Abstract
Motivation: Identifying drug–target interactions is an important task in drug discovery. To reduce heavy time and financial cost in experimental way, many computational approaches have been proposed. Although these approaches have used many different principles, their performance is far from satisfactory, especially in predicting drug–target interactions of new candidate drugs or targets.
Methods: Approaches based on machine learning for this problem can be divided into two types: feature-based and similarity-based methods. Learning to rank is the most powerful technique in the feature-based methods. Similarity-based methods are well accepted, due to their idea of connecting the chemical and genomic spaces, represented by drug and target similarities, respectively. We propose a new method, DrugE-Rank, to improve the prediction performance by nicely combining the advantages of the two different types of methods. That is, DrugE-Rank uses LTR, for which multiple well-known similarity-based methods can be used as components of ensemble learning.
Results: The performance of DrugE-Rank is thoroughly examined by three main experiments using data from DrugBank: (i) cross-validation on FDA (US Food and Drug Administration) approved drugs before March 2014; (ii) independent test on FDA approved drugs after March 2014; and (iii) independent test on FDA experimental drugs. Experimental results show that DrugE-Rank outperforms competing methods significantly, especially achieving more than 30% improvement in Area under Prediction Recall curve for FDA approved new drugs and FDA experimental drugs.
Availability: http://datamining-iip.fudan.edu.cn/service/DrugE-Rank
Contact: zhusf@fudan.edu.cn
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The identification of drug–target interactions is a crucial process in drug discovery, which can facilitate the understanding of drug action mechanism, disease pathology and drug side effect (Keiser et al., 2009; Lounkine et al., 2012; Nunez et al., 2012). Using biochemical experiments to identify these interactions is a reliable, yet expensive and time-consuming approach. In fact, the cost of developing a new FDA approved drug has doubled every 9 years since 1950, with only around 20 drugs approved by FDA per year (Scannell et al., 2012). Furthermore, there exist a huge number of unexplored compounds and human proteins, which makes it impossible to examine their interactions effectively by experimental approaches. For instance, there are over 35 million chemical compounds in PubChem database, and only less than 7000 have protein target information (Bolton et al., 2008). On the other hand, more than 140 000 human proteins are recorded in UniProtKB database (Boutet et al., 2007). It is estimated that the number of possible targets is between 6000 and 8000 (Overington et al., 2006), whereas only around 1300 known drugs are in DrugBank (Law et al., 2014). To reduce the huge time and financial cost of experimental approaches, many computational approaches have been proposed to select a small number of most promising candidate drug–target interactions for further experimental validation (Ding et al., 2014; Mousavian and Masoudi-Nejad, 2014; Yamanishi, 2013).
Traditional computational approaches for identifying drug–target interactions usually focus on a particular target of interest. These approaches can be divided into two categories, target-based methods (Lyne, 2002) and ligand-based methods (Acharya et al., 2011). The target-based methods rely on known 3D structure of targets and use docking techniques to simulate the interactions between targets and candidate drugs. However, the 3D structures of many important targets, especially G-protein coupled receptors and ion channels, are unavailable. On the other hand, ligand-based methods rely on the known interacting ligands of target proteins to define a pharmacophore model that describe the common characteristic of binding ligands. In this case, if the number of known binding ligands is very small, ligand-based methods cannot work well.
To overcome the limitation of traditional approaches and also realize large-scale prediction, machine learning has attracted lots of attention (Ding et al., 2014; Mousavian and Masoudi-Nejad, 2014; Yamanishi, 2013). This approach can use not only known drug–target interactions, but also various types of information on drug and targets, implementing the idea of integrating the chemical space of drugs and genomic space of targets. The methods of this approach can be roughly divided into two groups: feature-based methods (Nagamine and Sakakibara, 2007; Yabuuchi et al., 2011; Tabei et al., 2012) and similarity-based methods (Ding et al., 2014).
