary antibodies used were donkey anti-rabbit conjugated to horseradish peroxidase (HRP) (NA934, GE Healthcare) and sheep anti-mouse conjugated to HRP (NA931, GE Healthcare), both at 1:5000 dilutions. HRP conjugates were detected using Amersham ECL Prime reagent according to the manufacturer’s instructions.
Data availability
Strains (Table S1) are available upon request. Table S2 contains the full SGA data set and is available for download.
Results
Synthetic genetic array analysis of aneuploid yeast cells
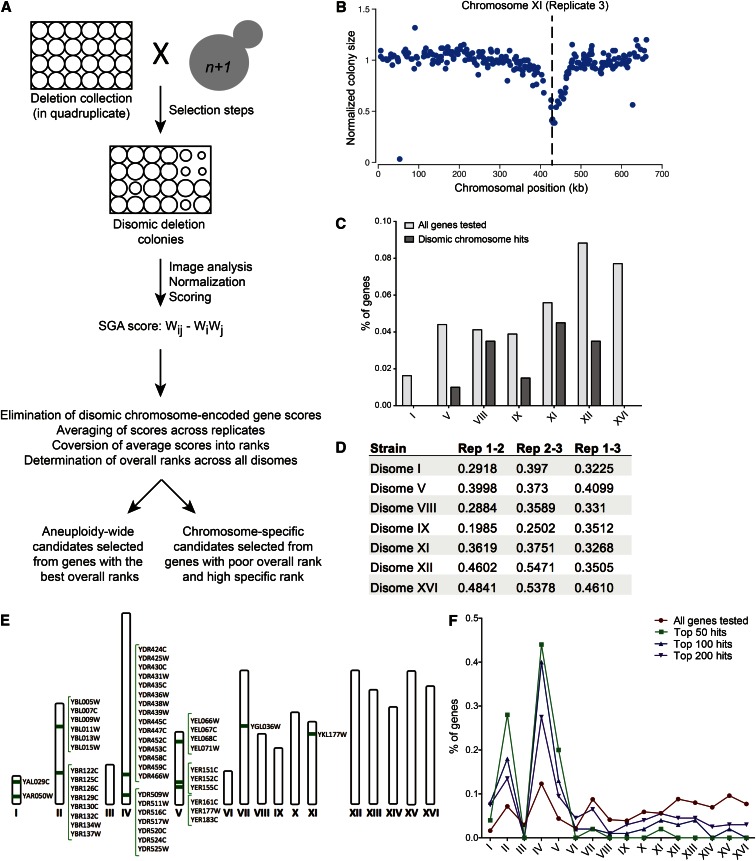
To characterize aneuploid cell biology on an unbiased, genome-wide scale, we conducted seven synthetic negative fitness screens in triplicate using disomic yeast strains as queries (Figure 1A). These seven strains, which have a haploid base ploidy and contain one extra copy of a single yeast chromosome (I, V, VIII, IX, XI, XII, or XVI), were selected to represent a range of disomic chromosome sizes and fitness defects. For each screen, quantification of colony sizes was performed using an approach developed for customized small-scale screens that includes image analysis and multiple normalization steps (Wagih et al. 2013). Scores are a reflection of the colony size, and thus the fitness, of each disomic deletion mutant, with scores less than −0.1 reflecting a synthetic fitness defect and scores greater than 0.1 indicating synthetic fitness enhancement. The technical success of each screen was assessed based on two metrics. First, we confirmed the presence of linkage between the selectable markers on the disomic chromosomes and candidate synthetic gene deletions on that chromosome (Figure 1B and Figure S1). This linkage derives from decreased recombination between the selectable marker and genes located nearby on the same chromosomal segment and should therefore be a marker of a technically successful screen (Baryshnikova et al. 2013). Second, we see a reduction in the number of synthetic negative fitness interactions between a given disome and gene deletions located on the disomic chromosome relative to all other chromosomes (Figure 1C). Upon deleting a gene located on the disomic chromosome, normal copy number of this gene has been restored rather than eliminated, and this is unlikely to impair disomic fitness in most cases. The screens conducted with disomes I, V, IX, XII, and XVI showed substantial depletion of synthetic negative fitness interactions of genes located on the disomic chromosome (Figure 1C). However, this was not the case for VIII and XI (Figure 1C), indicating that many of the negative interactions obtained in these screens are likely to be false positives. Reproducibility between replicates was within the range expected for this methodology, with correlation values ranging from r = 0.2 to 0.55 (Figure 1D). The results of all 21 screens can be found in Table S2.
Figure 1.
Synthetic lethal screen design, analysis, and quality control. (A) Schematic of the synthetic lethal screen. Disomes carrying appropriate selectable markers were crossed to the deletion collection as a 1536-spot array. After sporulation, progeny were pinned to a series of selective media to yield cells containing the disomic chromosome as well as the gene deletion of the parent strain. Images of the final plates were analyzed for pixel size, and normalization and scoring was performed as described (Wagih et al. 2013). The resultant SGA score is a standard multiplicative score where Wij is the fitness of the observed value, Wi is the median fitness of all the strains in the experiment, and Wj is the median fitness of the disomic strain. Scores were normalized for consistency across replicates and ranked both for each disome and overall. (B) Colony sizes of disomes harboring deletions of genes located on the disomic chromosome are shown for the screen of that disome. An example for disome XI is shown. The same analysis for the other disomic strains is shown in Figure S1. The dashed line marks the site of the selectable marker on the disomic chromosome. Genes located close to the site of the selectable marker on the disomic chromosome have smaller colony sizes due to lack of recombination between the gene and the selectable marker. (C) Depletion of synthetic negative growth defects between the disomes and deletions of genes located on the disomic chromosome. The percentage of all genes tested present on each disomic chromosome is plotted in light gray. The percentages of gene deletions identified among the top 200 potential synthetic negative fitness interactions for each disomic screen that are located on the disomic chromosome are plotted in dark gray. (D) Correlation values (r) for replicate screens using the same disomic query strain are shown. (E) Chromosomal locations of the top 50 candidate deletions with putative synthetic negative fitness defects with all disomic strains screened in the SGA are illustrated. (F) The percentages of genes on each chromosome found among the top 50, 100, or 200 candidate deletions for aneuploidy-wide synthetic fitness impairment are plotted. The distribution of all deletions tested (red line) is plotted for comparison.
To analyze the data, we first averaged scores across the three replicates and converted them into ranks to control for absolute score variability across screens. Ranks were both evaluated individually for each disome to identify disome-specific synthetic fitness defects and summed across screens to detect synthetic fitness defects common among many disomic strains analyzed. Before performing the aneuploidy-wide data analysis, scores for genes encoded on the disomic chromosome were removed from the data for that disome to exclude instances in which gene copy number had been restored to euploidy by the deletion rather than eliminated. Finally, we converted these summed ranks into an aneuploidy-wide rank, and the top candidates based on this rank are shown in Table S3.
