ABSTRACT
Although all 12 subtypes of human interferon alpha (IFN-α) bind the same receptor, recent results have demonstrated that they elicit unique host responses and display distinct efficacies in the control of different viral infections. The IFN-α2 subtype is currently in HIV-1 clinical trials, but it has not consistently reduced viral loads in HIV-1 patients and is not the most effective subtype against HIV-1 in vitro. We now demonstrate in humanized mice that, when delivered at the same high clinical dose, the human IFN-α14 subtype has very potent anti-HIV-1 activity whereas IFN-α2 does not. In both postexposure prophylaxis and treatment of acute infections, IFN-α14, but not IFN-α2, significantly suppressed HIV-1 replication and proviral loads. Furthermore, HIV-1-induced immune hyperactivation, which is a prognosticator of disease progression, was reduced by IFN-α14 but not IFN-α2. Whereas ineffective IFN-α2 therapy was associated with CD8+ T cell activation, successful IFN-α14 therapy was associated with increased intrinsic and innate immunity, including significantly higher induction of tetherin and MX2, increased APOBEC3G signature mutations in HIV-1 proviral DNA, and higher frequencies of TRAIL+ NK cells. These results identify IFN-α14 as a potent new therapeutic that operates via mechanisms distinct from those of antiretroviral drugs. The ability of IFN-α14 to reduce both viremia and proviral loads in vivo suggests that it has strong potential as a component of a cure strategy for HIV-1 infections. The broad implication of these results is that the antiviral efficacy of each individual IFN-α subtype should be evaluated against the specific virus being treated.
IMPORTANCE The naturally occurring antiviral protein IFN-α2 is used to treat hepatitis viruses but has proven rather ineffective against HIV in comparison to triple therapy with the antiretroviral (ARV) drugs. Although ARVs suppress the replication of HIV, they fail to completely clear infections. Since IFN-α acts by different mechanism than ARVs and has been shown to reduce HIV proviral loads, clinical trials are under way to test whether IFN-α2 combined with ARVs might eradicate HIV-1 infections. IFN-α is actually a family of 12 distinct proteins, and each IFN-α subtype has different efficacies toward different viruses. Here, we use mice that contain a human immune system, so they can be infected with HIV. With this model, we demonstrate that while IFN-α2 is only weakly effective against HIV, IFN-α14 is extremely potent. This discovery identifies IFN-α14 as a more powerful IFN-α subtype for use in combination therapy trials aimed toward an HIV cure.
INTRODUCTION
In 1957, Isaacs and Lindenmann discovered the potent antiviral effects of type I interferons (IFNs), and when C. Weissmann succeeded in cloning IFN genes into bacterial vectors in 1979, the scientific community believed that IFN would be an effective treatment for most viral infections. Thirty-five years later, only two viral infections, hepatitis B (HBV) and C (HCV) virus infections, are treated with interferon alpha (IFN-α). Interestingly, human IFN-α is not a single entity, but rather, the products of a multigene family encoding 12 IFN-α subtypes (1), all of which bind to the IFN-α/β receptor. Each subtype binds the receptor using distinctive contacts (2), thereby eliciting distinct signaling events (3, 4) and variable biological outcomes (5). While the unique affinity of the subtypes for each receptor subunit seems to contribute to differential downstream signaling, there appear to be additional factors influencing the unique outcomes elicited by individual subtypes that are currently not fully understood. Additionally, evolutionary selection of multiple functional IFN-α subtypes indicates that each subtype has essential and nonredundant functions (6). Evidence of this includes studies in the Friend retrovirus (FV) model, which showed that treatment with distinct IFN-α subtypes induced specific downstream responses that effectively controlled FV infections, whereas other subtypes induced responses that were ineffective (7, 8). Furthermore, murine herpesvirus or influenza virus infections were controlled by different subtypes than those inhibiting FV (9–13). Recent in vitro work demonstrated not only differential relative expression of IFN-α subtypes in human plasmacytoid dendritic cells (pDCs) after human immunodeficiency virus type 1 (HIV-1) exposure, but also variable HIV-1 antiviral potencies of IFN-α subtypes in gut lamina propria mononuclear cell (LPMC) cultures (14). Thus, effective antiviral therapy with IFN-α is quite specific to both the IFN-α subtype and the virus. This raises the critical question of whether the IFN-α2 subtype is the most efficacious subtype to be using against HIV-1 therapeutically.
IFN-α is rapidly induced and secreted by cells to limit virus replication through numerous and varied mechanisms, including the induction of intrinsic restriction factors (15–18) and the activation of innate and adaptive immune responders. In vitro studies with pDCs, which are the primary producers of IFN-α, indicated that the predominant IFN-α subtypes produced by pDCs in response to HIV-1 (IFN-α1, -2, and -5) have only weak anti-HIV-1 activities (14). Thus, HIV-1 predominantly induces IFN-α subtypes that are ineffective against HIV-1 in vitro. Analyses of IFN-α subtype profiles from peripheral blood mononuclear cells (PBMCs) collected from HIV-1-infected patients during different stages of disease also revealed that some of the subtypes shown to be most potent against HIV-1 in vitro are not produced until late-stage disease, when the immune system may be too damaged to respond effectively (14, 19). Thus, it is possible that the IFN-α subtypes most protective against HIV-1 are not induced in a timely manner and that therapeutic delivery of a more efficacious subtype(s) may be beneficial. This approach was previously successful in the FV mouse retrovirus model (8). One hesitation in using IFN-α as a therapeutic is that plasma IFN-α levels in HIV-1 patients have been shown to correlate with pathogenic immune activation (20) and possibly induction of apoptosis in CD4+ T cells (21). However, whether all IFN-α subtypes are associated with this chronic activation has not been delineated, and while studies suggest that there is dysregulation of the IFN-α response in pathogenic HIV/simian immunodeficiency virus (SIV) infections, a direct role for IFN-α in mediating chronic activation and disease progression has yet to be definitively proven (reviewed in reference 22).
Until now, IFN-α therapy in humans has focused almost exclusively on IFN-α2. While IFN-α2 is efficacious in the treatment of HBV and HCV, HIV-1 studies have shown much less promise (23–34). However, encouraging results from recent clinical studies (23, 26, 35) and new discoveries demonstrating IFN-α induction of antiretroviral restriction factors have led to a renewed interest in IFN-α therapy. IFN-α and antiretroviral (ARV) drugs work via different mechanisms, and IFN-α's unique capacity to reduce the proviral load combined with ARV therapy could lead to a functional cure for HIV-1. To this end, new IFN-α2 clinical trials have recently been approved. However, given the recently discovered widely divergent anti-HIV-1 activities of different IFN-α subtypes in vitro, we believe that a key to the success of such therapy is the identification of the IFN-α subtype that has the most potent anti-HIV-1 activity in vivo. These studies are now necessary to assess the complex immunomodulatory effects of different IFN-α subtypes that cannot be analyzed in vitro. Since the interactions between IFNs and their receptors are relatively species specific (36), and since the highly variable antiviral efficacies of IFN-α subtypes in disparate viral systems are now well recognized, it is critical to test the effects of human IFN-α subtypes in a human system infected with HIV-1. At this time, the only practical model to test human IFN-α subtypes against HIV-1 in vivo is the humanized mouse. We used triple-knockout bone marrow-liver-thymus (TKO-BLT) mice reconstituted with a human immune system. These mice have multilineage human hematopoietic cell reconstitution, including T cells, B cells, NK cells, dendritic cells, and monocytes/macrophages; they are susceptible to HIV infection; and they are able to mount HIV-specific B cell and T cell responses (37). All of the lymphocytes in these mice are of human origin, and the mice support and recapitulate the hallmarks of a bona fide human HIV-1 infection (37, 38). Since in vitro studies indicated that IFN-α14 was extremely potent against HIV-1 in comparison to the clinically approved IFN-α2, we compared the two subtypes in vivo.
MATERIALS AND METHODS
Ethics statement.
Fetal tissues for reconstitution of humanized mice were obtained through anonymous donations with informed written consent via Advanced Bioscience Resources. This research was conducted under NIH Office of Human Subjects Research Exemption 4980. All animal studies were performed under an animal study proposal approved by the Rocky Mountain Laboratories, NIAID, NIH Animal Care and Use Committee (number 2012-61) following all regulations and guidelines of the Public Health Service's Office of Laboratory Animal Welfare. Rocky Mountain Laboratories is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Humanized TKO-BLT mice.
C57BL/6 Rag2−/− γc−/− CD47−/− (TKO) mice were humanized using the BLT method, as previously described (37, 38). Briefly, 6- to 10-week-old mice received 5.0 Gy whole-body irradiation prior to transplantation of 17- to 22-week-gestation human thymus and liver under the kidney capsule, followed by intravenous injection of autologous liver-derived CD34+ hematopoietic progenitor cells. The animals in this study were housed under specific-pathogen-free conditions.
Recombinant IFN-α subtypes.
Human IFN-α subtype genes were optimized for expression in Escherichia coli. Isolated inclusion body proteins denatured with guanidine hydrochloride were refolded in arginine refolding buffer and purified by anion-exchange and size exclusion chromatography (39). Protein concentrations were determined using NanoDrop 2000c (Thermo Scientific, Wilmington, DE), and endotoxin levels were less than 0.0025 endotoxin units (EU)/ml (ToxinSensor; Genscript, Piscataway, NJ). These laboratory-produced proteins were used for all experiments except those shown in Fig. 1D. For the experiments in Fig. 1D, the IFN-α proteins were purchased from PBL Assay Science (Piscataway, NJ) as indicated below.
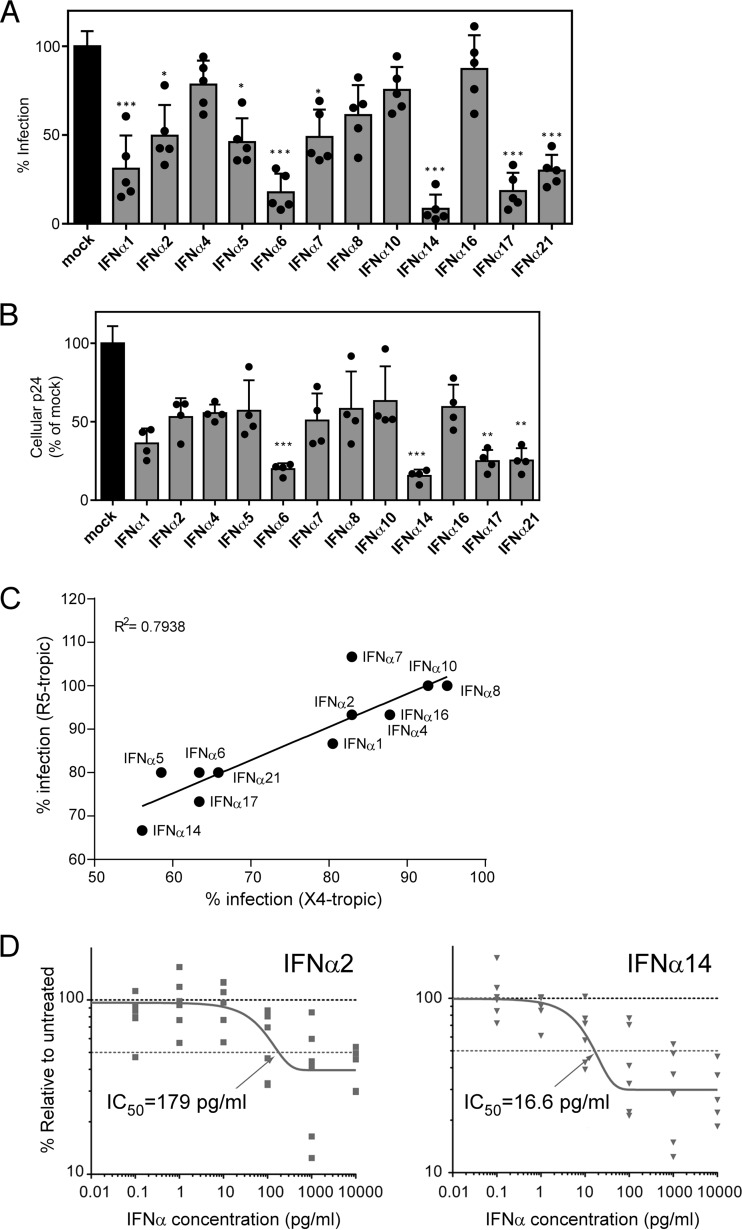
FIG 1.
