ABSTRACT
Glucose is the preferred carbon source for the yeast Saccharomyces cerevisiae. Glucose limitation activates Snf1 protein kinase, a key regulator of energy homeostasis that promotes utilization of alternative carbon sources and enforces energy conservation. Snf1 activation requires phosphorylation of its T-loop threonine (Thr210) by upstream kinases. When glucose is abundant, Snf1 is inhibited by Thr210 dephosphorylation. This involves the function of the type 1 protein phosphatase Glc7, which is targeted to Snf1 by a regulatory subunit, Reg1. The reg1 mutation causes increased Snf1 activity and mimics various aspects of glucose limitation, including slower growth. Reg2 is another Glc7 regulatory subunit encoded by a paralogous gene, REG2. Previous evidence indicated that the reg2 mutation exacerbates the Snf1-dependent slow-growth phenotype caused by reg1, suggesting a link between Reg2 and Snf1. Here, we explore this link in more detail and present evidence that Reg2 contributes to Snf1 Thr210 dephosphorylation. Consistent with this role, Reg2 interacts with wild-type Snf1 but not with nonphosphorylatable Snf1-T210A. Reg2 accumulation increases in a Snf1-dependent manner during prolonged glucose deprivation, and glucose-starved cells lacking Reg2 exhibit delayed Snf1 Thr210 dephosphorylation and slower growth recovery upon glucose replenishment. Accordingly, cells lacking Reg2 are outcompeted by wild-type cells in the course of several glucose starvation/replenishment cycles. Collectively, our results support a model in which Reg2-Glc7 contributes to the negative control of Snf1 in response to glucose refeeding after prolonged starvation. The competitive growth advantage provided by Reg2 underscores the evolutionary significance of this paralog for S. cerevisiae.
IMPORTANCE The ability of microorganisms to respond to stress is essential for their survival. However, rapid recovery from stress could be equally crucial in competitive environments. Therefore, a wise stress response program should prepare cells for quick recovery upon reexposure to favorable conditions. Glucose is the preferred carbon source for the yeast S. cerevisiae. Glucose depletion activates the stress response protein kinase Snf1, which functions to limit energy-consuming processes, such as growth. We show that prolonged glucose deprivation also leads to Snf1-dependent accumulation of Reg2 and that this protein helps to inhibit Snf1 and to accelerate growth recovery upon glucose replenishment. Cells lacking Reg2 are readily outcompeted by wild-type cells during glucose depletion/replenishment cycles. Thus, while prolonged glucose deprivation might seem to put yeast cells “on their knees,” concomitant accumulation of Reg2 helps configure the cells into a “sprinter's crouch start position” to spring into action once glucose becomes available.
INTRODUCTION
Glucose is a preferred source of carbon and energy for a wide variety of microorganisms. Although the yeast Saccharomyces cerevisiae can exist in many different environments, it is particularly known for its ability to flourish under glucose-rich conditions, a property exploited by humans for millennia to make wine (reviewed in references 1 and 2). Consistent with the importance of glucose for its growth and metabolism, S. cerevisiae has evolved a complex regulatory network that controls responses to glucose availability, with a central role played by a protein kinase called Snf1 (sucrose nonfermenting 1) (reviewed in reference 3).
Yeast Snf1 is a founding member of the highly conserved Snf1/AMP-activated protein kinase (AMPK) family (reviewed in reference 4). A unifying feature of Snf1/AMPK kinases is that they serve as the “fuel gauge” (5) of the eukaryotic cell: as energy levels decrease, these kinases are activated and function to balance the energy “budget” by reducing energy spending and by increasing energy “income.” In mammals, AMPK coordinates cell growth and proliferation with energy availability (6) and has been implicated in health conditions ranging from type 2 diabetes to cancer (7, 8). In yeast, lack of Snf1 leads to inability to utilize carbon sources that are less preferred than glucose, including sucrose, galactose, maltose, and starch (9–11). Genetic manipulation of Snf1 and/or its targets has been a useful strategy for achieving various practical goals in yeast biotechnology, such as improved maltose utilization (12), galactose utilization and ethanol production (13), fatty acid production (14, 15), and biomass yields (16). Thus, greater understanding of Snf1 regulation could help identify additional approaches for rational design of yeast strains.
When glucose is limiting, Snf1 is activated through the actions of three partially redundant protein kinases (Sak1, Tos3, and Elm1), which phosphorylate its conserved T-loop threonine residue (Thr210) (17–21). When glucose becomes abundant, Snf1 is inhibited by dephosphorylation, which involves the function of the PP2A-like protein phosphatase Sit4 and the type 1 protein phosphatase Glc7 (4, 22, 23).
The type 1 protein phosphatase Glc7 associates with various regulatory subunits that target it to appropriate substrates, thereby allowing it to have multiple cellular functions. One of these subunits, Reg1 (24), is known to play a major role in enabling Glc7 to recognize and dephosphorylate Thr210 of Snf1 (17, 25, 26). The reg1 mutation causes increased levels of phospho-Thr210-Snf1 and Snf1 kinase activity, thereby mimicking glucose limitation; indeed, cells lacking Reg1 exhibit constitutive (glucose-insensitive) expression of Snf1-dependent genes required for alternate carbon source utilization (17, 24, 27). In addition, the reg1 mutation confers a Snf1-dependent slow-growth phenotype (22, 28, 29). This slow growth, which is observed even in the presence of abundant glucose, likely reflects hyperactivation of an energy-saving function of Snf1. Genetic interactions of the reg1 mutation with respect to the slow-growth phenotype could identify additional regulators in the Snf1 pathway.
Reg2 is another regulatory subunit of Glc7 that shares homology with Reg1 (28, 30). In fact, REG1 and REG2 are a pair of paralogous genes that arose during an ancient whole-genome duplication event in an ancestral yeast species (31). Although the reg2 mutation alone does not cause any phenotypic defects comparable to those caused by reg1, it does exacerbate the slow-growth phenotype of the reg1 mutant; importantly, the slow growth of the reg1 reg2 double mutant depends on Snf1, suggesting that, like Reg1, Reg2 might be functionally related to Snf1 (28).
In this study, we addressed the possible role of Reg2 in the negative control of Snf1. We present evidence that Reg2 physically interacts with Snf1 and contributes to its Thr210 dephosphorylation in response to abundant glucose and that this effect depends on the ability of Reg2 to interact with Glc7. Reg2 accumulates during prolonged glucose deprivation in a Snf1-dependent manner, and glucose-starved cells lacking Reg2 exhibit a delay in Snf1 Thr210 dephosphorylation and growth recovery upon glucose replenishment. We also show that cells lacking Reg2 are outcompeted by wild-type cells during glucose starvation/replenishment cycles, supporting the evolutionary significance of Reg2 for S. cerevisiae.
MATERIALS AND METHODS
Strains and growth conditions.
The S. cerevisiae strains used in this study are listed in Table 1. Except for strain CTY10-5d (R. Sternglanz, SUNY, Stony Brook, NY), used for yeast two-hybrid assays where indicated, all the strains were in the Σ1278b genetic background and were descendants of strains MY1384 (MATa; prototroph), MY1401 (MATα ura3Δ leu2Δ his3Δ), and MY1402 (MATa ura3Δ leu2Δ trp1Δ) of the isogenic Sigma2000 series (Microbia, Cambridge, MA). To generate Σ1278b derivatives with reg2Δ::KanMX6, the marker sequence was amplified by PCR with primers flanking the REG2 open reading frame (ORF). The mutant allele was first introduced into a wild-type diploid (MKY324 × MKY341) by transformation; all yeast transformations were performed using standard methods (32). The genotype of the heterozygous REG2/reg2Δ::KanMX6 diploid was confirmed by PCR analysis of genomic DNA, and haploid reg2Δ::KanMX6 segregants were recovered from the heterozygous diploid by tetrad analysis. Construction of the reg1Δ::URA3, reg1Δ::KanMX6, and snf1Δ::KanMX6 derivatives was described previously (33, 34). The reg1Δ reg2Δ double mutant was constructed by crossing MMY9 (reg2Δ::KanMX6) and KBY247 (reg1Δ::URA3), followed by tetrad analysis.
