Abstract
Autosomal-dominant polycystic kidney disease (ADPKD) is a common, progressive, adult-onset disease that is an important cause of end-stage renal disease (ESRD), which requires transplantation or dialysis. Mutations in PKD1 or PKD2 (∼85% and ∼15% of resolved cases, respectively) are the known causes of ADPKD. Extrarenal manifestations include an increased level of intracranial aneurysms and polycystic liver disease (PLD), which can be severe and associated with significant morbidity. Autosomal-dominant PLD (ADPLD) with no or very few renal cysts is a separate disorder caused by PRKCSH, SEC63, or LRP5 mutations. After screening, 7%–10% of ADPKD-affected and ∼50% of ADPLD-affected families were genetically unresolved (GUR), suggesting further genetic heterogeneity of both disorders. Whole-exome sequencing of six GUR ADPKD-affected families identified one with a missense mutation in GANAB, encoding glucosidase II subunit α (GIIα). Because PRKCSH encodes GIIβ, GANAB is a strong ADPKD and ADPLD candidate gene. Sanger screening of 321 additional GUR families identified eight further likely mutations (six truncating), and a total of 20 affected individuals were identified in seven ADPKD- and two ADPLD-affected families. The phenotype was mild PKD and variable, including severe, PLD. Analysis of GANAB-null cells showed an absolute requirement of GIIα for maturation and surface and ciliary localization of the ADPKD proteins (PC1 and PC2), and reduced mature PC1 was seen in GANAB+/− cells. PC1 surface localization in GANAB−/− cells was rescued by wild-type, but not mutant, GIIα. Overall, we show that GANAB mutations cause ADPKD and ADPLD and that the cystogenesis is most likely driven by defects in PC1 maturation.
Introduction
Autosomal-dominant polycystic kidney disease (ADPKD) is one of the most common inherited disorders—it affects ∼1/1,000 individuals worldwide—and is characterized by progressive cyst development and expansion in the kidneys.1, 2 In ∼50% of affected individuals, ADPKD results in end-stage renal disease (ESRD), and 4%–10% of ESRD worldwide is due to ADPKD (see GeneReviews in Web Resources). ADPKD is caused by mutations in PKD1 (MIM: 601313) or PKD2 (MIM: 173910) (∼85% and ∼15% of mutation-resolved families, respectively).3, 4, 5, 6 Genotype-phenotype studies indicate an average age of 55.6 years for ESRD associated with truncating PKD1 mutations, 67.9 years for non-truncating PKD1 mutations, and 79.7 years for polycystic kidney disease 2 (PKD2 [MIM: 613095]).7 Larger kidneys (measured by the height-adjusted, MRI-determined total kidney volume [htTKV]) and an earlier decline in renal function (measured by the estimated glomerular filtration rate [eGFR]) are also associated with PKD1 (MIM: 173900).5, 8 The PKD1- and PKD2-associated proteins, polycystin 1 (PC1) and 2 (PC2), respectively, are membrane glycoproteins with the primary cilium as a likely functional site.9 PC1 is cleaved at the G-protein-coupled receptor proteolytic site (GPS), and this cleavage is essential for its function.10, 11 PC1 consists of two glycoforms of the N- and C-terminal (NT and CT, respectively) GPS-cleaved products: (1) mature, endoglycosidase H (EndoH) resistant (NTR and CTR) and (2) immature, EndoH sensitive (NTS and CTS).12, 13, 14 The level of the mature glycoforms is associated with disease severity, and forming a complex with PC2 is critical for PC1 maturation and surface and ciliary localization.12, 15, 16
In a number of comprehensive studies of the genes mutated in ADPKD, 7%–10% of families are genetically unresolved (GUR).5, 6, 7, 17 The typical finding of mild kidney disease in GUR cases5 suggests that few such families are explained by missed, fully penetrant mutations at the complex, segmentally duplicated PKD1 locus.3, 18 However, hypomorphic mutations at the existing loci—including those due to mosaicism,19, 20 phenocopies associated with autosomal-dominant tubulointerstitial kidney disease (ADTKD) loci,21, 22 and phenotypic and screening mistakes—most likely explain some unlinked and GUR families.23 Nevertheless, additional genetic heterogeneity seems possible.
Subjects affected by ADPKD have a ∼5× higher risk of developing intracranial aneurysms (ICAs).24 However, polycystic liver disease (PLD) is the most frequent extra-renal complication in ADPKD; a minority of mainly females develop severe PLD that requires surgical intervention.25, 26 Unlike severe PKD, severe PLD is not associated with a particular genic or PKD1 allelic type.25 Isolated, autosomal-dominant PLD (ADPLD [MIM: 174050]) is a separate inherited disorder where severe PLD can also occur, but renal cysts are absent or very rare.27 ADPLD is caused by mutations in PRKCSH (MIM: 177060), SEC63 (MIM: 608648), or LRP5.28, 29, 30, 31 PRKCSH encodes the β subunit of glucosidase II (GIIβ);32, 33 GII is an endoplasmic-reticulum (ER)-resident enzyme that catalyzes hydrolysis of the middle and innermost glucose residues of peptide-bound oligosaccharides, and it triggers quality-control assessment of glycoprotein folding through the calnexin and calreticulin cycle.34, 35, 36, 37, 38, 39 SEC63 facilitates the translocation of secreted or membrane proteins across the ER membrane.40, 41, 42, 43 Induced loss of Prkcsh or Sec63 in mouse kidneys results in PKD, and an interactive PC1-PC2 network with PC1 as the rate-limiting component has been proposed to maintain tubular differentiation.13 This suggests that PRKCSH mutations might act through PC1 depletion, although PC2 reduction has also been noted with Prkcsh depletion or loss.44, 45 There is strong evidence of further genetic heterogeneity in ADPLD, given that only ∼50% of cases have been explained by the known genes mutated in this disease.27, 46
In this study, we employed global and focused screening of GUR ADPKD- and ADPLD-affected families to identify a gene mutated in these disorders. Characterization of cells null for this gene links the pathogenesis to the maturation and localization of PC1 and PC2.
Subjects and Methods
Sample and Data Collection and Clinical Analysis
The relevant institutional review boards and ethics committees approved all studies, and participants gave informed consent. Blood samples for DNA isolation were collected from the proband and all available family members and were isolated by standard methods by the Mayo Biospecimens Accessioning and Processing Core or the Genkyst study. Clinical and imaging data were obtained by review of clinical records. Total kidney volume (TKV) and total liver volume (TLV) were measured from clinical MRI and computed-tomography (CT) imaging at the Mayo Translational PKD Center, which employed the stereology method with Analyze software or a semi-automated approach,47 and were adjusted for height (htTKV or htTLV). Enlarged kidneys or livers were defined as the mean + 2 SDs of the normal htTKV or htTLV adjusted for average height.26, 48 Kidney function was calculated from clinical serum creatinine measurements with the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) formula in adults49 and the Schwarz formula in the pediatric individual50 and expressed as mL/min/1.73 m2. Age at onset of high blood pressure was defined as when the affected individual started antihypertensive medication.
Whole-Exome Sequencing and Bioinformatics Analysis
Families were defined as GUR when no mutations were detected from Sanger sequencing or multiplex ligation-dependent probe amplification of PKD1 and PKD2. In addition, the Genkyst cohort, including families PK20016, PK20017, and P1174, were screened for HNF1B (MIM: 189907) mutations, and families with a possible ADPLD diagnosis, including P1073, M472, and M656, were screened for PRKCSH and SEC63 mutations.
Total genomic DNA was quantified with a Qubit 2.0 fluorometer (dsDNA BR Assay Kit, Thermo Fisher Scientific) and quality checked with a NanoDrop. Subsequently, 250 ng or 1 μg of DNA was sheared by sonication (Covaris E210, 150–200 bp) and purified by AMPure XP beads (Agencourt). The shearing efficiency was checked on a Agilent Bioanalyzer 2100 (DNA1000 assay). Whole-exome capture and Illumina library preparation were performed with the Agilent SureSelectXT Human All Exon V5+UTRs Kit on an Agilent Bravo workstation. The enriched library was sequenced with 101 bp paired-end reads on an Illumina HiSeq 2000 in the Mayo Medical Genome Facility. On average, three to four exomes per lane were multiplexed. Genome_GPS v.3.0.1 (Mayo Bioinformatics Core) was employed as a comprehensive secondary analysis pipeline for variant calling. In short, FASTQ files were aligned to the hg19 reference genome (UCSC Genome Browser) with Novoalign (V2.08.01) with the options -hdrhd off -v 120 -c 4 -i PE 425,80 -x 5 -r Random, and realignment and recalibration were performed with the Genome Analysis Toolkit (GATK) (3.3-0) Best Practices v.3 for each family separately. Overall, 75.6% of mapped reads aligned to the captured region, and 98.9% of the captured region was covered at 10× read depth. Multi-sample variant calling was performed with the GATK (3.3-0) Haplotype Caller, and variants were filtered with Variant Quality Score Recalibration for both SNVs and indels. Variant mining was performed with Golden Helix SNP & Variation Suite v.8 (SVS). All families were analyzed independently with the following filters: (1) quality filter of read depth ≥ 10× and genotype quality ≥ 20, (2) selection according to autosomal-dominant sample genotype pattern, (3) removal of Exome Aggregation Consortium (ExAC) Browser variants with a minor allele frequency > 0.1%, and (4) characterization of coding and non-coding SNVs within 14 bp of the splice site and subsequent removal of SNPs predicted to be neutral by one or more of six dbNSFP tools (SIFT, PolyPhen-2 HVAR, MutationTaster, Mutation Assessor, FATHMM, and FATHMM MKL) and dbscSNV (removal of SNVs with Ada and RF scores < 0.6). Table S1 lists the variants that remained after the SVS analysis.
