Abstract
A recent meta-analysis of multiple genome-wide association and follow-up endometrial cancer case-control datasets identified a novel genetic risk locus for this disease at chromosome 14q32.33. To prioritize the functional SNP(s) and target gene(s) at this locus, we employed an in silico fine-mapping approach using genotyped and imputed SNP data for 6,608 endometrial cancer cases and 37,925 controls of European ancestry. Association and functional analyses provide evidence that the best candidate causal SNP is rs2494737. Multiple experimental analyses show that SNP rs2494737 maps to a silencer element located within AKT1, a member of the PI3K/AKT/MTOR intracellular signaling pathway activated in endometrial tumors. The rs2494737 risk A allele creates a YY1 transcription factor-binding site and abrogates the silencer activity in luciferase assays, an effect mimicked by transfection of YY1 siRNA. Our findings suggest YY1 is a positive regulator of AKT1, mediating the stimulatory effects of rs2494737 increasing endometrial cancer risk. Identification of an endometrial cancer risk allele within a member of the PI3K/AKT signaling pathway, more commonly activated in tumors by somatic alterations, raises the possibility that well tolerated inhibitors targeting this pathway could be candidates for evaluation as chemopreventive agents in individuals at high risk of developing endometrial cancer.
Introduction
Endometrial cancer (MIM: 608089) (cancer of the lining of the uterine corpus) is the fourth most diagnosed cancer in women in Europe and North America.1, 2 To date, analyses of multiple genome-wide association study (GWAS) and follow-up datasets, comprising up to 7,737 endometrial cancer cases and 37,144 controls, have identified seven risk loci at genome-wide significance for this disease, including HNF1B (MIM: 189907),3, 4 CYP19A1 (MIM: 107910),5 and novel loci on chromosomes 13q22.1, 6q22.31, 8q24.21, 15q15.1, and 14q32.33.6 The lead SNP at the 14q32.33 locus, rs2498796, represents a single association signal located in the region of the AKT1 (MIM: 164730) oncogene.6 AKT1 is a member of the P13K/AKT/MTOR intracellular signaling pathway affecting cell survival and proliferation.7 This gene is of particular interest for endometrial cancer as increased PI3K/AKT/MTOR signaling is a common occurrence in endometrial tumors, and in aggressive subtypes in particular.8 Somatic alterations in one or more members of the PI3K/AKT/MTOR signaling pathway are common, with PTEN (MIM: 601728) as the most frequently altered gene.9 Moreover, high PIK3CA (MIM: 171834) copy number and elevated levels of phosphorylated AKT have been associated with aggressive disease.8, 10, 11 Our previous bioinformatic analysis indicated that rs2498796 and other SNPs in high linkage disequilibrium (LD) with this SNP might also regulate other nearby genes SIVA1 (MIM: 605567), ZBTB42 (MIM: 613915), ADSSL1 (MIM: 612498), and INF2 (MIM: 610982).6 Here, we detail in silico fine-mapping and bioinformatic investigation of an expanded set of genotyped and imputed SNPs at 14q32.33, derived from the meta-analysis dataset described above, and multiple laboratory analyses to identify the functional SNP(s) and target gene(s) increasing endometrial cancer risk at this locus.
Material and Methods
Previously, meta-analysis of data for 7,737 endometrial cancer cases and 37,144 controls of European ancestry from three GWAS datasets (ANECS, SEARCH, and NSECG) and two follow-up datasets (iCOGs and NSECG Phase 2) identified rs2498796 (OR = 1.12 for the minor A allele, 95% CI:1.07–1.17, p value = 3.55 × 10−8) as the top SNP representing a single association signal at the 14q32.33 endometrial cancer risk locus.6 For the current study we employed an in silico fine-mapping approach12 previously used to fine-map other endometrial cancer risk loci,4, 5, 13, 14 focussing on the 1Mb region surrounding rs2498796 (bases 104,743,220-105,743,220; NCBI build 37/hg19 assembly). The current analysis utilized genotyped and imputed SNP data for the three GWAS (ANECS, SEARCH, and NSECG) and the iCOGs follow-up datasets and included a total of 6,608 endometrial cancer cases and 37,925 controls (details of these datasets can be found in4, 5). The Cheng et al. analysis had included a total of 420 genotyped and imputed SNPs with minor allele frequencies (MAF) ≥1% and information scores ≥0.9 per dataset within the focal region.6 To expand the search for potentially functional SNPs, we considered all genotyped and imputed SNPs (N = 2,922) with MAF ≥1% and information scores ≥0.4 per dataset. As described previously4, regional imputation to the 1,000 Genomes v3 2012 release was conducted separately for each of the four datasets, based on inference panels of SNPs typed for each dataset, using IMPUTE v2.15 Association testing was performed separately for each dataset using frequentist tests with a logistic regression model in SNPTEST v2.16, and standard fixed effects meta-analysis using the beta estimates and standard errors per dataset conducted using METAL.17 The regional association plot was created using LocusZoom.18 Log-likelihood tests were used to determine the most likely causal SNPs by comparing the log-likelihoods obtained from the meta-analysis of our top SNPs (p < 10-6) with that of the most significantly associated SNP. SNPs with odds of 100:1 or better of being the top SNP were prioritized as potential causal candidates for bioinformatic and functional analyses.4, 19, 20 LD between SNPs was calculated from European Phase 3 1000 Genomes data and accessed from the National Cancer Institute LDlink tool.21
Bioinformatic Analysis
Bioinformatic analyses on SNPs prioritized by the log-likelihood tests were performed using publically available datasets from ENCODE22, which includes information such as the location of promoter and enhancer histone marks, open chromatin, bound proteins and altered motifs for the Ishikawa endometrial cancer cell line. Data from Hnisz et al.23 and PreSTIGE24 was accessed to identify the location of likely enhancers and their gene targets in a cell-specific context.
Expression Analyses
Expression quantitative trait locus (eQTL) analyses were conducted using uterine tissue-specific data (N = 70) generated by the Genotype-Tissue Expression Project (GTEx)25, and SNP (Affymetrix 6.0 arrays), RNA-seq and copy number (CNV) data for endometrial carcinoma samples (N = 526) and normal tissue samples adjacent to endometrial carcinoma (N = 29) obtained from restricted (SNP and RNA-Seq) and public (CNV) data portals of The Cancer Genome Atlas (TCGA).26 For the TCGA data, to investigate the expression of all AKT1 isoforms, including unannotated transcripts, unprocessed RNA-Seq FASTQ files were adapter trimmed using cutadapt (v1.8.1) and aligned to the Ensembl27 GRCh37 reference (version 70) using STAR28 (v2.4.2a). RNA-SeQC29 (v1.1.8.1) was used to assess sequencing quality for all aligned data. Gene and transcript counts were estimated using RSEM30 (v1.2.22). Genotypes for AKT1 region SNPs present in the 1000 Genomes v3 2012 dataset which were not present on the Affymetrix 6.0 arrays were imputed using MaCH31, 32 and minimac33, 34 software. eQTL analyses were performed on transcripts expressed in >80% of samples using Kruskal-Wallis tests adjusting for copy number and sequencing method, with Bonferroni corrected p values < 0.006 (0.05/8 transcripts per SNP) considered statistically significant.
