Abstract
Summary: In this article, we introduce a robust and efficient strategy for deriving global and allele-specific copy number alternations (CNA) from cancer whole exome sequencing data based on Log R ratios and B-allele frequencies. Applying the approach to the analysis of over 200 skin cancer samples, we demonstrate its utility for discovering distinct CNA events and for deriving ancillary information such as tumor purity.
Availability and implementation: https://github.com/xfwang/CLOSE
Contact: xuefeng.wang@stonybrook.edu or michael.krauthammer@yale.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
DNA copy number alternations (CNA) have been extensively studied in cancer. Copy number amplifications and homozygous deletions from CNA play an important role in the development of cancer. Genome-wide CNA detection of large number of tumor samples is a vital step to identify cancer driver events and to gain insights into cancer progression and prevention. Next-generation sequencing (NGS) technologies have quickly become the CNA analysis platform of choice due to higher resolution and sensitivity compared to traditional array-based approaches such as comparative genomic hybridization (CGH). However, NGS-specific CNA statistical and computational methods are currently not well established and are mostly based on the total copy number gains or losses as represented by the log ratio of sequence coverage in a tumor-normal pair. These relative measurements do not take into account tumor purity, clonality, as well as other valuable information accrued by NGS data. In this correspondence, we highlight a set of concepts and analytic tools that enables the fast and accurate dissection of whole-genome CNA profiles by integrating the totality of information available from NGS data. Based on whole exome sequencing (WES) data of 253 melanoma tumor-normal pairs, we demonstrate that our approaches allow for a more accurate estimation of copy number status and identification of otherwise undetectable CNA patterns.
Well-developed array-based CNA analytical tools are primarily based on the segmentation and smoothing of Log R Ratio (LRR) and B-allele frequency (BAF) (Wang et al., 2007), which provide orthogonal evidence for copy number status. LRR corresponds to the difference in the hybridization signal in a particular genome region between normal and tumor DNA, and BAF measures the relative contributions of the maternal and paternal alleles to this signal. Most NGS-based tools, however, rely solely on LRR due to the inherent challenges in assessing BAF. First, the density of observed single nucleotide variants (SNVs) is relatively sparse, especially in WES. Also, the accuracy of the BAF measurement is dependent on the sequence coverage, which can vary between gene regions and sequencing modalities. Finally, the BAF is sensitive to sequencing and mapping errors. Therefore, BAF information remains underutilized in NGS experiments, and BAF is often used in an ad hoc manner—in an attempt to validate the results obtained from LRR. In this paper, we propose a fast procedure that is based on both LRR and BAF and that works seamlessly with existing NGS pipelines. We show that the derived multivariate CNA profiles allow for an in-depth characterization of the somatic structural variations in cancer samples. A set of analysis scripts from vcf file preparation to the derivation of allele-specific copy numbers are provided in the pipeline package CLOSE (a toolkit for CNA/LOH analysis with Sequencing data) and its companion R package CLOSE-R.
