Abstract
Increasing incidence of inflammatory bowel diseases such as Crohn’s disease (CD) in developed nations is associated with changes to the environment, such as decreased prevalence of helminth colonization and alterations to the gut microbiota. We find that helminth infection protects mice deficient in the CD susceptibility gene Nod2 from intestinal abnormalities by inhibiting colonization with an inflammatory Bacteroides species. Colonization resistance to Bacteroides was dependent on type-2 immunity, which promoted the establishment of a protective microbiota enriched in Clostridiales. Additionally, we show that individuals from helminth-endemic regions harbor a similar protective microbiota, and that deworming treatment reduced Clostridiales and increased Bacteroidales. These results support a model of the hygiene hypothesis whereby certain individuals are genetically susceptible to the consequences of a changing microbial environment.
Dramatic increases in the incidence of inflammatory bowel disease (IBD) in the developed world point towards alterations in the environment, including changes to the gut microbiota (1) and decreased exposure to intestinal parasites such as helminths (2). Evidence supporting a central role of the microbiota in the pathogenesis of IBD has led to a growing interest in defining the symbiotic relationship between the host and specific microbial species (3). Symbiotic relationships described in insects that develop to defend against environmental hazards (defensive symbiosis) (4) may be applicable to host-microbiota interactions. For example, specific bacterial taxa found within the human gut microbiota likely mediate resistance to antibiotic-associated diarrhea caused by Clostridium difficile (5). Loss of beneficial members of the microbiota potentially contribute to chronic inflammatory diseases as well. Also, helminths and the gut microbiota have co-evolved with their mammalian hosts, but the mechanisms of these interactions and the consequence of decreased exposure to intestinal helminths remain unclear. Here, we find that helminths can reduce intestinal inflammatory responses by promoting expansion of protective bacterial communities that inhibit pro-inflammatory bacterial taxa.
We previously reported that mice deficient in Nod2 develop several small intestinal (SI) abnormalities in a manner dependent on a ubiquitous member of the gut microbiota, Bacteroides vulgatus (6). Consistent with the specific association between NOD2 variants and ileal Crohn’s disease (CD) (7), an IBD that affects the SI, the most striking abnormality was a SI goblet cell defect that resulted in a compromised mucus layer, allowing sustained colonization by B. vulgatus. We found that chronic infection of Nod2−/− mice with the parasitic worm Trichuris muris restored SI goblet cell numbers and morphology (Figure 1A, B, S1A, B). These changes were not detected in the colon, and wild-type (WT) mice infected with T. muris did not display non-specific goblet cell hyperplasia (Figure S1C). Elevated epithelial levels of the antimicrobial lectin Reg3β and interferon (IFN)γ+ CD8+ intraepithelial lymphocytes (IELs), inflammatory markers associated with goblet cell defects in Nod2−/− mice (6), were also reduced upon T. muris infection (Figure 1C−E, S1D, E, S2). Nod2−/− mice develop severe intestinal pathologies following SI injury induced by the non-steroidal anti-inflammatory drug (NSAID) piroxicam. T. muris infection prevented the intestinal bleeding and perforation, exaggerated weight loss, mucus depletion, splenomegaly, and bacterial translocation that were observed in uninfected Nod2−/− mice treated with piroxicam (Figure 1F, S3A–C, S4). Blind histology analysis confirmed reductions in specific pathologies such as abscesses, epithelial hyperplasia, villus blunting, and immune infiltrates (Figure 1G, H, S3D–J). These results indicate that T. muris infection ameliorates spontaneous and inducible intestinal defects in Nod2−/− mice.
Figure 1. Trichuris muris infection reverses intestinal abnormalities in Nod2−/− mice.