Inhibition of HIV-1 replication by IFN-α subtypes in vitro. (A and B) The infectivity of supernatants harvested from PBMCs exposed to X4-tropic HIV-1NL4-3_IRES_eGFP in the presence of different human IFN-α subtypes for 2 days was determined. (A) The supernatants were incubated with TZM-bl reporter cells, and β-galactosidase activity was compared to that of untreated supernatants after 3 days of incubation. (B) Additionally, the cells were lysed, and the cellular p24 protein content was analyzed by ELISA. Five healthy donors were used for the IFN-α inhibition assay, and the TZM-bl assay was measured in triplicate. Mean values plus standard errors of the mean (SEM) are shown, and individual donors are represented by dots. ***, P < 0.001; **, P < 0.01; *, P < 0.05; determined by one-way analysis of variance (ANOVA) with Dunnett's posttest. (C) The infectivities of IFN-α-treated X4- and R5-tropic viruses relative to untreated PBMCs were determined and compared using a Pearson correlation test. (D) Dose-response inhibition in mucosal immune cells. LPMCs (n = 6 donors) were infected with HIV-1BaL, resuspended with various doses of IFN-α2 and IFN-α14 (or mock infected), and evaluated for intracellular Gag p24+ expression in CD3+ CD8− cells at 4 dpi by flow cytometry. The data were normalized against mock infection for each donor. The dose-response curves were generated using a one-phase decay equation that was also used to evaluate the IC50.
Determination of IFN-α units.
Because the standard biological method to quantify interferons is with antiviral assays, we were concerned that the differential antiviral effects of the various interferon subtypes might produce aberrant results. Therefore, we developed a stable reporter cell line using human retinal pigment epithelial cells (ATCC CRL2302) transfected with a plasmid containing interferon-stimulated response element (ISRE) promoter/enhancer elements driving a luciferase reporter gene (pISRE; Stratagene) (40). Cells were grown for 24 h before adding serial dilutions of our recombinant IFN-α subtypes and commercially available IFN-α subtypes (PBL Assay Science, Piscataway, NJ) for 4 h. The cells were lysed with Bright Glo lysis buffer (Promega), and luciferase activity was measured 10 min later. Six experiments were done comparing the stated activities of commercially available IFN-α subtypes (PBL Assay Science, Piscataway, NJ) with relative light units (RLU) obtained from our ISRE assay; 500 U/ml (PBL units) corresponded to a mean of 7,610 RLU for IFN-α2 and 7,182 RLU for IFN-α14. The RLU for the IFN-α subtypes we synthesized were then converted to PBL units for the in vivo experiments. All the units given in the text correspond to PBL units. PBL determines the activities of interferons using a cytopathic inhibition assay on bovine kidney cells (MDBK) with vesicular stomatis virus (VSV) and on the human lung carcinoma cell line A549 with encephalomyocarditis virus (EMCV).
In vitro viral inhibition assay.
Coisogenic X4- and R5-tropic HIV-1 stocks were produced by transfection of HEK293T cells with X4- HIV-1NL4-3_IRES_eGFP and R5- HIV-1NL4-3_92TH014.12_IRES_eGFP, respectively. R5- HIV-1NL4-3_92TH014.12_IRES_eGFP contains an exchanged R5-tropic env V3 loop, as described previously (41, 42). One million phytohemagglutinin (PHA) (1 μg/ml)- and interleukin 2 (IL-2) (10 ng/ml)-stimulated PBMCs from 5 healthy donors were incubated with 350 μl of virus containing 250 ng of p24 antigen, with or without IFN-α subtypes (10 ng/ml), for 6 h at 37°C. The cells were washed twice with 1 ml of phosphate-buffered saline (PBS) and resuspended in complete RPMI 1640 medium supplemented with IL-2 (10 ng/ml) and IFN-α subtypes for 2 days at 37°C. The level of infectious HIV-1 in the supernatant was determined by TZM-bl assay 3 days postinfection in triplicate. Pelleted PBMCs were lysed before measuring cell-associated p24 by enzyme-linked immunosorbent assay (ELISA). Briefly, Nunc Immuno Maxi Sorb surface 96-well plates were coated with a mouse anti-p24 monoclonal antibody (MAK183; EXBIO) overnight. After blocking, Triton X-100-lysed supernatants or cells were transferred to the 96-well plates. The next day, the plates were washed and incubated with a polyclonal rabbit anti-HIV-1 p24 antibody (Eurogentec) for 1 h. Next, the plates were washed and incubated with a goat anti-rabbit antibody conjugated with horseradish peroxidase (Dianova; 111-035-008), followed by the addition of tetramethylbenzidine (TMB) peroxidase substrate. The reaction was stopped with 0.5 M H2SO4. The absorbance of each microplate well was determined using a microplate reader and calibrated against the absorbance of an HIV-1 p24 antigen standard or standard curve.
Dose-response curves in LPMCs.
Macroscopically normal human jejunum tissues that would otherwise be discarded were obtained from patients undergoing elective abdominal surgery. The patients signed a release form for the unrestricted use of these discarded tissues, and all protected information was deidentified to laboratory personnel. The protocol was given exempt research status by the Colorado Multiple Institutional Review Board at the University of Colorado. Human gut LPMCs from 6 different donors (each in duplicate) were obtained following tissue disaggregation and collagenase digestion (43, 44). The LPMCs were spinoculated with HIV-1BaL at 10 ng p24/106 cells (AIDS Research Reagent Program; catalog number 4984) and resuspended in different IFN-α2 or IFN-α14 concentrations, and the percentage of p24+ cells in the CD3+ CD8− cell fraction was evaluated at 4 days postinfection (dpi) by flow cytometry, as previously described (14). The recombinant IFN-α2 and IFN-α14 proteins were purchased from PBL Assay Science (catalog number 11002-1). Data were normalized against the mock-treated control for each donor, set as 100%. Normalized data from all 6 donors were averaged, and the 50% inhibitory concentration (IC50) and Vres (residual replication at maximal IFN-α concentrations) (45) were calculated using a one-phase decay equation in Prism 5.0.
HIV-1 challenge and IFN-α treatment.
HIV-1JR-CSF stocks were prepared by transfection of 293FT cells (Invitrogen, Grand Island, NY) with clone pYK-JRCSF, obtained from Irvin S. Y. Chen and Yoshio Koyanagi through the NIH AIDS Research and Reference Reagent Program. TZM-bl reporter cells (NIH AIDS Research and Reference Reagent Program, from John Kappes, Xiaoyun Wu, and Tranzyme Inc.) were used to determine stock concentrations. Experimental-group sizes were chosen to ensure adequate statistical power. Mice were assigned to groups to control as closely as possible for gender and the level of human reconstitution and for the level of p24 antigenemia at 5 weeks postinfection. The mice were infected intraperitoneally with 10,000 tissue culture infectious units (TCIU) of HIV-1JR-CSF. IFN-α subtypes (1.5 × 105 U/mouse) were injected either within 2 h of infection or after 5 weeks of infection and subsequently at 24-h intervals for 10 days. The interferon dose for treating the mice (0.14 million international units [MIU]/m2) was based on the dose of IFN-α2b used to treat humans for melanoma (20 MIU/m2) (46, 47) normalized to the body surface area (48).
Isolation of plasma and human leukocytes.
Blood samples were collected in EDTA and centrifuged to obtain plasma. Splenocytes were obtained by passage through 70-μm filters, followed by blood cell lysis with ACK (NH4Cl, 0.15 M; KHCO3, 10 mM; EDTA, 0.1 M). CD4+ splenocytes used for proviral quantification were preenriched using positive selection (Miltenyi, San Diego, CA).
Flow cytometry.
Splenocytes were analyzed by staining using CCR7-phycoerythrin (PE)-Cy7, Ki67-eFluor 488, CD107a-fluorescein isothiocyanate (FITC), CD45-V500, and CD3-V450 (BD Biosciences, San Jose, CA); HLA-DR and CD56-A700 Alexa Fluor 700 (Biolegend, San Diego, CA); CD45RA-PE, CD38-PE, CD4-PE-Cy7, CD8-allophycocyanin (APC)-eFluor 780, Lin (CD3, CD14, and CD19) peridinin chlorophyll protein (PerCP)-Cy5.5, and TRAIL-PE (eBioscience, San Diego, CA); and Granzyme B-PerCP (R&D Systems, Minneapolis, MN).
Quantification of plasma viremia, plasma cytokines, and proviral burden.
HIV-1 p24 levels in plasma were determined by ELISA (Advanced Bioscience Laboratories, Rockville, MD). Plasma cytokine levels were quantified using a Bio-Plex cytokine assay specific for human cytokines on the Bioplex-200 system. Analyses were done using Bio-Plex Manager software v6.1.1 (Bio-Rad, Hercules, CA).
HIV-1 RNA and HIV-1 DNA detection.
Plasma HIV-1 RNA was measured by using the Abbott RealTime HIV-1 assay on the m2000 system. The detection limit for an input volume of 200 μl plasma was 150 copies/ml. Genomic DNA was isolated with the QIAamp DNA minikit (Qiagen). Determination of proviral HIV-1 DNA was performed by quantitative real-time PCR on a per cell basis as detailed previously (49), using lysate from 5 × 106 CD4-enriched splenocytes.
Real-time PCR for antiretroviral interferon-stimulated genes (ISGs).
RNA was isolated from 5 × 106 splenocytes using the ZR-Duet DNA/RNA MiniPrep kit (Zymo Research, Irvine, CA). Real-time PCR analysis of mRNA expression was completed as previously described (50). Primers were tested on controls to ensure amplification of a single product. Data for each sample were calculated as the percent difference in the threshold cycle (CT) value (ΔCT = CT GAPDH − CT gene). Gene expression was plotted as a percentage of gene expression relative to that of GAPDH for each sample.
