Abstract
Protein phosphatase 2A (PP2A) plays a critical multi-faceted role in the regulation of the cell cycle. It is known to dephosphorylate over 300 substrates involved in the cell cycle, regulating almost all major pathways and cell cycle checkpoints. PP2A is involved in such diverse processes by the formation of structurally distinct families of holoenzymes, which are regulated spatially and temporally by specific regulators. Here, we review the involvement of PP2A in the regulation of three cell signaling pathways: wnt, mTOR and MAP kinase, as well as the G1→S transition, DNA synthesis and mitotic initiation. These processes are all crucial for proper cell survival and proliferation and are often deregulated in cancer and other diseases.
Keywords: Cancer, cell cycle, cell division, mitosis, phosphatase, PP2A
Introduction
Numerous proteins are involved in regulating the complex processes in cell division, and kinases and phosphatases are the primary regulators. Several kinases and phosphatases are well understood and reviewed in detail (Belle et al., 1990; Bononi et al., 2011; Fisher et al., 2012; Holt, 2012; Hunter, 1995; Mochida & Hunt, 2012). It is clear from early work that the initial focus on cell cycle regulation was kinases, and phosphatases were thought of merely as housekeeping enzymes. More recently, phosphatases are increasingly appreciated for their tight regulation and specific action on key players in the cell cycle (Janssens & Goris, 2001; Virshup & Shenolikar, 2009). One of the most versatile and important phosphatases involved in cell division is protein phosphatase 2A (PP2A). PP2A regulates every stage of the cell cycle in several critical pathways and, not surprisingly, has been widely implicated in tumor suppression (Eichhorn et al., 2009). As such, PP2A is being actively investigated as a therapeutic target (Sangodkar et al., 2015). This review is an attempt to aggregate the numerous substrates dephosphorylated by PP2A and discuss its regulatory activity in major pathways at each stage of the cell cycle.
Protein phosphatase 2A: a complex and diverse family of phosphatases
Background
Eukaryotic phosphatases can be divided into three super families: the serine/threonine phosphatases (PSPs), the tyrosine phosphatases (PTPs) and the dual specificity phosphatases (DSPs) [reviewed in (Shi, 2009; Hunter, 1995; Virshup & Shenolikar, 2009)]. There are around 100 PTPs, approximately equivalent to the number of tyrosine kinases in the genome. Over 400 serine/threonine kinases are expressed in the human proteome (Manning et al., 2002), exceeding that of PSPs by more than 10 fold. Serine/threonine phosphorylation constitutes more than 98% of total protein phosphorylation inside mammalian cells; however, the number of genes encoding PSPs (7) is very small. This controversy is reconciled by the fact that some of the PSPs form a large number of diverse oligomeric complexes. In particular, PP2A forms ~100 heterotrimeric holoenzymes and protein phosphatase 1 (PP1) forms ~400 heterodimeric holoenzymes. Some kinases also form oligomeric complexes, such as cell cycle dependent kinases (CDKs) and mTOR, on a scale much smaller than PP2A and PP1 though, underlying that complex and tight control of both kinases and phosphatases are important for cellular processes.
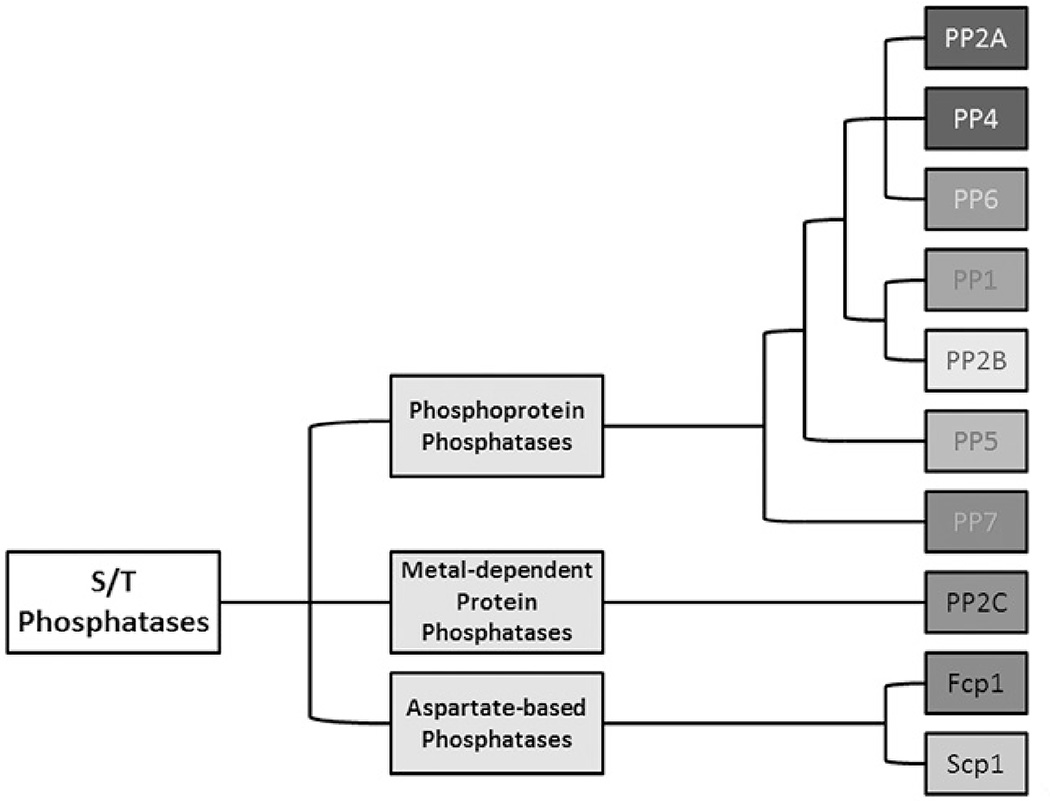
The PSPs are further divided into three families: phosphoprotein phosphatases (PPPs), metal-dependent protein phosphatases (PPMs) and aspartate-based phosphatases (Figure 1). The PPP family is the largest family of phosphatases, and many PPPs are involved in cell cycle regulation, including PP2A (Hunter, 1995; Shi, 2009; Virshup & Shenolikar, 2009). The PPP family phosphatases have a structurally conserved active site configuration. Two catalytic metal ions are coordinated by six conserved residues [two aspartate (D), one asparagine (N) and three histidine (H) residues], and a catalytic water molecule. Phosphate binding is coordinated by one conserved histidine and two arginine (R) residues. The dephosphorylation reaction proceeds via an SN2 mechanism with the activated water serving as a nucleophile to attack the phosphate group attached to Ser or Thr residues (Shi, 2009). PP2A is one of the most complex members in the PPP family, regulating diverse physiological and cellular processes such as neuronal stabilization, cardiac muscle function and the cell cycle. As such, it is implicated in many human diseases such as Alzheimer’s disease, cardiac disease and cancer (Eichhorn et al., 2009; Heijman et al., 2013; Kotlo et al., 2012; Martin et al., 2013). PP2A affects such variety of processes due to the formation of diverse heterotrimeric holoenzymes.
Figure 1.
Serine/threonine phosphatases are classified based on biochemical mechanism. They are divided into three families, the aspartate-based phosphatases, the metal-dependent protein phosphatases and the phosphoprotein phosphatases. The phosphoprotein phosphatases have similar active site configurations and require catalytic metal ions in the active site. PP2A is a member of this family. Adapted from Stanevich (2013). (see colour version of this figure at www.informahealthcare.com/bmg).
Regulation and activation of PP2A
Each PP2A holoenzyme is formed by a combination of three subunits: a common catalytic (C or PP2Ac) subunit containing the active site, a regulatory (B) subunit which confers substrate specificity and a common scaffolding (A) subunit that holds B and C together (Xu et al., 2006). There are two isoforms, α and β, for both A and C, and they share high sequence homology. The α isoform for each is expressed at a much higher level and is the predominant isoform studied in PP2A research. In addition to A and B subunits, cellular PP2Ac is also found associated with α4 protein and TOR Signaling Pathway Regulator-like (TIPRL) (Nakashima et al., 2013). Extensive efforts on understanding the structural and biochemical basis of PP2A regulation illuminated a linear pathway for the biogenesis of PP2A holoenzymes (Figure 2). The exact function of α4 on PP2Ac has been difficult to unravel. Our recent structural evidence suggests it preferentially binds to the partially folded PP2Ac and stabilizes it for stable latency (Jiang et al., 2013). α4 stabilizes PP2Ac in part by protecting it from ubiquitination by Midline 1 (MID1) and preventing its subsequent degradation (Liu et al., 2001; Short et al., 2002). This provides a pool of latent PP2Ac for the biogenesis of diverse heterotrimeric holoenzymes while simultaneously preventing the unregulated phosphatase activity of free PP2Ac and protecting cells from nontargeted dephosphorylation (Jiang et al., 2013).
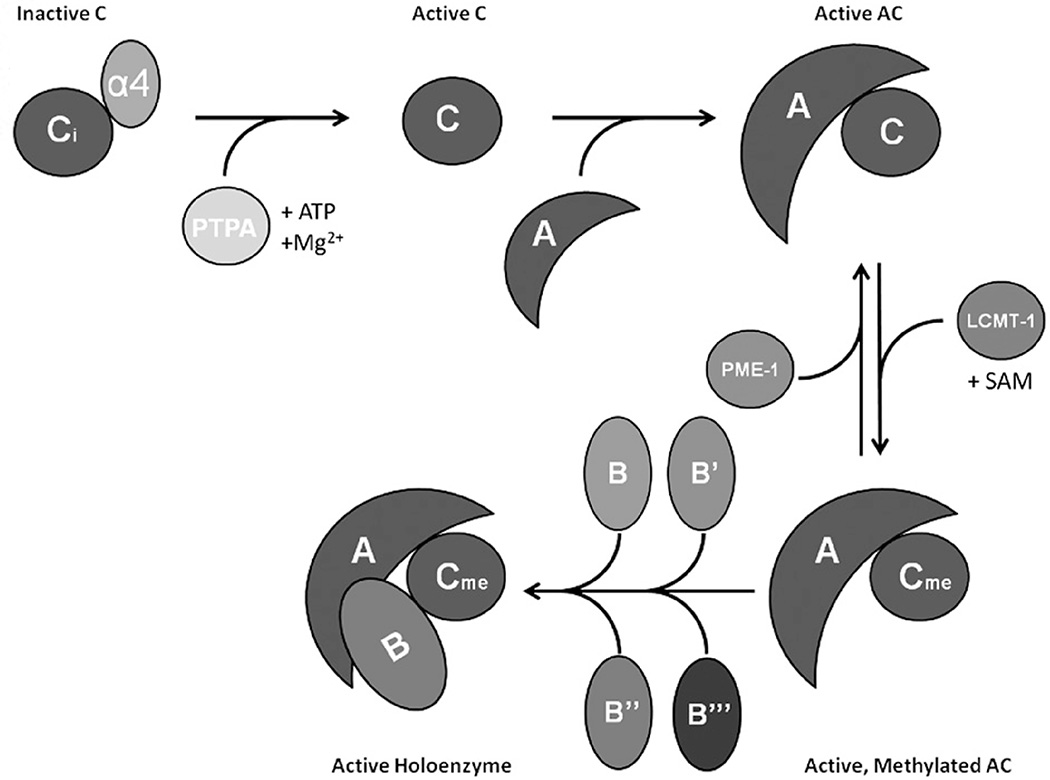
Figure 2.
PP2A biogenesis and holoenzyme assembly is regulated by unique factors. α4 protects inactive PP2Ac from ubiquitination by MID1. Activating metal ions are loaded by PTPA, and active PP2Ac binds to the scaffold subunit (A). The C-terminal tail of PP2Ac can be methylated by LCMT-1 and reversed by PME-1. Active, methylated PP2A-AC can then form holoenzymes with one B subunit. These available B subunits are divided into four families: B, B′, B″ and B‴, each with unique characteristics and regulation. (see colour version of this figure at www.informahealthcare.com/bmg).
Consequently, PP2A must be activated before being assembled into active holoenzymes. The phosphotyrosyl phosphatase activator (PTPA), now known as PP2A-specific phosphatase activator, plays a critical role in PP2A activation (Guo et al., 2014). PTPA stabilizes an active conformation of the active site and facilitates the loading of catalytic metal ions (Guo et al., 2014). PP2A together with PTPA forms a combined ATP-binding pocket, which orients the γ-phosphate of ATP to directly chelate catalytic metal ions. Following activation, the phosphatase active site catalyzes ATP hydrolysis. This is crucial for efficient loading of authentic catalytic metal ions and acquisition of pSer/Thr-specific phosphatase activity (Guo et al., 2014). Evidence suggests there is a Zn2+ ion in the active site and that ATP is required to load a Mg2+ ion into the second position to activate PP2A (Guo et al., 2014).
PP2Ac also undergoes post-translational modification on its unstructured carboxy-terminal tail (Janssens et al., 2008; Lee & Pallas, 2007); phosphorylation on T304 and Y307 and carboxymethylation on L309 (Low et al., 2014). The latter is reversibly controlled by PP2A-specific methyltransferase known as leucine carboxy methyltransferase (LCMT-1), and by PP2A-specific methylesterase 1 (PME-1) (Stanevich et al., 2011; Xing et al., 2008). PP2A methylation is essential for cellular function, and cells will undergo apoptosis in the absence of LCMT-1 (Lee & Pallas, 2007). Reduction in LCMT-1 expression or over-expression of PME-1 can promote transformation through Akt or S6K pathways (Jackson & Pallas, 2012). Methylation is crucial for the formation of stable heterotrimeric B/PR55 family holoenzymes inside cells (Longin et al., 2007), but it is not required for in vitro assembly nor is the carboxymethylated PP2Ac tail visible in the PP2A-Bα structure (Xu et al., 2008). Carboxymethylation is also not required for in vitro assembly of PP2A-B′ holoenzymes, but B′ holoenzyme structures show the carboxymethylated tail is situated in an area between the A–B interface with several negatively charged residues, suggesting a possible role of methylation in charge neutralization (Cho & Xu, 2007; Xu et al., 2006). Although PP2Ac carboxymethylation is not strictly required for holoenzyme assembly in vitro, it is clearly required for proper in vivo holoenzyme assembly (Lee & Pallas, 2007; Mumby, 2001). PP2Ac methylation also fluctuates during the cell cycle, indicating that regulation of PP2Ac methylation and holoenzyme assembly is required for cell cycle regulation (Janssens et al., 2008; Yu et al., 2001).
Structural diversity of holoenzymes
Protein phosphatase 2A (PP2A) can act on a wide range of substrates via its diverse holoenzymes, each containing a distinct B subunit from four families: the B (PR55), B′ (PR56), B″ (PR72) and B‴ (Striatin) (Shi, 2009). Currently, the identified regulatory subunits are encoded by 15 genes which can be alternatively spliced to yield 26 different B subunits (Eichhorn et al., 2009). A summary of subunit nomenclature can be found in Table 1. These subunits share sequence homology within each family, but have little-to-no sequence homology between the families (Eichhorn et al., 2009). No specific “recognition motif” has been identified for PP2A substrates, and the recognition is likely due to structural elements inherent to each subunit. The structure of the core dimer of PP2A revealed important insights on how holoenzyme assembly and activity are regulated (Xing et al., 2006). The core of PP2Ac contains two central β-sheets flanked by α-helices, with the loops connecting to the β-sheets forming the active site, and the active site loops harbor six conserved residues that chelate catalytic metal ions. The active site loops are highly dynamic (Guo et al., 2014; Jiang et al., 2013). As such, all holoenzyme (and core enzyme) structures solved to date required potent inhibitors such as microcystin LR (MCLR) or okadaic acid (OA) to stabilize the active site for crystallization (Wlodarchak et al., 2013; Xing et al., 2006; Xu et al., 2006,2008,2009).
Table 1.
A summary of PP2A subunit nomenclature.
| PP2A subunit family | Protein name/isoform | Other names |
|---|---|---|
| A | Aα | PR65α |
| Aβ | PR65β | |
| B/PR55 | Bα | B55α/PR55α |
| Bβ1 | B55β1/PR55β1 | |
| Bβ2 | B55β2/PR55β2 | |
| Bγ | B55γ/PR55γ | |
| Bδ | B55δ/PR55δ | |
| B′/PR61 | Bα | B56α/PR61α |
| Bβ | B56β/PR61β | |
| B′γ1 | B56γ1/PR61γ1 | |
| B′γ2 | B56γ2/PR61γ2 | |
| B′γ3 | B56γ3/PR61γ3 | |
| B′δ | B56δ1/PR61δ | |
| B′ε | B56ε/PR61ε | |
| B″/PR72 | B″α | PR130 |
| B″α | PR72 | |
| B″β | PR70 | |
| B″γ | G5PR | |
| B″′/Striatin | B‴ | Striatin |
| C | Cα | PP2Acα |
| Cβ | PP2Acβ |
There is a variety of abbreviations used for protein subunits in the PP2A field. Many regulatory subunits have splicing variants, such as Bβ and B′γ. Note that not all of them are shown. Table adapted from Van Kanegan & Strack (2009).