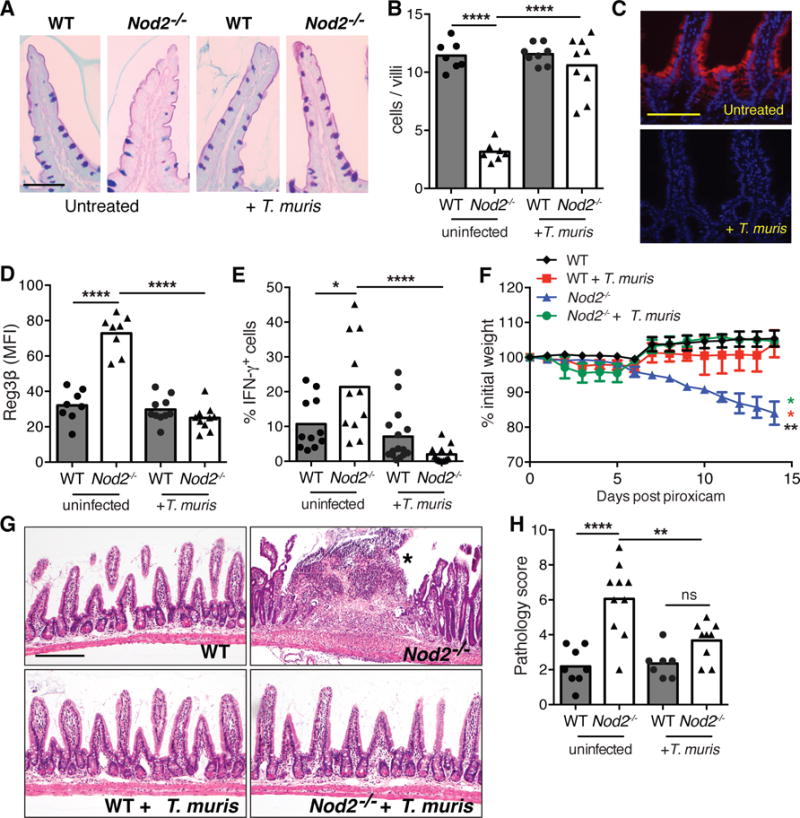
(A–B) PAS-Alcian blue stained small intestinal sections (A) and quantification of the number of goblet cells displaying normal morphology per villi (B) from uninfected and T. muris infected WT and Nod2−/− mice (n≥7 per genotype). (C–D) Immunofluorescence (IF) analysis of Reg3β in small intestine (C) and quantification of the mean fluorescence intensity (MFI) (D) of above mice (n≥8 per genotype). (E) Quantification of the proportion of CD8+ intra-epithelial lymphocytes (IELs) expressing IFN-γ by flow cytometry (n≥11 per genotype). (F–H) Quantification of weight loss (F), H&E-stained small intestinal sections (G), and quantification of pathology (48) (H), following piroxicam treatment of uninfected and T. muris infected WT and Nod2−/− mice. Asterisk denotes an abscess in (G). (n≥7 per genotype). *p<0.05, **p<0.01, and ****p<0.0001 by ANOVA with Holm-Sidak multiple comparisons test for (B), (D), (E), (F), and (H). Scale bar represents 50 μm in (A), 100 μm in (C) and (G). Data are represented as mean ± SEM in (F), each data point represents an individual mouse and bar denotes mean in (B), (D), (E), and (H), from at least two independent experiments.
Consistent with the dependence of these inflammatory pathologies on B. vulgatus (6), T. muris infection reduced bacterial burden to the limit of detection in the stool and SI tissue of Nod2−/− mice (Figure 2A, F). B. vulgatus inhibition was dependent on lymphocytes (Fig S5A–C), potentially reflecting goblet cell activation by type-2 cytokines (interleukin (IL)-4 and IL-13) produced by T helper (TH) cells during helminth infections. Indeed, we found increased phosphorylation of the type-2 transcription factor Stat6 in the SI epithelium of T. muris-infected Nod2−/− mice (Figure 2B, S5D). Also, T. muris infection only transiently inhibited B. vulgatus and did not restore goblet cells in Stat6−/− mice reconstituted with Nod2−/− bone marrow (Figure 2C, S5E). T. muris-infected Nod2−/− mice displayed a dominant TH2 response characterized by a >10-fold increase in IL-13+ CD4+ T cells in the lamina propria (Figure 2D, E, S5F, G). We confirmed these results with a second helminth, Heligmosomoides polygyrus, which induced an even greater TH2 response compared with T. muris, perhaps reflecting the distinct anatomical niches of these parasites (Figure 2H, D, S6C, D, S7B). H. polygyrus completely abolished tissue-associated B. vulgatus, restored goblet cells, and reduced IFNγ+ IELs in Nod2−/− mice (Figure 2F, 2G, S6A, B, S7A). Blocking IL-13 inhibited the effect of H. polygyrus on B. vulgatus and goblet cells, and administering recombinant IL-13 (rIL-13) or rIL-4 to Nod2−/− mice was sufficient to reproduce the effect of helminth infection (Figure 2I, J, K, L, S6E). RNA-seq analysis of intestinal tissues from rIL-13 treated Nod2−/− mice revealed a wound healing response characterized by expression of M2 macrophage genes (Figure 2M, S6F, Table S1). These results are consistent with the anti-inflammatory role of M2 macrophages in the gut (8, 9), and help explain how helminth infection ameliorates the exacerbated intestinal injury response in Nod2−/− mice. These results do not contradict the regulatory response induced by H. polygyrus in the colon (9, 10), because type-2 immunity and regulatory T cells can function concurrently to reduce inflammation (11).