Sanger Sequencing and Mutation Validation
Primers to amplify the 25 exons of GANAB (GenBank: NM_198335.3), plus ∼100 bp of flanking intervening sequences (IVSs), were designed with MacVector 12.0.6, and 50 ng of genomic DNA was used for PCR amplification (primers and PCR conditions are available upon request). Sanger sequencing was performed at Beckman Coulter Genomics according to standard approaches, and variants were identified with Mutation Surveyor software (SoftGenetics) and designated on the 25 exon GANAB isoform and corresponding protein (GenBank: NP_938149.2). When samples were available, segregation analysis was performed by sequence analysis of the mutated exonic fragment. Amino acid changes detected by Sanger sequencing were evaluated with the programs SIFT and Align GVGD, and the atypical splicing change in family M656 was evaluated with NNSPLICE 0.9 (Berkeley Drosophila Genome Project). The families affected by GANAB mutations originate from the US (M263, M641, 290100, M656, and M472), France (PK20016 and PK20017), Macedonia (P1174), and Spain (P1073). One family came from the HALT Progression of Polycystic Kidney Disease (HALT-PKD) cohort, two came from Genkyst, and the remainder came from the Mayo PKD Center population.
Using CRISPR/Cas9 to Generate Targeted GANAB-Mutated Cell Lines
Guide RNAs predicted to have the lowest off-targeting effect were cloned into pX330 (SpCas9) and verified by sequencing (see Table S2 for sequences). GANAB ex11-IVS12 was amplified by PCR (primer sequences are available on request), and the 550 bp genomic product was cloned into pCAG-EGxxFP51 with restriction sites BamHI and EcoRI. Each gRNA was then co-transfected with p-CAG-EG-GANAB(11-IVS-12)-FP in renal cortical tubular epithelial (RCTE) cells and scored after 24 hr for EGFP-positive cells. gRNA4 was selected as the most efficient cutter and used for generating cell lines. pX330-gRNA4 was transfected into wild-type (WT) RCTE cells by electroporation, and the cells were allowed to recover for 36 hr prior to splitting and re-seeding as single-cell suspensions in a 96-well plate. Cells were grown for 10 days for the establishment of single clone cell colonies, split in half, and re-seeded for screening. Screening was performed with genomic DNA extraction followed by amplification using GANAB ex11-IVS12 primers and a subsequent T7 mismatch assay. For this assay, PCR amplicons were denatured at 95°C for 2 min and cooled gradually to 25°C at a rate of −2°C per second. The reaction mixture was then subjected to T7 endonuclease (T7E1, New England BioLabs) for 20 min at 37°C and visualized in 2% agarose gels. Clones were additionally screened by western blotting (WB) for the α subunit of glucosidase II (GIIα) and Sanger sequencing.
Generating FLAG-GIIα Constructs with Missense Variants
C-terminal Myc-DDK-tagged GIIα (isoform 3 [GenBank: NM_198335.3]) was obtained from OriGene. Missense mutations were introduced by site-directed mutagenesis with Q5 high-fidelity polymerase (New England BioLabs); primer sequences are shown in Table S3.
WB Studies, Glycosylation Analysis, and Immunoprecipitation
For purification of crude membrane protein, cells were grown to confluence, washed with Dulbecco’s PBS (DPBS), scraped and reconstituted in LIS buffer (10 mM Tris HCl [pH 7.4], 2.5 mM MgCl2, and 1mM EDTA) plus protease inhibitors (Complete, Roche), and incubated on ice for 15 min. Homogenized total membrane lysates were prepared by repeated passage through a 25.5G needle and centrifugation at 4,600 RPM for 5 min. All procedures employed pre-chilled containers and were performed in the cold room to minimize protein loss to degradation. For immunoprecipitation (IP), the pellet of crude membrane protein (75 mg) was solubilized in IP buffer, and samples were pre-cleared with blank A/G agarose for 2 hr. Antibodies to the C terminus of PC1 (PC1-CT) or PC2 (YCE2) were added overnight and then incubated with 50 mL of packed washed A/G Agarose (Thermo) for 2 hr. The agarose was washed three times in IP buffer and once in ice-cold H2O, and the protein was eluted in either lithium dodecyl sulfate (LDS) plus tris(2-carboxyethyl)phosphine (TCEP) or agarose, split into three equal parts (untreated, EndoH, and PNGaseF), and subjected to deglycosylation analyses. Purified membrane protein and IP eluates were deglycosylated with EndoH and PNGaseF according to the manufacturer’s (New England BioLabs) instructions. Twenty-five micrograms of input and 100% of the IP were loaded per SDS-PAGE lane.
Surface Glycoprotein Labeling and Immunoprecipitation
Surface glycoprotein labeling of living cells was performed as previously described.15 Monolayer cells in 10 cm culture dishes were washed twice with ice-cold DPBS and then oxidized at 4°C in 1 mM NaIO4 containing DPBS (pH 7) for 30 min, quenched with ice-cold 1 mM glycerol in DPBS for 5 min, and washed twice with ice-cold DPBS. Oxime ligation was performed in the presence of 10 mM Aniline (Sigma-Aldrich) and 100 μM EZ-Link Alkoxyamine-PEG4-Biotin (Thermo Fisher Scientific) in 1% FBS supplemented with ice-cold DPBS buffer (pH 7.5) for 1 hr. Biotinylated cells were then washed three times in PBS and scraped. Reaction specificity was ensured with streptavidin-488 (Alexa Fluor) staining of a small sample of cells and visualized by fluorescent microscopy. Scraped cells were collected by centrifugation, subjected to purification of crude membrane protein, and solubilized in IP buffer (20 mM HEPES [pH 7.5], 137 mM NaCl, 1% NP-40, 10% (w/v) glycerol, 2 mM EDTA, and 2.5 mM MgCl2) supplemented with protease inhibitors (Roche) for 15 min. Non-solubilized protein was removed by centrifugation and discarded. Neutravidin agarose was washed in IP buffer three times, and 50 mL of packed agarose was added to solubilized membrane protein and agitated at 4°C for 2 hr. Samples were washed three times in IP buffer and once in ice-cold H2O. Protein was eluted in either LDS plus TCEP or agarose and subjected to deglycosylation analyses. Tris-acetate gels were washed with ultrapure water and stained with the ProteoSilver Silver Stain Kit (Sigma-Aldrich) according to the manufacturer’s instructions.
Transfection, Confocal Microscopy, Immunofluorescence, and Surface PC1 Labeling
RCTE cells were split at a ratio of 1:2 the day before electroporation and transfected at ∼80% confluency. Electroporation of RCTE cells was performed with the Bio-Rad Gene Pulser with a square-wave protocol: 110V, 25 ms pulse and 0.2 cm cuvettes (Bio-Rad) in electroporation buffer (20 mM HEPES, 135 mM KCl, 2 mM MgCl2, and 0.5% Ficoll 400 [pH 7.6]). TagGFP-PC2 and mCherry-PC1 used in this study have been previously described.15 RCTE cells were grown on glass coverslips, washed once with DPBS, fixed in 3.5% paraformaldehyde (PF) for 30 min, permeabilized with 0.1% Triton in DPBS (pH 7.5), washed again in PBS, and incubated in blocking buffer (10% normal goat serum, 1% BSA, and 0.1% Tween in PBS [pH 7.5]) for 30 min. After three PBS washes, primary antibodies were added to IF buffer (1% BSA, PBS [pH 7.5], and 0.1% Tween) for 2 hr at room temperature or overnight at 4°C with gentle agitation. After three PBS washes, conjugated secondary antibody (AlexaFluor, Invitrogen) was added for 1 hr. DAPI was added for 1 min to stain nuclei.
For surface labeling of mCherry-PC1, transfected RCTE cells were cooled at 4°C for 15 min, washed once in ice-cold PBS and pre-chilled mCherry antibody (BioVision), and incubated in 0.5% BSA in PBS for 30 min at 4°C. Cells were then fixed in 3.5% PF, and conjugated secondary antibody was added to IF buffer for 30 min. Confocal microscopy was performed with a Zeiss Axiovert equipped with Apotome.
For pH shift and SDS IF, the fixation and antigen-retrieval method was performed for visualizing endogenous PC2 in MEFs. For partially denaturing the protein, cells were grown to 100% confluency and serum starved for 48 hr, fixed in 3% PF (pH 7.5) for 15 min, fixed in 4% PF (pH 11]) in 100 mM borate buffer for 15 min, and then permeabilized in 5% SDS for 5 min. Subsequently, PC2 antibody (H280) staining was performed overnight at a 1:200 dilution at 4°C as described above.
Antibodies
The following antibodies were used: PC1-NT, 7e1252 (N-terminal, mouse monoclonal; 1/1,000 for WB); PC1-CT, EB08670 (C-terminal, goat; Everest Biotech; 1/250 for IP); PC2, H280 (rabbit; Santa Cruz; 1/5,000 for WB and 1/200 for IF); PC2, YCE2 (mouse monoclonal; Santa Cruz; 1/2,000 for WB and 1/500 for IF); EGFR (rabbit; BD Transduction labs; 1/1,000 for WB); acetylated α-tubulin (Invitrogen; 1/5,000 for IF); mCherry, 5993-100 (rabbit; BioVision; 1/1,000 for surface labeling); FLAG M2 (Sigma; 1/1,000 for IF); and GIIα, ab179805 (Abcam; 1/2,000 for WB).