The A-subunit consists of 15 Huntington-elongation-A-subunit-TOR (HEAT) repeats arranged in a horseshoe shape. PP2Ac binds to the ridge region of repeats 10–15 and the regulatory subunits bind to the ridge of the N-terminal repeats. The A-subunit can undergo a large degree of conformational changes, explaining how so many structurally diverse B subunits can form active holoenzymes with the same A–C dimer (Wlodarchak et al., 2013). The B′γ1 holoenzyme was the first holoenzyme structure solved (Xu et al., 2006). Similar to the A-subunit, the B′γ1 subunit is also a HEAT repeat protein. The structure of the Bα holoenzyme demonstrated a much wider conformation for the A-subunit than the B′ holoenzyme, with little interaction between the B and C subunits (Xu et al., 2008). The Bα subunit is a 7-bladed β-propeller with a hairpin that extends to interact with the side face of the N-terminal HEAT repeats of the A-subunit (Xu et al., 2008). Recently, the high-resolution structure of a B″ holoenzyme associated with PR70 and two structures of B″ family subunits in isolation were finally solved (Dovega et al., 2014; Wlodarchak et al., 2013). These structures show that the B″ subunits are distinct from other families and consist of a multi-domain arrangement with two prominent calcium binding EF hands and a hydrophobic interacting motif. One of the EF hands directly contacts the top ridge of the scaffold subunit and is important for A–B″ binding (Dovega et al., 2014; Wlodarchak et al., 2013). The N-terminal hydrophobic motif binds to the N-terminal side surface of the A-subunit while the C-terminal domain interacts with PP2Ac. These tripartite contacts between AC and PR70 force the A-subunit into a tight conformation, and this is required for enhanced substrate-specific dephosphorylation (Wlodarchak et al., 2013). These observations suggest that precise orientation of substrates is dependent on subtle structural features and compact conformation of the holoenzymes derived from large conformational changes in the A-subunit. Future structural and functional studies are required to illuminate these mechanisms in more detail.
Given the diversity of holoenzyme structures, it is no surprise that PP2A has been suggested or confirmed to dephosphorylate over 300 substrates (Table 2). Most of these substrates are involved in cell cycle regulation, and although some of PP2A-mediated dephosphorylation cause positive regulation of proliferation pathways, the majority of PP2A-mediated dephosphorylation events play a negative regulatory role. PP2A is implicated in a wide array of human diseases due to its prominent function in the cell cycle and many other essential cellular processes.
Table 2.
PP2A-interacting proteins identified by literature and database searches. Reviews, Pubmed and Biogrid (Chatr-Aryamontri et al., 2015) results are presented.
The first row is the abbreviated interacting protein, the second is B subunit if known, the third is dephosphorylation site if known, the fourth is interacting boundary if known and the last row indicates the reference to the work documenting the interaction. Abbreviations were used based on Biogrid entries using Uniprot naming rules. Historical/common names are given in parenthesis where standard abbreviations are not commonly used or not easily interpretable. All proteins mentioned are human, with the exception of important human viral proteins, which have virus names indicated in [brackets].
The cell cycle initiation: signaling pathways
The initiation of the cell cycle is controlled by many diverse and complex signaling pathways. There is a large and incredibly detailed body of information on each of these signaling pathways elsewhere. Presented here is a brief review on three critical signaling pathways, Wnt, mTOR and MAPK, with a focus on the role of PP2A in their regulation.
Wnt signaling pathway
The Wnt pathway is involved in the regulation of cell proliferation and polarity as well as embryonic development. It facilitates the initiation of the cell cycle by activating the transcription of critical promoters of cell division such as cyclin D1 and c-Myc (He et al., 1998; Rimerman et al., 2000). In the absence of Wnt signaling (Figure 3A), the protein β-catenin is degraded by the action of a complex composed of axin, adenomatous polyposis coli (APC), glycogen synthase kinase 3β (GSK3β) and casein kinase 1 (CK1) [reviewed in Clevers & Nusse (2012) and MacDonald et al. (2009)]. GSK3β and CK1 phosphorylate β-catenin, targeting it for ubiquitination and proteasomal degradation (Amit et al., 2002; van Noort et al., 2002). APC and axin have unique domains that bind to CK1 and GSK3β to serve as scaffolds to increase β-catenin phosphorylation, and these scaffolds are often found mutated in cancers (Dajani et al., 2003; Kinzler et al., 1991; Nishisho et al., 1991). Wnt signaling (Figure 3B) is activated when extracellular Wnt binds to the receptor frizzled and co-receptor LRP 5/6. An intracellular complex is then formed with the receptors disheveled, axin, CK1 and GSK3β, which then prevents the phosphorylation and subsequent degradation of β-catenin (Julius et al., 2000). β-Catenin can then accumulate in the nucleus and bind to TCF family transcription factors and activate Wnt responsive genes (Behrens et al., 1996). These include critical promoters of cell division such as cyclin D1 and c-Myc (He et al., 1998; Rimerman et al., 2000).
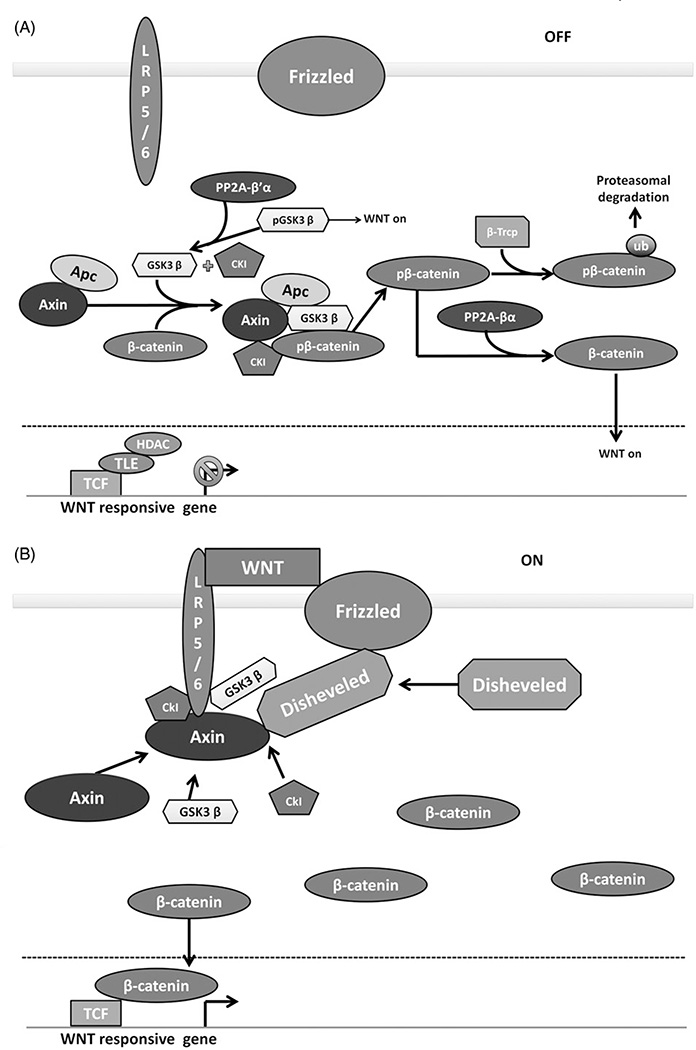
Figure 3.
PP2A positively and negatively regulates the Wnt signaling pathway. (A) Wnt OFF. In the absence of Wnt signaling, a complex of Axin, Apc, GSK3β and CK1 phosphorylate β-catenin, targeting it for proteasomal degradation. PP2A-B′α promotes β-catenin degradation by removing an inhibitory phosphorylation on GSK3β. PP2A-Bα can directly dephosphorylate β-catenin, promoting the activation of wnt responsive genes. (B) Wnt signaling ON. In the presence of Wnt ligand, Wnt receptors LRP5/6 and frizzled sequester the Axin, GSK3β and CK1, preventing the phosphorylation of β-catenin. β-Catenin accumulates and translocates to the nucleus, promoting the transcription of Wnt responsive genes. Figure adapted from MacDonald et al. (2009). (see colour version of this figure at www.informahealthcare.com/bmg).
β-Catenin is the central substrate in Wnt signaling, and its regulation is highly dependent on phosphorylation and dephosphorylation. The phosphorylation events are sequential, with CK1 phosphorylating S45 followed by GSK3β phosphorylating T41, S37 and S33 (Amit et al., 2002; van Noort et al., 2002). Phosphorylation at S37 and S33 allows the ubiquitin ligase β-transducin repeat containing protein (β-TRCP) to bind β-catenin and target it for degradation (Latres et al., 1999). In addition to phosphorylating β-catenin, CK1 and GSK3β can phosphorylate APC and axin which increases the affinity of β-catenin for these scaffolds (Ferrarese et al., 2007; Ha et al., 2004; Jho et al., 1999). These phosphorylation events are disrupted when the complex is perturbed by Wnt signaling. β-catenin, APC and axin can be dephosphorylated by phosphatases such as PP2A and PP1, and this event can also increase β-catenin levels. PP1 increases β-catenin levels by dephosphorylating axin which reduces its affinity for GSK3β (Luo et al., 2007). Unlike PP1, PP2A has a dual and opposing role in β-catenin regulation (Figure 3). The PP2A-Bα holoenzyme has been shown to directly interact with and dephosphorylate β-catenin to enhance Wnt signaling (Zhang et al., 2009). In addition to dephosphorylating the residues relevant to destruction, this holoenzyme can also dephosphorylate residues S552 and S675, the functionality of which has yet to be elucidated (Zhang et al., 2009). The B55α holoenzyme can also directly bind axin, likely through a different domain than the one that PP2Ac can bind (Zhang et al., 2009). In contrast, the PP2A B′α holoenzyme has been implicated in negative regulation of Wnt signaling (Figure 3). B′α can bind to the destruction complex through APC. Overexpression of B′α results in decreased β-catenin levels and the amino terminus of β-catenin being required for this effect (Seeling et al., 1999). In addition to β-catenin regulation, PP2A negatively regulates Wnt signaling through GSK3β both directly and indirectly (Figure 3). GSK3β is inhibited by phosphorylation on S9 by AKT (Leung-Hagesteijn et al., 2001). DNAJB6 with Heat Shock Cognate 40 (HSC40) can recruit PP2A to GSK3β where it can directly dephosphorylate S9 and activate GSK3β (Mitra et al., 2012), which targets more β-catenin for destruction (Kumar et al., 2012). PP2A also inhibits Protein Kinase B (AKT or PKB) (Kumar et al., 2012), which indirectly activates GSK3β by downregulating phosphorylation on S9. This pathway also provides an important intersection with the mTOR pathway, another critical cell cycle initiating pathway.
Mechanistic target of rapamycin (mTOR)
The mTOR pathway is involved in many diverse cellular processes. It is stimulated by amino acids, cellular metabolism, and growth factors, and results in increased growth, metabolism and biomolecule synthesis (Laplante & Sabatini, 2012). These are crucial for accumulating enough cellular components required for cell division. Due to the multifaceted role of mTOR in cell regulation, it is an intensely studied pathway with major implications in cancer, heart disease and even some neurological diseases such as autism (Chen et al., 2014; Fruman & Rommel, 2014; Laplante & Sabatini, 2012; Sciarretta et al., 2014;). Rapamycin was known to have toxic effects on yeast, and the responsible genes (DDR1&2/TOR1&2) were discovered in 1993, with the protein being discovered 1 year later (Brown et al., 1994; Cafferkey et al., 1993; Kunz et al., 1993). Two complexes are formed with the mTOR catalytic protein: mTORC1 and mTORC2 (Shimobayashi & Hall, 2014). Both complexes contain some shared as well as some unique components. The shared components are the tti1 and tel2 scaffolds, deptor and mLST8 (Shimobayashi & Hall, 2014). mTORC1 contains the unique proteins raptor and pras40, whereas mTORC2 contains rictor, mSin1 and protor1/2 (Laplante & Sabatini, 2012; Shimobayashi & Hall, 2014). Deptor is an inhibitor of both mTOR complexes and suppresses the function of S6 kinase 1 (S6K1), AKT and Serum and Glucocorticoid regulated kinase 1 (SGK1) (Peterson et al., 2009). Deptor is highly overexpressed in some multiple myelomas, and this overexpression can induce AKT function due to loss of feedback inhibition of phosphoinositide-3 kinase (PI3K) from mTORC1 (Peterson et al., 2009). Raptor and rictor help regulate substrate specificity to mTORC1 and mTORC2, respectively. Raptor binds with mTOR in the mTORC1 complex and is necessary for binding and phosphorylation of S6K1 and 4E-BP1, which in turn induce protein synthesis and proliferation (Kim et al., 2002; Nojima et al., 2003). GβL (also known as mLST8) is found in both mTORC2 and mTORC1 and is essential in stabilizing the interaction of mTOR with raptor (Kim et al., 2003; Laplante & Sabatini, 2012).
Both complexes are regulated by highly diverse processes. Wnt signaling can activate both mTOR complexes, as can stimulation by insulin (Inoki et al., 2006; Shimobayashi & Hall, 2014). Wnt can stimulate mTORC2 directly through the GTPase RAC1 and stimulates mTORC1 indirectly by inactivating glycogen synthase kinase 3β, which is necessary to activate an mTORC1 inhibitor, tuberous sclerosis complex 2 (TSC2) (Inoki et al., 2006; Shimobayashi & Hall, 2014). Insulin, long known to stimulate protein synthesis, activates mTOR through a central molecule, phosphatydilinositol-3,4,5-triphosphate, produced by PI3K (Hsu et al., 2011; Shimobayashi & Hall, 2014). This molecule can directly stimulate mTORC2 and can indirectly stimulate mTORC1 by activation of AKT which inhibits TSC2 (Klippel et al., 1997; Shimobayashi & Hall, 2014). Free amino acids activate mTORC1 via binding to RAS-related GTP binding protein (RAG) heterodimers, causing a global conformational change (Sancak et al., 2008). The RAG complex associated with amino acids can recruit mTORC1 to the lysosome where it is activated by binding to RAS homolog enriched in brain (RHEB) (Sancak et al., 2008; Shimobayashi & Hall, 2014).
Although the mTOR regulation pathways are well established, the downstream substrates of mTOR are not very well characterized. Mass spectrometric studies have identified 93 potential substrates in human embryonic kidney cells and 174 potential substrates in mouse embryonic fibroblasts (Hsu et al., 2011). Very few of these substrates have been studied in detail. The best known substrates of mTOR are the ribosomal S6 kinase (S6K) and eIF4e binding protein 1 (4E-BP) (Shimobayashi & Hall, 2014). Many proteins that are involved in growth and proliferation are encoded by mRNAs that have secondary structures in their 5′-UTR (untranslated region) that inhibit scanning by the 40S ribosomal subunit [reviewed in Ma & Blenis (2009)]. Phosphorylation of 4E-BP by mTORC1 can inhibit its binding to eIF4E, which is necessary to recruit the pre-initiation complex (Burnett et al., 1998; Ma & Blenis, 2009). An important component of the pre-initiation complex is eIF4B, which upon phosphorylation recruits eIF4A, a family of RNA helicases that facilitate efficient unwinding of secondary mRNA structures. eIF4B phosphorylation is mediated by S6K after phosphorylation and activation by mTORC1 (Burnett et al., 1998; Ma & Blenis, 2009). These mTOR substrates both work in concert to translate these structured mRNAs to enhance growth and proliferation. mTOR regulates autophagy by the phosphorylation of Unc 51-like kinase 1 (ULK1). Autophagy is promoted by ULK1 when activated by phosphorylation on S317 and S777 by AMP activated protein kinase (AMPK) (Kim et al., 2011). mTOR phosphorylates ULK1 on S757, which prevents AMPK phosphorylation and subsequent activation (Kim et al., 2011). Recently, LIPIN1, a protein that helps promote lipid biosynthesis, was identified as a potential mTOR substrate, but more work is needed to further characterize this substrate (Yuan et al., 2012).
There is evidence that PP2A can associate and dephosphorylate S6K, and the same report indicated that mTOR can inactivate PP2A, providing two modes of S6K activation (Peterson et al., 1999) (Figure 4). PP2A has also been shown to affect mTOR activity by regulating AKT. AKT, which inhibits TSC2, requires phosphorylation on T308 and S473 for activation, and the PP2A B′α holoenzyme has been shown to dephosphorylate AKT on T308, thereby inactivating it (Kuo et al., 2008) (Figure 4). Upstream of AKT, driven by insulin signaling, is insulin receptor substrate 1 (IRS1) which is necessary to transduce insulin receptor signaling to PI3K (Carlson et al., 2004). PP2A can dephosphorylate IRS1, leading to its stabilization and mTOR can inhibit PP2A activity toward IRS1 potentially directly phosphorylating IRS1 at S307, leading to IRS1 degradation (Carlson et al., 2004; Hartley & Cooper, 2002) (Figure 4).
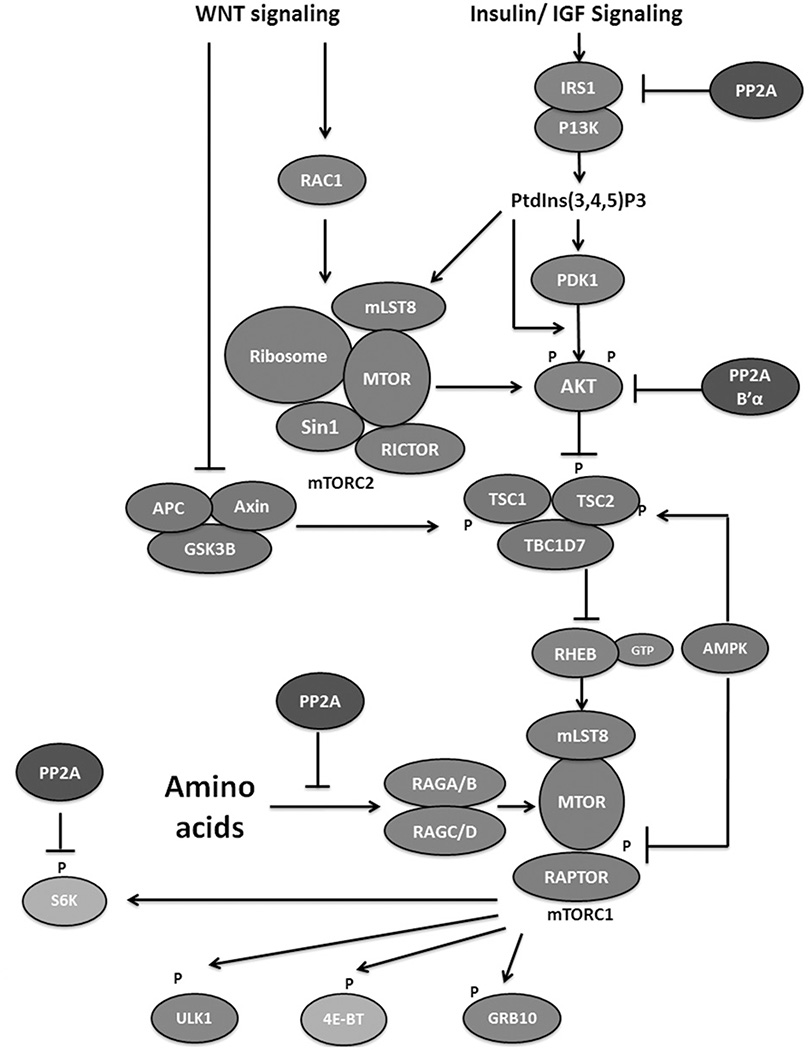
Figure 4.