Mutation analysis of proviral HIV-1 DNA.
Genomic DNA was extracted from lymph node samples using a Qiagen DNAEasy kit. Amplification of the gp41/nef region was performed by nested PCR using Phusion Taq (New England BioLabs). Following preamplification, nested PCR was performed using Illumina MiSeq-configured primers. Amplicons were sequenced by Illumina MiSeq as previously described (51). Sequences with >80% identity to the JR-CSF sequence (GenBank accession number M38429) were analyzed. Total mutations (including gaps) and GG→AG mutations were evaluated using custom Perl scripts (52).
Statistics.
Statistical calculations were performed with GraphPad Prism (GraphPad Software, La Jolla, CA) using the tests specified in the figure legends.
RESULTS
Antiviral activities of IFN-α subtypes on HIV-1 replication in PBMCs.
A recent in vitro study using human gut LPMCs showed that the antiviral effects of different IFN-α subtypes on HIV-1 suppression were not equal (14). To assess and confirm the best IFN-α subtypes to follow up with experiments in humanized mice, we conducted an in vitro screening assay. Activated PBMCs from five different healthy human donors were infected with HIV-1 in the presence or absence of 10 ng/ml of each of the 12 human IFN-α subtypes. HIV-1 infectivity in supernatants was measured 2 days later using a reporter cell line (Fig. 1A). Analysis of cell-associated HIV-1 was also assessed by p24 ELISA (Fig. 1B). The two assays gave similar results in that IFN-α2 only moderately suppressed HIV-1 compared to IFN-α14, which exhibited the most potent anti-HIV-1 activity of all the subtypes, a result that was consistent with the recent findings in LPMC cultures (14). Levels of suppression by the different subtypes were equivalent for both X4 and R5 viruses (Fig. 1C).
The relative potencies of commercially obtained IFN-α2 and IFN-α14 (PBL Assay Science) were next tested in a dose-response experiment using LPMCs infected with HIV-1BaL. The aggregate data from six different LPMC donors are shown in Fig. 1D. IFN-α14 was 10.8-fold more potent (in picograms per milliliter), 13.6-fold more potent (in units per milliliter), and 11.0-fold more potent (nanomolar) than IFN-α2. IFN-α14 also inhibited HIV-1 with greater potency than IFN-α2 at maximal IFN-α concentrations, as there was more residual HIV-1 replication (Vres) (45) in HIV-1-infected LPMCs treated with IFN-α2 (Vres = 40%) versus IFN-α14 (Vres = 30%). Based on these results, IFN-α14 was selected for in vivo studies in HIV-1-infected humanized mice using the clinically approved IFN-α2 subtype as the comparison control.
IFN-α14-mediated suppression of HIV-1 replication in humanized mice.
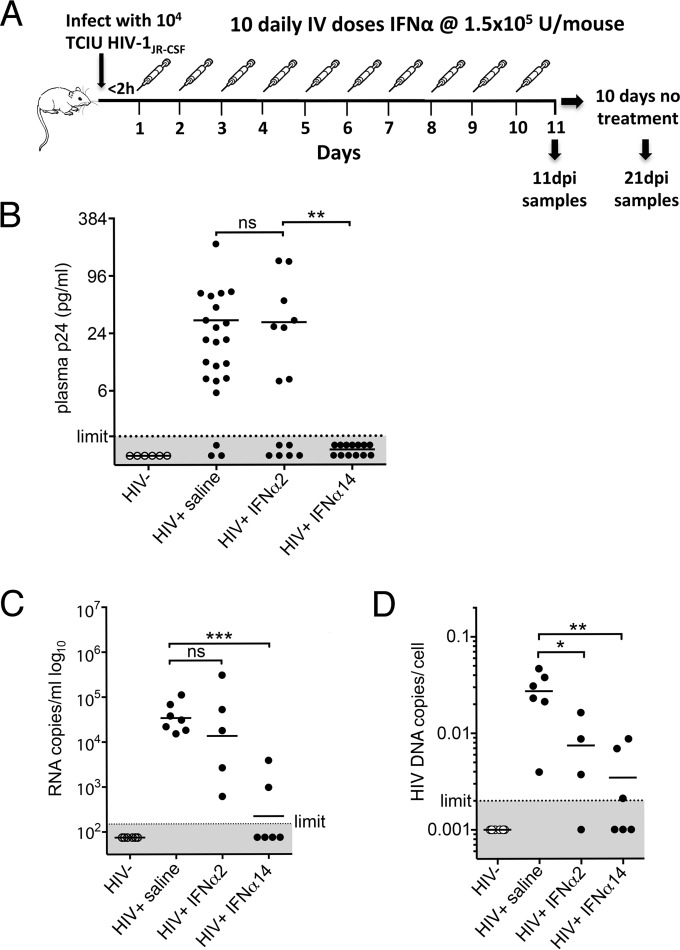
Recently described humanized TKO-BLT mice were used as the model for HIV-1 infection using R5-tropic HIV-1JR-CSF. TKO-BLT mice develop high levels of multilineage human hematopoietic cells with lymphoid tissues of human origin, are susceptible to HIV-1 infection, and recapitulate the classic hallmarks of human HIV-1 infection, including persistent infection and CD4+ T cell depletion (37, 38). To quantify the biological activities of our recombinantly produced IFN-α subtypes, we used an ISRE reporter assay normalized to commercially available IFN-α2 and IFN-α14. Since the dosage of IFN-α that can be used clinically is limited by undesirable side effects (53), we chose a high unit dose of IFN-α2 that is used to treat melanoma patients (46) (converted to the mouse equivalent [48]) to demonstrate the maximal efficacy that would be clinically achievable by IFN-α2 compared to the same unit dose of IFN-α14. To simulate postexposure prophylaxis, IFN-α was administered within 2 h of HIV-1 infection and continued daily for 10 days. The mice were euthanized at 11 dpi for analysis or were left untreated for an additional 10 days (to 21 dpi) to assess for viral rebound (Fig. 2A). Plasma samples from two separate cohorts of TKO-BLT mice reconstituted from separate human donors were tested for HIV-1 p24 at 11 dpi (1 day after the final IFN-α treatment). By chi-square analysis, there were significantly more p24-negative animals in the IFN-α14 group than in the IFN-α2 group (P = 0.0019). There was also a higher proportion of p24-negative animals in the IFN-α2 group than in the untreated controls (P = 0.0475), but the P value is above the Bonferroni-corrected statistical significance level (a P value of 0.025) required for multiple-comparison analyses. However, it is quite possible that larger group sizes would show that IFN-α2 had a significant effect. Regardless, the number of HIV p24-negative animals in the IFN-α14 group was significantly greater than in the IFN-α2 group by chi-square analysis (Fig. 2B).
FIG 2.
IFN-α14 suppression of HIV-1 replication in vivo. (A) The in vivo experimental design consisted of intraperitoneal infection of TKO-BLT mice with 104 TCIU of HIV-1JR-CSF, followed by intravenous administration of either IFN-α2, IFN-α14, or mock saline within 2 h of infection. Treatment was administered daily for 10 consecutive days, followed by sample collection either 24 h after the final injection (11 dpi) or after an additional 10-day period of no treatment (21 dpi). (B) HIV-1 p24 levels detected by ELISA in plasma collected from HIV-1JR-CSF-infected TKO-BLT mice 24 h after the final IFN treatment (11 dpi). The dots represent individual mice reconstituted from three separate human donors. The results comparing controls to IFN-α2 and IFN-α2 to IFN-α14 (detectable p24 versus undetectable p24) were analyzed by chi-square analysis with a Bonferroni correction for multiple comparisons (**, P = 0.0019; ns, not significant). (C) HIV-1 RNA copies per milliliter of plasma at 11 dpi, detected by quantitative PCR (qPCR). (D) HIV-1 proviral copies detected in CD4 cell-enriched TKO-BLT splenocytes at 11 dpi. One of the samples from the IFN-α2 group did not provide data, as the PCR was inhibited. (B to D) The horizontal lines denote means. (C and D) Statistical analyses were done by one-way ANOVA with Dunnett's posttest for multiple comparisons. ***, P < 0.001; **, P < 0.01; *, P < 0.05. Each dot represents a separate mouse.
A third cohort was infected and treated in the same manner so that sufficient plasma from a terminal bleed could be obtained to test for viremia using a highly sensitive RNA PCR assay. HIV-1 RNA levels were lower than the detection limit of 150 copies/ml of plasma in four of six mice treated with IFN-α14 and low in the other two (Fig. 2C). The mean reduction in HIV-1 RNA compared to the mock-treated controls was 154-fold. In contrast, treatment with IFN-α2 provided only a 2.5-fold reduction in plasma viral RNA levels. HIV-1 proviral loads in human CD4+ T cells isolated from spleens by magnetic-bead sorting were also analyzed. Interestingly, IFN-α2 treatment significantly reduced provirus levels in spleen cells, although the reduction (3.7-fold) was not as strong as with IFN-α14 treatment (7.9-fold) (Fig. 2D). Thus, a brief postexposure prophylaxis therapy with IFN-α14 effectively reduced both viremia and splenic proviral loads. In contrast, high-dose administration of IFN-α2 reduced proviral loads but had little effect on viremia.
HIV-1 relapse following cessation of therapy.
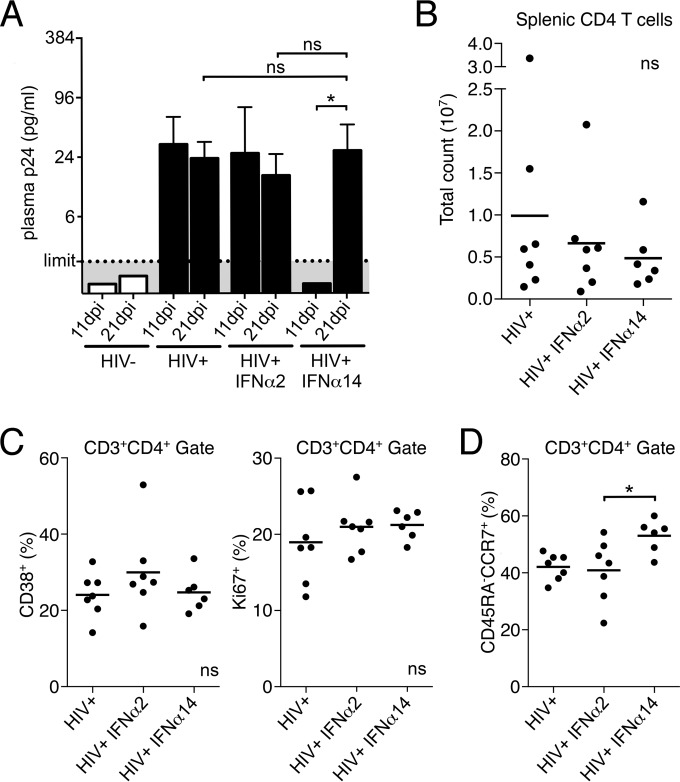
The undetectable viral RNA and provirus in some IFN-α14-treated animals suggested that treatment might have prevented establishment of latent infection. To examine that possibility, mice were treated as before and treatments were discontinued at day 10. The mice were then rested until 21 dpi, at which point they were tested for viral rebound. Even though all IFN-α14-treated mice were negative for HIV-1 p24 at 11 dpi, they all rebounded to the levels of untreated mice following 10 days without therapy (Fig. 3A). The results from a recent rhesus macaque study that treated SIV infection with human IFN-α2 suggested that IFN-α therapy might exacerbate aspects of the disease, such as T cell activation and depletion (54). However, examination of the HIV-1-infected humanized mice treated with human IFN-α showed similar CD4+ T cell counts (Fig. 3B) and levels of CD4+ T cell activation and proliferation (Fig. 3C) after viral rebound. Of note, the IFN-α14 group had significantly better preservation of CD4+ central memory T cells at 21 dpi (Fig. 3D). Thus, 10 days of IFN-α14 therapy did not clear infectious virus, but it did not increase CD4+ T cell depletion, and the central memory compartment was better maintained than in HIV-1-infected control mice. Thus, human IFN-α therapy in HIV-1-infected humanized mice did not exacerbate negative sequelae but instead preserved critical immune cell subsets by suppressing viral replication. Furthermore, the superior suppression of HIV-1 by IFN-α14 during the time of therapy suggested that it would be a more efficacious subtype than IFN-α2 for the treatment of HIV-1 infections.
