ABSTRACT
Many factors, such as the substrate and the growth phase, influence biosynthesis of secondary metabolites in microorganisms. Therefore, it is crucial to consider these factors when establishing a bioprospecting strategy. Mimicking the conditions of the natural environment has been suggested as a means of inducing or influencing microbial secondary metabolite production. The purpose of the present study was to determine how the bioactivity of Vibrionaceae was influenced by carbon sources typical of their natural environment. We determined how mannose and chitin, compared to glucose, influenced the antibacterial activity of a collection of Vibrionaceae strains isolated because of their ability to produce antibacterial compounds but that in subsequent screenings seemed to have lost this ability. The numbers of bioactive isolates were 2- and 3.5-fold higher when strains were grown on mannose and chitin, respectively, than on glucose. As secondary metabolites are typically produced during late growth, potential producers were also allowed 1 to 2 days of growth before exposure to the pathogen. This strategy led to a 3-fold increase in the number of bioactive strains on glucose and an 8-fold increase on both chitin and mannose. We selected two bioactive strains belonging to species for which antibacterial activity had not previously been identified. Using ultrahigh-performance liquid chromatography–high-resolution mass spectrometry and bioassay-guided fractionation, we found that the siderophore fluvibactin was responsible for the antibacterial activity of Vibrio furnissii and Vibrio fluvialis. These results suggest a role of chitin in the regulation of secondary metabolism in vibrios and demonstrate that considering bacterial ecophysiology during development of screening strategies will facilitate bioprospecting.
IMPORTANCE A challenge in microbial natural product discovery is the elicitation of the biosynthetic gene clusters that are silent when microorganisms are grown under standard laboratory conditions. We hypothesized that, since the clusters are not lost during proliferation in the natural niche of the microorganisms, they must, under such conditions, be functional. Here, we demonstrate that an ecology-based approach in which the producer organism is allowed a temporal advantage and where growth conditions are mimicking the natural niche remarkably increases the number of Vibrionaceae strains producing antibacterial compounds.
INTRODUCTION
Following the first era of discovery of bioactive compounds from natural sources, high-throughput screenings of compound libraries produced by combinatorial chemistry and rational drug design were preferred over natural product discovery (1). Disappointingly, the discovery rate of this approach was much lower than expected, and the lack of new leads triggered a return to the search for novel bioactive molecules from microorganisms (1, 2).
Recent progress in genome sequencing and mining has demonstrated a significant number and degree of diversity in microbial biosynthetic gene clusters. However, this potential can often not be unfolded and detected under standard laboratory conditions (3, 4), and, today, one challenge in the discovery of natural products is to elicit these silent/cryptic biosynthetic gene clusters. The one strain-many compounds method, where strains are cultivated under a range of growth conditions, has been suggested as a solution (5).
Secondary metabolites are likely to play many different roles in natural bacterial behavior, including antagonistic interactions and intercellular communication (6, 7). Hence, elicitation of the expression of silent biosynthetic gene clusters could rely on recreating the natural environmental conditions in the research laboratory (8–10). With this in mind, Seyedsayamdost (11) demonstrated that two previously silent biosynthetic gene clusters in Burkholderia thailandensis could be elicited by low concentrations of molecules of microbial origin. Also, antibacterial compounds have been shown to be produced by marine bacteria only when they were cultivated under conditions mimicking their natural intertidal environment (12–14).
Following the increasing interest in natural products from the marine environment during the last decades of the 20th century, several groups are now pursuing methods for the identification and production of natural product in marine microorganisms (15, 16). Our group took part in the global marine research expedition Galathea 3 (http://www.galathea3.dk), with the aim of, on a global scale, isolating marine bacteria with bioactivity potential. We cultured microorganisms on marine agar and subsequently screened all colonies for antagonism against the fish pathogen Vibrio anguillarum, which is very sensitive to antibacterial compounds produced by marine bacteria. We isolated approximately 300 bioactive Vibrionaceae strains (17). During rescreening, only 39 strains retained their antagonistic activity (18). We isolated the potent antibiotics holomycin and andrimid from Vibrio coralliilyticus and Photobacterium galatheae, respectively (9, 18), as well as modulators of virulence in Staphylococcus aureus, such as ngercheumicins F, G, H, and I (19), nigribactin (20), and solonamide B (21). However, we were challenged by the marked reduction in bioactivity during rescreening.
We reasoned that one cause for this loss of activity could be that significant secondary metabolite production mostly occurs during the late-exponential and the stationary phases of microbial growth, and we hypothesized that the biodiscovery rate could be increased if the producing organisms were allowed more time to grow before being exposed to the target organism. In the initial screening and isolation, colonies were allowed to grow for 3 to 5 days before being tested (17), but this temporal advantage was not given during the rescreening (18). We also questioned whether the use of naturally co-occurring substrates, such as mannose and chitin, would restore bioactivity. Mannose is ubiquitous in the marine environment, where it is commonly used by algae for protein glycosylation and production of extracellular polysaccharides (22, 23). Chitin is the most abundant organic molecule in the marine environment, being a component of the exoskeleton of crustaceans and zooplankton (24). It is a polysaccharide composed of N-acetylglucosamine (GlcNAc) units. Vibrionaceae bacteria are considered among the major actors in marine chitin catabolism, and the chitin utilization pathway is conserved within the family (25, 26). In Vibrio cholerae, chitin and derivatives can regulate the expression of genes involved in chitin metabolism (27) and also in biofilm formation and in virulence (28). In V. coralliilyticus, growth on chitin doubles the yield of the antibiotic andrimid compared to growth on glucose (9).
The aim of this study was to determine to which extent the use of substrates naturally present in the niche of isolation and the growth phase of the producer could restore (or induce) the biosynthesis of antibacterial compounds in a collection of 295 Vibrionaceae isolates. The number of antagonizing strains was greatly increased when the assay was performed on chitin and was up to 8-fold higher when the potential producers were given a temporal advantage over the target strain.
MATERIALS AND METHODS
Bacterial strains.
Two hundred ninety-five Vibrionaceae strains were isolated during the Danish Galathea 3 global research expedition (17). Strains were selected based on their abilities to inhibit the growth of Vibrio anguillarum and were identified as Vibrionaceae based on their 16S rRNA gene sequences (17). Species affiliation of strains producing antibacterial extracts (see below), which had not been previously assigned to a species by multilocus sequence analysis, was carried out by analysis of the fur gene (29). The fur gene sequences were retrieved from whole-genome sequences (WGS) or sequencing of PCR products, obtained as described elsewhere (29).