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| CTY10-5d | MATa gal4 gal80 URA3::lexAop-lacZ his3 leu2 ade2 trp1 | R. Sternglanz |
| MKY191 | MATα ura3Δ leu2Δ his3Δ | This laboratory |
| MKY323 | MATα ura3Δ | This laboratory |
| MKY324 | MATα ura3Δ | This laboratory |
| MKY339 | MATa ura3Δ leu2Δ his3Δ | This laboratory |
| MKY341 | MATa leu2Δ his3Δ trp1Δ | This laboratory |
| MKY343 | MATa ura3Δ leu2Δ his3Δ trp1Δ | This laboratory |
| MKY362 | MATa ura3Δ leu2Δ his3Δ trp1Δ snf1Δ::KanMX6 | This laboratory |
| MKY366 | MATα ura3Δ leu2Δ his3Δ snf1Δ::KanMX6 | This laboratory |
| MKY601 | MATa ura3Δ leu2Δ his3Δ reg1Δ::KanMX6 | This laboratory |
| KBY238 | MATα ura3Δ leu2Δ his3Δ reg1Δ::KanMX6 snf1Δ::KanMX6 | This laboratory |
| KBY266 | MATα ura3Δ leu2Δ his3Δ reg1Δ::KanMX6 | This laboratory |
| KBY247 | MATa ura3Δ leu2Δ his3Δ trp1Δ reg1Δ::URA3 | This study |
| MMY9 | MATα ura3Δ reg2Δ::KanMX6 | This study |
| MMY18 | MATα ura3Δ his3Δ trp1Δ reg2Δ::KanMX6 | This study |
| MMY40 | MATα leu2Δ trp1Δ reg1Δ::URA3 | This study |
| MMY41 | MATa ura3Δ his3Δ reg2Δ::KanMX6 | This study |
| MMY42 | MATa leu2Δ trp1Δ reg1Δ::URA3 reg2Δ::KanMX6 | This study |
| MMY128 | MATα ura3Δ his3Δ | This study |
The rich medium was yeast extract-peptone (YEP) supplemented with extra tryptophan (40 mg/liter) and adenine (20 mg/liter); synthetic complete (SC) medium lacking appropriate supplements was used to select for plasmids (32). Unless otherwise indicated, the media contained 2% glucose, and yeast cells were grown at 30°C.
Yeast two-hybrid assays.
All the plasmids expressing fusion proteins for the yeast two-hybrid experiments were based on vector pEG202, pACTII, or pVP16, all of which provide expression from the ADH1 promoter (35–37). Plasmids pIT469 (38) and pRJ79 (39) express LexA-Snf1 and VP16-Snf1 from vectors pEG202 and pVP16, respectively. Plasmids pRJ215 and pRJ217 express LexA-Snf1-K84R and LexA-Snf1-T210A, respectively, from vector pEG202 (40, 41). To construct pLexA-Glc7, a PCR fragment encompassing the GLC7 ORF was inserted at the BamHI site of pEG202. To construct pLexA-Reg2 and pGAD-Reg2, a PCR fragment encompassing the REG2 ORF was inserted at the BamHI sites of pEG202 and pACTII, respectively. Plasmids pLexA-Reg2-IF and pGAD-Reg2-IF are identical to plasmids pLexA-Reg2 and pGAD-Reg2, respectively, except that they carry mutations that change the Ile169 and Phe171 codons of Reg2 to Met and Ala codons, respectively; the mutations were introduced essentially as described previously (42) using a mutagenic primer that also creates silent diagnostic restriction sites overlapping the Met and Ala codons (NdeI and HaeIII, respectively).
The reporter strains were CTY10-5d, carrying an integrated lexAop-lacZ reporter, and MKY343, carrying a lexAop-lacZ reporter plasmid, pSH18-18, a derivative of pLR1Δ1 (43). The reporter strains were cotransformed with pairs of plasmids expressing the protein pairs being tested. The transformants were grown to mid-log phase with plasmid selection in SC lacking the appropriate supplements and containing 2% glucose and then shifted for 3 h to an otherwise identical medium containing 0.05% glucose. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units, as described previously (44).
Expression of the fusion proteins was confirmed by immunoblotting as follows. Representative transformants were grown under conditions identical to those used to measure β-galactosidase activity. Cell extracts were prepared using the boiling/alkaline lysis method (45). Proteins were separated by SDS-PAGE and analyzed by immunoblotting. LexA fusion proteins were detected with anti-LexA antibody (Millipore). The GAD-Reg2 and GAD-Reg2-IF fusion proteins contain a hemagglutinin (HA) epitope tag between the Gal4 activation domain (GAD) and Reg2 (or Reg2-IF) and were detected with the anti-HA antibody HA-7 (Sigma-Aldrich). VP16-Snf1 was detected using the anti-polyhistidine antibody H1029 (Sigma-Aldrich), which strongly recognizes Snf1 due to the presence of a natural tract of 13 consecutive histidines (45). Thr210-phosphorylated Snf1 proteins were detected with anti-phospho-Thr172-AMPK (Cell Signaling Technology) as described previously (45). Signals were detected by enhanced chemiluminescence using Pierce ECL2 or HyGlo (Denville Scientific).
Coimmunoprecipitation assays.
Plasmid pHA-Reg2 expresses N-terminal triple-HA-epitope-tagged Reg2 (HA-Reg2) and was constructed by inserting the REG2 coding sequence into the BamHI site of vector pSK134HA, which provides expression from the ADH1 promoter (34). Plasmids pSM14 and pMO16 (46) express untagged wild-type Snf1 and Snf1-T210A, respectively, from vector pRS316 (47). Cells of strain MKY362 (snf1Δ) were transformed with plasmid pSM14 expressing Snf1 or with plasmid pMO16 expressing Snf1-T210A and cotransformed with plasmid pHA-Reg2. The cells were grown to mid-log phase with plasmid selection in SC lacking leucine and uracil and containing 2% glucose and then shifted to an otherwise identical medium containing 0.05% glucose for 15 min, which is sufficient for Snf1 activation (22). Protein extracts were prepared, and immunoprecipitations (from 200 μg of protein per reaction) were performed essentially as described previously (40) in a buffer containing 0.1% Triton X-100 and 100 mM NaCl, followed by three washes in the presence of 250 mM NaCl; the antibody used for immunoprecipitation of Snf1 and Snf1-T210A was the anti-polyhistidine antibody H1029 (Sigma-Aldrich), which recognizes the natural polyhistidine tract in Snf1 (45). The immunoprecipitates were examined for the presence of HA-Reg2 by immunoblotting with anti-HA-peroxidase (Roche). The total levels of immunoprecipitated Snf1 and its Thr210 phosphorylation state were determined using anti-polyhistidine and anti-phospho-Thr172-AMPK antibodies, respectively (45). Signals were detected by enhanced chemiluminescence using Pierce ECL2 or HyGlo (Denville Scientific). The extracts were analyzed by immunoblotting similarly (10 μg protein per lane).
Analysis of regulation of Reg2 expression.
To analyze Reg2 expression from the native REG2 promoter, a low-copy-number plasmid, pReg2-L-HA, was used. This plasmid encodes a Reg2 protein C-terminally tagged with LexA followed by triple HA (here referred to as Reg2-L-HA) and was constructed as follows. First, the REG2 gene, including its 0.52-kb upstream regulatory sequence (the entire intergenic region between REG2 and its upstream neighbor RFS1), was amplified by PCR using wild-type genomic DNA as the template; the reverse primer used for this PCR also included a BspEI site and a sequence encoding a single HA tag. The resulting PCR fragment was then inserted into the BamHI site of the low-copy-number CEN-URA3 vector pRS316 (47), yielding plasmid pReg2-Bsp-HA. Next, the lexA ORF was amplified by PCR using plasmid pEG202 as the template; the reverse primer used for this PCR also included sequences for two consecutive HA tags. The resulting fragment was inserted into the BspEI site of plasmid pReg2-Bsp-HA, yielding pReg2-L-HA. The plasmid was then transformed into wild-type and mutant yeast strains, and the transformants were subjected to various culture conditions as specified below. Protein extracts were prepared using the boiling/alkaline lysis method (45), and Reg2-L-HA expression levels were analyzed by immunoblotting with the anti-HA antibody HA-7 as described above.
Mixed-culture competition assays.
Strains MKY323 (MATα ura3) and MMY9 (MATα ura3 reg2Δ::KanMX6) were grown to mid-log phase in YEP with 2% glucose. The cultures were adjusted to an optical density at 600 nm (OD600) of 0.15 with YEP-2% glucose medium, mixed together in equal proportions, and grown for 4 h to an OD600 of ∼0.6. The mixed-culture cells were then subjected to 10 cycles of glucose starvation/glucose replenishment. For glucose starvation, cells were shifted to YEP medium lacking glucose for 24 h. For glucose replenishment, the starved cells were shifted to YEP containing 2% glucose (at an initial OD600 of 0.15) and grown for 5 to 6 h; all incubations were at 30°C with shaking at 200 rpm. To determine the proportion of reg2Δ::KanMX6 cells, culture aliquots were plated on YEP with 2% glucose, and following colony formation, the number of kanamycin-resistant colonies was determined by replica plating onto YEP containing 2% glucose and 200 μg/ml kanamycin.
RESULTS
Reg2 interacts with Snf1 in the two-hybrid system.