FIG 3.
Cessation of IFN-α14 treatment. (A) HIV-1 p24 levels (means plus standard deviations [SD]) in plasma of mice 24 h after the final IFN injection (11 dpi) and after 10 additional days of no treatment (21 dpi). Limit, limit of detection within each assay. HIV−, n = 7; HIV+, n = 7; HIV+ IFN-α2, n = 7; HIV+ IFN-α14, n = 6. Paired t tests between 11 and 21 dpi were done for each treatment group; one-way ANOVA with Tukey's posttest was used to compare treatment groups at 21 dpi. (B) CD4+ T cell counts in spleens of mice at 21 dpi. (C and D) Frequencies of activated (CD38+) and proliferating (Ki67+) (C) and central memory (CD45RA− CCR7+) (D) CD4+ T cells at 21 dpi. The horizontal lines denote means. ns, not significant; *, P < 0.05 (one-way ANOVA with Tukey's posttest). Each dot represents a separate mouse.
IFN-α14 suppresses acute HIV-1 infection in humanized mice.
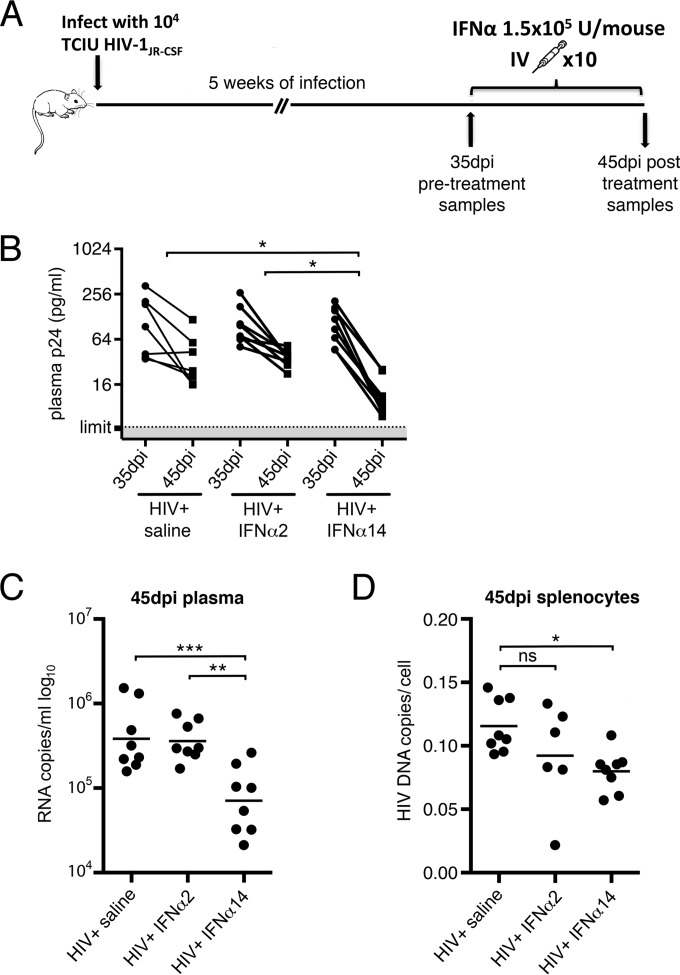
To investigate the efficacy of IFN-α14 in reducing acute HIV-1 infections, humanized mice were again infected with HIV-1JR-CSF, and the infections were allowed to progress for 5 weeks prior to initiation of treatment with either IFN-α2 or IFN-α14 (Fig. 4A). The mice were then tested for p24 levels and placed in treatment groups so that the mean plasma p24 levels of all the groups before the initiation of therapy were closely equivalent (Fig. 4B, 35 dpi). Again, the mice were treated daily for 10 days, followed by analysis the following day. HIV-1 plasma viremia was still in the acute phase at the start of IFN-α treatment, so plasma p24 antigen levels dropped over the next 10 days, even in mock-treated mice (Fig. 4B). Ten days of therapy with IFN-α2 did not produce a statistically significant reduction in p24 compared to mock-treated mice (Fig. 4B). In contrast, mice treated with IFN-α14 had 3.4-fold-lower p24 levels than mock-treated mice and 3-fold-lower levels than the IFN-α2-treated mice (Fig. 4B). Using the more sensitive plasma RNA quantification by PCR at the termination of the experiment, the mean plasma HIV-1 RNA levels were 5-fold (0.704 log10) lower in the IFN-α14 group than in the IFN-α2 group (Fig. 4C). IFN-α is unique among commonly prescribed antiretroviral drugs in its ability to reduce HIV-1 proviral loads later in infection (26, 35) and therefore is considered a promising candidate to be used in cure strategies. Indeed, analysis of proviral loads in human CD4+ T cells isolated from spleens of mice treated in acute infection revealed 30% lower proviral loads in the IFN-α14 group than in controls (Fig. 4D). Treatment with IFN-α2 reduced proviral loads by 20%, but that reduction did not reach the P value of 0.05 for statistical significance. Thus, IFN-α14 suppressed both plasma HIV-1 loads and levels of cellular provirus even during acute infection.
FIG 4.
IFN-α14 suppression of established HIV-1 infection. (A) The in vivo experimental design consisted of intraperitoneal infection of TKO-BLT mice with 104 TCIU of HIV-1JR-CSF, followed by 5 weeks of infection. At 35 dpi, plasma p24 levels were determined and the mice were assigned to similarly infected treatment groups for intravenous (IV) administration of IFN-α2, IFN-α14, or mock saline. Treatment was administered daily for 10 consecutive days, followed by sample collection 24 h after the final injection (45 dpi). (B) Levels of HIV-1 antigenemia as determined by p24 ELISA at the start (35 dpi) and 24 h after the final IFN injection (45 dpi). The percent reduction in plasma p24 was used to determine significance by one-way ANOVA with Tukey's posttest. (C) HIV-1 RNA copies per milliliter of plasma at 45 dpi, detected by qPCR. (D) HIV-1 proviral copies detected in CD4 cell-enriched TKO-BLT splenocytes at 45 dpi. The horizontal lines denote means. Statistical analyses were done by one-way ANOVA with Tukey's posttest. ns, not significant; ***, P < 0.001; **, P < 0.01; *, P < 0.05. Each dot represents a separate mouse.
Cytokine profile after IFN-α14 therapy.
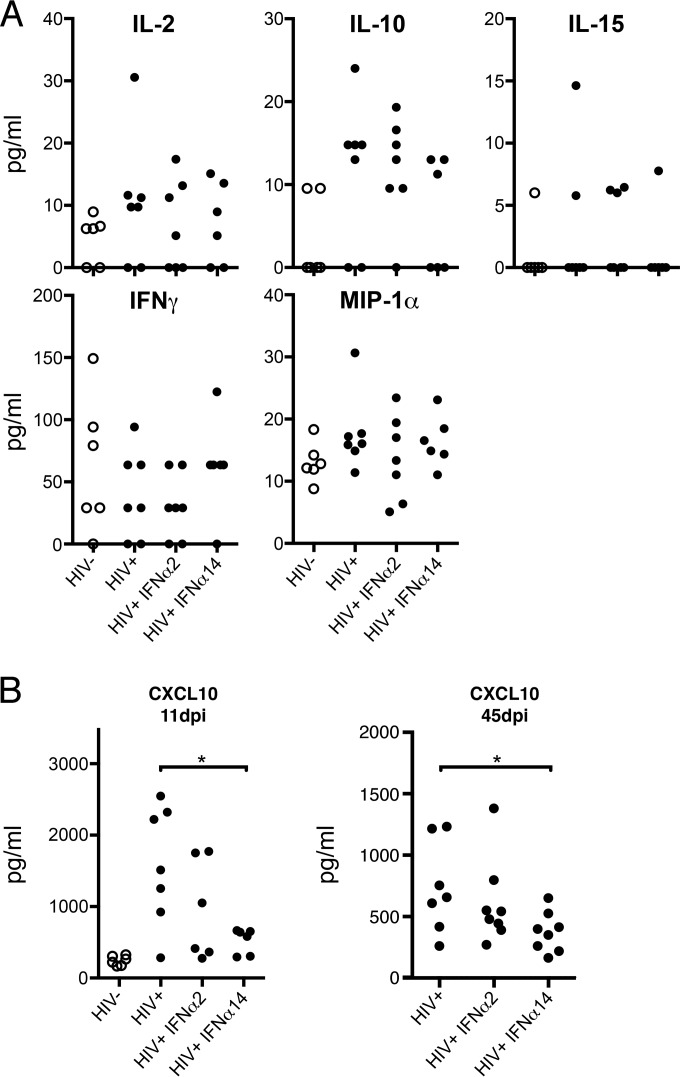
HIV-1 infection and the resultant IFN-α responses are known to induce the production of cytokines. Proinflammatory factors, such as IL-1β, tumor necrosis factor alpha (TNF-α), and CXCL10 (IP-10), appear to be detrimental to the host (55, 56), while MIP-1α is likely beneficial (57). To assess these responses in our model, we analyzed plasma samples at 11 dpi for the presence of 11 different human cytokines. The cytokines IL-1β, IL-4, IL-5, IL-6, and TNF-α were not detected above baseline levels, and no differences between treatment groups were found for IL-2, IL-10, IL-15, IFN-γ, or MIP-1α (Fig. 5A). Compared to uninfected controls, CXCL10 concentrations were similarly elevated in both IFN-α2-treated and mock-treated HIV-1-infected mice. The mice with the highest levels of p24 in plasma also had the highest levels of CXCL10 (data not shown). Thus, it appeared that HIV-1 infection induced CXCL10 rather than IFN-α2 treatments. CXCL10 levels in IFN-α14-treated mice were significantly lower than in mock-treated controls at 11 dpi (Fig. 5B). Cytokines from mice treated at 5 weeks postinfection were also analyzed. Again, there were no significant differences in plasma cytokine levels between any groups (data not shown), except for CXCL10. At 45 dpi, CXCL10 levels were generally lower than at 11 dpi and were again significantly reduced in IFN-α14-treated mice compared to controls (Fig. 5B). The combined results suggested that IFN-α14 therapy suppressed CXCL10 production either directly or through its ability to suppress HIV-1 loads.