Cell Lines
Endometrial cancer cell lines Ishikawa and EN-1078D (both heterozygous for the SNPs under investigation) were grown in DMEM medium with 10% FCS and antibiotics. Cell lines were maintained under standard conditions, routinely tested for Mycoplasma and short tandem repeat (STR) profiled to confirm cell line identity.
Chromatin Conformation Capture
Chromatin conformation capture (3C) libraries were generated using NcoI as described previously.35 3C interactions were quantitated by real-time PCR (qPCR) using primers designed within restriction fragments (Table S1). All qPCR was performed on a RotorGene 6000 using MyTaq HS DNA polymerase (Bioline) with the addition of 5 mM of Syto9, annealing temperature of 66°C and extension of 30 s. 3C analyses were performed in three independent 3C libraries from each cell line with each experiment quantified in duplicate. BAC clones covering the 14q32 region were used to create artificial libraries of ligation products in order to normalize for PCR efficiency. Data were normalized to the signal from the BAC clone library and, between cell lines, by reference to a region within GAPDH (MIM: 138400).35 All qPCR products were electrophoresed on 2% agarose gels, gel purified, and sequenced to verify the 3C product.
Electromobility Shift Assays
Gel shift assays were performed with Ishikawa and EN-1078D nuclear lysates and biotinylated oligonucleotide duplexes (Table S2). Nuclear lysates were prepared using the NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific) as per the manufacturer’s instructions. Total protein concentrations in nuclear lysates were determined by Bradford’s method. Duplexes were prepared by combining sense and antisense oligonucleotides in NEBuffer2 (New England Biolabs) and heat annealing at 80°C for 10 min and slow cooling to 25°C for 1 hr. Binding reactions were performed in binding buffer (10% [vol/vol] glycerol, 20 mM HEPES [pH 7.4], 1 mM DTT, protease inhibitor cocktail [Roche], 0.75 μg poly[dI:dC] [Sigma-Aldrich]) with 7.5 μg of nuclear lysate. For competition assays, binding reactions were pre-incubated with 1 pmol of competitor duplex (Table S3) at 25°C for 10 min before the addition of 10 fmol of biotinylated oligo duplex and a further incubation at 25°C for 15 min. For gel-supershift assays, 5 μg of rabbit polyclonal YY1 antibody (Santa Cruz H-414) or C/EBP antibody (Santa Cruz sc-150) was added immediately before probe addition. The rabbit pre-immune IgG (Santa Cruz sc-2027) was used as a negative control. Reactions were separated on 10% (WT/vol) Tris-Borate-EDTA (TBE) polyacrylamide gels (Bio-Rad) in TBE buffer at 160 V for 40 min. Duplex-bound complexes were transferred onto Zeta-Probe positively-charged nylon membranes (Bio-Rad) by semi-dry transfer at 25 V for 20 min then cross-linked onto the membranes under 254 nm ultra-violet light for 10 min. Membranes were processed with the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) as per the manufacturer’s instructions. Chemiluminescent signals were visualized with the C-DiGit blot scanner (LI-COR).
Plasmid Construction and Reporter Assays
Promoter-driven luciferase reporter constructs were generated by the insertion of PCR amplified fragments containing AKT1 canonical (can), AKT1 alternative (alt), or ZBTB42 promoters into the MluI and BglII sites of pGL3-Basic. A 2537 bp fragment containing a putative regulatory element (PRE) identified by bioinformatic analysis was generated by PCR and cloned into BamHI and SalI sites of the modified pGL3-promoter constructs (Table S4). The minor (risk-increasing) alleles of individual SNPs were introduced into the PRE sequences by overlap extension PCR or gBlocks (Integrated DNA Technologies). Sequencing of all constructs confirmed variant incorporation (Australian Genome Research Facility). Ishikawa and EN-1078D cells were transfected with equimolar amounts of luciferase reporter plasmids and 50 ng of pRL-SV40 transfection control plasmid with Lipofectamine 2000. The total amount of transfected DNA was kept constant at 600 ng for each construct by the addition of pUC19 as a carrier plasmid. Luciferase activity was measured 24 hr post-transfection by the Dual-Glo Luciferase Assay System. To correct for any differences in transfection efficiency or cell lysate preparation, we normalized Firefly luciferase activity to Renilla luciferase and measured the activity of each construct relative to the promoter alone construct, which had a defined activity of 1. Statistical significance was tested by log transforming the data and performing two-way ANOVA, followed by Dunnett’s multiple comparisons test in GraphPad Prism.
siRNA Silencing for Reporter Assays
Two Silencer Select siRNAs against YY1 (siYY1; s224779) and a Silencer Select nontargeting siRNA (siCON; 4390843) were purchased from Life Technologies (Thermo Fisher Scientific). For silencing, Ishikawa cells were co-transfected with the relevant luciferase reporter plasmids and 100 nM of either YY1 or non-targeting siRNAs with Lipofectamine 2000. Luciferase assays were performed as described above after 72 hr. qPCR was performed as described previously36 to validate YY1 knockdown.
Chromatin Immunoprecipitation
Ishikawa cells were cross-linked with 1% formaldehyde at 37°C for 10 min, rinsed once with ice-cold PBS containing 5% BSA and once with PBS, and harvested in PBS containing 1X protease inhibitor cocktail (Roche). Harvested cells were centrifuged for 2 min at 3,000 rpm. Cell pellets were resuspended in 0.35 mL of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, 1X protease inhibitor cocktail) and sonicated 3 times for 15 sec at 70% duty cycle (Branson SLPt) followed by centrifugation at 13,000 rpm for 15 min. Supernatants were collected and diluted in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1). Two micrograms of antibody was prebound for 6 hr to protein G Dynabeads (Life Technologies) and then added to the diluted chromatin for overnight immunoprecipitation. The magnetic bead-chromatin complexes were collected and washed six times in RIPA buffer (50 mM HEPES [pH 7.6], 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCl), then twice with TE buffer. To reverse the cross-linking, the magnetic bead complexes were incubated overnight at 65°C in elution buffer (1% SDS, 0.1 M NaHCO3). DNA fragments were purified using a QIAquick Spin Kit (Qiagen). For qPCR, 2.0 uL from a 100 uL immunoprecipitated chromatin extraction and 40 cycles of amplification were used. All PCR products were sequenced by Sanger sequencing (AGRF). Antibodies used were anti-NFκB p50 (06-886), anti-YY1 (sc-1703-X), and control IgG (sc-2027). ChIP primers are listed in Table S5.