Figure 2. Helminth infection inhibits Bacteroides vulgatus colonization through a type-2 immune response.
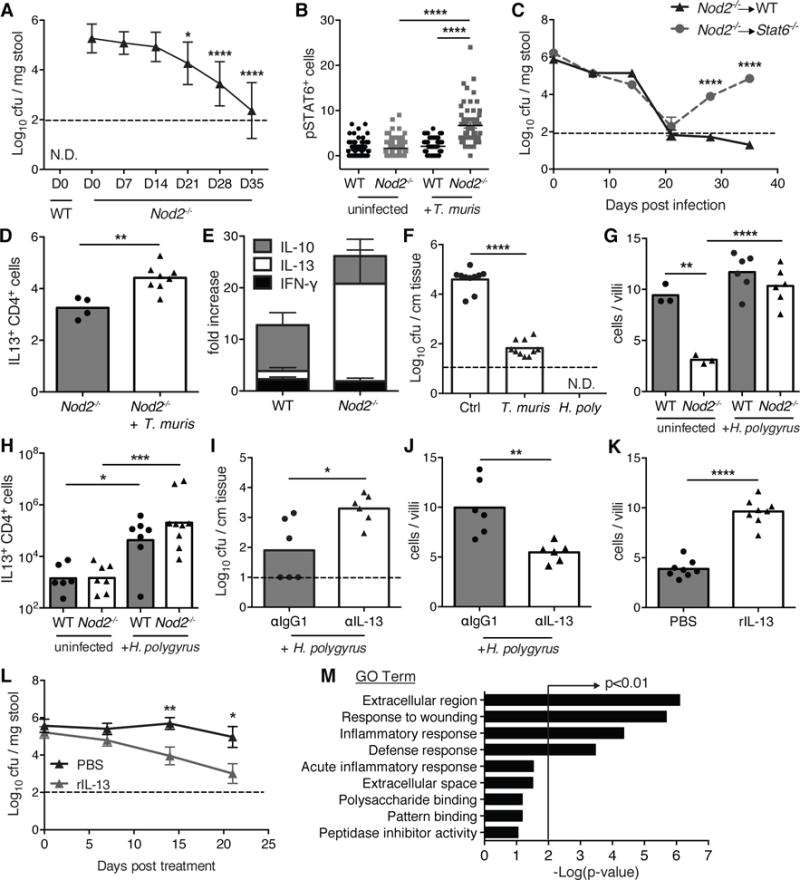
(A) Quantification of B. vulgatus colony forming units (cfu) in stool from T. muris infected WT and Nod2−/− mice (n≥10 per genotype). (B) Quantification of pSTAT6 staining in the small intestine of T. muris infected WT and Nod2−/− mice (n≥3 per genotype). (C) Quantification of B. vulgatus in stool from T. muris infected WT (Nod2−/− → WT) and Stat6−/− (Nod2−/− → Stat6−/−) mice reconstituted with Nod2−/− bone marrow (BM). Both WT and Stat6−/− chimeric mice were gavaged with B. vulgatus to ensure equal colonization before T. muris infection (n≥5 per genotype). (D) Quantification of the total number of small intestinal lamina propria CD4+ T cells expressing IL-13 in uninfected and T. muris infected Nod2−/− mice (n≥4 per genotype). (E) Fold-increase in the number CD4+ T cells producing IFN-γ, IL-13, or IL-10 in the small intestinal lamina propria of T. muris infected WT and Nod2−/− mice, normalized to uninfected mice (n≥4 per genotype). (F) Quantification of B. vulgatus associated with small intestinal tissue of uninfected, T. muris infected, and H. polygyrus infected Nod2−/− mice (n≥10 per genotype). (G–H) Quantification of goblet cells displaying normal morphology per villi (G) and total number of small intestinal lamina propria CD4+ T cells expressing IL-13 (H) in uninfected and H. polygyrus infected WT and Nod2−/− mice (n≥3 per genotype). (I–J) Quantification of B. vulgatus in small intestinal tissue (I), and goblet cells displaying normal morphology (J) in H. polygyrus infected Nod2−/− mice treated with antibody to IL-13 or isotype control (n=6 per genotype). (K–L) Quantification of goblet cells displaying normal morphology (K) and B. vulgatus in stool (L) in Nod2−/− mice treated with recombinant IL-13 or PBS (n=8 per genotype). (M) Pathway analysis based on GO terms of genes upregulated in Nod2−/− mice treated with recombinant IL-13 compared to PBS controls. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001 by ANOVA with Holm-Sidak multiple comparisons test for (A), (B), (G) and (H), and unpaired t-test for (C), (D), (F), and (I)−(L). Data are represented as mean ± SEM in (A), (B), (C), (E), and (L), each data point represents an individual mouse and bar denotes mean in (D), and (F)–(K), from at least two independent experiments.