FIG 5.
Plasma cytokine analysis. (A) Plasma was collected 24 h after the final IFN-α injection in both the acute- and established-infection experiments and assayed for human cytokine and chemokine levels using a custom 11-plex bead assay. Data from 11 days postinfection are shown. (B) Comparison of CXCL10 levels in plasma collected from mice at 11 dpi (one-way ANOVA with Dunnet's posttest; *, P < 0.05) or 45 dpi (unpaired t test; *, P < 0.05). Analyte levels were quantified in picograms per milliliter of TKO-BLT plasma using standards provided with the assay. Each dot represents a separate mouse.
Cellular immune responses in IFN-α-treated mice.
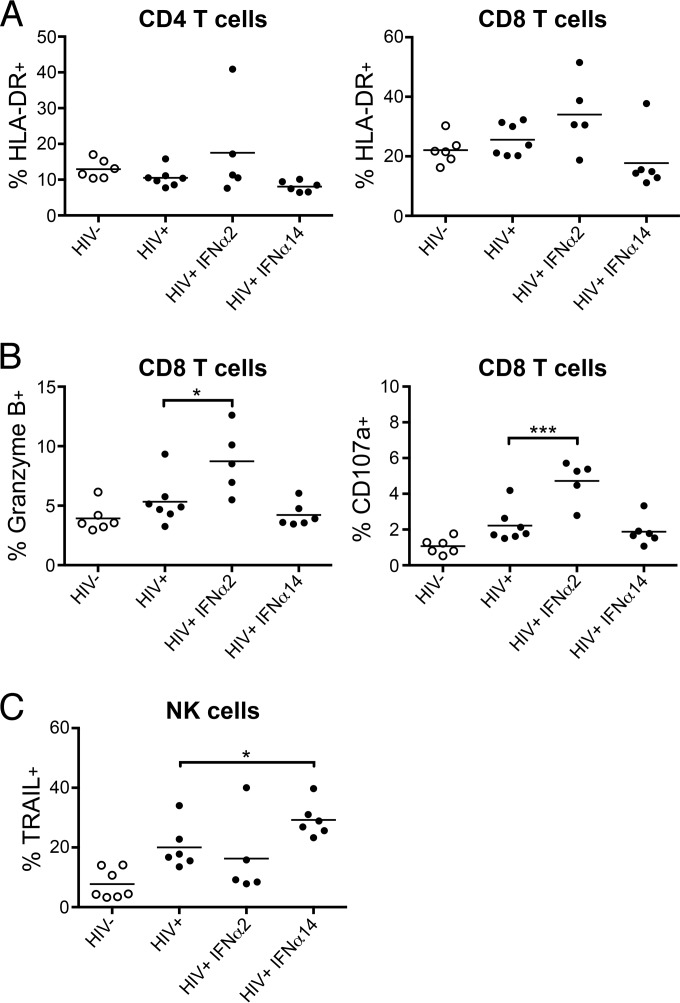
Since type I IFNs can stimulate lymphocyte responses, we tested for their effects on NK cells and T cells 1 day after termination of IFN-α therapy. HLA-DR expression is a marker of T cell activation, but we saw no increase in the frequency of HLA-DR-expressing CD4+ T cells (Fig. 6A, left). Compared to untreated controls, mice treated with IFN-α2 showed a slightly increased percentage of HLA-DR-positive CD8+ T cells (Fig. 6A, right). Although the HLA-DR increase was not statistically significant, it was associated with a significantly increased frequency of CD8+ T cells expressing two cytotoxicity-associated molecules, granzyme B and CD107a (Fig. 6B). These results are consistent with the known ability of IFN-α2 to activate CD8+ T cells (58). In contrast, IFN-α14 had no detectable effect on CD8+ T cells but was associated with significantly higher percentages of NK cells expressing the cytotoxic molecule TRAIL (Fig. 6C). Thus, treatment with different IFN-α subtypes activated unique downstream immune effectors with IFN-α2 treatment, resulting in activated CD8+ T cells, and with IFN-α14 treatment, producing more activated NK cells.
FIG 6.
IFN-α-induced changes in cell-mediated responses. Flow cytometry was used to analyze activation and functional markers on T cells and NK cells. (A) Frequency of activated (HLA-DR+) CD4 and CD8 T cells in spleens of mice from each treatment group at 11 dpi. (B) Frequency of splenic CD8+ T cells positive for cytoplasmic Granzyme B and membrane-bound CD107a at 11 dpi. Statistical analyses were done by one-way ANOVA with Dunnet's posttest. ***, P < 0.001; *, P < 0.05. (C) Percentages of splenic NK cells expressing the cytotoxic effector molecule TRAIL at 11 dpi. *, P < 0.05 (t test between HIV-1+ mock-treated and IFN-α14-treated groups). The horizontal lines denote the means. Each dot represents a separate mouse.
Subtype-specific induction of ISGs.
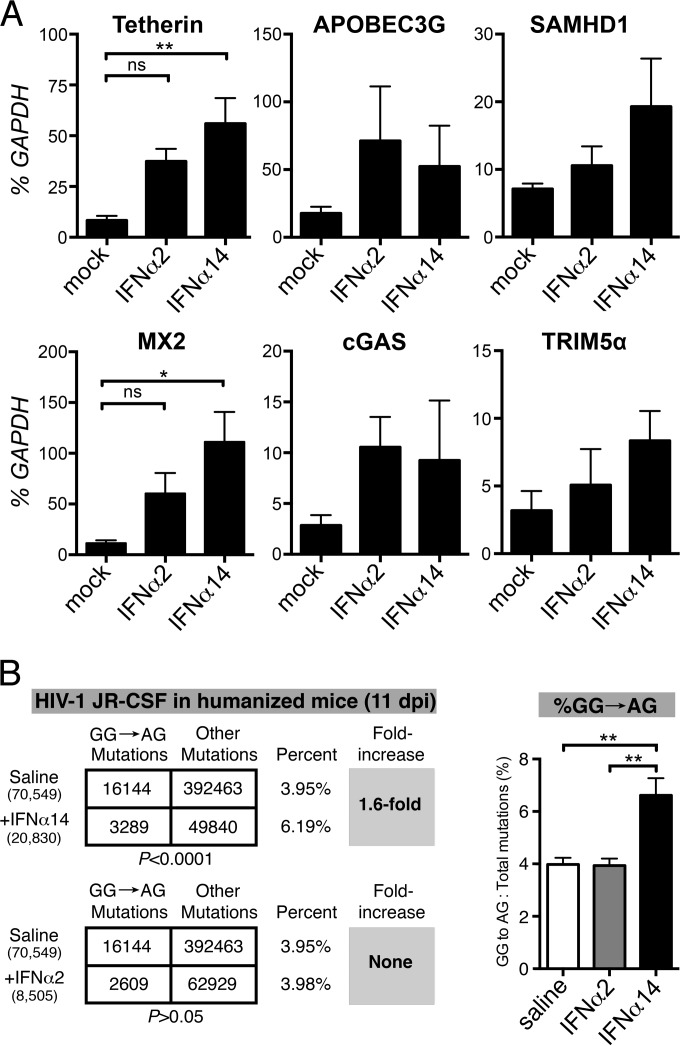
We next sought to determine if subtype efficacy might be associated with differential induction of ISGs. Since hundreds of genes are regulated by interferon (59), we focused on those previously associated with restricting HIV-1 replication. To analyze subtype-specific induction of ISG transcription, uninfected TKO-BLT mice were injected with either saline, IFN-α2, or IFN-α14, and splenocyte RNA was analyzed 6 h later. No significant upregulation of the innate viral sensor MB21D1/cGas or the viral restriction factors APOBEC3G, SAMHD1, or TRIM5α were observed (Fig. 7A). However, similar to the IFN-α subtype-specific ISG induction previously seen in vitro (14), IFN-α14, but not IFN-α2, induced statistically significant upregulation of BST2, which encodes the HIV restriction factor tetherin, and MX2, which encodes myxovirus resistance 2, also a potent inhibitor of HIV-1 (17, 18) (Fig. 7A). Thus, IFN-α14 upregulated in vivo transcription of two genes encoding proteins with demonstrated anti-HIV-1 activity.
FIG 7.
Induction of ISGs. (A) The transcription levels of six ISGs encoding HIV-1 sensors or restriction factors were determined in splenocytes harvested from HIV-1-negative TKO-BLT mice 6 h after intravenous injection with a single 1.5 × 105 U dose of the indicated IFN or mock saline control injection. RNA levels were determined by qPCR and are expressed as a percentage (means + SEM) of the human housekeeping GAPDH transcripts detected in the same sample. n = 5 mice per group. Statistical analyses were done by one-way ANOVA with Tukey's posttest. ns, not significant; **, P < 0.01; *, P < 0.05. (B) Evaluation of APOBEC3G signature mutations in IFN-α-treated mice. A 350-bp segment of the HIV-1 gp41/nef region was amplified from lymph node DNA from humanized mice treated with saline (n = 5), IFN-α14 (n = 4), and IFN-α2 (n = 3) at 11 dpi for next-generation sequencing. (Left) DNA sequences from all mice per cohort were pooled, and the relative numbers of GG→AG mutations relative to the total number of mutations were compared using a 2-by-2 contingency test with Yates' correction. The numbers of sequences analyzed are shown in parentheses, and the percentages of GG→AG mutations relative to the total number of mutations are shown. (Right) Percentages (means + SD) of GG→AG mutations relative to the total number of mutations computed per mouse. **, P < 0.01 (one-way ANOVA with Bonferroni's posttest).
Although APOBEC3G transcription was not upregulated by IFN-α14, it was reported in FV infection that IFN-α activation of critical APOBEC3 antiviral functions occurred at a posttranscriptional level (60). Additionally, treatment of HIV-1-infected LPMC cultures with specific IFN-α subtypes has been shown to result in increased APOBEC3 activity, but not increased transcription (14). Encapsidation of APOBEC3G into HIV-1 virions results in decreased reverse transcription and increased G-to-A hypermutation of the integrating provirus (61), causing both lower viral replication and potentially defective provirus incapable of producing infectious virus. Thus, a predominance of GG→AG mutations is a signature of APOBEC3G activity. To look for evidence of APOBEC3G signature mutations, we used a recently described (14, 52) next-generation sequencing approach to analyze lymph node samples from mice at 11 dpi for proviral HIV-1 GG→AG DNA mutations. Indeed, relative to mock treatment, IFN-α14 treatment, but not IFN-α2 treatment, significantly increased APOBEC3G signature mutations in HIV-1 (Fig. 7B).
