Abstract
DNA double-strand breaks (DSBs) pose a threat to genome stability and are repaired through multiple mechanisms. Rarely, telomerase, the enzyme that maintains telomeres, acts upon a DSB in a mutagenic process termed telomere healing. The probability of telomere addition is increased at specific genomic sequences termed sites of repair-associated telomere addition (SiRTAs). By monitoring repair of an induced DSB, we show that SiRTAs on chromosomes V and IX share a bipartite structure in which a core sequence (Core) is directly targeted by telomerase, while a proximal sequence (Stim) enhances the probability of de novo telomere formation. The Stim and Core sequences are sufficient to confer a high frequency of telomere addition to an ectopic site. Cdc13, a single-stranded DNA binding protein that recruits telomerase to endogenous telomeres, is known to stimulate de novo telomere addition when artificially recruited to an induced DSB. Here we show that the ability of the Stim sequence to enhance de novo telomere addition correlates with its ability to bind Cdc13, indicating that natural sites at which telomere addition occurs at high frequency require binding by Cdc13 to a sequence 20 to 100 bp internal from the site at which telomerase acts to initiate de novo telomere addition.
INTRODUCTION
Chromosomes in the budding yeast Saccharomyces cerevisiae, as in all eukaryotes, terminate with specialized nucleoprotein structures called telomeres. S. cerevisiae telomeric DNA consists of ∼250 to 350 bp of TG1–3/AC1–3 repeats and a short (∼10-bp) terminal G-rich 3′ overhang (1). Because the conventional DNA replication machinery cannot fully replicate chromosome ends, telomeres shorten with each cell division cycle. In most eukaryotes, telomere shortening is counteracted by the enzyme telomerase, a ribonucleoprotein complex that uses an intrinsic RNA subunit as the template for telomeric DNA synthesis. Associated with telomeric DNA are proteins that protect chromosome ends from nucleolytic resection and prevent chromosome end-to-end fusions by distinguishing natural chromosome ends from ends generated by DNA double-strand breaks (DSBs) (2). These protective functions make telomeres essential for the maintenance of genome integrity and cell viability.
In S. cerevisiae, optimal telomere length requires a balance between positive and negative regulatory mechanisms mediated by telomere-binding proteins, including Cdc13 and Rap1 (3). Cdc13, a telomere sequence-specific single-stranded DNA (ssDNA) binding protein, recruits telomerase to telomeres during the S/G2 phase of the cell cycle through interaction with Est1, a subunit of the telomerase holoenzyme (reviewed in reference 3). Rap1 binds to the double-stranded telomeric repeat, forming a telomere length-regulatory complex through interactions of its C-terminal domain with Rif1 and Rif2 (4, 5). Regulation occurs through a counting mechanism in which telomere length is inversely proportional to the number of Rif1 and Rif2 molecules present at a telomere (6, 7).
Cells experience insults to their genome from endogenous and exogenous sources, including reactive oxygen species, radiation, and chemical mutagens (8). DSBs resulting from these sources pose an enormous threat to genome stability and cell viability, since failure to repair DSBs can cause chromosome rearrangements and/or chromosome loss. Eukaryotic cells utilize two main pathways for DSB repair: a homologous recombination (HR) pathway, which uses a sister chromatid or homologous chromosome as the template for DSB repair, and a nonhomologous end-joining (NHEJ) pathway, in which broken DNA ends are directly ligated (9). Inaccurate repair of DSBs can give rise to gross chromosomal rearrangements (GCRs), large-scale changes in chromosome structure that include interstitial deletions, chromosome end-to-end fusions, and translocations (10). Direct action of telomerase at DSBs results in yet another type of GCR, de novo telomere addition, in which all genetic information distal to the DSB is lost (11).
In yeast, either Cdc13 or Rap1 can stimulate de novo telomere addition. Cdc13 appears to facilitate telomerase recruitment to DSBs in a manner similar to its role at endogenous telomeres (12). Following DSB induction by homothallic switching (HO) endonuclease, Cdc13 and Est1 are both recruited to an artificial telomere seed (∼80-bp TG tract) inserted adjacent to the HO site, and artificial tethering of Cdc13 adjacent to the break is sufficient to stimulate de novo telomere addition (12). Est1 recruitment depends on its interaction with Cdc13, although the converse is not true (12). In contrast to its negative regulatory role at endogenous telomeres, Rap1 stimulates de novo telomere addition at artificial chromosomal (13, 14) or extrachromosomal termini containing short telomere-like sequences (15). While informative about the mechanisms of telomerase recruitment, the vast majority of the studies referenced here utilize artificial sequences to facilitate the formation of de novo telomeres. Cdc13 and/or Rap1 could stimulate de novo telomere addition at endogenous intrachromosomal TG-rich sequences with the potential to bind one or both proteins. However, a potential role for Cdc13 or Rap1 at such sequences has not been directly addressed.
Given that de novo telomere addition at intrachromosomal TG-rich sequences has the potential to influence genome stability, we sought to identify the cis- and trans-acting factors required for de novo telomere addition at such endogenous sequences. Here, we investigate the requirements for de novo telomere addition at an 84-bp site of repair-associated telomere addition, located 35 kb from the left telomere of chromosome V (SiRTA 5L-35). As previously described (16), the majority of telomere additions at SiRTA 5L-35 occur within a 23-bp TG-rich sequence, which we refer to as the Core sequence.
We find that a separate TG-rich sequence located centromere proximal to the Core, and which itself is infrequently targeted for telomere addition, is required for high levels of telomere addition at the Core (referred to here as the “Stim” sequence). Using two different approaches to differentially target Cdc13 or Rap1 to SiRTA 5L-35, we show that it is likely the ability of Cdc13 to bind the Stim sequence that promotes telomere addition at SiRTA 5L-35. SiRTA 5L-35 therefore has a bipartite structure in which Cdc13 binding to an upstream sequence stimulates telomere addition at a neighboring target site. Finally, we report the identification of a new SiRTA located 44 kb from the left telomere of chromosome IX and show that SiRTA 9L-44 has a bipartite structure similar to that of SiRTA 5L-35.
MATERIALS AND METHODS
Yeast strains and plasmids.
Table S1 in the supplemental material contains a complete list of the strains used in this study. Unless otherwise noted, strains were grown in yeast extract-peptone-dextrose (YEPD) at 30°C. In strains utilized for HO cleavage assays (YKF1310 and YKF1308), the HMRa sequence on chromosome III was replaced with nat, which confers resistance to nourseothricin. All gene deletions were generated by one-step gene replacement with a selectable marker and were confirmed by PCR. hxt13::URA3 disruptions were created by using a plasmid in which the KpnI-SphI restriction fragment from HXT13 was replaced with URA3 or by amplifying the hxt13::URA3 locus from an existing strain using primers hxt13::URA3 F and hxt13::URA3 R (see Table S2 in the supplemental material). The HO cleavage site was integrated on chromosomes VII and IX by one-step gene replacement using plasmid pJH2017 (HOcs::HPH; gift of J. Haber) as the template with selection for hygromycin B resistance. URA3 was integrated on chromosome VII by one-step gene replacement at the PAU11 locus and on chromosome IX by one-step gene replacement at the SOA1 locus. Primers utilized for PCR are listed in Table S2 in the supplemental material.
Strain YKF1409 (rad52::LEU2) was generated by transformation with BamHI-digested plasmid pSM20, which contains the rad52::LEU2 disruption allele (17).
Mutations in SiRTA 5L-35 (see Fig. 2 to 4) were introduced by two-step gene replacement (18). DNA fragments containing each mutation were generated by PCR using gene splicing by overlap extension (gene SOEing) (19). Met-NPR2 For primer was used with a reverse primer containing the desired mutation to generate fragment I, and NPR2 mid-Rev primer was used with a forward primer containing the same mutation to generate fragment II (see Table S2 in the supplemental material). Fragments I and II were extended by mutually primed synthesis using the Met NPR2 For and Mid NPR2 Rev primers to generate a final PCR product that was cleaved with HindIII and XbaI for ligation into pRS306. The integration plasmid was linearized with BamHI or BclI prior to transformation and selection for Ura+ integrants. To facilitate the identification of strains containing the desired mutations following selection on medium containing 5-fluoroorotic acid (5-FOA), two-step integration was first used to create strain YKF1366, in which SiRTA 5L-35 is deleted. Subsequent strains were created by reintroducing mutated versions of SiRTA 5L-35 into strain YKF1366 by two-step integration. The resulting 5-FOAr isolates were screened by PCR, and candidates were sequenced to confirm the presence of the desired mutations.
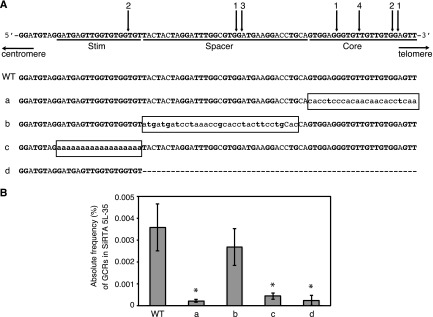
FIG 2.