Results
WES Identifies GANAB as a Gene Mutated in ADPKD
After PKD1 and PKD2 mutation analysis for base-pair and larger rearrangements, we identified 327 GUR families out of ∼3,600 screened. These GUR families originated from the HALT-PKD cohort (64) (an ADPKD clinical trial) and the Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) (16) and Genkyst (124) ADPKD observational studies. In addition, 123 Mayo PKD Center GUR families affected by ADPKD or mild renal cystic disease and PLD were included. Although a firm clinical diagnosis of ADPKD was made in 247 GUR pedigrees, in 80 pedigrees the disease presentation was more atypical (cystic kidney disease without kidney enlargement), although the vast majority exceeded the defined ultrasound or MRI criteria for an ADPKD diagnosis.53, 54 In the seven remaining families, the disease presentation was more consistent with ADPLD, although all but one had some renal cysts. PRKCSH and SEC63 screening was also performed in these families before inclusion. Six multiplex GUR ADPKD-affected families were screened by WES, and standard screening methods were employed to identify mutated, dominant genes (see Subjects and Methods); the detected genes and variants are listed in Table S1. A missed PKD1 mutation was identified in one family (M560).
From this WES analysis, one candidate gene, GANAB, encoding the catalytic subunit of glucosidase II (GIIα), appeared most promising given that PRKCSH encodes the non-catalytic β subunit of this enzyme, and mutations in this gene cause ADPLD. GANAB (in chromosomal region 11q12.3; genomic size 21.9 kb) has two splice forms shown by in silico and RT-PCR analysis to be approximately equally expressed in the human kidneys and liver (Figure S1): isoform 3 (GenBank: NM_198335.3) has 966 aa (∼110 kDa), 25 exons, and 2,898 bp of coding sequence, and isoform 2 (GenBank: NM_198334.2) has 944 aa (∼107 kDa), 24 exons (in-frame skipping of exon 6), and 2,832 bp of coding sequence.36, 38 We employed the larger splice form for mutation screening and designation and functional studies. ExAC Browser exome data (60,706 individuals) list five loss-of-function (LoF) GANAB mutations out of an expected 49.7 (probability of LoF intolerance [pLI] = 1.0; see Subjects and Methods), a figure similar to that for PKD2 (7/37.6; pLI = 1.0) and consistent with GANAB mutations’ causing dominant disease.55, 56
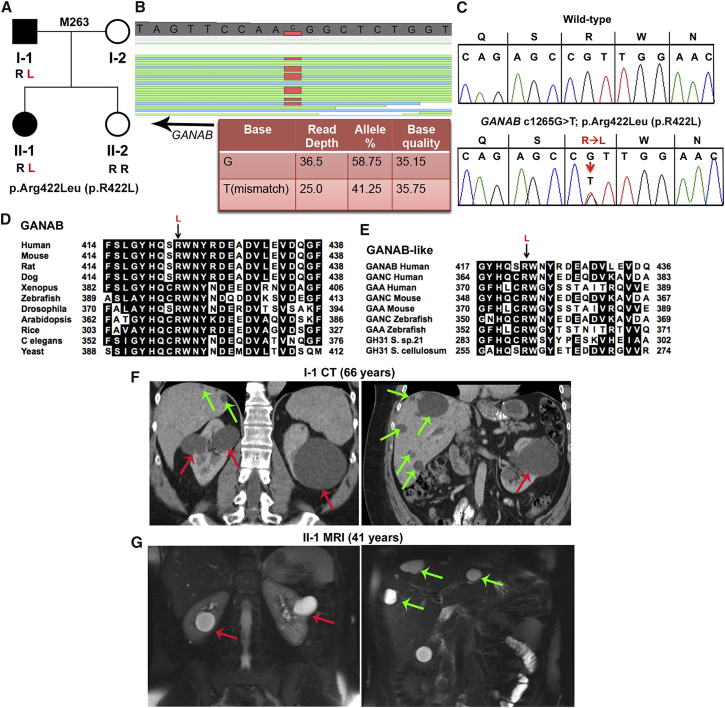
The GANAB missense variant, c.1265G>T (p.Arg422Leu), was detected by WES in family M263 and found in the affected father and daughter, but not the unaffected daughter (Figures 1A–1C). In silico analysis, including conservation in multi-sequence alignments (MSAs) of proteins orthologous to yeast and related glucosidases showed that this variant is highly predicted to be pathogenic (Figures 1D and 1E). Both of the affected individuals had mild kidney and significant liver cystic disease (Figures 1F and 1G and Table 1).
Figure 1.
WES Analysis Reveals GIIα p.Arg422Leu (c.1265G>T) in Family M263 as the Likely Pathogenic Variant
(A) Pedigree of family M263 shows that the two affected individuals (I-1 and II-1, shaded in black) have the GIIα p.Arg422Leu (p.R422L) missense variant resulting from c.1256G>T in exon 12, but the unaffected daughter (II-2, for whom no cysts were detected on ultrasound at 30 years of age) does not.
(B) GenomeBrowse (SVS, Golden Helix) view of the WES results from II-1 show GANAB variant c.1265G>T (reverse strand), and details of the reads are tabulated below.
(C) Sanger sequencing confirmation of heterozygous GIIα variant p.Arg422Leu (p.R422L) (c.1256G>T) in II-1. The WT is shown for comparison.
(D) MSA of GIIα orthologous proteins shows invariance of Arg422 from humans to yeast. In silico analysis of the likely pathogenicity of GIIα p.Arg422Leu (p.R422L) shows variant scores (SIFT = 0.00, Align GVGD = C65) characteristic of a highly likely pathogenic mutation.
(E) MSA of related glucosidases GANC (neutral alpha-glucosidase C) and GAA (lysosomal alpha-glucosidase) of various eukaryotic species and prokaryotic GH31 (glycosyl hydrolase family 31) shows invariant conservation of GIIα Arg422.
(F) CT scan with contrast of kidneys and liver of individual I-1 at 66 years shows a few large kidney cysts (red arrows) and multiple scattered liver cysts (green arrows).
(G) T2-weighted MRI of II-1 at 41 years shows a few kidney (red arrows) and liver (green arrows) cysts.
Table 1.
Clinical Presentation of Kidney and Liver Disease in the 20 Affected Individuals from Nine Families with GANAB Mutations
| Family | GANAB Mutation | Subject | Sex | eGFRa(Age in Years) | HBP (Age in Yearsb) |
Radiologic Presentation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Kidneys |
Liver |
||||||||||||
| Type | Agec | Cysts | Volumed | Figure | Cysts | Volumee | Figure | ||||||
| M263f | c.1265G>T (p.Arg422Leu) | I-1 | M | 78 (66) | N (67) | CT | 66 | ∼10 bilateral cysts (largest 11 cm) | 302g | 1F | >50 scattered cysts (largest 6 cm) | 1,226 | 1F |
| II-1 | F | 91 (42) | N (43) | MRI | 41 | ∼10 bilateral cysts (largest 3 cm) | 211 | 1G | >20 scattered cysts (largest 3 cm) | 835 | 1G | ||
| M641 | c.1914_1915delAG (p.Asp640Glnfs∗77) | II-1 | F | 86 (51) | Y (40) | CT | 55 | ∼15 bilateral cysts (largest 10 cm) | 822g | S3A | no liver cysts detected | 1,505h | S3A |
| II-2 | F | 104 (46) | N (50) | CT | 45 | ∼10 bilateral cysts (largest 6 cm) | 318g | 2B | ∼20 scattered cysts (largest 2 cm) | 764 | 2B | ||
| 290100 | c.1914_1915delAG (p.Asp640Glnfs∗77) | I-1 | M | 78 (65) | N (65) | MRI | 58 | ∼8 bilateral cysts (largest 2 cm) | 227 | 2E | >30 scattered cysts (largest 3 cm) | 1,255 | 2E |
| II-1 | M | 87 (25) | Y (13) | MRI | 24 | ∼12 bilateral cysts (largest 2.5 cm) | 259 | 2D | none | 832 | 2D | ||
| P1174 | c.1214C>G (p.Thr405Arg) | I-1 | M | NAi | N (61) | US | 55 | 3 cysts in the left kidney | NE | S3B | 1 cyst (1.5 cm) | NE | – |
| II-1 | F | NAi | N (35) | US | 29 | 2 cysts in the right kidney | NE | 2H | NA | NA | – | ||
| III-1 | M | 122 (9) | N (9) | MRI | 9 | ∼5 bilateral cysts (largest 2 cm) | 116 | 2G | none | 492 | – | ||
| M656 | c.2690+2_+7del | I-1 | M | NAi | Y (55) | CT∗ | 67 | multiple small cysts | NE | S3C | none | NE | S3C |
| II-1 | M | 84 (39) | N (39) | US | 44 | multiple cysts reported | NA | – | multiple cysts reported | NA | – | ||
| II-2 | F | 77 (50) | N (50) | US | 52 | ∼5–10 bilateral cysts (largest 2 cm) | NE | S3D | >20 scattered cysts (largest 5 cm) | NE | S3D | ||
| II-3 | M | 95 (49) | Y (35) | MRI | 43 | >30 bilateral cysts (largest 3 cm) | SE | 2L | >20 scattered cysts (largest 1 cm) | NE | 2L | ||
| PK20016 | c.39−1G>C | II-1 | M | 90 (53) | Y (45) | CT∗ | 52 | ∼20 bilateral cysts (largest 10 cm) | 665g | 3B | ∼20 scattered cysts (largest 2 cm) | 1,449h | 3B |
| PK20017 | c.2176C>T (p.Arg726∗) | II-1 | F | 77 (78) | Y (53) | US | 78 | ∼40 bilateral cysts (largest 3 cm) | NE | 3D | ∼20 scattered cysts (largest 1.5 cm) | NA | 3D |
| P1073 | c.2515C>T (p.Arg839Trp) | I-2 | F | NA | NA | US | (78) | unknown | NA | – | multiple cysts reported | NA | – |
| II-1 | F | 86 (50) | N | CT∗ | 43 | ∼8 bilateral cysts (largest 1 cm) | 196 | 3F | severe PLD, transplant at 43 years | 4,641h | 3F | ||
| II-2 | M | NA | NA | US | (44) | unknown | NA | – | multiple cysts reported | NA | – | ||
| M472 | c.152_153delGA (p.Arg51Lysfs∗21) | II-1 | F | 82 (58) | N (58) | MRI | 58 | ∼20 cysts (largest 1 cm) | 223 | 3I | severe PLD, resections at 47 and 48 years | 1,749j,h | 3I |
| II-2k | F | NA | N (50) | MRI | 50 | ∼16 bilateral cysts (largest 6 cm) | 305g | S3E | >50 scattered cysts (largest 5 cm) | 1,249h | S3E | ||
Abbreviations are as follows: CT, computed tomography; CT∗, contrast-enhanced CT; eGFR, estimated glomerular filtration rate; HBP, high blood pressure; N, no; NA, not available; NE, not enlarged; SE, slightly enlarged; US, ultrasound; and Y, yes.