Results
Association and Likelihood Testing at the 14q32.33 Endometrial Cancer Risk Locus Prioritizes Three SNPs for Follow-Up
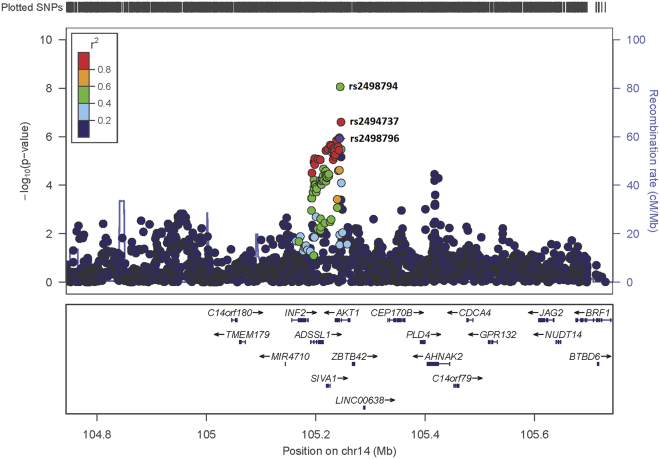
A total of 2,922 SNPs with MAF ≥1% and information scores ≥0.4 per endometrial cancer dataset were included in the fine-mapping analysis, representing 76.6% of the SNPs with a MAF ≥1% in the 1000 Genomes 2012 reference panel in this region (hg19 chr14: 104,743,220-105,743,220; Table S6). Considering SNPs correlated (r2 > 0.2) with rs2498796, the previously reported top endometrial cancer risk SNP at this locus,6 coverage was good, with >94% of correlated SNPs in the same 1000 Genomes reference panel represented in each dataset.
Association and log-likelihood tests prioritized two SNPs for bioinformatic and functional follow-up: rs2498794 (OR = 1.13, 95% CI 1.09–1.17, p value 8.7 × 10-9) and rs2494737 (OR = 1.13, 95% CI 1.08–1.17, p value 2.5 × 10-7; Table 1 and Table S7). No other SNP was significant at p < 1 × 10-4 in analyses conditioning on rs2498794 or rs2494737 (r2 to each other 0.54, Table S7), confirming the single association signal at this locus (Figure 1). As SNP rs2498796 (p value 1.2 × 10-6 in the current analysis, r2 to rs2498794 0.43, and to rs2494737 0.83; Table 1) was the original endometrial cancer risk SNP reported for this locus, it was also included in the bioinformatic and functional analyses detailed below. Neither rs2498794 nor rs2494737 had been reported in our previous genome-wide analysis because of the more stringent imputation threshold used in that study.
Table 1.
Association of the Top Candidate Causal SNPs at Chromosome 14q32.33 with Endometrial Cancer Risk
| SNP | Position (Build 19) | Minor allele | Common allele | MAFa | OR (95% CI)b | p value | r2to rs2498796 | Likelihood ratioc |
|---|---|---|---|---|---|---|---|---|
| rs2498794 | 105245251 | G | A | 0.48 | 1.13 (1.09–1.17) | 8.7 × 10−9 | 0.43 | 1 |
| rs2494737 | 105246325 | A | T | 0.30 | 1.13 (1.08–1.17) | 2.5 × 10−7 | 0.83 | 26 |
| rs2498796 | 105243220 | A | G | 0.30 | 1.11 (1.07–1.16) | 1.2 × 10−6 | -- | 120 |
Minor (risk-increasing) allele frequency.
OR for the effect allele.
Ratio of the likelihood of rs2498794 to the likelihood of this SNP.
Figure 1.
Regional Association Plot for the 14q32.33 Endometrial Cancer Risk Locus
The location (Build 19) and –log10 p value of the original top SNP at this locus, rs2498796,6 is shown in purple; all other SNPs are shown in colors corresponding to their r2 (linkage disequilibrium) value with rs2498796.
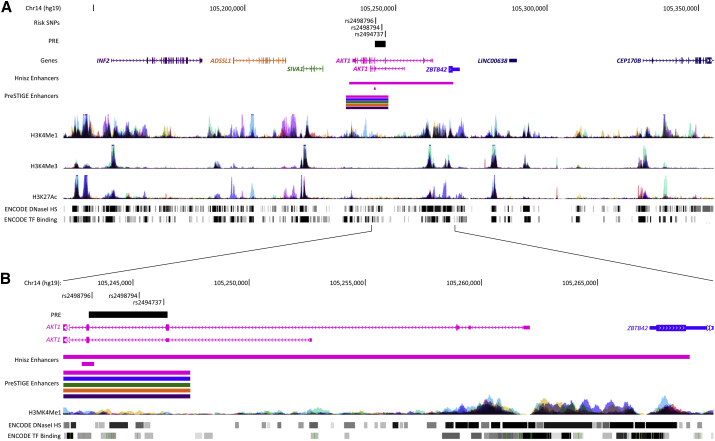
The Top Candidate SNPs Fall within a Putative Regulatory Element that Frequently Interacts with AKT1 and ZBTB42 Promoter Regions
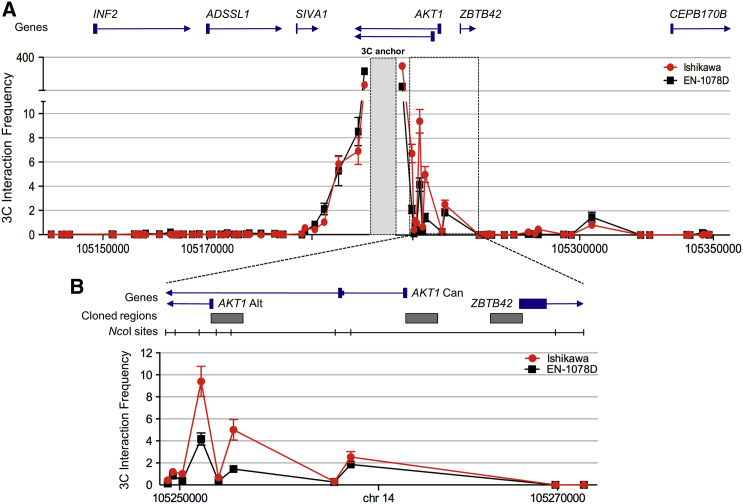
Analysis of cis enhancer-gene interactions using data from Hnisz et al.23 and PreSTIGE24 identified AKT1, ZBTB42, SIVA1, ADSSL1, and INF2 as potential candidate target genes of a PRE located in the region containing the top candidate SNPs (Figure 2). To determine the target gene(s) of the PRE, we performed chromosome conformation capture (3C) using an anchor primer within the PRE and primers within restriction fragments spanning all protein coding gene promoters within 2Mb of the PRE. The results showed that the PRE frequently interacted with a canonical and alternative promoter of AKT1 and the ZBTB42 promoter in both Ishikawa and EN-1078D endometrial cancer cells (Figures 3A and 3B). To assess any potential impact of SNP rs2494737 on chromatin looping, we performed allele-specific 3C in heterozygous Ishikawa cell lines. A primer was designed to incorporate the rs2494737 into the 3C PCR products, which were then Sanger sequenced. The sequence profiles indicate that the cancer risk- and non-risk-associated rs2494737 alleles form loops with the AKT1 and ZBTB42 promoters with equal efficiencies (Figure S1). No significant interactions were detected between the PRE and other flanking genes including SIVA1, ADSSL1, INF2, and CEPB170B (Figure 3A and Figure S2).