Preparation of colloidal chitin.
Colloidal chitin was prepared following a modified version of the method published by Hsu and Lockwood (30). Ten grams of practical-grade shrimp shell chitin (Sigma C9213) was added to 400 ml of 37% HCl at 4°C and stirred at this temperature for 6 h. The solution was poured into 4 liters of cold H2O and incubated overnight at 4°C before it was neutralized with solid NaOH. After centrifugation (6,000 × g for 10 min), the supernatant was discarded and the chitin pellet was suspended in 500 ml of H2O and autoclaved. The concentration of colloidal chitin was calculated from the dry weight of a subsample.
Screening of Vibrionaceae strains for antibacterial activity.
Square petri dishes containing 20 g/liter sea salts (Sigma S9883), 3 g/liter Casamino Acids (BD 223050), 15 g/liter agar (AppliChem A0949), and either 2 g/liter colloidal chitin or 2 g/liter mannose were prepared. As the control, the same was done, with the same medium used in the original screening procedure (30 g/liter instant ocean, 3 g/liter Casamino Acids, 4 g/liter glucose, 10 g/liter agar) (17). Bacterial strains were grown overnight, aerated (200 rpm) at room temperature in half-strength yeast tryptone sea salts (YTSS) (31). One microliter of each culture was spotted onto the three media. On each plate, 35 strains were spotted in rows, where the distance between two strains was 20 mm horizontally and 15 mm vertically. Each plate was produced three times. On one plate, 1 μl of an overnight culture of the target strain, Vibrio anguillarum 90-11-287, grown in half-strength YTSS, was spotted simultaneously at a distance of 5 mm from the potential producers of antimicrobial compounds. On the second copy of each plate, an identical process was performed after 24 h; on a third plate, after 48 h. Plates were incubated at 25°C and examined 24 or 48 h after the target strain had been spotted. A biological replicate was performed for the isolates that were bioactive in the first screening.
In silico analysis of the distribution of chiS and the (GlcNAc)2 operon.
The chiS (VC0622) gene and the (GlcNAc)2 operon (VC0611-VC0620) of Vibrio cholerae were searched against a custom-built genome database using MultiGeneBlast (32). For the preparation of the database, genome sequences were downloaded from the GenBank database.
Extraction of bioactive compounds from liquid cultures.
All strains showing a consistent bioactivity were grown aerated (200 rpm) in 10 ml of 2% Sigma sea salts solution with 0.3% Casamino Acids and 0.2% colloidal chitin (sea salt broth and chitin medium [SSBC]) for 48 h at 25°C. Cultures were extracted with an equal volume of high-performance liquid chromatography (HPLC)-grade ethyl acetate (EtOAc) for 20 min. The organic phase was transferred to fresh vials and evaporated until dry under a stream of nitrogen. Extracts were dissolved in 250 μl methanol (MeOH) and stored at −20°C until further analysis. The activity of the extracts against Vibrio anguillarum 90-11-287 was tested in a well diffusion agar (WDA) assay (33).
Genome sequencing and bioinformatics analysis.
High-purity DNA was obtained for Vibrio furnissii S0821 and Vibrio fluvialis S1110 by repeated phenol-chloroform-isoamyl alcohol purification followed by RNase treatment and DNA precipitation, as described previously (34). Quantification was performed on a NanoDrop spectrometer (Saveen Werner, Sweden) and a Qubit 2.0 analyzer (Invitrogen, United Kingdom). Construction of 500-bp libraries and 100-bp paired-end sequencing of genomes were performed by the Beijing Genome Institute (Hong Kong) on a HiSeq2000. Sequencing data were assembled to contigs in CLC genomic workbench (CLC Bio, Aarhus, Denmark) using the de novo assembly algorithm. The draft genomes of strains S0821 and S1110 were annotated with the NCBI prokaryotic genome annotation pipeline (35) and submitted to antiSMASH 2.0 (36) and BAGEL3 (37) for analysis of biosynthetic gene clusters.
Ultrahigh-performance liquid chromatography–high-resolution mass spectrometry.
Ultrahigh-performance liquid chromatography–high-resolution mass spectrometry (UHPLC-HRMS) was performed on an Agilent Infinity 1290 UHPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a diode array detector. Separation was obtained on an Agilent Poroshell 120 phenyl-hexyl column (2.1 × 250 mm, 2.7 μm) with a linear gradient consisting of H2O (A) and acetonitrile (B), both buffered with 20 mM formic acid, starting at 10% for B and increased to 100% in 15 min, where it was held for 2 min, returned to 10% in 0.1 min, and remained for 3 min (0.35 ml/min, 60°C). An injection volume of 1 μl was used. Mass spectroscopy (MS) detection was performed on either an Agilent 6545 QTOF MS equipped with Agilent dual jet stream electrospray ion source with a drying gas temperature of 160°C, gas flow of 13 liters/min, sheath gas temperature of 300°C, and flow of 16 liters/min, or an Agilent 6550 QTOF MS equipped with Agilent dual jet stream electrospray ion source with a drying gas temperature of 250°C, gas flow of 8 liters/min, sheath gas temperature of 300°C, and flow of 12 liters/min. Capillary voltage was set to 4,000 V, and nozzle voltage, to 500 V. Mass spectra were recorded as centroid data at an m/z of 85 to 1,700, and MS/MS was done at 10, 20, and 40 eV fragmentation energy, scanning an m/z of 30 to 1,700. The acquisition was 10 spectra/s. Lock mass solution in 70:30 MeOH:H2O was infused in the second sprayer using an extra pump at a flow of 15 μl/min using a 1:100 splitter. The solution contained 1 μM tributylamine (Sigma-Aldrich) and 10 μM hexakis(2,2,3,3-tetrafluoropropoxy)phosphazene (Apollo Scientific, Ltd., Cheshire, United Kingdom) as lock masses. The [M + H]+ ions (m/z, 186.2216 and 922.0098, respectively) of both compounds were used.
Influence of culture conditions on bioactivity and characterization of the antibacterial compound.
