ABSTRACT
The second messenger cyclic diguanylate (c-di-GMP) is an important regulator of motility in many bacterial species. In Pseudomonas aeruginosa, elevated levels of c-di-GMP promote biofilm formation and repress flagellum-driven swarming motility. The rotation of P. aeruginosa's polar flagellum is controlled by two distinct stator complexes, MotAB, which cannot support swarming motility, and MotCD, which promotes swarming motility. Here we show that when c-di-GMP levels are elevated, swarming motility is repressed by the PilZ domain-containing protein FlgZ and by Pel polysaccharide production. We demonstrate that FlgZ interacts specifically with the motility-promoting stator protein MotC in a c-di-GMP-dependent manner and that a functional green fluorescent protein (GFP)-FlgZ fusion protein shows significantly reduced polar localization in a strain lacking the MotCD stator. Our results establish FlgZ as a c-di-GMP receptor affecting swarming motility by P. aeruginosa and support a model wherein c-di-GMP-bound FlgZ impedes motility via its interaction with the MotCD stator.
IMPORTANCE The regulation of surface-associated motility plays an important role in bacterial surface colonization and biofilm formation. c-di-GMP signaling is a widespread means of controlling bacterial motility, and yet the mechanism whereby this signal controls surface-associated motility in P. aeruginosa remains poorly understood. Here we identify a PilZ domain-containing c-di-GMP effector protein that contributes to c-di-GMP-mediated repression of swarming motility by P. aeruginosa. We provide evidence that this effector, FlgZ, impacts swarming motility via its interactions with flagellar stator protein MotC. Thus, we propose a new mechanism for c-di-GMP-mediated regulation of motility for a bacterium with two flagellar stator sets, increasing our understanding of surface-associated behaviors, a key prerequisite to identifying ways to control the formation of biofilm communities.
INTRODUCTION
Cyclic diguanylate (c-di-GMP) is a ubiquitous bacterial second messenger responsible for regulating a range of cellular processes, including motility and biofilm formation (1). In general, low intracellular c-di-GMP levels are associated with motile lifestyles, while elevated levels of c-di-GMP promote surface attachment and sessile lifestyles (2, 3). c-di-GMP is synthesized from two molecules of GTP by diguanylate cyclases (DGCs) and degraded by c-di-GMP-specific phosphodiesterases (PDEs) (1, 4). Many DGCs and PDEs involved in motility regulation have been characterized, but the mechanisms by which c-di-GMP regulates motility are poorly understood in Pseudomonas aeruginosa and in other bacterial species.
To regulate numerous biological functions, c-di-GMP binds to specific effector proteins or RNA (reviewed in reference 5). Recent studies have focused on identifying these c-di-GMP effectors and their mechanisms for regulating c-di-GMP-dependent processes. One class of effectors is the PilZ domain-containing protein family, which is characterized by conserved c-di-GMP binding motifs RXXXR and D/NXSXXG (6, 7). PilZ domain-containing proteins typically bind c-di-GMP and, in the c-di-GMP-bound state, influence cellular processes, including polysaccharide production, virulence, biofilm formation, and motility control (8–14). The PilZ domain-containing protein YcgR of Escherichia coli and its homologs in Salmonella enterica and Bacillus subtilis have been shown to bind to c-di-GMP and to inhibit cellular motility in response to c-di-GMP (15–20). Evidence suggests that these PilZ domain proteins impede flagellar function by directly interacting with parts of the flagellar motor. In E. coli, interactions have been demonstrated between YcgR and three different flagellar motor-associated proteins, MotA, FliG, and FliM (15, 17, 18). In B. subtilis, evidence suggests that the YcgR homolog YpfA (now called DgrA) (20) interacts with MotA (16). The P. aeruginosa genome encodes seven PilZ domain-containing proteins that have been shown to bind to c-di-GMP and an eighth PilZ domain protein that lacks c-di-GMP binding, but no link between these proteins and flagellar motility has been established in this organism (12, 21–23).
We have previously reported a connection between c-di-GMP-dependent repression of swarming and the activity of flagellar stator proteins (24). Stator proteins form the ion-translocating channels that are necessary for generating torque to power flagellar rotation (25, 26). P. aeruginosa and its relatives are distinguished from many other flagellated bacteria in that they have two sets of proton-dependent stators, MotAB and MotCD (27, 28). Our previous studies have shown that these stators play distinct roles in the control of surface-associated swarming motility—one set of stators promotes swarming motility (MotCD), and a second set (MotAB) prevents swarming motility. From this work, we suggested a model by which P. aeruginosa controls swarming motility in response to c-di-GMP via a unique stator-swapping mechanism between these distinct MotAB and MotCD stator complexes (24). Specifically, MotCD was more likely to be found colocalized with the motor as c-di-GMP levels decreased, thereby presumably enabling surface motility (24).
Here we tested the importance of PilZ domain proteins in the control of P. aeruginosa swarming motility and demonstrate that the PilZ domain protein PA14_20700, here named FlgZ after the P. fluorescens and P. putida homologs (29), and the Pel polysaccharide contribute to c-di-GMP-mediated swarming repression. We provide evidence that FlgZ interacts directly with stator protein MotC but does not interact with MotA. Both the function of FlgZ in swarming repression and its ability to interact with MotC depend on c-di-GMP binding. Furthermore, we show that the localization of a green fluorescent protein (GFP)-FlgZ fusion to the pole of the cell is increased at high c-di-GMP levels and depends on the presence of MotCD. Thus, we suggest that FlgZ functions to repress swarming motility in response to c-di-GMP by specifically targeting the function of MotCD, the swarming-promoting stator set, by preventing the engagement of MotCD with the rotor.
MATERIALS AND METHODS
Strains and media.
Bacterial strains used in this study are listed in Table S1 in the supplemental material. P. aeruginosa PA14 and E. coli S17-1 λpir and BTH101 were routinely grown in lysogeny broth (LB) or on LB solidified with 1.5% agar. When antibiotic selection was appropriate for P. aeruginosa, gentamicin (Gm) was used at 30 μg/ml. For E. coli selections, Gm was used at 10 μg/ml, carbenicillin (Cb) at 50 μg/ml, kanamycin (Kan) at 50 μg/ml, and nalidixic acid (Nal) at 20 μg/ml.