In feature-based methods, drugs and targets are represented by feature vectors, which are derived from their properties, such as drug fingerprints and the sequence descriptors of targets. Currently, the most high performance approach in this direction uses ‘Learning To Rank (LTR)’ (Agarwal et al., 2010; Rathke et al., 2010; Zhang et al., 2015), a rather new paradigm in machine learning. In principle, LTR is very powerful. For example, in binary classification, positive examples ranked lower and negative examples ranked higher are more penalized, while examples are rather treated equally in any classification method (Li, 2011). LTR has been originally developed in information retrieval for ranking web pages, according to multiple users’ queries, meaning that LTR is considered for ‘multilabel classification’, in which each instance is labeled by multiple labels (Liu, 2009). Identifying drug–target interactions can also be multilabel classification, in the sense that drugs can be interacted (labeled) by multiple targets. In fact, statistics from DrugBank (before March 2014) shows 4.58 interacting targets on average for each FDA approved small molecule drug, and 4.5 interacting FDA approved small molecule drugs on average for each target protein. So each drug (target) is labeled around 4.5 times on average, and LTR is perfectly fitted to drug–target interaction prediction.
In similarity-based methods, different types of similarity among drugs (e.g. chemical structure similarity) and among targets (e.g. protein sequence similarity) can be used to learn models to predict the interaction between drug and targets. A common assumption of these methods is that similar drugs will interact with similar targets and vice versa. A distinct advantage of similarity-based methods is that this assumption can be explicitly incorporated into the prediction model for identifying novel drug–target interactions. Following the pioneering work of (Yamanishi et al., 2008), a lot of similarity-based methods have been developed by using different techniques, such as kernel learning (Bleakley and Yamanishi, 2009; Jacob and Vert, 2008; Mei et al., 2013; van Laarhoven et al., 2011; van Laarhoven and Marchiori, 2013), matrix factorization (Gönen, 2012; Zheng et al., 2013), regularized least squares (Xia et al., 2010) and multilabel k-nearest neighbor (Shi et al., 2015). These methods are thought to be reasonable and valid, because the drug and target similarities generate the chemical space of drugs and genomic space of targets, respectively, and the methods try to understand the connection between the two spaces. This point leads to the high-performance of similarity-based methods.
We address the problem of predicting drug–target interactions for new drugs or new targets, which is especially challenging (Ding et al., 2014), due to three main reasons. First, new drugs or targets do not have known interactions, which makes the training of prediction models difficult. Second, the connections among different drugs or targets are not well considered in the existing computational models. Third, given a new drug (or target), there are many possible interacting targets (or drugs). Until December 2015, there are at least approximately 1200 FDA approved small molecule drugs and also at least 1300 target human proteins in DrugBank (Law et al., 2014). To this end, we propose a new method, DrugE-Rank, to take the most advantage of both the feature-based and similarity-based machine learning methods. First, DrugE-Rank uses LTR, which provides currently the most powerful performance and is totally suitable for this problem. Second, in the framework of LTR, DrugE-Rank performs an ensemble learning, that is integrating the prediction by diverse cutting-edge techniques. It has adopted different prediction methods as well as regular inputs (i.e. features of drug and targets). Third, in the ensemble framework, LTR has only the top promising drugs (or targets) suggested by each component methods as the input, instead of considering all possible chemical compounds. This manner avoids using irrelevant chemical compounds and eventually reduces computational burden heavily.
We examined the performance of DrugE-Rank by using manually annotated drug–target interactions in DrugBank (Law et al., 2014). We generated three different datasets from DrugBank: (i) FDA approved drugs appeared in DrugBank before March 2014; (ii) new targets and FDA approved targets appeared in DrugBAnk after March 2014; and (iii) FDA experimental drugs. We first compared the performance of DrugE-Rank with competing methods by cross-validation on the first dataset. Experimental results on this cross-validation show that DrugE-Rank outperformed all competing methods, being statistically significant. The improvement when having new drug was especially promising, where DrugE-Rank achieved an Area under Prediction Recall curve (AUPR) of 0.4917, 27.3% higher than that by the best component method (an AUPR of 0.3864). The performance of DrugE-Rank was further validated by two different independent tests using the second and third datasets. For example, for the new FDA approved drugs appearing in DrugBank after March 2014, DrugE-Rank achieved an AUPR of 0.2031, 60.8% improvement against that by the best component method (an AUPR of 0.1263). Also we analyzed top predicted drugs and targets from a variety of viewpoints, giving several insights on the data of drug–target interactions.