The reduction of B. vulgatus in the presence of helminths could be mediated indirectly through alterations to the gut microbiota downstream of the type-2 response. Cohousing mice allows for coprophagic transmission of microbial populations without transfer of parasites because the worms are not sexually mature until ~35 days post infection and eggs require several weeks for germination (12). We found that uninfected Nod2−/− mice cohoused with T. muris-infected Nod2−/− mice showed a similar decrease in B. vulgatus colonization (Figure 3A, S8A). This reduction in B. vulgatus levels was not observed in uninfected Nod2−/− mice when they were instead cohoused with T. muris-infected WT mice (Figure S8B, C). 16S rDNA sequencing analysis of stool samples indicated that the alterations to microbial community compositions are different for T. muris-infected WT and Nod2−/− mice (Fig 3B), which may reflect different intestinal responses between WT and Nod2−/− mice (Figure 2E). Whereas there is reduced alpha diversity in infected WT mice, as previously reported (13, 14), Nod2−/− mice increased their alpha diversity at Day 21 post infection (Fig. S8D). The most significantly reduced bacterial taxa in infected Nod2−/− mice were Prevotella and Bacteroides genus (belonging to the order Bacteroidales), and the Lachnospiraceae family of the order Clostridiales were the most significantly increased (Figure 3C). The increase in Clostridiales was less evident in WT mice (Figure 3B), potentially explaining why cohousing Nod2−/− mice with T. muris-infected WT mice was ineffective in reducing B. vulgatus burden. The expansion of Clostridiales was also observed in the stool of uninfected Nod2−/− mice treated with rIL-13 or rIL-4 (Figure 3D, S8E). The expansion of Clostridiales was even more pronounced among tissue-associated bacteria in the SI following T. muris or H. polygyrus infection (Figure S8F, G). Thus, helminth infection and type-2 cytokines inhibit B. vulgatus and expand Clostridiales strains.
Figure 3. Inhibition of Bacteroides vulgatus is associated with expansion of Clostridiales following helminth infection.