Saccharomyces cerevisiae strain InvSc1 (Invitrogen) was used for constructing plasmids via in vivo homologous recombination (30). InvSc1 was grown in yeast extract-peptone-dextrose (1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose). Synthetic defined medium lacking uracil was used to select for plasmid-harboring yeast.
Construction of mutant strains and plasmids.
Table S2 in the supplemental material lists all plasmids used in this study. Primers used in plasmid and mutant construction are listed in Table S3. In-frame deletion mutants were constructed via allelic exchange as previously described (30). Integrants were isolated on LB medium supplemented with Gm and Nal followed by sucrose counterselection. Resolved integrants were confirmed by PCR and sequencing.
Point mutations made to flgZ plasmids were generated using a modified protocol for in vitro site-directed mutagenesis (31). Briefly, forward and reverse oligonucleotide primers were designed to contain mismatches for generating the desired point mutation. These primers were first used separately to amplify the parental vector using Phusion DNA polymerase (NEB) for four cycles. Products from the forward and reverse reactions were then combined and amplified for an additional 18 cycles. The parental, nonmutagenized plasmid was digested using DpnI before transformation of the products in E. coli.
Swarming motility assays.
Swarming motility was tested by inoculating 2 μl of overnight cultures onto M8 minimal salts medium supplemented with 0.53% agar and glucose (0.2%), MgSO4 (1 mM), and Casamino Acids (0.5%), as reported previously (32). Arabinose was used at 0.2% where indicated for expression plasmids with the PBAD promoter. Swarm assays were incubated at 37°C for 16 to 19 h. Quantification of swarm zones was performed using ImageJ software (National Institutes of Health) (33).
Free-swimming capillary assay.
To perform a zero-flow, free-swimming capillary assay, 100 μl of mid-log-phase P. aeruginosa culture (optical density at 600 nm [OD600] between 0.6 and 1.0) was pelleted at 1,000 × g for 1 min and gently resuspended in 500 μl motility buffer (liquid medium of the same composition as the swarm plates but lacking agar). The resuspended cells were incubated at room temperature (RT) for 20 min to allow recovery of motility, before 50 μl were drawn into a 0.2-by-2-mm glass capillary tube by capillary action. The ends of the tube were sealed with silicon grease to prevent evaporation and flow, the tube was attached to a glass slide, and bacteria were visualized in phase contrast at ×20 magnification at the center of the tube for 1 to 2 min with a video frame rate of 20 ms.
The image segmentation software Tracker (34) was used to detect cells and form tracks, using the “Threshold” algorithms with the following parameters: upper threshold, 0.52; lower threshold, 0.48; minimal cluster size, 12. To exclude wrong connections of tracks, a cutoff value of 90 μm/s per single frame was applied. More than 400 tracks from three videos per strain with a minimal track length of 50 frames were analyzed, and swimming speeds of all detected bacteria, as well as of bacteria classified as motile (swimming speed, >10 μm/s), were compared.
Western blot analysis of protein level.
Determination of the levels of wild-type (WT) and mutant proteins in whole-cell extracts was performed as reported previously (24) and is summarized as follows. Cells were cultured on 0.53% swarm medium for 16 to 19 h. Cells were harvested from swarm medium using ethanol-washed plastic coverslips and then centrifuged for 1 min at room temperature (RT). Supernatants were removed, and cell pellets were stored at −80°C prior to further processing. To generate cell lysates, cells were resuspended in buffer (200 mM Tris-HCl [pH 7.5], 1 mM EDTA, 2 mM MgCl2, cOmplete protease inhibitors [Roche Diagnostics Corp., Indianapolis, IN]), Benzonase nuclease (Novagen, San Diego, CA) was added to reach a final concentration of ∼50 units/ml, and the reaction mixture was lysed by French press or bead beating. For Western blotting, whole-cell lysates were mixed with a final concentration of 1× SDS and 100 mM dithiothreitol (DTT). Samples were boiled for 5 min and resolved by SDS-PAGE using Any kD gels or 12% polyacrylamide gels (Bio-Rad). Proteins were transferred to a nitrocellulose membrane and probed with an anti-penta-His antibody (Qiagen, Valencia, CA). Nitrocellulose membranes were washed with Tris-buffered saline–Tween (TBS-Tween) and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 30 min at RT. Proteins were detected using ECL Plus Western blotting substrate (Pierce).
c-di-GMP measurements.
Cells were collected from swarm plates after incubation at 37°C for 18 h. Nucleotide extraction was performed as previously described (35, 36). c-di-GMP measurement analysis was performed by liquid-chromatography mass spectrometry (LC-MS/MS) at the Mass Spectrometry Facility at Michigan State University.
Bacterial two-hybrid analysis.
Protein-protein interactions were examined using a bacterial adenylate cyclase two-hybrid (BATCH) system obtained from Euromedex (Souffelweyersheim, France) as previously described (37, 38). In this assay, full-length proteins of interest are fused to either the T18 or T25 fragment of Bordetella pertussis adenylate cyclase and then coexpressed in E. coli BTH101 cells. Interaction between the two hybrid proteins functionally reconstitutes the catalytic domain of adenylate cyclase, leading to cyclic AMP (cAMP) synthesis and transcriptional activation of the lac operon.
In this study, each of genes motA, motC, fliG, and fliM was cloned into pKNT25, pKT25, pUT18, and pUT18C vectors. All vectors containing the motA gene include a C-terminal 6×His tag. We tested pairwise interactions with T18 and T25 fusions in both orientations. Transformants were 10-fold serially diluted and spotted (2 μl) on LB agar containing Cb, Kan, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside) (0.5 mM) and incubated for 40 h at 30°C. The efficiency of the interactions was determined via β-galactosidase activity assays, as previously described (39).
Immunoprecipitation assays.
Cells were collected from swarm plates as described above. Swarm plates contained 0.2% arabinose to induce the expression of plasmid-based genes. Whole-cell lysates were prepared in lysis buffer containing 20 mM Tris (pH 8), 10 mM MgCl2, cOmplete EDTA-free Protease Inhibitor Cocktail (Roche), Benzonase nuclease, and 10 mM imidazole. Each immunoprecipitation mixture contained 500 μl lysate, 40 μl ProBond nickel-chelating resin (Invitrogen), and 0.8% Thesit (Sigma) to solubilize membranes. Immunoprecipitation mixtures were incubated at 4°C for 75 min. When indicated, they contained 5 μM c-di-GMP. The nickel resin was washed in lysis buffer once for 15 min at 4°C and then three times at RT with gentle shaking prior to SDS-PAGE analysis. Immunoprecipitation mixtures containing 5 μM c-di-GMP were washed with lysis buffer containing 5 μM c-di-GMP. For Western blotting, proteins were transferred to a nitrocellulose membrane and probed with an antihemagglutinin (anti-HA) antibody (Covance, Princeton, NJ).