Figure 2.
Regulatory Landscape at the 14q32.33 Endometrial Cancer Risk Locus
(A) The location of the candidate SNPs are represented by black ticks, and the PRE is shown as a black box. Gene structures are depicted with exons (vertical boxes) joined by introns (lines). The subset of enhancers predicted in Hnisz et al.23 and PreSTIGE24 which overlap the candidate causal SNPs are shown as colored bars, where the color matches its predicted gene target. Regions showing histone binding (H3K4Me1, indicative of regulatory regions; H3K4Me3, indicative of promoters; and H3K27Ac, indicative of active enhancers), DNAseI hypersensitivity (indicative of open chromatin, with darker shading indicating stronger experimental signal) and transcription factor (TF) binding in multiple ENCODE cell lines are indicated at the bottom of the panel.
(B) Zoomed-in view of the location of candidate SNPs, PRE, and nearby gene promoter regions.
Figure 3.
Candidate Causal SNPs Are Located within a PRE that Interacts with the AKT1 and ZBTB42 Promoter Regions
(A) 3C interaction profiles between the PRE and local genes in Ishikawa and EN-1078D endometrial cancer cell lines. The 3C anchor (which contains the PRE) is shown as a grey box and significant interactions are outlined.
(B) Zoomed-in view of significant interactions. AKT1 Can and AKT1 Alt denote a canonical and alternative AKT1 promoter (prom) region, respectively. 3C libraries were generated with NcoI. Graphs represent three biological replicates. Error bars represent SD. Regions cloned into reporter gene constructs are shown as grey horizontal boxes.
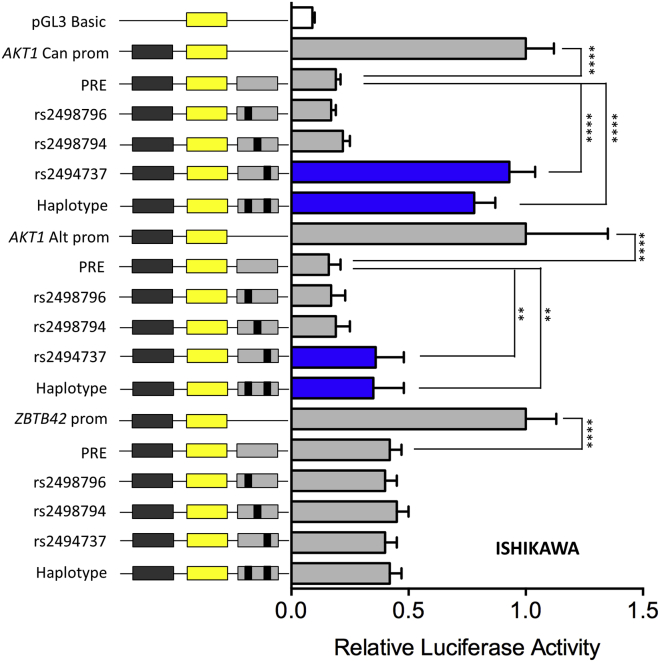
SNP rs2494737 Affects the Regulatory Capability of the PRE on AKT1 Promoter Regions
The regulatory capability of the PRE, combined with the effects of candidate SNPs, was examined in luciferase reporter assays in Ishikawa and EN-1078D cell lines. PRE constructs containing the reference (common, protective) alleles of the three candidate SNPs significantly reduced their associated target gene promoter activities, suggesting that the PRE can act as a transcriptional silencer (Figure 4 and Figure S3). Inclusion of the minor (risk-increasing) allele of rs2494737 significantly increased the canonical and alternative AKT1 promoter activities in both cell lines, but had no effect on the ZBTB42 promoter. In contrast, inclusion of the minor (risk-increasing) alleles of SNPs rs2498796 and rs2498794 had no significant effects on AKT1 or ZBTB42 promoter activities (Figure 4 and Figure S3).
Figure 4.
The Risk Allele of SNP rs2494737 Enhances AKT1 Promoter Activity
Luciferase reporter assays following transient transfection of Ishikawa endometrial cancer cell lines. The putative regulatory element (PRE) containing the major SNP alleles were cloned downstream of target gene promoter-driven luciferase constructs. AKT1 can and AKT1 alt denote a canonical and alternative AKT1 promoter (prom) region, respectively. Minor (risk-increasing) SNP alleles were engineered into the constructs and are designated by the rs ID of the corresponding SNP. Haplotype denotes a construct that contains the minor alleles of rs2498796 and rs2494737. Error bars denote 95% confidence intervals from three independent experiments performed in duplicate. P values were determined by 2-way ANOVA followed by Dunnett’s multiple comparisons test (∗∗p < 0.01, ∗∗∗∗p < 0.0001).