PP2A negatively regulates the mTOR signaling pathway. The mTOR complexes are colored blue. Proteins involved in mTOR inhibition and activation are colored red and green, respectively with PP2A in purple. Downstream factors inhibiting and stimulating growth are colored magenta and teal, respectively. Growth factors stimulate the mTOR pathway via inhibiting the function of the TSC complex that inhibits mTOR activation. Wnt signaling can inhibit the TSC complex or directly stimulate mTORC2. Amino acids can also stimulate mTOR activity. PP2A inhibits the mTOR pathway by inhibiting IRS1 in the insulin signaling pathway or MAP4K3 in the amino acid pathway, or by inhibiting AKT function. PP2A can also reverse mTOR phosphorylation of S6K. Figure adapted from Shimobayshi & Hall (2014). (see colour version of this figure at www.informahealthcare.com/bmg).
Multiple reports have indicated that mTOR can negatively regulate PP2A activity, and most of this negative regulation supports mTOR activation through insulin signaling and PI3K. In contrast, PP2A can negatively regulate mTOR when amino acids are not present. MAP4K3 can signal to activate mTOR when amino acids are present, and autophosphorylation on S170 is necessary for this activation (Yan et al., 2010). When amino acids are withdrawn, PP2A dephosphorylates S170 and prevents mTOR activation by this pathway (Yan et al., 2010) (Figure 4).
The role of PP2A in mTOR activation is further regulated by its regulatory proteins. TIPRL can overcome amino acid withdrawal and stimulate mTOR activation by inhibiting PP2A phosphatase activity. However, the yeast homolog, TIP41, has a negative effect on mTOR activation by binding to TAP42 (Nakashima et al., 2013). In contrast to observations in yeast, the association between PP2Ac and α4, the mammalian homolog of TAP42, is not dependent on mTOR, indicating that the functions of TIPRL and α4 in mTOR signaling are not conserved and remain to be deciphered (Yoo et al., 2008).
The mTOR pathway is a critical pathway to initiate cell growth, and the regulation of this pathway is exceedingly complex, involving many feedback loops and antagonistic partnerships, especially with PP2A. mTOR has been intensely investigated and is frequently targeted for potential treatment of diseases such as cancer. Nevertheless, there are still important gaps in our understanding of mTOR substrates, regulation and crosstalk with other signaling pathways. Further study will deepen our understanding of growth signaling and possibly lead to significant drug development.
Mitogen activated protein kinase signaling pathway
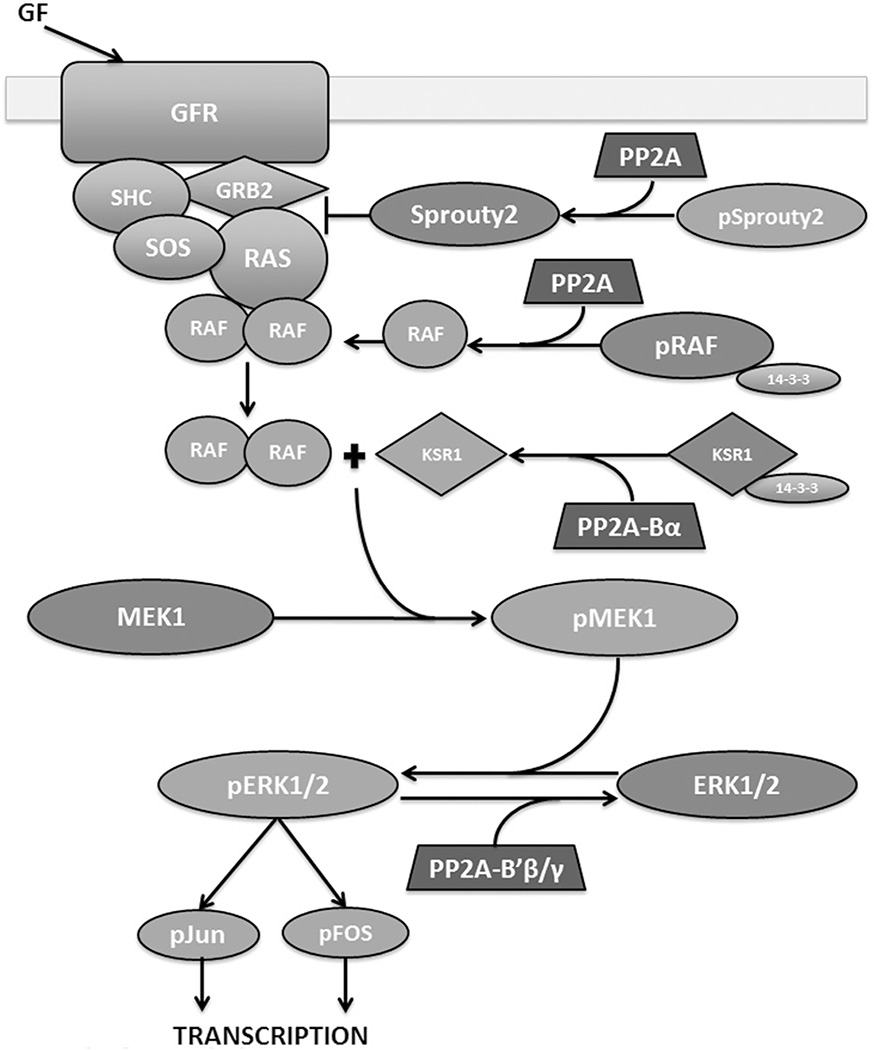
Mitogen-activated protein (MAP) kinase pathways help regulate many cellular functions such as proliferation, differentiation and apoptosis. There are four families of MAP kinases: ERK1/2, ERK5, JNK and p38 [reviewed in Hommes et al. (2003), Imajo et al. (2006), McCubrey et al. (2007), Meloche & Pouyssegur (2007)]. When activated, MAP kinases phosphorylate downstream substrates to induce cellular responses. MAPKs are activated by upstream kinases (MAPK kinases), and those MAPKKs are activated by further upstream kinases, MAPKK kinases (MAPKKKs) (Imajo et al., 2006). These kinases are activated by cellular growth signals, cytokines or stress signals. ERK5, JNK and p38 generally have pro-apoptotic functions and are activated by stresses, whereas ERK1/2 promotes proliferation and transformation (Imajo et al., 2006; Meloche & Pouyssegur, 2007; Wu, 2007). The ERK1/2 pathway was the first MAPK pathway discovered and is the best studied. Growth factors such as epidermal growth factor (EGF) or fibroblast growth factor (FGF) bind to their respective receptors and recruit a complex of SRC homology-2-containing protein (SHC), growth factor receptor bound protein 2 (GRB2) and son of sevenless (SOS) (Egan et al., 1993) (Figure 5). This complex changes Ras conformation, disrupting GDP interaction and promoting GTP association which activates Ras and recruits Raf (MAPKKK) to the membrane bound complex (Freedman et al., 2006; Milburn et al., 1990), where Raf is activated by dimerization and phosphorylation (Rajakulendran et al., 2009). Raf then phosphorylates and activates MEK1, which subsequently phosphorylates and activates ERK1/2 (Crews et al., 1992; Wu et al., 1996) (Figure 5). ERK1/2 phosphorylates transcription factors Jun and Fos which can then translocate to the nucleus and bind DNA to initiate transcription of genes involved in cell cycle regulation such as AP-1, which in turn can promote cyclin D1 expression (Monje et al., 2005; Weber et al., 1997). ERK1/2 can also phosphorylate and stabilize c-Myc, which can then enhance its transcriptional activity toward cell cycle promoting genes such as cyclin D1 and CDC25A (Mathiasen et al., 2012; Meloche & Pouyssegur, 2007).
Figure 5.
PP2A positively and negatively regulates the MAPK signaling pathway. Growth factors stimulate a complex of proteins: SHC, GRB2 and SOS to assemble on a growth factor receptor. This complex activates Ras which starts a signal cascade from activation of Raf, to activation of MEK, ERK and eventually the transcription factors that activate the transcription of growth related genes. PP2A can activate Raf by dephosphorylating S259 and causing 14–3-3 release. PP2A-Bα dephosphorylates S392 of KSR1, which leads to dissociation of 14–3-3 from KSR1, essential for MEK1 activation. PP2A negatively regulates MAPK upstream by activating Sprouty2, which inhibits GRB2 and subsequent RAS complex formation. PP2A-B′β/B′γ can directly dephosphorylate ERK1/2 downstream of the signaling cascade, thereby inactivating it. Factors promoting cell division are shown in green and those opposing cell division are shown in red. Figure adapted from McCubrey et al. (2007). (see colour version of this figure at www.informahealthcare.com/bmg).
The ERK1/2 MAPK pathway and its downstream substrates are also regulated by the action of phosphatases. There are at least 11 MAPK phosphatases (MKPs), which are split into three families based on cellular localization. There is significant cross-activity between the MKPs and all four of the MAPK pathways, and many of the MKPs, such as MKP-1 and MKP-3, have an overall transforming effect and are implicated in many cancers [reviewed in Wu (2007)]. MKP-1 has been the best studied of the MKPs, and is implicated in a variety of cancers. One of the mechanisms implicated in maintaining cell survival is its ability to prevent stress-induced apoptosis by preferentially dephosphorylating p38 and JNK, inactivating two critical stress-induced apoptotic pathways in the cell (Franklin & Kraft, 1997; Franklin et al., 1998).
Protein phosphatase 2A (PP2A) appears to have a role primarily in negative regulation of the ERK MAPK pathway (Figure 5). SHC is an important member of the complex that binds growth receptors and activates Ras (Egan et al., 1993). PP2A can bind to the phospho-tyrosine binding domain of SHC and negatively regulate Ras activation (Ugi et al., 2002). After growth factor stimulation, T317 phosphorylation of SHC can dissociate PP2A and allow downstream activation (Ugi et al., 2002). It is currently unknown whether PP2A actively dephosphorylates SHC or which regulatory subunit is responsible for PP2A’s inhibitory effect (Ugi et al., 2002). PP2A can also directly inactivate ERK by dephosphorylation (Letourneux et al., 2006) (Figure 5). This is mediated by the B′β and B′γ subunits, which can also be phosphorylated by ERK if IEX-1 is expressed, thus reversing PP2A mediated inactivation (Letourneux et al., 2006). Sprouty2 is an inhibitor of FGF stimulated ERK activation. Sprouty2 is normally phosphorylated and cannot bind Grb2, and phosphorylation on T55 allows c-Cbl to bind and target sprouty2 for degradation by the proteasome (Lao et al., 2007). Upon FGF stimulation, sprouty2 is dephosphorylated by PP2A, which exposes the Grb2 binding motif on the C-terminus (Lao et al., 2007) (Figure 5). When bound to Grb2, ras is unable to be recruited to the complex and be activated, thus downregulating ERK activation by FGF (Egan et al., 1993). PP2A binds to sprouty2 between residues 50–60, competing with c-Cbl binding and thus activating and protecting sprouty2 (Lao et al., 2007).
PP2A can also positively regulate ERK MAPK signaling. EGF receptors are targeted for ubiquitination and degradation by c-Cbl, which requires phosphorylation on various residues (McCubrey et al., 2007). This interaction is disrupted upon recruitment of SRC homology 2 domain containing inositol polyphosphate phosphatase, SHIP2 (Zwaenepoel et al., 2010). PR130, a PP2A regulatory subunit from the B″ family, forms a holoenzyme which can form a complex with SHIP2 and is required for SHIP2-mediated stabilization of EGFR (Zwaenepoel et al., 2010). Mapping studies indicate that the catalytic domain of SHIP2 interacts with the EF hands of PR130, and mutation in this region disrupts the stabilizing effect of PR130 holoenzyme on EGFR (Zwaenepoel et al., 2010). It is currently unknown whether catalytic activity is required for this effect and whether PP2A-PR130 dephosphorylates EGFR, SHIP2 or other associated targets. Downstream of growth factor receptor, the kinase suppressor of ras (KSR1) is a critical positive regulator of ras signaling (Ory et al., 2003). It is a necessary scaffold to transduce the activation signal from ras-1 to MEK to ERK. PP2A-Bα holoenzyme is associated with the KSR1 complex and is required for MEK activation (Ory et al., 2003). When phosphorylated, S392 of KSR1 associates with 14–3-3 protein and remains cytoplasmic. PP2A-Bα directly dephosphorylates KSR1 at S392 which is then freed from 14–3-3 and can translocate to the membrane, an event required for MEK activation (Ory et al., 2003) (Figure 5). Similarly, PP2A and PP1 have been shown to positively regulate Raf-1 activity by dephosphorylating S259, allowing 14–3-3 release from Raf-1 and membrane translocation (Jaumot & Hancock, 2001) (Figure 5). PP2A-B′β and -B′δ can also positively regulate MAPK signaling in neuronal PC12 cells through action on TrkA. PP2A enhances autophosphorylation of TrkA likely by dephosphorylating an inhibitory Ser/Thr, allowing Ras activation by TrkA and sustained MAPK signaling (Van Kanegan & Strack, 2009).
Taken together, the role of PP2A in regulation of MAPK pathway is complex and other as-yet-unidentified regulatory proteins may be involved. Signaling scaffolding proteins, such as KSR1, are crucial for coordinating spatiotemporal control of the function of kinases, phosphatases and other signaling molecules. The role of PP2A in both positive and negative regulation of MAPK are likely crucial for fine-tuning and precise control of this pathway. There is also crosstalk with many of these pathways as well as other cell cycle promoting pathways, highlighting the importance of phosphatases in regulating the initiation of the cell cycle and the complexity by which they do so.
Cell cycle progression: Rb and the G1-S transition
In G1 phase, cell cycle initiation pathways, such as those mentioned above, initiate growth and transcription of factors, such as cyclin D1, that control cell cycle progression. Before the cell can transition from G1 to synthesis (S) phase, it must pass through a critical cell checkpoint to ensure that the cell is ready for DNA synthesis. The master regulator of this checkpoint, and first discovered tumor suppressor protein, is the retinoblastoma tumor suppressor protein (Rb).
Rb phosphorylation
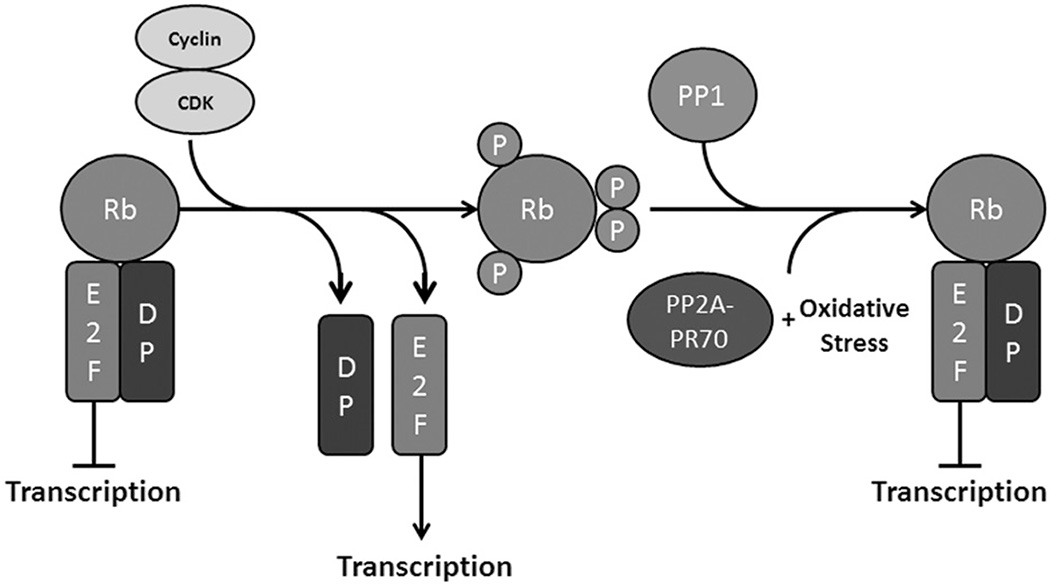
Retinoblastoma tumor suppressor protein (Rb) is an approximately 105 kDa protein consisting of three functional domains: an N-terminal structured region, a two-part central pocket region and a C-terminal unstructured region (Harbour & Dean, 2000). There are two other proteins structurally and functionally related to Rb: p107 and p130. Together, these proteins make up the pocket protein family and have all been implicated in diverse cellular processes such as cell cycle progression, apoptosis, senescence, differentiation, and angiogenesis [reviewed in Indovina et al. (2013)]. These proteins bind and inactivate E2F transcription factors, with Rb binding E2F 1–3, and p107/p130 binding E2F4 & 5 (Figure 6) (Indovina et al., 2013; Kolupaeva & Janssens, 2013). E2F 1–3 are transcriptional activators and mostly express cell cycle genes such as cyclins E and A, and CDC25 [reviewed in Harbour & Dean (2000), Indovina et al. (2013), Nevins (2001)]. E2F4 & 5 are transcriptional repressors and are involved in maintaining genomic stability and redundant functions with Rb [reviewed in Dominguez-Brauer et al. (2010) and Plesca et al. (2007)]. The pocket proteins bind to E2F transcription factors along with their dimerization partners (DPs), preventing their translocation to the nucleus and transcriptional activation (Rubin, 2013). The pocket proteins are phosphorylated by cyclin/CDK holoenzymes on numerous residues, weakening the interaction between them and the E2Fs, causing dissociation and E2F transcriptional activation [reviewed in Kolupaeva & Janssens (2013) and Rubin (2013)] (Figure 6). Rb is phosphorylated by cyclin D/CDK4 and cyclin E/CDK2 in G1, and numerous phosphorylation events gradually lead to the release of E2F transcription factors (Brown et al., 1999). One possible mechanism for this gradual release is due to the association of E2F with multiple domains of Rb (Rubin et al., 2005). The pocket domain alone is not sufficient for E2F dimerization, as the C-terminal region of Rb adopts a stable conformation upon association with E2F1-DP and increases the binding of the complex more than 36 fold (Rubin et al., 2005). Loss of this interaction pre-disposes Rb-E2F to dissociate, and this region is phosphorylated by cyclin D/CDK4/6 early in G1, thus providing a model for how sequential phosphorylation events dissociate Rb-E2F (Rubin et al., 2005). Rb levels do not change throughout the cell cycle, indicating that phosphorylation events need to be reversed to reset Rb after cell division (Kolupaeva & Janssens, 2013).