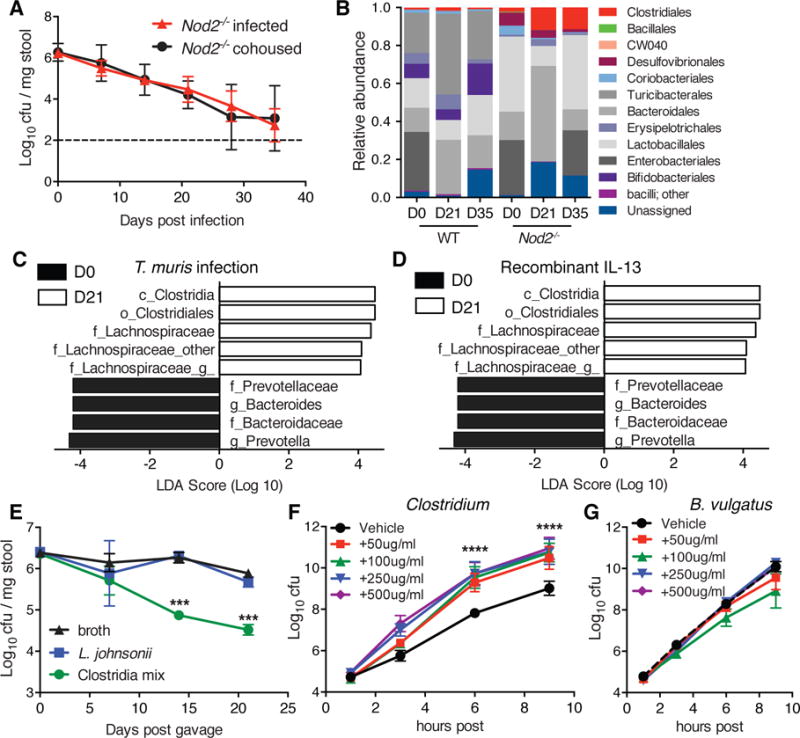
(A) Quantification of B. vulgatus in stool harvested from uninfected and T. muris infected Nod2−/− mice co-housed for the duration of the experiment (n≥4). (B) Relative abundance of taxonomic groups in response to T. muris infection in the stool of WT and Nod2−/− mice as determined by 16S sequencing (n≥5 per genotype). (C) Supervised analysis of 16S sequencing data with LDA effect size (LEfSe) comparing Nod2−/− mice at D0 and D21 post infection with T. muris using an LDA threshold score of 4 (n≥5). (D) LEfSE analysis to determine alterations to the stool microbiota after recombinant IL-13 treatment of Nod2−/− mice using an LDA threshold score of 4 (n≥5). (E) Quantification of B. vulgatus in stool harvested from Nod2−/− mice gavaged with sterile broth, L. johnsonii, or a mix of 17 Clostridiales and Erysipelotrichales strains (n≥3). (F–G) Quantification of Clostridium species (Clostridiales #28) (F) or B. vulgatus (G) in the presence of varying concentrations of pig intestinal mucin or vehicle in the culture media. ***p<0.001, ****p<0.0001 by ANOVA with Holm-Sidak multiple comparisons test for (E), and (F). Data are represented as mean ± SEM from at least two independent experiments.
To determine if Clostridia can directly inhibit B. vulgatus, we inoculated Nod2−/− mice with a mixture of clusters IV, XIVa, and XVIII Clostridiales and Erysipelotrichales strains isolated from human feces (15). Repetitive gavaging of Nod2−/− mice with this mixture, but not sterile broth or an equivalent number of Lactobacillus johnsonii (a host-interactive commensal bacterium (16)), led to a decrease in B. vulgatus over time (Figure 3E). Increased mucus production by goblet cells may alter the intestinal environment to favor Clostridiales, because we found that the addition of mucin to anaerobic cultures accelerates the growth of all three representative Clostridia strains tested but not B. vulgatus (Figure 3F, G, S8H, I). Hence, our results indicate that in Nod2−/− mice, the mucus response associated with type-2 immunity during helminth infection expands Clostridia strains that can inhibit colonization of B. vulgatus.
IBD is less prevalent in regions where helminth colonization is endemic. We previously found that helminth-colonized individuals among indigenous populations in Malaysia, known as the Orang Asli, have higher microbial diversity than negative individuals (17). We compared rural Orang Asli of the Temuan subtribe from a village 40km away, with individuals living in urbanized Kuala Lumpur (96% versus 5.3% of individuals colonized by intestinal helminths, respectively) (Table S2). People living in Kuala Lumpur predominantly cluster in a group driven by abundance of a single Bacteroides OTU (TaxID 3600504), which is less abundant in the Orang Asli (Figure 4A, B). In contrast, the helminth-positive Orang Asli falls into a second group characterized by Faecalibacterium and Prevotella (Figure 4A). This division between urban and rural populations in microbiota dominances is observed in other Asian countries (18).
Figure 4. Helminth colonization in humans is associated with a decrease in Bacteroidales and an increase in Clostridiales.