High rates of telomere addition at SiRTA 5L-35 require two separable sequences. (A) Top schematic, sequence of SiRTA 5L-35 with arrows indicating sites of de novo telomere addition. The most 3′ chromosomal nucleotide with identity to the cloned de novo telomere is indicated. Individual cloned sequences are shown in Fig. S4 in the supplemental material. Numbers above arrows indicate the number of independent telomere addition events mapped to each site. Stim, Spacer, and Core are defined in the text. Bottom schematic, mutations created in SiRTA 5L-35. Uppercase letters represent unchanged nucleotides, lowercase letters enclosed in box represent mutated nucleotides, and the dashed line indicates deleted nucleotides. (B) Core and Stim sequences contribute to the formation of GCR events within SiRTA 5L-35. The frequency (%) at which GCR events occur within SiRTA 5L-35 following induction of HO endonuclease expression on medium containing galactose is shown for the WT strain and for the mutant strains as depicted in panel A. Averages from three independent replicates are shown with standard deviations. Mutants marked with an asterisk are significantly different from WT (P < 0.05) by analysis of variance (ANOVA) with post hoc Tukey's honestly significant difference (HSD).
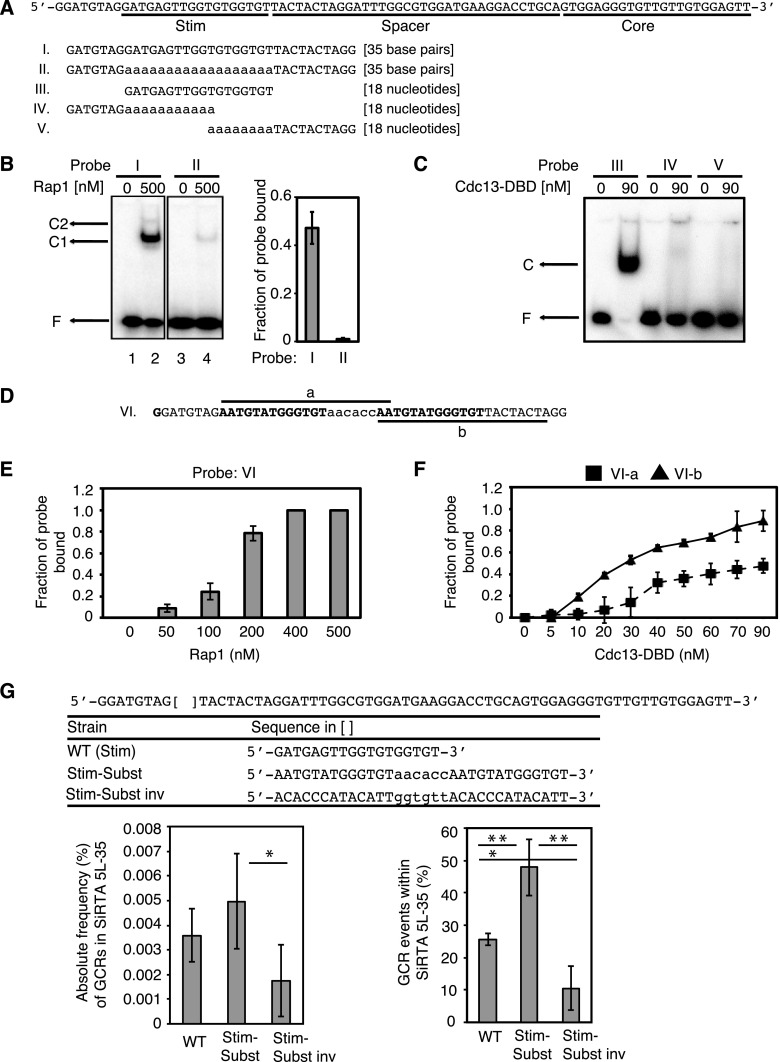
FIG 4.
The Stim sequence of SiRTA 5L-35 binds Cdc13 and Rap1 in vitro and can be functionally replaced with a sequence that binds both proteins. (A) Top schematic, sequence of SiRTA 5L-35 as described for Fig. 2A. Bottom schematic, sequences of probes utilized for Cdc13-DBD and Rap1 binding assays. Mutated bases are shown in lowercase letters. Probes are identified with Roman numerals, and this numbering is maintained throughout the figures. Probes I and II are double stranded; probes III to V are single stranded. (B) Binding of recombinant Rap1 to probe I (WT SiRTA 5L-35) and probe II [Stim poly(A) mutation, Fig. 2A]. The mobility of free probe (F) and two bound complexes (C1 and C2) is indicated. The fraction of probe bound when utilizing 500 nM Rap1 is quantified on the right from three independent experiments. Error bars represent standard deviations. (C) Binding of recombinant Cdc13-DBD to probes III, IV, and V, as indicated. Probe III contains the WT SiRTA 5L-35 sequence. Probes IV and V contain the Stim poly(A) mutation from Fig. 2A. This sequence was tested in two pieces to avoid the formation of secondary structure. The mobility of free probe (F) and bound complex (C) is indicated. (D) The sequence shown was designed to contain two predicted Rap1 binding sites (bold) separated by a linker sequence (lowercase letters). Probe VI is double stranded and is utilized in data shown in panel E. Probes VI-a and VI-b are single-stranded sequences as indicated and are utilized in data shown in panel F. (E) Binding of the indicated concentration of recombinant Rap1 to probe VI. (F) Binding of the indicated concentration of recombinant Cdc13-DBD to probes VI-a or VI-b. For panels E and F, the average fraction of probe bound was determined from three independent experiments. Error bars represent standard deviations. (G) The SiRTA 5L-35 Stim sequence (indicated by brackets) was replaced at the endogenous locus on chromosome V with the three sequences depicted here. Stim-Subst is identical to the sequence of Probe VI. Stim-Subst inv is the reverse complement of the Stim-Subst sequence. Average results and standard deviations from three HO cleavage assays are shown as the absolute frequency (%) of GCR formation within SiRTA 5L-35 (left graph) or the percentage of total GCR events that occur within SiRTA 5L-35 (right graph). Samples with statistically different values by ANOVA with post hoc Tukey's HSD are indicated (*, P < 0.05; **, P < 0.01).
For other mutations (described in Fig. 5 and 6), a portion of SiRTA 5L-35 including the Stim sequence was replaced with URA3 in strain YKF1323 by one-step gene replacement to generate strain YKF1585. Sequences to be integrated were generated by PCR using the HS forward and HS reverse primers and transformed into strain YKF1585. After allowing cells to recover for 24 to 48 h on rich medium, cells were replica plated to medium containing 5-FOA. Mutations were confirmed by sequencing.
FIG 5.
The rate of de novo telomere addition at SiRTA 5L-35 correlates with the ability of the SiRTA Stim sequence to bind Cdc13. (A) Probes utilized for Cdc13-DBD and Rap1 binding assays. Probes I, III, and VI are identical to those described for Fig. 4. Probes followed by “Rap1 EMSA” are double stranded. Those indicated with “Cdc13 EMSA” are single stranded. (B) Binding of the indicated concentration of recombinant Rap1 to double-stranded probes shown in panel A. (C) Binding of the indicated concentration of recombinant Cdc13-DBD to single-stranded probes shown in panel A. In panels B and C, the average fraction of probe bound was determined from three independent experiments. Error bars represent standard deviations. (D) The SiRTA 5L-35 Stim sequence (indicated by brackets) was replaced at the endogenous locus on chromosome V with the sequences depicted here. Average results and standard deviations for three HO cleavage assays are shown as absolute frequency (%) of GCR formation within SiRTA 5L-35 (left graph) or percentage of total GCR events within SiRTA 5L-35 (right graph). Averages indicated with 2 asterisks are significantly different (P < 0.01) by unpaired Student's t test.
FIG 6.
Artificial recruitment of Cdc13 to the SiRTA 5L-35 stimulatory site increases the rate of GCR formation. (A) The SiRTA 5L-35 Stim sequence was replaced with two tandem copies of the Gal4 upstream activating sequence (2× UAS) or with a string of adenines [poly(A); identical sequence to that of mutation d in Fig. 2A]. (B) The rate of GCR formation within SiRTA 5L-35 is shown for strains containing either the 2× UAS or poly(A) sequences integrated in place of SiRTA 5L-35 Stim. Cells are transformed with pRS314 (empty vector) or with pRS414 expressing either CDC13 or RAP1 as N-terminal fusions with the Gal4 DNA binding domain (GBD). Values for the three rightmost columns are maximum estimates (see Materials and Methods). Error bars indicate standard deviations for three independent experiments. Averages indicated with 2 asterisks are significantly different (P < 0.01) by ANOVA with post hoc Tukey's HSD.
SiRTA 5L-35 spacerΔ was integrated on chromosome VII, and mutations were introduced at SiRTA 9L-44 by two-step gene replacement essentially as described above. Table S2 in the supplemental material contains the sequences of primers used for strain construction.
Plasmid pAB180 (pRS414-ADHpromoter-GBD-CDC13) was a gift from A. Bianchi. pAB180-Rap1 was created by replacing the CDC13 open reading frame in pAB180 with the full-length RAP1 open reading frame at the NcoI and AatII sites. All amplified regions were confirmed by sequencing.
GCR assays.
For spontaneous GCR assays, cells cultured overnight in synthetic drop-out medium lacking uracil (SD-Ura) were used to inoculate YEPD cultures (approximately 30 per strain). YEPD cultures were grown overnight to saturation (approximately 24 h) and plated onto medium containing 5-FOA and canavanine (Can) to isolate GCRs (5-FOAr Canr colonies). Only one colony was analyzed from each plate to ensure independence. The approximate location and nature of GCR events were determined by multiplex PCR (20) (see Fig. S1 in the supplemental material) and Southern blotting (see below).