2 Methods and results
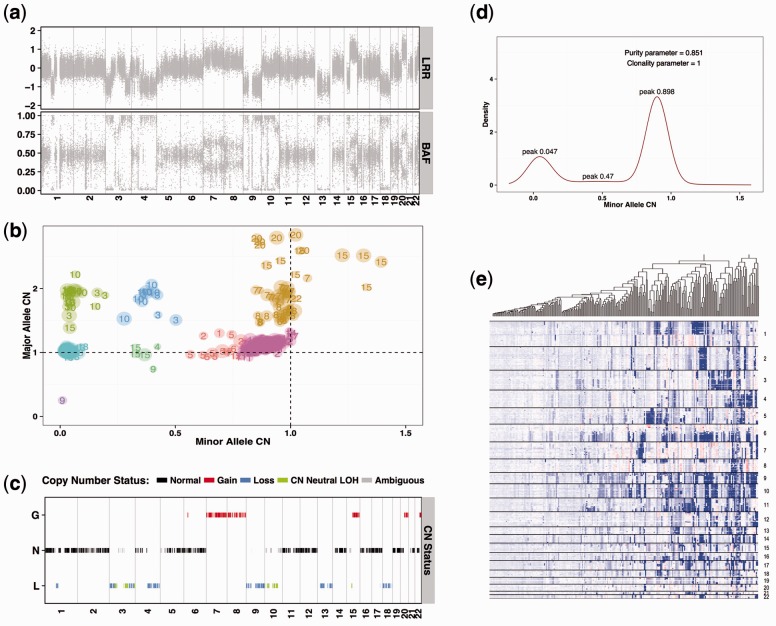
A conceptual overview of our strategy is illustrated in Figure 1. NGS sequencing data is first segmented into LRR regions using existing programs, and we only use the BAF of identified variant positions that are heterozygous in the paired normal tissues in the subsequent joint modelling steps. The average or median LRR and BAF values are calculated for each segment. For convenience and as a mean to reduce data sparsity, the segmental BAF values are further converted into the LAF (lesser allele frequency) measures according the formula as described in the Supplementary Methods. We can then apply a joint likelihood model at the segmental level for integrating information from both LRR and LAF data for deriving allele-specific copy number (ASCN) estimates. ASCN is a 2 dimensional representation of copy number data that encodes the status of the A and B alleles (i.e. the maternal and paternal alleles) (LaFramboise et al., 2005; Van Loo et al., 2010). To ensure reliable estimation, it is important to exclude outlier and less informative (normal) regions. We assign more weight to regions with LAF smaller than 0.25, because they are much less prone to sequencing related bias and represent important anchor points for recognition of copy number alterations. We derive sample purity and ploidy based on the traditional full likelihood model and through a position-based approach that explores distances from canonical coordinates (Li and Li, 2014).
Fig. 1.
Genome-wide CNA profile of melanoma samples. (a) LRR and BAF calls from WES of matched normal and tumor samples. (b) Bivariate genome-wide allele-specific copy number profile (detailed in Supplementary Materials) and nonparametric Bayesian clustering based on Chinese Restaurant Process (CRP). (c) Predicted copy number status. (d) Density estimation of minor allele copy number estimation by the Dirichlet Process mixture of normal distributions. (e) Hierarchical clustering of 253 melonoma samples based on the minor allele copy number estimates
Importantly, we derived a generalized LRR-LAF mapping function (Equation 8 in the Supplementary Methods) that can be used when the sample ploidy parameter is not given or cannot be estimated with enough certainty. In this case, the ASCN estimates become relative measures, instead of absolute ones. Relative ASCN estimates are still sufficient in determining copy number status (Chen et al., 2014). This function was used recursively in CLOSE for calculating canonical coordinates or lines under different settings. It provides a unified systematic method for determining copy number status, as well as a basis for further exploring clonal mixtures with a tumor.
Both LRR and LAF measurements exhibit strong variability (Bao et al., 2014; Carter et al., 2012) with baseline values that are often shifted from null (0.5 for LAF) depending on sample purity. As a robust alternative to the model- or likelihood- based method for copy number detection, we propose to call absolute or relative ASCN based on nonparametric Bayesian clustering with the Chinese Restaurant Process (CRP). The cluster that is closest to the baseline coordinates corresponds to normal regions, and other copy number alternation regions can be inferred accordingly. The derived CNA profile (Fig. 1b) greatly facilitates genome-wide data exploration by grouping samples into different CNA-sets. Importantly, the use of ASCN allows the detection of copy number neutral LOH (loss of heterozygosity) regions. These regions do no change their total DNA copy numbers and have been a blind spot for LRR-based tools. Therefore, CLOSE-R currently implements three methods for copy number inference: (i) the proposed CRP or clustering based method; (ii) the likelihood- or model-based method; and, for comparison, (iii) the joint segmentation model of LRR and LAF modified based on the R package falcon (Chen et al., 2014). Based on a group of randomly selected samples, we compared the total and allele specific copy number estimates obtained from these three methods (Supplementary Fig. S2) and found consistent results across different samples. We also compared the global parameter estimation implemented in CLOSE (based on the modified likelihood approach) with a similar package Sequenza (Favero et al., 2015); they returned very close estimates for purity even at different ploidy levels (Supplementary Table S1 and Supplementary Fig. S3).