Figure 6.
Rb phosphorylation promotes transcription of E2F responsive genes. Rb normally binds E2F transcription factors and their dimerization partners. When phosphorylated by cyclin/CDK heterodimers, Rb loses affinity for E2F and free E2F is allowed to promote transcription. Rb is dephosphorylated at the end of mitosis, allowing reassociation with E2F. Normally, PP1 dephosphorylates Rb at the end of mitosis, but PP2A-PR70 can dephosphorylate Rb under oxidative stress conditions. (see colour version of this figure at www.informahealthcare.com/bmg).
Phosphatases in Rb regulation
The specific roles of kinases in Rb phosphorylation have been well established; however, the role of phosphatases in Rb regulation continues to be discovered. Protein phosphatase 1 and PP2A are the primary phosphatases that regulate Rb (and p107/130) function (Kolupaeva & Janssens, 2013). PP1 is responsible for complete dephosphorylation of Rb after mitosis, whereas PP2A functions throughout the cell cycle, dephosphorylating Rb and p107/130 in response to various stimuli [reviewed in Kolupaeva & Janssens (2013) and Kurimchak & Grana (2012)].
Protein phosphatase 1 (PP1) and PP2A are both known to dephosphorylate Rb, and PP1 appears to compete for the same CDK docking sites (Alberts et al., 1993; Hirschi et al., 2010). Such competitive interaction has also been suggested between CDKs and PP2A for p107 (Kolupaeva et al., 2013), and when CDKs are elevated they outcompete phosphatases causing an irreversible cell cycle progression signal switch. CDKs compete with phosphatases and switch the signal toward hyperphosphorylation and irreversible progression of the cell cycle (Garriga et al., 2004). In support of the competition hypothesis, specific overexpression of Bα induces p107 dephosphorylation (Jayadeva et al., 2010). Bα can directly associate with p107 but has little affinity for pRb, therefore, additional holoenzymes may be required to mediate cell cycle arrest by pocket protein activation (Jayadeva et al., 2010).
In contrast to constitutive competitive interactions, extracellular signaling or stress factors can induce PP2A-modulated dephosphorylation of the pocket proteins with no significant changes in CDK activity or PP2A expression (Cicchillitti et al., 2003). Sustained FGF signaling can arrest cell growth in chondrocytes, which is the opposite to the effect in most other cell types (Kolupaeva et al., 2013). FGF signaling leads to Bα dephosphorylation, increasing Bα affinity for p107, and chondrocytes have a large constitutively expressed Bα population (Kolupaeva et al., 2013). Phosphorylation of Bα allows increased association of PP2A-Bα holoenzymes with p107 and subsequent dephosphorylation and cell cycle arrest (Kolupaeva et al., 2013). The extracellular factor all-trans-retinoic acid (ATRA) appears to induce PP2A-specific dephosphorylation of p130 (Purev et al., 2011). Upon ATRA treatment, PP2A can bind to and dephosphorylate p130, protecting it from ubiquitination and degradation (Purev et al., 2011). PP2A can also mediate p130’s translocation to the nucleus due to dephosphorylation of S1080 and T1097, exposing the NLS and allowing binding by importins α and β (Purev et al., 2011; Soprano et al., 2006). Under oxidative stress conditions, the PP2A-PR70 holoenzyme can dephosphorylate Rb, and this activity is dependent on Ca2+ stimulation (Magenta et al., 2008) (Figure 6). One potential underlying mechanism is that oxidative stress induces an influx of Ca2+, which stimulates PR70 holoenzyme formation and results in specific Rb dephosphorylation by PP2A-PR70, although further investigation is needed.
The regulation of pocket proteins by dephosphorylation is complex and results from the interplay between competition with CDKs and specific mitogenic or stress stimuli. There may also be crosstalk between various signaling pathways in this process, as pocket proteins are central effectors through which many pathways funnel. S phase induction by Rb represents a commitment by the cell to DNA synthesis and phosphatases continue to be important in regulating this process.
DNA synthesis and regulation of the origin recognition complex
Once the cell passes the G1-S checkpoint, it is committed to the process of synthesizing DNA. The genome in eukaryotes is far too large for synthesis to proceed in a linear fashion from one end to another, so synthesis proceeds from discrete origins of replication. In yeast, these origins are defined by specific DNA sequences; however, human origins are likely defined by DNA structural features [reviewed in Hyrien et al. (2013)]. Excluding DNA recognition, the origin recognition complex functions in a conserved manner and is highly regulated.
ORC assembly and regulation
The origin recognition complex (ORC) is a large protein complex that binds to DNA at the origins of synthesis and recruits all of the proteins required to unzip and polymerize DNA [reviewed in Bell (2002) and Duncker et al. (2009)]. There are and ORC proteins (ORC1–6) that bind to DNA at the origins [referred to as autonomously replicating sequences (ARS) in yeast] (Duncker et al., 2009). These proteins all bind and hydrolyze ATP, and ATPase activity is required for their assembly and recruitment of other complex members (Bell & Stillman, 1992). In late G1/early S, cell division control 6 protein (Cdc6) binds to the ORC proteins and is the critical component for further ORC assembly (Liang et al., 1995). Cdc6 facilitates the loading of Cdt1 and ORC6 to ORC1–5 which then facilitate the loading of the mini-chromosome maintenance proteins (MCM2–7) (Nishitani et al., 2000). Cdc6 also has ATPase activity, and hydrolysis of ATP leads to conformational changes, which increases the binding affinity of the MCM proteins for the complex (Shin et al., 2003). The MCM proteins are helicases, and when properly bound they begin to unwind DNA for replication. The MCM proteins are subsequently phosphorylated by Dbf4/Cdc7 which allows the recruitment of RPA and Cdc45 to the unzipped origin, facilitating the loading of DNA polymerase which then synthesizes new DNA (Sheu & Stillman, 2006; Tanaka & Nasmyth, 1998).
Regulation of Cdc6 by phosphorylation
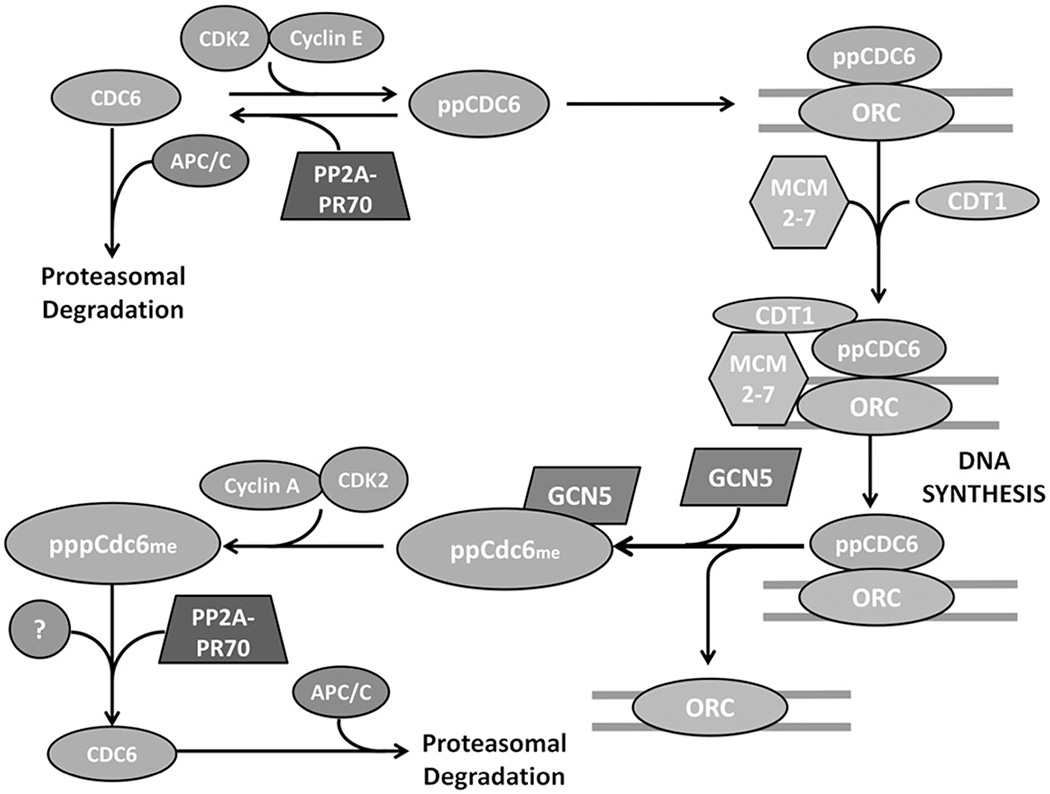
For error free cell division, DNA must be synthesized once and only once. One of the chief ways the cell regulates this process is by allowing the origins to fire only once. This restriction is accomplished by the tight regulation of Cdc6. In the absence of phosphorylation near its N-terminal destruction motifs, the anaphase-promoting complex/cyclosome (APC/C) targets Cdc6 for ubiquitination and proteasomal degradation (Mailand & Diffley, 2005). In G1, Cdc6 is phosphorylated by cyclin E/CDK2 on S54 and S74, protecting it from degradation and allowing it to be transported into the nucleus and to bind to the ORC (Jallepalli et al., 1997; Mailand & Diffley, 2005) (Figure 7). After origin firing in early S phase, Cdc6 is acetylated by general control non-derepressible 5 (GCN5) and phosphorylated by cyclin A/CDK2 on S106 (Paolinelli et al., 2009; Petersen et al., 1999). These modifications tag Cdc6 for nuclear export where it is degraded in the cytoplasm. Cdc6 degradation can only happen when the protective residues are dephosphorylated, and PP2A-PR70 has been shown to dephosphorylate Cdc6 in vivo (Davis et al., 2008). Our recent study showed that Cdc6 is specifically dephosphorylated by PP2A-PR70 holoenzyme and not others (Wlodarchak et al., 2013). PP2A-PR70 binds Cdc6 near the phosphorylated residues, likely due to a charge recognition pattern, and a compact holoenzyme conformation is critical for optimal enzymatic activity (Wlodarchak et al., 2013). The in vivo timing and location of PP2A-PR70 dephosphorylation is not fully characterized, but it likely occurs after origin firing to prevent re-assembly and possibly before origin assembly to regulate synthesis (Figure 7).
Figure 7.
Cdc6 is necessary for assembly of the pre-replication complex and subsequent DNA synthesis. In G0, Cdc6 is ubiquinated by the anaphase promoting complex/cyclosome (APC/C) and degraded by the proteasome. In G1, Cyclin E/CDK2 phosphorylates Cdc6 on S54 and S74, protecting it from degradation. Cdc6 is translocated into the nucleus where it binds the origin recognition complex and is required to recruit Cdt1 and MCM2–7 and form the prereplication complex. After firing of the origins, Cdc6 is methylated by GCN5 causing its dissociation from the ORC. Cdc6 is then phosphorylated on S106 by Cyclin A/CDK2 and translocated to the cytoplasm. PP2A-PR70 is thought to dephosphorylate Cdc6 either at this point in G2 and/or in G1, ensuring Cdc6 destruction and regulating DNA synthesis. Figure adapted from Mumby (2009). (see colour version of this figure at www.informahealthcare.com/bmg).
Mitosis: PP2A as a gatekeeper from mitotic entry to mitotic exit
Inhibition of PP2A is required for mitotic entry
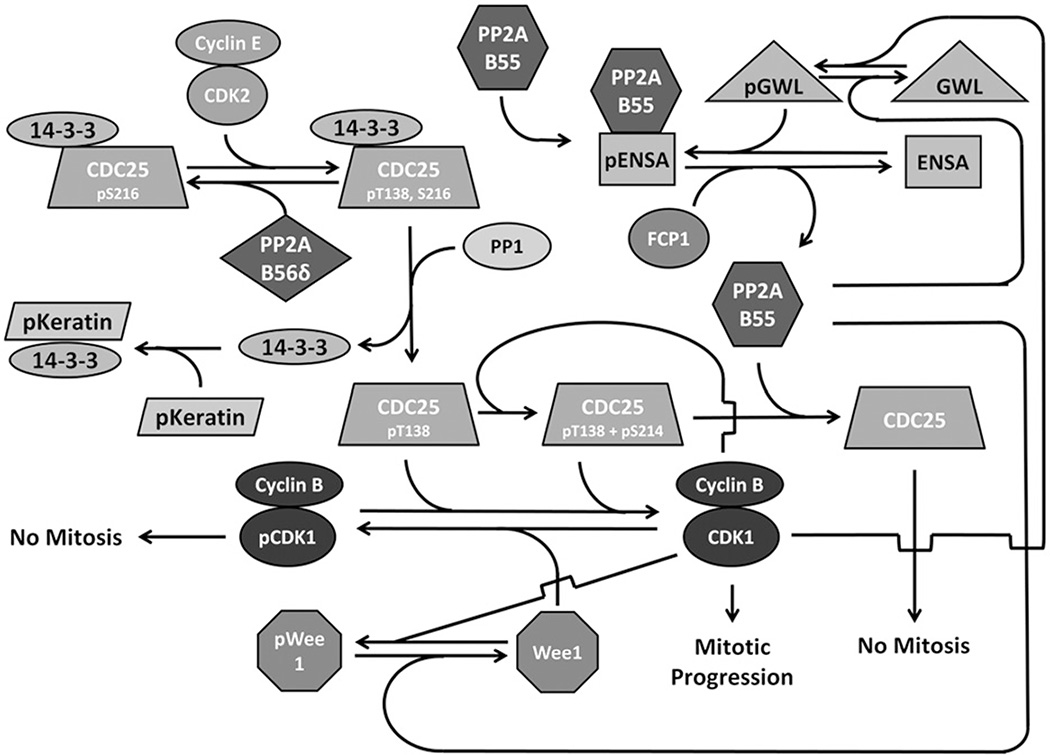
The transition from G2 to M phase is elicited by many factors and pathways, but one of the most critical events is the activation of CDK1, which is concomitant with the inactivation of PP2A-B55 holoenzyme (Mochida et al., 2009). The role of CDK1 was discovered over 40 years ago, but the complex regulatory pathways in which it is involved continue to be studied (Fisher et al., 2012). CDK1 is kept inactive by phosphorylation of S14 and Y15 by Wee1 and Myt1 (Mueller et al., 1995). During the G2→M transition, CDK1 is activated by a group of dual-specificity phosphatases, Cdc25a, b and c (herein collectively referred to as CDC25), which themselves are subject to a complex regulatory network involving several kinases and phosphatases (Lammer et al., 1998) [reviewed in Johnson & Kornbluth (2012)]. Before mitotic entry, CDC25 is phosphorylated on S216 by CaMKII and can also be phosphorylated by Chk1 to arrest the cell cycle (Hutchins et al., 2003). This allows 14–3-3 protein to associate with CDC25 and prevent its nuclear translocation (Margolis et al., 2006a). Chk1 also phosphorylates PP2A-B56δ on S37 which subsequently enhances its activity toward pT138 of CDC25, keeping CDC25 inactive (Margolis et al., 2006a). At the end of G2, CDK2-cyclin E phosphorylates CDC25 T138, decreasing the affinity of 14–3-3 to CDC25 (Margolis et al., 2006a). The decreased affinity allows gradual 14–3-3 dissociation, and the free 14–3-3 becomes bound in a phospho-keratin sink (Margolis et al., 2006a). The re-exposed S216 can then be dephosphorylated by PP1, preventing 14–3-3 re-association (Margolis et al., 2003,2006b). The now active CDC25 can dephosphorylate pT14 and pY15 of CDK2, subsequently activating it (Gautier et al., 1991). Once CDK1 is active, it can phosphorylate CDC25 at S214, enhancing the affinity of CDC25 for PP1 and causing activation of additional CDK1, leading to rapid mitotic progression (Margolis et al., 2006b) (Figure 8).