Although we performed 21 technically successful screens, there were various potential sources of false positives inherent in these screens. Indeed, when sorted by chromosomal position, the distribution of the deletions that exhibit synthetic fitness defects with multiple disomic strains is conspicuously nonrandom. Among the top 50 deletions that exhibited synthetic fitness defects across aneuploid strains, all but 4 were found in clusters of at least three genes (Figure 1E). However, as we expand the list of candidate genes further, to 100 or 200 candidates, the distribution progressively approaches that of all genes tested in these screens, reducing the clustering of the data substantially (Figure 1F). One potential contributor to this clustering of gene deletions exhibiting synthetic fitness defects with multiple disomes is the previously described neighboring gene effect (Baryshnikova and Andrews 2012; Ben-Shitrit et al. 2012). This phenomenon is due to artifacts introduced during the construction of the deletion collection. Deletion of some genes in the deletion collection has unanticipated effects on the expression of neighboring genes. This results in multiple strains with adjacent deletions behaving similarly in screens due to the fact that the same gene is disrupted in each of these different strains. Such artifacts have a particular impact on our assessment of top aneuploidy-wide candidates, as rankings in this group of synthetic interactions depend not only on the strength of the interactions but also on how broadly they occur across strains, placing more weight on weaker, but common synthetic negative interactions. The clustering of candidate synthetic gene deletions may also be due to differences between the disomic strains (W303 background) and the deletion collection (S288C) in that W303 may harbor one or more genetic variants that cause aneuploidy tolerance. For example, we observed synthetic negative interactions for a stretch of genes on chromosome IV that spans 78.4 kb and includes 15 of the 50 most highly ranked aneuploidy-wide candidates (Figure 1E). Observing a collinear set of negative interactions derived from an SGA screen suggests that the W303 parental strain may carry a genetic variant within this chromosome IV segment that can act as a suppressor of the fitness defect associated with disome aneuploidy. The SGA technology could be used to map these strain-specific suppressors in this way (Jorgensen et al. 2002). We conclude that neighboring gene effects, aneuploidy-tolerating alleles in the W303 strain background, and known errors in the deletion collection including widespread aneuploidy enrich for false positives among deletion candidates that exhibit synthetic fitness defects with multiple disomes.
Karyotype-specific synthetic negative fitness interactions inform disomic cell biology
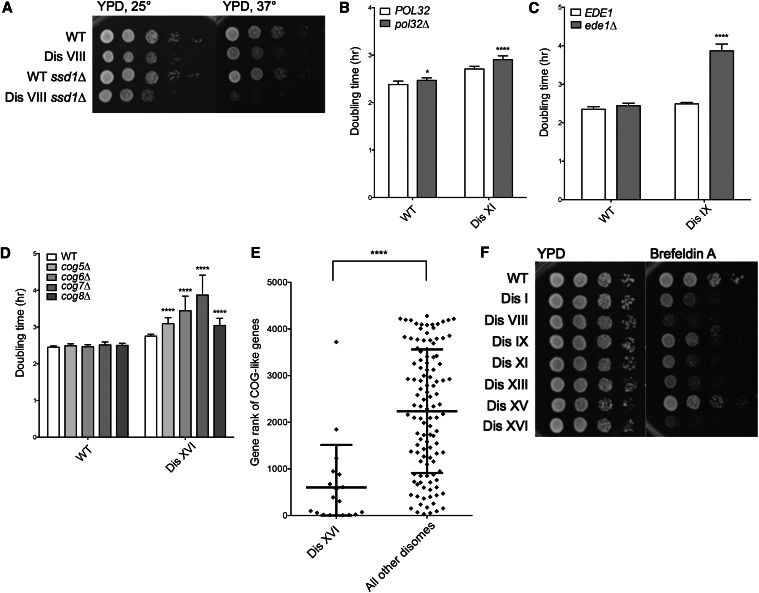
We first analyzed gene deletions that showed putative synthetic fitness defects with individual disomes, presumably due to karyotype-specific impacts on their biology. To perform this analysis, the top 50 candidates by rank were determined for each disome, and those with poor overall ranks across the disomes were prioritized for analysis, as these should represent deletions that specifically impair proliferation of a particular disome (Table 1). To validate the SGA results, we generated de novo gene deletions and introduced these deletions into previously characterized disomes (Torres et al. 2007). Doubling time measurements of strains obtained in this manner showed that 7 of 11 (63.6%) deletions chosen for this analysis exhibited synthetic fitness defects with a specific disomy (Table 1). The degrees of these synthetic fitness defects were variable. For example, deletion of SSD1 conferred a mild fitness defect on disome VIII (Figure 2A), as did deletion of POL32 on disome XI (Figure 2B). In contrast, other deletions had a severe impact on the fitness of individual disomes. For example, deletion of EDE1 dramatically increased the doubling time of disome IX cells (Figure 2C). This synthetic interaction was specific because deletion of EDE1 had little or no effect on the fitness of wild-type cells and disomes VIII, XI, or XVI (Figure S2A).
Table 1. Validation of deletions that impair fitness of specific disomes.
| Gene | Disome | Disome-specific rank | Disome-wide rank | Confirmed? |
|---|---|---|---|---|
| NPL4 | V | 23 | 1366 | No |
| SSD1 | VIII | 12 | 1691 | Yes (37°) |
| EDE1 | IX | 3 | 2774 | Yes |
| HIR1 | IX | 20 | 1352 | No |
| SLT2 | IX; XI | 18; 16 | 2559 | No |
| POL32 | XI | 21 | 1621 | Yes |
| CBF1 | XVI | 7 | 2557 | No |
| COG5 | XVI | 44 | 2289 | Yes |
| COG6 | XVI | 66 | 3375 | Yes |
| COG7 | XVI | 20 | 2092 | Yes |
| COG8 | XVI | 2 | 1554 | Yes |
Figure 2.
Karyotype-specific synthetic negative interactions. (A) Cultures of wild-type and disome VIII strains either wild type or deleted for SSD1 were grown overnight in YPD, and 10-fold serial dilutions were plated on YPD plates at the indicated temperatures. (B–D) Doubling times of disomes either wild type or deleted for the indicated genes were determined by growing cells in YPD at 25° and taking OD600 measurements every 15 min over 24 hr. SD is shown. *P < 0.01, ****P < 0.00001; Student’s t-test. (E) Genes that exhibited similar negative synthetic interactions in SGA analyses as the COG5–8 genes were identified using the DRYGIN online database (Koh et al. 2009). The distribution of specific ranks of these genes (CBF1, TLG2, ARL1, IRS4, SYS1, GYP1, RIC1, YPT6, VPS51, VPS63, ERV14, RUD3, YLR269C, GET2, COY1, IMH1, OCA1, ENT4, SEC22) in the disome XVI SGA screen are plotted, as well as the distribution of the ranks of these genes across the other disome SGA screens (****P < 0.0001, Mann–Whitney test). (F) Cultures of wild-type and disomic strains were grown overnight in YPD, and 10-fold serial dilutions were plated on YPD plates with or without 200 μg/ml Brefeldin A. Plates were incubated at 30° for 2 days before images were taken.
Deletion of four genes (COG5–8) located on three separate chromosomes, which encode components of the conserved oligomeric Golgi (COG) complex, exhibited synthetic negative interactions with disome XVI (Figure 2D). The COG complex is involved in protein glycosylation and trafficking (Whyte and Munro 2001; Ram et al. 2002; Suvorova 2002; Smith and Lupashin 2008). COG5–8 encode components of one lobe of this complex (Fotso et al. 2005). Individual deletion of each of these genes increased the doubling time of disome XVI by between 17 and 67 min, while having close-to-no effect on the growth of wild-type cells and little-to-no impact on other disomes (Figure S2B).