Expressed in mL/min/1.73 m2 on the basis of the last data available; obtained with the CKD-EPI formula in adults49 and the Schwarz formula in the pediatric individual.50
Age at HBP diagnosis or blood-pressure measurement.
Age at imaging examination. Values in parentheses are the present age if images are not available.
Kidney volume measurement or estimate (NE or SE). Values are the measured height-adjusted total kidney volume (htTKV) in mL/m.
Liver volume measurement or estimate (NE). Values are the measured height-adjusted total liver volume (htTLV) in mL/m.
The mutation in this pedigree was first identified by whole-exome sequencing.
Enlarged values: mean + 2 SDs of normal male and female htTKV.48
Enlarged values: mean + 2 SDs of normal male and female htTLV.26
Kidney function was reported to be within the normal range.
htTLV after resections.
A sample was unavailable, and so the GANAB mutation was not confirmed.
Identification of Further ADPKD-Affected Families with GANAB Mutations
Given that mutations in GANAB might account for disease in families with unresolved ADPKD, we analyzed our remaining GUR cohort of 321 families by Sanger sequencing of the coding region. From this analysis, we identified eight additional families affected by GANAB mutations: three frameshift, two splicing, one nonsense, and two missense mutations (Table 1). Data from the ExAC Browser showed that none of the GANAB variants, except c.152_153delGA (reported once), have been reported in the 60,706 unrelated individuals sequenced as part of various disease-specific and population genetic studies.55 Additionally, none of these variants were seen in the NHLBI Exome Sequence Project (ESP) Exome Variant Server.
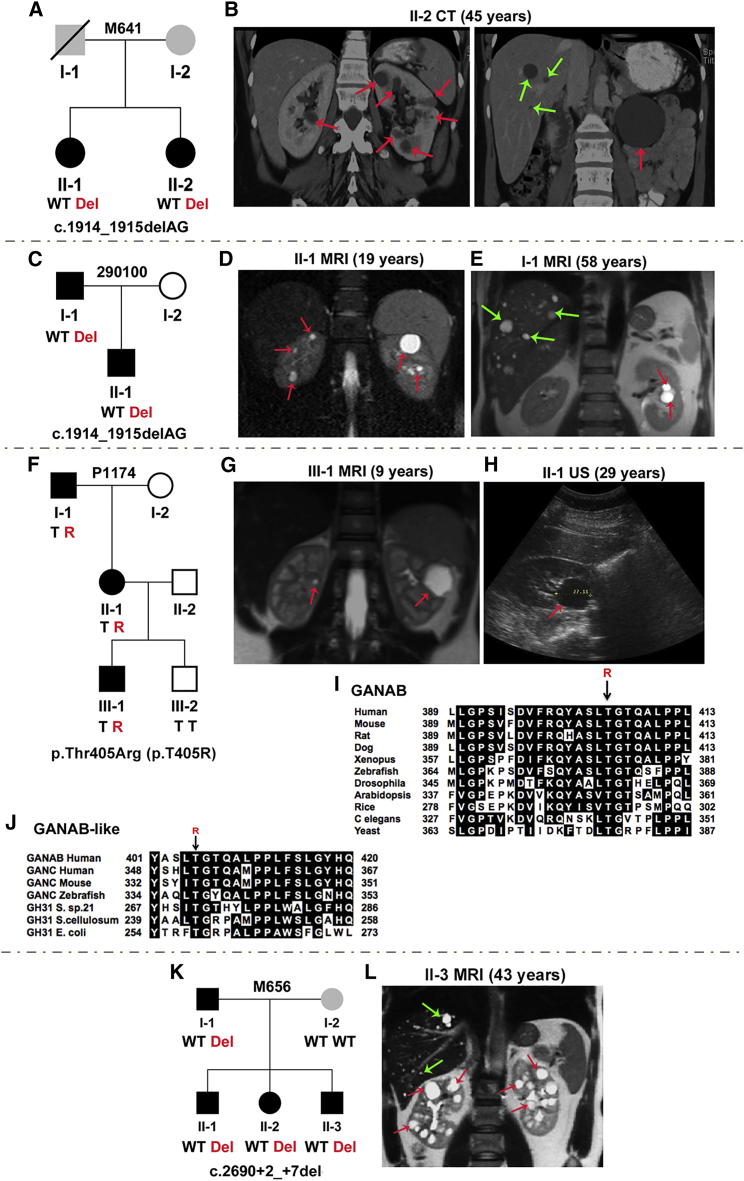
The GANAB frameshift mutation, c.1914_1915delAG (p.Asp640Glnfs∗77), was found in two families, M641 and 290100 (Figures 2A–2E, Figures S2A, S2B, and S3A, and Table 1). In M641, two sisters had relatively mild PKD with variable liver cysts. The family history was unclear: the father died at 75 years with renal cell cancer but no reported cysts, and there was no information on the mother. Both sisters had ICAs (see the Figure 2 legend for details), and the father was reported to have a ruptured aneurysm. In family 290100, the father and son both had mild PKD, and multiple liver cysts were present in the father, but not the son.
Figure 2.
Characterization of GANAB Mutations in Four ADPKD-Affected Families
(A) Pedigree of family M641 shows c.1914_1915delAG (p.Asp640Glnfs∗77) (exon 17) in two affected siblings. Both sisters had basilar tip aneurysms, and II-2 also had two aneurysms detected on the left middle cerebral artery. The affected status of the parents is unclear (gray shading); I-1 had renal cell carcinoma and a ruptured aneurysm, but no reported PKD.
(B) CT scan of kidneys and liver from II-2 shows multiple kidney and occasional liver cysts.
(C) Pedigree of family 290100 shows c.1914_1915delAG in the father and son.
(D and E) MRI shows a few kidney but no hepatic cysts in II-1 (D) and a few kidney and scattered liver cysts in I-1 (E).
(F) Pedigree of family P1174 shows p.Thr405Arg (p.T405R) (c.1214C>G, exon 11) in three affected individuals; III-2, for whom no cysts were detected by ultrasound at 5 years, did not have the variant.
(G and H) MRI of III-1 shows bilateral kidney cysts (G), and ultrasound (US) of II-1 shows a single large renal cyst (H).
(I) MSA of GIIα orthologs shows that Thr405 is invariant across species. In silico mutation analysis highly predicts p.Thr405Arg (p.T405R) to be pathogenic (SIFT = 0.00, Align GVGD = C65).
(J) MSA of GANAB-like proteins shows invariant conservation of this residue.
(K) Pedigree of family M656 shows c.2690+2_+7del (IVS20) in four affected members. Splicing predictions show complete loss of the donor site. The mother (I-2), who does not have the GANAB mutation, has ∼5 kidney and ∼30 liver cysts, but no detected PKD1, PKD2, PRKCSH, or SEC63 mutations.
(L) MRI of II-3 shows small hepatic andmultiple renal cysts. Red and green arrows indicate kidney and liver cysts, respectively. Where multiple cysts are present, only representative cysts are highlighted.