For HO cleavage GCR assays, cells were grown in SD-Ura medium (with 2% raffinose) to an optical density at 600 nm (OD600) of ∼0.3 to 1. Cells were plated on yeast extract-peptone medium with 2% galactose (YEPG), and a dilution was plated on YEPD to determine total cell number. After 3 days, colonies were counted and galactose-resistant colonies were transferred to SD medium containing 5-FOA to isolate GCR events. At least 100 galactose-resistant (Galr) colonies were individually plated on medium containing 5-FOA to determine the rate of URA3 loss. If necessary, additional colonies were obtained by replica plating. The approximate location and nature of GCR events was determined by multiplex PCR (20; see also Fig. S2 in the supplemental material) and Southern blotting (see below).
The absolute frequency at which GCR events occur within a SiRTA is calculated by multiplying the rate of survival on galactose (colonies on galactose/colonies on glucose, corrected for dilution factor) by the fraction of Galr colonies capable of growth on medium containing 5-FOA and the fraction of Galr 5-FOAr colonies in which the GCR event occurred within the SiRTA as measured by PCR. Approximately 30 Galr 5-FOAr colonies were analyzed for each experiment, and the averages and standard deviations presented are derived from a minimum of three independent experiments. In a few cases, rates shown are upper estimates because no 5-FOA-resistant colonies were detected among 100 colonies analyzed or no events were obtained within the SiRTA sequence among ∼30 Galr 5-FOAr colonies. These cases are indicated in the figure legends when applicable.
Southern blotting.
For the characterization of GCR events, approximate locations determined by multiplex PCR were used to design restriction enzyme digests and probes for Southern blot analysis. Information about the restriction enzymes and primers utilized are available upon request. Digested fragments were separated on a 0.7% agarose gel, transferred to nylon membrane (Hybond N+), and probed with [32P]dCTP-labeled telomeric DNA. Radioactive membranes were exposed to Phosphor screens (Molecular Dynamics), and screens were scanned with Typhoon TRIO variable mode imager (GE Healthcare). Telomere addition was determined by visualization of the characteristic smear generated by the heterogeneous telomere sequence at a size consistent with the PCR result.
Protein purification.
Plasmid pET21a-Cdc13-DBD-His6 (21) was a gift from Deborah Wuttke (University of Colorado, Boulder, CO). Escherichia coli BL21(DE3) transformed with pET21a-Cdc13-DBD-His6 was grown at 37°C in LB medium with ampicillin (50 mg/liter) to an OD600 of ∼0.6. Cdc13-DBD-His6 expression was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM at 22°C for approximately 6 h, after which cells were harvested by centrifugation. Cells were resuspended in buffer A (25 mM HEPES–NaOH, 10% glycerol, 300 mM NaCl, 0.01% NP-40, 20 mM imidazole, 1 mM benzamidine, 0.2 mM phenylmethylsulfonyl fluoride), and cell lysis was achieved by incubation with lysozyme solution (1 mg/ml lysozyme, 1 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 10 mM NaCl, 0.5% Triton X-100), followed by sonication for 60 s (two 30-s cycles) using a Branson 450 Sonifier (power setting 3, 70% duty cycle). Supernatant was separated from cellular debris by centrifugation, and Cdc13-DBD-His6 was purified from supernatant with Ni-nitrilotriacetic acid (Ni-NTA)–agarose beads. Beads were washed in buffer A, and protein was eluted from beads in buffer B (25 mM HEPES–NaOH, 10% glycerol, 300 mM NaCl, 0.01% NP-40, 200 mM imidazole, 1 mM benzamidine, 0.2 mM phenylmethylsulfonyl fluoride). Eluted protein was dialyzed overnight in buffer C (25 mM HEPES–NaOH, 10% glycerol, 300 mM NaCl). Protein concentration was estimated by polyacrylamide gel electrophoresis, using bovine serum albumin as a standard. Purification of Rap1 has been previously described (22).
EMSA.
To generate electrophoretic mobility shift assay (EMSA) probes, oligonucleotide pairs were mixed in equimolar ratios, boiled in 1× annealing buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA), and slowly cooled to room temperature. Probes were radiolabeled with T4 polynucleotide kinase or by fill-in synthesis with Klenow polymerase. For Rap1 EMSAs, each 20-μl EMSA reaction mixture contained binding buffer (20 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol [DTT], 70 mM KCl, 1 mM EDTA, 5% glycerol, 2.5 ng/μl bovine serum albumin [BSA]), 1.5 μg herring sperm DNA (Sigma-Aldrich), labeled probe (30 to 50 nM), and the indicated amounts of Rap1. Reaction products were separated by 5% native polyacrylamide gel electrophoresis (PAGE) in 0.5× Tris-borate-EDTA (TBE) at 100 V for 45 min. Gels were fixed in 10% acetic acid–20% methanol and then exposed to Phosphor screens. Screens were scanned using the Typhoon TRIO Variable Mode Imager, and results were analyzed with ImageQuant TL 7.0 software (GE Heathcare). For Cdc13 EMSAs, each 20-μl EMSA reaction mixture contained binding buffer (50 mM Tris-HCl [pH 7.5], 1 mM DTT, 75 mM KCl, 75 mM NaCl, 0.1 mM EDTA, 15% glycerol, 1 mg/ml BSA), labeled probe (62.5 nM), and the indicated amount of Cdc13-DBD protein. Reaction products were separated on 6% native polyacrylamide gels (containing 5% glycerol) in 1× TBE buffer at 200 V for 30 min. Subsequent steps were as described for Rap1 above.
Cloning and sequencing of de novo telomeres.
Cloning of telomeric DNA was performed as previously described (23) with minor modifications. Blunting of genomic DNA ends was done with T4 DNA polymerase (New England BioLabs) in the presence of 0.1 mM deoxynucleoside triphosphates (dNTPs). Sequences for “ds oligo 1” and “ds oligo 2” (used to create the double-stranded oligonucleotide that was ligated to the blunted DNA ends) can be found in Table S2 in the supplemental material. Telomere PCR was performed with ds oligo 2 and a primer internal to the de novo telomere (see Table S2 in the supplemental material). PCR products were separated in 2% agarose gels, purified, and ligated into pGEM-T Easy vector (Promega) or pDrive Vector (Qiagen). Sequencing of inserts was carried out by GenHunter (Nashville, TN) or Genewiz (South Plainfield, NJ) using the primer M13F (−20) (see Table S2 in the supplemental material).
RESULTS
An internal telomere-like sequence on chromosome V in S. cerevisiae is a target of de novo telomere addition.
In S. cerevisiae, a short sequence within the NPR2 gene on the left arm of chromosome V (chrV-L) incurs a high frequency of de novo telomere addition relative to that observed in flanking sequences (16). To determine the magnitude of this effect in our strain, we utilized an assay (24) in which CAN1 and URA3 are utilized to select GCR events within a 12-kb region on chrV-L between CAN1 and the first essential gene (Fig. 1A). Independent liquid cultures were plated on medium containing canavanine (Can) and 5-fluoroorotic acid (5-FOA), drugs toxic to cells expressing CAN1 and URA3, respectively. A single surviving colony was isolated from each culture for analysis. As previously shown (16, 24), nearly all cells resistant to both drugs lost DNA sequences distal to CAN1. The approximate location of each rearrangement was determined by multiplex PCR using primer pairs spanning the 12-kb region (see Fig. S1 in the supplemental material), and each chromosome rearrangement was subsequently classified as “telomere addition” or “other” using Southern blot analysis to detect the characteristic “smear” generated by heterogeneous telomeric repeats.
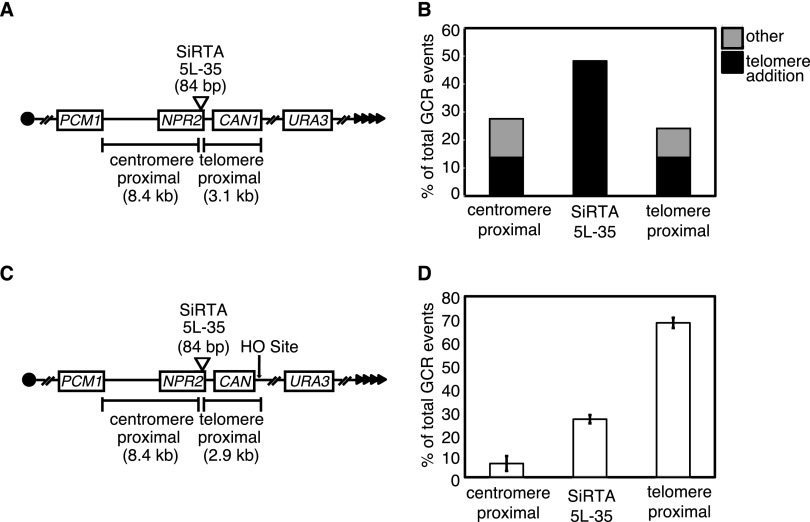
FIG 1.
SiRTA 5L-35 incurs a high frequency of de novo telomere addition relative to flanking sequences. (A) Schematic of chromosome V GCR assay system. Filled triangles represent the terminal telomeric repeats. Throughout the figures, chromosome arms are diagrammed with the telomere to the right. This convention places the 3′ terminus upon which telomerase directly acts on the top strand of DNA. (B) Distribution of spontaneously occurring GCR events in the WT strain. GCR events were mapped by multiplex PCR (see Fig. S1 in the supplemental material) to one of the three regions indicated in panel A. The type of event (telomere addition or “other”) was determined by Southern blotting. A total of 29 events were analyzed. The enrichment of GCR events within the 84-bp SiRTA relative to the expected frequency (assuming a random distribution of GCR events across the 11.5-kb target region) was significant by Fisher's exact test (P < 0.001). (C) Schematic of the chromosome V HO-inducible GCR assay system. Expression of the HO endonuclease is induced by growth on medium containing galactose, and the site of HO cleavage is indicated (arrow). (D) Distribution of HO endonuclease-induced GCR events in the WT strain. Data are from three independent experiments and ∼30 to 40 GCRs per experiment. Error bars represent standard deviations.