Fluorescence microscopy and data processing.
P. aeruginosa bacteria for fluorescence images were picked from the edges of swarm plate colonies, as previously described (24, 32), and resuspended in motility buffer (liquid medium of the same composition as the swarm plates but lacking the agar). Two streaks of bacteria were resuspended in 800 μl motility buffer and centrifuged (3,750 × g, 3 min). The bacterial pellet was gently resuspended in 25 to 50 μl motility buffer to ensure similar bacterial densities. A 1.5-μl volume of the resuspension was placed on a microscope slide layered with a pad of 1.5% agarose in motility buffer and covered with a coverslip. Fluorescence microscopy was performed as previously described (40) with minor modifications: a Deltavision Spectris optical sectioning microscope (Applied Precision) equipped with a UPlanSApo 100 × 1.40 oil objective (Olympus) combined with ×1.6 auxiliary magnification was used, resulting in a pixel size of 101.31 nm. An Evolve electron-multiplying charge-coupled-device (EMCCD) camera (Photometrics) was used to take differential interference contrast (DIC) and fluorescence photomicrographs. For fluorophore visualization, a GFP/hsGFP filter set (excitation [Ex], 475/28 nm; emission [Em], 522/44 nm) and an EM gain of 50 was used. A stack of 20 DIC frames (Δz = 100 nm) were taken with 0.005-s exposures. Fluorescence frames corresponding to the center of the bacterium were acquired for 12 s (0.4-s exposure time [t]; Δt = 1 s). To minimize the effect of cytosolic autofluorescence and to identify stable fluorescent foci, the first three frames were discarded, and data from the remaining frames were averaged. The fluorescence background of each averaged image was determined and maximum and minimum (max/min) intensities were set to 9,200 units above the average value and 800 units below the average, respectively, using ImageJ (33). Polar fluorescent spots were then identified by eye in a blinded analysis. For each strain, more than 2,000 bacteria in 11 fields of view from three independent experiments were analyzed.
RESULTS
FlgZ and Pel mediate c-di-GMP-dependent repression of swarming motility in a mutant that makes high levels of c-di-GMP.
In P. aeruginosa, deletion of the bifA gene, which encodes a c-di-GMP-degrading phosphodiesterase, results in elevated intracellular c-di-GMP levels and repression of swarming motility (41). We have previously demonstrated that the flagellar stator protein MotA and pilin-associated protein PilY1 contribute to swarming repression in the ΔbifA mutant (24, 42). We expected that, in addition to these factors, a c-di-GMP-binding effector would likely be involved in swarming repression in response to high c-di-GMP levels. The P. aeruginosa genome encodes eight PilZ domain-containing proteins, and seven of these PilZ proteins have been shown to bind to c-di-GMP. One PilZ domain protein, PA14_20700, has a YcgR domain also found in the YcgR protein, a c-di-GMP-responsive flagellar motility control protein of E. coli and S. enterica (23). It was recently shown that an ortholog of PA14_20700 in P. fluorescens participates in biofilm formation and that another ortholog in P. putida is able to repress swimming motility when expressed from a plasmid (29). On the basis of knowledge of these roles for similar proteins, we focused on PA14_20700 as a potential candidate for c-di-GMP-mediated swarming repression. Orthologs of PA14_20700 in P. fluorescens and P. putida are called flgZ, as the gene is positioned downstream of the flgMN genes, so we adopt that nomenclature for P. aeruginosa and refer to PA14_20700 as the flgZ gene (29).
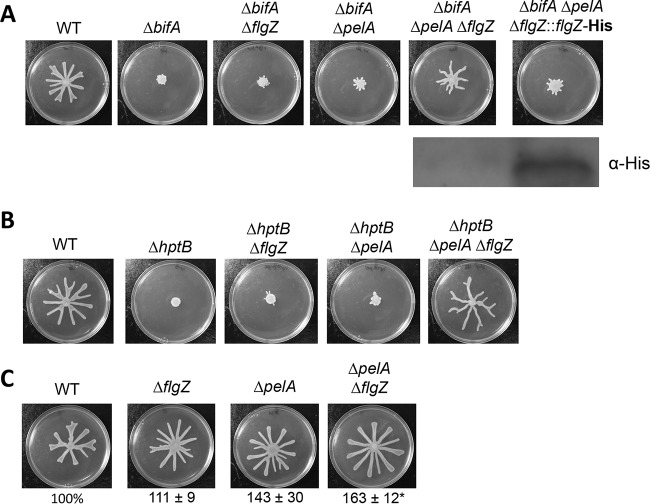
To investigate whether FlgZ participates in repression of swarming motility under conditions of elevated c-di-GMP levels in P. aeruginosa, we constructed a ΔbifA ΔflgZ double mutant and examined its swarming phenotype. As shown in Fig. 1A, deletion of the flgZ gene did not restore swarming to the ΔbifA mutant. In addition to its inability to swarm, another hallmark of the ΔbifA mutant is increased production of the Pel polysaccharide component of extracellular polymeric substances (EPS), which causes cells on swarm medium to appear wrinkly (41). We noted that the ΔbifA ΔflgZ mutant cells have a wrinkly appearance, indicating that Pel production remains high in this mutant. Pel plays important roles in cell aggregation, and we hypothesized that elevated levels of Pel may be inhibiting cell mobility and masking any contributions of FlgZ to regulating flagellum-mediated swarming motility in the ΔbifA mutant. Similarly, other extracellular polysaccharides have been shown to impair cell motility in S. enterica (19), B. subtilis (43), and Listeria monocytogenes (44).
FIG 1.