Extracts from the cultures V. furnissii S0821 and V. fluvialis S1110 were analyzed by UHPLC-HRMS, as described above. Extracts from the strains grown in SSBC supplemented with 0.1 g/liter ferric citrate were also prepared and analyzed. For the bioassay-guided fractionation, 50 cultures of strain S0821 grown in 10 ml SSBC for 48 h were extracted with an equal volume of EtOAc, and extracts were pooled and evaporated until dryness under nitrogen occurred. Portions of the pooled S0821 culture extracts were fractionated by mixed-mode anion exchange solid-phase extraction (SPE) on an Oasis Max cartridge (Waters, Milford, MA) (30 μm, 30 mg, 1 ml). The sample was dissolved in 400 μl of 3:1 H2O:MeOH containing 2% ammonium hydroxide and then directly loaded onto a conditioned SPE column. The column was sequentially eluted with 2 ml of 3:1 H2O:MeOH (F1), 2 ml of 1:1 H2O:MeOH (F2), 2 ml of MeOH (F3), 1 ml of H2O and 1 ml of 3:1 H2O:MeOH containing 1% formic acid (F4), 2 ml of 1:1 H2O:MeOH containing 1% formic acid (F5), and, finally, 2 ml of MeOH containing 1% formic acid (F6). The fractions were dried under a stream of nitrogen before being resuspended in 200 μl MeOH. Fractions were tested for antibacterial activity in a WDA assay and for siderophore activity in a chrome azurol S (CAS) assay (38). Extracts were mixed with CAS solution in a 1:1 ratio, and the color change from blue to orange, indicating siderophore activity, was checked after 15 min and 24 h.
Nucleotide sequence accession numbers.
Sequence data generated in this study were deposited in GenBank under accession numbers LKHS00000000 (WGS of strain S0821), LKHR00000000 (WGS of strain S1110), and KT952522 to KT952526 (fur gene sequences of strains S1162, S1732, S2054, S2056, and S2150, respectively).
RESULTS
Screening of strains for antibacterial activity.
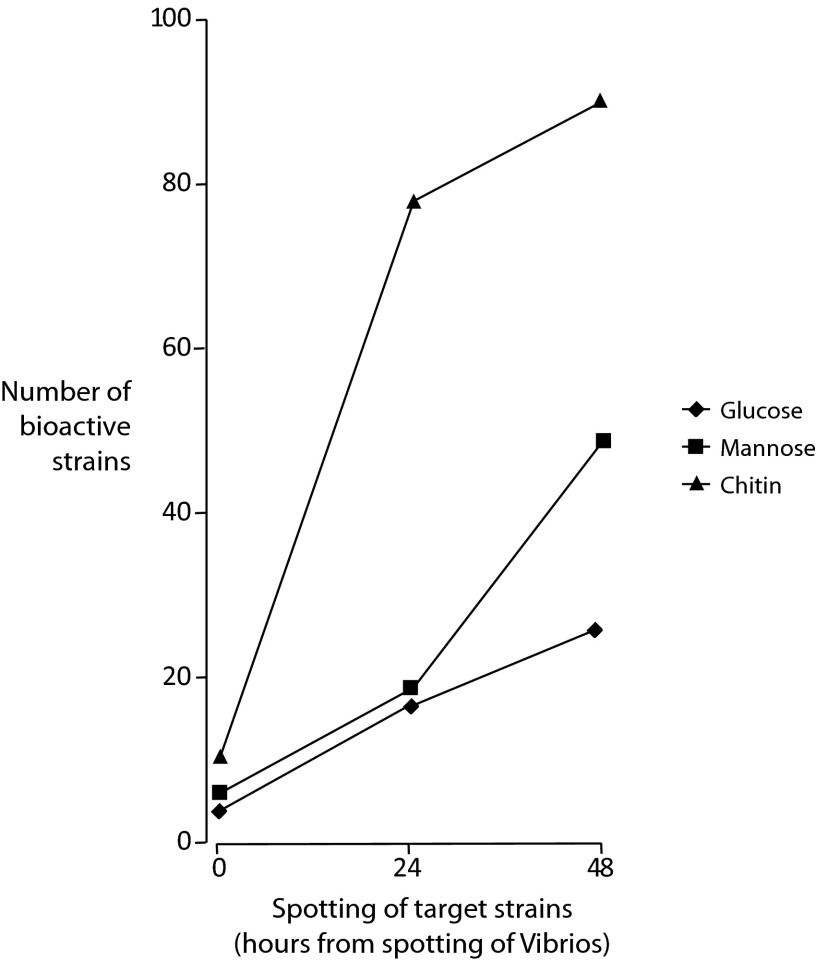
Of the 295 Vibrionaceae strains, 4 isolates antagonized V. anguillarum when grown on the glucose medium, 6 when grown on the mannose medium, and 11 when grown on the chitin medium, using a procedure where the potential producers were not given any temporal advantage over the target strain. When the target strain was spotted 24 h after the potential bioactive strains, 6 isolates were bioactive on glucose, 19 on mannose, and 78 on chitin. Finally, 26, 49, and 91 strains were bioactive on glucose, mannose, and chitin, respectively, when the target strain was spotted with a 48-h delay (Fig. 1). Examples of one plate and of the behavior of one strain (V. furnissii S0821) over time on the mannose- and the chitin-based media are shown in Fig. S1 in the supplemental material.
FIG 1.
Number of bioactive Vibrionaceae strains (of 295 in total) on glucose (rhombus), mannose (square), and chitin (triangle) allowing 0, 24, and 48 h pregrowth of the potential producer before exposing the target strain, Vibrio anguillarum.
Ethyl acetate extracts from the 91 antagonizing strains grown in chitin-containing liquid medium for 48 h were tested in a well diffusion assay against V. anguillarum. Extracts from V. coralliilyticus (strains S2043, S2052, S2054, S2056, and S4053), Vibrio nigripulchritudo (S2601, S2600, and S2604), V. fluvialis (S1110 and S1162), V. furnissii (S0821), and two Vibrio sp. (S1732 and S2150) inhibited the growth of V. anguillarum (Table 1). The strongest inhibition (i.e., the largest inhibition zone) was observed in extracts from the V. coralliilyticus strains. The extracts from the V. furnissii and V. fluvialis strains were moderately growth inhibitory based on the size of the clearing zone. The remaining extracts exhibited weak antibacterial activity.
TABLE 1.