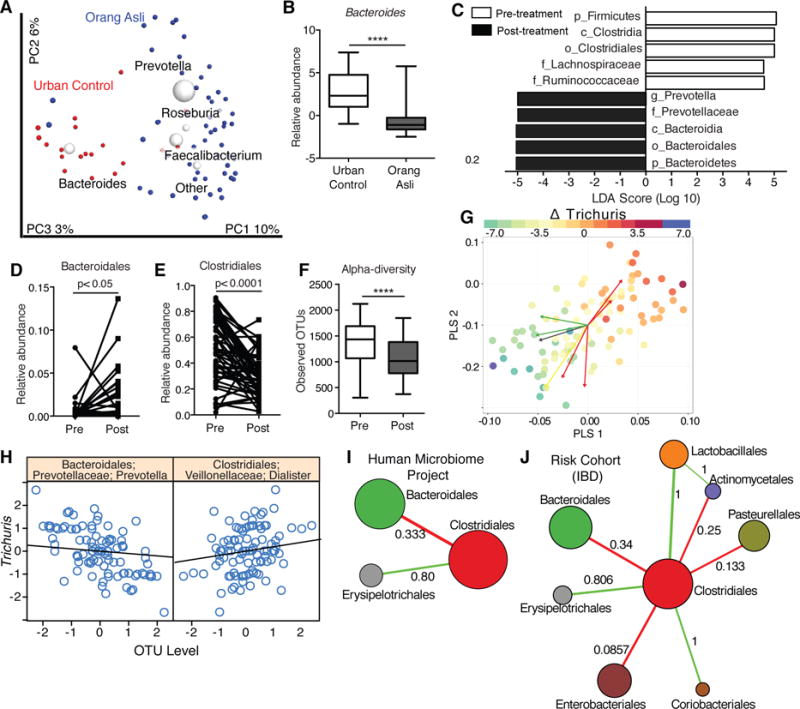
(A) Beta diversity plots of gut microbiota from urban controls in Kuala Lumpur (red dots) or the Orang Asli (blue dots). (B) Relative abundance of a dominant Bacteroides OTU in the Orang Asli and urban controls. (C–F) Supervised LEfSE analysis (C), relative abundance of Bacteroidales (D) and Clostridiales (E), and alpha diversity as Observed OTUs (F) of the Orang Asli stool microbiota pre and post treatment with Albendazole. (n= 19 for urban controls and 55 Orang Asli. n = 53 for deworming experiments). (G) Partial Least Squares regression biplots examining within subject variances with repeated measures design to identify bacterial taxa associated with Trichuris trichiura worm burden (intensity of spots). Red arrows are Clostridiales taxa and green arrows are Bacteroidales taxa. (H) Specific OTUs identified to be positively (Dialister) or negatively (Prevotella) associated with changes to T. trichiura egg burdens. (I–J) Microbial network inference demonstrating an antagonistic relationship between Clostridiales and Bacteroidales communities from the Human Microbiome Project (I) and the pediatric IBD RISK cohort (J). The node diameter is proportional to the geometric mean of the OTU’s relative abundance. Numerical values on the edges represent the fraction of edges that are either majority positive (Green) or majority negative (Red). Also see Figure S10. ****p<0.0001 by unpaired t-test in (B), and paired t-test in (D)–(F).
To control for factors other than helminth colonization (e.g., diet), we analyzed stool samples collected from the Orang Asli before and after deworming treatment with Albendazole (Figure S9A, B, Table S3). Alpha-diversity of microbial communities was significantly reduced following treatment (Figure 4F, S9C, D). By LEfSe, Clostridiales was the most significantly reduced order, whereas Bacteroidales (Prevotella) was significantly expanded post treatment (Figure 4C–E, S9E). Utilizing the egg burden data, we combined Centered Log-Ratio (CLR) transformation with Partial Least Square (PLS) regression to examine within subject changes, incorporating a repeated measures design (48). The resulting model showed that changes in Trichuris trichiura egg burden post treatment within individuals are strongly associated with a small set of bacterial taxa, independently of age and gender (Figure 4G, S10A–C, Table S4). Specifically, Dialister and Coprococcus are two members of the order Clostridiales positively associated with changes in egg burden, whereas the Bacteroidales species Prevotella and another OTU are negatively associated (Figure 4H, S10D). Individuals without reduced egg burden did not show these changes in the microbiome, indicating that these findings are unlikely to be due to non-specific effects of Albendazole treatment (Figure S10E–G). Overall, these data support our hypothesis that helminth infection promotes the expansion of Clostridiales communities that outcompete Bacteroidales communities, although the TH2 response was not examined here. Finally, we applied a method (SPIEC-EASI) for inference of microbial ecological networks (19) to publicly available human microbiome datasets consisting of healthy USA residents (Human Microbiome Project and American Gut Project) and pediatric IBD patients (RISK cohort) (20–22) and found that the antagonistic relationship between Clostridiales and Bacteroidales is the most consistently observed negative relationship (Figure 4I, J, S11).