A competitive advantage of the CRP approach is the straightforward ability to explore tumor cellularity and subclonal cancer architecture using the allele specific copy numbers. As illustrated in Figure 1d, the minor allele copy number (minCN) approximately follows a mixture of one, two or three Gaussian distributions. The spacing between the central point of the first cluster of minCN and that of the third, if existent, corresponds to the concentration ratio of the tumor sample. The peak in the middle, if existent, is useful for estimating aneuploidy or subclonality. The deviation of the main mode (of the density plot in Fig. 1d) of the minor allele copy number from one captures the systematic bias caused by sequencing and mapping errors. The Dirichlet Process fit can be successfully applied to identify these peaks given a purity measure is as low as 0.5. The abstracted features from the CNA profile aid in conducting the integrative analysis of multiple cancer samples. We performed three sets of hierarchical clustering of all of 253 available melanoma samples, based on minCN, and both minor and major allele copy numbers, respectively (Fig. 1e, and Supplementary Fig. S4). The two-channel clustering shows major copy number events across the genome, including losses in chromosomes 9 and 10, and gains of 1q, 7p, 7q and 8q (left panel, Supplementary Fig. S4). A clustering plot based on majCN reveals a set of melanomas, including YUMOKI, YURDE and YUWALI, with a distinct chromosome 7 gain (right panel, top, in Supplementary Fig. S4), and a tight grouping of melanoma samples from the same patients. The combined clustering does not reveal the chromosome 7 finding and fails to group samples from the same patient (see YUCLAT and YUZEST). Because a few samples in our dataset were also submitted to TCGA, we were able to compare our results to the publicly available copy-number segmentation results based on array data (Affymetrix SNP6), showing considerable consistency between the results (Supplementary Fig. S5).
Taken together, this study describes a timely and lightweight solution for cancer CNA detection that is scalable to large-scale sequencing studies. A similar approach can be applied for accurate CNA detection in high-depth single-cell sequencing, and the solution is extensible to whole genome sequencing. A remaining challenge is the joint identification of unknown parameters such as ploidy and cellularity—in which parameters may not be identifiable without imposing additional assumptions. Nevertheless, we hope that our solution offers a path forward for deconvolving the complexities of the cancer genome, and stimulates further interest in allele-specific CNA profiling based on LRR and BAF.
3 Conclusions
We propose a lightweight approach for assessing WES-based global DNA copy number abberations that (i) replicates copy number calls from exisiting analysis methodologies; (ii) derives tumor ploidy and cellularity; and (iii) provides information for cross-tumor integrative analysis.
Supplementary Material
Acknowledgements
The authors would like to thank Bo Li for helpful discussion.
Funding
M.K. was supported by the National Cancer Institute (Yale SPORE in skin cancer, P50 CA121974). M.C. was supported in part by the National Institutes of Health (NIH) grants R01 CA082659 and P01CA142538. X.W. was supported in part by the NIH grant P20 CA192994.
Conflict of Interest: none declared.
References
- Bao L., et al. (2014) AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data. Bioinformatics, 30, 1056–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., et al. (2014) Allele-specific copy number profiling by next-generation DNA sequencing. Nucleic Acids Res., 43:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S.L., et al. (2014) Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol., 30, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero F., et al. (2015) Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol., 26: 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFramboise T., et al. (2005) Allele-specific amplification in cancer revealed by SNP array analysis. PLoS Comput. Biol., 1, e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Li J. (2014) A general framework for analyzing tumor subclonality uing SNP array and DNA sequencing data. Genome Biol., 15, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo P., et al. (2010) Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. USA, 107, 16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., et al. (2007) PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res., 17, 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.