The observed synthetic effects offer genetic evidence for a protein transport defect in disome XVI cells. To further test this possibility, we used the online Data Repository of Yeast Genetic Interactions, a databank of all SGA analyses performed to date (DRYGIN) (Koh et al. 2009), to generate a list of gene deletions whose own SGA results correlated positively with those of screens using COG5–8 deletions as queries (Table S4). These COG-like genes are therefore genetically similar to COG5–8. We then analyzed the specific ranks of the deletion of these COG-like genes (excluding those encoded on chromosome XVI) in the disome XVI SGA screen as well as across the other six screens (Figure 2E and Figure S2C). We observed a strong bias for COG-like genes to be ranked among the deletions that exhibited the highest degree of negative fitness defects with disome XVI, while showing no deviation from the average rank distribution for the other disomes. Furthermore, correlation analyses between our disomic SGA screens and the repository of previously performed SGA screens revealed that the disome XVI SGA results are highly correlated with SGA screens employing deletions in genes involved in vesicle-mediated transport (Bonferroni-corrected P = 3.78 × 10−9) (Costanzo et al. 2010).
Consistent with the hypothesis that disomy of chromosome XVI confers defects in protein transport, we observed that disome XVI cells are highly sensitive to Brefeldin A, a drug that targets protein trafficking by inhibiting the assembly of coatomer on COPII vesicles, thereby blocking transport between the ER and Golgi (Figure 2F) (Shah and Klausner 1993; Dinter and Berger 1998). Although this phenotype is strongest in disome XVI, other disomes that are not sensitive to the deletion of genes encoding COG complex components also exhibit increased sensitivity to Brefeldin A. This finding suggests that perturbations in protein transport are widespread among disomic strains but this phenomenon is related to the COG complex only in disome XVI cells. To date, we have not been able to identify a single gene located on chromosome XVI the increased dosage of which causes synthetic growth defects with deletions of genes encoding COG complex subunits. This observation raises the possibility that increased copy number of multiple genes is responsible for the severe synthetic fitness defect between disomy XVI and loss of COG complex function. We conclude that SGA technology can successfully identify specific aspects of disome cell biology that are likely governed by particular dosage imbalances of genes encoded on the chromosomes in excess, exemplified by the specific vesicle transport defects in disome XVI cells.
Chromosome-specific synthetic fitness defect explained by a single gene dosage change
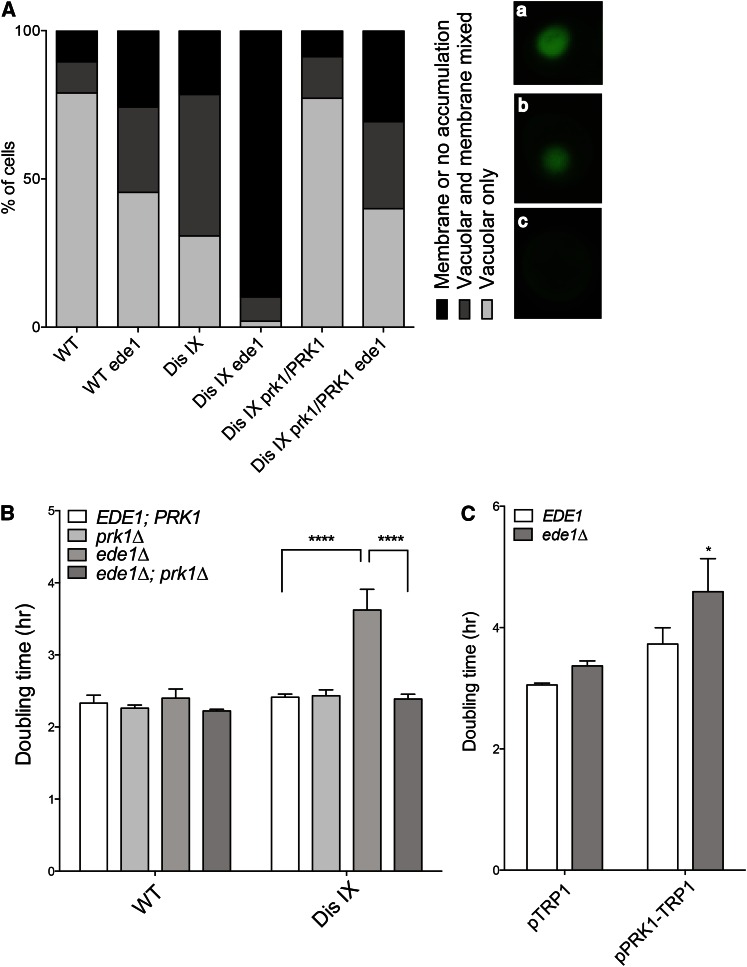
The most severe karyotype-specific synthetic interaction that we identified was caused by deletion of EDE1, an early endocytic gene, in disome IX. The doubling time of disome IX increased by >80 min upon EDE1 deletion, while wild-type growth slowed by only 5 min (Figure 2C). This effect is karyotype-specific, as the same gene deletion had little-to-no effect on other disomes tested (Figure S2A).
EDE1 encodes a coat protein that is recruited to the sites of endocytic patches on the plasma membrane early in the process of endocytosis initiation (Gagny et al. 2000; Goode et al. 2015). Known phenotypes of ede1∆ cells include gross defects in endocytosis, which can be assayed by following the internalization and vacuolar accumulation of the fluorescent dye Lucifer yellow (Dulic et al. 1991). Consistent with their synthetic negative fitness defect with deletion of EDE1, we find that cells disomic for chromosome IX, but not other disomic strains, exhibit defects in endocytosis (Figure 3A and Figure S2D). Only 31% of disome IX cells fully internalized LY after 1 hr, in comparison to 79% of wild-type cells. Furthermore, the endocytosis defect of disome IX cells was exacerbated by deletion of EDE1; only 10% of disomic cells lacking EDE1 showed any accumulation at all of LY in the vacuole, in contrast to 90% of wild-type cells (Figure 3A).
Figure 3.
The growth defect of disome IX cells lacking EDE1 is due to excess PRK1. (A) Cells were incubated with Lucifer yellow dye for 1 hr at room temperature, washed three times, and imaged. The distribution of internalization phenotypes is shown. n > 100 cells per strain. Images on the right show the different classes of Lucifer yellow localization: (a) vacuolar accumulation, (b) mixed vacuolar and membrane localization, and (c) membrane or no localization. (B and C) Doubling times were determined as described in Figure 2 in YPD medium (B) or synthetic complete medium without tryptophan (C). SD is shown. *P < 0.01, ****P < 0.00001; Student’s t-test.
As Ede1 has a well-defined cellular function, we next determined whether the specific synthetic endocytosis defect conferred on disome IX cells by deletion of EDE1 could be explained by the dosage imbalance of a known chromosome IX-encoded endocytic gene. We identified a chromosome IX-encoded protein kinase, Prk1, that negatively regulates various endocytic proteins (Zeng and Cai 1999, 2005). PRK1 is known to be a dosage-sensitive gene the overexpression of which causes actin abnormalities and lethality (Zeng and Cai 1999; Makanae et al. 2012). We deleted one copy of PRK1 in disome IX cells and determined its effects on the synthetic growth and endocytosis defects observed in disome IX ede1∆ cells. Remarkably, restoration of haploid PRK1 dosage suppressed the growth defect of disome IX ede1∆ cells, restoring their growth rate to that of disome IX EDE1 cells (Figure 3B). Importantly, deleting PRK1 did not affect the proliferative abilities of disome IX cells harboring a wild-type EDE1 allele (Figure 3B).