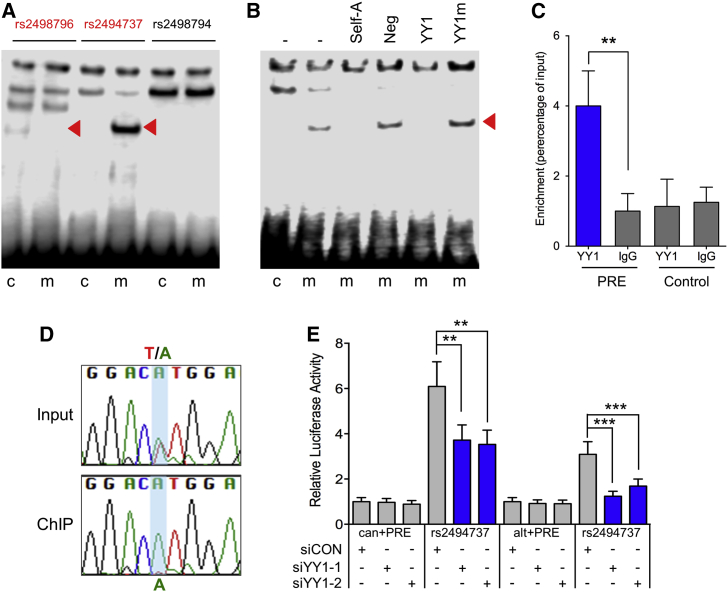
The Risk Allele of SNP rs2494737 Binds the YY1 Transcription Factor
We used bioinformatic analyses and functional studies to examine DNA-protein interactions for the three candidate SNPs. In silico prediction tools including HaploReg37 and Alibaba238 predicted all three SNPs to alter transcription factor (TF) binding (Table S8 and Figure S4). We performed electrophoretic mobility shift assays (EMSAs) to assess binding of TFs to the common (protective) and minor (risk-increasing) alleles of each of these SNPs and showed allele-specific protein binding for rs2494737 and rs2498796 (Figure 5A and Figure S5). Competition with TF binding sites suggested that YY1 binds to the minor (risk-increasing) allele of rs2494737 and NF-κB binds to the common allele of rs2498796 (Figure 5B and Figure S5). No other predicted TFs were able to compete for binding at either site, including CEBPA, AP2, and CREB (Figure S5). Supershift assays using anti-YY1 antiserum indicated that the protein binding the minor allele of rs2494737 is likely to be YY1 (Figure S6). Chromatin immunoprecipitation (ChIP) in heterozygous Ishikawa cells confirmed occupancy of YY1 binding in vivo and showed it is preferentially recruited to the minor A (risk-increasing) allele of rs2494737 (Figures 5C and 5D and Figure S7). The importance of YY1 binding was confirmed in cotransfection assays that showed that two independent siRNAs against YY1 repressed the promoter activation in the presence of the minor A allele of rs2494737 (Figure 5E and Figure S8). We found no evidence of CEBPA binding to the rs2494737 site or NF-κB binding to the rs2498796 site in vivo.
Figure 5.
The Risk Allele of rs2494737 Demonstrates Allele-Specific YY1 Binding
(A) EMSAs to detect allele-specific binding of nuclear proteins. Oligonucleotides were incubated with Ishikawa nuclear extracts. Red arrowheads show bands of different mobility detected between the common (C) and minor (risk-increasing) (m) alleles for the candidate causal SNPs.
(B) Oligonucleotides for SNP rs2494737 were incubated with Ishikawa nuclear extracts. Red arrowhead indicates the band that was competed for complex formation on the minor (m) allele. Competitor oligonucleotides are listed above each panel and were used at 100-fold molar excess: (−) no competitor; (Neg) a non-specific competitor; (YY1) consensus binding site; (YY1m) an identical oligonucleotide but with a mutated binding site.
(C) ChIP–qPCR on SNP rs2494737 in heterozygous Ishikawa cell lines. ChIP assays were performed with YY1 antibody or non-immune IgG, a region 3.2kb upstream of the predicted YY1-binding site served as a negative control (Control). Graphs represent two biological replicates. Error bars denote SD. P-values were determined with a two-tailed t-test (∗∗p < 0.01).
(D) Sanger sequencing of the PCR fragment generated using primers flanking SNP rs2494737 following YY1 ChIP-qPCR and the input DNA controls.
(E) Luciferase assays in Ishikawa cells shows the effect of YY1 siRNA silencing on the activity of the AKT1 canonical (can) and alternative (alt) promoter regions with the PRE containing the reference T allele (can+PRE; alt+PRE) or the risk A allele (rs2494737). Error bars denote 95% confidence intervals from three independent experiments performed in duplicate. P values were determined by two-way ANOVA followed by Dunnett’s multiple comparisons test (∗∗p < 0.01, ∗∗∗p < 0.001). The level of YY1 silencing is shown in Figure S8.
Gene-Expression Analysis in Uterine Tissue
Association between SNPs in the AKT1 region and AKT1 mRNA expression was investigated in both normal and endometrial tumor tissue. In the GTEx dataset, with the three candidate SNPs as input, rs2497896 was associated with increased AKT1 expression in normal uterine tissue (sample N = 70, p = 0.01, Figure S9), but no eQTL effect was detected for rs2494737 or rs2498794, suggesting a stochastic effect due to the reasonably small sample size as these SNPs are in moderate to high LD with each other. Performing an eQTL search for AKT1 in uterine tissue returned no results. However, including all GTEx tissues revealed all three candidate SNPs, and others in moderate to high LD, to be highly significantly associated with AKT1 expression in thyroid tissue (N = 278; rs2494737 = 3.6 × 10−14, rs2498796 = 5.10×10−25, and rs2498794=6.1 × 10−19; Table S7), indicating these SNPs are eQTLs for AKT1 in some cellular contexts. In the TCGA datasets, no SNP in the AKT1 region was associated with differential AKT1 expression of any isoform in normal endometrial tissue (N = 29) or in endometrial tumors (N = 526; Figure S10).
Discussion
In the largest association study for endometrial cancer to date, a recent meta-analysis of five GWAS and follow-up datasets revealed the presence of one multi-variant haplotype at the 14q32.33 chromosomal locus associated with the risk of this cancer.6 In consideration of the fact that our genotyping platforms were not specifically designed for fine-mapping of this region, we conducted in silico fine-mapping of the 14q32 region using SNPs with imputation scores down to 0.4. We identified two SNPs as most likely to be the causal SNPs increasing endometrial cancer risk in this region: SNPs rs2498794 and rs2494737, in moderate and high LD, respectively, with the original hit at this locus rs2498796.6 Multiple laboratory analyses then confirmed that rs2494737 has a functional impact on the AKT1 oncogene, a gene of potential biological relevance to endometrial cancer risk as other PI3K pathway mutations have been detected in precursor lesions of complex atypical endometrial hyperplasia.39
Our fine-mapping, together with multiple lines of bioinformatic and experimental evidence indicate that rs2494737 is the functional SNP most likely to be relevant for endometrial cancer at the 14q32 risk locus. However, additional bioinformatic analyses indicated multiple regulatory elements across the region that contained several less significantly risk-associated SNPs. Additionally, our SNP coverage of the region was not complete, although ∼98% of SNPs in at least moderate LD (r2 > 0.2), and 100% of SNPs in high LD (r2 > 0.8), with rs2498796 in the 1000 Genomes 2012 panel were also present in our datasets, and imputed to high quality scores (>0.71). Therefore, we cannot rule out the possibility that additional SNPs exert effects on AKT1 expression via alternative mechanisms. For example, at the well-characterized 8q24 risk locus, multiple risk-associated enhancers interact with MYC in a tissue-specific manner.40 Furthermore, a few recent studies have indicated that risk-associated SNPs might also influence epigenetic features,41, 42 adding yet another layer of complexity to the control of gene expression.