Antibacterial activity of 13 ethyl acetate extracts against V. anguillarum shown as the diameter of clearing zones
| Strain | Species | Inhibitiona of V. anguillarum |
|---|---|---|
| S0821 | V. furnissii | ++ |
| S1110 | V. fluvialis | ++ |
| S1162 | V. fluvialis | ++ |
| S1732 | Vibrio sp. | + |
| S2043 | V. coralliilyticus | +++ |
| S2052 | V. coralliilyticus | +++ |
| S2054 | V. coralliilyticus | +++ |
| S2056 | V. coralliilyticus | ++ |
| S2150 | Vibrio sp. | + |
| S2600 | V. nigripulchritudo | + |
| S2601 | V. nigripulchritudo | + |
| S2604 | V. nigripulchritudo | + |
| S4053 | V. coralliilyticus | +++ |
Clearing zone diameters: +, between 1 and 15 mm; ++, between 16 and 25 mm; +++, over 25 mm.
Distribution of chiS and the (GlcNAc)2 operon.
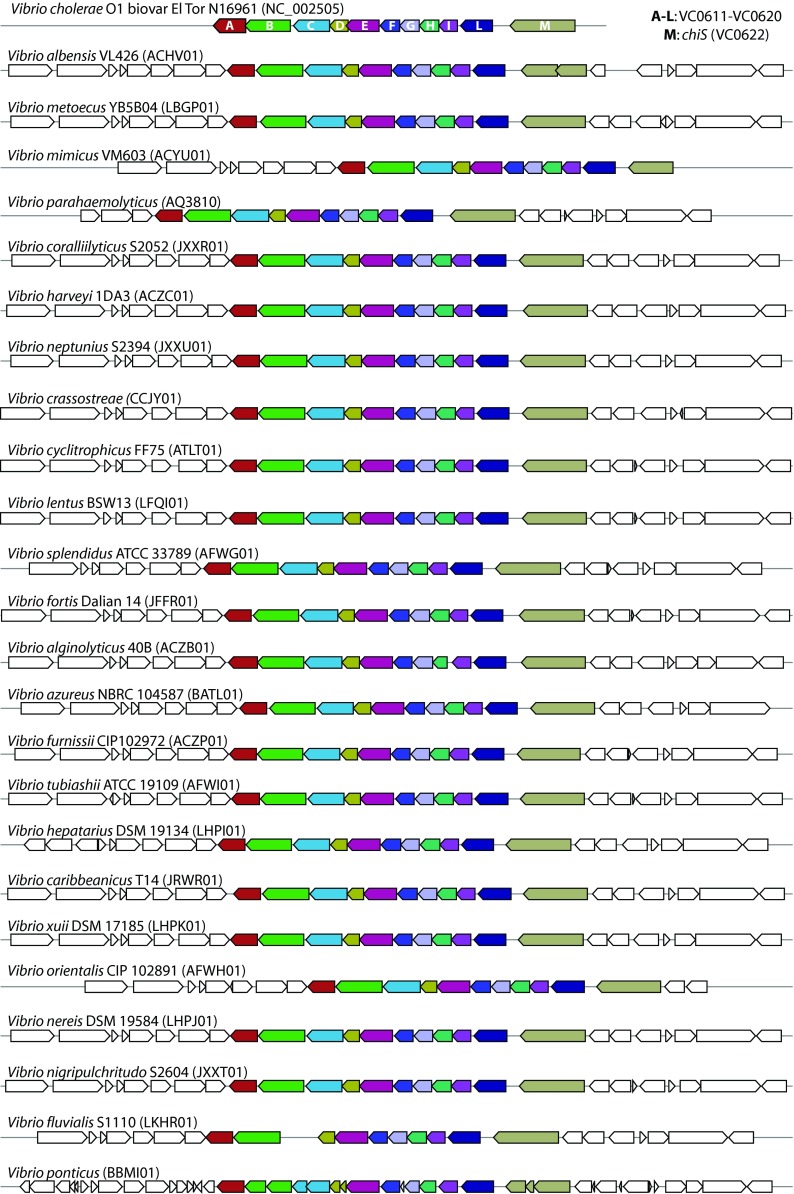
Given the pronounced increase in bioactivity when chitin was used as growth substrate, we speculated that this could be due to a simple substrate change (e.g., catabolite repression) or to a direct involvement of chitin in the regulation. Since chitin is indeed involved in regulation of phenotypes in Vibrio species (27, 39–41), we addressed the possible chitin-dependent regulation of secondary metabolism in Vibrionaceae, possibly through the chitin catabolic cascade sensor histidine kinase (ChiS) regulatory system (see Discussion). Therefore, we investigated the distribution of the chiS gene and of the (GlcNAc)2 operon in 33 genomes of vibrio species belonging to 8 of the 17 proposed Vibrio clades (42) and to 3 of the 4 proposed Photobacterium clades (42). In total, 22 Vibrio and 11 Photobacterium genomes were included in the analysis. This choice was driven by the quantity and the quality of the publicly available genome sequences. MultiGeneBlast-based analysis showed that the chiS gene and the complete (GlcNAc)2 operon are widely distributed in both Vibrio and Photobacterium species, both being present in all analyzed species (Fig. 2 and 3).
FIG 2.
Distribution of the chiS gene and of the (GlcNAc)2 operon among Vibrio spp. GenBank/EMBL/DDBJ accession numbers of the used genomes are indicated in parentheses.
FIG 3.
Distribution of the chiS gene and of the (GlcNAc)2 operon (VC0611-VC0620) among Photobacterium spp. GenBank/EMBL/DDBJ accession numbers of the used genomes are indicated in parentheses.
Genome mining of Vibrio furnissii and Vibrio fluvialis.
Contig-based draft genomes of V. furnissii S0821 and V. fluvialis S1110 were obtained by assembling the sequencing data in CLC genomics workbench. The genome size was 5.0 Mb for V. furnissii S0821 and was 4.5 Mb for V. fluvialis S1110. The antiSMASH analysis of the genomes found six putative biosynthetic gene clusters in V. furnissii S0821 and five in V. fluvialis S1110 (Table 2). Due to the phylogenetic relatedness of V. furnissii and V. fluvialis (42) and the similarity of the antiSMASH results for the two strains, we thought it likely that the antibacterial activity of the two extracts could be due to the same compound(s).
TABLE 2.