Elevated dosage of PRK1 not only is required for the growth defect of disome IX ede1∆ cells, but also is sufficient. The doubling time of wild-type cells expressing a CEN plasmid harboring PRK1 under the control of its own promoter was increased by an average of 52 min upon deletion of EDE1 (Figure 3C). In fact, these analyses had to be conducted in medium selecting for the plasmid, as the plasmid was highly unstable and prone to loss in rich medium. Thus, duplication of PRK1 in disome IX cells is both necessary and sufficient to confer a fitness defect on cells lacking EDE1. Consistent with this result, restoration of haploid PRK1 dosage in disome IX ede1∆ cells also suppressed their endocytosis defect (Figure 3A). We conclude that some severe karyotype-specific synthetic interactions can be explained by the change in dosage of a single disome-encoded gene. This result underscores the complexity of aneuploid cell biology, as the phenotypes of aneuploid cells depend in part on the identity of the imbalanced chromosome(s) while also exhibiting known general aneuploid phenotypes elicited by many different aneuploid karyotypes (Torres et al. 2007; Thorburn et al. 2013).
SGA analysis identifies gene deletions detrimental to many different disomes
In addition to chromosome-specific synthetic interactions, we identified genes the deletion of which impairs the fitness of many different disomic strains. We ranked deletions according to the severity of the synthetic fitness defect that they exhibited with each disome, summed these ranks across strains, and assigned an aneuploidy-wide rank. To validate the aneuploidy-wide synthetic interactions revealed by the screen, we chose a subset of 11 genes with ranks ranging from 1 to 152 (Table S3). We created de novo deletions of the chosen genes, introduced them into the disomic strains, and analyzed their fitness by calculating their doubling times in rich medium (YPD) at 25°. This analysis revealed that, of the 11 candidates, 5 deletions (45.5%) exhibited the synthetic growth defects observed in the SGA analysis across many disomic strains (Table 2). As mentioned above, this modest validation percentage can be explained in part by artifacts stemming from the neighboring gene effect and/or the presence of potential aneuploidy-tolerating alleles in W303 (Baryshnikova and Andrews 2012). Consistent with this hypothesis, none of the genes the deletion of which was confirmed to exhibit a synthetic fitness defect with the disomes clustered in the same genomic region as any other confirmed gene deletions.
Table 2. Validation of deletions that impair fitness of multiple disomes.
| Gene | Rank | Disome I | Disome V | Disome IX | Disome XI | Disome XIV | Disome II | Disome XIII | Disome XVI | Disome XII | Disome VIII | Disome XV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FLO1 | 1 | NE | ND | ND | NE | ND | ND | ND | NE | ND | NE | ND |
| SHE3 | 3 | NE | ND | NE | NE | ND | NA | NE | NE | ND | NE | ND |
| UBP3* | 11 | NE | NA | NE | −− | −− | −− | NE | −− | −− | − | −− |
| SNX41 | 12 | ND | ND | − | NE | ND | ND | ND | NE | ND | − | ND |
| TSA2 | 15 | NE | ND | NE | NE | ND | ND | NE | NE | ND | NE | ND |
| CYM1 | 30 | NE | ND | NE | NE | ND | ND | ND | NE | ND | NE | ND |
| PPN1* | 43 | − | NE | − | −− | −− | NE | − | − | NE | − | − |
| TPS1* | 48 | − | ND* | −− | −− | NE | NA | −− | −− | − | NE | NE |
| SIR1 | 111 | ND | NE | NE | NE | ND | ND | NE | NE | ND | NE | ND |
| MNN10* | 115 | NE | − | − | −− | ND* | NE | −− | −− | −− | − | −− |
| RVS167* | 152 | −− | −− | −− | −− | − | NE | NE | − | ND* | − | NE |
–, negative synthetic effect; – –, strong negative synthetic effect; NE, no effect; ND*, no data because this strain was found to have lost its extra chromosome in at least one case; ND, no data; NA, gene on disomic chromosome so not tested. (*) indicates confirmed gene deletions with synthetic negative fitness interactions across disomic strains.
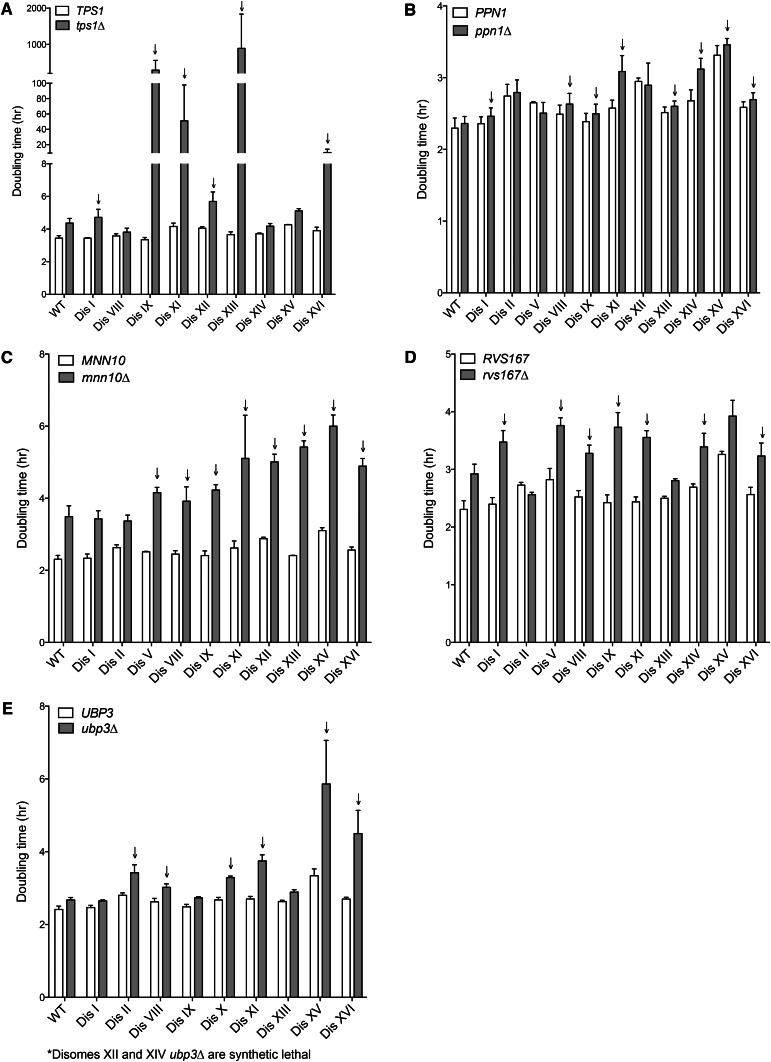
Among the deletions identified as conferring a proliferation defect on multiple disomes was that of TPS1, which encodes a subunit of trehalose-6-phosphate synthase, an enzyme critical for trehalose production (Bell et al. 1992). Deletion of this gene caused an extreme proliferation defect in disomes IX, XI, XIII, and XVI and impaired fitness in multiple other disomes (Figure 4A). Trehalose confers stress resistance to yeast cells (D’Amore et al. 1991; Singer and Lindquist 1998), and this function may be important for the survival of several disomic strains, which are known to experience diverse cellular stresses (Torres et al. 2007; Sheltzer et al. 2011; Oromendia et al. 2012; Dephoure et al. 2014; Blank et al. 2015).
Figure 4.
Deletion of MNN10, PPN1, RSV167, TPS1, and UBP3 decreases proliferation rate of many disomic yeast strains. Doubling times of the indicated strains were determined as described in Figure 2. Cells were grown in either YPD medium (B–E) or YEP containing 2% raffinose 2% galactose (A) as deletion of TPS1 causes lethality in glucose. Arrows indicate disomes the doubling time of which increases more upon gene deletion than does that of wild type.