Three PCR primers were designed to detect GCR formation within an 84-bp TG-rich sequence encompassing the telomere addition hot spot within NPR2 (see Fig. S1 in the supplemental material, PCR products 4 and 5). To reflect the propensity for de novo telomere addition, we refer to this sequence as the site of repair-associated telomere addition 35 kb from the left telomere of chromosome V (SiRTA 5L-35). Similar to what was seen in previous reports (16), 48.3% (14 of 29) of GCR events occurred in SiRTA 5L-35. The remaining events were split evenly between the regions centromere and telomere proximal to SiRTA 5L-35 (Fig. 1B). In every case, GCR events within SiRTA 5L-35 involved de novo telomere addition, whereas GCRs in the flanking regions consisted of both telomere additions and other chromosome rearrangements (Fig. 1B). Given that the 84-bp SiRTA 5L-35 comprises less than 1% of the total region analyzed, this sequence incurs a remarkably high frequency of de novo telomere addition.
To eliminate concern that telomere addition arises from a propensity for DNA breakage within or near this region, we utilized a second approach in which a site-specific DNA break is introduced ∼3 kb distal to SiRTA 5L-35. The strain utilized (25) contains a single recognition site for the yeast homothallic switching endonuclease (HO endonuclease) within the CAN1 gene (Fig. 1C). Homologous sequences on chromosome III are deleted to eliminate repair by gene conversion. The HO endonuclease gene is expressed from a galactose-inducible promoter such that growth on medium containing galactose results in a DSB ∼3 kb distal to SiRTA 5L-35 (Fig. 1C). Most cells accurately repair the DSB, resulting in a cleavage-repair cycle that culminates in cell death. However, approximately 0.1% of cells survive as a result of mutations at the HO site that prevent further cleavage. These cells have incurred small insertions or deletions or have lost all DNA distal to the HO site. The latter are identified by selection on 5-FOA for cells lacking the distal URA3 marker (Fig. 1C). We refer to cells that survive on galactose and have lost the URA3 marker (Galr 5-FOAr colonies) as GCR events. The propensity for GCR formation to occur at SiRTA 5L-35 is expressed in two ways: (i) as the overall rate at which GCR formation occurs within SiRTA and (ii) as the fraction of GCR events that occur within SiRTA. In both cases, values are the averages (with standard deviations) for at least three independent experiments with 25 to 35 GCR events analyzed per experiment.
By PCR analysis (see Fig. S2A in the supplemental material), 25.7% ± 1.9% of GCR events following HO cleavage occurred within SiRTA 5L-35, 68.3% ± 2.3% occurred between the HO cleavage site and SiRTA 5L-35, and the remainder occurred in the centromere-proximal region between SiRTA 5L-35 and the first essential gene (Fig. 1D). We analyzed a subset of events (69 from two independent assays) by Southern blotting. Of 18 events that mapped to SiRTA 5L-35, 17 (94.4%) involved de novo telomere addition (see Fig. S2B in the supplemental material). Events that occurred distal to SiRTA 5L-35 fell into two classes. The majority of distal events (36 of 48) occurred at or immediately adjacent to the HO cleavage site (within 60 bp), and of those, 75.0% involved de novo telomere addition. In contrast, only 12 events occurred within the 3 kb separating the HO site from SiRTA 5L-35 and 58.3% of those events involved de novo telomere addition (see Fig. S2B in the supplemental material). De novo telomere addition events at SiRTA 5L-35 are mediated by telomerase since deletion of RAD52 to eliminate recombination-mediated telomere maintenance (26) did not reduce the fraction of GCR events occurring at SiRTA 5L-35 (see Fig. S3 in the supplemental material).
A sequence internal to the direct target of telomere addition is required for high levels of de novo telomere addition at SiRTA 5L-35.
To identify sequences required to direct high levels of de novo telomere addition at SiRTA 5L-35, we cloned and sequenced 14 telomere addition events following HO cleavage. Telomere addition occurred at seven different sites, four of which were used more than once (Fig. 2A). All 14 events were independent, as reflected in the divergent telomere sequences added to each (see Fig. S4 in the supplemental material). Of the 14 telomere additions, 8 (57.1%) occurred within the original 23-bp TG-rich sequence defined by Stellwagen et al. (16). Interestingly, very few events occurred at the centromere-proximal end of SiRTA 5L-35, despite the telomere-like nature of that sequence (Fig. 2A).
To determine which sequences contribute to the high rate of de novo telomere addition, we created a series of mutations, diagrammed in Fig. 2A. Mutation of the 23-bp TG-rich sequence in which telomere addition frequently occurs (each base mutated to its complement) decreased the overall frequency of GCR events at SiRTA 5L-35 16-fold (Fig. 2B, mutation a) and reduced the percentage of total GCR events mapping to SiRTA 5L-35 from 25.7% ± 1.9% to 4.8% ± 2.5% (see Fig. S5 in the supplemental material). In contrast, mutation of the neighboring sequence (mutation b) did not significantly change the overall rate of GCR formation (Fig. 2B), with 20.3% ± 3.7% of the GCR events mapping to SiRTA 5L-35 (see Fig. S5 in the supplemental material). To address a potential role for the TG-rich sequence at the centromere-proximal end of SiRTA 5L-35, we mutated this 18-bp region to adenine (mutation c). Although this sequence is rarely the direct target of telomere addition, the effect on the rate of GCR formation within SiRTA 5L-35 was nearly as pronounced as that seen when the target sequences themselves were mutated (Fig. 2B; compare mutations a and c) and only 5.6% ± 2.1% of total GCR events mapped to SiRTA 5L-35 (see Fig. S5 in the supplemental material). Furthermore, only 3 of 5 of those events involved de novo telomere addition (data not shown). While this sequence enhances telomere addition at SiRTA 5L-35, it is insufficient to support high levels of de novo telomere addition since a strain containing only the centromere-proximal sequences (mutation d) underwent a low rate of GCR formation within SiRTA 5L-35 with a small fraction of GCR events at SiRTA 5L-35 (4.4% ± 1.9%) (Fig. 2B; see also Fig. S5 in the supplemental material).
We conclude that high levels of telomere addition at SiRTA 5L-35 require a bipartite structure in which one sequence serves as the primary, direct target of telomere addition (the SiRTA Core; defined by mutation a), while the other sequence (SiRTA Stim; defined by mutation c) stimulates telomere addition within or near the Core sequence. The spacer between these sites makes no sequence-specific contribution to the stimulation of de novo telomere addition.
The Core and Stim sequences of SiRTA 5L-35 are sufficient to stimulate high levels of de novo telomere addition at an ectopic site.
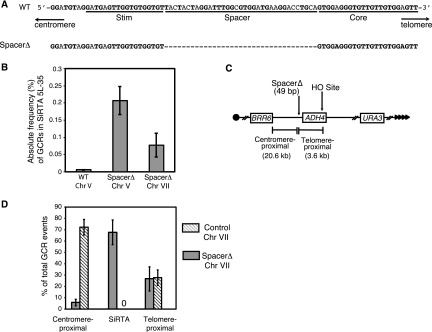
Given that the spacer between the SiRTA 5L-35 Core and Stim sequences could be mutated with little effect on telomere addition, we tested the effect of deleting this sequence (Fig. 3A). Strikingly, the overall rate of GCR formation within SiRTA 5L-35 following HO cleavage increased 36-fold compared to the wild-type SiRTA 5L-35 (spacerΔ) (Fig. 3B), and 74.2% ± 15.6% of total GCR events occurred within SiRTA 5L-35. Analysis by Southern blotting of 35 GCR events from one representative assay showed that all 28 events within SiRTA 5L-35 spacerΔ involved de novo telomere addition (data not shown). In addition, 5 of the remaining 7 events, originally classified as telomere proximal by PCR, actually involved telomere addition within 100 bp of the spacerΔ variant of SiRTA 5L-35. No events of this type were observed among 69 GCR events characterized in the wild-type (WT) strain.
FIG 3.
The SiRTA Stim and Core sequences are sufficient to stimulate de novo telomere addition at an ectopic location. (A) Top schematic, sequence of SiRTA 5L-35 as described for Fig. 2A. Bottom schematic, spacerΔ mutation created in SiRTA 5L-35. The dashed line indicates deleted nucleotides. (B) The absolute frequency (% total cells) of GCR formation at SiRTA 5L-35 is shown for the spacerΔ variant at its endogenous location on chromosome V and at an ectopic site on chromosome VII. Data for WT SiRTA 5L-35 are shown for comparison (same as Fig. 2B). Values are the averages from three independent experiments with standard deviations. (C) Schematic of the modified left arm of chromosome VII. Sizes of the regions between the integrated spacerΔ sequence and either the HO cleavage site (telomere proximal) or the most distal essential gene (BRR6; centromere proximal) are indicated. (D) The percentage of GCR events occurring in each indicated region on chromosome VII is shown for the experimental strain (SiRTA 5L-35 spacerΔ) and a control strain (no integration). In the control strain, no GCR events were observed in the 219-bp region that is replaced by the spacerΔ variant in the experimental strain. Values are averages from three independent experiments with standard deviations.
