Gene-environment interactions, where the biological effect of an exposure depends on an individual’s genotype, are widely held to be ubiquitous—and rightly so, considering epidemiologists have long abandoned the paradigm of ascribing disease to either “nature” or “nurture” (if indeed they ever thought of etiology in unifactoral terms) and now seek to understand the joint action of both “nature” and “nurture.” However, statistical interactions, where a quantitative measure of exposure effect differs according to genotype, are far from ubiquitous in epidemiologic studies of human disease (1). The small number of replicated gene-environment interactions in human observational studies stands in sharp contrast to the widespread evidence for gene-environment interaction from experimental studies in model organisms (2). This discrepancy is a puzzle. Is there something fundamentally different about the biology of human complex traits? Are there limitations to how gene-environment interactions have been studied in humans? Or both?
Stenzel et al. (3) discuss two important methodological challenges facing epidemiologic studies of gene-environment interactions: the lack of exposure variability in standard designs and exposure measurement error. Both of these factors can lead to loss of power to detect gene-environment interactions. Stenzel et al. show that for rare binary exposures oversampling exposed individuals in case-control studies can improve power relative to sampling cases and controls without regard to exposure. They consider designs that oversample exposed cases and controls equally or that only oversample cases. The advantage of oversampling exposed individuals declines and eventually disappears as exposure misclassification increases.
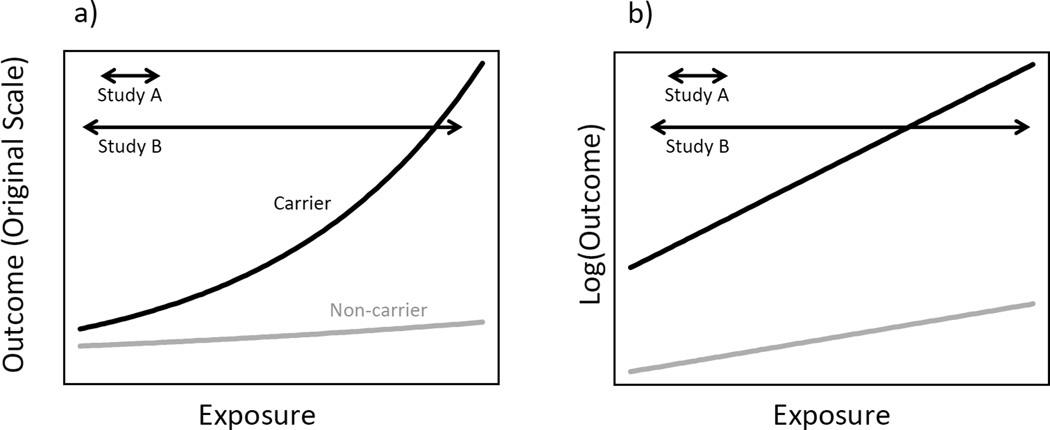
Stenzel et al. consider a binary exposure and binary outcome, but the intuition behind the increase in power from oversampling exposed individuals is perhaps better conveyed by a continuous outcome and continuous exposure. Figure 1 illustrates the range of gene-environment effects captured by two studies: Study A, which only samples a small range of exposure, and Study B, which samples a broad range. The difference in exposure range could be due to an exposure-driven sampling design—for example, if both studies have been conducted in the same base population but Study B has oversampled the extremes of the exposure distribution—or the difference could be caused by differences in the base populations between the two studies. In either case, it is clear that Study B captures more variability in the exposure and hence more variability in the gene-environment interaction term, leading to greater power, regardless of how the outcome is scaled. In fact, on the original scale, the interaction is extremely subtle across the range sampled by Study A; the interaction only becomes apparent when more extreme exposures are considered.
Figure 1.
Mean outcome (a) and log mean outcome (b) as a function of exposure and genotype. Arrows denote range of exposure captured by two hypothetical studies.