The synthetic fitness defect conferred by tps1∆ on the disomes underscores the sensitivity to various stresses conferred by aneuploidy; however, the reasons for other synthetic defects identified by the screen are less obvious. Deletion of PPN1 caused a subtle decrease in proliferative potential in disomes I, VIII, IX, XI, XIII, XIV, XV, and XVI (Figure 4B). PPN1 encodes an endo- and exopolyphosphatase that is important for the hydrolysis and utilization of long-chain polyphosphates (Sethuraman et al. 2001; Pestov et al. 2005). Why its deletion impairs the fitness of multiple disomes more than that of wild type is at present not understood; however, polyphosphate has a plethora of proposed cellular roles, including phosphate metabolism, cation chelation, and stress tolerance (Kornberg et al. 1999). The synthetic fitness defect caused by the PPN1 deletion in multiple disomes represents an interesting avenue for further investigation.
Remarkably, many of the genes the deletion of which conferred a fitness defect on multiple disomes were involved in protein trafficking and membrane-related processes. For example, deletion of MNN10, which encodes a Golgi mannosyltransferase subunit (Jungmann et al. 1999), caused striking fitness defects in 8 of 10 disomes analyzed (Figure 4C). MNN10 functions in a complex with four other proteins, deletions of two of which were tested in the SGA screen. Deletion of HOC1 also exhibited fitness defects in the SGA when introduced into multiple disomes, with an overall rank of 60 across disomic strains (Table S2). Deletion of MNN11 also caused a fitness decrease in multiple disomes, with a rank of 271 (Table S2). Together, these data indicate that many disomic strains exhibit increased sensitivity to perturbations in mannosyltransferase function.
RVS167 encodes an actin-associated endo- and exocytosis protein (Youn et al. 2010; Smaczynska-de Rooij et al. 2011). Its deletion substantially increased the doubling time of all but disomes II, XIII, and XV (Figure 4D). Remarkably, in two disomes where a decrease in fitness was not observed upon deletion of RVS167, the proliferation defect conferred by deletion of RVS167 was in fact suppressed. Disome II and disome XIII cells lacking RVS167 grew faster than wild-type cells lacking the gene (Figure 4D). This finding indicates that the two chromosomes harbor genes that suppress the adverse effects of deleting RVS167 on cellular fitness. Finally, deletion of UBP3, which encodes a deubiquitinase (Baxter and Craig 1998), impaired the fitness of almost all disomes examined and in two cases caused synthetic lethality (Figure 4E). This ubiquitin protease has also been implicated in vesicle transport, among other processes (Cohen et al. 2003; Ossareh-Nazari et al. 2010).
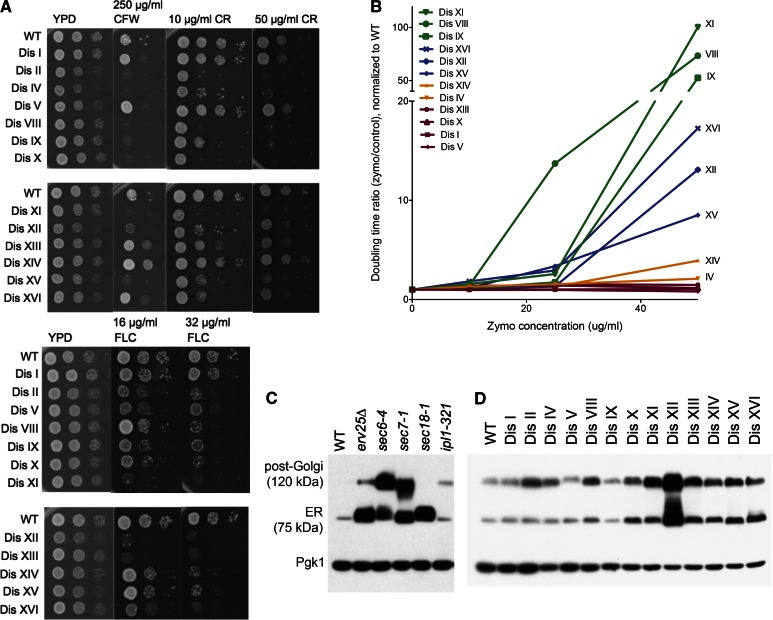
Taken together, genes the deletion of which exhibited synthetic negative interactions with one or more disomes were significantly enriched for the gene ontology (GO) term “vesicle-mediated transport” (Bonferroni-corrected P = 1.2 × 10−4). This observation indicates that protein trafficking and membrane defects may be recurrent phenotypes of aneuploid cells. Indeed, the vast majority of the disomes exhibited phenotypes consistent with protein transport defects. Cell-wall formation relies on protein trafficking; thus, cells defective in this process exhibit sensitivity to cell-wall-damaging agents such as Calcofluor white (CFW) or Congo red (CR) (Ram and Klis 2006). All but disomes I, V, and XIV exhibited a decreased ability to form colonies on medium containing CFW or CR (Figure 5A). Moreover, every disome except disome I was more sensitive to the antifungal drug fluconazole, which inhibits Erg11 and leads to ergosterol depletion and thus cell-membrane defects and accumulation of toxic sterol intermediates (Kontoyiannis 2000; Abe et al. 2009). Furthermore, most disomes were highly sensitive to addition of the yeast cell-wall lytic enzyme zymolyase to the growth medium (Figure 5B), indicating pervasive cell-membrane and cell-wall defects.
Figure 5.
Protein-trafficking defects are widespread among disomic yeast strains. (A) Wild-type and disomic cells were grown in YPD overnight. Tenfold dilutions were spotted on YPD plates containing the indicated concentrations of CFW, CR, or fluconazole (FLC). Plates were incubated for 2 days at 30° before images were taken. (B) Zymolyase sensitivity was assayed by performing doubling time measurements in YPD ± the indicated concentrations of zymolyase (20T), as described in Figure 2. The ratio of doubling time in the presence of zymolyase to doubling time without the enzyme is plotted as a function of zymolyase concentration, normalized to wild type. (C and D) Cells were grown to mid-log phase in YPD medium at room temperature, and total protein was extracted by trichloroacetic acid precipitation. In the case of the ipl1-321 strain, the culture was shifted to 30° for 3 hr before sampling. Samples were run on SDS-PAGE, and Ccw14p mobility was analyzed by Western blot analysis. The ER and post-Golgi forms of the protein are indicated. (C) Known secretory mutants display accumulation of the protein at predictable points in the secretory pathway and are shown for comparison. Pgk1 is shown as loading control.