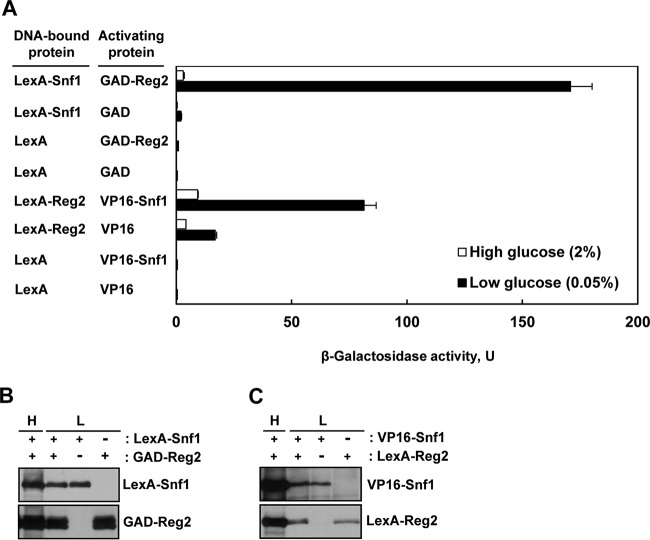
We first tested whether, like Reg1, Reg2 physically interacts with Snf1. To address this possibility, we first used the yeast two-hybrid system. We constructed a fusion protein in which the Gal4 activation domain (GAD) is fused at the amino terminus of Reg2 (GAD-Reg2) and tested the ability of GAD-Reg2 to activate transcription of an integrated lexAop-lacZ reporter gene when coexpressed with a LexA-Snf1 fusion protein (38) in the reporter strain CTY10-5d. Under high-glucose conditions (2% glucose), reporter activation in cells expressing LexA-Snf1 and GAD-Reg2 was not substantially higher than in the vector controls, but under glucose-limiting conditions (0.05% glucose), LexA-Snf1 and GAD-Reg2 interacted strongly (Fig. 1A), and the increased interaction could not be attributed to increased fusion protein levels (Fig. 1B).
FIG 1.
Reg2 and Snf1 interact in the two-hybrid system. (A) Transformants of CTY10-5d with plasmids expressing the indicated protein pairs were grown to mid-log phase in selective SC medium lacking histidine and leucine and containing 2% glucose and then shifted for 3 h to an otherwise identical medium containing 0.05% glucose. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units. The values are averages for 4 to 8 transformants. The error bars indicate standard errors. (B and C) Expression of the fusion proteins during growth in 2% glucose (H, high glucose) and after shift to 0.05% glucose (L, low glucose). Transformants of CTY10-5d expressing the indicated proteins (+) or carrying the corresponding vectors (−) were grown as for panel A, and expression of the fusion proteins was analyzed by immunoblotting as described in Materials and Methods.
We also constructed a LexA-Reg2 fusion and examined its ability to interact with a VP16-Snf1 fusion (39). Similarly, the interaction was stronger under glucose-limiting conditions (Fig. 1A) without an increase in the levels of the interaction partners (Fig. 1C).
Thus, these experiments indicated that Reg2 and Snf1 interact in the two-hybrid system in a glucose-regulated manner.
Mutation of the Snf1 activation loop threonine abolishes interaction with Reg2.
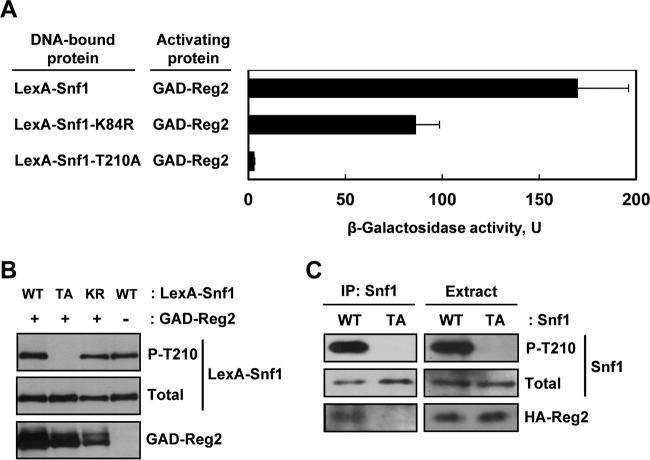
To determine if the Reg2-Snf1 interaction depends on the Thr210 phosphorylation state or catalytic activity of Snf1, we used yeast two-hybrid assays to compare the ability of GAD-Reg2 to interact with LexA-Snf1 and with two mutant derivatives: nonphosphorylatable LexA-Snf1-T210A and kinase-dead LexA-Snf1-K84R with a mutation in the ATP-binding site (48).
Under high-glucose conditions, the mean β-galactosidase activities were low for all the combinations examined (2.5 U or lower); under glucose-limiting conditions, GAD-Reg2 strongly interacted with LexA-Snf1 (170 U) and with kinase-dead LexA-Snf1-K84R (86 U), but there was no significant interaction with nonphosphorylatable LexA-Snf1-T210A (<3 U) (Fig. 2A). The observed interaction defect between GAD-Reg2 and LexA-Snf1-T210A could not be attributed to reduced expression of the fusion proteins, as determined by immunoblot analysis (Fig. 2B). Furthermore, this defect could not be attributed to the lack of catalytic activity, since GAD-Reg2 still interacted quite strongly with the kinase-dead LexA-Snf1-K84R.
FIG 2.
Mutation of the Snf1 activation loop threonine abolishes interaction with Reg2. (A) Wild-type strain MKY343 carrying a lexAop-lacZ reporter (plasmid pSH18-18) and expressing the indicated protein pairs was grown to mid-log phase in selective SC medium lacking uracil, histidine, and leucine and containing abundant (2%) glucose and then shifted for 3 h to an otherwise identical medium containing limiting (0.05%) glucose. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units. Shown are the results for limiting glucose; the values are averages for five transformants, and the error bars indicate standard errors. Under glucose-rich conditions, the values were 2.5 U or lower. (B) Representative transformants were cultured under glucose-limiting conditions as for panel A, and the levels of LexA-Snf1 phosphorylated at Thr210 (P-T210), total LexA-Snf1 (Total), and GAD-Reg2 were confirmed by immunoblotting as described in Materials and Methods. WT, LexA-Snf1; TA, LexA-Snf1-T210A; KR, LexA-Snf1-K84R. The cells also expressed GAD-Reg2 (GAD-Reg2, +) or carried the corresponding vector pACTII (GAD-Reg2, −). (C) Coimmunoprecipitation assays. Cells of strain MKY362 (snf1Δ) transformed with combinations of plasmids expressing HA-Reg2 and wild-type Snf1 (WT) or nonphosphorylatable Snf1-T210A (TA) were grown to mid-log phase in selective SC medium lacking leucine and uracil and containing abundant (2%) glucose and then shifted to an otherwise identical medium containing limiting (0.05%) glucose for 15 min. Following the shift to limiting glucose, protein extracts were prepared, and Snf1 was immunoprecipitated with anti-polyhistidine antibody. The immunoprecipitates (IP: Snf1) and extracts (Extract) were examined by immunoblotting for the presence of phospho-Thr210-Snf1 (P-T210), total Snf1 protein (Total), and HA-Reg2.
We further examined the interaction between Reg2 and nonphosphorylatable Snf1-T210A by coimmunoprecipitation. We coexpressed HA-Reg2 and either Snf1 or Snf1-T210A in a snf1Δ strain. Cells were cultured in limiting glucose (to promote Thr210 phosphorylation), protein extracts were prepared, and the Snf1 and Snf1-T210A proteins were immunoprecipitated with anti-polyhistidine antibody. The immunoprecipitates were then examined for the presence of HA-Reg2 by immunoblotting. HA-Reg2 coimmunoprecipitated with Snf1, but not with Snf1-T210A (Fig. 2C).
Collectively, these experiments indicated that Thr210 of Snf1 is critical for its interaction with Reg2, suggesting that like Reg1 (25), Reg2 interacts with the phosphorylated form of Snf1.
The reg2Δ mutation does not result in constitutive Thr210 phosphorylation of Snf1 during exponential growth on high glucose.
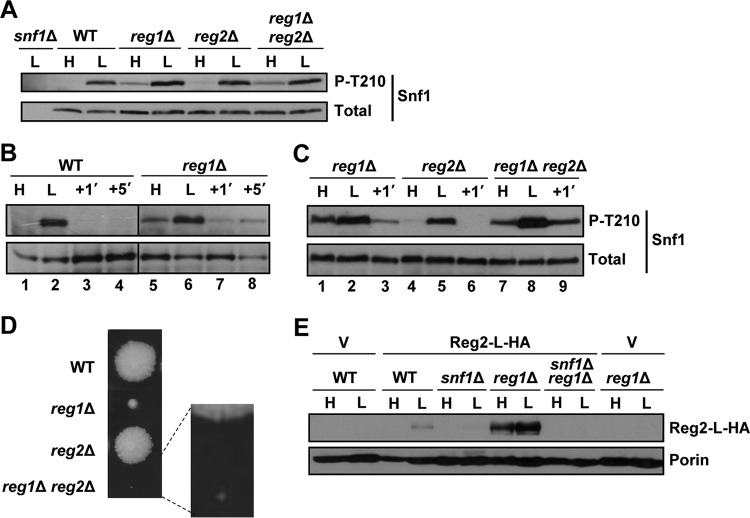
To investigate the role of Reg2 in Snf1 regulation, we first tested for possible effects of the reg2Δ mutation on the Thr210 phosphorylation status of Snf1. Wild-type and mutant cells were grown to mid-log phase in the presence of abundant (2%) glucose and shifted to limiting (0.05%) glucose for 1 h, and the Thr210 phosphorylation status of Snf1 was determined by immunoblotting as described previously (45). The levels of phospho-Thr210-Snf1 in the reg2Δ mutant were comparable to those in the wild type under both conditions (Fig. 3A). Thus, unlike the reg1Δ mutation (17) (see Fig. 7), the reg2Δ mutation did not lead to any discernible difference in the Thr210 phosphorylation status of Snf1 during exponential growth in high glucose.