We took advantage of this remarkably high level of de novo telomere addition to ask whether the SiRTA 5L-35 Core and Stim sequences are sufficient to confer this property to an ectopic site. A 49-bp sequence containing the SiRTA 5L-35 Core and Stim sequences (SiRTA-spacerΔ) was integrated within nonessential sequences on chromosome VII-L, ∼20 kb from the first essential gene (BRR6). The galactose-inducible HO cleavage cassette was placed 3 kb telomere proximal to the ectopic SiRTA sequence, and URA3 was integrated to monitor the rate of terminal deletion (Fig. 3C). A strain containing only the HO cleavage site and URA3 marker served as a control.
In the control strain, no GCR events were observed within a 542-bp sequence corresponding to the insertion site and 72.2% ± 6.9% of the GCR events occurred centromere-proximal to this location (Fig. 3D). In striking contrast, 67.6% ± 10.8% of total GCR events in the experimental strain occurred within SiRTA-spacerΔ and only 5.7% ± 2.8% mapped to the centromere-proximal region (Fig. 3D). Southern analysis was conducted on 35 events from a single experiment. Of 22 events mapped by PCR to the SiRTA-spacerΔ sequence, 20 (91%) involved telomere addition. Furthermore, of 12 events that mapped telomere proximal to the SiRTA-spacerΔ sequence, all but one involved telomere addition within 100 bp of SiRTA-spacerΔ. Therefore, in this subset of 35 GCR events, 91.4% involved de novo telomere addition within or immediately adjacent to the SiRTA-spacerΔ insert. The overall rate of GCR formation within SiRTA-spacerΔ was modestly (2.7-fold) lower on chromosome VII than at the endogenous location on chromosome V but still much higher than that observed for the endogenous SiRTA 5L-35 (Fig. 3B).
Taken together, these results indicate that the SiRTA 5L-35 Core and Stim sequences are sufficient to support de novo telomere addition following a distal chromosome break. Reducing the spacing between the stimulatory and core sequences dramatically increases the rate of de novo telomere addition. Interestingly, sequences located within approximately 100 bp of the SiRTA nucleate telomere addition when the spacer sequence is deleted, most likely because the stimulatory sequence is now closer to these sites.
Sequences that bind Rap1 and Cdc13 stimulate de novo telomere addition at SiRTA 5L-35.
We reasoned that the enhancing properties of SiRTA Stim may arise from one or more proteins bound at that site. Given the TG-rich nature of this sequence, we investigated the ability of Rap1 and Cdc13 to bind the SiRTA Stim sequence in vitro. Rap1 is a double-stranded DNA binding protein that binds at high frequency within the endogenous telomeric repeat (27) but binds additional internal chromosomal sites as a transcription factor (28). Cdc13 binds the single-stranded overhang at the yeast telomere (29–33) and could bind at SiRTA Stim following resection of a DSB at a distal site.
Electrophoretic mobility shift assays (EMSAs) were performed using recombinant Rap1 and the DNA binding domain of Cdc13 (Cdc13-DBD; amino acids 497 to 694) to monitor binding to double-stranded or single-stranded target DNAs, respectively. The DNA binding domain of Cdc13 alone closely mimics the binding specificity of the full-length protein (21). Indeed, the endogenous SiRTA Stim sequence is bound by both Rap1 (Fig. 4A and B, probe I) and by Cdc13-DBD (Fig. 4A and C, probe III). As predicted, the poly(A) mutation that disrupts SiRTA Stim function (Fig. 2) reduces binding by Rap1 and Cdc13-DBD (Fig. 4A to C, probes II, IV, and V), consistent with one or both of these proteins playing a role in the stimulation of de novo telomere addition.
To address whether binding by Rap1 and/or Cdc13 is sufficient to stimulate de novo telomere addition, we designed a sequence predicted to contain two tandem Rap1 binding sites and to have the ability to bind Cdc13. By EMSA, this sequence (Fig. 4D, Stim-Subst) binds Rap1 with higher affinity than the endogenous SiRTA Stim sequence (Fig. 4B and E, compare binding to probes I and VI) and also binds Cdc13-DBD (Fig. 4F, probes VIa and VIb). The Stim-Subst sequence was integrated in place of the SiRTA Stim sequence on chromosome V, and the frequency of GCR events at SiRTA 5L-35 was measured. Consistent with either Rap1 and/or Cdc13 playing a role in the stimulation of telomere addition, this artificial sequence stimulated GCR events at SiRTA 5L-35 at a rate equivalent to the endogenous sequence and increased the fraction of GCR events occurring within SiRTA 5L-35 (Fig. 4G).
To verify that the Stim-Subst sequence is not the direct target of telomere addition, we designed a reverse primer within the SiRTA spacer sequence that generates a PCR product only if a de novo telomere is added within or near the SiRTA Core sequence (see Fig. S6 in the supplemental material). Consistent with our sequencing results in the WT strain, in which 12 of 14 (85.7%) telomere addition events were located distal to SiRTA Stim (Fig. 2A), 17 of 20 (85.0%) previously uncharacterized events generated a PCR product in this assay (see Fig. S6 in the supplemental material). Importantly, PCR product was obtained in 34 of 35 (97.1%) SiRTA GCR events in the Stim-Subst strain (see Fig. S6 in the supplemental material), indicating that Stim-Subst enhances the probability of telomere addition within the SiRTA Core.
To test whether stimulation is orientation dependent, we inverted the Stim-Subst sequence. Inversion reduced the rate of GCR formation at SiRTA 5L-35 2.8-fold and reduced the fraction of GCR events within SiRTA 5L-35 from 47.8% ± 7.1% to 10.4% ± 6.8% (Fig. 4G). Rap1 is expected to retain binding to the Stim-Subst sequence regardless of orientation. In contrast, the ability of Cdc13 to bind requires orientation-dependent exposure of its single-stranded TG-rich binding site during resection from a distal double-strand break, suggesting that Cdc13 may be the functionally relevant protein in this context.
Binding of Cdc13 within SiRTA Stim is sufficient to drive de novo telomere addition at the neighboring Core sequence.
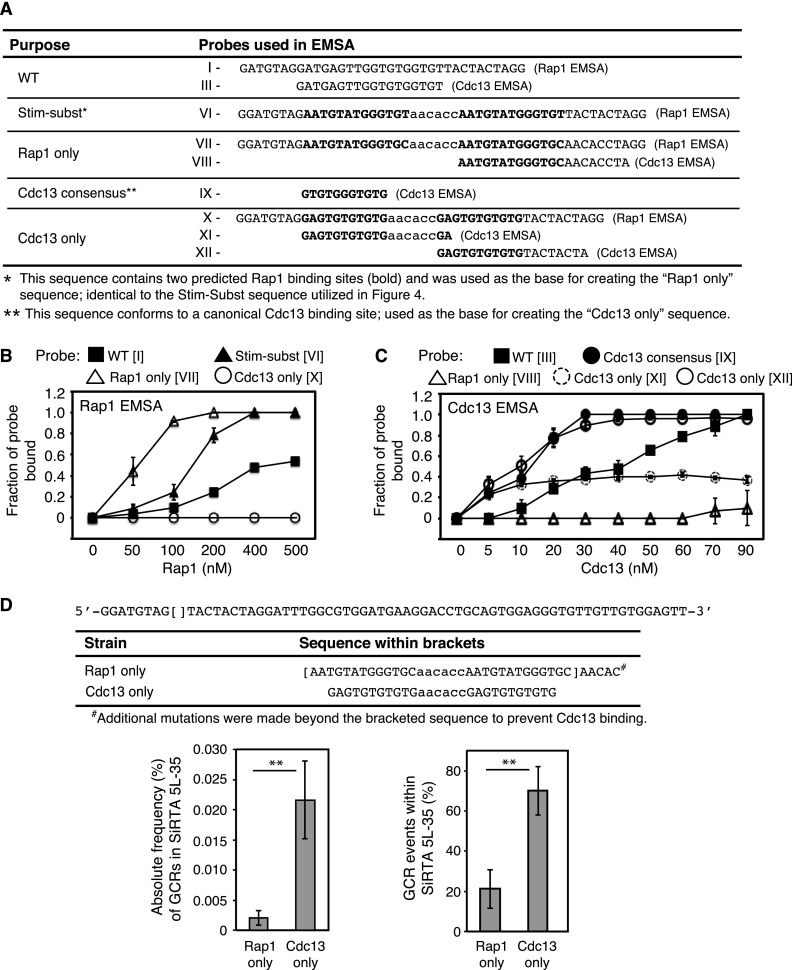
To distinguish the effects by Rap1 and Cdc13, we designed sequences to support differential binding. To create a sequence that binds Rap1 but not Cdc13, we began with the Stim-Subst sequence containing two sites predicted to bind Rap1 separated by an AC-rich spacer sequence (Fig. 4D and 5A, probe VI). Since the 5′ portion of this sequence binds weakly to Cdc13-DBD (Fig. 4F, probe VIa), we mutated only the most 3′ residue in this repeat, a change predicted to retain Rap1 association but disrupt Cdc13 binding. At the second Rap1 site, we mutated both that same 3′ nucleotide and several nucleotides that lie adjacent to the defined Stim sequence (Fig. 5A, probe VII). As shown in Fig. 5B, the engineered “Rap1 only” sequence binds Rap1 with higher affinity than either the original SiRTA-Subst sequence or the endogenous SiRTA Stim. Mutations in the second Rap1 binding site essentially eliminate Cdc13-DBD binding to this sequence (compare Fig. 4F, probe VIb, with Fig. 5C, probe VIII).