Potential for the production of secondary metabolites from V. furnissii S0821 and V. fluvialis S1110 based on antiSMASH and Cluster Finder algorithms
| Algorithm | Type of cluster | No. of clusters |
Similarity (%)a | |
|---|---|---|---|---|
| V. furnissii S0821 | V. fluvialis S1110 | |||
| AntiSMASH | Hserlactone | 1 | 1 | |
| Ectoine | 1 | 1 | 66/66 ectoine BGCb | |
| NRPSc | 1 | 1 | 72/72 vibriobactin BGC | |
| Arylpolyene | 1 | 1 | 90/75 APEd BGC | |
| Bacteriocin | 1 | 1 | ||
| Other | 1 | 0 | 5% lipopolysaccharide BGC | |
| Cluster Finder | Saccharide | 2 | 3 | –e |
| Putative | 8 | 9 | —f | |
| Fatty acid | 2 | 2 | ||
| Saccharide-fatty acid | 1 | 1 | ||
Percentages on the left and right sides of each slash refer to V. furnissii S0821 and to V. fluvialis S1110, respectively.
BGC, biosynthetic gene cluster.
NRPS, nonribosomal peptide synthetase.
APE, arylpolyene.
For V. furnissii S0821, two clusters with 29% gene similarity to the O- and K-antigen BGC; for V. fluvialis S1110, two clusters with 3% and 18% gene similarity to the O- and K-antigen BGC.
For V. furnissii S0821, one cluster with 4% gene similarity to the xantholipin BGC; for V. fluvialis S1110, one cluster with 14% gene similarity to the O-antigen BGC and one cluster with 36% gene similarity to the vibrioferrin BGC.
Both genomes harbored a biosynthetic gene cluster for the production of the quorum-sensing auto-inducer molecules, acyl-homoserine lactones (AHLs), and biosynthetic gene clusters with a relatively high gene similarity to those for the biosynthesis of ectoine, vibriobactin, and aryl polyenes. A cluster for bacteriocin production was identified in both strains, but the bacteriocin prediction tool BAGEL3 was not consistent with the antiSMASH results. Although BAGEL3 did predict the presence of one bacteriocin gene cluster, it differed from the one predicted by antiSMASH. A BLAST-based homology search using the bacteriocin amino acid sequences predicted in the two genomes (see Table S1 in the supplemental material) as queries revealed a high similarity (E value, 0; homology, >98%) with endopeptidases from the M23 superfamily involved in cell wall biogenesis.
Investigation of the antibacterial compound produced by Vibrio furnissii and Vibrio fluvialis.
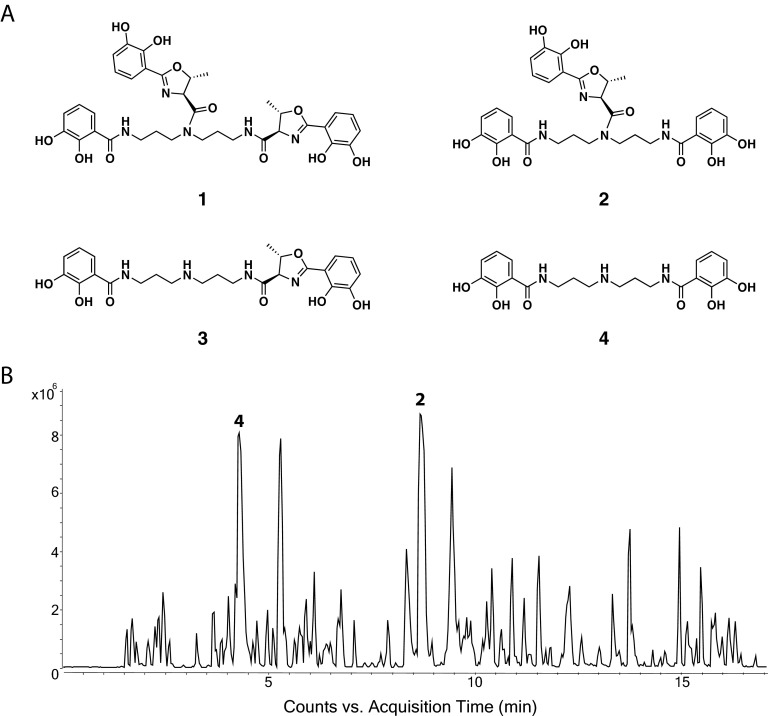
There are no reports in the literature describing antibacterial compounds in V. furnissii and V. fluvialis. Given the importance of these two species as human pathogens (43, 44), we focused on these strains to determine the nature of the compound(s) responsible for the activity. Working under the hypothesis that these closely related species likely produced similar antimicrobial compounds, the bioactive extracts were dereplicated through a two-phase approach; first, extracts were compared with extracts from cultures of related strains, which did not display bioactivity in the well diffusion assay. Compounds that were found in both the active and inactive strains were assumed to not be responsible for the observed antibacterial activity. The remaining unassigned compounds were further dereplicated by searching for all known compounds produced by Vibrio species found in AntiBase 2012, MarinLit 2012, and the Dictionary of Natural Products. Analysis of the dereplicated UHPLC-HRMS data revealed the presence of an abundant compound with ions at an m/z of 623.2342 [M + H]+ and 645.2158 [M + Na]+ in extracts from cultures of V. furnissii S0821 and V. fluvialis S1110, which was tentatively identified as the siderophore fluvibactin based on the accurate mass (mass deviation, 0.96 ppm). Subsequent MS/MS analysis, comparison with the literature UV spectrum, and isolation and nuclear magnetic resonance analysis confirmed this assignment (see Fig. S2 to S5 and Tables S2 and S3 in the supplemental material). The UHPLC-HRMS analysis also found another abundant ion with an m/z of 404.1818, which was assigned to the known compound 4 [N,N-bis-(2,3-dihydroxybenzoyl)-norspermidine] (mass deviation, 0.49 ppm) (Fig. 4A and B).
FIG 4.
(A) Structures of vibriobactin (1) (54), fluvibactin (2) (55), compound 3 (3), and compound 4 (4). (B) UHPLC-HRMS total ion chromatogram of the culture extract from V. furnissii. The peaks assigned to fluvibactin (2) and compound 4 are highlighted.