To examine protein trafficking in the disomes directly, we assessed the maturation of the inner cell-wall mannoprotein Ccw14p. In wild-type cells, Ccw14p is expressed as a GPI-linked protein that is rapidly transported by the secretory pathway to the cell surface, where it becomes covalently linked to the cell-wall glucan, causing the protein to become insoluble (Mrsa et al. 1999). Thus, in wild-type cell extracts only the small fraction of Ccw14p in transit in the secretory pathway is soluble and therefore detectable by Western blotting (Figure 5C, WT). However, in cells that have defects in protein trafficking, transport intermediates accumulate that can readily be detected by Western blot analysis. Two features of accumulation of Ccw14p transport intermediates provide a simple and versatile assay for defects in the secretory pathway. First, because Ccw14p transits the secretory pathway so rapidly in wild-type cells, even a modest kinetic delay in the secretory pathway will allow the accumulation of the corresponding intermediate to be detected. Second, because Ccw14p receives additional mannose residues in the Golgi, the intermediate form of Ccw14p that accumulates in cells with an ER-to-Golgi transport defect (e.g., erv25Δ or sec18-1) can be distinguished from the form that accumulates in cells with a Golgi transport defect (e.g., sec7-1) and from the form resulting from a defect in secretory vesicle fusion with the plasma membrane (e.g., sec6-4) (Moukadiri et al. 1997; C. A. Kaiser laboratory, unpublished data) (Figure 5C). These features allow modest, nonlethal defects in the secretory pathway to be detected and classified according to the step in the pathway that is affected. Examination of the disomes for accumulation of soluble transport intermediates of Ccw14p revealed that a number of them had significant protein-trafficking defects. We observed clear ER-to-Golgi trafficking defects in disomes X, XI, XII (note CCW14 is encoded on chromosome XII), XIII, XIV, XV, and XVI (Figure 5D). Post-Golgi trafficking defects were even more prevalent; disomes II, IV, VIII, X, XI, XII, XIII, XIV, XV, and XVI all accumulated the 120-kDa form of the protein (Figure 5D). These results cannot be explained by increased expression of the CCW14 transcripts in the disomes (Torres et al. 2007) and correlate well with the sensitivity of the disomes to Brefeldin A (Figure 2F). Together these findings indicate that protein transport defects are a frequent occurrence among disomic yeast strains. Whether protein transport-associated processes are especially sensitive to gene dosage changes, thereby leading to protein transport defects in many different disomic yeast strains, will be an important question to address in the future.
To determine whether the protein-trafficking defects observed in the disomes are also present in cells harboring more complex aneuploid karyotypes, we examined Ccw14p maturation in an ipl-321 mutant (Biggins et al. 1999). IPL1 encodes a protein essential for accurate chromosome segregation (Chan and Botstein 1993). When cells harboring the temperature-sensitive ipl1-321 allele are grown at a semipermissive temperature (30°), this mutant missegregates chromosomes at a high frequency (Oromendia et al. 2012). After growth at 30° for 3 hr, 30% of ipl1-321 cells have missegregated a GFP-marked chromosome IV. The 120-kDa form of Ccw14p also accumulated in ipl1-321 mutant cells when grown for 3 hr at the semipermissive temperature, indicating that cells harboring random aneuploidies also exhibit defects in post-Golgi trafficking (Figure 5C). We conclude that protein-trafficking defects are widespread among aneuploid yeast strains. The molecular basis for this phenotype remains to be elucidated, but our work has successfully identified new and unanticipated phenotypes of aneuploid cells, including a pathway the function of which is broadly compromised across aneuploid strains.
Discussion
Synthetic genetic array analysis of disomic yeast informs aneuploid cell biology
The unbiased, genome-wide investigation into the biology of aneuploid cells described here has elucidated novel aspects of aneuploid cell biology. We were able to identify at least 12 gene deletions that cause either disome-specific or aneuploidy-wide fitness defects despite a substantial background of false-positive synthetic negative interactions. The high number of false positives likely stems from a few main sources. First, the screen was performed with disomic strains that were hybrids between W303 and S288C because the disomes were created in the W303 background, whereas the deletion collection is of the S288C background. Validations of potential synthetic deletions were not performed in the hybrid background but rather in the pure W303 background. Thus, deletions that exhibited a negative impact only on disomic strains of the hybrid background would not have been confirmed in this manner. It is interesting to note, however, that this difference in strain backgrounds may be able to explain some of the clustering of candidate synthetic gene deletions. The W303 parental strain may carry genetic variants within these regions that can act as suppressors of the fitness defect associated with many different disomies. Moreover, false-positive genetic interactions can also be revealed in SGA screens because of the deletion collection itself. The deletion collection was generated in a high-throughput manner and thus is prone to some level of inaccuracy (Giaever et al. 2002; Ben-Shitrit et al. 2012; Giaever and Nislow 2014). Indeed, expression profiling of a subset of the deletion collection revealed aneuploidy to be a feature of 8% of deletion strains (Hughes et al. 2000). Finally, the deletion collection suffers from an artifact known as the neighboring gene effect, in which the activity of a gene of interest is often impacted by the deletion of neighboring genes (Baryshnikova and Andrews 2012; Ben-Shitrit et al. 2012). We believe that this latter artifact can account for some of the false-positive negative synthetic interactions, especially among the deletions that affect the fitness of multiple disomes that were identified as small clusters. In fact, 3 of the genes in the list of 50, the negative fitness effects of which on multiple disomes were confirmed, were all accompanied by at least two neighboring gene candidates. Regardless of these complications, we were able to detect seven chromosome-specific synthetic negative interactions and at least five deletions that confer a growth defect on multiple disomes and to identify a new aneuploidy-compromised pathway.
Protein transport pathways are prone to dosage sensitivity and exhibit synthetic fitness defects with individual disomes
We identified a strong, specific synthetic fitness defect between the deletion of any nonessential member of the COG complex and disomy XVI. The COG complex is involved in retrograde transport within the Golgi apparatus, recycling Golgi-resident proteins to their appropriate cisternal locations (Suvorova 2002; Ungar et al. 2006; Willett et al. 2013). Although deletion of any of the COG5–8 genes had little-to-no effect on the proliferation of wild-type cells, each deletion significantly slowed proliferation of disome XVI cells. This suggests a dosage imbalance with a specific gene or genes encoded on chromosome XVI or a broader defect in vesicle trafficking in this strain that is exacerbated by deletion of these factors. Consistent with the latter possibility, we found that disome XVI cells exhibit severe trafficking defects in multiple assays. This strain is sensitive to Brefeldin A, and furthermore, disome XVI cells exhibit a significant defect in the transport of Ccw14p from the ER to the Golgi. Owing to these observations and the fact that we have thus far been unable to identify a single gene on chromosome XVI the excess of which confers heightened sensitivity to deleting genes encoding COG complex subunits, we favor the hypothesis that both chromosome-specific gene effects and bulk aneuploid proteotoxicity contribute to the trafficking phenotypes of this strain; however, the precise reason why disome XVI cells are so defective in protein transport remains to be determined.
Another karyotype-specific synthetic fitness defect that we investigated in detail was the negative genetic interaction between the deletion of the endocytic factor EDE1 and disomy IX. We were able to explain this chromosome-specific genetic interaction as the result of duplication of a single gene encoded on chromosome IX, PRK1, which encodes a regulatory kinase that phosphorylates and inactivates multiple proteins involved in endocytosis. Deleting one of the two copies of PRK1 in disome IX cells abolished the synthetic interaction between the deletion of EDE1 and disomy IX and restored endocytosis activity in these cells.
In wild-type haploid yeast cells, very few genes cause significant proliferation defects when present in two copies instead of one (Sopko et al. 2006; Makanae et al. 2012; Bonney et al. 2015). Our results indicate that dosage changes can, however, have a profound impact on biological pathways that are already compromised because of genetic perturbations. An extra copy of PRK1 has a severe impact on cells deleted for EDE1. Similarly, synergistic effects between specific aneuploidies and environmental conditions that impact the activity of certain biological pathways have previously been documented. For example, particular disomies have been shown to enhance or suppress the growth of yeast cells grown under adverse environmental conditions (Pavelka et al. 2010; Chen et al. 2012, 2015). Therefore, single-gene amplifications can have significant phenotypic impacts on aneuploid cells, and SGA screens are a useful method to identify these karyotype-specific vulnerabilities.