Figure 8.
PP2A negatively regulates the cell cycle through CDC25 and Wee1. In G2, Greatwall, Fcp and Wee1 are dephosphorylated, keeping PP2A-B55 active and CDK1 inactive. CDC25 is phosphorylated at S216, allowing 14–3-3 association, holding it inactive. At the transition from G2 to M, CDC25 is phosphorylated at T138, weakening 14–3-3 binding and allowing dissociation with subsequent binding to a phospho-keratin pool. The now exposed S216 can be dephosphorylated by PP1 activating CDC25. Active CDC25 dephosphorylates CDK1 at T14 and Y15 thereby activating it. The active CDK1 can then phosphorylate several substrates required for mitotic progression. In addition, CDK1 participates in several positive feedback loops. It phosphorylates Wee1, preventing direct inactivation, and it phosphorylates CDC25 at S214, increasing its affinity for PP1 and allowing for more CDC25 activation. Furthermore, CDK1 can prevent CDC25 inactivation by PP2A-B55 by phosphorylating Greatwall, which in turn phosphorylates ENSA, which binds to PP2A-B55 and keeps it inactive. CDK1 also phosphorylates FCP1, preventing it from dephosphorylating ENSA and releasing PP2A-B55. Figure adapted from Johnson & Kornbluth (2012) and Hegarat et al. (2014). (see colour version of this figure at www.informahealthcare.com/bmg).
Protein phosphatase 2A (PP2A)-B55 holoenzyme provides additional mechanism for the complex CDK1 regulation. Before mitotic initiation, PP2A-B55 dephosphorylates Wee1 and Greatwall kinase, keeping both inactive (Harvey et al., 2011; Hegarat et al., 2014). It can also dephosphorylate and subsequently inactivate CDC25 at mitotic exit (Forester et al., 2007; Johnson & Kornbluth, 2012). In addition, cyclin A-CDK2 begins to phosphorylate Greatwall kinase at T194 and activate it at the G2→M transition (Hegarat et al., 2014). Greatwall phosphorylates ENSA that subsequently binds to and inhibits PP2A-B55, preventing CDC25 repression (Mochida et al., 2010). As more CDK1 is activated, a positive feedback loop ensures PP2A-B55 inactivation. CDK1 also phosphorylates Greatwall and FCP1, keeping a majority of ENSA phosphorylated and bound to PP2A-B55 (Hegarat et al., 2014). Furthermore, CDK1 inactivates Wee1 and, without the antagonistic effect of PP2A-B55, ensures its activation (Watanabe et al., 2005). This complex network of regulation and positive feedback loops serve to inactivate PP2A while activating CDK1, rapidly driving entry into mitosis.
PP2A in reorganization of cellular structures during mitosis
Protein phosphatase 2A (PP2A) holoenzymes play a critical role in regulating reorganization of cellular structures during mitosis, including nuclear envelope breakdown, rearrangement of intracellular organelles, such as the endo plasmic reticulum and the Golgi apparatus, assembly of mitotic chromosomes, assembly of the mitotic spindle and attachment of cytoplasmic microtubules to kinetochores, which are crucial for proper partitioning of cellular materials into emerging daughter cells during cytokinesis. A significant amount of knowledge on cellular reorganization during mitosis had been reviewed (Wurzenberger & Gerlich, 2011). Here, we primarily focus on the function of PP2A in these critical processes.
Nuclear envelop breakdown and reassembly is tightly coordinated with mitotic phosphorylation and dephosphorylation (reviewed in Guttinger et al. (2009)). Nuclear envelop breakdown was facilitated by CDK1-dependent phosphorylation of lamin proteins and subsequent disassembly of the nuclear lamina (Peter et al., 1990), and phosphorylation of nucleoporins-mediated disassembly of nuclear pore complexes (Laurell et al., 2011). PP2A (Schmitz et al., 2010) and its closely related PP1 (Thompson et al., 1997) play a role in nuclear envelope reassembly during mitotic exit with unclear mechanisms. It remains to be determined whether lamin and nucleoporins are the specific substrates of PP2A. Disassembly and reassembly of the Golgi apparatus is also driven by mitotic phosphorylation and dephosphorylation. While phosphorylation of Golgi matrix protein GM130 induces disassembly (Wei & Seemann, 2009), PP2A-mediated dephosphorylation of GM130 induces Golgi reassembly during mitotic exit, which involves PP2A-B55α holoenzyme (Lowe et al., 2000; Schmitz et al., 2010).
Mitotic chromosomal assembly is regulated by condensin I and condensin II, which belongs to a class of conserved condensin complexes that play essential roles in mitotic chromosome condensation by collaborating with other chromosomal components (Hagstrom & Meyer, 2003; Jessberger, 2002). PP2A interacts with condensin II and plays an essential role in targeting condensin II to chromosomes (Takemoto et al., 2009). Intriguingly, this process does not require the phosphatase activity of PP2A (Takemoto et al., 2009). Chromatin decondensation requires PP1 and its regulatory subunits Repo-Man (recruits PP1 onto mitotic chromatin at anaphase protein) and PNUTS (phosphatase 1 nuclear targeting subunit) (Landsverk et al., 2005; Vagnarelli et al., 2006). The role of PP2A in this process is less characterized. Nonetheless, a recent study showed that a midzone-associated Aurora B gradient monitors chromosome position along the division axis and to prevent premature chromosome decondensation by retaining Condensin I until effective separation of sister chromatids is achieved (Afonso et al., 2014). Both PP1 and PP2A phosphatases counteract this gradient and promoted chromosome decondensation (Afonso et al., 2014).
Proper kinetochore–microtubule attachments are tightly controlled by Aurora B-mediated phosphorylation and PP2A/PP1-mediated dephosphorylation. Aurora B phosphorylates multiple substrates at the kinetochore to destabilize and correct erroneous kinetochore–microtubule attachments (Welburn et al., 2010). While PP1 is considered the major phosphatase counteracting Aurora B (Carmena et al., 2012), the PP2A-B56α holoenzyme also plays a critical role in stabilizing kinetochore–microtubule attachments by counteracting Aurora B phosphorylation (Foley et al., 2011). Pseudokinase BUBR1 seems to play a critical role in integration of kinase and phosphatase activities to ensure proper formation of stable kinetochore–microtubule attachments (Suijkerbuijk et al., 2012). Phosphorylation of a conserved KARD domain n BUBR1 by PLK1 (polo-like kinase 1) promotes direct interaction of BUBR1 with the PP2A-B56α phosphatase (Suijkerbuijk et al., 2012), a potential mechanism for the recruitment of PP2A-B56α to the inner kinetochore prior to microtubule attachment (Foley et al., 2011). Removal of BUBR1 from mitotic cells or inhibition of PLK1 reduces PP2A-B56α kinetochore binding (Suijkerbuijk et al., 2012), suggesting that PLK1 and BUBR1 cooperate to stabilize kinetochore–microtubule interactions by regulating kinetochore localization of the PP2A-B56α holoenzyme.
PP2A in spindle checkpoint, regulation of APC/C-CDC20 and mitotic exit
The rise in APC/C-CDC20 activity initiates mitotic exit by targeting several mitotic determinants for degradation, resulting in the formation and separation of two interphase daughter cells. APC/C is kept inactive by the spindle assembly checkpoint until all chromosomes attach to microtubules originating from opposite spindle poles [reviewed in Musacchio & Salmon (2007)]. The early mitotic inhibitor 1 (Emi1) and 2 (Emi2) play a critical role in inhibition of APC/C, and PP2A-B56 holoenzymes was found to promote the inhibitory activity of Emi2 and maintain the spindle assembly checkpoint, which was antagonized by the activity of CDK1 (Tischer et al., 2012). CDK1-mediated phosphorylation of APC/C inhibitory proteins primes its own inactivation. At mitotic exit, APC/C-CDC20 induces proteasomal destruction of cyclin B, and inactivates mitotic CDK1 (Sullivan & Morgan, 2007). Inactivation of CDK1 is expected to lead to reactivation of PP2A-B55 to mediate dephosphorylation of CDK1 substrates at mitotic exit.
Adenomatous polyposis coli (APC)/C-CDC20 also mediates the degradation of securin to initiate chromosome segregation. Securin inhibits the protease separase; removal of securin allows separase to cleave the sister chromatid cohesion 1 (SCC1) subunit of the cohesin complex (Sullivan & Morgan, 2007). Cohesin function is also regulated by PLK1 and PP2A. PLK1 promotes dissociation of cohesin from chromosome arms by phosphorylating the cohesin subunit SA2 during prometaphase (Sumara et al., 2002). Shugoshin 1 recruits PP2A–B56 to protect SA2 against PLK1-mediated phosphorylation and thereby maintains a pool of persistent cohesion, and prevents premature separation of sister chromatids (Kitajima et al., 2006; Tang et al., 2006; Tanno et al., 2010; Xu et al., 2009).
With limited information on specific substrates targeted by diverse PP2A holoenzymes, the above knowledge likely merely represents a small fraction of PP2A function during mitosis. More questions need to be addressed regarding how PP2A holoenzyme activity is temporally and spatially controlled for tight regulation of numerous events during mitosis.
Concluding remarks
The cell cycle harbors complex and intricate processes and may be the most studied aspect in biology. This highlights its importance in understanding the origins of most of human disease and what can be done to intervene for therapeutic purposes. Great strides have been made in understanding the complex players of the cell cycle, and the importance of regulation by reversible phosphorylation cannot be underestimated.
The role of PP2A in regulating the cell cycle is only beginning to be investigated. It is involved in most major cell cycle initiation pathways as well as in regulating major checkpoints during cell cycle phase transitions. PP2A is implicated in dephosphorylating many more cell cycle pathway substrates than could be discussed here (Table 2), further highlighting its importance to properly functioning cells. The kinases involved in regulating the cell cycle typically exert their action through transcription-level changes and/or regulation of protein stability, whereas the cellular level of PP2A scaffold and catalytic subunits are stable throughout the cell cycle. Although PP2A expression differs from that leading to “canonical” kinase regulation, PP2A is one of the primary cell cycle regulating enzymes due to the dynamic nature of its holoenzyme assembly, activation and inhibition. As discussed, it is a major target in several key pathways, both for protein activation and inactivation, and it is frequently targeted for inhibition due to its antagonistic effects in these pathways. Although kinases may have taken center stage in the study of cell cycle regulation, the intricate connectivity between these kinases and PP2A shows its importance in the tight regulation of these processes.
In addition to its wide involvement in cellular processes, PP2A is a complex group of enzymes, and its assembly and activity are highly regulated making it challenging to study. No PP2A-substrate binding consensus sequence has been identified yet, indicating that structural and biochemical information is required to understand the mechanisms by which PP2A regulates substrate dephosphorylation. Currently, high-throughput assays are being explored to identify substrates and characterize protein–protein interactions significantly faster than traditional methods. These large scale assays combined with structural and biochemical characterization will provide an unprecedented amount of information to the PP2A field and possibly identify new targets of cell cycle regulation. These new targets may be incredibly useful in developing drugs or biomarkers for preventing, diagnosing or treating human disease, and more refined knowledge on existing PP2A-substrate interactions may help improve current treatments.
Acknowledgments
Declaration of interest
This project is supported by NIH RO1 GM096060–01 (Y. Xing). N. Wlodarchak is supported by NIH T32 (CA009135) and the Hartwell Foundation (133-PRJ24TF).
References
- Abdelmohsen K, Srikantan S, Yang X, et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DG, Coffee RL, Jr, Zhang H, et al. Positive regulation of Raf1-MEK1/2-ERK1/2 signaling by protein serine/threonine phosphatase 2A holoenzymes. J Biol Chem. 2005;280:42644–42654. doi: 10.1074/jbc.M502464200. [DOI] [PubMed] [Google Scholar]
- Adler HT, Nallaseth FS, Walter G, Tkachuk DC. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J Biol Chem. 1997;272:28407–28414. doi: 10.1074/jbc.272.45.28407. [DOI] [PubMed] [Google Scholar]
- Afonso O, Matos I, Pereira AJ, et al. Feedback control of chromosome separation by a midzone Aurora B gradient. Science. 2014;345:332–336. doi: 10.1126/science.1251121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JH, Mcavoy T, Rakhilin SV, et al. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci USA. 2007a;104:2979–2984. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JH, Sung JY, Mcavoy T, et al. The B″/PR72 subunit mediates Ca2+-dependent dephosphorylation of DARPP-32 by protein phosphatase 2A. Proc Natl Acad Sci USA. 2007b;104:9876–9881. doi: 10.1073/pnas.0703589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichem A, Kalveram B, Spinnenhirn V, et al. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci. 2012;125:4576–4585. doi: 10.1242/jcs.107789. [DOI] [PubMed] [Google Scholar]
- Alberts AS, Thorburn AM, Shenolikar S, et al. Regulation of cell cycle progression and nuclear affinity of the retinoblastoma protein by protein phosphatases. Proc Natl Acad Sci USA. 1993;90:388–392. doi: 10.1073/pnas.90.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado-Kristensson M, Andersson T. Protein phosphatase 2A regulates apoptosis in neutrophils by dephosphorylating both p38 MAPK and its substrate caspase 3. J Biol Chem. 2005;280:6238–6244. doi: 10.1074/jbc.M409718200. [DOI] [PubMed] [Google Scholar]
- Amit S, Hatzubai A, Birman Y, et al. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammosova T, Washington K, Debebe Z, et al. Dephosphorylation of CDK9 by protein phosphatase 2A and protein phosphatase-1 in Tat-activated HIV-1 transcription. Retrovirology. 2005;2:47. doi: 10.1186/1742-4690-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle MI, Komiyama NH, Delaney A, et al. The SH3 domain of postsynaptic density 95 mediates inflammatory pain through phosphatidylinositol-3-kinase recruitment. EMBO Rep. 2010;11:473–478. doi: 10.1038/embor.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold HK, Sears RC. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol. 2006;26:2832–2844. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold HK, Zhang X, Daniel CJ, et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J. 2009;28:500–512. doi: 10.1038/emboj.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asencio C, Davidson IF, Santarella-Mellwig R, et al. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 2012;150:122–135. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]
- Avdi NJ, Malcolm KC, Nick JA, Worthen GS. A role for protein phosphatase-2A in p38 mitogen-activated protein kinase-mediated regulation of the c-Jun NH(2)-terminal kinase pathway in human neutrophils. J Biol Chem. 2002;277:40687–40696. doi: 10.1074/jbc.M204455200. [DOI] [PubMed] [Google Scholar]
- Baroja ML, Vijayakrishnan L, Bettelli E, et al. Inhibition of CTLA-4 function by the regulatory subunit of serine/threonine phosphatase 2A. J Immunol. 2002;168:5070–5078. doi: 10.4049/jimmunol.168.10.5070. [DOI] [PubMed] [Google Scholar]
- Bauman AL, Apparsundaram S, Ramamoorthy S, et al. Cocaine and antidepressant-sensitive biogenic amine transporters exist in regulated complexes with protein phosphatase 2A. J Neurosci. 2000;20:7571–7578. doi: 10.1523/JNEUROSCI.20-20-07571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Sotnikova TD, Marion S, et al. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell. 2005;122:261–273. doi: 10.1016/j.cell.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Behrens J, Von Kries JP, Kuhl M, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Belle R, Cormier P, Poulhe R, et al. Protein phosphorylation during meiotic maturation of Xenopus oocytes: cdc2 protein kinase targets. Int J Dev Biol. 1990;34:111–115. [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennin DA, Don AS, Brake T, et al. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B’ subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J Biol Chem. 2002;277:27449–27467. doi: 10.1074/jbc.M111693200. [DOI] [PubMed] [Google Scholar]
- Bishop JD, Nien WL, Dauphinee SM, Too CK. Prolactin activates mammalian target-of-rapamycin through phosphatidylinositol 3-kinase and stimulates phosphorylation of p70S6K and 4E-binding protein-1 in lymphoma cells. J Endocrinol. 2006;190:307–312. doi: 10.1677/joe.1.06368. [DOI] [PubMed] [Google Scholar]
- Bisson N, James DA, Ivosev G, et al. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011;29:653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- Bobulescu IA, Quinones H, Gisler SM, et al. Acute regulation of renal Na+/H+ exchanger NHE3 by dopamine: role of protein phosphatase 2A. Am J Physiol Renal Physiol. 2010;298:F1205–F1213. doi: 10.1152/ajprenal.00708.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bononi A, Agnoletto C, De Marchi E, et al. Protein kinases and phosphatases in the control of cell fate. Enzyme Res. 2011;2011:329098. doi: 10.4061/2011/329098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsotto M, Cavarec L, Bouillot M, et al. PP2A-Bgamma subunit and KCNQ2 K + channels in bipolar disorder. Pharmacogenomics J. 2007;7:123–132. doi: 10.1038/sj.tpj.6500400. [DOI] [PubMed] [Google Scholar]
- Breitman M, Zilberberg A, Caspi M, Rosin-Arbesfeld R. The armadillo repeat domain of the APC tumor suppressor protein interacts with Striatin family members. Biochim Biophys Acta. 2008;1783:1792–1802. doi: 10.1016/j.bbamcr.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Brown VD, Phillips RA, Gallie BL. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol. 1999;19:3246–3256. doi: 10.1128/mcb.19.5.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, et al. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferkey R, Young PR, Mclaughlin MM, et al. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Gerrits B, Marra G, et al. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J Biol Chem. 2007;282:2976–2986. doi: 10.1074/jbc.M609989200. [DOI] [PubMed] [Google Scholar]