DISCUSSION
This study presents new evidence that IFN-α subtypes elicit differential anti-HIV-1 activity in vivo, with much more potent responses induced by IFN-α14 than IFN-α2 when administered at the same dose. The results were consistent between different assays and between cohorts of mice reconstituted from different donors. Ten days of IFN-α14 therapy either as a postexposure prophylactic or in the treatment of acute HIV-1 infections produced significant drops in antigenemia, viremia, and proviral loads. In contrast, IFN-α2 did not control antigenemia and viremia effectively despite the administration of a dose known to be at the upper limit of clinical tolerability. Interestingly, proviral load decreases were observed following IFN-α2 treatments, although the decrease during acute infection was not significant or as great as that observed with IFN-α14. Decreases may have resulted from the killing of infected cells by IFN-α2-induced cytolytic T lymphocytes (CTL), as shown in Fig. 6B. Additionally, the remaining proviruses in IFN-α2-treated mice may have been more replication competent than the APOBEC3G-mutated provirus detected in IFN-α14-treated mice. Interestingly, the superior efficacy of IFN-α14 therapy was associated, not with CD8+ CTL activity, but rather, with the induction of multiple mechanisms of intrinsic and innate immunity. That said, it is not currently known whether any or all of the associated mechanisms were responsible for the in vivo control of HIV-1 in this study. In agreement with recent work in human LPMC cultures (14), we found that in vivo, IFN-α14, but not IFN-α2, significantly upregulated the transcription of two intrinsic restriction factors with well-established anti-HIV-1 activity, MX2 and tetherin (Fig. 7A). We also found evidence of increased APOBEC3G-mediated hypermutation of the HIV-1 provirus in IFN-α14- but not IFN-α2-treated mice (Fig. 7B). Finally, IFN-α14 therapy resulted in an expanded population of TRAIL-expressing NK cells. Given that IFN-α induces hundreds of genes, it is not surprising to find that multiple potential mechanisms of protection were induced in IFN-α14-treated mice. The ability of IFN-α14 to induce a multipronged attack on HIV-1 is one of the aspects that makes it so appealing as a therapeutic, especially since the potential mechanisms of protection involved are different from those employed by current antiretroviral drugs and could therefore potentiate their effects.
HIV-1 has evolved mechanisms to evade the antiviral activities of both tetherin and APOBEC3G, but no antagonist of MX2 has yet been described. MX2 is a key interferon-inducible inhibitor of HIV-1 that blocks at a postentry/early preintegration step of the HIV-1 replication cycle in vitro (17, 18). Expression of MX2 by target cells renders them resistant to HIV-1 infection, so the induction of MX2 by IFN-α14 could play a significant role in preventing infection and spread in vivo. The results presented here demonstrate the first in vivo association between MX2 expression and HIV-1 resistance. Also upregulated by IFN-α14 was tetherin, which inhibits the release of virions from infected cell membranes and is antagonized by HIV-1 Vpu (62). The extent of tetherin induction has been shown to correlate with reduced viral loads in HCV/HIV-1-coinfected patients treated with ribavirin and IFN-α2 (16). In addition to its direct antiviral effects, tetherin has also been associated with improved antiretroviral NK cell responses (63, 64). In the current study, IFN-α14 treatment induced expression of the cytotoxic molecule TRAIL on NK cells, which has been associated with the control of HCV in patients treated with pegylated IFN-α2 and ribavirin (65). In addition, IFN-α-induced NK cell activation and increased TRAIL expression correlated with retroviral control in a mouse model (8), and NK cell responses have been shown to play an important role in HIV-1 immunity (66). APOBEC3G is an intrinsic HIV restriction factor (67, 68), which upon incorporation into newly assembled virions restricts HIV-1 replication in the next target cell by physically impeding reverse transcription and/or inducing lethal G-to-A hypermutations (68–72), primarily converting tryptophans (TGG) to stop codons (TGA). Retrovirus restriction by APOBEC3 is evolutionarily conserved, and deletion of APOBEC3 in mice abrogated the therapeutic effect of IFN-α during FV infection (60). We identified a significant increase in APOBEC3G signature mutations in IFN-α14-treated but not IFN-α2-treated mice. Overall, multiple mechanisms were associated with IFN-α14-induced control of HIV-1, including increased transcription (MX2 and tetherin) and activity (APOBEC3G) of intrinsic immune factors and activation of innate immunity (TRAIL+ NK cells).
There is evidence from both macaques and HIV-1 patients that treatment with IFN-α might lead to detrimental effects from increased immune hyperactivation and dysfunction (54, 73). Thus, it was important to determine whether IFN-α therapy elevated levels of plasma CXCL10, a hallmark of HIV-1-induced immune dysregulation (55, 56). CXCL10 has been reported to be inducible by IFN-α (74, 75) and is produced by HIV-1-infected macrophages and dendritic cells as a chemoattractant to recruit activated T cells to sites of infection (76). As CD4+ T cells are major targets for HIV-1, such recruitment accelerates their infection and loss (76). Thus, CXCL10 levels are predictive of both HIV-1 loads (77) and disease progression (78). In this study, elevated CXCL10 was indeed observed in HIV-1-infected controls and in HIV-1-infected, IFN-α2-treated animals. In contrast, IFN-α14-treated mice, which had only very low levels of HIV-1 infection, also had low levels of CXCL10. Thus, elevated CXCL10 was associated with HIV-1 viremia rather than IFN-α treatment. Importantly, IFN-α14-treated mice also had significantly better preservation of central memory CD4+ T cells, indicative of less HIV-1-induced immunopathology. These results are consistent with studies in SIV-infected African green monkeys (AGM) and sooty mangabeys (SM), demonstrating that administration of high-dose rhesus macaque IFN-α2 did not impede the resolution of immune activation and ISG expression that is specific to these nonpathogenic models (79), nor did it exacerbate disease (80, 81). They are also consistent with studies in SIV-infected macaques showing that blockade of the IFN receptor accelerated disease progression (54), indicating that endogenous type I IFN responses are beneficial and impede SIV pathology. This is in contrast to studies where macaques receiving human IFN-α2 displayed accelerated disease progression despite downregulation of ISG expression and levels of immune activation similar to or lower than those of placebo controls (54).
It is becoming apparent that subtle differences in the amino acid sequences of IFN-α subtypes can produce a significant effect on the binding affinity for the IFN-α/β receptor, downstream signaling events, and antiviral efficacy (2–5, 82). Given that rhesus macaque IFN-α2 sequences compared to human and SM/AGM IFN-α2 are only 92 and 98% identical, respectively, future studies in macaques should consider using species-specific IFNs rather than human IFNs (83). In fact, IFN-α14 may not be the most effective subtype in suppressing SIV, and a complete in vitro analysis of all subtypes is warranted before in vivo studies are initiated. Additionally, a recent study in mice chronically infected with LCMV (84) suggested that persistent IFN-β, but not IFN-α, responses are associated with immune dysregulation and virus chronicity. This result supports two pivotal concepts in our study: (i) different type I IFN subtypes can mediate distinct biological effects despite binding to the same receptor and (ii) type I IFNs may be useful drugs for the treatment of chronic viral infections if one determines the optimal subtype for the particular virus.
Although therapy of HIV-1 patients with IFN-α2 has produced decreases in viremia in some studies (24–31), the use of IFN-α2 has been based more upon its availability as a pharmaceutical-grade protein and its efficacy in treating HCV and HBV infections than upon evidence that it is the most efficacious subtype against HIV-1. Recent in vitro work (14), combined with these new in vivo results, suggest that, based on differential expression patterns, restriction factor induction, mobilization of immune effectors, and ultimate anti-HIV-1 potency, not all subtypes play redundant roles in HIV-1 infection, and therefore, they should be individually evaluated as anti-HIV-1 therapeutics. Extrapolation of the results in humanized mice to HIV-1 infections in humans suggests that IFN-α14 might prove more effective than IFN-α2, which is currently in clinical HIV-1 trials. Such IFN studies would most likely be done in patients undergoing standard antiretroviral therapy, something that we have not yet tested in our model. Thus, it remains to be seen whether IFN-α14 will be similarly effective in that scenario.
It should be noted that these humanized mice do not have an entirely intact human immune system. For example, the lymphoid organ structure is not completely normal because the T cells and B cells are interspersed rather than delineated into zones (37). The impact of differences between these chimeric animals and intact animals on the current results is not known. However, since HIV does not infect mouse cells, no direct effect of human IFN-α interaction with mouse cells could impact HIV-1 levels. Furthermore, the lack of mouse lymphocytes and NK cells in this model precludes any indirect effects from cytokines or chemokines produced by these cells. TKO-BLT mice do contain murine myeloid cells that could potentially respond to human IFN-α. Most cytokines produced by mouse macrophages, including macrophage colony-stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and IL-6, are not reactive with human receptors (85). However, IL-1β has weak reactivity with the human receptor (86), as does mouse TNF-α. Thus, there remains the possibility of an indirect effect on HIV-1-infected human cells. That said, the current results from our in vivo studies comparing IFN-α2 and IFN-α14 are highly consistent with the in vitro results in Fig. 1, as well as results from the recent IFN-α subtype study using human LPMCs infected with HIV-1 (14). Studies are under way to determine if latent HIV infections can be established in this model using antiretroviral therapy. Since IFN-α14 functions via different mechanisms than antiretroviral drugs, it will be of great interest to test a combination of the two in an attempt to eliminate the replication-competent HIV-1 reservoir, thereby producing a functional cure. A clinical study of this nature is under way with IFN-α2 (https://clinicaltrials.gov/ct2/show/NCT02227277). Our results suggest that IFN-α14 may provide even stronger benefits in such a trial. The results from our study have implications for the use of IFN-α as an antiviral therapeutic for other viruses, as well. Although IFN-α2 is approved for the treatment of HBV, HCV, and AIDS-related Kaposi's sarcoma, it will be important in the future to determine if IFN-α2 is the most effective and safe subtype for these and other viruses.
ACKNOWLEDGMENTS
We thank Martin McCarter (University of Colorado) for the gut tissue samples, Stephanie Dillon for advice on LPMC cultures, Barbara Bleekmann for excellent technical assistance, and M. A. Joyce for critical reading of the manuscript.