FlgZ and Pel polysaccharide contribute to swarming motility repression. (A) Top panel: representative swarm plates of the indicated strains. Bottom panel: Western blot probed with anti-His antibody to detect FlgZ-His expression in the ΔbifA ΔpelA ΔflgZ::flgZ-His strain in which the flgZ gene deletion is complemented by allelic replacement, resulting in expression of a His epitope-tagged FlgZ protein. (B) Representative swarm plates of the indicated strains. (C) Representative swarm plates of the indicated strains. The values below the swarm plates indicate the percentages (means ± standard errors of the means [SEM] of the results determined in three independent experiments performed with six plates each) of plate surface coverage of the mutant strains relative to that of the WT strain (set at 100%). Significance was determined by analysis of variance and Dunnett's posttest comparison for differences relative to the WT. *, P < 0.05 (compared to WT).
To remove the potential influence of Pel on swarming motility, we introduced a mutation in the pelA gene to eliminate a critical function required for Pel production (45). Deleting the pelA gene in the bifA mutant did not restore swarming; however, when flgZ and pelA were both deleted in a ΔbifA mutant background, swarming motility was largely restored compared to that of the wild-type strain (Fig. 1A). As reported previously, the ΔbifA ΔpelA mutant was unable to swarm (41) (Fig. 1A). We complemented the ΔbifA ΔflgZ ΔpelA mutant by allelic replacement of the flgZ deletion with a His-tagged wild-type flgZ allele (Fig. 1A). The complemented ΔbifA ΔpelA ΔflgZ::flgZ-His strain did not swarm, indicating that FlgZ participates in swarming inhibition in the ΔbifA ΔpelA mutant. Confirming the ability of the FlgZ-His protein to complement the flgZ deletion, we could detect the FlgZ-His protein by Western blotting performed on cells harvested from a swarm agar plate (Fig. 1A, bottom panel). To examine whether FlgZ had an impact on swimming motility in addition to its effects on swarming, we performed a zero-flow, free-swimming capillary assay to compare the single-cell motilities of wild-type (WT) and ΔflgZ mutant cells. We found that the ΔflgZ mutant swims slightly faster than the WT (see Fig. S1 in the supplemental material), indicating that FlgZ plays a role in inhibiting both swimming and swarming.
FlgZ is just one of eight PilZ domain-containing proteins in P. aeruginosa, while E. coli and S. enterica have only two PilZ proteins each. It is possible that additional PilZ domain proteins in P. aeruginosa play roles in c-di-GMP-responsive swarming motility repression. To examine whether other PilZ domain proteins participate in swarming motility repression, we constructed deletions of each PilZ domain protein-encoding gene in ΔbifA and ΔbifA ΔpelA mutant backgrounds. As shown in Fig. S2A in the supplemental material, deletions of each of the seven remaining PilZ domain proteins did not restore swarming to the ΔbifA mutant. When introduced into the ΔbifA ΔpelA mutant, only a mutation in pilZ resulted in a small increase in swarming but did not do so to the extent observed in the ΔbifA ΔpelA ΔflgZ mutant (see Fig. S2B). These results indicate that FlgZ is the main c-di-GMP effector protein contributing to swarming motility repression in the ΔbifA ΔpelA mutant, although PilZ may also play a minor role in this process. While PilZ does not appear to bind c-di-GMP (12, 23), this protein participates in biogenesis of type IV pili (46), and we suspect that the defect in the function of type IV pili is responsible for the impact on swarming (35). Given the substantial role of FlgZ in regulating cell surface motility, we focus on the contribution of this protein for the remainder of the work presented here.
We next confirmed that the increased motility observed in flgZ mutant strains was not the result of a decrease in c-di-GMP levels. We used mass spectrometry to quantify intracellular c-di-GMP levels in wild-type P. aeruginosa PA14 as well as in the ΔflgZ, ΔpelA, ΔpelA ΔflgZ, ΔbifA, ΔbifA ΔflgZ, ΔbifA ΔpelA, and ΔbifA ΔpelA ΔflgZ mutants. In most cases, c-di-GMP levels did not change significantly with the introduction of the flgZ mutation (see Fig. S3 in the supplemental material). However, the ΔbifA ΔflgZ ΔpelA mutant had significantly higher c-di-GMP levels than the ΔbifA ΔpelA mutant (see Fig. S3) despite the fact that the triple mutant showed increased motility versus the double mutant; this result indicates that swarming in the ΔbifA ΔflgZ ΔpelA mutant is not due to decreased c-di-GMP levels.
To assess whether FlgZ and Pel contribute to swarming motility repression under conditions of high levels of c-di-GMP generally, and not specifically in response to the absence of bifA, we used another strain with high c-di-GMP levels which carries a mutation in the hptB gene. The hptB gene encodes a histidine phosphotransfer protein involved in a complex regulatory cascade (47–49). The ΔhptB mutant of P. aeruginosa PAK has been demonstrated to produce hyperbiofilms and to show elevated Pel polysaccharide production and elevated c-di-GMP levels (48, 50). By mass spectrometry analysis, we found that, in the P. aeruginosa PA14 strain, the ΔhptB mutant produces 4.3-fold more c-di-GMP than the wild type, while the ΔbifA mutant produces 12.8-fold more c-di-GMP than the wild type. Mirroring our observations in the ΔbifA mutant background, the ΔhptB single mutant and the ΔhptB ΔpelA and ΔhptB ΔflgZ double mutants were unable to swarm, but swarming was restored in a ΔhptB ΔflgZ ΔpelA triple mutant (Fig. 1B). Notably, swarming motility appeared to be fully restored to wild-type levels in the ΔhptB ΔflgZ ΔpelA mutant in contrast to the partial restoration observed for the ΔbifA ΔflgZ ΔpelA mutant. We suspect that this discrepancy is due to the difference in c-di-GMP levels measured for the ΔhptB and ΔbifA mutants.
In addition to the roles of FlgZ and Pel in motility repression in two distinct high-c-di-GMP mutant backgrounds, we found that a ΔpelA ΔflgZ mutant hyperswarmed compared to the wild-type strain (Fig. 1C). The ability of flgZ and pelA mutations to alleviate swarming repression in two high-c-di-GMP backgrounds and in the wild-type background suggests that FlgZ's impact on swarming is not due to the response to any particular mutation but is instead a general function of FlgZ.
FlgZ's impact on swarming motility control depends on its conserved c-di-GMP binding motif.