FIG 3.
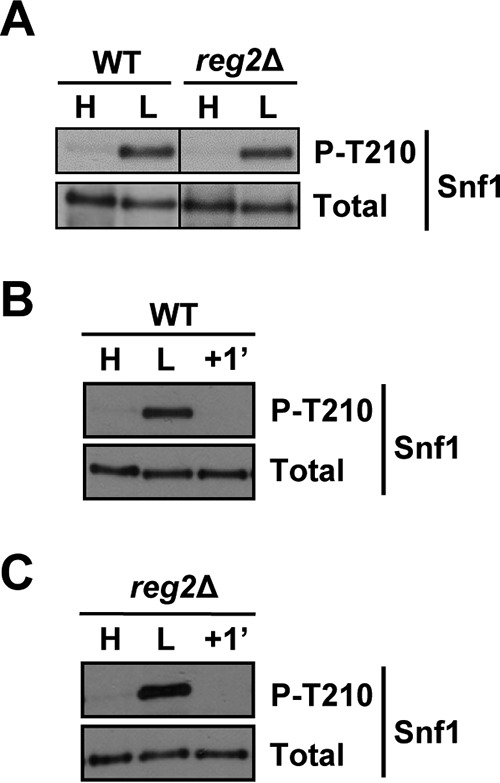
Lack of Reg2 does not lead to constitutive accumulation of phospho-Thr210-Snf1. (A) Cells of the indicated genotypes were grown to mid-log phase in YEP containing 2% glucose (H, high glucose) followed by a shift for 1 h to YEP containing 0.05% glucose (L, low glucose). (B and C) Cells were grown as for panel A, and the glucose-limited cells were then replenished with abundant (2%) glucose for 1 min (+1′). The levels of phospho-Thr210-Snf1 (P-T210) and total Snf1 protein (Total) were analyzed by immunoblotting. The strains were MMY128 (WT) and MMY18 (reg2Δ).
FIG 7.
Combined effects of reg1Δ and reg2Δ. (A to C) Cells of the indicated genotypes were grown to mid-log phase in YEP containing 2% glucose (H, high glucose) and shifted for 1 h to YEP containing 0.05% glucose (L, low glucose); where indicated, the glucose-limited cells were then replenished with abundant (2%) glucose for 1 min (+1′) or 5 min (+5′). The levels of phosphorylated Thr210 (P-T210) and total Snf1 protein (Total) were analyzed by immunoblotting. The strains were MMY128 (WT), MMY40 (reg1Δ), MMY41 (reg2Δ), MMY42 (reg1Δ reg2Δ), and MKY362 (snf1Δ). (D) A representative tetratype tetrad from a cross between strains MMY9 (reg2Δ) and KBY247 (reg1Δ). Segregants were micromanipulated onto YEP containing 2% glucose and grown for 4 days. The centers of the colonies are ∼5 mm apart. (E) Cells of the indicated genotypes transformed with vector pRS316 (V) or pReg2-L-HA (Reg2-L-HA) were grown in SC medium lacking uracil and containing 2% glucose (H, high glucose) and then shifted for 1 h to an otherwise identical medium containing 0.05% glucose (L, low glucose). The levels of tagged Reg2 expressed from its native promoter (Reg2-L-HA) were analyzed by immunoblotting; Por1 was used as a loading control (Porin) and detected using an anti-Por1 antibody (Invitrogen). The strains were MKY191 (WT), MKY366 (snf1Δ), KBY266 (reg1Δ), and KBY238 (snf1Δ reg1Δ).
It remained possible that the reg2Δ mutant differs from the wild type with respect to rapid Snf1 dephosphorylation in response to glucose replenishment. To test this possibility, glucose-limited wild-type and reg2Δ cells were prepared as described above and then replenished with glucose for 1 min. However, no obvious dephosphorylation defect was observed in the mutant (Fig. 3B and C).
Overexpression of Reg2 leads to reduced phospho-Thr210-Snf1 during growth in abundant glucose.
We next examined the effects of Reg2 overexpression. To this end, we used LexA-Reg2, which is expressed from the ADH1 promoter of the multicopy vector pEG202 (35). Overexpression of LexA-Reg2 resulted in a significant reduction of the levels of phospho-Thr210-Snf1 both in the wild type and in the reg1Δ mutant during growth in abundant glucose (Fig. 4A). Consistent with previous results (28), in control experiments, we observed that overexpression of LexA-Reg2 improved the growth of the reg1Δ mutant (Fig. 4B).
FIG 4.
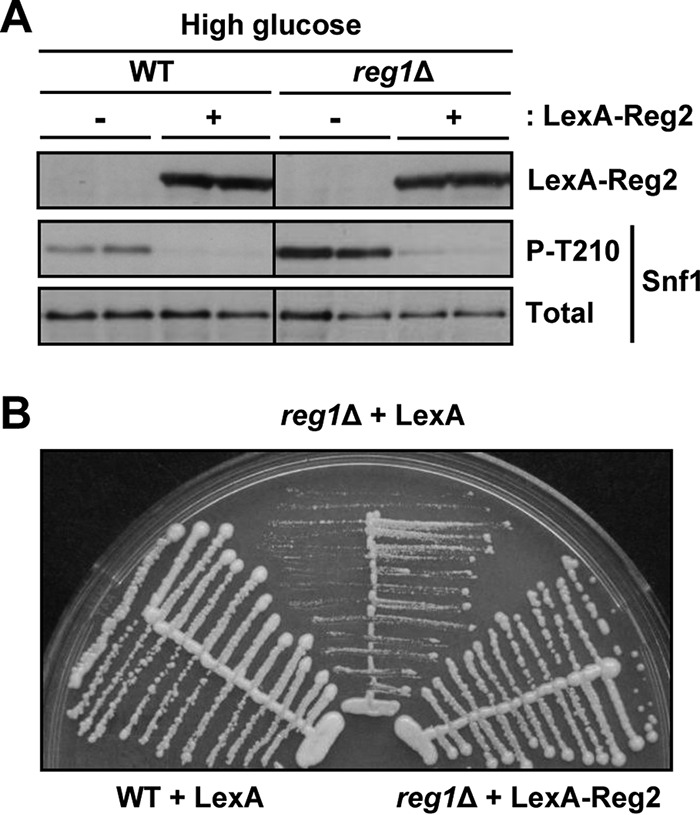
Effects of Reg2 overexpression under glucose-rich conditions. (A) Overexpression of Reg2 during growth in abundant glucose reduces phospho-Thr210-Snf1 levels. Strains of the indicated genotypes carrying plasmid pLexA-Reg2 (LexA-Reg2, +) or the corresponding vector pEG202 (LexA-Reg2, −) were grown to mid-log phase in selective SC medium lacking histidine and containing 2% glucose. The levels of LexA-Reg2, phospho-Thr210-Snf1 (P-T210), and total Snf1 protein (Total) were analyzed by immunoblotting. (B) Overexpression of LexA-Reg2 suppresses the slow growth caused by reg1Δ. Cells of the indicated genotypes carrying vector pEG202 (LexA) or plasmid pLexA-Reg2 (LexA-Reg2) were streaked on SC medium lacking histidine and containing 2% glucose and grown for 3 days. The strains were MKY343 (WT) and KBY247 (reg1Δ).
In contrast to cells grown in high glucose, wild-type cells with LexA-Reg2 exhibited no discernible reduction in the phospho-Thr210 level in limiting glucose (Fig. 5A). It should be noted that, as we often observe with proteins expressed from the vector pEG202, the LexA-Reg2 protein level was somewhat reduced in limiting glucose relative to abundant glucose (Fig. 5B), but it seems unlikely that this reduction alone could be responsible for the lack of any apparent effect on Snf1. We conclude that Reg2 overexpression negatively affects Snf1, at least in the presence of abundant glucose.
FIG 5.