In this study, Clostridiales are an example of defensive symbionts with an antagonistic interaction with another common commensal bacteria (Bacteroidales), which we consistently observed in all human gut microbiome datasets. Bacteroidales are pathogenic only in susceptible Nod2 deficient hosts and this competition reverses disease pathologies. Many CD patients do not carry NOD2 variants, and hence may not respond to helminths, which have failed in clinical trials. Helminths may be beneficial only in patients with NOD2 variants or have pro-inflammatory Bacteroidales species. We propose that certain individuals may be more susceptible to deleterious consequences of a changing microbial environment and an understanding of the contribution of genetic and environmental factors towards the development of inflammatory diseases is essential to devise therapeutic strategies that consider the heterogeneity of etiologies.
Supplementary Material
Acknowledgments
We thank D. Artis for seed stock of T. muris, H. Silva for assistance with flow cytometry panels, B. Zeck and L. Chiriboga for assistance with immunohistochemistry, and NYUSoM Flow Cytometry and Cell Sorting Center, Genomics Core, and Immunohistochemistry core (supported in part by National Institute of Health (NIH) grants P30CA016087 and UL1 TR00038). The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. RNA-seq data has been made publicly available and can be accessed using the GEO accession number GSE76504. Fastq files and corresponding mapping files for 16S-sequencing data are available on request. Clostridia strains derived from human microbiota are available from K. Honda under a material transfer agreement with RIKEN. This work was supported by NIH grants DK103788 (K.C. and P.L.), DK093668 (K.C.), HL123340 (K.C.), AI093811 (P.L.), AI007180 (Z.K. and M.J.B.), DK090989 (Z.K. and M.J.B.), AI107588 (W.C.G); Broad Medical Research Program (P.L.); Kevin and Marsha Keating Family Foundation (P.L.); The MCJ Amelior Foundation (W.C.G); NIH/NCATS UL1 TR000038 (K.C. and P.L.); philanthropic support from Bernard Levine (K.C. and P.L.); and UM.C/625/HIR/MOE/MED/23 (Y.L. and P.L). K.C. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
Supplementary Materials
Materials and Methods
Figs. S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11
References (23–47)
References and notes
- 1.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstock JV, Elliott DE. Helminths and the IBD hygiene hypothesis. Inflamm Bowel Dis. 2009;15:128–133. doi: 10.1002/ibd.20633. [DOI] [PubMed] [Google Scholar]
- 3.Kamada N, Nunez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology. 2014;146:1477–1488. doi: 10.1053/j.gastro.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science. 2010;329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 5.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014;41:311–324. doi: 10.1016/j.immuni.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleynen I, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2015 doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter MM, et al. In vitro-derived alternatively activated macrophages reduce colonic inflammation in mice. Gastroenterology. 2010;138:1395–1405. doi: 10.1053/j.gastro.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 9.Ziegler T, et al. A novel regulatory macrophage induced by a helminth molecule instructs IL-10 in CD4+ T cells and protects against mucosal inflammation. J Immunol. 2015;194:1555–1564. doi: 10.4049/jimmunol.1401217. [DOI] [PubMed] [Google Scholar]
- 10.Hang L, et al. Heligmosomoides polygyrus bakeri infection activates colonic Foxp3+ T cells enhancing their capacity to prevent colitis. J Immunol. 2013;191:1927–1934. doi: 10.4049/jimmunol.1201457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaiss MM, et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity. 2015;43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klementowicz JE, Travis MA, Grencis RK. Trichuris muris: a model of gastrointestinal parasite infection. Seminars in immunopathology. 