Two recent studies of the effect of the interaction between FTO rs9939609 genotype and physical activity on body mass index provide a concrete example of the scenario in Figure 1. A study in largely sedentary European and North American populations required a very large sample size (218,166) to detect a small, nominally significant interaction effect between this SNP and physical activity: the per-minor allele increase in odds of obesity decreased by 6% in the physically active group relative to the physically inactive (p=0.001) (4). On the other hand, a study in India that captured a much broader range of physical activity (from sedentary city dwellers to very active rural farmworkers) identified a qualitatively similar interaction (the minor allele was associated with increased waist size in the least active subjects but not in the most active; p=0.008) in a much smaller sample size (1,129) (5).
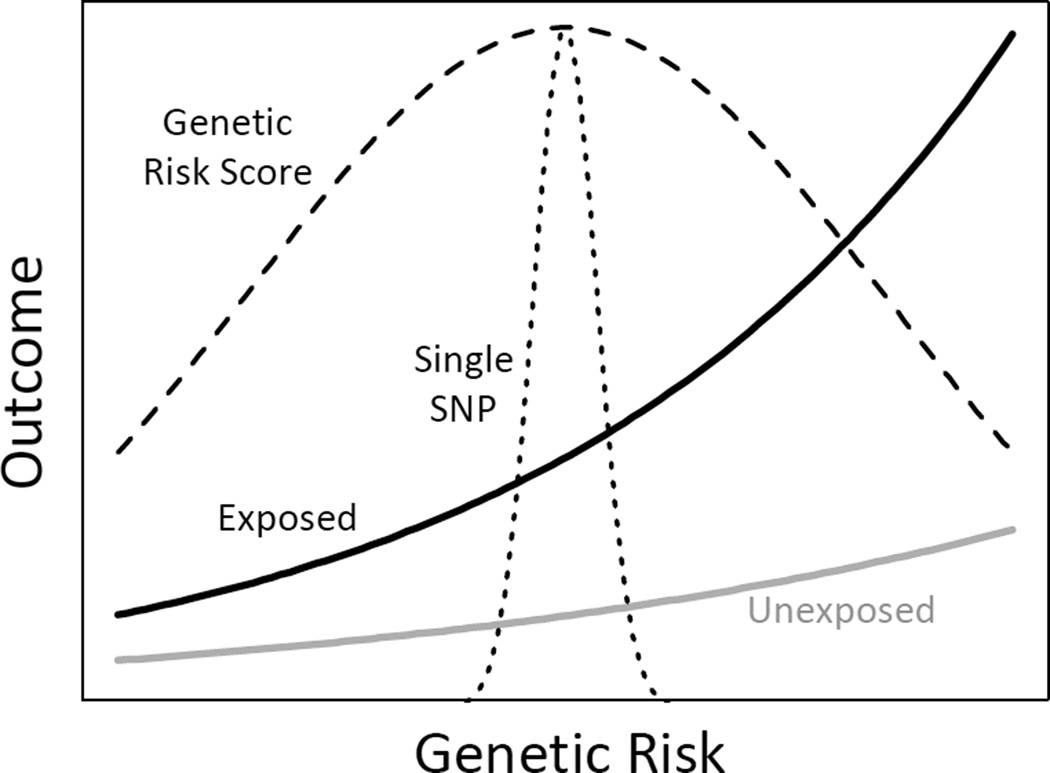
Recent advances in our understanding of common genetic markers associated with a broad range of human traits and diseases enable us to turn this idea around: we might be able to increase power detect gene-environment interactions by increasing the range genetic susceptibility under study (6). Figure 2 contrasts an analysis that focuses on a single nucleotide polymorphism (SNP) with an analysis that considers a genetic risk score, for example a multi-SNP genetic instrument for body mass index, as might be used in a Mendelian randomization study (7). In this situation, by capturing more of the relevant genetic variability, the SNP score increases power to detect gene-environment interaction. This power increase is contingent on the true joint gene-environment effects having the form displayed in Figure 2, or at least on most SNPs in the score having gene-environment interaction effects in the same direction, but there is already some evidence supporting interaction effects of this type (8–11).