REFERENCES
- 1.Hardy MP, Owczarek CM, Jermiin LS, Ejdeback M, Hertzog PJ. 2004. Characterization of the type I interferon locus and identification of novel genes. Genomics 84:331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, Nolan GP, Piehler J, Schreiber G, Garcia KC. 2011. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146:621–632. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaks E, Gavutis M, Uze G, Martal J, Piehler J. 2007. Differential receptor subunit affinities of type I interferons govern differential signal activation. J Mol Biol 366:525–539. doi: 10.1016/j.jmb.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 4.Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, Moolchan K, Pestka S, Schreiber G. 2011. Binding and activity of all human alpha interferon subtypes. Cytokine 56:282–289. doi: 10.1016/j.cyto.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Cull VS, Tilbrook PA, Bartlett EJ, Brekalo NL, James CM. 2003. Type I interferon differential therapy for erythroleukemia: specificity of STAT activation. Blood 101:2727–2735. doi: 10.1182/blood-2002-05-1521. [DOI] [PubMed] [Google Scholar]
- 6.Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, Sironi M, Tichit M, Bouchier C, Casanova JL, Barreiro LB, Quintana-Murci L. 2011. Evolutionary genetic dissection of human interferons. J Exp Med 208:2747–2759. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlach N, Gibbert K, Alter C, Nair S, Zelinskyy G, James CM, Dittmer U. 2009. Anti-retroviral effects of type I IFN subtypes in vivo. Eur J Immunol 39:136–146. doi: 10.1002/eji.200838311. [DOI] [PubMed] [Google Scholar]
- 8.Gibbert K, Joedicke JJ, Meryk A, Trilling M, Francois S, Duppach J, Kraft A, Lang KS, Dittmer U. 2012. Interferon-alpha subtype 11 activates NK cells and enables control of retroviral infection. PLoS Pathog 8:e1002868. doi: 10.1371/journal.ppat.1002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Austin BA, James C, Silverman RH, Carr DJ. 2005. Critical role for the oligoadenylate synthetase/RNase L pathway in response to IFN-beta during acute ocular herpes simplex virus type 1 infection. J Immunol 175:1100–1106. doi: 10.4049/jimmunol.175.2.1100. [DOI] [PubMed] [Google Scholar]
- 10.Austin BA, James CM, Harle P, Carr DJ. 2006. Direct application of plasmid DNA containing type I interferon transgenes to vaginal mucosa inhibits HSV-2 mediated mortality. Biol Proced Online 8:55–62. doi: 10.1251/bpo118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James CM, Abdad MY, Mansfield JP, Jacobsen HK, Vind AR, Stumbles PA, Bartlett EJ. 2007. Differential activities of alpha/beta IFN subtypes against influenza virus in vivo and enhancement of specific immune responses in DNA vaccinated mice expressing haemagglutinin and nucleoprotein. Vaccine 25:1856–1867. doi: 10.1016/j.vaccine.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 12.Tsang SL, Leung PC, Leung KK, Yau WL, Hardy MP, Mak NK, Leung KN, Fung MC. 2007. Characterization of murine interferon-alpha 12 (MuIFN-alpha12): biological activities and gene expression. Cytokine 37:138–149. doi: 10.1016/j.cyto.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Yeow WS, Lai CM, Beilharz MW. 1997. The in vivo expression patterns of individual type I interferon genes in murine cytomegalovirus infections. Antiviral Res 34:17–26. doi: 10.1016/S0166-3542(96)01018-2. [DOI] [PubMed] [Google Scholar]
- 14.Harper MS, Guo K, Gibbert K, Lee EJ, Dillon SM, Barrett BS, McCarter MD, Hasenkrug KJ, Dittmer U, Wilson CC, Santiago ML. 2015. Interferon-alpha subtypes in an ex vivo model of acute HIV-1 infection: expression, potency and effector mechanisms. PLoS Pathog 11:e1005254. doi: 10.1371/journal.ppat.1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan N, Chen ZJ. 2012. Intrinsic antiviral immunity. Nat Immunol 13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillai SK, Abdel-Mohsen M, Guatelli J, Skasko M, Monto A, Fujimoto K, Yukl S, Greene WC, Kovari H, Rauch A, Fellay J, Battegay M, Hirschel B, Witteck A, Bernasconi E, Ledergerber B, Gunthard HF, Wong JK, Swiss HIV Cohort Study. 2012. Role of retroviral restriction factors in the interferon-alpha-mediated suppression of HIV-1 in vivo. Proc Natl Acad Sci U S A 109:3035–3040. doi: 10.1073/pnas.1111573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, Hue S, Barclay WS, Schulz R, Malim MH. 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann C, Taubert D, Jung N, Fatkenheuer G, van Lunzen J, Hartmann P, Romerio F. 2009. Preferential upregulation of interferon-alpha subtype 2 expression in HIV-1 patients. AIDS Res Hum Retroviruses 25:577–581. doi: 10.1089/aid.2008.0238. [DOI] [PubMed] [Google Scholar]
- 20.Hardy GA, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, Mudd J, Schacker T, Funderburg NT, Pilch-Cooper HA, Debernardo R, Rabin RL, Lederman MM, Harding CV. 2013. Interferon-alpha is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One 8:e56527. doi: 10.1371/journal.pone.0056527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraietta JA, Mueller YM, Yang G, Boesteanu AC, Gracias DT, Do DH, Hope JL, Kathuria N, McGettigan SE, Lewis MG, Giavedoni LD, Jacobson JM, Katsikis PD. 2013. Type I interferon upregulates Bak and contributes to T cell loss during human immunodeficiency virus (HIV) infection. PLoS Pathog 9:e1003658. doi: 10.1371/journal.ppat.1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle T, Goujon C, Malim MH. 2015. HIV-1 and interferons: who's interfering with whom? Nat Rev Microbiol 13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moron-Lopez S, Gomez-Mora E, Salgado M, Ouchi D, Puertas MC, Urrea V, Navarro J, Jou A, Perez M, Tural C, Clotet B, Montaner LJ, Blanco J, Crespo M, Martinez-Picado J. 2016. Short-term treatment with IFNalpha diminishes expression of HIV-1 and reduces CD4+ T-cell activation in HIV/HCV-coinfected patients on antiretroviral therapy. J Infect Dis 213:1008–1012. doi: 10.1093/infdis/jiv521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdel-Mohsen M, Deng X, Liegler T, Guatelli JC, Salama MS, Ghanem Hel D, Rauch A, Ledergerber B, Deeks SG, Gunthard HF, Wong JK, Pillai SK. 2014. Effects of alpha interferon treatment on intrinsic anti-HIV-1 immunity in vivo. J Virol 88:763–767. doi: 10.1128/JVI.02687-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asmuth DM, Murphy RL, Rosenkranz SL, Lertora JJ, Kottilil S, Cramer Y, Chan ES, Schooley RT, Rinaldo CR, Thielman N, Li XD, Wahl SM, Shore J, Janik J, Lempicki RA, Simpson Y, Pollard RB, AIDS Clinical Trials Group A5192 Team. 2010. Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon alfa-2a in HIV-1-monoinfected participants: a phase II clinical trial. J Infect Dis 201:1686–1696. doi: 10.1086/652420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azzoni L, Foulkes AS, Papasavvas E, Mexas AM, Lynn KM, Mounzer K, Tebas P, Jacobson JM, Frank I, Busch MP, Deeks SG, Carrington M, O'Doherty U, Kostman J, Montaner LJ. 2013. Pegylated interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J Infect Dis 207:213–222. doi: 10.1093/infdis/jis663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brockmeyer NH, Poffhoff A, Bader A, Hochdorfer B, Schlottmann R, Rasokat H, Altmeyer P, Kreuter A. 2006. Treatment of condylomata acuminata with pegylated interferon alfa-2b in HIV-infected patients. Eur J Med Res 11:27–32. [PubMed] [Google Scholar]
- 28.Dianzani F, Rozera G, Abbate I, D'Offizi G, Abdeddaim A, Vlassi C, Antonucci G, Narciso P, Martini F, Capobianchi MR. 2008. Interferon may prevent HIV viral rebound after HAART interruption in HIV patients. J Interferon Cytokine Res 28:1–3. doi: 10.1089/jir.2007.0076. [DOI] [PubMed] [Google Scholar]
- 29.Frissen PH, de Wolf F, Reiss P, Bakker PJ, Veenhof CH, Danner SA, Goudsmit J, Lange JM. 1997. High-dose interferon-alpha2a exerts potent activity against human immunodeficiency virus type 1 not associated with antitumor activity in subjects with Kaposi's sarcoma. J Infect Dis 176:811–814. doi: 10.1086/517309. [DOI] [PubMed] [Google Scholar]
- 30.Haas DW, Lavelle J, Nadler JP, Greenberg SB, Frame P, Mustafa N, St Clair M, McKinnis R, Dix L, Elkins M, Rooney J. 2000. A randomized trial of interferon alpha therapy for HIV type 1 infection. AIDS Res Hum Retroviruses 16:183–190. doi: 10.1089/088922200309278. [DOI] [PubMed] [Google Scholar]
- 31.Tavel JA, Huang CY, Shen J, Metcalf JA, Dewar R, Shah A, Vasudevachari MB, Follmann DA, Herpin B, Davey RT, Polis MA, Kovacs J, Masur H, Lane HC. 2010. Interferon-alpha produces significant decreases in HIV load. J Interferon Cytokine Res 30:461–464. doi: 10.1089/jir.2009.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Cruz E, Lang JM, Frissen J, Furner V, Chateauvert M, Boucher CA, Dowd P, Stevens J. 1995. Zidovudine plus interferon-alpha versus zidovudine alone in HIV-infected symptomatic or asymptomatic persons with CD4+ cell counts > 150 x 10(6)/L: results of the Zidon trial. Zidon Study Group. AIDS 9:1025–1035. [PubMed] [Google Scholar]
- 33.Krown SE, Aeppli D, Balfour HH Jr. 1999. Phase II, randomized, open-label, community-based trial to compare the safety and activity of combination therapy with recombinant interferon-alpha2b and zidovudine versus zidovudine alone in patients with asymptomatic to mildly symptomatic HIV infection. HIV Protocol C91-253 Study Team. J Acquir Immune Defic Syndr Hum Retrovirol 20:245–254. doi: 10.1097/00042560-199903010-00005. [DOI] [PubMed] [Google Scholar]
- 34.Boue F, Reynes J, Rouzioux C, Emilie D, Souala F, Tubiana R, Goujard C, Lancar R, Costagliola D. 2011. Alpha interferon administration during structured interruptions of combination antiretroviral therapy in patients with chronic HIV-1 infection: INTERVAC ANRS 105 trial. AIDS 25:115–118. doi: 10.1097/QAD.0b013e328340a1e7. [DOI] [PubMed] [Google Scholar]
- 35.Sun H, Buzon MJ, Shaw A, Berg RK, Yu XG, Ferrando-Martinez S, Leal M, Ruiz-Mateos E, Lichterfeld M. 2014. Hepatitis C therapy with interferon-alpha and ribavirin reduces CD4 T-cell-associated HIV-1 DNA in HIV-1/hepatitis C virus-coinfected patients. J Infect Dis 209:1315–1320. doi: 10.1093/infdis/jit628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumaran J, Wei L, Kotra LP, Fish EN. 2007. A structural basis for interferon-alpha-receptor interactions. FASEB J 21:3288–3296. doi: 10.1096/fj.07-8585com. [DOI] [PubMed] [Google Scholar]