We took a similar approach to create a sequence capable of binding Cdc13 and not Rap1. Here, the starting sequence was 11 bases shown to support strong association with Cdc13 (31) (Fig. 5A, probe IX). The second base of this sequence (known to have little or no effect on Cdc13 binding [34]) was mutated to reduce similarity with the Rap1 binding consensus, and two of these sites were placed in tandem. As shown in Fig. 5B, this “Cdc13 only” sequence shows no detectable binding to Rap1 (probe X). Binding of this sequence to Cdc13 was measured using two probes that monitor binding to the 5′ (probe XI) or 3′ (probe XII) repeat. The 3′ repeat shows similar affinity for Cdc13-DBD as the 11-base consensus sequence (Fig. 5C, compare probes IX and XII), and both of these probes are bound more strongly than is the endogenous SiRTA Stim (Fig. 5C, probe III). The 5′ repeat is also bound by Cdc13-DBD, although more weakly than the 3′ repeat (Fig. 5C, probe XI). In conclusion, these in vitro binding analyses demonstrate that binding by Rap1 and Cdc13 can be separated, allowing us to test the specific effects of these proteins on SiRTA 5L-35 function.
The sequences defined above were integrated in place of the endogenous SiRTA Stim sequence on chromosome V (Fig. 5D). Integration of the “Rap1 only” sequence reduced the overall frequency of GCR formation at SiRTA 5L-35 to about one-half that of the Stim-Subst sequence upon which it is based, a rate similar to that observed for the Stim-Subst inverted sequence (compare Fig. 5D with Fig. 4G). In contrast, the sequence containing only binding sites for Cdc13 resulted in a GCR rate 8-fold higher than that of the endogenous sequence; 70.0% ± 9.8% of all GCR events occurred within SiRTA 5L-35 (Fig. 5D), and all of those events were the result of de novo telomere addition (data not shown). We again used a PCR-based strategy to determine where telomere addition occurred within SiRTA 5L-35. Despite the telomere-like nature of the “Cdc13 only” sequence, 50 of 63 (79.4%) de novo telomeres were added at least 20 bp distal to that site (see Fig. S6 in the supplemental material).
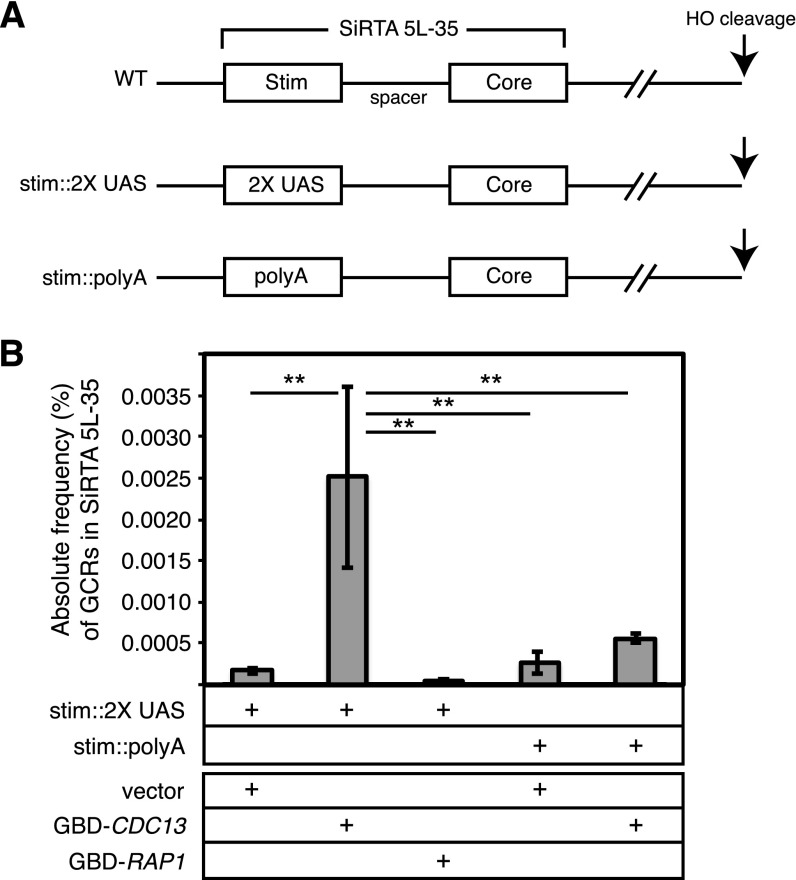
To confirm the ability of Cdc13 to stimulate de novo telomere addition, we replaced the SiRTA Stim sequence with two copies of the Gal4 upstream activating sequence (stim::2XUAS), which is recognized by the Gal4 DNA binding domain (35) (GBD) (Fig. 6A). Into this strain, we introduced either an empty vector or a plasmid expressing a fusion of GBD with full-length Cdc13 or Rap1. As expected, the strain containing stim::2XUAS and empty vector supported a rate of GCR formation at SiRTA 5L-35 indistinguishable from that of a strain containing the stim::poly(A) mutation (Fig. 6B; comparable to mutation c in Fig. 2). Expression of the GBD-Rap1 fusion protein in the stim::2XUAS strain failed to increase the rate of GCR formation at SiRTA 5L-35 (Fig. 6B), suggesting that Rap1 plays either no role or a minor role in stimulating telomere addition following a DSB.
In contrast, expression of the GBD-Cdc13 fusion protein in the stim::2XUAS strain increased the rate of GCR formation at SiRTA 5L-35 nearly 15-fold relative to the strain containing vector only (Fig. 6B). This increase is largely attributable to the recruitment of GBD-Cdc13 to the 2XUAS sequences on chromosome V since expression of the fusion protein in the stim::poly(A) strain had no significant effect on GCR formation at SiRTA 5L-35 compared to the same strain containing the empty vector (Fig. 6B).
Taken together, these results are consistent with a model in which resection of the 5′ strand following a DSB exposes one or more sites at which Cdc13 is able to associate with the SiRTA Stim sequence. Such binding is required to increase the rate at which the more telomere-proximal Core sequence is capable of nucleating de novo telomere addition and explains why the telomere-like SiRTA 5L-35 Core sequence alone is insufficient to maintain a high rate of GCR formation at that site.
A second SiRTA on chromosome IX also has a bipartite structure.
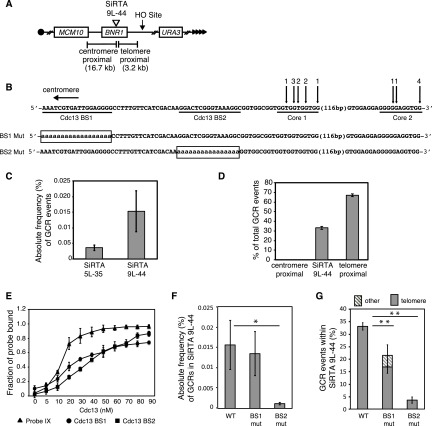
In searching for additional sequences with the hallmarks of a SiRTA, we identified a TG-rich sequence within the BNR1 gene. This site is located ∼16 kb distal to the first essential gene, MCM10, on chromosome IX-L. We integrated the HO recognition site ∼3 kb distal to this sequence (Fig. 7A) in a strain that expresses the HO endonuclease under galactose regulation and placed URA3 on the distal arm, allowing us to select GCR events as described above for SiRTA 5L-35. Our PCR strategy was designed to capture GCR events occurring in the most prominent TG-rich sequence (Fig. 7B, Core 1). However, Southern blotting revealed a second cluster of de novo telomere addition events ∼130 bp distal to the original sequence (Fig. 7B, Core 2; see also Fig. S7 in the supplemental material). These two sequences result in a combined frequency of GCR formation of 0.015% ± 0.006%, approximately three times higher than the rate observed at SiRTA 5L-35 (Fig. 7C). We subsequently refer to this site as SiRTA 9L-44 (SiRTA, 44 kb from the left telomere of chromosome IX). Similar to SiRTA 5L-35, 33.0% ± 1.4% of the GCR events obtained on chromosome IX occurred within SiRTA 9L-44 (including both Core sequences) (Fig. 7D). Interestingly, although Core 1 lies centromere proximal to Core 2 (and therefore will be rendered single-stranded after Core 2 in response to a distal DSB), 28.2% ± 5.2% of all GCR events occurred within Core 1, while 4.8% ± 4.4% occurred in Core 2 (data not shown), suggesting that Core 1 is more efficiently targeted. No GCR events occurred within the ∼16-kb region between SiRTA 9L-44 and the first essential gene (Fig. 7D).
FIG 7.