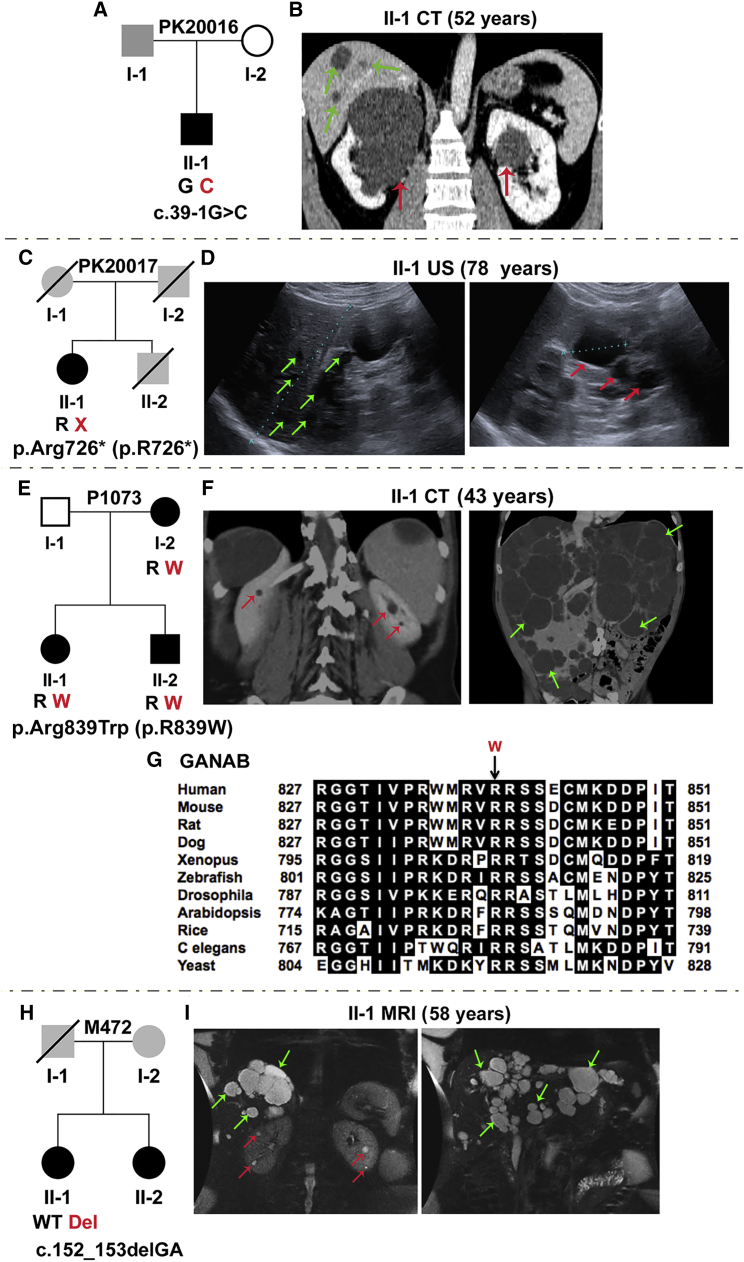
In family P1174, the GANAB missense variant, c.1214C>G (p.Thr405Arg), was found in three generations, including in individual III-1, in whom renal cysts were detected incidentally in infancy (Figures 2F–2H, Figures S2C and S3B, and Table 1). The substituted residue is invariant in orthologs and conserved in related proteins (Figures 2I and 2J). The GANAB splicing mutation, c.2690+2_+7del, was found in four affected family members in M656 (Figures 2K and 2L, Figures S2D, S3C, and S3D, and Table 1). All members (apart from II-3, who had multiple kidney cysts) had no to mild cystic liver disease and mild kidney disease. Of note, the mother (without the GANAB mutation) had multiple cysts of unresolved etiology. PK20016 had the splicing mutation c.39−1G>C, and PK20017 had the nonsense mutation c.2176C>T (p.Arg726∗), and both had only one known affected family member (Figures 3A–3D, Figures S2E and S2F, Table 1). Both individuals had multiple kidney cysts and a few liver cysts.
Figure 3.
Characterization of GANAB Variants in Four Families, Including ADPLD-Affected P1073 and M472
(A) Pedigree of family PK20016 shows the splicing mutation c.39−1G>C (IVS1). The family history is unclear because samples were unavailable, but one large renal cyst was reported in I-1.
(B) CT scan of II-1 shows bilateral kidney cysts and occasional hepatic cysts.
(C) Pedigree of family PK20017 shows p.Arg726∗ (p.R726∗) (c.2176C>T, exon 18), in the proband, II-1. A lack of DNA samples and clinical information precluded determining the family history. II-2 died at 55 years from a ruptured intracranial aneurysm, but his PKD status was unknown.
(D) Ultrasound examination of II-1 shows several liver (left), and kidney cysts (right).
(E) Pedigree of family P1073 shows p.Arg839Trp (p.R839W) (c.2515C>T, exon 22) in three affected individuals.
(F) CT scan of II-1 shows very few kidney cysts (left) but severe PLD (right). Gross images of the liver of this subject have been published.57
(G) MSA of GANAB (GIIα) orthologs shows invariant conservation of Arg839 across species. In silico mutation analysis highly predicts p.Arg839Trp (p.R839W) to be pathogenic (SIFT = 0.00, Align GVGD = C65).
(H) Pedigree of family M472 shows c.152_153delGA (p.Arg51Lysfs∗21) (p.R51fs; exon 3) in II-1.
(I) MRI of II-1 shows a few renal cysts (left) but significant PLD; this image was subsequent to earlier resections (Table 1). No sample was available from II-2, but imaging also showed predominant PLD (Figure S3E). Unavailable parental DNA samples and limited clinical information precluded determining the family history, but I-1 was reported to have had a cerebral hemorrhage. Red and green arrows indicate kidney and liver cysts, respectively. Where multiple cysts are present, only representative cysts are highlighted.
Identification of Families Affected by GANAB Mutations and ADPLD
Unlike the described ADPKD-affected families, families P1073 and M472 had a possible diagnosis of ADPLD, although a few renal cysts were present. P1073 had the missense mutation c.2515C>T (p.Arg839Trp), which is at an invariant site in orthologs and segregates in three affected family members (Figures 3E–3G, Figure S2G, and Table 1). As indicated, there were very few renal cysts but multiple liver cysts, and the daughter (II-1) required a liver transplant at 43 years. M472 individual II-1, who had the GANAB frameshift mutation c.152_153delGA (p.Arg51Lysfs∗21) (Figures 3H and 3I, Figure S2H, and Table 1), had a few small kidney cysts but severe PLD that required partial liver resections. A similar phenotype with less severe PLD was seen in II-2 (Figure S3E), although a sample was unavailable for mutation analysis.
Characterization of the Effect of GANAB Loss on PC1 and PC2 Maturation and Localization
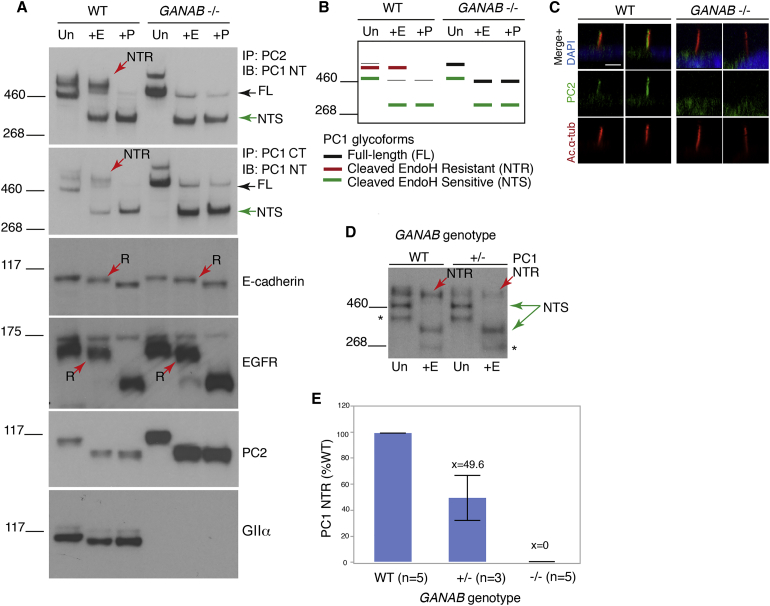
From the genetic studies, mutations in GANAB were shown to be a cause of ADPKD and ADPLD, so we next explored the mechanism of pathogenesis by cellular analysis. CRISPR/Cas9 targeting of GANAB exon 12 in human RCTE cells generated clones with biallelic frameshift mutations (null; clone C6) or a single in-frame deletion (heterozygous; E4) (Figures S4A–S4D). Analysis of the polycystin complex immunocaptured with PC2 or PC1-CT antibodies in GANAB−/− cells showed that the PC1 N-terminal, mature product (PC1-NTR) was absent (Figures 4A and 4B). In contrast, full-length (GPS-uncleaved) PC1, PC1-NTS, and PC2 were elevated, indicating that GIIα plays a major role in PC1 maturation. We previously showed an interdependence of PC1 and PC2 for localization, including to cilia,15 and so to assess localization of the polycystin complex, we analyzed PC2. Ciliary localization of PC2 was completely absent in GANAB−/− cells, although cilia formed normally (Figure 4C and Figure S4E).15, 16 Given that affected individuals harbored just one GANAB mutation, we assayed GANAB+/− cells and found a proportional, ∼50%, depletion of PC1-NTR (Figures 4D and 4E). Analysis of the maturation of other membrane proteins (epidermal growth factor receptor [EGFR] and E-cadherin) showed that they were not (or only mildly) affected by loss of GIIα (Figure 4A). Further analysis of terminally glycosylated proteins in GANAB−/− cells did not suggest a global disruption of surface-localized proteins (Figures S4F–S4H).
Figure 4.
GIIα Is Required for PC1 ER Exit and Maturation
(A) Deglycosylation analysis of WT and GANAB−/− RCTE membrane protein either untreated (Un) or treated with EndoH (+E) or PNGaseF (+P). IP was used to enrich the PC1 complex with C-terminal PC1 (PC1-CT) or PC2 (YCE2) antibodies and immunodetected with the N-terminal PC1 (PC1-NT) antibody (7e12). Complete loss of the mature PC1 glycoform15 (NTR; red arrow) was observed in GANAB−/− cells, and full-length PC1 (FL) and PC1-NTS became more abundant. Mature E-cadherin and EGFR were not or only marginally affected by GANAB loss; the EndoH-resistant protein (R, red arrow) persisted. No EndoH-resistant form of PC2 was noted,15 but the protein was upregulated in GANAB−/− cells. Loss of WT GIIα was confirmed in GANAB−/− cells with the use of a C-terminal antibody.
(B) Schematic representation of the observed PC1 banding pattern in the WT and GANAB−/− cells shown in (A).
(C) Confocal z stack rendering of primary cilia in confluent WT and GANAB−/− cells in which acetylated α-tubulin (Ac.α-tub) and PC2 were detected shows no cilia PC2 signal in GANAB−/− cells. Nuclei were stained with DAPI, and 100 ciliated cells were analyzed in three independent experiments. The scale bar represents 10 μm.