- 37.Lavender KJ, Pang WW, Messer RJ, Duley AK, Race B, Phillips K, Scott D, Peterson KE, Chan CK, Dittmer U, Dudek T, Allen TM, Weissman IL, Hasenkrug KJ. 2013. BLT-humanized C57BL/6 Rag2-/-gammac-/-CD47-/- mice are resistant to GVHD and develop B- and T-cell immunity to HIV infection. Blood 122:4013–4020. doi: 10.1182/blood-2013-06-506949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavender KJ, Messer RJ, Race B, Hasenkrug KJ. 2014. Production of bone marrow, liver, thymus (BLT) humanized mice on the C57BL/6 Rag2(-/-)gammac(-/-)CD47(-/-) background. J Immunol Methods 407:127–134. doi: 10.1016/j.jim.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalie E, Jaitin DA, Abramovich R, Schreiber G. 2007. An interferon alpha2 mutant optimized by phage display for IFNAR1 binding confers specifically enhanced antitumor activities. J Biol Chem 282:11602–11611. doi: 10.1074/jbc.M610115200. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, Jonjic S, Koszinowski U, Hengel H. 2005. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-{gamma} signaling and antiviral responses. J Exp Med 201:1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindler M, Munch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Muller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, Bailes E, Roques P, Sodora DL, Silvestri G, Sharp PM, Hahn BH, Kirchhoff F. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 42.Papkalla A, Munch J, Otto C, Kirchhoff F. 2002. Nef enhances human immunodeficiency virus type 1 infectivity and replication independently of viral coreceptor tropism. J Virol 76:8455–8459. doi: 10.1128/JVI.76.16.8455-8459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dillon SM, Manuzak JA, Leone AK, Lee EJ, Rogers LM, McCarter MD, Wilson CC. 2012. HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli. J Immunol 189:885–896. doi: 10.4049/jimmunol.1200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steele AK, Lee EJ, Manuzak JA, Dillon SM, Beckham JD, McCarter MD, Santiago ML, Wilson CC. 2014. Microbial exposure alters HIV-1-induced mucosal CD4+ T cell death pathways ex vivo. Retrovirology 11:14. doi: 10.1186/1742-4690-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P. 2013. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daud A, Soon C, Dummer R, Eggermont AM, Hwu WJ, Grob JJ, Garbe C, Hauschild A. 2012. Management of pegylated interferon alpha toxicity in adjuvant therapy of melanoma. Expert Opin Biol Ther 12:1087–1099. doi: 10.1517/14712598.2012.694421. [DOI] [PubMed] [Google Scholar]
- 47.Payne MJ, Argyropoulou K, Lorigan P, McAleer JJ, Farrugia D, Davidson N, Kelly C, Chao D, Marshall E, Han C, Wellman S, Middleton MR. 2014. Phase II pilot study of intravenous high-dose interferon with or without maintenance treatment in melanoma at high risk of recurrence. J Clin Oncol 32:185–190. doi: 10.1200/JCO.2013.49.8717. [DOI] [PubMed] [Google Scholar]
- 48.Reagan-Shaw S, Nihal M, Ahmad N. 2008. Dose translation from animal to human studies revisited. FASEB J 22:659–661. [DOI] [PubMed] [Google Scholar]
- 49.Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. 2008. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc 3:1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- 50.Butchi NB, Woods T, Du M, Morgan TW, Peterson KE. 2011. TLR7 and TLR9 trigger distinct neuroinflammatory responses in the CNS. Am J Pathol 179:783–794. doi: 10.1016/j.ajpath.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halemano K, Guo K, Heilman KJ, Barrett BS, Smith DS, Hasenkrug KJ, Santiago ML. 2014. Immunoglobulin somatic hypermutation by APOBEC3/Rfv3 during retroviral infection. Proc Natl Acad Sci U S A 111:7759–7764. doi: 10.1073/pnas.1403361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrett BS, Guo K, Harper MS, Li SX, Heilman KJ, Davidson NO, Santiago ML. 2014. Reassessment of murine APOBEC1 as a retrovirus restriction factor in vivo. Virology 468–470:601–608. doi: 10.1016/j.virol.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pestka S. 2007. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem 282:20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- 54.Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, Hill BJ, Timmer JK, Reiss E, Yarden G, Darko S, Contijoch E, Todd JP, Silvestri G, Nason M, Norgren RB Jr, Keele BF, Rao S, Langer JA, Lifson JD, Schreiber G, Douek DC. 2014. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rychert J, Strick D, Bazner S, Robinson J, Rosenberg E. 2010. Detection of HIV gp120 in plasma during early HIV infection is associated with increased proinflammatory and immunoregulatory cytokines. AIDS Res Hum Retroviruses 26:1139–1145. doi: 10.1089/aid.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Norris PJ, Pappalardo BL, Custer B, Spotts G, Hecht FM, Busch MP. 2006. Elevations in IL-10, TNF-alpha, and IFN-gamma from the earliest point of HIV type 1 infection. AIDS Res Hum Retroviruses 22:757–762. doi: 10.1089/aid.2006.22.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cocchi F, DeVico AL, Yarchoan R, Redfield R, Cleghorn F, Blattner WA, Garzino-Demo A, Colombini-Hatch S, Margolis D, Gallo RC. 2000. Higher macrophage inflammatory protein (MIP)-1alpha and MIP-1beta levels from CD8+ T cells are associated with asymptomatic HIV-1 infection. Proc Natl Acad Sci U S A 97:13812–13817. doi: 10.1073/pnas.240469997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curtsinger JM, Valenzuela JO, Agarwal P, Lins D, Mescher MF. 2005. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol 174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- 59.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harper MS, Barrett BS, Smith DS, Li SX, Gibbert K, Dittmer U, Hasenkrug KJ, Santiago ML. 2013. IFN-alpha treatment inhibits acute Friend retrovirus replication primarily through the antiviral effector molecule Apobec3. J Immunol 190:1583–1590. doi: 10.4049/jimmunol.1202920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol 337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 62.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 63.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, Chen BK. 2014. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J Virol 88:6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li SX, Barrett BS, Heilman KJ, Messer RJ, Liberatore RA, Bieniasz PD, Kassiotis G, Hasenkrug KJ, Santiago ML. 2014. Tetherin promotes the innate and adaptive cell-mediated immune response against retrovirus infection in vivo. J Immunol 193:306–316. doi: 10.4049/jimmunol.1400490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stegmann KA, Bjorkstrom NK, Veber H, Ciesek S, Riese P, Wiegand J, Hadem J, Suneetha PV, Jaroszewicz J, Wang C, Schlaphoff V, Fytili P, Cornberg M, Manns MP, Geffers R, Pietschmann T, Guzman CA, Ljunggren HG, Wedemeyer H. 2010. Interferon-alpha-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology 138:1885–1897. doi: 10.1053/j.gastro.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 66.Jost S, Altfeld M. 2013. Control of human viral infections by natural killer cells. Annu Rev Immunol 31:163–194. doi: 10.1146/annurev-immunol-032712-100001. [DOI] [PubMed] [Google Scholar]
- 67.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 68.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 69.Harris RS, Sheehy AM, Craig HM, Malim MH, Neuberger MS. 2003. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat Immunol 4:641–643. doi: 10.1038/ni0703-641. [DOI] [PubMed] [Google Scholar]
- 70.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krisko JF, Martinez-Torres F, Foster JL, Garcia JV. 2013. HIV restriction by APOBEC3 in humanized mice. PLoS Pathog 9:e1003242. doi: 10.1371/journal.ppat.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cha L, Berry CM, Nolan D, Castley A, Fernandez S, French MA. 2014. Interferon-alpha, immune activation and immune dysfunction in treated HIV infection. Clin Transl Immunology 3:e10. doi: 10.1038/cti.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, Gladue RP, Lin W, Boyd JG, Moser B, Wood DE, Sahagan BG, Neote K. 1998. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med 187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeremski M, Dimova RB, Benjamin S, Penney MS, Botfield MC, Talal AH. 2015. Intrahepatic and peripheral CXCL10 expression in hepatitis C virus-infected patients treated with telaprevir, pegylated interferon, and ribavirin. J Infect Dis 211:1795–1799. doi: 10.1093/infdis/jiu807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Foley JF, Yu CR, Solow R, Yacobucci M, Peden KW, Farber JM. 2005. Roles for CXC chemokine ligands 10 and 11 in recruiting CD4+ T cells to HIV-1-infected monocyte-derived macrophages, dendritic cells, and lymph nodes. J Immunol 174:4892–4900. doi: 10.4049/jimmunol.174.8.4892. [DOI] [PubMed] [Google Scholar]
- 77.Gray CM, Hong HA, Young K, Lewis DA, Fallows D, Manca C, Kaplan G. 2013. Plasma interferon-gamma-inducible protein 10 can be used to predict viral load in HIV-1-infected individuals. J Acquir Immune Defic Syndr 63:e115–e116. doi: 10.1097/QAI.0b013e3182930ea8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiao Y, Zhang T, Wang R, Zhang H, Huang X, Yin J, Zhang L, Xu X, Wu H. 2012. Plasma IP-10 is associated with rapid disease progression in early HIV-1 infection. Viral Immunol 25:333–337. doi: 10.1089/vim.2012.0011. [DOI] [PubMed] [Google Scholar]
- 79.Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, Francella N, Sidahmed A, Smith AJ, Cramer EM, Zeng M, Masopust D, Carlis JV, Ran L, Vanderford TH, Paiardini M, Isett RB, Baldwin DA, Else JG, Staprans SI, Silvestri G, Haase AT, Kelvin DJ. 2009. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest 119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vanderford TH, Slichter C, Rogers KA, Lawson BO, Obaede R, Else J, Villinger F, Bosinger SE, Silvestri G. 2012. Treatment of SIV-infected sooty mangabeys with a type-I IFN agonist results in decreased virus replication without inducing hyperimmune activation. Blood 119:5750–5757. doi: 10.1182/blood-2012-02-411496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacquelin B, Petitjean G, Kunkel D, Liovat AS, Jochems SP, Rogers KA, Ploquin MJ, Madec Y, Barre-Sinoussi F, Dereuddre-Bosquet N, Lebon P, Le Grand R, Villinger F, Muller-Trutwin M. 2014. Innate immune responses and rapid control of inflammation in African green monkeys treated or not with interferon-alpha during primary SIVagm infection. PLoS Pathog 10:e1004241. doi: 10.1371/journal.ppat.1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vazquez N, Schmeisser H, Dolan MA, Bekisz J, Zoon KC, Wahl SM. 2011. Structural variants of IFNalpha preferentially promote antiviral functions. Blood 118:2567–2577. doi: 10.1182/blood-2010-12-325027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Villinger F, Brar SS, Mayne A, Chikkala N, Ansari AA. 1995. Comparative sequence analysis of cytokine genes from human and nonhuman primates. J Immunol 155:3946–3954. [PubMed] [Google Scholar]
- 84.Ng CT, Sullivan BM, Teijaro JR, Lee AM, Welch M, Rice S, Sheehan KC, Schreiber RD, Oldstone MB. 2015. Blockade of interferon beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe 17:653–661. doi: 10.1016/j.chom.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Q, Khoury M, Chen J. 2009. Expression of human cytokines dramatically improves reconstitution of specific human-blood lineage cells in humanized mice. Proc Natl Acad Sci U S A 106:21783–21788. doi: 10.1073/pnas.0912274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang JJ, Newton RC, Rutledge SJ, Horuk R, Matthew JB, Covington M, Lin Y. 1988. Characterization of murine IL-1 beta. Isolation, expression, and purification. J Immunol 140:3838–3843. [PubMed] [Google Scholar]