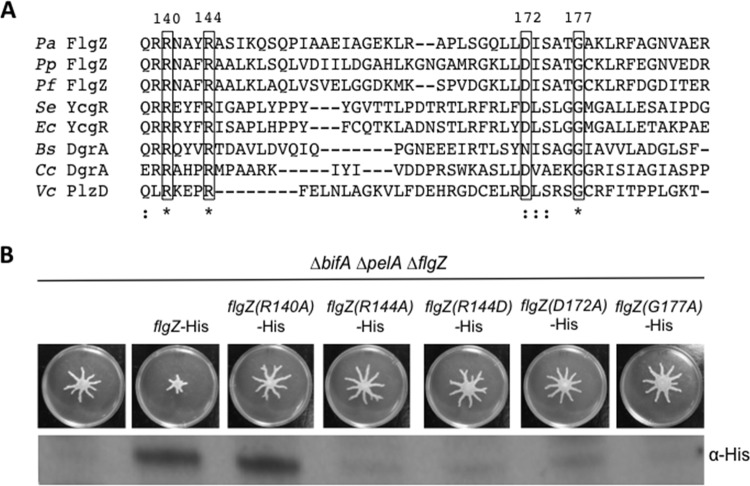
Amino acid residues of the conserved motifs RXXXR and D/NXSXXG are critical for c-di-GMP binding in PilZ domain-containing proteins, including E. coli YcgR (8), P. aeruginosa Alg44 (12), Caulobacter crescentus DgrA (10), Vibrio cholerae PlzD (7), and Borrelia burgdorferi PlzA (13). The PilZ domain of P. aeruginosa FlgZ contains these conserved residues, as demonstrated by sequence alignment with PilZ domain-containing proteins of other species (Fig. 2A). To test whether FlgZ's role in swarming repression requires this c-di-GMP binding motif, we generated strains in which we complemented the ΔbifA ΔflgZ ΔpelA mutant with a His-tagged variant of FlgZ carrying single amino acid substitutions in the PilZ domain and tested the ability of these mutant FlgZ proteins to restore swarming repression. A R140A substitution resulted in the production of a detectable FlgZ protein that was no longer able to repress swarming (Fig. 2B). The R144A, R144D, D172A, and G177A mutant variants of FlgZ-His each resulted in a protein that could not be detected by Western blotting (Fig. 2B). This result suggests that the conserved R140 residue required for c-di-GMP binding is critical for FlgZ's role in repressing swarming in response to c-di-GMP.
FIG 2.
Conserved residues in the c-di-GMP binding domain are important for FlgZ stability and function. (A) Multiple-sequence alignment of the predicted c-di-GMP binding region of FlgZ in P. aeruginosa (Pa) with orthologs from P. putida (Pp), P. fluorescens (Pf), S. enterica (Se), E. coli (Ec), and B. subtilis (Bs) along with other PilZ domain-containing proteins from C. crescentus (Cc) and V. cholerae (Vc). The sequence alignment was generated by Clustal Omega (59, 60) using the complete PilZ domain of each protein as predicted by SMART (61, 62). A portion of the alignment is shown here. Clustal Omega determined conservation of residues. ★, a fully conserved residue; :, a residue with strongly similar properties. The boxed conserved residues were targeted for site-directed mutagenesis. Numbers correspond to the amino acid residues in the P. aeruginosa FlgZ full-length protein. (B) Top panel: representative swarm assays of the indicated strains. Bottom panel: protein levels determined using Western blotting and anti-His antibody to detect expression of the wild-type strain and mutant FlgZ-His variants.
FlgZ interacts with the flagellar stator protein MotC.
PilZ domain proteins in other bacteria have been shown to influence motility via direct interactions with one or more components of the flagellar motor. Thus, we hypothesized that FlgZ may impact swarming motility by a similar mechanism. We employed a bacterial adenylate cyclase two-hybrid assay in E. coli to probe for interactions between FlgZ and components of the P. aeruginosa flagellar motor: MotA, MotC, FliG, and FliM. MotA and MotC are components of ion-translocating stators, and FliG and FliM are the rotor component and switch protein of the motor, respectively. The motor requires MotC, FliG, and FliM for functional swarming motility (51), whereas MotA reduces swarming motility in P. aeruginosa.
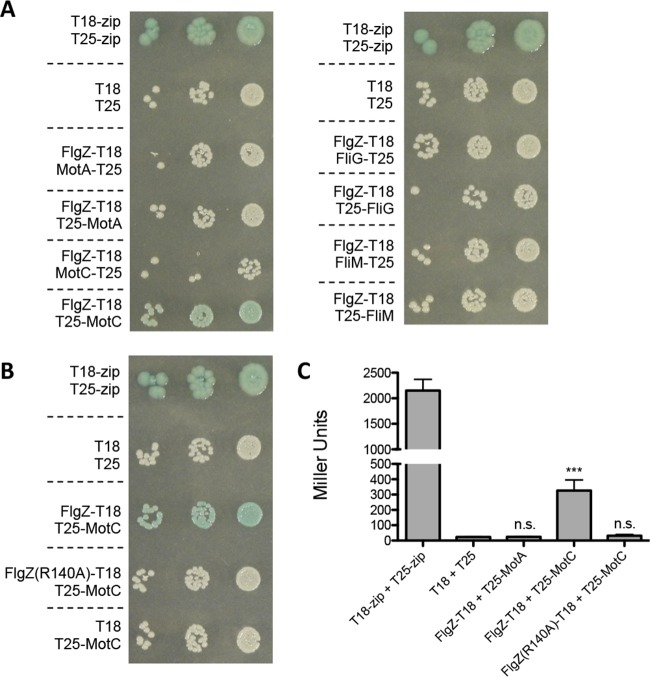
Full-length proteins were fused to either the T18 or T25 subunit of adenylate cyclase, and these hybrid proteins were coexpressed in E. coli BTH101 to test each potential interaction. An interaction between hybrid proteins can be detected as blue coloring on medium containing the substrate X-Gal. As shown in Fig. 3A, FlgZ was found to interact with MotC but not with MotA, FliG, or FliM. To investigate whether the c-di-GMP binding region of FlgZ is important for the interaction between FlgZ and MotC, we next tested the point mutant FlgZ (R140A) in the bacterial two-hybrid assay. As shown in Fig. 3B, FlgZ (R140A) was not able to interact with MotC, indicating that residue R140 is critical both for FlgZ's function in swarming motility repression and for the FlgZ-MotC interaction. As an additional negative control, we cotransformed the pUT18 empty vector with T25-MotC and saw no blue coloring, as expected (Fig. 3B). The strength of these interactions was quantified using β-galactosidase assays, and strains coexpressing FlgZ and MotC fusion proteins showed significantly higher β-galactosidase levels than the negative control (Fig. 3C). Mutating R140 in the FlgZ protein resulted in a significant reduction in interactions with MotC to a level observed for the FlgZ-MotA interaction (Fig. 3C).