- Carlson CJ, White MF, Rondinone CM. Mammalian target of rapamycin regulates IRS-1 serine 307 phosphorylation. Biochem Biophys Res Commun. 2004;316:533–539. doi: 10.1016/j.bbrc.2004.02.082. [DOI] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SF, Sucher NJ. An NMDA receptor signaling complex with protein phosphatase 2A. J Neurosci. 2001;21:7985–7992. doi: 10.1523/JNEUROSCI.21-20-07985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GI, Tisayakorn S, Jorgensen C, et al. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J Biol Chem. 2008;283:29273–29284. doi: 10.1074/jbc.M803443200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Yang CC, Hsieh CL, et al. A functional genomic approach reveals the transcriptional role of EDD in the expression and function of angiogenesis regulator ACVRL1. Biochim Biophys Acta. 2013;1829:1309–1319. doi: 10.1016/j.bbagrm.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Chen J, Alberts I, Li X. Dysregulation of the IGF-I/PI3K/AKT/mTOR signaling pathway in autism spectrum disorders. Int J Dev Neurosci. 2014;35:35–41. doi: 10.1016/j.ijdevneu.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, et al. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Chuang JY, Wang SA, Yang WB, et al. Sp1 phosphorylation by cyclin-dependent kinase 1/cyclin B1 represses its DNA-binding activity during mitosis in cancer cells. Oncogene. 2012;31:4946–4959. doi: 10.1038/onc.2011.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchillitti L, Fasanaro P, Biglioli P, et al. Oxidative stress induces protein phosphatase 2A-dependent dephosphorylation of the pocket proteins pRb, p107, and p130. J Biol Chem. 2003;278:19509–19517. doi: 10.1074/jbc.M300511200. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Colland F, Jacq X, Trouplin V, et al. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JL, Wilder PJ, Gilmore JM, et al. The SOX2-interactome in brain cancer cells identifies the requirement of MSI2 and USP9X for the growth of brain tumor cells. PLoS One. 2013;8:e62857. doi: 10.1371/journal.pone.0062857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews CM, Alessandrini A, Erikson RL. The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science. 1992;258:478–480. doi: 10.1126/science.1411546. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Roel G, Eichhorn PJ, et al. PR72, a novel regulator of Wnt signaling required for Naked cuticle function. Genes Dev. 2005;19:376–386. doi: 10.1101/gad.328905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Roel G, Eichhorn PJ, et al. PR130 is a modulator of the Wnt-signaling cascade that counters repression of the antagonist Naked cuticle. Proc Natl Acad Sci USA. 2006;103:5397–5402. doi: 10.1073/pnas.0507237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossthwaite AJ, Ciruela A, Rayner TF, Cooper DM. A direct interaction between the N terminus of adenylyl cyclase AC8 and the catalytic subunit of protein phosphatase 2A. Mol Pharmacol. 2006;69:608–617. doi: 10.1124/mol.105.018275. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Barwacz CA, Cribbs JT, Strack S. Unfolding-resistant translocase targeting: a novel mechanism for outer mitochondrial membrane localization exemplified by the Bbeta2 regulatory subunit of protein phosphatase 2A. J Biol Chem. 2005;280:27375–27382. doi: 10.1074/jbc.M503693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, et al. Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. EMBO J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Yan Z, Martinez B, Mumby MC. Protein phosphatase 2A is targeted to cell division control protein 6 by a calcium-binding regulatory subunit. J Biol Chem. 2008;283:16104–16114. doi: 10.1074/jbc.M710313200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehde S, Rohaly G, Schub O, et al. Two immunologically distinct human DNA polymerase alpha-primase subpopulations are involved in cellular DNA replication. Mol Cell Biol. 2001;21:2581–2593. doi: 10.1128/MCB.21.7.2581-2593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobierzewska A, Giltiay NV, Sabapathi S, et al. Protein phosphatase 2A and neutral sphingomyelinase 2 regulate IRAK-1 protein ubiquitination and degradation in response to interleukin-1beta. J Biol Chem. 2011;286:32064–32073. doi: 10.1074/jbc.M111.238030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge-Kafka KL, Bauman A, Mayer N, et al. cAMP-stimulated protein phosphatase 2A activity associated with muscle A kinase-anchoring protein (mAKAP) signaling complexes inhibits the phosphorylation and activity of the cAMP-specific phosphodiesterase PDE4D3. J Biol Chem. 2010;285:11078–11086. doi: 10.1074/jbc.M109.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohoney KM, Guillerm C, Whiteford C, et al. Phosphorylation of p53 at serine 37 is important for transcriptional activity and regulation in response to DNA damage. Oncogene. 2004;23:49–57. doi: 10.1038/sj.onc.1207005. [DOI] [PubMed] [Google Scholar]
- Dominguez-Brauer C, Brauer PM, Chen YJ, et al. Tumor suppression by ARF: gatekeeper and caretaker. Cell Cycle. 2010;9:86–89. doi: 10.4161/cc.9.1.10350. [DOI] [PubMed] [Google Scholar]
- Dovega R, Tsutakawa S, Quistgaard EM, et al. Structural and biochemical characterization of human PR70 in isolation and in complex with the scaffolding subunit of protein phosphatase 2A. PLoS One. 2014;9:e101846. doi: 10.1371/journal.pone.0101846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Chesnokov IN, Mcconkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan SE, Giddings BW, Brooks MW, et al. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature. 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Creyghton MP, Wilhelmsen K, et al. A RNA interference screen identifies the protein phosphatase 2A subunit PR55gamma as a stress-sensitive inhibitor of c-SRC. PLoS Genet. 2007;3:e218. doi: 10.1371/journal.pgen.0030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan GH, Yang W, Sai J, Richmond A. Phosphorylationin-dependent association of CXCR2 with the protein phosphatase 2A core enzyme. J Biol Chem. 2001;276:16960–16968. doi: 10.1074/jbc.M009292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarese A, Marin O, Bustos VH, et al. Chemical dissection of the APC Repeat 3 multistep phosphorylation by the concerted action of protein kinases CK1 and GSK3. Biochemistry. 2007;46:11902–11910. doi: 10.1021/bi701674z. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Howard MJ, Mcdaid JR, et al. PKA, PKC, and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue-specific transcriptional regulation. Mol Cell. 2003;12:1225–1237. doi: 10.1016/s1097-2765(03)00425-8. [DOI] [PubMed] [Google Scholar]
- Fisher D, Krasinska L, Coudreuse D, Novak B. Phosphorylation network dynamics in the control of cell cycle transitions. J Cell Sci. 2012;125:4703–4711. doi: 10.1242/jcs.106351. [DOI] [PubMed] [Google Scholar]
- Foerster S, Kacprowski T, Dhople VM, et al. Characterization of the EGFR interactome reveals associated protein complex networks and intracellular receptor dynamics. Proteomics. 2013;13:3131–3144. doi: 10.1002/pmic.201300154. [DOI] [PubMed] [Google Scholar]
- Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester CM, Maddox J, Louis JV, et al. Control of mitotic exit by PP2A regulation of Cdc25C and Cdk1. Proc Natl Acad Sci USA. 2007;104:19867–19872. doi: 10.1073/pnas.0709879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin CC, Kraft AS. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-activated protein kinase in U937 cells. J Biol Chem. 1997;272:16917–16923. doi: 10.1074/jbc.272.27.16917. [DOI] [PubMed] [Google Scholar]
- Franklin CC, Srikanth S, Kraft AS. Conditional expression of mitogen-activated protein kinase phosphatase-1, MKP-1, is cytoprotective against UV-induced apoptosis. Proc Natl Acad Sci USA. 1998;95:3014–3019. doi: 10.1073/pnas.95.6.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman TS, Sondermann H, Friedland GD, et al. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc Natl Acad Sci USA. 2006;103:16692–16697. doi: 10.1073/pnas.0608127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz A, Brayer KJ, Mccormick N, et al. Phosphorylation of serine 526 is required for MEKK3 activity, and association with 14-3-3 blocks dephosphorylation. J Biol Chem. 2006;281:6236–6245. doi: 10.1074/jbc.M509249200. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DX, Kuo YL, Liu BY, et al. Human T-lymphotropic virus type I tax activates I-kappa B kinase by inhibiting I-kappa B kinase-associated serine/threonine protein phosphatase 2A. J Biol Chem. 2003;278:1487–1493. doi: 10.1074/jbc.M210631200. [DOI] [PubMed] [Google Scholar]
- Fuhrer DK, Yang YC. Complex formation of JAK2 with PP2A, P13K, and Yes in response to the hematopoietic cytokine interleukin-11. Biochem Biophys Res Commun. 1996;224:289–296. doi: 10.1006/bbrc.1996.1023. [DOI] [PubMed] [Google Scholar]
- Galea MA, Eleftheriou A, Henderson BR. ARM domain-dependent nuclear import of adenomatous polyposis coli protein is stimulated by the B56 alpha subunit of protein phosphatase 2A. J Biol Chem. 2001;276:45833–45839. doi: 10.1074/jbc.M107149200. [DOI] [PubMed] [Google Scholar]
- Garriga J, Jayaraman AL, Limon A, et al. A dynamic equilibrium between CDKs and PP2A modulates phosphorylation of pRB, p107 and p130. Cell Cycle (Georgetown, Tx) 2004;3:1320–1330. doi: 10.4161/cc.3.10.1183. [DOI] [PubMed] [Google Scholar]
- Gautier J, Solomon MJ, Booher RN, et al. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- Gil-Bernabe AM, Romero F, Limon-Mortes MC, Tortolero M. Protein phosphatase 2A stabilizes human securin, whose phosphorylated forms are degraded via the SCF ubiquitin ligase. Mol Cell Biol. 2006;26:4017–4027. doi: 10.1128/MCB.01904-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jonnalagadda JC, Douglas P, et al. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J, Hwang J, Carrier KJ, et al. Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 2011;12:54. doi: 10.1186/1471-2091-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz J, Probst A, Mistl C, et al. Distinct role of protein phosphatase 2A subunit Calpha in the regulation of E-cadherin and beta-catenin during development. Mech Dev. 2000;93:83–93. doi: 10.1016/s0925-4773(00)00267-7. [DOI] [PubMed] [Google Scholar]
- Grant MM. Identification of SUMOylated proteins in neuroblastoma cells after treatment with hydrogen peroxide or ascorbate. BMB Rep. 2010;43:720–725. doi: 10.5483/BMBRep.2010.43.11.720. [DOI] [PubMed] [Google Scholar]
- Greco TM, Yu F, Guise AJ, Cristea IM. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10:M110.004317. doi: 10.1074/mcp.M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold-Prenner I, Kamibayashi C, Maruoka EM, et al. Physical and functional interactions between type I transforming growth factor beta receptors and Balpha, a WD-40 repeat subunit of phosphatase 2A. Mol Cell Biol. 1998;18:6595–6604. doi: 10.1128/mcb.18.11.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Stanevich V, Wlodarchak N, et al. Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone. Cell Res. 2014;24:190–203. doi: 10.1038/cr.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Song E, Ma S, et al. Proteomics strategy to identify substrates of LNX, a PDZ domain-containing E3 ubiquitin ligase. J Proteome Res. 2012;11:4847–4862. doi: 10.1021/pr300674c. [DOI] [PubMed] [Google Scholar]
- Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- Ha NC, Tonozuka T, Stamos JL, et al. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Hagstrom KA, Meyer BJ. Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- Hall DD, Feekes JA, Arachchige Don AS, et al. Binding of protein phosphatase 2A to the L-type calcium channel Cav1.2 next to Ser1928, its main PKA site, is critical for Ser1928 dephosphorylation. Biochemistry. 2006;45:3448–3459. doi: 10.1021/bi051593z. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–E67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- Hartley D, Cooper GM. Role of mTOR in the degradation of IRS-1: regulation of PP2A activity. J Cell Biochem. 2002;85:304–314. doi: 10.1002/jcb.10135. [DOI] [PubMed] [Google Scholar]
- Harvey SL, Enciso G, Dephoure N, et al. A phosphatase threshold sets the level of Cdk1 activity in early mitosis in budding yeast. Mol Biol Cell. 2011;22:3595–3608. doi: 10.1091/mbc.E11-04-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashigasako A, Machide M, Nakamura T, Matsumoto K. Bidirectional regulation of Ser-985 phosphorylation of c-met via protein kinase C and protein phosphatase 2A involves c-Met activation and cellular responsiveness to hepatocyte growth factor. J Biol Chem. 2004;279:26445–26452. doi: 10.1074/jbc.M314254200. [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Hegarat N, Vesely C, Vinod PK, et al. PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit. PLoS Genet. 2014;10:e1004004. doi: 10.1371/journal.pgen.1004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijman J, Dewenter M, El-Armouche A, Dobrev D. Function and regulation of serine/threonine phosphatases in the healthy and diseased heart. J Mol Cell Cardiol. 2013;64:90–98. doi: 10.1016/j.yjmcc.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Heikkinen PT, Nummela M, Leivonen SK, et al. Hypoxiaactivated Smad3-specific dephosphorylation by PP2A. J Biol Chem. 2010;285:3740–3749. doi: 10.1074/jbc.M109.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi A, Cecchini M, Steinhardt RC, et al. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol. 2010;17:1051–1057. doi: 10.1038/nsmb.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Bunemann M, et al. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat Methods. 2005;2:171–176. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Bottger F, Stemmann O, Taylor SS. Protein phosphatase 2A and separase form a complex regulated by separase autocleavage. J Biol Chem. 2007;282:24623–24632. doi: 10.1074/jbc.M702545200. [DOI] [PubMed] [Google Scholar]
- Holt LJ. Regulatory modules: coupling protein stability to phopshoregulation during cell division. FEBS Lett. 2012;586:2773–2777. doi: 10.1016/j.febslet.2012.05.045. [DOI] [PubMed] [Google Scholar]
- Hommes DW, Peppelenbosch MP, Deventer SJHV. Mitogen activated protein (MAP) kinase signal transduction pathways and novel anti-inflammatory targets. Gut. 2003;52:144–151. doi: 10.1136/gut.52.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn V, Thelu J, Garcia A, et al. Functional interaction of Aurora-A and PP2A during mitosis. Mol Biol Cell. 2007;18:1233–1241. doi: 10.1091/mbc.E06-12-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HK, Forsburg SL, John UP, et al. Isolation and characterization of the Pin1/Ess1p homologue in Schizosaccharomyces pombe. J Cell Sci. 2001;114:3779–3788. doi: 10.1242/jcs.114.20.3779. [DOI] [PubMed] [Google Scholar]
- Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- Hutchins JR, Dikovskaya D, Clarke PR. Regulation of Cdc2/cyclin B activation in Xenopus egg extracts via inhibitory phosphorylation of Cdc25C phosphatase by Ca(2+)/calmodulin-dependent protein [corrected] kinase II. Mol Biol Cell. 2003;14:4003–4014. doi: 10.1091/mbc.E03-02-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Rappailles A, Guilbaud G, et al. From simple bacterial and archaeal replicons to replication N/U-domains. J Mol Biol. 2013;425:4673–4689. doi: 10.1016/j.jmb.2013.09.021. [DOI] [PubMed] [Google Scholar]
- Imajo M, Tsuchiya Y, Nishida E. Regulatory mechanisms and functions of MAP kinase signaling pathways. IUBMB Life. 2006;58:312–317. doi: 10.1080/15216540600746393. [DOI] [PubMed] [Google Scholar]
- Indovina P, Marcelli E, Casini N, et al. Emerging roles of RB family: new defense mechanisms against tumor progression. J Cell Physiol. 2013;228:525–535. doi: 10.1002/jcp.24170. [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- Ito A, Kataoka TR, Watanabe M, et al. A truncated isoform of the PP2A B56 subunit promotes cell motility through paxillin phosphorylation. EMBO J. 2000;19:562–571. doi: 10.1093/emboj/19.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JB, Pallas DC. Circumventing cellular control of PP2A by methylation promotes transformation in an Akt-dependent manner. Neoplasia. 2012;14:585–599. doi: 10.1593/neo.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Brown GW, Muzi-Falconi M, et al. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Jaumot M, Hancock JF. Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14-3-3 interactions. Oncogene. 2001;20:3949–3958. doi: 10.1038/sj.onc.1204526. [DOI] [PubMed] [Google Scholar]
- Jayadeva G, Kurimchak A, Garriga J, et al. B55alpha PP2A holoenzymes modulate the phosphorylation status of the retinoblastoma-related protein p107 and its activation. J Biol Chem. 2010;285:29863–29873. doi: 10.1074/jbc.M110.162354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R. The many functions of SMC proteins in chromosome dynamics. Nat Rev Mol Cell Biol. 2002;3:767–778. doi: 10.1038/nrm930. [DOI] [PubMed] [Google Scholar]
- Jho E, Lomvardas S, Costantini F. A GSK3beta phosphorylation site in axin modulates interaction with beta-catenin and Tcf-mediated gene expression. Biochem Biophys Res Commun. 1999;266:28–35. doi: 10.1006/bbrc.1999.1760. [DOI] [PubMed] [Google Scholar]
- Jiang L, Stanevich V, Satyshur KA, et al. Structural basis of protein phosphatase 2A stable latency. Nat Commun. 2013;4:1699. doi: 10.1038/ncomms2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Kornbluth S. Phosphatases driving mitosis: pushing the gas and lifting the brakes. Prog Mol Biol Transl Sci. 2012;106:327–341. doi: 10.1016/B978-0-12-396456-4.00008-0. [DOI] [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, et al. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius MA, Schelbert B, Hsu W, et al. Domains of axin and disheveled required for interaction and function in wnt signaling. Biochem Biophys Res Commun. 2000;276:1162–1169. doi: 10.1006/bbrc.2000.3607. [DOI] [PubMed] [Google Scholar]
- Junttila MR, Puustinen P, Niemela M, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130:51–62. doi: 10.1016/j.cell.2007.04.044. [DOI] [PubMed] [Google Scholar]