It is especially interesting to note that certain cellular processes and molecular functions are more susceptible to dosage alterations. For example, protein trafficking is repeatedly found enriched in dosage sensitivity data sets, potentially helping to explain our COG5–8 complex result (Sopko et al. 2006; Makanae et al. 2012). Our SGA analyses also revealed genes involved in this process to exhibit synthetic interactions with diverse aneuploid karyotypes. Why protein trafficking and membrane-related processes are more sensitive to gene dosage alterations remains to be established.
Protein trafficking: a novel weakness of aneuploid cells
In addition to karyotype-specific synthetic genetic effects, we were able to identify genes the deletion of which impaired the fitness of multiple disomic strains. We identified five genes the deletion of which increases the doubling time of the majority of disomes tested. Due to the roles of multiple of these proteins in protein trafficking and localization to membranous structures including the vacuole, Golgi, and plasma membrane, we explored the status of both protein trafficking and the cell wall, the integrity of which depends heavily on this process. Interestingly, the majority of disomes exhibited defects in both cell-wall integrity and protein trafficking.
Although defects in protein transport and cell-wall stability are pervasive in aneuploid cells, different disomes exhibited different phenotypes with respect to specificity and severity. These findings indicate that protein-trafficking processes are highly sensitive to changes in gene dosage. Consistent with this idea is the finding that mutations in cell-wall components such as mannoproteins have previously been demonstrated to be dosage-sensitive, with both deletion and overexpression of these proteins causing significant cell-wall defects (Moukadiri et al. 1997). Furthermore, screens designed to globally identify dosage-sensitive genes have classified protein transport and cell-wall-related genes as enriched in these data sets (Sopko et al. 2006; Makanae et al. 2012). It is plausible that the reason why protein-trafficking defects are so pervasive among the disomic strains is because gene-specific dosage imbalances enhance mild protein-trafficking defects that are the result of increased stress on the proteostasis pathways imposed by aneuploidy. However, compromising proteasome function by chemical or genetic means alone is not sufficient to cause protein-trafficking defects (S. E. Dodgson, unpublished observations), indicating that simply increasing proteotoxic load is not sufficient to impair vesicle-mediated transport. However, synergistic effects may exist. That is, the additional proteins produced in aneuploid cells may impose stress on the protein transport machinery that is highly specific and depends on an array of precise protein interactions that are highly sensitive to dosage imbalances of their components. Regardless, we have discovered a biological pathway broadly compromised in aneuploid cells and, strikingly, our SGA data set has the potential to uncover additional unanticipated consequences of genome imbalance.
Aneuploidy-wide synthetic gene deletions open new avenues of investigation for cancer therapeutics
The vast majority of solid tumors and hematopoietic malignancies are highly aneuploid (Beroukhim et al. 2010). Although these cancer cells do not exhibit the proliferation defects associated with their unbalanced karyotypes [perhaps because they have acquired mutations that allow them to tolerate this level of aneuploidy and proliferate uncontrollably (Torres et al. 2010; Gordon et al. 2012)], they may retain aneuploidy-induced vulnerabilities such as increased sensitivity to proteotoxic agents and metabolic challenge (Williams et al. 2008; Tang et al. 2011; Santaguida et al. 2015). These general vulnerabilities of aneuploid cells, now including potential protein-trafficking defects, could represent novel therapeutic targets independent of cancer cell karyotype.
In this work, we have demonstrated the utility of an additional avenue for identifying novel cancer therapeutics. When specific karyotype changes of the tumor are known, individual chromosome-specific vulnerabilities could be exploited. A case in point is disome XVI. Disome XVI cells are especially sensitive to genetic and chemical interference with vesicle transport. Many tumor types harbor characteristic chromosome gains or losses. For example, 46% of Ewing sarcomas exhibit a gain of chromosome 8, and 33% have gained chromosome 12 (Maurici et al. 1998). Chromosome 7 gain is a recurrent characteristic of both spontaneous colorectal cancer and many glioblastomas (Lopez-Gines et al. 2005; Grade et al. 2006), and monosomy of chromosome 3 is a frequent karyotype aberration in ocular melanoma (Jovanovic et al. 2013), among other examples (reviewed in Gordon et al. 2012). Targeting chromosome-characteristic sensitivities could augment traditional therapies in these cancer types with distinguishing karyotype changes. As most basic cellular processes are highly conserved from yeast to humans, budding yeast is the ideal model system to discover such karyotype-specific vulnerabilities.
Acknowledgments
We thank Frank Solomon and members of the Amon lab for suggestions and critical reading of this manuscript. This work was supported by the National Institutes of Health (NIH) (grant GM056800 to A.A.). Work in the Boone lab is supported by grant R01HG005853 from the NIH and grants from the Canadian Institutes for Health Research (now the Foundation Scheme program). S.E.D. was supported by a Massachusetts Institute of Technology School of Science Fellowship in Cancer Research. A.A. is also an investigator of the Glenn Foundation for Biomedical Research.
Footnotes
Communicating editor: S. Biggins
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185660/-/DC1.
Literature Cited
- Abe F., Usui K., Hiraki T., 2009. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 48: 8494–8504. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A., Andrews B., 2012. Neighboring-gene effect: a genetic uncertainty principle. Nat. Methods 9: 341–343. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A., Costanzo M., Kim Y., Ding H., Koh J., et al. , 2010. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat. Methods 7: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A., VanderSluis B., Costanzo M., Myers C. L., Cha R. S., et al. , 2013. Global linkage map connects meiotic centromere function to chromosome size in budding yeast. G3 (Bethesda) 3: 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter B. K., Craig E. A., 1998. Isolation of UBP3, encoding a de-ubiquitinating enzyme, as a multicopy suppressor of a heat-shock mutant strain of S. cerevisiae. Curr. Genet. 33: 412–419. [DOI] [PubMed] [Google Scholar]
- Bell W., Klaassen P., Ohnacker M., Boller T., Herweijer M., et al. , 1992. Characterization of the 56-kDa subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur. J. Biochem. 209: 951–959. [DOI] [PubMed] [Google Scholar]
- Ben-Shitrit T., Yosef N., Shemesh K., Sharan R., Ruppin E., et al. , 2012. Systematic identification of gene annotation errors in the widely used yeast mutation collections. Nat. Methods 9: 373–378. [DOI] [PubMed] [Google Scholar]
- Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., et al. , 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., et al. , 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13: 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank H. M., Sheltzer J. M., Meehl C. M., Amon A., 2015. Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast. Mol. Biol. Cell 26: 1440–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney M. E., Moriya H., Amon A., 2015. Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage-sensitive genes. Genes Dev. 29: 898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrejon F., Gomez A., Sanz M., Durán A., Roncero C., 2006. The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot. Cell 5: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Botstein D., 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135: 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Bradford W. D., Seidel C. W., Li R., 2012. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Mulla W. A., Kucharavy A., Tsai H.-J., Rubinstein B., et al. , 2015. Targeting the adaptability of heterogeneous aneuploids. Cell 160: 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Stutz F., Belgareh N., Haguenauer-Tsapis R., Dargemont C., 2003. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat. Cell Biol. 5: 661–667. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amore T., Crumplen R., Stewart G. G., 1991. The involvement of trehalose in yeast stress tolerance. J. Ind. Microbiol. 7: 191–196. [Google Scholar]