FIG 3.
Detection of interaction between FlgZ and MotC by bacterial two-hybrid analysis. (A and B) Full-length flgZ (A) and flgZ (R140A) (B), as well as the flagellar motor genes motA, motC, fliG, and fliM, were cloned into vector pKNT25, pKT25, pUT18, or pUT18C and cotransformed into E. coli BTH101 cells. The coexpressed fusion protein combinations for each transformation are indicated on the left. The transformants were 10-fold serial diluted, spotted (2 μl) on LB agar containing Cb, Kan, X-Gal, and IPTG, and then incubated for 40 h at 30°C. Cells cotransformed with empty vectors served as negative controls, and cells cotransformed with leucine zipper vectors provided by the manufacturer (T18-zip and T25-zip) served as positive controls. The degradation of X-Gal (blue) indicates a positive protein-protein interaction. (C) Bacterial two-hybrid interactions were quantified by measuring β-galactosidase activity in transformants grown in LB broth supplemented with Cb and Kan overnight at 30°C. The data represent results of three independent experiments performed with three or four biological replicates each, and values are reported as means ± SEM. Significance was determined by analysis of variance and Dunnett's posttest comparison for differences relative to the negative control (T18 + T25). n.s., not significant; ***, P < 0.001 (the positive control [T18-zip + T25-zip] was not included in the statistical analysis).
To further substantiate the evidence for physical interaction between FlgZ and MotC, we assessed the ability of FlgZ and MotC to coprecipitate. To probe this interaction, FlgZ was C-terminally HA tagged in P. aeruginosa strains carrying overexpression plasmid pMotA-His or pMotC-His. These His-tagged versions of MotA and MotC have been used previously by our group and are functional in swarming assays (24). We used a nickel resin to enrich for MotA-His or MotC-His and tested for FlgZ-HA coprecipitation with and without the addition of 5 μM c-di-GMP. Our results indicate that FlgZ interacts with MotC in the presence of 5 μM c-di-GMP and does not interact with MotC in the absence of c-di-GMP (Fig. 4). FlgZ does not interact with MotA regardless of the presence or absence of c-di-GMP. In strains lacking His-tagged MotA or MotC, only background bands were detected (Fig. 4, far right lanes).
FIG 4.
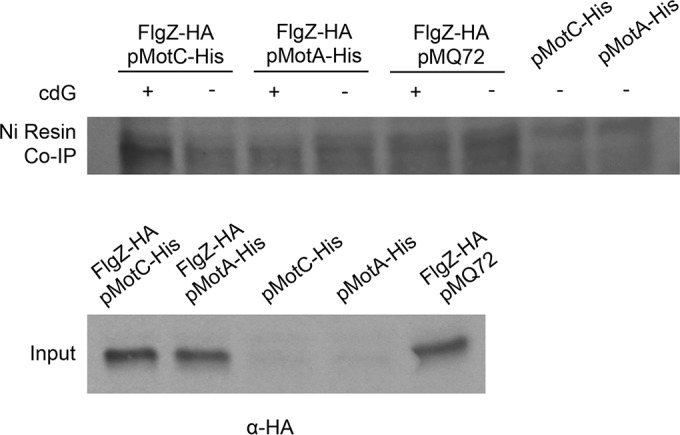
Immunoprecipitation analysis to assess FlgZ and MotC interaction. Immunoprecipitations (Co-IP) with a nickel-chelating resin were performed with cell lysates expressing FlgZ-HA and MotA-His or MotC-His. Western blots of precipitate (top panel) and input (bottom panel) were probed with anti-HA. Immunoprecipitations were performed with and without 5 μM c-di-GMP (cdG), as indicated.
FlgZ localization is consistent with an interaction between FlgZ and MotCD.
Our data suggest that there is a c-di-GMP-dependent interaction between FlgZ and MotC. To assess whether c-di-GMP levels and the presence of MotCD stator sets influence subcellular localization of FlgZ, we replaced the flgZ gene with a gfp-flgZ fusion at its native chromosomal locus. We complemented the ΔbifA ΔflgZ ΔpelA mutant by allelic replacement of the flgZ deletion with gfp-flgZ to show that this GFP fusion does not interfere with the function of FlgZ in swarming motility (see Fig. S4 in the supplemental material).
Previous work suggested that FlgZ-like proteins in P. fluorescens, E. coli, and S. enterica localize to the flagellar basal body and that this localization is enhanced in mutants with high c-di-GMP levels (15, 18, 29). In P. aeruginosa cells harvested from a swarm agar plate, we observed that GFP-FlgZ could indeed localize to the pole (Fig. 5). Furthermore, the percentage of cells with polarly localized GFP-FlgZ significantly increased in the ΔbifA mutant (Fig. 5A), which produces more c-di-GMP than the wild-type strain (see Fig. S3 in the supplemental material). This finding indicates that increased levels of c-di-GMP impact localization of FlgZ.
FIG 5.
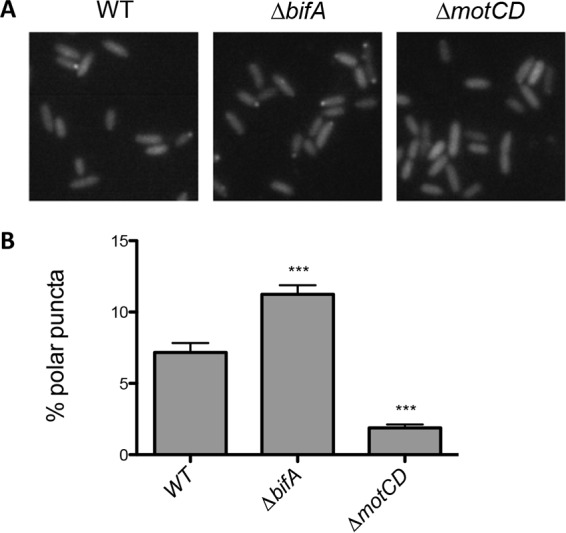
Localization of GFP-FlgZ is impacted by c-di-GMP levels and MotCD. (A) Representative images of WT, ΔbifA, and ΔmotCD strains expressing GFP-FlgZ. (B) For each strain, more than 2,000 bacteria from three independent experiments were analyzed, and values are reported as means ± SEM. Significance was determined by analysis of variance and Dunnett's posttest comparison for differences relative to WT. ***, P < 0.001.