These compounds (fluvibactin and compound 4) were not detected when extracts were prepared from V. furnissii S0821 and V. fluvialis S1110 grown in chitin medium supplemented with 0.1 g/liter of ferric citrate (see Fig. S6 in the supplemental material). These extracts were not inhibitory against V. anguillarum 90-11-287 (see Fig. S7). The bioactive extract was then divided into fractions by mixed-mode anion exchange SPE. Only the extract fraction containing the putative fluvibactin was inhibitory to V. anguillarum (Table 3; see also Fig. S8). A chrome azurol S assay performed on the same fraction confirmed the siderophoric nature of the compound (Table 3). The use of anion-exchange chromatography allowed for the separation of fluvibactin from N-(3-oxo-decanoyl-L)-homoserine lactone (O-C10-HSL), which coeluted under the reverse-phase conditions used for UHPLC-HRMS analysis. Fractions containing the AHL (F3) (see Fig. S6) did not show bioactivity, and the AHL was also found to be present in nonbioactive iron-supplemented cultures (see Fig. S7). O-C10-HSL was identified based on accurate mass, retention time, and the characteristic homoserine fragment ion at an m/z of 102.0549 (45).
TABLE 3.
Siderophore (column CAS assay) and antibacterial activity of the raw extract from a culture of V. furnissii S0821 and of the six derived fractions (F1 to F6)a
| Sample | CAS assay | Inhibition zone (mm) |
|---|---|---|
| Raw extract | Yellow | 20 |
| F1 | Blue | 0 |
| F2 | Blue | 0 |
| F3 | Blue | 0 |
| F4 | Blue | 0 |
| F5 | Dark orange | 9 |
| F6 | Yellow | 23 |
| Blank | Blue | 0 |
The addition of a siderophore to the CAS solution causes a change in color from dark blue to orange-yellow. Activity against V. anguillarum is measured as the diameter of inhibition zones.
DISCUSSION
We investigated to what extent culture parameters could affect (restore or induce) the production of antibacterial compounds in a collection of marine Vibrionaceae whose members were initially isolated based on their ability to antagonize the fish pathogen V. anguillarum. However, in later rescreenings, only approximately 10% of them retained the activity. With the use of substrates typical to the natural niche of isolation, and allowing potential producer strains to reach a late growth phase, we could restore the bioactivity in one-third of the strains. Allowing V. fluvialis and V. furnissii to reach late growth phases before exposure to the target strain led to the identification of the siderophore fluvibactin as the entity responsible for antibacterial activity.
Different carbon sources can lead to significantly different profiles in microbial secondary metabolism (5, 9, 46). In our investigation, we used three molecules (glucose, mannose, and chitin) that are abundant in the marine environment (22, 24) as the substrates for marine Vibrionaceae. The numbers of bioactive (antibacterial) strains were nearly 2- and 3.5-fold higher when mannose and chitin, respectively, were used as carbon sources compared to glucose.
The high efficacy of chitin in restoring (or inducing) the production of antibacterial compounds in the tested strains is in agreement with the ecology and lifestyle of Vibrionaceae that are adapted to live in marine niches richer in this polysaccharide than in other carbohydrates (25, 26). Indeed, vibrios are well known for their associations with chitin-rich biotic surfaces, such as zooplankton (24, 47). Chitinase genes and the chitin utilization pathway are conserved in Vibrionaceae (25, 26), and natural competence is induced by chitin in Vibrio vulnificus (48) and V. cholerae (49). In V. cholerae, chitin affects also chitin catabolism (27), biofilm formation, and virulence (28, 50).
Chitin-dependent regulation of secondary metabolism mediated by the transcriptional regulator DasR occurs in the soil bacterium Streptomyces coelicolor A3 (2) (51). In vibrios, one possible mechanism for a similar regulation could be through the two-component histidine kinase sensor, ChiS, which has been characterized in Vibrio cholerae and is activated by chitin-derived oligosaccharides (27). In the proposed model, a putative cognate receptor regulates the expression of target genes involved in the above-mentioned phenomena (27). Hunt and colleagues (25) suggested that genes with high homology to chiS (VC0622) and to some of the genes from the downstream (GlcNAc)2 operon (VC0611-VC0613 and VC0616-VC0619) are widespread among Vibrionaceae. However, their genome analysis included a limited number (i.e., 10) of species, possibly due to the low availability of genome sequences at the time the study was conducted. We performed a broader analysis and showed that, indeed, both chiS and the complete (GlcNAc)2 operon (VC0611-VC0620), which were detected in all analyzed genomes, are very conserved and maintain their topological organization in Vibrio and Photobacterium species (Fig. 2 and 3). The (GlcNAc)2 operon includes the gene encoding the periplasmic (GlcNAc)2-binding protein which inactivates ChiS when chitin is not present in the environment (27). Given its importance in V. cholerae, such a degree of conservation of genes hypothesized to be involved in the ChiS regulatory system in Vibrionaceae indicates that chitin could serve a regulatory role in the whole family. Certainly, chitin-dependent regulation of phenomena, such as biofilm formation and biosynthesis of antibacterial compounds, would be advantageous during competition for nutrients with other microorganisms in the marine environment.
Although the use of chitin restored or induced the production of antibacterial compounds in approximately one-third of the isolates, this approach was not effective with the majority of the strains, even when they were allowed longer times before exposure to the target strain. Induction of silent/cryptic biosynthetic gene clusters has been achieved by exposing bacteria or fungi to small molecules produced by naturally co-occurring microorganisms (11, 52, 53). Similarly, it is likely that molecules that were present in the local seawater used to prepare the medium for the original screening/isolation procedure or that were produced by strains that were tested on the same plate (co-cultivated) might have elicited the biosynthesis of antibacterial compounds. Co-cultivation could therefore also be a strategy to be used to induce the production of antibacterial compounds in our strain collection.
Extracts of cultures from V. furnissii S0821 and V. fluvialis S1110 had antibacterial activity against V. anguillarum. The bioactivity was present in all tested media; however, on chitin, the antagonistic activity could be observed earlier than on the other media (data not shown). Genome analysis of the strains provided a list of four biosynthetic gene clusters potentially responsible for the biosynthesis of the antibacterial compound. Three of them (AHL, ectoine, and arylpolyenes) could be excluded as the cause of bioactivity through testing of pure standards in WDA and analysis of UV/Vis spectra of the extracts (see Fig. S8 and S9 in the supplemental material).