- Kanemitsu MY, Jiang W, Eckhart W. Cdc2-mediated phosphorylation of the gap junction protein, connexin43, during mitosis. Cell Growth Differ. 1998;9:13–21. [PubMed] [Google Scholar]
- Kawabe T, Muslin AJ, Korsmeyer SJ. HOX11 interacts with protein phosphatases PP2A and PP1 and disrupts a G2/M cell-cycle checkpoint. Nature. 1997;385:454–458. doi: 10.1038/385454a0. [DOI] [PubMed] [Google Scholar]
- Keen JC, Zhou Q, Park BH, et al. Protein phosphatase 2A regulates estrogen receptor alpha (ER) expression through modulation of ER mRNA stability. J Biol Chem. 2005;280:29519–29524. doi: 10.1074/jbc.M505317200. [DOI] [PubMed] [Google Scholar]
- Kiely PA, O’Gorman D, Luong K, et al. Insulin-like growth factor I controls a mutually exclusive association of RACK1 with protein phosphatase 2A and beta1 integrin to promote cell migration. Mol Cell Biol. 2006;26:4041–4051. doi: 10.1128/MCB.01868-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Nilbert MC, Su LK, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Klippel A, Kavanaugh WM, Pot D, Williams LT. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V, Daempfling L, Basilico C. The B55alpha regulatory subunit of protein phosphatase 2A mediates fibroblast growth factor-induced p107 dephosphorylation and growth arrest in chondrocytes. Mol Cell Biol. 2013;33:2865–2878. doi: 10.1128/MCB.01730-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V, Janssens V. PP1 and PP2A phosphatases–cooperating partners in modulating retinoblastoma protein activation. FEBS J. 2013;280:627–643. doi: 10.1111/j.1742-4658.2012.08511.x. [DOI] [PubMed] [Google Scholar]
- Kotlo K, Johnson KR, Grillon JM, et al. Phosphoprotein abundance changes in hypertensive cardiac remodeling. J Proteomics. 2012;77:1–13. doi: 10.1016/j.jprot.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn MP, Egger-Adam D, Wodarz A. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev Cell. 2009;16:901–908. doi: 10.1016/j.devcel.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Kray AE, Carter RS, Pennington KN, et al. Positive regulation of IkappaB kinase signaling by protein serine/threonine phosphatase 2A. J Biol Chem. 2005;280:35974–35982. doi: 10.1074/jbc.M506093200. [DOI] [PubMed] [Google Scholar]
- Kristensen AR, Gsponer J, Foster LJ. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods. 2012;9:907–909. doi: 10.1038/nmeth.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger KM, Daaka Y, Pitcher JA, Lefkowitz RJ. The role of sequestration in G protein-coupled receptor resensitization. Regulation of beta2-adrenergic receptor dephosphorylation by vesicular acidification. J Biol Chem. 1997;272:5–8. doi: 10.1074/jbc.272.1.5. [DOI] [PubMed] [Google Scholar]
- Kumar A, Pandurangan AK, Lu F, et al. Chemopreventive sphingadienes downregulate Wnt signaling via a PP2A/Akt/GSK3beta pathway in colon cancer. Carcinogenesis. 2012;33:1726–1735. doi: 10.1093/carcin/bgs174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Kuo YC, Huang KY, Yang CH, et al. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–1892. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- Kurimchak A, Grana X. PP2A holoenzymes negatively and positively regulate cell cycle progression by dephosphorylating pocket proteins and multiple CDK substrates. Gene. 2012;499:1–7. doi: 10.1016/j.gene.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer C, Wagerer S, Saffrich R, et al. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J Cell Sci. 1998;111:2445–2453. doi: 10.1242/jcs.111.16.2445. [DOI] [PubMed] [Google Scholar]
- Landsverk HB, Kirkhus M, Bollen M, et al. PNUTS enhances in vitro chromosome decondensation in a PP1-dependent manner. Biochem J. 2005;390:709–717. doi: 10.1042/BJ20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao DH, Yusoff P, Chandramouli S, et al. Direct binding of PP2A to Sprouty2 and phosphorylation changes are a prerequisite for ERK inhibition downstream of fibroblast growth factor receptor stimulation. J Biol Chem. 2007;282:9117–9126. doi: 10.1074/jbc.M607563200. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E, Chiaur DS, Pagano M. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of beta-catenin. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, et al. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Lechward K, Zolnierowicz S, Hemmings BA. Eukaryotic translation termination factor 1 associates with protein phosphatase 2A and targets it to ribosomes. Biochemistry (Mosc) 1999;64:1373–1381. [PubMed] [Google Scholar]
- Lee JA, Pallas DC. Leucine carboxyl methyltransferase-1 is necessary for normal progression through mitosis in mammalian cells. J Biol Chem. 2007;282:30974–30984. doi: 10.1074/jbc.M704861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Kim DU, Lee MY, Choi KY. Identification of proteins interacting with the catalytic subunit of PP2A by proteomics. Proteomics. 2007;7:206–214. doi: 10.1002/pmic.200600480. [DOI] [PubMed] [Google Scholar]
- Lees JA, Buchkovich KJ, Marshak DR, et al. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneux C, Rocher G, Porteu F. B56-containing PP2A dephosphorylate ERK and their activity is controlled by the early gene IEX-1 and ERK. EMBO J. 2006;25:727–738. doi: 10.1038/sj.emboj.7600980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Hagesteijn C, Mahendra A, Naruszewicz I, Hannigan GE. Modulation of integrin signal transduction by ILKAP, a protein phosphatase 2C associating with the integrin-linked kinase, ILK1. EMBO J. 2001;20:2160–2170. doi: 10.1093/emboj/20.9.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V, Ryan CE, Piwnica-Worms H. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol Cell Biol. 2006;26:7529–7538. doi: 10.1128/MCB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Cai X, Shouse GP, et al. A specific PP2A regulatory subunit, B56gamma, mediates DNA damage-induced dephosphorylation of p53 at Thr55. EMBO J. 2007;26:402–411. doi: 10.1038/sj.emboj.7601519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Makkinje A, Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996;271:11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- Li S, Wang L, Berman MA, et al. RNAi screen in mouse astrocytes identifies phosphatases that regulate NF-kappaB signaling. Mol Cell. 2006;24:497–509. doi: 10.1016/j.molcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Liedtke CM, Wang X, Smallwood ND. Role for protein phosphatase 2A in the regulation of Calu-3 epithelial Na+K+2Cl−, type 1 co-transport function. J Biol Chem. 2005;280:25491–25498. doi: 10.1074/jbc.M504473200. [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Lin SS, Bassik MC, Suh H, et al. PP2A regulates BCL-2 phosphorylation and proteasome-mediated degradation at the endoplasmic reticulum. J Biol Chem. 2006;281:23003–23012. doi: 10.1074/jbc.M602648200. [DOI] [PubMed] [Google Scholar]
- Liu J, Prickett TD, Elliott E, et al. Phosphorylation and microtubule association of the Opitz syndrome protein mid-1 is regulated by protein phosphatase 2A via binding to the regulatory subunit alpha 4. Proc Natl Acad Sci USA. 2001;98:6650–6655. doi: 10.1073/pnas.111154698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Eisenman RN. Regulation of c-Myc protein abundance by a protein phosphatase 2A-glycogen synthase kinase 3β-negative feedback pathway. Genes Cancer. 2012;3:23–36. doi: 10.1177/1947601912448067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longin S, Zwaenepoel K, Louis JV, et al. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J Biol Chem. 2007;282:26971–26980. doi: 10.1074/jbc.M704059200. [DOI] [PubMed] [Google Scholar]
- Louis JV, Martens E, Borghgraef P, et al. Mice lacking phosphatase PP2A subunit PR61/B’delta (Ppp2r5d) develop spatially restricted tauopathy by deregulation of CDK5 and GSK3beta. Proc Natl Acad Sci USA. 2011;108:6957–6962. doi: 10.1073/pnas.1018777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C, Quistgaard EM, Kovermann M, et al. Structural basis for PTPA interaction with the invariant C-terminal tail of PP2A. Biol Chem. 2014;395:881–889. doi: 10.1515/hsz-2014-0106. [DOI] [PubMed] [Google Scholar]
- Lowe M, Gonatas NK, Warren G. The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J Cell Biol. 2000;149:341–356. doi: 10.1083/jcb.149.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Peterson A, Garcia BA, et al. Protein phosphatase 1 regulates assembly and function of the beta-catenin degradation complex. EMBO J. 2007;26:1511–1521. doi: 10.1038/sj.emboj.7601607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Arnold HK, Lilly MB, et al. Negative regulation of Pim-1 protein kinase levels by the B56beta subunit of PP2A. Oncogene. 2007;26:5145–5153. doi: 10.1038/sj.onc.1210323. [DOI] [PubMed] [Google Scholar]
- Ma OK, Sucher NJ. Molecular interaction of NMDA receptor subunit NR3A with protein phosphatase 2A. Neuroreport. 2004;15:1447–1450. doi: 10.1097/01.wnr.0000132773.41720.2d. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Macdonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenta A, Fasanaro P, Romani S, et al. Protein phosphatase 2A subunit PR70 interacts with pRb and mediates its dephosphorylation. Mol Cell Biol. 2008;28:873–882. doi: 10.1128/MCB.00480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mao L, Yang L, Arora A, et al. Role of protein phosphatase 2A in mGluR5-regulated MEK/ERK phosphorylation in neurons. J Biol Chem. 2005;280:12602–12610. doi: 10.1074/jbc.M411709200. [DOI] [PubMed] [Google Scholar]
- Margolis SS, Perry JA, Forester CM, et al. Role for the PP2A/B56delta phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell. 2006a;127:759–773. doi: 10.1016/j.cell.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SS, Perry JA, Weitzel DH, et al. A role for PP1 in the Cdc2/Cyclin B-mediated positive feedback activation of Cdc25. Mol Biol Cell. 2006b;17:1779–1789. doi: 10.1091/mbc.E05-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SS, Walsh S, Weiser DC, et al. PP1 control of M phase entry exerted through 14-3-3-regulated Cdc25 dephosphorylation. EMBO J. 2003;22:5734–5745. doi: 10.1093/emboj/cdg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein LY, Mclaughlin PJ, Stanton JB, et al. Bestrophin interacts physically and functionally with protein phosphatase 2A. J Biol Chem. 2002;277:30591–30597. doi: 10.1074/jbc.M204269200. [DOI] [PubMed] [Google Scholar]
- Martin L, Latypova X, Wilson CM, et al. Tau protein phosphatases in Alzheimer’s disease: the leading role of PP2A. Ageing Res Rev. 2013;12:39–49. doi: 10.1016/j.arr.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Mathiasen DP, Egebjerg C, Andersen SH, et al. Identification of a c-Jun N-terminal kinase-2-dependent signal amplification cascade that regulates c-Myc levels in ras transformation. Oncogene. 2012;31:390–401. doi: 10.1038/onc.2011.230. [DOI] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcconnell JL, Gomez RJ, Mccorvey LR, et al. Identification of a PP2A-interacting protein that functions as a negative regulator of phosphatase activity in the ATM/ATR signaling pathway. Oncogene. 2007;26:6021–6030. doi: 10.1038/sj.onc.1210406. [DOI] [PubMed] [Google Scholar]
- Mccubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilleur MA, Akpovi CD, Pelletier RM, Vitale ML. Tumor necrosis factor-alpha-induced anterior pituitary folliculostellate TtT/GF cell uncoupling is mediated by connexin 43 dephosphorylation. Endocrinology. 2007;148:5913–5924. doi: 10.1210/en.2007-0767. [DOI] [PubMed] [Google Scholar]
- Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- Merrill RA, Slupe AM, Strack S. N-terminal phosphorylation of protein phosphatase 2A/Bbeta2 regulates translocation to mitochondria, dynamin-related protein 1 dephosphorylation, and neuronal survival. FEBS J. 2013;280:662–673. doi: 10.1111/j.1742-4658.2012.08631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Tong L, Devos AM, et al. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Mitra A, Menezes ME, Pannell LK, et al. DNAJB6 chaperones PP2A mediated dephosphorylation of GSK3beta to downregulate beta-catenin transcription target, osteopontin. Oncogene. 2012;31:4472–4483. doi: 10.1038/onc.2011.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Shima H, Tanuma N, et al. Protein phosphatase type 2A, PP2A, is involved in degradation of gp130. Mol Cell Biochem. 2005;269:183–187. doi: 10.1007/s11010-005-3089-x. [DOI] [PubMed] [Google Scholar]
- Mochida S, Hunt T. Protein phosphatases and their regulation in the control of mitosis. EMBO Rep. 2012;13:197–203. doi: 10.1038/embor.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009;28:2777–2785. doi: 10.1038/emboj.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Maslen SL, Skehel M, Hunt T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science. 2010;330:1670–1673. doi: 10.1126/science.1195689. [DOI] [PubMed] [Google Scholar]
- Monje P, Hernandez-Losa J, Lyons RJ, et al. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J Biol Chem. 2005;280:35081–35084. doi: 10.1074/jbc.C500353200. [DOI] [PubMed] [Google Scholar]
- Moreno CS, Park S, Nelson K, et al. WD40 repeat proteins striatin and S/G2 nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J Biol Chem. 2000;275:5257–5263. doi: 10.1074/jbc.275.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- Mumby M. A new role for protein methylation: switching partners at the phosphatase ball. Sci STKE. 2001:1. doi: 10.1126/stke.2001.79.pe1. 2001. [DOI] [PubMed] [Google Scholar]
- Mumby M. Lab Webpage [Online] Dallas (TX): UT Southwestern; 2009. [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Naji S, Ambrus G, Cimermancic P, et al. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol Cell Proteomics. 2012;11:M111.015313. doi: 10.1074/mcp.M111.015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Tanimura-Ito K, Oshiro N, et al. A positive role of mammalian Tip41-like protein, TIPRL, in the amino-acid dependent mTORC1-signaling pathway through interaction with PP2A. FEBS Lett. 2013;587:2924–2929. doi: 10.1016/j.febslet.2013.07.027. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, et al. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Stevens C, Phillips AM, et al. TLE1 modifies the effects of NOD2 in the pathogenesis of Crohn’s disease. Gastroenterology. 2011;141:972–981. e1–e2. doi: 10.1053/j.gastro.2011.05.043. [DOI] [PubMed] [Google Scholar]
- Nishisho I, Nakamura Y, Miyoshi Y, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Masuda HT, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010;12:719–727. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- Oberg EA, Nifoussi SK, Gingras AC, Strack S. Selective proteasomal degradation of the B’β subunit of protein phosphatase 2A by the E3 ubiquitin ligase adaptor Kelch-like 15. J Biol Chem. 2012;287:43378–43389. doi: 10.1074/jbc.M112.420281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oellerich T, Bremes V, Neumann K, et al. The B-cell antigen receptor signals through a preformed transducer module of SLP65 and CIN85. EMBO J. 2011;30:3620–3634. doi: 10.1038/emboj.2011.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kamibayashi C, Serrano M, et al. p53-dependent association between cyclin G and the B′ subunit of protein phosphatase 2A. Mol Cell Biol. 1996;16:6593–6602. doi: 10.1128/mcb.16.11.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Li H, Jensen MR, et al. Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol Cell. 2002;9:761–771. doi: 10.1016/s1097-2765(02)00504-x. [DOI] [PubMed] [Google Scholar]
- Olah J, Vincze O, Virok D, et al. Interactions of pathological hallmark proteins: tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J Biol Chem. 2011;286:34088–34100. doi: 10.1074/jbc.M111.243907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory S, Zhou M, Conrads TP, et al. Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr Biol. 2003;13:1356–1364. doi: 10.1016/s0960-9822(03)00535-9. [DOI] [PubMed] [Google Scholar]
- Palkowitsch L, Leidner J, Ghosh S, Marienfeld RB. Phosphorylation of serine 68 in the IkappaB kinase (IKK)-binding domain of NEMO interferes with the structure of the IKK complex and tumor necrosis factor-alpha-induced NF-kappaB activity. J Biol Chem. 2008;283:76–86. doi: 10.1074/jbc.M708856200. [DOI] [PubMed] [Google Scholar]
- Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat Struct Mol Biol. 2009;16:412–420. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- Paroni G, Cernotta N, Dello Russo C, et al. PP2A regulates HDAC4 nuclear import. Mol Biol Cell. 2008;19:655–667. doi: 10.1091/mbc.E07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Doree M, et al. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990;60:791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sorensen CS, et al. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci USA. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman MR, Barr RK, Gliddon BL, et al. A critical role for the protein phosphatase 2A B’a regulatory subunit in dephosphorylation of sphingosine kinase 1. Int J Biochem Cell Biol. 2011;43:342–347. doi: 10.1016/j.biocel.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Plesca D, Crosby ME, Gupta D, Almasan A. E2F4 function in G2: maintaining G2-arrest to prevent mitotic entry with damaged DNA. Cell Cycle. 2007;6:1147–1152. doi: 10.4161/cc.6.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Brautigan DL. Cytokine activation of p38 mitogen-activated protein kinase and apoptosis is opposed by alpha-4 targeting of protein phosphatase 2A for site-specific dephosphorylation of MEK3. Mol Cell Biol. 2007;27:4217–4227. doi: 10.1128/MCB.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purev E, Soprano DR, Soprano KJ. PP2A interaction with Rb2/p130 mediates translocation of Rb2/p130 into the nucleus in alltrans retinoic acid-treated ovarian carcinoma cells. J Cell Physiol. 2011;226:1027–1034. doi: 10.1002/jcp.22418. [DOI] [PubMed] [Google Scholar]
- Qian J, Beullens M, Lesage B, Bollen M. Aurora B defines its own chromosomal targeting by opposing the recruitment of the phosphatase scaffold Repo-Man. Curr Biol. 2013;23:1136–1143. doi: 10.1016/j.cub.2013.05.017. [DOI] [PubMed] [Google Scholar]