(D) Immunoblot of PC1-NT in WT and GANAB+/− cells shows a reduced level of PC1-NTR in GANAB+/− cells. Asterisks indicate a non-specific product.15
(E) Quantification of PC1-NTR shows a reduction to ∼50% (p < 0.001, Student’s t test) in heterozygous cells and a complete loss in homozygous, GANAB−/− cells.
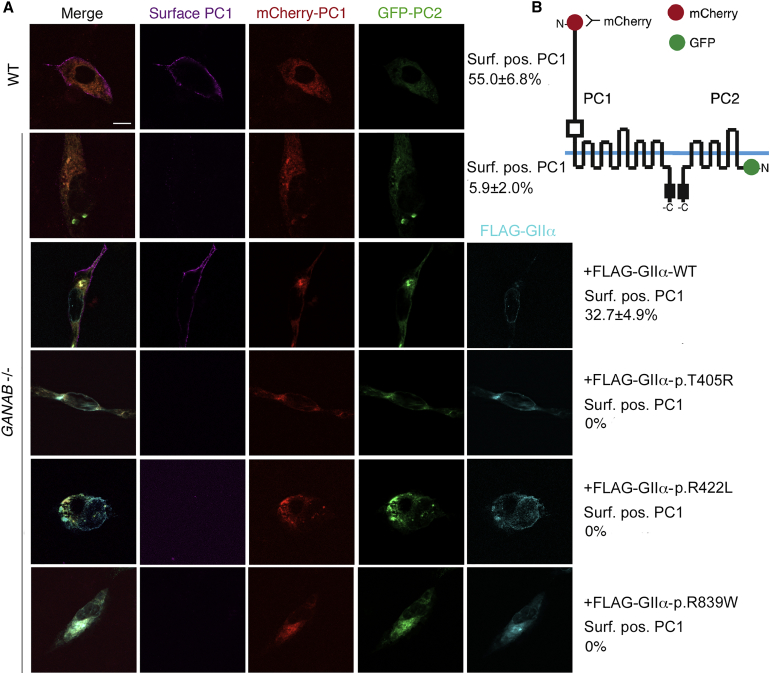
Functional Analysis of GANAB Mutations and Variants
To determine the effect of GANAB loss on PC1 localization by immunofluorescence and to test the pathogenicity of detected GIIα variants, we co-transfected WT and GANAB−/− cells with tagged PC1 and PC2 constructs (mCherry-PC1-V5 and GFP-PC2; Figures 5A and 5B).15 GIIα loss prevented efficient surface localization of tagged PC1, which was restored by co-expression of WT FLAG-GIIα (Figure 5A). Identified GIIα missense variants (p.Thr405Arg, p.Arg422Leu, and p.Arg839Trp), expressed as FLAG-GIIα constructs, failed to rescue PC1 surface localization in GANAB−/− cells (Figure 5A), whereas three other variants considered likely to be neutral (c.284A>G [p.Gln95Arg], c.760A>G [p.Thr254Ala], and c.991C>T [p.Arg331Cys]) restored surface localization (Figure S5).
Figure 5.
Surface Localization of PC1 Requires WT GIIα and Is Disrupted by GANAB Missense Mutations
(A) WT and GANAB−/− cells were co-transfected with WT tagged PC1 and PC2, mCherry-PC1, and TagGFP-PC2 and examined for surface mCherry-PC1 labeling. Co-transfected cells were screened for live cell-surface PC1 signal and quantified as the percentage of surface-positive PC1 cells out of the total co-transfected cells. Surface PC1 was detected on 55.0% ± 6.8% of WT cells but only 5.9% ± 2.0% of GANAB−/− cells (p ≤ 0.0001), and the level was rescued to 32.7% ± 4.9% by co-transfection with the WT GANAB (FLAG-GIIα) plasmid. Co-transfection with the newly identified putative GANAB missense mutations cloned in FLAG-GIIα, p.Thr405Arg (p.T405R), p.Arg422Leu (p.R422L), and p.Arg839Trp (p.R839W) did not rescue PC1 surface localization (bottom three panels; all p < 0.0001 versus WT rescue). Student’s t test was performed to determine significance in at least 100 triple-transfected cells analyzed among three independent experiments. The scale bar represents 20 μm.
(B) Diagram of the constructs.
Discussion
The combination of the human genetic studies and cellular analysis of GANAB-null cells showed that mutations in GANAB cause ADPKD and ADPLD, that loss or reduction of GIIα is associated with maturation and localization defects of PC1 and PC2, and that the identified mutations cannot rescue the PC1 localization defect.
The renal phenotype associated with GANAB mutations is consistently mild without renal insufficiency, such that any kidney enlargement is due to a few large cysts. The phenotype caused by GANAB mutations is more similar to PKD2 than to PKD1 but is apparently even milder.58 One individual had more severe PKD (M656 member II-3), and here an undetermined cystogenic influence from the mother might be significant. Applying imaging diagnostic criteria developed in PKD1- and PKD2-affected families53, 54 might be unreliable in families affected by GANAB mutations even in those with a largely renal phenotype given the disease mildness and the variability within families. The significance of the vascular disease, noted in families M641, PK20017, and M472, is presently unclear. Only in M641 do the definitely affected sisters have ICAs; the vascular phenotypes in three other individuals in these families are not proven to be linked to GANAB mutations.
The liver disease is variable and ranges from no cysts to severe PLD requiring surgical intervention. The highly variable PLD phenotype is characteristic of ADPKD and ADPLD. Allelic effects do not appear to explain this variability, although some evidence of familial clustering of severe PLD suggests that genetic modifiers play a significant role.25, 46 It is interesting that GANAB mutations were identified as the cause of the disease in two of the seven ADPLD-affected families studied, suggesting that the mutation-detection rate might be higher in ADPLD-affected families. Although there might be ascertainment bias because ADPKD was the diagnosis in the vast majority of screened families, the overall phenotype appears to involve more renal disease than described in PRKCSH.59 The reason for this is unclear given that they are subunits of the same protein, and further analysis of GANAB in ADPLD and PRKCSH in ADPKD populations is required. We show that phenotypes consistent with mild ADPKD and ADPLD involving a few renal cysts can be caused by GANAB mutations. Rather than considering ADPKD and ADPLD to be strictly separate diseases, we suggest recording the full range of phenotypes associated with each gene in which mutations are associated with ADPKD and/or ADPLD.
GANAB mutations account for ∼3% of GUR ADPKD-affected families (∼0.3% total ADPKD), although given that many GUR cases most likely involve PKD1 or PKD2 mutations that were missed (see Introduction and Paul et al.23), they are probably responsible for a much greater proportion of missing genetic causes of ADPKD. In addition, because of the mild phenotype, these mutations are most likely underdiagnosed and, in particular, might be more common in families with mild PKD and significant PLD.
Defects in glycosylation and protein trafficking underlie a large number of human diseases,60, 61 but the interesting aspect here is the specificity of the phenotype associated with disruption of a step in a process important for many proteins. GII functions in the early cargo-recruitment steps of the calnexin and calreticulin cycle, which facilitates the quality control and maturation of transmembrane glycoproteins.34 GANAB−/− cells and S. pombe mutants are viable without growth defects,62, 63 and we have demonstrated a lack of global, surface glycoprotein deficiencies in GANAB−/− cells, which suggests that endomannosidase activity and other chaperones and folding-assisting proteins can generally compensate for this loss, at least in non-stress conditions. The complexity due to protein size and extensive N-linked glycosylation might underlie the critical dependence of PC1 on GII and the calnexin and calreticulin cycle for achieving native folding. At this stage, it is unclear whether the enrichment of PLD associated with GII deficiency indicates that the liver is particularly vulnerable to reduction of this enzyme. In contrast to that described for SEC63 deficiency,15, 62, 64, 65 the defect we observed in GANAB−/− cells was complete disruption of PC1 maturation, which increased ER accumulation of cleaved PC1 while only marginally affecting GPS cleavage. In RCTE cells, GANAB heterozygosity was associated with a ∼50% reduction of PC1-NTR, a level predisposing to cyst development in association with stochastic, renal injury and/or somatic events.12, 15, 66, 67 However, fully understanding quantitatively how PC1 maturation is influenced by GII dosage will require further studies, which could provide insights into a therapeutic role for cellular and molecular chaperones in ADPKD.
Acknowledgments
We thank the families and coordinators for involvement in the study, the Exome Aggregation Consortium, Tatyana Masyuk, and other HALT Progression of Polycystic Kidney Disease (HALT-PKD) and Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) investigators: Drs. Grantham, Yu, and Winklehofer (Kansas Medical Center), Bae, Abebe, and Landsittel (University of Pittsburgh), Schrier and Brosnahan (University of Colorado Denver), Perrone and Miskulin (Tufts University), Braun (Cleveland Clinic), Steinman (Beth Israel Deaconess Medical Center), Mrug (University of Alabama at Birmingham), Rahbari-Oskoui (Emory University), Bennett (Legacy Health, Portland), Flessner (National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK]), Moore (Carolinas HealthCare, Charlotte), and Czarnecki (Brigham and Women’s Hospital). This study received support from NIDDK grant DK058816, the Mayo PKD Translational Center (DK090728), an American Heart Association postdoctoral fellowship (B.P.), the Mayo Clinic Nephrology Training Grant (T32DK007013 to V.G.G.), an American Society of Nephrology (ASN) Foundation Kidney Research Fellowship (E.C.-L.G.) and Ben J. Lipps Research Fellowship (K.H.), the Mayo Graduate School (E.K.D.), the Zell Family Foundation, and Robert M. and Billie Kelley Pirnie. The CRISP and HALT-PKD studies were supported by NIDDK cooperative agreements (DK056943, DK056956, DK056957, DK056961, DK062410, DK062408, DK062402, DK082230, DK062411, and DK062401), National Center for Research Resources General Clinical Research Centers, and National Center for Advancing Translational Sciences Clinical and Translational Science Awards. The Genkyst cohort was supported by National Plans for Clinical Research, Groupement Interrégional de Recherche Clinique et d’Innovation (GIRCI Grand Ouest), and the French Society of Nephrology.