Publicly available enhancer data from multiple cell types indicate that the region harboring rs2494737 might target a number of genes in the 14q32 region, some of which are highly plausible endometrial cancer candidate genes. Our 3C analyses show that in endometrial tumor cells the rs2494737 region specifically targets AKT1 and ZBTB42, while luciferase assays showed that the rs2494737 minor A (risk-increasing) allele affects only AKT1, increasing the activity of both the canonical and an alternative promoter. Therefore, we expect the causal risk allele to result in increased expression of one or more AKT1 isoforms in vivo. Although we observed no significant effect of the rs2494737 minor allele on overall or isoform-specific AKT1 expression in normal uterine or endometrial tumor tissue, there are multiple possible explanations. One reason could be that the risk allele affects AKT1 expression in endometrial epithelial cells that represent only a fraction of the total cells in a normal uterine sample, which is composed of substantially more endometrial stromal cells as well as underlying myometrium. Any effect on expression might also occur only in specific cellular contexts. Further, the lack of association in the normal tissue sample sets examined might also be due to low power, with only 47% and 23% power to detect an effect of a SNP (MAF 0.3) explaining even 5% of the variance in AKT1 expression in the GTEx and TCGA datasets, respectively. We had 99.9% power to detect the same effect in the larger (N = 526) endometrial tumor dataset, although here any eQTL effect might be difficult to detect due to the overall increase in AKT1 expression seen in endometrial tumor cells in general. The apparent discrepancy between eQTL results and our in vitro findings is not unprecedented: functional SNPs in CCND1 (MIM: 168461)36 and MYC (MIM: 190080)43 show no association with gene expression in human tumor cells, although one MYC region SNP (rs6983267) has been demonstrated to have a functional effect in vivo.43
AKT1, a serine/threonine kinase highly expressed in the endometrium,44 regulates many processes including cell metabolism, proliferation, survival, growth, and angiogenesis45 and is already of considerable interest as a potential therapeutic target for endometrial cancer.9, 46 Activation of the PI3K/AKT/mTOR intracellular signaling pathway, of which AKT1 is a member, occurs in numerous cancers9, 45 and up to 80% of endometrial tumors.47 This pathway activation has been linked to somatic mutations and copy-number alterations in various PI3K/AKT/mTOR pathway genes, including inactivating mutations and deletion of the PTEN tumor-suppressor gene and activating mutations or amplifications in the PIK3R1 (MIM: 171833) and/or PIK3CA genes.48 AKT1 mutations are rare, with an oncogenic c.49G>A (p.Glu17Lys) (NM_005163.2) mutation occurring in only ∼2% of endometrial tumors,10, 11 and activation is thought to result from the concomitant loss or activation of upstream pathway proteins.48
Our results indicate a possible additional mechanism whereby the presence of a common SNP allele results in increased AKT1 transcriptional activity mediated through YY1 and results in an increased risk of endometrial cancer. YY1 is found at elevated levels in numerous cancers, including breast (MIM: 114480), prostate (MIM: 176807), and cervical cancers (MIM: 603956),49 and was recently demonstrated to be over-expressed particularly in early stage (I and II) endometrial tumors, indicating this transcription factor could be a molecular marker of early tumor development.50 Of note, YY1 knockdown using small inferring RNA (siRNA) and small hairpin RNA (shRNA) reduced YY1 protein levels, decreased cell proliferation and reduced cell motility of the AN3CA endometrial cancer cell line, while siYY1 injected directly into xenograft tumors in mice delayed endometrial tumor growth.50 Although these data would suggest YY1 is a potential therapeutic target, transcription factors are notoriously hard to target with small molecules. The data presented here suggests that elevated levels of YY1 are oncogenic in part through upregulation of AKT1 expression, which is a signaling pathway that is more amenable to drug targeting.51
Activation of AKT1 requires translocation to the plasma membrane followed by phosphorylation of the Thr308 and Ser473 residues: high levels of p-AKT1 are a marker of poor prognosis in endometrial and other cancers.8, 52, 53 A large number of inhibitors targeting mTOR and/or PI3K have been tested in early clinical trials in multiple tumor types, however, toxicity issues have complicated their ongoing development and many have not been taken forward into large phase III trials. Several AKT inhibitors are also in development, and initial clinical activity recently reported in several different solid tumor types including breast, lung, and gynaecological tumors carrying the AKT1 c.49G>A (p.Glu17Lys) hotspot mutation.54, 55 There is a current emphasis on reducing systemic toxicities by optimizing scheduling as well as evaluating nanoparticles to target these agents to tumors and reduce systemic exposure,56 nonetheless it is unlikely that AKT inhibitors developed for treatment of metastatic disease will have an acceptable toxicity profile to be used as chemopreventive agents.
A promising alternative might be the re-positioning of the type 2 diabetes drug metformin. This drug has multiple mechanisms of action targeting both metabolism, by decreasing circulating glucose levels, as well as altering intracellular signaling by activating AMPK.57 Activation of AMPK has been shown to inhibit mTOR, a downstream effector of PI3K/AKT signaling. Metformin is currently being evaluated in the adjuvant treatment of endometrial cancer58 as well as large chemoprevention trials (e.g. the Diabetes Prevention Program Outcomes Study). It would be interesting to determine the outcome analyses from these large-scale chemoprevention trials if patients were genotyped and retrospectively stratified based on their germline AKT1 risk alleles. Perhaps the ability of metformin to blunt mTOR signaling would be reflected in a greater decrease in endometrial cancer incidence in the AKT1 SNP carriers treated with metformin.
Although our data indicate that AKT1 is the likely target gene, it is possible that these SNPs also exert functional effects through long-range control of other genes under different conditions of cell activation or in other cell types, including nearby SIVA1, ZBTB42, ADSSL1, and INF2. Notably, SIVA1 is reported to activate and suppress apoptosis, a process dysregulated in cancer. Among other roles, SIVA1 can inhibit p53 tumor suppressor functions and is mutated in up to 90% of aggressive endometrial tumors.26, 59 ZBTB42 (zinc finger and BTB domain containing 42) is a poorly characterized member of the C2H2 zinc finger protein family60. It is highly expressed in subsynaptic nuclei in skeletal muscles underlying the neuromuscular junctions,60 and might be involved in muscle development.61 ADSSL1 (Adenylosuccinate Synthase Like 1) is a muscle isozyme that is selectively deleted in carcinogen-induced mouse lung adenocarcinomas.62 While INF2 (Inverted Formin 2) encodes a member of the diaphanous-related formin family, which is involved in remodelling the actin and microtubule cytoskeltons.63 Mutations in this gene are reported to cause a form of autosomal-dominant focal and segmental glomerulosclerosis and Charcot-Marie-Tooth disease.64, 65
In conclusion, we have identified a common SNP allele associated with endometrial cancer risk that functions to increase AKT1 expression through YY1-mediated transcription. Identification of an endometrial cancer risk allele within a member of the PI3K/AKT signaling pathway, more commonly activated in tumors by somatic alterations, raises the possibility that well-tolerated inhibitors targeting this pathway could be candidates for evaluation as chemopreventive agents in individuals at high risk of developing endometrial cancer.