2 Related work
Feature-based learning is a general framework of machine learning, especially classification, for which various approaches have been proposed and successfully used in a wide variety of applications. LTR is a rather new paradigm in machine learning, which is motivated by information retrieval, where with respect to users’ queries, more relevant web pages should be ranked higher (Li, 2011). LTR is powerful for binary classification in the sense that lower ranked positive examples and higher ranked negatives are penalized more, while general classification methods treat all examples equally. Already a variety of methods for LTR have been proposed. One of such method is LambdaMART (Burges, 2010), which has been used in many applications (Liu et al., 2015b). Currently, LTR has proven to be a powerful approach not only in information retrieval, but also in many other applications, even in computational biology, such as protein remote homology detection (Liu et al., 2015a), peptide identification from proteomics data (Qeli et al., 2014) and biomedical semantic indexing (Liu et al., 2015b). In particular, recently LTR has been applied to ligand-based virtual screening (Agarwal et al., 2010; Rathke et al., 2010; Zhang et al., 2015). Initially, a pair-wise LTR method, SVMRank, was rather straightforwardly applied to rank chemical structures for drug discovery, showing higher performance than support vector regression (SVR) (Agarwal et al., 2010). Then this method with SVMRank was outperformed by StructRank (Rathke et al., 2010). Also in a more application-oriented manner, six already available software/algorithms of LTR were compared for ligand-based virtual screening (Zhang et al., 2015). In this line of work, we emphasize that our proposed method, DrugE-Rank, has several unique points, to outperform the existing work: (i) DrugE-Rank can use ensemble learning or information from multiple component methods, for which cutting-edge similarity-based machine learning methods can be used. In addition, in training, we filter candidate drugs (or targets) by using component methods, and this step works well to remove irrelevant drugs (false positives) and as a result reduce the entire computational cost. (ii) The focus of DrugE-Rank is on predicting new drugs (or new targets), while the existing work are for ligand-based virtual screening (to rank the candidate chemical compounds), where the target is not necessarily new. (iii) DrugE-Rank uses all known drug–target interactions in DrugBank, meaning that this work is a thorough study, while the data used in existing work are interactions of a single target or a small number of targets only.
One uniqueness of the problem of predicting drug–target interactions is that given data are not only interactions between drugs and targets, but also similarities between drugs (and those of targets), which can be thought to represent the chemical and genomic spaces, respectively. This is a well-advocated paradigm in chemogenomics. Thus, similarity-based approaches are well accepted by pharmacologists and relevant methods have been widely proposed, achieving high predictive performance (Ding et al., 2014). DrugE-Rank uses cutting-edge similarity-based methods as components of ensemble learning. See Section 3.3 for their schemes which are explained as component methods of DrugE-Rank.
3 Methods: DrugE-Rank
3.1. Notations
We use to represent drug set, and for target set. Y is the drug–target interaction matrix, where Yij = 1 if there is interaction between di and tj, otherwise Yij = 0. Let be a binary vector, called interaction profile of drug di, where the j-th element of is 1 if drug di interacts with target tj; otherwise 0. Similarly let be a binary vector, called interaction profile of target tj. is the genomic similarity matrix of targets, and is genomic similarity between ti and tj in ; is the chemical similarity matrix of drugs, and is the genomic similarity between di and dj in ; is the feature vector of target t, and is the feature matrix of T. is the feature vector of drug d, and is the feature matrix of D.
3.2. Overview
The framework of DrugE-Rank is to predict drugs (or targets) given a new target (or a new drug). For simplicity, in this section, we explain our method by the case that a new target is given (and then ranking drugs). We note that the reverse case can be easily explained by replacing drugs and targets with targets and drugs, respectively. Given a new target, each target can be viewed as an instance, and all drugs can be viewed as labels. That is, given a new target , identifying relevant drugs is to predict the labels of instance t, which can be considered as a multilabel classification, since not only one label (drug) but also multiple labels (drugs) are considered. To solve this problem, DrugE-Rank uses LTR, which is originally developed in information retrieval for ranking web pages with respect to user’ query. That is, in information retrieval, t and D can be a query and a set of web pages, respectively, and predicting drugs interacting with t can be solved by ranking drugs in D by LTR.
The procedure of DrugE-Rank has four steps. Before explaining the detail of the four steps, we will briefly explain the six component methods of DrugE-Rank.