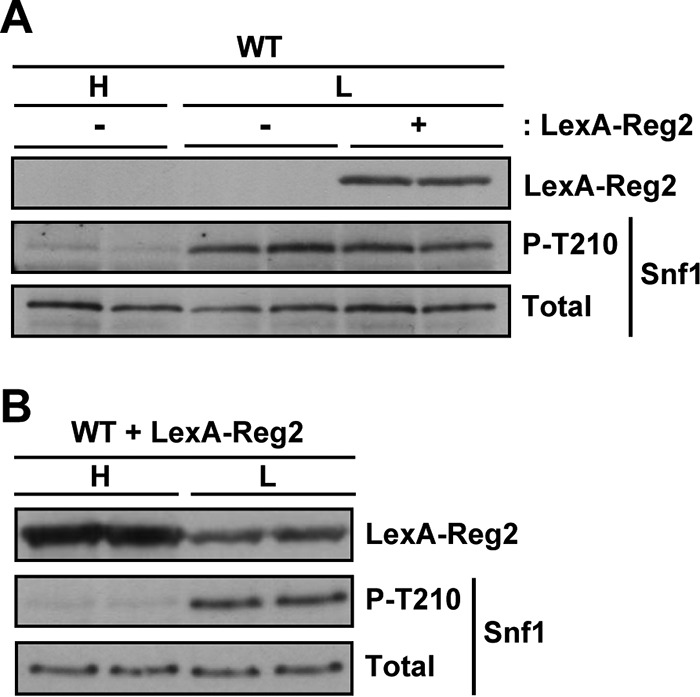
Overexpression of Reg2 has no significant effect on the Thr210 phosphorylation status of Snf1 in low glucose. (A) Wild-type cells (MKY343) transformed with pLexA-Reg2 (LexA-Reg2, +) or with the corresponding vector pEG202 (LexA-Reg2, −) were grown to mid-log phase in SC medium lacking histidine and containing 2% glucose (H, high glucose) and then shifted to an otherwise identical medium containing 0.05% glucose (L, low glucose). The levels of LexA-Reg2, phospho-Thr210-Snf1 (P-T210), and total Snf1 protein (Total) were analyzed by immunoblotting. (B) Expression of LexA-Reg2 in 2% glucose (H, high glucose) and in 0.05% glucose (L, low glucose). Cells of MKY343 (WT) transformed with pLexA-Reg2 were grown as for panel A.
The RVxF motif of Reg2 is required for its interaction with Glc7.
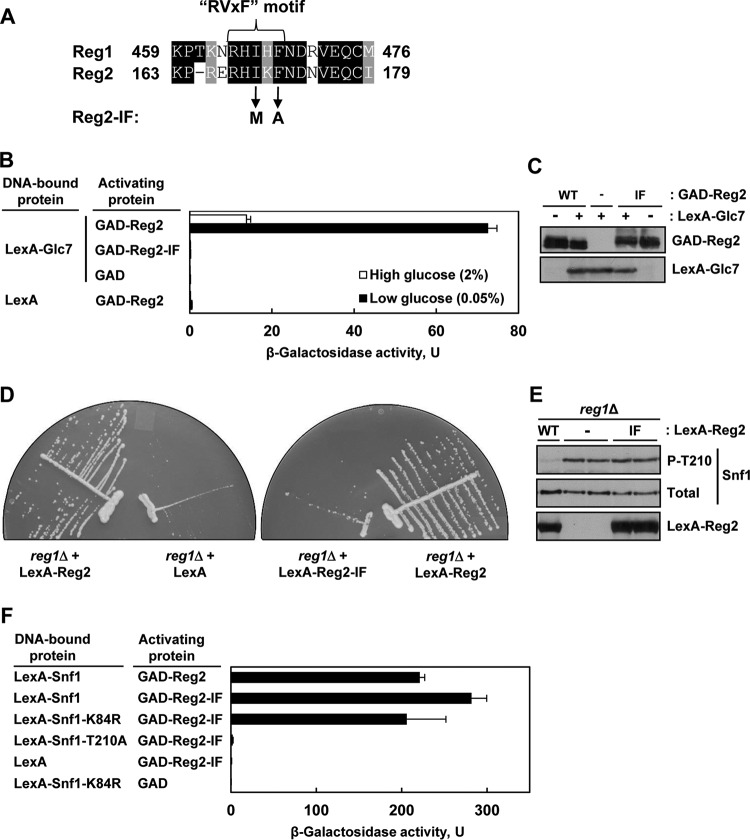
Reg1 and Reg2 both possess variants of the so-called RVxF motif found in many eukaryotic type 1 protein phosphatase-interacting proteins (RHIHF and RHIKF, respectively) (Fig. 6A). It was previously shown that the RVxF motif of Reg1 is required for its interaction with Glc7 (42, 49–51). To determine the role of the apparent RVxF motif of Reg2, we mutated the two critical hydrophobic residues, isoleucine (Ile169) and phenylalanine (Phe171), to methionine and alanine, respectively, by analogy to prior studies with Reg1 (42, 51). Since the Reg1 double mutant (I466M F468A) was previously called Reg1-IF (42), we similarly refer to the equivalent Reg2 double mutant (I169M F171A) as Reg2-IF.
FIG 6.
Mutation of the RVxF motif in Reg2 affects its ability to interact with Glc7 and to dephosphorylate Snf1. (A) Alignment of the RVxF motifs of Reg1 and Reg2. The amino acid identities and similarities are shaded black and gray, respectively. The arrows indicate the changes introduced in the Reg2-IF mutant. (B) Reg2-IF does not interact with Glc7 in the two-hybrid system. Transformants of CTY10-5d with plasmids expressing the indicated protein pairs were grown to mid-log phase in selective SC medium lacking histidine and leucine and containing 2% glucose and then shifted for 3 h to an otherwise identical medium containing 0.05% glucose. β-Galactosidase activity was assayed in the permeabilized cells and expressed in Miller units. The values are averages for 4 to 8 transformants. The error bars indicate standard errors. (C) Representative transformants of CTY10-5d expressing GAD-Reg2 (GAD-Reg2, WT) or GAD-Reg2-IF (GAD-Reg2, IF), carrying the corresponding vector (GAD-Reg2, −) and simultaneously expressing LexA-Glc7 (LexA-Glc7, +), or carrying the corresponding vector (LexA-Glc7, −) were grown as for panel B, and expression of the fusion proteins was confirmed by immunoblotting as described in Materials and Methods. (D) Overexpression of wild-type Reg2, but not the Reg2-IF mutant, improves growth of reg1Δ cells. reg1Δ cells of strain KBY247 carrying vector pEG202 (LexA), plasmid pLexA-Reg2 (LexA-Reg2), or plasmid pLexA-Reg2-IF (LexA-Reg2-IF) were streaked on SC medium lacking histidine and containing 2% glucose and grown for 3 days. (E) Overexpression of LexA-Reg2-IF during growth in abundant glucose does not reduce phospho-Thr210-Snf1 levels. reg1Δ cells of strain KBY247 carrying plasmid pLexA-Reg2 (LexA-Reg2, WT), pLexA-Reg2-IF (LexA-Reg2, IF), or the corresponding vector pEG202 (LexA-Reg2, −) were grown to mid-log phase in selective SC medium lacking histidine and containing 2% glucose. The levels of LexA-tagged proteins (LexA-Reg2), phospho-Thr210-Snf1 (P-T210), and total Snf1 protein (Total) were analyzed by immunoblotting. (F) Reg2-IF interacts with Snf1 in the yeast two-hybrid system. Transformants of CTY10-5d with plasmids expressing the indicated protein pairs were grown as for panel B. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units. Shown are the results for limiting glucose; the values are averages for 4 to 8 transformants, and the error bars indicate standard errors.
Consistent with previous results (28), wild-type Reg2 interacted strongly with Glc7 in the yeast two-hybrid system (Fig. 6B). In contrast, the Reg2-IF mutant exhibited a severe Glc7 interaction defect. Immunoblot analysis showed similar levels of the LexA-Glc7, GAD-Reg2, and GAD-Reg2-IF fusion proteins in both cases (Fig. 6C). These results strongly suggest that the RVxF motif of Reg2 is required for its interaction with Glc7.
In support of a functional defect, overexpression of Reg2-IF (as LexA-Reg2-IF) was unable to restore the growth of the reg1Δ mutant to the wild-type level (Fig. 6D) and to reduce the levels of phospho-Thr210-Snf1 in this mutant during growth on high glucose (Fig. 6E).
It was previously reported that the RVxF motif of Reg1 is required for interaction not only with Glc7 but also with Snf1, since Reg1-IF was unable to interact with either (42). We therefore also tested Reg2-IF for interaction with Snf1. The observed interaction pattern was similar to that of wild-type Reg2, as GAD-Reg2-IF interacted with LexA-Snf1 and kinase-dead LexA-Snf1-K84R under low-glucose conditions, but it did not interact with nonphosphorylatable LexA-Snf1-T210A (Fig. 6F). Thus, these results provide no evidence that the RVxF motif of Reg2 is required for its interaction with Snf1.
Combined effects of reg1Δ and reg2Δ.
The above-described overexpression and interaction experiments provided evidence that Reg2 is involved in the negative control of Snf1. Since the reg2Δ single mutation did not affect the Thr210 phosphorylation status of Snf1 in our earlier experiments (Fig. 3), we next examined whether reg2Δ could affect Snf1 when combined with reg1Δ.