The remaining predicted biosynthetic gene cluster had 72% gene similarity to the biosynthetic gene cluster for the catechol siderophore produced by V. cholerae vibriobactin (54). Due to the phylogenetic relatedness of V. furnissii and V. fluvialis with V. cholerae (42), we hypothesized that the identified biosynthetic gene cluster encodes the fluvibactin nonribosomal peptide synthetase. Fluvibactin is a siderophore produced by V. fluvialis (55), which differs from vibriobactin only in that it contains a single l-threonine residue rather than two (Fig. 4). Siderophores similar to fluvibactin can inhibit bacterial and fungal growth (20, 56), and catechol iron chelators have also been suggested to protect bacteria from oxidative stress (57, 58). Hence, beside the competitive advantage during surface colonization due to the antibacterial activity of fluvibactin, producers of this compound might as well be protected from oxidative stress, which is a prevalent phenomenon in the marine environment (59).
Conclusion.
We have shown that a rational choice of substrates typical of the niche of isolation of microorganisms is a valid cultivation strategy to enhance the numbers of bioactive strains in a screening step. Our results suggest a role of chitin in the production of secondary metabolism in Vibrionaceae. The genomes of members of this family of bacteria harbor great potential for chitin catabolism. Hence, genomic studies could help to predict which substrates other families of microorganisms might prefer and, possibly, lead to the elicitation of biosynthetic gene clusters. Also, allowing the potential producing strain a temporal advantage (reaching stationary phase) is an important aspect to consider when designing a screening strategy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jette Melchiorsen for assistance with DNA extraction and the CAS assay. We thank Agilent Technologies for the thought leader donation of the UHPLC-qTOF system.
S.G. was supported by an early stage researchers grant from the People program (Marie Curie Actions) of the European Union's seventh framework program, FP7-People-2012-ITN, under grant agreement 317058, BacTory. C.P. was supported by a grant from the Villum foundation (VKR023285). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
This work was carried out as part of the Galathea 3 expedition under the auspices of the Danish Expedition Foundation. This is Galathea 3 contribution no. P116.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00730-16.
REFERENCES
- 1.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 2.Harvey AL, Edrada-Ebel R, Quinn RJ. 2015. The reemergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 3.Machado H, Sonnenschein EC, Melchiorsen J, Gram L. 2015. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genomics 16:158. doi: 10.1186/s12864-015-1365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helfrich EJ, Reiter S, Piel J. 2014. Recent advances in genome-based polyketide discovery. Curr Opin Biotechnol 29:107–115. doi: 10.1016/j.copbio.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Bode HB, Bethe B, Hoefs R, Zeeck A. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3:619–627. doi:. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien J, Wright GD. 2011. An ecological perspective of microbial secondary metabolism. Curr Opin Biotechnol 22:552–558. doi: 10.1016/j.copbio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Wietz M, Duncan K, Patin NV, Jensen PR. 2013. Antagonistic interactions mediated by marine bacteria: the role of small molecules. J Chem Ecol 39:879–891. doi: 10.1007/s10886-013-0316-x. [DOI] [PubMed] [Google Scholar]
- 8.Vizcaino MI, Guo X, Crawford JM. 2014. Merging chemical ecology with bacterial genome mining for secondary metabolite discovery. J Ind Microbiol Biotechnol 41:285–299. doi: 10.1007/s10295-013-1356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wietz M, Månsson M, Gram L. 2011. Chitin stimulates production of the antibiotic andrimid in a Vibrio coralliilyticus strain. Environ Microbiol Rep 3:559–564. doi: 10.1111/j.1758-2229.2011.00259.x. [DOI] [PubMed] [Google Scholar]
- 10.Palková Z. 2004. Multicellular microorganisms: laboratory versus nature. EMBO Rep 5:470–476. doi: 10.1038/sj.embor.7400145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seyedsayamdost MR. 2014. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A 111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan L, Boyd KG, Grant Burgess J. 2002. Surface attachment induced production of antimicrobial compounds by marine epiphytic bacteria using modified roller bottle cultivation. Mar Biotechnol 4:356–366. doi: 10.1007/s10126-002-0041-x. [DOI] [PubMed] [Google Scholar]
- 13.Yan L, Boyd KG, Adams DR, Burgess JG. 2003. Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl Environ Microbiol 69:3719–3727. doi: 10.1128/AEM.69.7.3719-3727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkar S, Saha M, Roy D, Jaisankar P, Das S, Gauri Roy L, Gachhui R, Sen T, Mukherjee J. 2008. Enhanced production of antimicrobial compounds by three salt-tolerant actinobacterial strains isolated from the Sundarbans in a niche-mimic bioreactor. Mar Biotechnol 10:518–526. doi: 10.1007/s10126-008-9090-0. [DOI] [PubMed] [Google Scholar]
- 15.Davidson B. 1995. New dimensions in natural products research: cultured marine microorganisms. Curr Opin Biotechnol 6:284–291. doi: 10.1016/0958-1669(95)80049-2. [DOI] [Google Scholar]
- 16.Proksch P, Edrada RA, Ebel R. 2002. Drugs from the seas—current status and microbiological implications. Appl Microbiol Biotechnol 59:125–134. doi: 10.1007/s00253-002-1006-8. [DOI] [PubMed] [Google Scholar]
- 17.Gram L, Melchiorsen J, Bruhn JB. 2010. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar Biotechnol 12:439–451. doi: 10.1007/s10126-009-9233-y. [DOI] [PubMed] [Google Scholar]
- 18.Wietz M, Mansson M, Gotfredsen CH, Larsen TO, Gram L. 2010. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar Drugs 8:2946–2960. doi: 10.3390/md8122946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kjaerulff L, Nielsen A, Mansson M, Ingmer H, Gram L, Thomas O, Gotfredsen CH. 2013. Identification of four new agr quorum sensing-interfering cyclodepsipeptides from a marine Photobacterium. Mar Drugs 11:5051–5162. doi: 10.3390/md11125051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen A, Mansson M, Wietz M, Varming A, Phipps R, Larsen T, Gram L, Ingmer H. 2012. Nigribactin, a novel siderophore from Vibrio nigripulchritudo, modulates Staphylococcus aureus virulence gene expression. Mar Drugs 10:2584–2595. doi: 10.3390/md10112584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen A, Mansson M, Bojer MS, Gram L, Larsen TO, Novick RP, Frees D, Frøkiær H, Ingmer H. 2014. Solonamide B inhibits quorum sensing and reduces Staphylococcus aureus mediated killing of human neutrophils. PLoS One 9:e84992. doi: 10.1371/journal.pone.0084992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popper ZA, Michel G, Hervé C, Domozych DS, Willats WGT, Tuohy MG, Kloareg B, Stengel DB. 2011. Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol 62:567–590. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- 23.Aluwihare LI, Repeta DJ, Chen RF. 2002. Chemical composition and cycling of dissolved organic matter in the mid-Atlantic bight. Deep Sea Res Part II Top Stud Oceanogr 49:4421–4437. doi: 10.1016/S0967-0645(02)00124-8. [DOI] [Google Scholar]