Acknowledgments
The QIMR Berghofer groups were supported by a Rio Tinto Ride to Conquer Cancer (RTCC)/Weekend to End Women’s Cancers (WEWC) Grant and NHMRC project grants 1058415 to S.L.E. and 1031333 to A.B.S. A.M.D acknowledges a CRUK grant C8197/A16565. I.T. acknowledges core funding to the Wellcome Trust Centre for Human Genetics from the Wellcome Trust (090532/Z/09/Z). A.B.S. is supported by an NHMRC Senior Research Fellowship (1061779). The funders have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: June 2, 2016
Footnotes
Supplemental Data includes ten figures, eight tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.04.012.
Web Resources
1000 Genomes, http://www.1000genomes.org
ENCODE, https://www.encodeproject.org/
GTEx Portal, http://www.gtexportal.org/home/
National Cancer Institute LDlink tool, http://analysistools.nci.nih.gov/LDlink/
OMIM, http://www.omim.org/
PreSTIGE, http://genetics.case.edu/prestige/
QTL Genetic Power Calculator, http://pngu.mgh.harvard.edu/∼purcell/gpc/
The Cancer Genome Atlas, http://cancergenome.nih.gov/
Supplemental Data
References
- 1.Ferlay J.S.I., Ervik M., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. International Agency for Research on Cancer; Lyon, France: 2013. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Spurdle A.B., Thompson D.J., Ahmed S., Ferguson K., Healey C.S., O’Mara T., Walker L.C., Montgomery S.B., Dermitzakis E.T., Fahey P., Australian National Endometrial Cancer Study Group. National Study of Endometrial Cancer Genetics Group Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat. Genet. 2011;43:451–454. doi: 10.1038/ng.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Painter J.N., O’Mara T.A., Batra J., Cheng T., Lose F.A., Dennis J., Michailidou K., Tyrer J.P., Ahmed S., Ferguson K., National Study of Endometrial Cancer Genetics Group (NSECG) CHIBCHA Consortium. Australian National Endometrial Cancer Study Group (ANECS) RENDOCAS. Australian Ovarian Cancer Study (AOCS) GENICA Network Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum. Mol. Genet. 2015;24:1478–1492. doi: 10.1093/hmg/ddu552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson D.J., O’Mara T.A., Glubb D.M., Painter J.N., Cheng T., Folkerd E., Doody D., Dennis J., Webb P.M., Gorman M. CYP19A1 fine-mapping and Mendelian randomisation: estradiol is causal for endometrial cancer. Endocr. Relat. Cancer. 2015;23:77–91. doi: 10.1530/ERC-15-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng T.H., Thompson D.J., O’Mara T.A., Painter J.N., Glubb D.M., Flach S., Lewis A., French J.D., Freeman-Mills L., Church D., National Study of Endometrial Cancer Genetics Group (NSECG) Australian National Endometrial Cancer Study Group (ANECS) RENDOCAS. CHIBCHA Consortium. AOCS Group Five endometrial cancer risk loci identified through genome-wide association analysis. Nat. Genet. 2016 doi: 10.1038/ng.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 8.Salvesen H.B., Carter S.L., Mannelqvist M., Dutt A., Getz G., Stefansson I.M., Raeder M.B., Sos M.L., Engelsen I.B., Trovik J. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc. Natl. Acad. Sci. USA. 2009;106:4834–4839. doi: 10.1073/pnas.0806514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slomovitz B.M., Coleman R.L. The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin. Cancer Res. 2012;18:5856–5864. doi: 10.1158/1078-0432.CCR-12-0662. [DOI] [PubMed] [Google Scholar]
- 10.Shoji K., Oda K., Nakagawa S., Hosokawa S., Nagae G., Uehara Y., Sone K., Miyamoto Y., Hiraike H., Hiraike-Wada O. The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br. J. Cancer. 2009;101:145–148. doi: 10.1038/sj.bjc.6605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen Y., Shalmon B., Korach J., Barshack I., Fridman E., Rechavi G. AKT1 pleckstrin homology domain E17K activating mutation in endometrial carcinoma. Gynecol. Oncol. 2010;116:88–91. doi: 10.1016/j.ygyno.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Barrett J.H., Taylor J.C., Bright C., Harland M., Dunning A.M., Akslen L.A., Andresen P.A., Avril M.F., Azizi E., Bianchi Scarrà G., GenoMEL Consortium Fine mapping of genetic susceptibility loci for melanoma reveals a mixture of single variant and multiple variant regions. Int. J. Cancer. 2015;136:1351–1360. doi: 10.1002/ijc.29099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvajal-Carmona L.G., O’Mara T.A., Painter J.N., Lose F.A., Dennis J., Michailidou K., Tyrer J.P., Ahmed S., Ferguson K., Healey C.S., National Study of Endometrial Cancer Genetics Group (NSECG) Australian National Endometrial Cancer Study Group (ANECS) RENDOCAS. Australian Ovarian Cancer Study (AOCS) GENICA Network Candidate locus analysis of the TERT-CLPTM1L cancer risk region on chromosome 5p15 identifies multiple independent variants associated with endometrial cancer risk. Hum. Genet. 2015;134:231–245. doi: 10.1007/s00439-014-1515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Mara T.A., Glubb D.M., Painter J.N., Cheng T., Dennis J., Attia J., Holliday E.G., McEvoy M., Scott R.J., Ashton K., Australian National Endometrial Cancer Study Group (ANECS) National Study of Endometrial Cancer Genetics Group (NSECG) RENDOCAS. AOCS Group Comprehensive genetic assessment of the ESR1 locus identifies a risk region for endometrial cancer. Endocr. Relat. Cancer. 2015;22:851–861. doi: 10.1530/ERC-15-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 17.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Udler M.S., Tyrer J., Easton D.F. Evaluating the power to discriminate between highly correlated SNPs in genetic association studies. Genet. Epidemiol. 2010;34:463–468. doi: 10.1002/gepi.20504. [DOI] [PubMed] [Google Scholar]
- 21.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sal lari R., Lupien M., Markowitz S., Scacheri P.C. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium G.T., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., Benz C.C., Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flicek P., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchsberger C., Abecasis G.R., Hinds D.A. minimac2: faster genotype imputation. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghoussaini M., Edwards S.L., Michailidou K., Nord S., Cowper-Sal Lari R., Desai K., Kar S., Hillman K.M., Kaufmann S., Glubb D.M., Australian Ovarian Cancer Management Group. Australian Ovarian Cancer Management Group Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat. Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.French J.D., Ghoussaini M., Edwards S.L., Meyer K.B., Michailidou K., Ahmed S., Khan S., Maranian M.J., O’Reilly M., Hillman K.M., GENICA Network. kConFab Investigators Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2015;44:D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. (Gedrukt) 2002;2:S1–S15. [PubMed] [Google Scholar]