A site at which de novo telomere addition occurs at high frequency on chromosome IX (SiRTA 9L-44) has an organization similar to that of SiRTA 5L-35. (A) Schematic of the left arm of chromosome IX. Sizes of the regions between SiRTA 9L-44 and either the HO cleavage site (telomere proximal) or the most distal essential gene (MCM10; centromere proximal) are indicated. (B) Top schematic, sequence of SiRTA 9L-44 with vertical arrows indicating sites of de novo telomere addition. The sequence is oriented with the telomere to the right so that the DNA strand depicted is the direct 3′ primer upon which telomerase acts. In each case, the most 3′ chromosomal nucleotide with identity to the cloned de novo telomere is indicated. Numbers above arrows indicate the number of independent telomere addition events mapped to each site. Sequences predicted to bind Cdc13 are underlined (Cdc13 BS1 and Cdc13 BS2). Bottom schematic, mutations created in SiRTA 9L-44. Uppercase letters represent unchanged nucleotides; lowercase letters enclosed in box represent mutated nucleotides. (C) The absolute frequency (%) of GCR events within SiRTA 5L-35 or SiRTA 9L-44 is shown. (D) The percentage of GCR events occurring in each indicated region on chromosome IX is shown. No GCR events were observed in the region centromere proximal to SiRTA 9L-44. Values in panels C and D are averages for three independent experiments with standard deviations. (E) Binding of the indicated concentration of recombinant Cdc13-DBD to single-stranded probe IX (Fig. 5A) or single-stranded probes corresponding to the underlined sequences in panel B. The average fraction of probe bound was determined from three independent experiments. Error bars represent standard deviations. (F) The BS1 mutant (mut) and BS2 mut sequences shown in panel B were inserted at the endogenous SiRTA 9L-44 locus on chromosome IX, and the absolute frequency (%) of de novo formation within SiRTA 9L-44 was determined. (G) The percentage of total GCR events that occur within SiRTA 9L-44 for WT and the indicated mutant strains is shown. GCR events involving de novo telomere addition were identified by Southern blotting; any event that does not involve telomere addition is classified as “other.” Results in panels F and G are the averages and standard deviations for three independent experiments. Samples with statistically different values by ANOVA with post hoc Tukey's HSD are indicated (*, P < 0.05; **, P < 0.01).
Given that we identified a stimulatory sequence within SiRTA 5L-35 capable of binding to Cdc13 (Fig. 2 and 4), we sought to identify similar sequence(s) that may contribute to telomere addition at SiRTA 9L-44. Using EMSA, we identified two sites that bind Cdc13-DBD in vitro (Cdc13 BS1 and Cdc13 BS2) (Fig. 7B and E). Mutation of Cdc13 BS1 to polyadenine had no effect on the overall frequency of GCR events at SiRTA 9L-44 relative to WT (Fig. 7F), although the fraction of GCR events that occurred within SiRTA 9L-44 was significantly reduced (from 33.0% ± 1.4% to 21.4% ± 6.2% [Fig. 7G]). This reduction occurred specifically at Core 1, since the fraction of events at Core 2 remained similar to WT (4.8% ± 4.4% in WT versus 7.5% ± 1.5% in BS1). Of the GCR events that occurred at SiRTA 9L-44 when Cdc13 BS1 was mutated, 4.6% ± 4.2% were not de novo telomere addition events, a phenomenon never observed in WT cells (Fig. 7G). Together, these results suggest a minor contribution by Cdc13 BS1 to the high rate of de novo telomere addition at SiRTA 9L-44.
In contrast, mutation of Cdc13 BS2 to polyadenine strongly reduced the overall rate of GCR formation at SiRTA 9L-44 (∼18-fold) (Fig. 7F). Only 3.5% ± 1.3% of GCR events occurred within SiRTA 9L-44 (Fig. 7G), and none were within Core 2. Cdc13-DBD binds similarly to BS1 and BS2 in vitro, suggesting that the efficacy of these sequences in stimulating telomere addition correlates poorly with the strength of binding by Cdc13 (compare Fig. 7E, F, and G). The functional difference between these sequences may be explained, at least in part, by their differing proximity to the sites of de novo telomere addition at Core 1 and Core 2.
In conclusion, we identified a SiRTA on chromosome IX that bears striking similarities to the SiRTA on chromosome V. Both SiRTAs contain TG-rich tracts within which telomere addition occurs, and the high rate of de novo telomere addition at these SiRTAs relative to neighboring sequences can be attributed to stimulatory sequences that lie centromere proximal to the major sites of telomere addition. Finally, EMSA analysis shows that Cdc13 binds these stimulatory sequences in vitro. Thus, increased telomerase activity at SiRTAs is likely achieved, at least in part, by the association of Cdc13 with one or more stimulatory sequences upstream of the sites of actual de novo telomere addition.
DISCUSSION
Endogenous sites of de novo telomere addition can affect genome stability and have been associated with cancer (36) and congenital disorders (37–39). While S. cerevisiae provides a useful model system to study mechanisms of de novo telomere addition, most studies utilize artificial sequences to stimulate telomere formation. The goal of this study was to examine naturally occurring sites at which de novo telomere addition is greatly favored and to identify cis- and trans-acting factors contributing to this property. We characterized two genomic sites (SiRTAs) at which de novo telomere addition occurs at a remarkably (at least 200-fold) increased rate compared to neighboring sequences. Zakian and colleagues reported a third hot spot of de novo telomere addition on chromosome VII at a location 50 kb internal to an HO cleavage site (40). Unlike the two sequences studied here, the chromosome VII site lies internal to the last essential gene on the chromosome arm (a disomic strain lacking RAD52 was used to maintain viability and limit homologous recombination). Given the ease with which these sites have been found, it seems likely that additional SiRTAs remain to be identified.
While many assays of de novo telomere addition utilize short telomeric tracts placed immediately adjacent (within 20 to 30 nucleotides [nt]) to the HO cleavage site (for examples, see references 12 and 41 to 43), the natural SiRTAs described here are located several kilobases internal to the induced break, requiring extensive resection prior to telomere addition. In our assays, we also observe telomere addition at or very close to the HO cut site (see Fig. S2 in the supplemental material). This propensity is at least partially a consequence of the TGTT-3′ overhang produced by HO endonuclease cleavage. When the recognition site is inverted to generate the complementary ACAA-3′ overhang, de novo telomere addition at the HO site is reduced and the fraction of events at SiRTA 5L-35 is increased (data not shown). Given these observations, studies of telomere addition at endogenous sites located at a distance from an induced break may more faithfully capture the mechanism of repair of a random chromosomal break than models in which a telomeric seed sequence is intentionally placed adjacent to the HO cleavage site.
We observe that both SiRTA 5L-35 and 9L-44 contain sequences that enhance telomerase action but act rarely, if ever, as the direct target of telomerase action (SiRTA Stim sequences). Because the telomere-binding proteins Cdc13 and Rap1 stimulate de novo telomere addition at artificial sequences in S. cerevisiae (12–15, 43), we hypothesized that one or both of these proteins could be responsible for the enhancing activity of the Stim sequence. Although Cdc13 and Rap1 bind similar sequences, we were able to design artificial sequences that bind with great preference to one protein as measured in vitro. Using this approach, we found that a sequence designed to facilitate Cdc13 binding is much more effective in the stimulation of de novo telomere addition than one binding primarily Rap1 (Fig. 5D). Importantly, artificial recruitment of GBD-Cdc13 to SiRTA 5L-35 (Fig. 6) led to high frequencies of telomere addition, suggesting that Cdc13 binding, and not the TG-rich sequences per se, stimulates de novo telomere addition. Replacement of Stim with the “Rap1 only” sequence did not reduce telomere addition as dramatically as the replacement of Stim with a poly(A) sequence or with the Gal4 UAS (compare Fig. 5D with Fig. 2B and 6B), so binding by Rap1 may also contribute to the stimulation of de novo telomere addition. Consistent with the proposal that binding by Cdc13 is important for the stimulatory effect of the Stim sequence, we find that high levels of telomere addition at SiRTA 9L-44 require a sequence capable of binding Cdc13 (Fig. 7B, BS2) and that this sequence stimulates telomere addition over a distance of more than 100 bp. Rap1 does not bind to the BS2 sequence in vitro (data not shown), suggesting that Cdc13 is sufficient to stimulate de novo telomere addition at this SiRTA.
The stimulation of de novo telomere addition by telomere-like sequences located internal to the site of telomerase action has been previously observed in both artificial and natural contexts. For example, de novo telomeres generated following DNA cleavage by HO endonuclease near a TG-rich seed sequence are frequently added at the 3′ overhang of the HO endonuclease target site rather than within the telomeric seed itself (23). At the SiRTA on chromosome VII reported by Mangahas et al. (40), telomere addition occurs at several closely spaced sequences located 37 to 49 bp distal to a 35-bp GT dinucleotide repeat. This sequence matches the GXGT(T/G)7 consensus for Cdc13 binding (34), consistent with the GT dinucleotide repeat acting as a Stim sequence in the manner that we report here for SiRTAs 5L-35 and 9L-44. However, as is the case at SiRTA 5L-35, the TG dinucleotide tract on chromosome VII may also bind Rap1 (44), so a contribution of Rap1 to Stim function cannot be ruled out.
Kramer and Haber reported that de novo telomere addition occurs ∼15 to 100 bp distal to an ectopic tract of 13 T2G4 repeats (Tetrahymena thermophila telomeric sequence), and a similar phenomenon was reported for plasmid substrates (45, 46). The Tetrahymena telomeric sequence contains the GXGT(T/G)7 consensus sequence for Cdc13 binding (34), although a (T2G4)3 oligonucleotide was shown to compete poorly with the yeast telomeric sequence for Cdc13 binding in vitro (29). This short sequence is predicted to contain only a single Cdc13 binding site, while the tract of 13 repeats utilized by Kramer and Haber has multiple potential binding sites (45). Combined with our observation that the Stim sequences in both SiRTA 5L-35 and 9L-44 bind Cdc13 with lower affinity than an optimized sequence from the yeast telomere (Fig. 5C and 7E), it is reasonable to suggest that the stimulatory effect observed by Kramer and Haber is due to Cdc13 binding to the ectopic T2G4 tract.