3.3 Component methods
We select six well-known, cutting-edge similarity-based methods as component methods: k-nearest neighbor (k-NN), Bipartite Local Model with support vector classification (BLM-svc) (Bleakley and Yamanishi, 2009), Bipartite Local Model with support vector regression (BLM-svr) (Bleakley and Yamanishi, 2009), Laplacian regularized least squares (LapRLS) (Xia et al., 2010), Network-based Laplacian regularized least squares (NetLapRLS) (Xia et al., 2010), Weighted Nearest Neighbor-based Gaussian Interaction Profile classifier (WNN-GIP) (van Laarhoven et al., 2011; van Laarhoven and Marchiori, 2013).
(i) k-NN
Given a new target tnew, we select a set of top k most similar targets, M by using . Then the interaction profile of tnew can be computed as follows: k-NN uses the closest instances (targets) to estimate interaction profile of a given new target, meaning that k-NN only uses local information of drug–target interactions.
(ii) BLM-svc
Given a new target tnew, to predict the interaction between tnew and drug d, BLM generates a prediction model by regarding each of other targets as one instance. That is, each instance (target) has one binary label showing if the corresponding target is interacting with drug d. The prediction model of BLM-svc is support vector classification, for which the kernel is generated from drug similarities .
(iii) BLM-svr
BLM-svr uses the framework of BLM-svc, just by replacing svc with svr.
(iv) LapRLS
LapRLS minimizes the squared loss between Yand F (interaction score matrix:parameter) with a regularized term of using and F. This minimization leads to an analytical solution, by which Fcan be obtained easily by a rule consisting of and Y.
(v) NetLapRLS
NetLapRLS is an extension of LapRLS by incorporating the network information of drug–target interaction into the regularizer under the same regularized least squares framework.
(vi) WNN-GIP
Gaussian interaction profile (GIP) generates a Gaussian kernel for drug pairs using the similarity of interaction profiles of drugs and also a Gaussian kernel for target pairs from those of targets. From these kernels, GIP then generates a kernel between drug–target pairs, which is used for predicting drug–target interactions by regularized least squares. KNN-GIP is a slight extension of GIP by using k-NN for computing Gaussian interaction profiles.
3.4 Entire procedure of DrugE-Rank
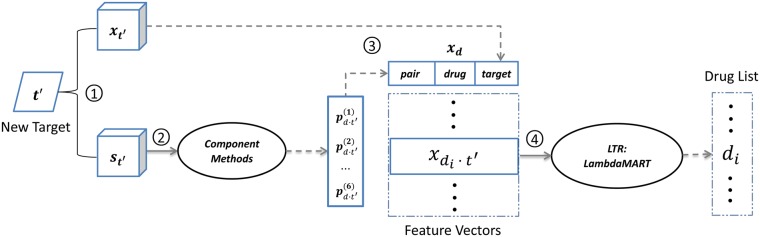
Figure 1 shows the entire procedure of DrugE-Rank, with four steps.
Fig. 1.
A four-step procedure of DrugE-Rank for predicting drugs’ interactions with a given target : (1) DrugE-Rank first computes feature vectors of target and arbitrary drug d and also similarity of targets and that of drugs, (2) DrugE-Rank runs six component methods by using similarities computed in Step 1 and the returning scores are generated as pair features, (3) A feature vector for drug d is generated from drug and target features, generated in Step 1 and pair features in Step 2, (4) Finally, the features are input into LTR and the resultant ranked drug list for can be returned
3.4.1. Step 1: preprocessing data
For a given new target tnew, we generate 147-dimension feature vector (target feature vector or TFV) by Composition, Transition and Distribution (CTD), which is extensively used and sequence-derived physicochemical features. We obtain this data from a web server, PROFEAT (Rao et al., 2011). The genomic similarity between tnew and a target t in T, that is , is computed by normalized Smith–Waterman score on sequence similarity (Yamanishi et al., 2008).
For the drug side, each drug d is represented by a 36-dimension GD (general descriptors) feature vector (drug feature vector or DFV), which is generated by an open-source Chemoinformatics software, RDKit.1 The GD features are, for each chemical compound, a set of physical molecular properties, such as van der Waals surface area, molar refractivity, log P (octanol/water), and partial charge. The chemical similarity between d and drug in D, that is , is calculated by Tanimoto coefficient over 2D chemical substructures of drugs.
Table 1 summarizes the information on drug and target feature vectors.
Table 1.