Published: June 2, 2016
Footnotes
Supplemental Data include five figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.05.004.
Accession Numbers
The accession number for the cDNA sequence reported in this paper is GenBank: NM_198335.3.
Web Resources
ADPKD Mutation Database, http://pkdb.mayo.edu
Align GVGD, http://agvgd.iarc.fr
Berkeley Drosophila Genome Project NNSPLICE 0.9, http://www.fruitfly.org/seq_tools/splice.html
ExAC Browser, http://exac.broadinstitute.org
GeneReviews, Harris, P.C., and Torres, V.E. (2015). Polycystic Kidney Disease, Autosomal Dominant, http://www.ncbi.nlm.nih.gov/books/NBK1246/
GeneTests, https://www.genetests.org
NCBI Nucleotide, http://www.ncbi.nlm.nih.gov/nuccore
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
SIFT, http://sift.jcvi.org
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Torres V.E., Harris P.C., Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 2.Harris P.C., Torres V.E. Polycystic kidney disease. Annu. Rev. Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The European Polycystic Kidney Disease Consortium The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;77:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki T., Wu G., Hayashi T., Xenophontos S.L., Veldhuisen B., Saris J.J., Reynolds D.M., Cai Y., Gabow P.A., Pierides A. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 5.Heyer C.M., Sundsbak J.L., Abebe K.Z., Chapman A.B., Torres V.E., Grantham J.J., Bae K.T., Schrier R.W., Perrone R.D., Braun W.E., HALT PKD and CRISP Investigators Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2016 doi: 10.1681/ASN.2015050583. Published online January 28, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audrézet M.P., Cornec-Le Gall E., Chen J.M., Redon S., Quéré I., Creff J., Bénech C., Maestri S., Le Meur Y., Férec C. Autosomal dominant polycystic kidney disease: comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum. Mutat. 2012;33:1239–1250. doi: 10.1002/humu.22103. [DOI] [PubMed] [Google Scholar]
- 7.Cornec-Le Gall E., Audrézet M.P., Chen J.M., Hourmant M., Morin M.P., Perrichot R., Charasse C., Whebe B., Renaudineau E., Jousset P. Type of PKD1 mutation influences renal outcome in ADPKD. J. Am. Soc. Nephrol. 2013;24:1006–1013. doi: 10.1681/ASN.2012070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grantham J.J., Torres V.E., Chapman A.B., Guay-Woodford L.M., Bae K.T., King B.F., Jr., Wetzel L.H., Baumgarten D.A., Kenney P.J., Harris P.C., CRISP Investigators Volume progression in polycystic kidney disease. N. Engl. J. Med. 2006;354:2122–2130. doi: 10.1056/NEJMoa054341. [DOI] [PubMed] [Google Scholar]
- 9.Ong A.C., Harris P.C. A polycystin-centric view of cyst formation and disease: the polycystins revisited. Kidney Int. 2015;88:699–710. doi: 10.1038/ki.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian F., Boletta A., Bhunia A.K., Xu H., Liu L., Ahrabi A.K., Watnick T.J., Zhou F., Germino G.G. Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc. Natl. Acad. Sci. USA. 2002;99:16981–16986. doi: 10.1073/pnas.252484899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei W., Hackmann K., Xu H., Germino G., Qian F. Characterization of cis-autoproteolysis of polycystin-1, the product of human polycystic kidney disease 1 gene. J. Biol. Chem. 2007;282:21729–21737. doi: 10.1074/jbc.M703218200. [DOI] [PubMed] [Google Scholar]
- 12.Hopp K., Ward C.J., Hommerding C.J., Nasr S.H., Tuan H.F., Gainullin V.G., Rossetti S., Torres V.E., Harris P.C. Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J. Clin. Invest. 2012;122:4257–4273. doi: 10.1172/JCI64313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedeles S.V., Tian X., Gallagher A.R., Mitobe M., Nishio S., Lee S.H., Cai Y., Geng L., Crews C.M., Somlo S. A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat. Genet. 2011;43:639–647. doi: 10.1038/ng.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurbegovic A., Kim H., Xu H., Yu S., Cruanès J., Maser R.L., Boletta A., Trudel M., Qian F. Novel functional complexity of polycystin-1 by GPS cleavage in vivo: role in polycystic kidney disease. Mol. Cell. Biol. 2014;34:3341–3353. doi: 10.1128/MCB.00687-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gainullin V.G., Hopp K., Ward C.J., Hommerding C.J., Harris P.C. Polycystin-1 maturation requires polycystin-2 in a dose-dependent manner. J. Clin. Invest. 2015;125:607–620. doi: 10.1172/JCI76972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H., Xu H., Yao Q., Li W., Huang Q., Outeda P., Cebotaru V., Chiaravalli M., Boletta A., Piontek K. Ciliary membrane proteins traffic through the Golgi via a Rabep1/GGA1/Arl3-dependent mechanism. Nat. Commun. 2014;5:5482. doi: 10.1038/ncomms6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossetti S., Consugar M.B., Chapman A.B., Torres V.E., Guay-Woodford L.M., Grantham J.J., Bennett W.M., Meyers C.M., Walker D.L., Bae K., CRISP Consortium Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2007;18:2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 18.Loftus B.J., Kim U.-J., Sneddon V.P., Kalush F., Brandon R., Fuhrmann J., Mason T., Crosby M.L., Barnstead M., Cronin L. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- 19.Consugar M.B., Wong W.C., Lundquist P.A., Rossetti S., Kubly V.J., Walker D.L., Rangel L.J., Aspinwall R., Niaudet W.P., Ozen S., CRISP Consortium Characterization of large rearrangements in autosomal dominant polycystic kidney disease and the PKD1/TSC2 contiguous gene syndrome. Kidney Int. 2008;74:1468–1479. doi: 10.1038/ki.2008.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan A.Y., Blumenfeld J., Michaeel A., Donahue S., Bobb W., Parker T., Levine D., Rennert H. Autosomal dominant polycystic kidney disease caused by somatic and germline mosaicism. Clin. Genet. 2015;87:373–377. doi: 10.1111/cge.12383. [DOI] [PubMed] [Google Scholar]
- 21.Heidet L., Decramer S., Pawtowski A., Morinière V., Bandin F., Knebelmann B., Lebre A.S., Faguer S., Guigonis V., Antignac C., Salomon R. Spectrum of HNF1B mutations in a large cohort of patients who harbor renal diseases. Clin. J. Am. Soc. Nephrol. 2010;5:1079–1090. doi: 10.2215/CJN.06810909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckardt K.U., Alper S.L., Antignac C., Bleyer A.J., Chauveau D., Dahan K., Deltas C., Hosking A., Kmoch S., Rampoldi L. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management--A KDIGO consensus report. Kidney Int. 2015;88:676–683. doi: 10.1038/ki.2015.28. [DOI] [PubMed] [Google Scholar]
- 23.Paul B.M., Consugar M.B., Ryan Lee M., Sundsbak J.L., Heyer C.M., Rossetti S., Kubly V.J., Hopp K., Torres V.E., Coto E. Evidence of a third ADPKD locus is not supported by re-analysis of designated PKD3 families. Kidney Int. 2014;85:383–392. doi: 10.1038/ki.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirson Y., Chauveau D., Torres V. Management of cerebral aneurysms in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2002;13:269–276. doi: 10.1681/ASN.V131269. [DOI] [PubMed] [Google Scholar]
- 25.Chebib F.T., Jung Y., Heyer C.M., Irazabal M.V., Hogan M.C., Harris P.C., Torres V.E., El-Zoghby Z.M. Effect of genotype on the severity and volume progression of polycystic liver disease in ADPKD. Nephrol. Dial. Transplant. 2016 doi: 10.1093/ndt/gfw008. Published online February 29, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogan M.C., Abebe K., Torres V.E., Chapman A.B., Bae K.T., Tao C., Sun H., Perrone R.D., Steinman T.I., Braun W. Liver involvement in early autosomal-dominant polycystic kidney disease. Clin. Gastroenterol. Hepatol. 2015;13 doi: 10.1016/j.cgh.2014.07.051. 155–164.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoevenaren I.A., Wester R., Schrier R.W., McFann K., Doctor R.B., Drenth J.P., Everson G.T. Polycystic liver: clinical characteristics of patients with isolated polycystic liver disease compared with patients with polycystic liver and autosomal dominant polycystic kidney disease. Liver Int. 2008;28:264–270. doi: 10.1111/j.1478-3231.2007.01595.x. [DOI] [PubMed] [Google Scholar]
- 28.Drenth J.P., te Morsche R.H., Smink R., Bonifacino J.S., Jansen J.B. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat. Genet. 2003;33:345–347. doi: 10.1038/ng1104. [DOI] [PubMed] [Google Scholar]