- Dephoure N., Hwang S., O’Sullivan C., Dodgson S. E., Gygi S. P., et al. , 2014. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3: e03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinter A., Berger E. G., 1998. Golgi-disturbing agents. Histochem. Cell Biol. 109: 571–590. [DOI] [PubMed] [Google Scholar]
- Dulic V., Egerton M., Elguindi I., Raths S., Singer B., et al. , 1991. Yeast endocytosis assays. Methods Enzymol. 194: 697–710. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Cope M. J., Goode B. L., Wendland B., Drubin D. G., 2001. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3: 687–690. [DOI] [PubMed] [Google Scholar]
- Fotso P., Koryakina Y., Pavliv O., Tsiomenko A. B., Lupashin V. V., 2005. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J. Biol. Chem. 280: 27613–27623. [DOI] [PubMed] [Google Scholar]
- Gagny B., Wiederkehr A., Dumoulin P., Winsor B., Riezman H., et al. , 2000. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J. Cell Sci. 113(Pt 18): 3309–3319. [DOI] [PubMed] [Google Scholar]
- Giaever G., Nislow C., 2014. The yeast deletion collection: a decade of functional genomics. Genetics 197: 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Eskin J. A., Wendland B., 2015. Actin and endocytosis in budding yeast. Genetics 199: 315–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. J., Resio B., Pellman D., 2012. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13: 189–203. [DOI] [PubMed] [Google Scholar]
- Grade M., Becker H., Liersch T., Ried T., Ghadimi B. M., 2006. Molecular cytogenetics: genomic imbalances in colorectal cancer and their clinical impact. Cell. Oncol. 28: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., Roberts C. J., Dai H., Jones A. R., Meyer M. R., et al. , 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25: 333–337. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Nelson B., Robinson M. D., Chen Y., Andrews B., et al. , 2002. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics 162: 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic P., Mihajlovic M., Djordjevic-Jocic J., Vlajkovic S., Cekic S., et al. , 2013. Ocular melanoma: an overview of the current status. Int. J. Clin. Exp. Pathol. 6: 1230–1244. [PMC free article] [PubMed] [Google Scholar]
- Jungmann J., Rayner J. C., Munro S., 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 274: 6579–6585. [DOI] [PubMed] [Google Scholar]
- Koh J. L. Y., Ding H., Costanzo M., Baryshnikova A., Toufighi K., et al. , 2009. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res. 38: D502–D507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D. P., 2000. Modulation of fluconazole sensitivity by the interaction of mitochondria and erg3p in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 46: 191–197. [DOI] [PubMed] [Google Scholar]
- Kornberg A., Rao N. N., Ault-Riché D., 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68: 89–125. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lopez-Gines C., Cerda-Nicolas M., Gil-Benso R., Pellin A., Lopez-Guerrero J. A., et al. , 2005. Association of chromosome 7, chromosome 10 and EGFR gene amplification in glioblastoma multiforme. Clin. Neuropathol. 24: 209–218. [PubMed] [Google Scholar]
- Makanae K., Kintaka R., Makino T., Kitano H., Moriya H., 2012. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res. 23: 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurici D., Perez-Atayde A., Grier H. E., Baldini N., Serra M., et al. , 1998. Frequency and implications of chromosome 8 and 12 gains in Ewing sarcoma. Cancer Genet. Cytogenet. 100: 106–110. [DOI] [PubMed] [Google Scholar]
- Moukadiri I., Armero J., Abad A., Sentandreu R., Zueco J., 1997. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 179: 2154–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrsa V., Ecker M., Strahl-Bolsinger S., Nimtz M., Lehle L., et al. , 1999. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 181: 3076–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S. I., Hassold T. J., Hunt P. A., 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13: 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia A. B., Dodgson S. E., Amon A., 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 26: 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Cohen M., Dargemont C., 2010. The Rsp5 ubiquitin ligase and the AAA-ATPase Cdc48 control the ubiquitin-mediated degradation of the COPII component Sec23. Exp. Cell Res. 316: 3351–3357. [DOI] [PubMed] [Google Scholar]
- Pavelka N., Rancati G., Zhu J., Bradford W. D., Saraf A., et al. , 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov N. A., Kulakovskaya T. V., Kulaev I. S., 2005. Effects of inactivation of the PPN1 gene on exopolyphosphatases, inorganic polyphosphates and function of mitochondria in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 5: 823–828. [DOI] [PubMed] [Google Scholar]
- Ram A. F. J., Klis F. M., 2006. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 1: 2253–2256. [DOI] [PubMed] [Google Scholar]
- Ram R. J., Li B., Kaiser C. A., 2002. Identification of Sec36p, Sec37p, and Sec38p: components of yeast complex that contains Sec34p and Sec35p. Mol. Biol. Cell 13: 1484–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Vasile E., White E., Amon A., 2015. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 29: 2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman A., Rao N. N., Kornberg A., 2001. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98: 8542–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N., Klausner R. D., 1993. Brefeldin A reversibly inhibits secretion in Saccharomyces cerevisiae. J. Biol. Chem. 268: 5345–5348. [PubMed] [Google Scholar]
- Sheltzer J. M., Blank H. M., Pfau S. J., Tange Y., George B. M., et al. , 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. A., Lindquist S., 1998. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1: 639–648. [DOI] [PubMed] [Google Scholar]
- Smaczynska-de Rooij I. I., Allwood E. G., Mishra R., Booth W. I., Aghamohammadzadeh S., et al. , 2011. Yeast dynamin Vps1 and amphiphysin Rvs167 function together during endocytosis. Traffic 13: 317–328. [DOI] [PubMed] [Google Scholar]
- Smith R. D., Lupashin V. V., 2008. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 343: 2024–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., et al. , 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21: 319–330. [DOI] [PubMed] [Google Scholar]
- Stingele S., Stoehr G., Peplowska K., Cox J. U. R., Mann M., et al. , 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova E. S., 2002. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157: 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.-C., Williams B. R., Siegel J. J., Amon A., 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn R. R., Gonzalez C., Brar G. A., Christen S., Carlile T. M., et al. , 2013. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol. Biol. Cell 24: 1274–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H. Y., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong A. H. Y., Boone C., 2012. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313: 171–192. [DOI] [PubMed] [Google Scholar]
- Torres E. M., Sokolsky T., Tucker C. M., Chan L. Y., Boselli M., et al. , 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- Torres E. M., Dephoure N., Panneerselvam A., Tucker C. M., Whittaker C. A., et al. , 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D., Oka T., Krieger M., Hughson F. M., 2006. Retrograde transport on the COG railway. Trends Cell Biol. 16: 113–120. [DOI] [PubMed] [Google Scholar]
- Wagih O., Usaj M., Baryshnikova A., VanderSluis B., Kuzmin E., et al. , 2013. SGAtools: one-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 41: W591–W596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B. A., Cleveland D. W., 2006. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18: 658–667. [DOI] [PubMed] [Google Scholar]
- Whyte J. R., Munro S., 2001. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev. Cell 1: 527–537. [DOI] [PubMed] [Google Scholar]
- Willett R., Ungar D., Lupashin V., 2013. The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem. Cell Biol. 140: 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. R., Prabhu V. R., Hunter K. E., Glazier C. M., Whittaker C. A., et al. , 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.-Y., Friesen H., Kishimoto T., Henne W. M., Kurat C. F., et al. , 2010. Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis. Mol. Biol. Cell 21: 3054–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Cai M., 1999. Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J. Cell Biol. 144: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Cai M., 2005. Prk1p. Int. J. Biochem. Cell Biol. 37: 48–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains (Table S1) are available upon request. Table S2 contains the full SGA data set and is available for download.