Drug and target feature vectors
| DFV | TFV | |
|---|---|---|
| Name | General descriptor | CTD |
| Dimension | 36 | 147 |
| Calculation | RDKit | PROFEAT |
| Description | PEOE_VSA1 - PEOE_VSA14, SMR_VSA1 - SMR_VSA10, SlogP_VSA1 - SlogP_VSA12 | Composition, transition and distribution |
3.4.2. Step 2: running component methods
We run six component methods to predict the score of drug–target interaction between any drug d and the given new target tnew, for i-th component, using the similarity between new target tnew and another target t, that is , as the input. The obtained score is further normalized to be a value between 0 and 1 as follows:
where maxi and mini are the maximum and minimum prediction scores of i-th component method. This means, for a given new target tnew and a drug d, we can have a 6-dimension vector (pair feature vector or PFV), where the i-th element of this vector is .
3.4.3. Step 3: setting up feature vector
We generate the final feature vector for any pair of drug d and a newly given target tnew, by concatenating the following three types of features:
TFV: target feature vector
As described in Step 1, target tnew is represented by a 147-dimension feature vector, .
DFV: drug feature vector
Also as mentioned in Step 1, each drug d is represented by a 36-dimension feature vector, xd.
PFV: pair feature vector
As described in Step 2, each drug–target pair can be represented by a 6-dimension feature vector , showing the strength of interaction between d and tnew.
Thus, each drug–target pair is represented by 189 features, which are the input of Step 4 for ranking drugs to interact with tnew.
3.4.4 Step 4: ranking drugs by LTR
We use LambdaMART (Burges, 2010) for training LTR. Given a new target tnew, we can have the feature vector of a drug, according to Step 3, by which for drugs, we can generate multiple feature vectors, which are given as the input to the trained LTR. A ranked list of drugs is then returned as the final prediction result.
We note that when training LTR, we do not use all possible drug–target pairs, and instead for each target, we use only top K drugs highly predicted to be interacting with the target by each component method. In fact drug–target interaction is highly sparse data, by which false positives can occur easily. So this manner of using only top K drugs is highly effective to avoid this type of false positives and eventually reduce the computational cost in training LTR.
4 Experiments
4.1. Data
Drug–target interaction (binary) data are obtained from DrugBank, a high-quality human annotated database (Law et al., 2014), in which drugs are labeled with protein targets. In the data of DrugBank, we used only small molecules (FDA approved or experimental drug) and human proteins for targets. We first focused on FDA approved drugs and then conducted preliminary data analysis over the drug–target interactions with FDA approved drugs. In total, this data consists of 1242 drugs, 1324 targets and 5701 interactions (0.4% of all possible drug–target pairs).
4.1.1. Promiscuity of drugs and targets
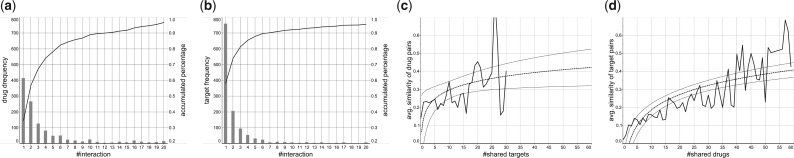
Figure 2 shows the distribution of the number of (a) drugs and (b) targets when they are classified by the number of interactions. The result shows that more than 95% drugs/targets are involved in less than 20 interactions, meaning the sparseness and unbalancedness of the data, while in both drugs and targets, around half of them have more than one interactions, implying high promiscuity of those drugs and targets.
Fig. 2.
Analysis on drug–target interactions with FDA-approved drugs consisting of 5701 known interactions between 1242 FDA approved drugs and 1324 human protein targets. (a) #interactions versus #drugs, (b) #interactions versus #targets, (c) #shared targets versus average similarity between drugs and (d) #shared drugs versus average similarity between targets. In (c) and (d), the dashed lines show the trend lines, being fitted on a logarithmic function, and the two outside lines show the confidence interval of the estimation ((c): P = 0.046, R2 = 0.19 and (d): P = 0.001, )
4.1.2. Correlation between #shared drugs/targets and similarity
A hypothesis of similarity-based methods is that similar drugs will interact with similar or the same targets, and vice versa. Figure 2 shows the average similarity between (c) drugs due to the number of shared targets and (d) targets due to the number of shared drugs (both by the solid lines). The dashed lines show trend lines, which indicates the rather positive correlation between the similarity and the number of shared drugs/targets, confirming the hypothesis of similarity-based methods,that is component methods in DrugE-Rank.