- 24.Gooday GW. 1990. The ecology of chitin degradation. Adv Microb Ecol 11:387–430. doi: 10.1007/978-1-4684-7612-5_10. [DOI] [Google Scholar]
- 25.Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riemann L, Azam F. 2002. Widespread N-acetyl-d-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl Environ Microbiol 68:5554–5562. doi: 10.1128/AEM.68.11.5554-5562.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Roseman S. 2004. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc Natl Acad Sci U S A 101:627–631. doi: 10.1073/pnas.0307645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederiksen RF, Paspaliari DK, Larsen T, Storgaard BG, Larsen MH, Ingmer H, Palcic MM, Leisner JJ. 2013. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology 159:833–847. doi: 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 29.Machado H, Gram L. 2015. The fur gene as a new phylogenetic marker for Vibrionaceae species identification. Appl Environ Microbiol 81:2745–2752. doi: 10.1128/AEM.00058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu SC, Lockwood JL. 1975. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González JM, Whitman WB, Hodson RE, Moran MA. 1996. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol 62:4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medema MH, Takano E, Breitling R. 2013. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol 30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hjelm M, Bergh Ø, Riaza A, Nielsen J, Melchiorsen J, Jensen S, Duncan H, Ahrens P, Birkbeck H, Gram L. 2004. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst Appl Microbiol 27:360–371. doi: 10.1078/0723-2020-00256. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 35.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Ciufo S, Li W. 2013. Prokaryotic genome annotation pipeline: the NCBI handbook, 2nd ed National Center for Biotechnology Information, Rockville, MD. [Google Scholar]
- 36.Medema MH, Blin K, Cimermancic P, De Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. 2011. AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. 2013. BAGEL3: automated identification of genes encoding bacteriocins and (non)bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–W453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 39.Sun S, Tay QXM, Kjelleberg S, Rice SA, McDougald D. 2015. Quorum sensing-regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J 9:1812–1820. doi: 10.1038/ismej.2014.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG. 2011. The N-acetyl-d-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol Microbiol 82:894–903. doi: 10.1111/j.1365-2958.2011.07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo Scrudato M, Blokesch M. 2012. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet 8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AR, Mino S, Nakagawa S, Sawabe T, Kumar R, Fukui Y, Satomi M, Matsushima R, Thompson FL, Gomez-Gil B, Christen R, Maruyama F, Kurokawa K, Hayashi T. 2013. Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol 4:414. doi: 10.3389/fmicb.2013.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramamurthy T, Chowdhury G, Pazhani GP, Shinoda S. 2014. Vibrio fluvialis: an emerging human pathogen. Front Microbiol 5:91. doi: 10.3389/fmicb.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruzzo C, Huq A, Colwell RR, Donelli G. 2005. Pathogenic Vibrio species in the marine and estuarine environment, p 217–252. In Belkin S, Colwell RR (ed), Oceans and health: pathogens in the marine environment. Springer, New York, NY. [Google Scholar]
- 45.Rasmussen BB, Nielsen KF, Machado H, Melchiorsen J, Gram L, Sonnenschein EC. 2014. Global and phylogenetic distribution of quorum sensing signals, acyl homoserine lactones, in the family of Vibrionaceae. Mar Drugs 12:5527–5546. doi: 10.3390/md12115527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sánchez S, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Avalos M, Guzmán-Trampe S, Rodríguez-Sanoja R, Langley E, Ruiz B. 2010. Carbon source regulation of antibiotic production. J Antibiot (Tokyo) 63:442–459. doi: 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- 47.Heidelberg JF, Heidelberg KB, Colwell RR. 2002. Bacteria of the gamma-subclass Proteobacteria associated with zooplankton in Chesapeake Bay. Appl Environ Microbiol 68:5498–5507. doi: 10.1128/AEM.68.11.5498-5507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gulig PA, Tucker MS, Thiaville PC, Joseph JL, Brown RN. 2009. USER friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl Environ Microbiol 75:4936–4949. doi: 10.1128/AEM.02564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto S, Mitobe J, Ishikawa T, Wai SN, Ohnishi M, Watanabe H, Izumiya H. 2014. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a noncanonical transmembrane regulator in Vibrio cholerae. Mol Microbiol 91:326–347. doi: 10.1111/mmi.12462. [DOI] [PubMed] [Google Scholar]
- 51.Nazari B, Kobayashi M, Saito A, Hassaninasab A, Miyashita K, Fujiia T. 2013. Chitin-induced gene expression in secondary metabolic pathways of Streptomyces coelicolor A3(2) grown in soil. Appl Environ Microbiol 79:707–713. doi: 10.1128/AEM.02217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroeckh V, Scherlach K, Nutzmann H-W, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A 106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. 2013. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4:e00459-13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griffiths GL, Sigel SP, Payne SM, Neilands JB. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem 259:383–385. [PubMed] [Google Scholar]
- 55.Yamamoto S, Okujo N, Fujita Y, Saito M, Yoshida T, Shinoda S. 1993. Structures of two polyamine-containing catecholate siderophores from Vibrio fluvialis. J Biochem 113:538–544. [DOI] [PubMed] [Google Scholar]
- 56.Bergeron RJ, Elliott GT, Kline SJ, Ramphal R, St James L. 1983. Bacteriostatic and fungostatic action of catecholamide iron chelators. Antimicrob Agents Chemother 24:725–730. doi: 10.1128/AAC.24.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adler C, Corbalán NS, Seyedsayamdost MR, Pomares MF, de Cristóbal RE, Clardy J, Kolter R, Vincent PA. 2012. Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS One 7:e46754. doi: 10.1371/journal.pone.0046754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adler C, Corbalan NS, Peralta DR, Pomares MF, De Cristóbal RE, Vincent PA. 2014. The alternative role of enterobactin as an oxidative stress protector allows Escherichia coli colony development. PLoS One 9:e84734. doi: 10.1371/journal.pone.0084734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lesser MP. 2006. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol 68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.