2012;34:815–828. doi: 10.1007/s00281-012-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm JB, et al. Chronic Trichuris muris Infection Decreases Diversity of the Intestinal Microbiota and Concomitantly Increases the Abundance of Lactobacilli. PLoS One. 2015;10:e0125495. doi: 10.1371/journal.pone.0125495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houlden A, et al. Chronic Trichuris muris Infection in C57BL/6 Mice Causes Significant Changes in Host Microbiota and Metabolome: Effects Reversed by Pathogen Clearance. PLoS One. 2015;10:e0125945. doi: 10.1371/journal.pone.0125945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 16.Pridmore RD, et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci U S A. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SC, et al. Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl Trop Dis. 2014;8:e2880. doi: 10.1371/journal.pntd.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama J, et al. Diversity in gut bacterial community of school-age children in Asia. Scientific reports. 2015;5:8397. doi: 10.1038/srep08397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtz ZD, et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11:e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gevers D, et al. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10:e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AmGut, American Gut Project. Technical report [Google Scholar]
- 23.Gundra UM, et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood. 2014;123:e110–122. doi: 10.1182/blood-2013-08-520619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO. A guide for managers of control programmes. Geneva: World Health Organization; 1998. Guidelines for the evaluation of soil-transmitted helminthiasis and schistosomiasis at community level. [Google Scholar]
- 25.Wakelin D. Acquired immunity to Trichuris muris in the albino laboratory mouse. Parasitology. 1967;57:515–524. doi: 10.1017/s0031182000072395. [DOI] [PubMed] [Google Scholar]
- 26.Else KJ, Wakelin D, Wassom DL, Hauda KM. MHC-restricted antibody responses to Trichuris muris excretory/secretory (E/S) antigen. Parasite Immunol. 1990;12:509–527. doi: 10.1111/j.1365-3024.1990.tb00985.x. [DOI] [PubMed] [Google Scholar]
- 27.Anthony RM, et al. Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat Med. 2006;12:955–960. doi: 10.1038/nm1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.E. Aronesty. (2011).
- 33.Caporaso J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. 623–656. [Google Scholar]
- 35.Chao A. Nonparametric estimation of the number of classes in a population. Scandinavian Journal of statistics. 1984:265–270. [Google Scholar]
- 36.Chen J, Bittinger K, Charlson E, Hoffmann C. Associating microbiome composition with environmental covariates using generalized UniFrac distances. 2012 doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. The ISME journal. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aitchison J. The statistical analysis of compositional data. Chapman and Hall; London: 1986. New York. [Google Scholar]
- 41.Chung D, Keles S. Sparse partial least squares classification for high dimensional data. Stat Appl Genet Mol Biol. 2010;9:Article 17. doi: 10.2202/1544-6115.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chun H, Keles S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J R Stat Soc Series B Stat Methodol. 2010;72:3–25. doi: 10.1111/j.1467-9868.2009.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim-Anh Lê Cao DR, Robert-Granié Christèle, Besse Philippe. A sparse PLS for variable selection when integrating omics data. Statistical applications in genetics and molecular biology. 2008;7 doi: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 44.Liu H, Roeder K, Wasserman L. Stability Approach to Regularization Selection (StARS) for High Dimensional Graphical Models. Adv Neural Inf Process Syst. 2010;24:1432–1440. [PMC free article] [PubMed] [Google Scholar]
- 45.Liquet B, Le Cao KA, Hocini H, Thiebaut R. A novel approach for biomarker selection and the integration of repeated measures experiments from two assays. BMC Bioinformatics. 2012;13:325. doi: 10.1186/1471-2105-13-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim-Anh Le Cao IG. Sebastien Dejean with key contributors Florian Rohart, Benoit Gautier, contributions from Pierre Monget, Jeff Coquery, FangZou Yao, and Benoit Liquet, mixOmics: Omics Data Integration Project. 2015 [Google Scholar]
- 47.Grencis RK. Immunity to helminths: resistance, regulation, and susceptibility to gastrointestinal nematodes. Annu Rev Immunol. 2015;33:201–225. doi: 10.1146/annurev-immunol-032713-120218. [DOI] [PubMed] [Google Scholar]
- 48.Materials and Methods
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