4.1.3. Generating subsets
We divided the entire DrugBank data into five subsets, named by Data-1 to Data-5, where Data-1 to Data-4 have FDA approved drugs and Data-5 has experimental drugs. Data-1 and Data-3 are distinct from Data-2 and Data-4 by the time (March, 2014) when drugs appear in DrugBank. Similarly, Data-1 and Data-2 are distinct from Data-3 and Data-4 by when (also March, 2014) targets appear in DrugBank. Table 2 shows the summary of this classification of subsets, and Table 3 shows the information on these datasets.
Table 2.
Binary drug–target interaction datasets from DrugBank
| FDA drugs (before 03/2014) | FDA drugs (after 03/2014) | Experimental drugs | |
|---|---|---|---|
| Targets (before 03/2014) | Data-1 | Data-2 | Data-5 |
| Targets (after 03/2014) | Data-3 | Data-4 |
Table 3.
Information on data subsets
| Data-1 | Data-2 | Data-3 | Data-4 | Data-5 | |
|---|---|---|---|---|---|
| #Targets | 1200 | 1200 | 124 | 124 | 1324 |
| #Drugs | 1178 | 64 | 1178 | 64 | 1753 |
| #Interactions | 5400 | 149 | 147 | 5 | 2094 |
| Interaction rate (%) | 0.382 | 0.194 | 0.101 | – | 0.09 |
| #Interactions per target | 4.5 | – | 1.19 | – | 1.58 |
| #Interactions per drug | 4.58 | 2.33 | – | – | 1.19 |
4.2. Experimental procedures
4.2.1. Competing methods
We compared DrugE-Rank with six component methods which are cutting-edge similarity-based machine learning methods for predicting drug–target interactions. Also we used two feature-based machine learning methods, that is random forest (RF) and gradient boosting decision tree (GBDT) as competing methods, where only the DFV and TFV were used for RF and GBDT. Furthermore, within DrugE-Rank, we tested three different sets of feature vectors including: (i) DFV and TFV, (ii) PFV and (iii) all features (= DFV, TFV and PFV), to check the contribution of the feature vectors to the performance improvement. We note that DrugE-Rank with DFV and TFV only is substantially equivalent to the existing work of using LTR, which is one of the most powerful feature-based machine learning approaches.
4.2.2. Evaluation criteria
Drug–target interactions are really sparse, by which false positives should be punished more in evaluation. Thus, we use AUPR, instead of AUC (Area Under the receiver operator characteristics curve), which treats all examples equally (Davis and Goadrich, 2006). Note that precision is the ratio of the correctly predicted positives to all predicted positives, and recall is the ratio of predicted positives to all positives.
4.2.3. Three types of experimental settings
Cross-validation over Data-1
We conducted 10 times 5-fold cross-validation over Data-1, thinking about new drugs or new targets, meaning repeating randomly dividing drugs or targets of Data-1 into five parts (one for testing, one for training LTR and the rest for training component models) for 10 times. Paired t-test was used to confirm the statistical significance of performance differences.
Independent test on Data-2 + Data-3 (new targets or new FDA approved drugs (appearing in DrugBank after March 2014))
We used Data-1 for training and Data-2 and Data-3 for testing. In training, all targets (or drugs) in Data-1 were randomly divided into five subsets, four out of the five for training component models and the rest one for training LTR. We repeated this procedure 10 times and reported the average prediction performance.
Independent test on Data-5 (interactions with experimental drugs)
We used all Data-1 to Data-4 for training and Data-5 for testing. The procedure is the same as the above independent test setting.
4.3. Parameter setting
4.3.1. Choosing parameter values
For k-NN, k was selected from {5,7,9,11,13,15,17} to maximize AUPR under cross-validation. BLM-svc, BLM-svr, LapRLS, NetLapRLS and WNN-GIP were implemented exactly following their publications, where LibSVM was used for BLM-svc and BLM-svr, with regularization coefficient C of 1. RF, GBDT and LambdaMART were implemented by RankLib.2 The default parameter setting was used for LapRLS and NetLapRLS. The decay rate T of WNN-GIP was selected from {0.7, 0.8, 0.85, 0.9, 0.95} by 5fold cross-validation to maximize AUPR. For RF, the number of bags was selected from {8,16,32,64,128} to maximize AUPR in cross-validation. Also for LambdaMART and GBDT, the number of trees and the number of leaves for each tree were selected from {8,16,32,64,128} and {4,8,16}, respectively, in a similar manner.