- 39.Hayes M.P., Wang H., Espinal-Witter R., Douglas W., Solomon G.J., Baker S.J., Ellenson L.H. PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin. Cancer Res. 2006;12:5932–5935. doi: 10.1158/1078-0432.CCR-06-1375. [DOI] [PubMed] [Google Scholar]
- 40.Ahmadiyeh N., Pomerantz M.M., Grisanzio C., Herman P., Jia L., Almendro V., He H.H., Brown M., Liu X.S., Davis M. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. USA. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voisin S., Almén M.S., Zheleznyakova G.Y., Lundberg L., Zarei S., Castillo S., Eriksson F.E., Nilsson E.K., Blüher M., Böttcher Y. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:103. doi: 10.1186/s13073-015-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell C.G., Finer S., Lindgren C.M., Wilson G.A., Rakyan V.K., Teschendorff A.E., Akan P., Stupka E., Down T.A., Prokopenko I., International Type 2 Diabetes 1q Consortium Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS ONE. 2010;5:e14040. doi: 10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sur I.K., Hallikas O., Vähärautio A., Yan J., Turunen M., Enge M., Taipale M., Karhu A., Aaltonen L.A., Taipale J. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 44.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 45.Vivanco I., Sawyers C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 46.Salvesen H.B., Haldorsen I.S., Trovik J. Markers for individualised therapy in endometrial carcinoma. Lancet Oncol. 2012;13:e353–e361. doi: 10.1016/S1470-2045(12)70213-9. [DOI] [PubMed] [Google Scholar]
- 47.Cheung L.W., Hennessy B.T., Li J., Yu S., Myers A.P., Djordjevic B., Lu Y., Stemke-Hale K., Dyer M.D., Zhang F. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fabi F., Asselin E. Expression, activation, and role of AKT isoforms in the uterus. Reproduction. 2014;148:R85–R95. doi: 10.1530/REP-14-0270. [DOI] [PubMed] [Google Scholar]
- 49.Castellano G., Torrisi E., Ligresti G., Malaponte G., Militello L., Russo A.E., McCubrey J.A., Canevari S., Libra M. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle. 2009;8:1367–1372. doi: 10.4161/cc.8.9.8314. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y., Zhou L., Lu L., Wang L., Li X., Jiang P., Chan L.K., Zhang T., Yu J., Kwong J. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013;32:3432–3442. doi: 10.1038/onc.2012.360. [DOI] [PubMed] [Google Scholar]
- 51.Courtney K.D., Corcoran R.B., Engelman J.A. The PI3K pathway as drug target in human cancer. J. Clin. Oncol. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gungorduk K., Ertas I.E., Sahbaz A., Ozvural S., Sarica Y., Ozdemir A., Sayhan S., Gokcu M., Yilmaz B., Sanci M. Immunolocalization of ERK1/2 and p-AKT in normal endometrium, endometrial hyperplasia, and early and advanced stage endometrioid endometrial adenocancer and their prognostic significance in malignant group. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014;179:147–152. doi: 10.1016/j.ejogrb.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 53.Zhu Z., Yu W., Fu X., Sun M., Wei Q., Li D., Chen H., Xiang J., Li H., Zhang Y. Phosphorylated AKT1 is associated with poor prognosis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015;34:95. doi: 10.1186/s13046-015-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyman, D.M., Smyth, L., Bedard, P.L., Oza, A., Dean, E., Armstrobg, A., Lima, J., Bando, H., Kabos, P., Perez-Fidalgo, J.A., et al. (2015). AZD5363, a catalytic pan-Akt inhibitor, in Akt1 E17K mutation positive advanced solid tumors. Molecular Targets and Cancer Therapeutics Meeting Boston Clinical trial number NCT01226316.
- 55.Eathiraj, S., Schwartz, B., Yu, Y., Wick, M.J., Hall, T., Chai, F., Sachdev, J., and Abbadessa, G. (2015). Targeting PI3K pathway dependent endometrial tumors with allosteric AKT inhibitors, ARQ 092 and ARQ 751 Molecular Targets and Cancer Therapeutics Meeting Boston.
- 56.Cadzow L., Lam M.H., Wang H., DeWitt D., Pucci V., Mo J.-R., Lewies-Clark E., Ferguson H., Gindy M., Low S. Molecular Targets and Cancer Therapeutics Meeting Boston; 2015. Accurins improve the pharmacokinetics, pharmacodynamics, tolerability and anti-tumor activity of the AKT inhibitor MK-2206. [Google Scholar]
- 57.Rena G., Pearson E.R., Sakamoto K. Molecular mechanism of action of metformin: old or new insights? Diabetologia. 2013;56:1898–1906. doi: 10.1007/s00125-013-2991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porta C., Paglino C., Mosca A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front. Oncol. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X., Zha M., Zhao X., Jiang P., Du W., Tam A.Y., Mei Y., Wu M. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat. Commun. 2013;4:1551. doi: 10.1038/ncomms2533. [DOI] [PubMed] [Google Scholar]
- 60.Devaney S.A., Mate S.E., Devaney J.M., Hoffman E.P. Characterization of the ZBTB42 gene in humans and mice. Hum. Genet. 2011;129:433–441. doi: 10.1007/s00439-010-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel N., Smith L.L., Faqeih E., Mohamed J., Gupta V.A., Alkuraya F.S. ZBTB42 mutation defines a novel lethal congenital contracture syndrome (LCCS6) Hum. Mol. Genet. 2014;23:6584–6593. doi: 10.1093/hmg/ddu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller J.C., Blake D.C., Jr., Herzog C.R. Adenylosuccinate synthetase 1 gene is a novel target of deletion in lung adenocarcinoma. Mol. Carcinog. 2009;48:1116–1122. doi: 10.1002/mc.20563. [DOI] [PubMed] [Google Scholar]
- 63.Chesarone M.A., DuPage A.G., Goode B.L. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 64.Barua M., Brown E.J., Charoonratana V.T., Genovese G., Sun H., Pollak M.R. Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int. 2013;83:316–322. doi: 10.1038/ki.2012.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyer O., Nevo F., Plaisier E., Funalot B., Gribouval O., Benoit G., Huynh Cong E., Arrondel C., Tete M.J., Montjean R. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.