We constructed reg1Δ reg2Δ double-mutant cells by genetic crossing and used immunoblotting to monitor the levels of phospho-Thr210-Snf1. Cells were grown in the presence of abundant glucose and then shifted to limiting glucose. In this experimental setting, however, the levels of phospho-Thr210-Snf1 in the reg1Δ reg2Δ double mutant were similar to those in the reg1Δ single mutant (Fig. 7A).
In the course of our experiments with the reg1Δ single mutant, we noted that glucose readdition to glucose-limited cells results in a rapid (1-min) reduction of phospho-Thr210-Snf1 to a level that was discernibly below that observed during exponential growth in abundant glucose (Fig. 7B, compare lanes 5 and 7). The effect was transient, as the level of phospho-Thr210-Snf1 increased again 5 min after glucose readdition (Fig. 7B, compare lanes 7 and 8). We considered the possibility that this rapid/transient dip in phospho-Thr210-Snf1 upon glucose replenishment might involve Reg2. We compared the Thr210 phosphorylation states of Snf1 in the reg1Δ single, reg2Δ single, and reg1Δ reg2Δ double mutants in response to readdition of glucose. Unlike with the reg1Δ single mutant, there was no extra dip in the reg1Δ reg2Δ double mutant at 1 min after glucose addition (Fig. 7C, compare lanes 7 and 9), suggesting that Reg2 might contribute to the rapid dephosphorylation of Snf1 upon glucose replenishment.
We note that these results still do not provide a conclusive explanation for the slow growth of the reg1Δ reg2Δ double mutant (28) (Fig. 7D). It is known that this phenotype is Snf1 dependent (28), but as described above, there was no obvious increase in phospho-Thr210-Snf1 in the reg1Δ reg2Δ double-mutant cells relative to reg1Δ single mutant cells during exponential growth in abundant glucose. It is possible that the slow growth of the double mutant is not directly related to Reg2's role in the negative control of Snf1 itself; instead, it could be related to a role in dephosphorylation of a Snf1 target involved in growth control, as was proposed previously (28). However, we cannot exclude the possibility that immunoblot analysis simply does not have sufficient resolution to capture potentially relevant effects of reg2Δ on Snf1 that are either transient or small in comparison to the effects of reg1Δ yet sufficient to translate into larger phenotypic effects.
The lack of an obvious phenotype produced by the reg2Δ single mutation yet the presence of a strong growth defect produced by its combination with reg1Δ might be related to differential expression of the Reg2 protein in REG1 versus reg1Δ cells. We therefore compared the levels of a C-terminally tagged Reg2 protein expressed from the native REG2 promoter in REG1 and in reg1Δ cells. We observed that Reg2 is not detectably expressed in the wild type grown on high glucose (Fig. 7E) and is expressed only weakly after a 1-h shift to low glucose (Fig. 7E). In contrast, Reg2 was expressed at much higher levels in reg1Δ cells, under both high- and low-glucose conditions (Fig. 7E).
In a previous microarray study, REG2 was listed among genes positively regulated by Snf1 at the mRNA level (52). To directly address the requirement for Snf1 for Reg2 protein expression, we examined the effects of the snf1Δ mutation in the REG1 and reg1Δ backgrounds. We observed a Reg2 expression defect in the snf1Δ mutant after a 1-h shift to low glucose (Fig. 7E). Moreover, we did not detect any Reg2 protein in the reg1Δ snf1Δ double mutant (Fig. 7E), indicating that even in the absence of Reg1, Reg2 protein expression remains Snf1 dependent.
Reg2 plays a role in Snf1 inactivation after prolonged glucose starvation.
We considered the possibility that the reg2Δ mutation failed to produce detectable effects on its own (in REG1 cells), because Reg2 protein expression is quite low under the experimental conditions employed so far. In other words, the reg2Δ single mutation could produce an effect under conditions where Reg2 is naturally expressed at a sufficiently high level. Since Reg2 expression showed an increase in response to short-term glucose limitation (1 h) (Fig. 7E), we reasoned that its expression might increase even further in response to a longer starvation time. Indeed, we detected a higher Reg2 protein level after depriving cells of glucose for 24 h (Fig. 8A), and expression still remained Snf1 dependent (Fig. 8B).
FIG 8.
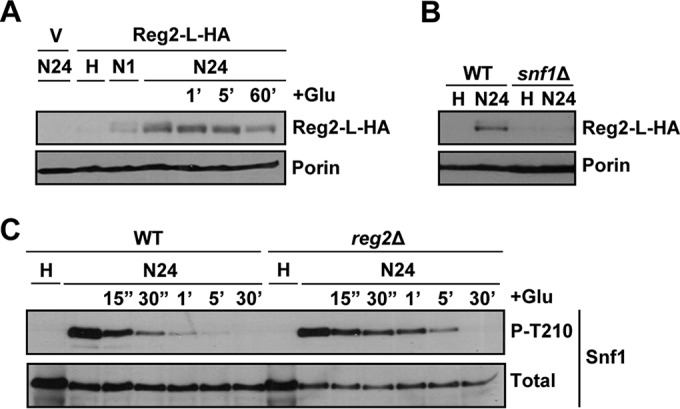
Reg2 accumulation increases after prolonged glucose deprivation in a Snf1-dependent manner. (A) reg2Δ cells transformed with the vector pRS316 (V) or pReg2-L-HA (Reg2-L-HA) were grown to mid-log phase in selective SC medium lacking uracil and containing 2% glucose (H, high glucose) and then shifted to an otherwise identical medium containing no glucose for either 1 h (N1) or 24 h (N24). After 24 h of glucose starvation, the cultures were replenished with 2% glucose (+Glu) for either 1 min (1′), 5 min (5′), or 60 min (60′). (B) Cells of the indicated genotypes carrying pReg2-L-HA (Reg2-L-HA) were grown to mid-log phase in selective SC medium lacking uracil and containing 2% glucose (H, high glucose) and then shifted to an otherwise identical medium containing no glucose for 24 h (N24). In panels A and B, the levels of tagged Reg2 expressed from the native REG2 promoter (Reg2-L-HA) and Por1 protein (Porin) were detected by immunoblotting as for Fig. 7E. (C) Cells of the indicated genotypes were grown to mid-log phase in YEP medium containing 2% glucose (H, high glucose) and shifted to an otherwise identical medium containing no glucose for 24 h (N24). Following 24 h in no glucose, the cells were replenished with 2% glucose (+Glu) for either 15 s (15″), 30 s (30″), 1 min (1′), 5 min (5′), or 30 min (30′). The levels of phospho-Thr210-Snf1 (P-T210) and total Snf1 protein (Total) were analyzed by immunoblotting. The strains were MMY9 (reg2Δ), MKY323 (WT), and MKY366 (snf1Δ).
When we added glucose back to wild-type and reg2Δ mutant cells prestarved for 24 h, we finally detected a difference between the cells. Specifically, Snf1 Thr210 dephosphorylation occurred more slowly in the reg2Δ mutant, as there was a clear difference in phospho-Thr210-Snf1 levels at 30 s, 1 min, and 5 min postreplenishment (Fig. 8C); in subsequent experiments, we also detected differences at later time points (Fig. 9B) (see below). These results strongly suggested that Reg2 has an important role in the rapid dephosphorylation of Snf1 when glucose is added back after prolonged starvation.
FIG 9.