Figure 2.
Figure 2. Mean outcome as a function of exposure and cumulative genetic risk. Dashed lines denote scaled densities for genetic risk captured by a single SNP or a multi-SNP genetic risk score.
The discussion of exposure misclassification in Stenzel et al. raises philosophical and increasingly important practical issues. On a philosophical level, the exposures we can measure are rarely if ever the etiologically relevant exposures. Some degree of model misspecification and exposure misclassification is inevitable. But on a practical level, many of the exposures we can measure and on which we could intervene are too expensive to measure directly in extremely large sample sizes. Instead, epidemiologic studies rely on inexpensive proxies—a practice which is only likely to increase, as epidemiologists incorporate different streams of Big Data into their studies (12). The results of Stenzel et al. suggest that the utility of designs that sample from a larger cohort based on an exposure proxy in order to identify who to genotype depends on the accuracy of the proxy. We suspect that the same caution applies to designs where the proxy is used to identify a subset of subjects whose exposures will be measured using a more expensive “gold standard” technology.
To return to the questions raised above: the relative lack of compelling gene-environment interactions in human observational studies is likely due to both the types of traits studied and how they are studied. Complex diseases result from the interplay of multiple biological processes affected by multiple genes and exposures. Even if an underlying intermediate trait exhibits strong gene-environment interaction, this interaction effect can be washed out at the disease level. At the same time, limited variability in exposure has likely also contributed to lack of power in human gene-environment interaction studies. Stenzel et al. demonstrate that thoughtful design can overcome this limitation. Design and analysis strategies that increase variability in sampled exposures and genetic susceptibilities deserve further consideration.
References
- 1.Hutter CM, Mechanic LE, Chatterjee N, Kraft P, Gillanders EM. Gene-environment interactions in cancer epidemiology: a National Cancer Institute Think Tank report. Genet Epidemiol. 2013;37(7):643–657. doi: 10.1002/gepi.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aschard H, Lutz S, Maus B, et al. Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Human genetics. 2012;131(10):1591–1613. doi: 10.1007/s00439-012-1192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenzel S, Ahn J, Boonstra P, Gruber S, Mukhejee B. The impact of exposure-biased sampling designs on detection of gene-environment interactions in case-control studies with potential exposure misclassification. Eur J Epidemiol. 2015 doi: 10.1007/s10654-014-9908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilpelainen TO, Qi L, Brage S, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS medicine. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore SC, Gunter MJ, Daniel CR, et al. Common genetic variants and central adiposity among Asian-Indians. Obesity. 2012;20(9):1902–1908. doi: 10.1038/oby.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aschard H. A theoretical perspective on interaction effects in genetic association studies. doi: 10.1002/gepi.21989. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, Kraft P. Methodological challenges in mendelian randomization. Epidemiology. 2014;25(3):427–435. doi: 10.1097/EDE.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi Q, Chu AY, Kang JH, et al. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367(15):1387–1396. doi: 10.1056/NEJMoa1203039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindstrom S, Schumacher F, Siddiq A, et al. Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers--results from BPC3. PLoS ONE. 2011;6(2):e17142. doi: 10.1371/journal.pone.0017142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langenberg C, Sharp SJ, Franks PW, et al. Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med. 2014;11(5):e1001647. doi: 10.1371/journal.pmed.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Z, Shrubsole MJ, Li G, et al. Interaction of cigarette smoking and carcinogen-metabolizing polymorphisms in the risk of colorectal polyps. Carcinogenesis. 2013;34(4):779–786. doi: 10.1093/carcin/bgs410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mooney SJ, Westreich DJ, El-Sayed AM. Commentary: epidemiology in the era of big data. Epidemiology. 2015;26(3):390–394. doi: 10.1097/EDE.0000000000000274. [DOI] [PMC free article] [PubMed] [Google Scholar]