- 29.Li A., Davila S., Furu L., Qian Q., Tian X., Kamath P.S., King B.F., Torres V.E., Somlo S. Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am. J. Hum. Genet. 2003;72:691–703. doi: 10.1086/368295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davila S., Furu L., Gharavi A.G., Tian X., Onoe T., Qian Q., Li A., Cai Y., Kamath P.S., King B.F. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat. Genet. 2004;36:575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- 31.Cnossen W.R., te Morsche R.H., Hoischen A., Gilissen C., Chrispijn M., Venselaar H., Mehdi S., Bergmann C., Veltman J.A., Drenth J.P. Whole-exome sequencing reveals LRP5 mutations and canonical Wnt signaling associated with hepatic cystogenesis. Proc. Natl. Acad. Sci. USA. 2014;111:5343–5348. doi: 10.1073/pnas.1309438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai K., Hirai M., Minoshima S., Kudoh J., Fukuyama R., Shimizu N. Isolation of cDNAs encoding a substrate for protein kinase C: nucleotide sequence and chromosomal mapping of the gene for a human 80K protein. Genomics. 1989;5:309–315. doi: 10.1016/0888-7543(89)90063-3. [DOI] [PubMed] [Google Scholar]
- 33.Trombetta E.S., Simons J.F., Helenius A. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 1996;271:27509–27516. doi: 10.1074/jbc.271.44.27509. [DOI] [PubMed] [Google Scholar]
- 34.Xu C., Ng D.T. Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 2015;16:742–752. doi: 10.1038/nrm4073. [DOI] [PubMed] [Google Scholar]
- 35.D’Alessio C., Dahms N.M. Glucosidase II and MRH-domain containing proteins in the secretory pathway. Curr. Protein Pept. Sci. 2015;16:31–48. doi: 10.2174/1389203716666150213160438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treml K., Meimaroglou D., Hentges A., Bause E. The alpha- and beta-subunits are required for expression of catalytic activity in the hetero-dimeric glucosidase II complex from human liver. Glycobiology. 2000;10:493–502. doi: 10.1093/glycob/10.5.493. [DOI] [PubMed] [Google Scholar]
- 37.Tannous A., Pisoni G.B., Hebert D.N., Molinari M. N-linked sugar-regulated protein folding and quality control in the ER. Semin. Cell Dev. Biol. 2015;41:79–89. doi: 10.1016/j.semcdb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelletier M.F., Marcil A., Sevigny G., Jakob C.A., Tessier D.C., Chevet E., Menard R., Bergeron J.J., Thomas D.Y. The heterodimeric structure of glucosidase II is required for its activity, solubility, and localization in vivo. Glycobiology. 2000;10:815–827. doi: 10.1093/glycob/10.8.815. [DOI] [PubMed] [Google Scholar]
- 39.Deprez P., Gautschi M., Helenius A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol. Cell. 2005;19:183–195. doi: 10.1016/j.molcel.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 40.Rothblatt J.A., Deshaies R.J., Sanders S.L., Daum G., Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadler I., Chiang A., Kurihara T., Rothblatt J., Way J., Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J. Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deshaies R.J., Sanders S.L., Feldheim D.A., Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- 43.Skowronek M.H., Rotter M., Haas I.G. Molecular characterization of a novel mammalian DnaJ-like Sec63p homolog. Biol. Chem. 1999;380:1133–1138. doi: 10.1515/BC.1999.142. [DOI] [PubMed] [Google Scholar]
- 44.Gao H., Wang Y., Wegierski T., Skouloudaki K., Pütz M., Fu X., Engel C., Boehlke C., Peng H., Kuehn E.W. PRKCSH/80K-H, the protein mutated in polycystic liver disease, protects polycystin-2/TRPP2 against HERP-mediated degradation. Hum. Mol. Genet. 2010;19:16–24. doi: 10.1093/hmg/ddp463. [DOI] [PubMed] [Google Scholar]
- 45.Hofherr A., Wagner C., Fedeles S., Somlo S., Köttgen M. N-glycosylation determines the abundance of the transient receptor potential channel TRPP2. J. Biol. Chem. 2014;289:14854–14867. doi: 10.1074/jbc.M114.562264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waanders E., te Morsche R.H., de Man R.A., Jansen J.B., Drenth J.P. Extensive mutational analysis of PRKCSH and SEC63 broadens the spectrum of polycystic liver disease. Hum. Mutat. 2006;27:830. doi: 10.1002/humu.9441. [DOI] [PubMed] [Google Scholar]
- 47.Kline T.L., Korfiatis P., Edwards M.E., Warner J.D., Irazabal M.V., King B.F., Torres V.E., Erickson B.J. Automatic total kidney volume measurement on follow-up magnetic resonance images to facilitate monitoring of autosomal dominant polycystic kidney disease progression. Nephrol. Dial. Transplant. 2016;31:241–248. doi: 10.1093/ndt/gfv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheong B., Muthupillai R., Rubin M.F., Flamm S.D. Normal values for renal length and volume as measured by magnetic resonance imaging. Clin. J. Am. Soc. Nephrol. 2007;2:38–45. doi: 10.2215/CJN.00930306. [DOI] [PubMed] [Google Scholar]
- 49.Levey A.S., Stevens L.A., Schmid C.H., Zhang Y.L., Castro A.F., 3rd, Feldman H.I., Kusek J.W., Eggers P., Van Lente F., Greene T., Coresh J., CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz G.J., Haycock G.B., Edelmann C.M., Jr., Spitzer A. A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics. 1976;58:259–263. [PubMed] [Google Scholar]
- 51.Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A., Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ong A.C.M., Harris P.C., Davies D.R., Pritchard L., Rossetti S., Biddolph S., Vaux D.J.T., Migone N., Ward C.J. Polycystin-1 expression in PKD1, early-onset PKD1, and TSC2/PKD1 cystic tissue. Kidney Int. 1999;56:1324–1333. doi: 10.1046/j.1523-1755.1999.00659.x. [DOI] [PubMed] [Google Scholar]
- 53.Pei Y., Obaji J., Dupuis A., Paterson A.D., Magistroni R., Dicks E., Parfrey P., Cramer B., Coto E., Torra R. Unified criteria for ultrasonographic diagnosis of ADPKD. J. Am. Soc. Nephrol. 2009;20:205–212. doi: 10.1681/ASN.2008050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei Y., Hwang Y.H., Conklin J., Sundsbak J.L., Heyer C.M., Chan W., Wang K., He N., Rattansingh A., Atri M. Imaging-based diagnosis of autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2015;26:746–753. doi: 10.1681/ASN.2014030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.(ExAC), E.A.C. (2016). Exome Aggregation Consortium (ExAC) Cambridge, MA (URL: http://exac.broadinstitute.org).
- 56.Lek M., Karczewski K., Minikel E., Samocha K., Banks E., Fennell T., O’Donnell-Luria A., Ware J., Hill A., Cummings B. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urribarri A.D., Munoz-Garrido P., Perugorria M.J., Erice O., Merino-Azpitarte M., Arbelaiz A., Lozano E., Hijona E., Jiménez-Agüero R., Fernandez-Barrena M.G. Inhibition of metalloprotease hyperactivity in cystic cholangiocytes halts the development of polycystic liver diseases. Gut. 2014;63:1658–1667. doi: 10.1136/gutjnl-2013-305281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris P.C., Bae K., Rossetti S., Torres V.E., Grantham J.J., Chapman A., Guay-Woodford L., King B.F., Wetzel L.H., Baumgarten D. Cyst number but not the rate of cystic growth is associated with the mutated gene in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2006;17:3013–3019. doi: 10.1681/ASN.2006080835. [DOI] [PubMed] [Google Scholar]
- 59.Cnossen W.R., Drenth J.P. Polycystic liver disease: an overview of pathogenesis, clinical manifestations and management. Orphanet J. Rare Dis. 2014;9:69. doi: 10.1186/1750-1172-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hung M.C., Link W. Protein localization in disease and therapy. J. Cell Sci. 2011;124:3381–3392. doi: 10.1242/jcs.089110. [DOI] [PubMed] [Google Scholar]
- 61.Hennet T., Cabalzar J. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction. Trends Biochem. Sci. 2015;40:377–384. doi: 10.1016/j.tibs.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 62.D’Alessio C., Fernández F., Trombetta E.S., Parodi A.J. Genetic evidence for the heterodimeric structure of glucosidase II. The effect of disrupting the subunit-encoding genes on glycoprotein folding. J. Biol. Chem. 1999;274:25899–25905. doi: 10.1074/jbc.274.36.25899. [DOI] [PubMed] [Google Scholar]
- 63.Reitman M.L., Trowbridge I.S., Kornfeld S. A lectin-resistant mouse lymphoma cell line is deficient in glucosidase II, a glycoprotein-processing enzyme. J. Biol. Chem. 1982;257:10357–10363. [PubMed] [Google Scholar]
- 64.Fedeles S.V., So J.S., Shrikhande A., Lee S.H., Gallagher A.R., Barkauskas C.E., Somlo S., Lee A.H. Sec63 and Xbp1 regulate IRE1α activity and polycystic disease severity. J. Clin. Invest. 2015;125:1955–1967. doi: 10.1172/JCI78863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore S.E., Spiro R.G. Demonstration that Golgi endo-α-D-mannosidase provides a glucosidase-independent pathway for the formation of complex N-linked oligosaccharides of glycoproteins. J. Biol. Chem. 1990;265:13104–13112. [PubMed] [Google Scholar]
- 66.Gallagher A.R., Germino G.G., Somlo S. Molecular advances in autosomal dominant polycystic kidney disease. Adv. Chronic Kidney Dis. 2010;17:118–130. doi: 10.1053/j.ackd.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takakura A., Contrino L., Zhou X., Bonventre J.V., Sun Y., Humphreys B.D., Zhou J. Renal injury is a third hit promoting rapid development of adult polycystic kidney disease. Hum. Mol. Genet. 2009;18:2523–2531. doi: 10.1093/hmg/ddp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.