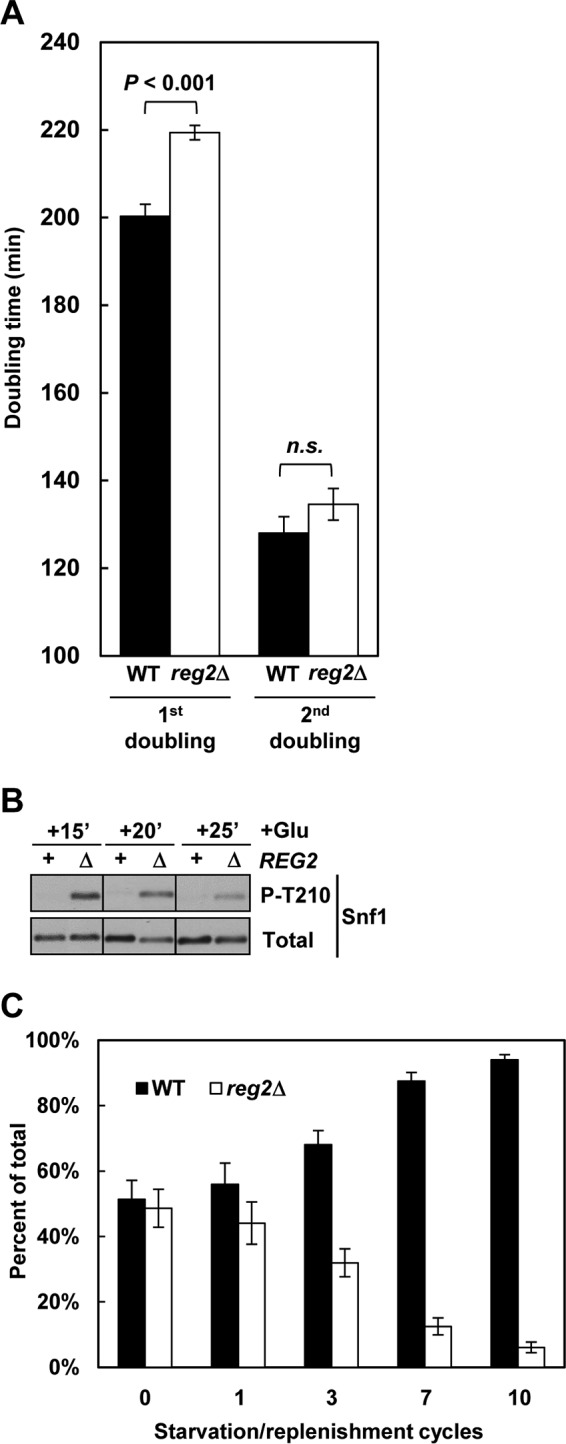
Cells lacking Reg2 exhibit delayed recovery from prolonged glucose deprivation. (A) Wild-type and reg2Δ cells were grown to mid-log phase in YEP containing 2% glucose and then shifted to YEP without glucose for 24 h. After this starvation period, the cultures were shifted (at an initial OD600 of 0.15) to YEP containing 2% glucose, and the OD600 of the cultures was monitored. The values are averages for three cultures of each genotype. The error bars indicate standard errors. (B) Wild-type (REG2, +) and reg2Δ (REG2, Δ) cells were grown to mid-log phase in YEP medium containing 2% glucose and shifted to YEP containing no glucose for 24 h, and glucose was then added at 2% (+Glu) for either 15 min (+15′), 20 min (+20′), or 25 min (+25′). The levels of phosphorylated Thr210 (P-T210) and total Snf1 protein (Total) were analyzed by immunoblotting. (C) Wild-type and reg2Δ cells were grown to mid-log phase in YEP medium containing 2% glucose, mixed in equal proportions, and subjected to 10 cycles consisting of a 24-h glucose starvation phase and a 5- to 6-h glucose replenishment phase, and the proportion of reg2Δ cells was determined as described in Materials and Methods. In all the experiments, the strains were MKY323 (WT) and MMY9 (reg2Δ).
Starved cells lacking Reg2 exhibit a growth recovery delay following glucose replenishment.
Since increased Snf1 activity can cause slower growth, we next compared the abilities of glucose-starved wild-type and reg2Δ mutant cells to resume growth upon glucose replenishment. When 2% glucose was added back to cells prestarved for 24 h, the reg2Δ mutant required ∼20 min longer than the wild type to undergo the first doubling (ca. 220 min versus 200 min, respectively) (Fig. 9A), but there was no statistically significant difference in their second doubling times (ca. 130 min) (Fig. 9A). The initial 20-min growth delay of the reg2Δ mutant also coincided with higher levels of phospho-Thr210-Snf1, which could be detected at 15, 20, and even 25 min after glucose readdition (Fig. 9B).
Although the 20-min growth recovery delay of the reg2Δ mutant may seem modest, if repeated over several starvation/replenishment cycles, it could translate into a substantial competitive disadvantage. To address this point, we performed mixed-culture competition assays. Wild-type and reg2Δ cells were grown to mid-log phase in glucose-rich medium, mixed in equal proportions, and subjected to repetitive cycles of 24-h glucose starvation followed by glucose replenishment. The proportion of the mutant cells in the culture progressively declined, dropping below 10% after 10 such cycles (Fig. 9C). Thus, these results underscore the evolutionary significance of the REG2 gene.
DISCUSSION
REG1 and REG2 of S. cerevisiae are a pair of paralogous genes dating back to an ancient whole-genome duplication event in an ancestral yeast species (53). In the course of evolution, most duplicate gene copies were lost, and more than 80% of the protein-coding genes in modern S. cerevisiae do not have a paralog (for a recent review, see reference 54). It was previously noted that many of the preserved gene pairs function in energy metabolism and/or its regulation (55). It was further proposed that this functional bias, together with the freedom of the paralogs to acquire a degree of specialization, may have been beneficial for the ability of S. cerevisiae to outcompete other species by rapid glucose fermentation (56). Rapid growth in glucose is associated with reduced Snf1 activity, emphasizing the importance of Snf1-inhibiting mechanisms. As a regulatory subunit of Glc7 protein phosphatase, the role of Reg1 in Snf1 inhibition has been firmly established (4), but the possible contribution of Reg2 to this process has not been fully understood.
In this study, we present evidence that Reg2 also contributes to the negative control of Snf1. First, like Reg1, Reg2 strongly interacts with Snf1; moreover, the results obtained suggest that this interaction requires that Snf1 be phosphorylated at Thr210, as would be expected if Snf1 is a dephosphorylation target for Reg2-Glc7. Second, Reg2 overexpression not only rescues the Snf1-dependent slow-growth phenotype of the reg1Δ mutant (28), but is also accompanied by a reduction in phospho-Thr210-Snf1. Third, in cells experiencing prolonged glucose deprivation, Reg2 is needed for faster Snf1 Thr210 dephosphorylation upon glucose replenishment.
Although Reg2 is substantially smaller than Reg1 (338 versus 1,014 amino acids, respectively), both proteins contain variants of the so-called RVxF motif found in many eukaryotic type 1 protein phosphatase-interacting proteins. It was previously shown that the RVxF motif of Reg1 is required for its interaction with Glc7 (42, 49–51). In this study, we present evidence that the RVxF motif of Reg2 is similarly required for Glc7 binding. This further supports the notion that, like Reg1, Reg2 is a bona fide regulatory subunit of Glc7.
Interestingly, the RVxF motif of Reg1 was also found to be required for Reg1's interaction not only with Glc7 but also with Snf1 (42). Based on these findings, a model was proposed in which the regulation of Snf1 by Reg1 occurs via a complex dynamic competition between Glc7 and Snf1 for Reg1 binding. Our results provide no evidence for such a model for Reg2, since an equivalent mutation of its RVxF motif did not abolish its interaction with Snf1. It remains possible that a similar binding competition could occur via an as yet unknown region of Reg2 other than its RVxF motif. However, it is also possible that the functional cycle of Reg2 is simpler than that of Reg1 and it does not require such competition.
Evidence indicates that the Reg1 protein is expressed at similar levels under glucose-rich and glucose-limiting conditions (42). In contrast, REG2 gene transcription (52, 57) and Reg2 protein abundance (this study) increase in response to glucose limitation in a Snf1-dependent manner. Furthermore, Reg2 protein expression increases even further following long-term glucose deprivation. We propose a model in which Snf1-dependent Reg2 accumulation during prolonged glucose deprivation prepares cells to more quickly dephosphorylate and inactivate Snf1 upon glucose replenishment (Fig. 10).
FIG 10.
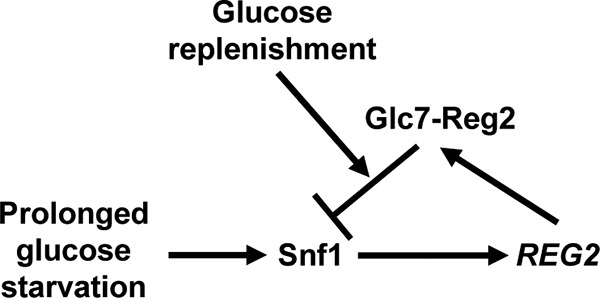
Proposed model. During steady-state growth in high glucose, Snf1 is inactive and REG2 is not expressed. Prolonged glucose starvation leads to Snf1-dependent accumulation of Reg2. Once glucose becomes abundant, Reg2 contributes to the rapid dephosphorylation and inactivation of Snf1.
We also show that Reg2 contributes to faster growth recovery when glucose is added back to cells after prolonged glucose deprivation. It seems logical to assume that at least part of the underlying mechanism involves Snf1 Thr210 dephosphorylation, but it seems possible, and even likely, that Reg2-Glc7 also dephosphorylates other growth-related targets. As proposed previously (28), it seems likely that some additional growth-related targets of Reg2-Glc7 could actually be phosphorylation substrates of Snf1 itself. In this regard, it has been recently reported that Snf1 phosphorylates and negatively regulates adenylate cyclase (58) and that it also inhibits the progrowth TORC1 pathway in response to glucose starvation (59, 60), raising the possibility that Reg2-Glc7 might target components of the Ras-cAMP-PKA and/or TORC1 pathway. It certainly also remains possible that some relevant targets of Reg2-Glc7 could lie outside the Snf1 pathway. Further studies will be required to address these various possibilities.
Regardless of whether the role of Reg2-Glc7 is limited to dephosphorylation and inactivation of Snf1 itself or extends to other targets, it is clear that the function provided by Reg2 bears evolutionary significance. The results presented in this study show that cells lacking Reg2 are readily outcompeted by wild-type cells in the course of a few glucose starvation/replenishment cycles. The ability of starved yeast cells to rapidly activate metabolism and growth following nutrient replenishment is crucial for competing with microbial neighbors, and the REG2 gene was likely preserved to help fulfill this function.