Given that we have demonstrated that FlgZ interacts with MotCD, we hypothesized that the polar localization of FlgZ might be at least in part dependent upon MotCD. To test this hypothesis, we introduced a ΔmotCD mutation into our strain expressing the functional GFP-FlgZ from its endogenous locus on the chromosome. We found significantly fewer cells with polar localization of GFP-FlgZ in the ΔmotCD mutant than in the wild type (Fig. 5B). This finding is consistent with our hypothesis that FlgZ interacts with the flagellar machinery via a direct interaction with MotC.
MotCD stator overexpression restores swarming motility in high-c-di-GMP backgrounds lacking Pel.
We previously demonstrated that the MotCD stator set is required for swarming motility and that there is a decrease in polar localization of MotD under conditions of high levels of c-di-GMP (24). We hypothesized that overexpression of MotCD would increase the presence of this stator at the flagellar motor and that this increase in MotCD levels could restore swarming to high-c-di-GMP backgrounds by overcoming the FlgZ-mediated swarming repression mechanism.
We found that, similarly to the impact of deleting the flgZ gene, overexpression of MotCD partially restores swarming motility to the ΔbifA ΔpelA mutant (Fig. 6). However, MotCD overexpression does not restore swarming motility in the ΔbifA mutant in which elevated Pel polysaccharide is produced. This result is consistent with our hypothesis that the effect of c-di-GMP on flagellar motor function can be masked by the contribution of Pel polysaccharide.
FIG 6.
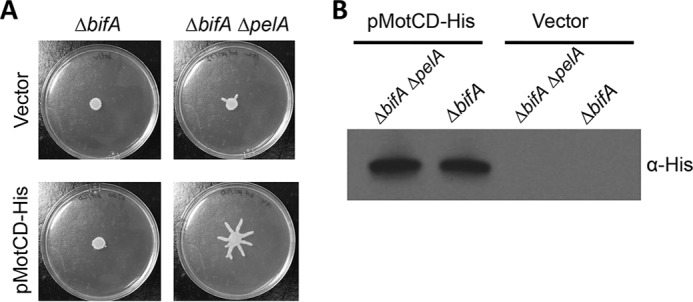
MotCD overexpression restores c-di-GMP-inhibited swarming only in the absence of Pel. (A) Representative swarm plates of the indicated strains carrying either an empty vector or a MotCD-His-expressing plasmid (pMotCD-His). Swarm plates contained 0.2% arabinose. (B) Protein levels determined by the use of Western blotting and anti-His antibody to detect expression of pMotCD-His.
DISCUSSION
The second messenger c-di-GMP coordinately regulates motility and extracellular polysaccharide synthesis to allow bacteria to efficiently transition between motile and sessile lifestyles. Regulation via c-di-GMP can occur at the level of transcription as well as posttranscriptionally and posttranslationally (52). In P. aeruginosa, c-di-GMP regulates the biosynthesis of both flagellar components and extracellular polysaccharide through the transcription factor FleQ (53). Posttranscriptional regulation of extracellular polysaccharide production is controlled by another c-di-GMP binding protein, PelD (54). Here we describe an additional layer of motility regulation: the first PilZ domain protein in P. aeruginosa with a role in flagellum-dependent swarming motility control.
Deletion mutants lacking the genes encoding each of the eight PilZ proteins of P. aeruginosa revealed that c-di-GMP-mediated repression of swarming motility is largely restored by the deletion of both flgZ and pelA genes. This finding indicates that inhibition of motility can be controlled by FlgZ and highlights a role for Pel polysaccharide in this process. The finding that Pel polysaccharide plays a role in motility repression in the absence of c-di-GMP effector protein FlgZ, particularly in the ΔbifA ΔflgZ and ΔhptB ΔflgZ mutants, aligns with previous findings indicating that extracellular polysaccharides impair motility in other species, including S. enterica (19), B. subtilis (43), and L. monocytogenes (44). However, P. aeruginosa is different from these and other organisms using c-di-GMP-dependent motility control in that eliminating either the c-di-GMP binding protein or the exopolysaccharide singly has no significant impact on motility in the high-c-di-GMP background.
It has been previously demonstrated that FlgZ binds c-di-GMP. We show that mutants with an amino acid substitution in the conserved c-di-GMP binding motif of FlgZ behave like a flgZ null mutant, suggesting that the PilZ domain of FlgZ is required for swarming motility inhibition and, consequently, that c-di-GMP binding is critical for FlgZ's function in swarming motility control. We measured global levels of c-di-GMP in flgZ mutants to ensure that the increased motility of the ΔbifA ΔflgZ ΔpelA mutant was not due to an overall decrease in c-di-GMP levels. Unexpectedly, we found that the ΔbifA ΔflgZ ΔpelA mutant has elevated levels of c-di-GMP compared to the ΔbifA ΔpelA mutant despite the enhanced swarming motility of the ΔbifA ΔflgZ ΔpelA triple mutant; we currently do not understand the basis for this observation.
P. aeruginosa has two stator sets to power a single flagellar motor, distinguishing it from many organisms that use PilZ proteins to control motility. We hypothesize that c-di-GMP-bound FlgZ inhibits swarming motility by specifically targeting the stator set that promotes swarming motility—MotCD. This hypothesis is supported by results of bacterial two-hybrid assays, coimmunoprecipitation experiments, and localization studies, all of which indicate a protein-protein interaction between FlgZ and MotC. These experiments also demonstrated that the presence of c-di-GMP and that of the conserved PilZ domain of FlgZ, which are necessary for motility inhibition, are also required for FlgZ-MotC interaction.