- Rajakulendran T, Sahmi M, Lefrancois M, et al. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- Rajgopal A, Young DW, Mujeeb KA, et al. Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J Cell Biochem. 2007;100:1509–1517. doi: 10.1002/jcb.21137. [DOI] [PubMed] [Google Scholar]
- Ricotta D, Hansen J, Preiss C, et al. Characterization of a protein phosphatase 2A holoenzyme that dephosphorylates the clathrin adaptors AP-1 and AP-2. J Biol Chem. 2008;283:5510–5517. doi: 10.1074/jbc.M707166200. [DOI] [PubMed] [Google Scholar]
- Rimerman RA, Gellert-Randleman A, Diehl JA. Wnt1 and MEK1 cooperate to promote cyclin D1 accumulation and cellular transformation. J Biol Chem. 2000;275:14736–14742. doi: 10.1074/jbc.m910241199. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci. 2013;38:12–19. doi: 10.1016/j.tibs.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin SM, Gall AL, Zheng N, Pavletich NP. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell. 2005;123:1093–1106. doi: 10.1016/j.cell.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Ruvolo VR, Kurinna SM, Karanjeet KB, et al. PKR regulates B56(alpha)-mediated BCL2 phosphatase activity in acute lymphoblastic leukemia-derived REH cells. J Biol Chem. 2008;283:35474–35485. doi: 10.1074/jbc.M800951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina AA, Chen W, Arroyo JD, et al. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell. 2007;129:969–982. doi: 10.1016/j.cell.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstag Y, Nebl G. Interaction of cofilin with the serine phosphatases PP1 and PP2A in normal and neoplastic human T lymphocytes. Adv Enzyme Regul. 2003;43:197–211. doi: 10.1016/s0065-2571(02)00031-6. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangodkar J, Farrington C, Mcclinch K, et al. All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. Q1FEBS J. 2015 doi: 10.1111/febs.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraf A, Oberg EA, Strack S. Molecular determinants for PP2A substrate specificity: charged residues mediate dephosphorylation of tyrosine hydroxylase by the PP2A/B’ regulatory subunit. Biochemistry. 2010;49:986–995. doi: 10.1021/bi902160t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Satoh J, Kawana N, Yamamoto Y. Pathway analysis of ChIP-Seq-based NRF1 target genes suggests a logical hypothesis of their involvement in the pathogenesis of neurodegenerative diseases. Gene Regul Syst Bio. 2013;7:139–152. doi: 10.4137/GRSB.S13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz AL, Srayko M, Dammermann A, et al. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–564. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GK, Gu F, Crump CM, et al. The phosphorylation state of an autoregulatory domain controls PACS-1-directed protein traffic. EMBO J. 2003;22:6234–6244. doi: 10.1093/emboj/cdg596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeling JM, Miller JR, Gil R, et al. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- Seth A, Sheth P, Elias BC, Rao R. Protein phosphatases 2A and 1 interact with occludin and negatively regulate the assembly of tight junctions in the CACO-2 cell monolayer. J Biol Chem. 2007;282:11487–11498. doi: 10.1074/jbc.M610597200. [DOI] [PubMed] [Google Scholar]
- Shanley TP, Vasi N, Denenberg A, Wong HR. The serine/threonine phosphatase, PP2A: endogenous regulator of inflammatory cell signaling. J Immunol. 2001;166:966–972. doi: 10.4049/jimmunol.166.2.966. [DOI] [PubMed] [Google Scholar]
- Shern JF, Sharer JD, Pallas DC, et al. Cytosolic Arl2 is complexed with cofactor D and protein phosphatase 2A. J Biol Chem. 2003;278:40829–40836. doi: 10.1074/jbc.M308678200. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- Shin JH, Grabowski B, Kasiviswanathan R, et al. Regulation of minichromosome maintenance helicase activity by Cdc6. J Biol Chem. 2003;278:38059–38067. doi: 10.1074/jbc.M305477200. [DOI] [PubMed] [Google Scholar]
- Short KM, Hopwood B, Yi Z, Cox TC. MID1 and MID2 homoand heterodimerise to tether the rapamycin-sensitive PP2A regulatory subunit, alpha 4, to microtubules: implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biol. 2002;3:1. doi: 10.1186/1471-2121-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtrichman R, Sharf R, Barr H, et al. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc Natl Acad Sci USA. 1999;96:10080–10085. doi: 10.1073/pnas.96.18.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano KJ, Purev E, Vuocolo S, Soprano DR. Rb2/p130 and protein phosphatase 2A: key mediators of ovarian carcinoma cell growth suppression by all-trans retinoic acid. Oncogene. 2006;25:5315–5325. doi: 10.1038/sj.onc.1209679. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanevich V. PhD Thesis. Madison, WI: University of Wisconsin-Madison; 2013. Protein phosphatase 2A methylation and stable latency. [Google Scholar]
- Stanevich V, Jiang L, Satyshur KA, et al. The structural basis for tight control of PP2A methylation and function by LCMT-1. Mol Cell. 2011;41:331–342. doi: 10.1016/j.molcel.2010.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, et al. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Sun L, Stoecklin G, Van Way S, et al. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-alpha mRNA. J Biol Chem. 2007;282:3766–3777. doi: 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- Sung U, Jennings JL, Link AJ, Blakely RD. Proteomic analysis of human norepinephrine transporter complexes reveals associations with protein phosphatase 2A anchoring subunit and 14-3-3 proteins. Biochem Biophys Res Commun. 2005;333:671–678. doi: 10.1016/j.bbrc.2005.05.165. [DOI] [PubMed] [Google Scholar]
- Surpili MJ, Delben TM, Kobarg J. Identification of proteins that interact with the central coiled-coil region of the human protein kinase NEK1. Biochemistry. 2003;42:15369–15376. doi: 10.1021/bi034575v. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Suzuki K. Regulation of protein phosphatase 2A-mediated recruitment of IQGAP1 to beta1 integrin by EGF through activation of Ca2+/calmodulin-dependent protein kinase II. J Cell Physiol. 2006;208:213–219. doi: 10.1002/jcp.20657. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shibata H, Shimakawa M, et al. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J Biol Chem. 1999;274:17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- Takemoto A, Maeshima K, Ikehara T, et al. The chromosomal association of condensin II is regulated by a noncatalytic function of PP2A. Nat Struct Mol Biol. 2009;16:1302–1308. doi: 10.1038/nsmb.1708. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shu H, Qi W, et al. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Tanno Y, Kitajima TS, Honda T, et al. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010;24:2169–2179. doi: 10.1101/gad.1945310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao GZ, Toivola DM, Zhou Q, et al. Protein phosphatase-2A associates with and dephosphorylates keratin 8 after hyposmotic stress in a site- and cell-specific manner. J Cell Sci. 2006;119:1425–1432. doi: 10.1242/jcs.02861. [DOI] [PubMed] [Google Scholar]
- Thompson LJ, Bollen M, Fields AP. Identification of protein phosphatase 1 as a mitotic lamin phosphatase. J Biol Chem. 1997;272:29693–29697. doi: 10.1074/jbc.272.47.29693. [DOI] [PubMed] [Google Scholar]
- Tischer T, Hormanseder E, Mayer TU. The APC/C inhibitor XErp1/Emi2 is essential for Xenopus early embryonic divisions. Science. 2012;338:520–524. doi: 10.1126/science.1228394. [DOI] [PubMed] [Google Scholar]
- Tran H, Bustos D, Yeh R, et al. HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC-axin interaction. J Biol Chem. 2013;288:3753–3767. doi: 10.1074/jbc.M112.415240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzcinska AM, Girstun A, Piekielko A, et al. Potential protein partners for the N-terminal domain of human topoisomerase I revealed by phage display. Mol Biol Rep. 2002;29:347–352. doi: 10.1023/a:1021237021338. [DOI] [PubMed] [Google Scholar]
- Tu LC, Yan X, Hood L, Lin B. Proteomics analysis of the interactome of N-myc downstream regulated gene 1 and its interactions with the androgen response program in prostate cancer cells. Mol Cell Proteomics. 2007;6:575–588. doi: 10.1074/mcp.M600249-MCP200. [DOI] [PubMed] [Google Scholar]
- Turowski P, Myles T, Hemmings BA, et al. Vimentin dephosphorylation by protein phosphatase 2A is modulated by the targeting subunit B55. Mol Biol Cell. 1999;10:1997–2015. doi: 10.1091/mbc.10.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugi S, Imamura T, Ricketts W, Olefsky JM. Protein phosphatase 2A forms a molecular complex with Shc and regulates Shc tyrosine phosphorylation and downstream mitogenic signaling. Mol Cell Biol. 2002;22:2375–2387. doi: 10.1128/MCB.22.7.2375-2387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P, Hudson DF, Ribeiro SA, et al. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol. 2006;8:1133–1142. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kanegan MJ, Strack S. The protein phosphatase 2A regulatory subunits B′beta and B’delta mediate sustained TrkA neurotrophin receptor autophosphorylation and neuronal differentiation. Mol Cell Biol. 2009;29:662–674. doi: 10.1128/MCB.01242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort M, Meeldijk J, van der Zee R, et al. Wnt signaling controls the phosphorylation status of beta-catenin. J Biol Chem. 2002;277:17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Keskitalo S, van Drogen A, et al. The protein interaction landscape of the human CMGC kinase group. Cell Rep. 2013a;3:1306–1320. doi: 10.1016/j.celrep.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Sacco R, Stukalov A, et al. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods. 2013b;10:307–314. doi: 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- Varlamov O, Kalinina E, Che FY, Fricker LD. Protein phosphatase 2A binds to the cytoplasmic tail of carboxypeptidase D and regulates post-trans-Golgi network trafficking. J Cell Sci. 2001;114:311–322. doi: 10.1242/jcs.114.2.311. [DOI] [PubMed] [Google Scholar]
- Vastiau A, Cao L, Jaspers M, et al. Interaction of the protein phosphatase 2A with the regulatory domain of the cystic fibrosis transmembrane conductance regulator channel. FEBS Lett. 2005;579:3392–3396. doi: 10.1016/j.febslet.2005.04.079. [DOI] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, Hijmans EM, Bernards R. Functional interaction between a novel protein phosphatase 2A regulatory subunit, PR59, and the retinoblastoma-related p107 protein. Oncogene. 1999;18:515–524. doi: 10.1038/sj.onc.1202316. [DOI] [PubMed] [Google Scholar]
- Wainszelbaum MJ, Liu J, Kong C, et al. TBC1D3, a hominoid-specific gene, delays IRS-1 degradation and promotes insulin signaling by modulating p70 S6 kinase activity. PLoS One. 2012;7:e31225. doi: 10.1371/journal.pone.0031225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PY, Weng J, Anderson RG. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- Wang W, Huang J, Wang X, et al. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Iwasaki J, et al. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc Natl Acad Sci USA. 2005;102:11663–11668. doi: 10.1073/pnas.0500410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Raben DM, Phillips PJ, Baldassare JJ. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D1 in G1 phase. Biochem J. 1997;326:61–68. doi: 10.1042/bj3260061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JH, Seemann J. Mitotic division of the mammalian Golgi apparatus. Semin Cell Dev Biol. 2009;20:810–816. doi: 10.1016/j.semcdb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal RS, Anderson KA, Means AR, Wadzinski BE. A signaling complex of Ca2+calmodulin-dependent protein kinase IV and protein phosphatase 2A. Science. 1998;280:1258–1261. doi: 10.1126/science.280.5367.1258. [DOI] [PubMed] [Google Scholar]
- Westphal RS, Coffee RL, Jr, Marotta A, et al. Identification of kinase-phosphatase signaling modules composed of p70 S6 kinase-protein phosphatase 2A (PP2A) and p21-activated kinase-PP2A. J Biol Chem. 1999;274:687–692. doi: 10.1074/jbc.274.2.687. [DOI] [PubMed] [Google Scholar]
- Wlodarchak N, Guo F, Satyshur KA, et al. Structure of the Ca2+dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation. Cell Res. 2013;23:931–946. doi: 10.1038/cr.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NT, Mesquita RD, Sweet M, et al. Charting the landscape of tandem BRCT domain-mediated protein interactions. Sci Signal. 2012;5:rs6. doi: 10.1126/scisignal.2002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GS. Role of mitogen-activated protein kinase phosphatases (MKPs) in cancer. Cancer Metastasis Rev. 2007;26:579–585. doi: 10.1007/s10555-007-9079-6. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Hansen DV, Guo Y, et al. Control of Emi2 activity and stability through Mos-mediated recruitment of PP2A. Proc Natl Acad Sci USA. 2007;104:16564–16569. doi: 10.1073/pnas.0707537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Noh SJ, Zhou G, et al. Selective activation of MEK1 but not MEK2 by A-Raf from epidermal growth factor-stimulated Hela cells. J Biol Chem. 1996;271:3265–3271. doi: 10.1074/jbc.271.6.3265. [DOI] [PubMed] [Google Scholar]
- Wu Z, Chen Y, Yang T, et al. Targeted ubiquitination and degradation of G-protein-coupled receptor kinase 5 by the DDB1-CUL4 ubiquitin ligase complex. PLoS One. 2012;7:e43997. doi: 10.1371/journal.pone.0043997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol. 2011;12:469–482. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- Xie Y, Avello M, Schirle M, et al. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 2013;288:2976–2985. doi: 10.1074/jbc.M112.430066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Deng X. Protein phosphatase 2A enhances the proapoptotic function of Bax through dephosphorylation. J Biol Chem. 2006;281:18859–18867. doi: 10.1074/jbc.M512543200. [DOI] [PubMed] [Google Scholar]
- Xing H, Hong Y, Sarge KD. Identification of the PP2A-interacting region of heat shock transcription factor 2. Cell Stress Chaperones. 2007;12:192–197. doi: 10.1379/CSC-249R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Li Z, Chen Y, et al. Structural mechanism of demethylation and inactivation of protein phosphatase 2A. Cell. 2008;133:154–163. doi: 10.1016/j.cell.2008.02.041. [DOI] [PubMed] [Google Scholar]
- Xing Y, Xu Y, Chen Y, et al. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell. 2006;127:341–353. doi: 10.1016/j.cell.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Xu P, Raetz EA, Kitagawa M, et al. BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biol Open. 2013;2:479–486. doi: 10.1242/bio.20134051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Chen Y, Zhang P, et al. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xing Y, Chen Y, et al. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- Xu Z, Cetin B, Anger M, et al. Structure and function of the PP2A-shugoshin interaction. Mol Cell. 2009;35:426–441. doi: 10.1016/j.molcel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Williams BR. The B56alpha regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Mol Cell Biol. 2000;20:5285–5299. doi: 10.1128/mcb.20.14.5285-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Hinoi T, Michiue T, et al. Inhibition of the Wnt signaling pathway by the PR61 subunit of protein phosphatase 2A. J Biol Chem. 2001;276:26875–26882. doi: 10.1074/jbc.M100443200. [DOI] [PubMed] [Google Scholar]
- Yan L, Mieulet V, Burgess D, et al. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- Yang CS, Xin HW, Kelley JB, et al. Ligand binding to the androgen receptor induces conformational changes that regulate phosphatase interactions. Mol Cell Biol. 2007;27:3390–3404. doi: 10.1128/MCB.02411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Fan GH, Wadzinski BE, et al. Protein phosphatase 2A interacts with and directly dephosphorylates RelA. J Biol Chem. 2001;276:47828–47833. doi: 10.1074/jbc.M106103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatim A, Benne C, Sobhian B, et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48:445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong FM, Hombauer H, Wendt KS, et al. Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr Biol. 2003;13:2058–2064. doi: 10.1016/j.cub.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Miller WT. Protein phosphatase 2A interacts with the Src kinase substrate p130(CAS) Oncogene. 2001;20:6057–6065. doi: 10.1038/sj.onc.1204735. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Reich NC, Miller WT. Involvement of protein phosphatase 2A in the interleukin-3-stimulated Jak2-Stat5 signaling pathway. J Interferon Cytokine Res. 2001;21:369–378. doi: 10.1089/107999001750277844. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Jimenez RH, Sanders JA, et al. The alpha4-containing form of protein phosphatase 2A in liver and hepatic cells. J Cell Biochem. 2008;105:290–300. doi: 10.1002/jcb.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XX, Du X, Moreno CS, et al. Methylation of the protein phosphatase 2A catalytic subunit is essential for association of Balpha regulatory subunit but not SG2NA, striatin, or polyomavirus middle tumor antigen. Mol Biol Cell. 2001;12:185–199. doi: 10.1091/mbc.12.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Pino E, Wu L, et al. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J Biol Chem. 2012;287:29579–2988. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yang J, Liu Y, et al. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem. 2009;284:22649–22656. doi: 10.1074/jbc.M109.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Kosik KS, Meigs TE, et al. Galpha12 directly interacts with PP2A: evidence FOR Galpha12-stimulated PP2A phosphatase activity and dephosphorylation of microtubule-associated protein, tau. J Biol Chem. 2004;279:54983–54986. doi: 10.1074/jbc.C400508200. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel K, Goris J, Erneux C, et al. Protein phosphatase 2A PR130/B″alpha1 subunit binds to the SH2 domain-containing inositol polyphosphate 5-phosphatase 2 and prevents epidermal growth factor (EGF)-induced EGF receptor degradation sustaining EGF-mediated signaling. FASEB J. 2010;24:538–547. doi: 10.1096/fj.09-140228. [DOI] [PubMed] [Google Scholar]