ACKNOWLEDGMENTS
We thank M. Carlson for reagents and M. Orlova and V. Strogolova for technical assistance and comments on the manuscript.
This work was supported by National Science Foundation grant MCB-0818837 to S.K.
REFERENCES
- 1.Pretorius IS. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729. [DOI] [PubMed] [Google Scholar]
- 2.Goddard MR, Greig D. 2015. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res 15:fov009. doi: 10.1093/femsyr/fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santangelo GM. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedbacker K, Carlson M. 2008. SNF1/AMPK pathways in yeast. Front Biosci 13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie DG, Carling D. 1997. The AMP-activated protein kinase: fuel gauge of the mammalian cell? Eur J Biochem 246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 6.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Hardie DG. 2007. AMP-activated protein kinase as a drug target. Annu Rev Pharmacol Toxicol 47:185–210. doi: 10.1146/annurev.pharmtox.47.120505.105304. [DOI] [PubMed] [Google Scholar]
- 8.Fogarty S, Hardie DG. 2010. Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim Biophys Acta 1804:581–591. doi: 10.1016/j.bbapap.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Carlson M, Osmond BC, Botstein D. 1981. Mutants of yeast defective in sucrose utilization. Genetics 98:25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celenza JL, Carlson M. 1984. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol 4:49–53. doi: 10.1128/MCB.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuchin SV, Kartasheva NN, Benevolensky SV. 1993. Genes required for derepression of an extracellular glucoamylase gene, STA2, in the yeast Saccharomyces. Yeast 9:533–541. doi: 10.1002/yea.320090510. [DOI] [PubMed] [Google Scholar]
- 12.Zhang CY, Bai XW, Lin X, Liu XE, Xiao DG. 2015. Effects of SNF1 on maltose metabolism and leavening ability of baker's yeast in lean dough. J Food Sci 80:M2879–M2885. doi: 10.1111/1750-3841.13137. [DOI] [PubMed] [Google Scholar]
- 13.Ostergaard S, Olsson L, Johnston M, Nielsen J. 2000. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat Biotechnol 18:1283–1286. doi: 10.1038/82400. [DOI] [PubMed] [Google Scholar]
- 14.Shi S, Chen Y, Siewers V, Nielsen J. 2014. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. mBio 5:e01130-14. doi: 10.1128/mBio.01130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi JW, Da Silva NA. 2014. Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J Biotechnol 187:56–59. doi: 10.1016/j.jbiotec.2014.07.430. [DOI] [PubMed] [Google Scholar]
- 16.Raab AM, Hlavacek V, Bolotina N, Lang C. 2011. Shifting the fermentative/oxidative balance in Saccharomyces cerevisiae by transcriptional deregulation of Snf1 via overexpression of the upstream activating kinase Sak1p. Appl Environ Microbiol 77:1981–1989. doi: 10.1128/AEM.02219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCartney RR, Schmidt MC. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem 276:36460–36466. [DOI] [PubMed] [Google Scholar]
- 18.Hong SP, Leiper FC, Woods A, Carling D, Carlson M. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci U S A 100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCartney RR, Rubenstein EM, Schmidt MC. 2005. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr Genet 47:335–344. doi: 10.1007/s00294-005-0576-2. [DOI] [PubMed] [Google Scholar]
- 20.Nath N, McCartney RR, Schmidt MC. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol Cell Biol 23:3909–3917. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJ, Schmidt MC, Hardie DG. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol 13:1299–1305. doi: 10.1016/S0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz A, Xu X, Carlson M. 2011. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci U S A 108:6349–6354. doi: 10.1073/pnas.1102758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz A, Liu Y, Xu X, Carlson M. 2012. Heterotrimer-independent regulation of activation-loop phosphorylation of Snf1 protein kinase involves two protein phosphatases. Proc Natl Acad Sci U S A 109:8652–8657. doi: 10.1073/pnas.1206280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu J, Carlson M. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J 14:5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludin K, Jiang R, Carlson M. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, McCartney RR, Chandrashekarappa DG, Mangat S, Schmidt MC. 2011. Reg1 protein regulates phosphorylation of all three Snf1 isoforms but preferentially associates with the Gal83 isoform. Eukaryot Cell 10:1628–1636. doi: 10.1128/EC.05176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong SP, Momcilovic M, Carlson M. 2005. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase α as Snf1-activating kinases in yeast. J Biol Chem 280:21804–21809. doi: 10.1074/jbc.M501887200. [DOI] [PubMed] [Google Scholar]
- 28.Frederick DL, Tatchell K. 1996. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol Cell Biol 16:2922–2931. doi: 10.1128/MCB.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett L, Orlova M, Maziarz M, Kuchin S. 2012. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 11:119–128. doi: 10.1128/EC.05061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, Tatchell K, Liu S, Michels CA. 2000. Protein phosphatase type-1 regulatory subunits Reg1p and Reg2p act as signal transducers in the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. Mol Gen Genet 263:411–422. doi: 10.1007/s004380051185. [DOI] [PubMed] [Google Scholar]
- 31.Byrne KP, Wolfe KH. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose MD, Winston F, Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 33.Kuchin S, Vyas VK, Carlson M. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol Cell Biol 22:3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orlova M, Kanter E, Krakovich D, Kuchin S. 2006. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 5:1831–1837. doi: 10.1128/EC.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golemis EA, Serebriiskii I, Gjuris J, Brent R. 1997. Current protocols in molecular biology, vol 3. Wiley, New York, NY. [Google Scholar]
- 36.Legrain P, Dokhelar MC, Transy C. 1994. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res 22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vojtek AB, Hollenberg SM, Cooper JA. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205–214. doi: 10.1016/0092-8674(93)90307-C. [DOI] [PubMed] [Google Scholar]
- 38.Kuchin S, Treich I, Carlson M. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc Natl Acad Sci U S A 97:7916–7920. doi: 10.1073/pnas.140109897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang R, Carlson M. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev 10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 40.Treitel MA, Kuchin S, Carlson M. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol 18:6273–6280. doi: 10.1128/MCB.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuchin S, Vyas VK, Kanter E, Hong SP, Carlson M. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabba S, Mangat S, McCartney R, Schmidt MC. 2010. PP1 phosphatase-binding motif in Reg1 protein of Saccharomyces cerevisiae is required for interaction with both the PP1 phosphatase Glc7 and the Snf1 protein kinase. Cell Signal 22:1013–1021. doi: 10.1016/j.cellsig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West RW Jr, Yocum RR, Ptashne M. 1984. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol 4:2467–2478. doi: 10.1128/MCB.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vyas VK, Kuchin S, Carlson M. 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlova M, Barrett L, Kuchin S. 2008. Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast 25:745–754. doi: 10.1002/yea.1628. [DOI] [PubMed] [Google Scholar]
- 46.Orlova M, Ozcetin H, Barrett L, Kuchin S. 2010. Roles of the Snf1-activating kinases during nitrogen limitation and pseudohyphal differentiation in Saccharomyces cerevisiae. Eukaryot Cell 9:208–214. doi: 10.1128/EC.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estruch F, Treitel MA, Yang X, Carlson M. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alms GR, Sanz P, Carlson M, Haystead TA. 1999. Reg1p targets protein phosphatase 1 to dephosphorylate hexokinase II in Saccharomyces cerevisiae: characterizing the effects of a phosphatase subunit on the yeast proteome. EMBO J 18:4157–4168. doi: 10.1093/emboj/18.15.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanz P, Alms GR, Haystead TA, Carlson M. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol 20:1321–1328. doi: 10.1128/MCB.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dombek KM, Voronkova V, Raney A, Young ET. 1999. Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol Cell Biol 19:6029–6040. doi: 10.1128/MCB.19.9.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young ET, Dombek KM, Tachibana C, Ideker T. 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem 278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- 53.Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 54.Wolfe KH. 2015. Origin of the yeast whole-genome duplication. PLoS Biol 13:e1002221. doi: 10.1371/journal.pbio.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuepfer L, Sauer U, Blank LM. 2005. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15:1421–1430. doi: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conant GC, Wolfe KH. 2007. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol 3:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lutfiyya LL, Iyer VR, DeRisi J, DeVit MJ, Brown PO, Johnston M. 1998. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150:1377–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicastro R, Tripodi F, Gaggini M, Castoldi A, Reghellin V, Nonnis S, Tedeschi G, Coccetti P. 2015. Snf1 phosphorylates adenylate cyclase and negatively regulates protein kinase A-dependent transcription in Saccharomyces cerevisiae. J Biol Chem 290:24715–24726. doi: 10.1074/jbc.M115.658005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hughes Hallett JE, Luo X, Capaldi AP. 2015. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 4:e09181. doi: 10.7554/eLife.09181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes Hallett JE, Luo X, Capaldi AP. 2014. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 198:773–786. doi: 10.1534/genetics.114.168369. [DOI] [PMC free article] [PubMed] [Google Scholar]