By impacting flagellar motor function in response to c-di-GMP, FlgZ can allow P. aeruginosa to regulate swarming motility after flagellar assembly is complete, thus enabling cells to quickly adapt to changing environmental conditions. The mechanism by which FlgZ's interaction with MotC may be altering MotCD engagement with the rotor will be the focus of future studies. Interestingly, evidence for stator exchange in the context of a dual-stator system also comes from studies in Shewanella oneidensis, in which sodium-dependent PomAB and proton-dependent MotAB drive rotation of a single flagellar rotor (55). In S. oneidensis, sodium levels drive stator dynamics by influencing the efficiency of incorporation of MotAB into the motor (56); however, the mechanism of stator switching in this organism is not yet clear. We propose that, due to the dynamic nature of the stator proteins with respect to the flagellar motor (57, 58), in the case of P. aeruginosa, one possibility is that, under conditions of high c-di-GMP levels, FlgZ interacts with MotCD stators to prevent MotCD engagement with the flagellar rotor, leaving room for increased incorporation of MotAB stators, which cannot drive swarming motility. Perhaps analogous mechanisms function to promote stator exchange in other organisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R37 AI83256-06 and Human Frontiers in Science Program grants to G.A.O. A.E.B. was supported by the NSF Graduate Research Fellowship.
We thank Chris Jones at the University of Oxford for assistance with analysis of the free-swimming capillary assay and Lijun Chen at the Michigan State University Mass Spectrometry Facility for quantitative analysis of c-di-GMP.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00196-16.
REFERENCES
- 1.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 2.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 4.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 7.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J 26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 9.Ko M, Park C. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol 303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 10.Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A 104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem 282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 13.Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, Schwartz I, Marconi RT. 2010. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol Med Microbiol 58:285–294. doi: 10.1111/j.1574-695X.2009.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect Immun 79:1815–1825. doi: 10.1128/IAI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Chai Y, Guo JH, Losick R. 2012. Evidence for cyclic Di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol 194:5080–5090. doi: 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 18.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zorraquino V, Garcia B, Latasa C, Echeverz M, Toledo-Arana A, Valle J, Lasa I, Solano C. 2013. Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J Bacteriol 195:417–428. doi: 10.1128/JB.01789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, Mukherjee S, Matthews PM, Hammad LA, Kearns DB, Dann CE III. 2013. Functional characterization of core components of the Bacillus subtilis cyclic-di-GMP signaling pathway. J Bacteriol 195:4782–4792. doi: 10.1128/JB.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Düvel J, Bertinetti D, Möller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jänsch L, Herberg FW, Häussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods 88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol 86:1424–1440. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchma SL, Delalez NJ, Filkins LM, Snavely EA, Armitage JP, O'Toole GA. 2015. Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J Bacteriol 197:420–430. doi: 10.1128/JB.02130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blair DF. 2003. Flagellar movement driven by proton translocation. FEBS Lett 545:86–95. doi: 10.1016/S0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- 26.Blair DF, Berg HC. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 27.Doyle TB, Hawkins AC, McCarter LL. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol 186:6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toutain CM, Zegans ME, O'Toole GA. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol 187:771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, González de Heredia E, Baena I, Martín-Martín I, Rivilla R, Martín M. 2014. Identification of flgZ as a flagellar gene encoding a PilZ domain protein that regulates swimming motility and biofilm formation in Pseudomonas. PLoS One 9:e87608. doi: 10.1371/journal.pone.0087608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O'Toole GA. 2015. Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. mBio 6:e01978-15. doi: 10.1128/mBio.01978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha DG, Kuchma SL, O'Toole GA. 2014. Plate-based assay for swarming motility in Pseudomonas aeruginosa. Methods Mol Biol 1149:67–72. doi: 10.1007/978-1-4939-0473-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood TM, Yates CA, Wilkinson DA, Rosser G. 2012. Simplified multitarget tracking using the PHD filter for microscopic video data. IEEE Trans Circuits Syst Video Technol 22:702–713. doi: 10.1109/TCSVT.2011.2177937. [DOI] [Google Scholar]
- 35.Kuchma SL, Griffin EF, O'Toole GA. 2012. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J Bacteriol 194:5388–5403. doi: 10.1128/JB.00899-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J Bacteriol 193:4685–4698. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battesti A, Bouveret E. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Diepold A, Kudryashev M, Delalez NJ, Berry RM, Armitage JP. 2015. Composition, formation, and regulation of the cytosolic C-ring, a dynamic component of the type III secretion injectisome. PLoS Biol 13:e1002039. doi: 10.1371/journal.pbio.1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuchma SL, Ballok AE, Merritt JH, Hammond JH, Lu W, Rabinowitz JD, O'Toole GA. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J Bacteriol 192:2950–2964. doi: 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttenplan SB, Blair KM, Kearns DB. 2010. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet 6:e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen LH, Koseoglu VK, Guvener ZT, Myers-Morales T, Reed JM, D'Orazio SE, Miller KW, Gomelsky M. 2014. Cyclic di-GMP-dependent signaling pathways in the pathogenic Firmicute Listeria monocytogenes. PLoS Pathog 10:e1004301. doi: 10.1371/journal.ppat.1004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51:675–690. [DOI] [PubMed] [Google Scholar]
- 46.Alm RA, Bodero AJ, Free PD, Mattick JS. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol 178:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhuwan M, Lee HJ, Peng HL, Chang HY. 2012. Histidine-containing phosphotransfer protein-B (HptB) regulates swarming motility through partner-switching system in Pseudomonas aeruginosa PAO1 strain. J Biol Chem 287:1903–1914. doi: 10.1074/jbc.M111.256586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bordi C, Lamy MC, Ventre I, Termine E, Hachani A, Fillet S, Roche B, Bleves S, Mejean V, Lazdunski A, Filloux A. 2010. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol 76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CT, Huang YJ, Chu PH, Hsu JL, Huang CH, Peng HL. 2006. Identification of an HptB-mediated multi-step phosphorelay in Pseudomonas aeruginosa PAO1. Res Microbiol 157:169–175. doi: 10.1016/j.resmic.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 50.Moscoso JA, Jaeger T, Valentini M, Hui K, Jenal U, Filloux A. 2014. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J Bacteriol 196:4081–4088. doi: 10.1128/JB.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 52.Wolfe AJ, Visick KL. 2008. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol 190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, Narberhaus F, Thormann KM. 2009. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol 71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- 56.Paulick A, Delalez NJ, Brenzinger S, Steel BC, Berry RM, Armitage JP, Thormann KM. 2015. Dual stator dynamics in the Shewanella oneidensis MR-1 flagellar motor. Mol Microbiol 96:993–1001. doi: 10.1111/mmi.12984. [DOI] [PubMed] [Google Scholar]
- 57.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I, Doerks T, Bork P. 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.