How might the bipartite structure of SiRTA 5L-35 facilitate de novo telomere addition? Genetic and biochemical analyses suggest that Cdc13 protects telomeres from extensive 5′ strand degradation (47–50). One possibility is that Cdc13 bound to the Stim sequence inhibits resection past SiRTA, thereby providing a grace period to allow for the formation of a stable telomere at the Core sequence. However, inhibition of resection alone does not account for the increased efficacy of Stim when it is brought in close proximity to Core (Fig. 3B). Another possibility is that Cdc13 must be bound to both the Stim and Core sequences to stably recruit telomerase to the SiRTA. This proposal is congruent with the results of Hirano and Sugimoto that show greatly increased stimulation of telomerase recruitment and de novo telomere addition when two tandem Cdc13 binding sites are placed adjacent to an HO-induced break, compared to the effect of a single site (43).
In addition to its C-terminal oligonucleotide/oligosaccharide binding (OB) fold domain (also known as its DNA-binding domain), Cdc13 contains an N-terminal OB fold that is involved in Cdc13 dimerization (51). The minimal binding site for both full-length Cdc13 and its isolated DBD is an 11-mer TG1–3 sequence. However, as demonstrated by EMSA, the Cdc13 N-terminal OB fold, which exists as a stable dimer in solution, does not bind to this 11-mer but binds with stronger affinity to single-stranded DNA (ssDNA) sequences at least 37 nucleotides long (51). These observations support a model in which each Cdc13 monomer binds to a separate site on a single molecule of ssDNA, with optimal binding depending on the distance between the two sites. Finally, mutations that disrupt Cdc13 dimerization in vitro cause telomere shortening when introduced in vivo (51). We propose that dimerization between Cdc13 monomers bound to the Stim and Core sequences is required to stably recruit telomerase to the SiRTA. This model additionally accounts for the increased efficacy of Stim when it is juxtaposed with Core, since in that context, the distance between the two monomers may be more optimally suited to stable telomerase recruitment.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Haber, Deborah Wuttke, and Alessandro Bianchi for generously providing us with strains and plasmids. We are grateful to Shyam Murali and Ilham Eli for strain construction.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00095-16.
REFERENCES
- 1.Osterhage JL, Friedman KL. 2009. Chromosome end maintenance by telomerase. J Biol Chem 284:16061–16065. doi: 10.1074/jbc.R900011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn EH. 1991. Structure and function of telomeres. Nature 350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 3.Wellinger RJ, Zakian VA. 2012. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191:1073–1105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy CF, Sussel L, Shore D. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 5.Wotton D, Shore D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 6.Marcand S, Gilson E, Shore D. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 7.Levy DL, Blackburn EH. 2004. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24:10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoeijmakers JH. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 9.Aylon Y, Kupiec M. 2004. DSB repair: the yeast paradigm. DNA Repair (Amst) 3:797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Kolodner RD, Putnam CD, Myung K. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 11.Pennaneach V, Putnam CD, Kolodner RD. 2006. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol 59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- 12.Bianchi A, Negrini S, Shore D. 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell 16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Ray A, Runge KW. 1998. The C terminus of the major yeast telomere binding protein Rap1p enhances telomere formation. Mol Cell Biol 18:1284–1295. doi: 10.1128/MCB.18.3.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossi S, Bianchi A, Damay P, Shore D. 2001. Telomere formation by Rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol Cell Biol 21:8117–8128. doi: 10.1128/MCB.21.23.8117-8128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lustig AJ, Kurtz S, Shore D. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 16.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schild D, Calderon IL, Contopoulo R, Mortimer RK. 1983. Cloning of yeast recombination repair genes and evidence that several are non-essential genes, p 417–427. In Friedberg EC, Bridges BA (ed), Cellular responses to DNA damage. Alan R. Liss, New York, NY. [Google Scholar]
- 18.Scherer S, David RW. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A 76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 20.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. 2013. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson EM, Halsey WA, Wuttke DS. 2002. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res 30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, Adkins CJ, Cartwright BR, Friedman KL. 2008. Yeast Est2p affects telomere length by influencing association of Rap1p with telomeric chromatin. Mol Cell Biol 28:2380–2390. doi: 10.1128/MCB.01648-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bairley RCB, Guillaume G, Vega LR, Friedman KL. 2011. A mutation in the catalytic subunit of yeast telomerase alters primer-template alignment while promoting processivity and protein-DNA binding. J Cell Sci 124:4241–4252. doi: 10.1242/jcs.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Kolodner RD. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23:81–85. [DOI] [PubMed] [Google Scholar]
- 25.Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. 2010. Sgs1 and Exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet 6:e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundblad V, Blackburn EH. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347–360. doi: 10.1016/0092-8674(93)90234-H. [DOI] [PubMed] [Google Scholar]
- 27.Conrad MN, Wright JH, Wolf AJ, Zakian VA. 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63:739–750. doi: 10.1016/0092-8674(90)90140-A. [DOI] [PubMed] [Google Scholar]
- 28.Shore D, Nasmyth K. 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51:721–732. doi: 10.1016/0092-8674(87)90095-X. [DOI] [PubMed] [Google Scholar]
- 29.Lin J-J, Zakian VA. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci U S A 93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nugent CI, Hughes TR, Lue NF, Lundblad V. 1996. Cdc13p: a single-strand telomeric DNA binding protein with a dual role in yeast telomere maintenance. Science 274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 31.Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc Natl Acad Sci U S A 97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourns BD, Alexander MK, Smith AM, Zakian VA. 1998. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol 18:5600–5608. doi: 10.1128/MCB.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukamoto Y, Taggart AKP, Zakian VA. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol 11:1328–1335. doi: 10.1016/S0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 34.Eldridge AM, Halsey WA, Wuttke DS. 2006. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry 45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giniger E, Varnum SM, Ptashne M. 1985. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 36.Fouladi B, Sabatier L, Miller D, Pottier G, Murnane JP. 2000. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia 2:540–554. doi: 10.1038/sj.neo.7900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kostiner DR, Nguyen H, Cox VA, Cotter PD. 2002. Stabilization of a terminal inversion duplication of 8p by telomere capture from 18q. Cytogenet Genome Res 98:9–12. doi: 10.1159/000068536. [DOI] [PubMed] [Google Scholar]
- 38.Fortin F, Beaulieu Bergeron M, Fetni R, Lemieux N. 2009. Frequency of chromosome healing and interstitial telomeres in 40 cases of constitutional abnormalities. Cytogenet Genome Res 125:176–185. doi: 10.1159/000230002. [DOI] [PubMed] [Google Scholar]
- 39.Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, Bonati MT, Giglio S, Guerrini R, Osimani S, Marelli S, Zucca C, Grasso R, Borgatti R, Mani E, Motta C, Molteni M, Romano C, Greco D, Reitano S, Baroncini A, Lapi E, Cecconi A, Arrigo G, Patricelli MG, Pantaleoni C, D'Arrigo S, Riva D, Sciacca F, Dalla Bernardina B, Zoccante L, Darra F, Termine C, Maserati E, Bigoni S, Priolo E, Bottani A, Gimelli S, Bena F, Brusco A, di Gregorio E, Bagnasco I, Giussani U, Nitsch L, Politi P, Martinez-Frias ML, Martínez-Fernández ML, Martínez Guardia N, Bremer A, Anderlid BM, Zuffardi O. 2011. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid syndrome. PLoS Genet 7:e1002173. doi: 10.1371/journal.pgen.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mangahas JL, Alexander MK, Sandell LL, Zakian VA. 2001. Repair of chromosome ends after telomere loss in Saccharomyces. Mol Biol Cell 12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Durocher D. 2010. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev 24:502–515. doi: 10.1101/gad.1869110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeyre C, Shore D. 2012. Anticheckpoint pathways at telomeres in yeast. Nat Struct Mol Biol 19:307–313. doi: 10.1038/nsmb.2225. [DOI] [PubMed] [Google Scholar]
- 43.Hirano Y, Sugimoto K. 2007. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell 18:2026–2036. doi: 10.1091/mbc.E06-12-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer KM, Haber JE. 1993. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev 7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 46.Murray AW, Claus TE, Szostak JW. 1988. Characterization of two telomeric DNA processing reactions in Saccharomyces cerevisiae. Mol Cell Biol 8:4642–4650. doi: 10.1128/MCB.8.11.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garvik B, Carson M, Hartwell L. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15:6128–6138. doi: 10.1128/MCB.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grandin N, Damon C, Charbonneau M. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO j 20:6127–6139. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ngo HP, Lydall D. 2010. Survival and growth of yeast without telomere capping by Cdc13 in the absence of Sgs1, Exo1, and Rad9. PLoS Genet 6:e1001072. doi: 10.1371/journal.pgen.1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greetham M, Skordalakes E, Lydall D, Connolly BA. 2015. The telomere binding protein Cdc13 and the single-stranded DNA binding protein RPA protect telomeric DNA from resection by exonucleases. J Mol Biol 427:3023–3030. doi: 10.1016/j.jmb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell MT, Smith JS, Mason M, Harper S, Speicher DW, Johnson FB, Skordalakes E. 2010. Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol Cell Biol 30:5325–5334. doi: 10.1128/MCB.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.