Component methods were used in DrugE-Rank, and also for comparing their performances with that of DrugE-Rank. Please note that data are different between the above two cases. For example, in DrugE-Rank, only a part of data was used, like in 5-fold cross-validation over Data-1, three among five folds for training component methods, one for training LTR and the last one for performance evaluation, while in performance comparison, the same last one for performance evaluation and the rest are for training the component method.
4.3.2. Selecting K, the number of labels
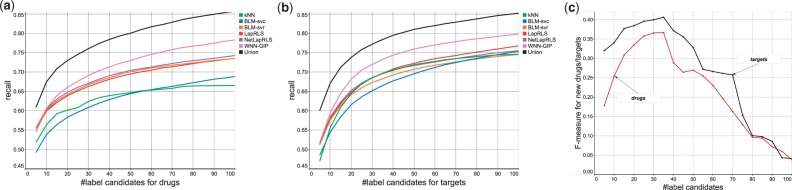
As shown by data analysis, drug–target interactions are very sparse and likely to cause false positives, meaning that reducing possible labels would be very useful. So we did a preliminary experiment to decide the number of K, that is possible number of labels or candidate drugs (for a given new target), by dividing Data-1 into five parts, one for training LTR, one for prediction and the rest for training component models. After prediction, we checked the precision and recall of top K prediction results by component models and the union of their prediction results, changing K = 5 to 100. Fig. 3(a) and (b) show that the recall values obtained by the results of this preliminary experiment. The union of the results obtained by component models reached 0.75 by integrating top 30, that is around 3% of all possible labels (drugs). This is much higher than that of each component model, implying that focusing on most confidently predicted drugs/targets is very useful. Finally, we computed F-measure, the harmonic mean of precision and recall, for predicting new drugs or targets. Fig. 3(c) shows the F-measure against the number of label candidates, indicating that the F-measure can be maximized for K = 35, which we used for all experiments in this article.
Fig. 3.
Selecting the number of labels by 10 × 5-fold cross-validation over Data-1. (a) Recall values by the prediction of component models and the union of their results, changed by the candidate drugs. (b) Also recall values, changed by the candidate targets. (c) F-measure values for predicting drugs/targets, obtained by the union of prediction results
4.4. Experimental results
4.4.1. Cross-validation over Data-1
Table 4 shows the performance results (AUPR) by six similarity-based methods, two feature-based methods and DrugE-Rank with three different sets of features. Among six similarity-based methods, BLM-svr was the best in both new drugs and new targets, being especially significant in new target prediction. On the other hand, feature-based methods gave worse results than BLM-svr. For example, for predicting new drugs, BLM-svr achieved an AUPR of 0.3864, outperforming an AUPR of 0.3418 by RF and of 0.3328 by GBDT. DrugE-Rank, when all features were used, achieved the highest AUPR of 0.4917 for predicting new drugs and 0.5906 for new targets, being statistically significant against all other cases except DrugE-Rank with pair features. In particular, the AUPR of 0.4917 for new drugs is around 27% higher than that by BLM-svr, the best similarity-based prediction method. On the other hand, DrugE-Rank with drug and target features only showed comparative performance with BLM-svr, being much worse than DrugE-Rank with pair features or all features. This result indicates that pair features are much more important than drug and target features. Also integrating pair features into drug and target features is important, confirming the usefulness of our idea.
Table 4.
AUPR for 10 × 5-fold cross-validation over Data-1 (P-values of paired t-test against DrugE-Rank with all features)
| Method | Drug-based interactions | Target-based interactions |
|---|---|---|
| k-NN | ||
| BLM-svc | ||
| BLM-svr | ||
| LapRLS | ||
| NetLapRLS | ||
| WNN-GIP | ||
| RF | ||
| GBDT | ||
| DrugE-Rank | ||
| (DFV and TFV) | ||
| DrugE-Rank | ||
| (PFV only) | ||
| DrugE-Rank | ||
| (all features) |
The AUPRs achieved by the best model, DrugE-Rank (all features), are highlighted in bold face.
Furthermore, by using the 10 trained models of DrugE-Rank, we predicted unknown drug–target interactions (for Data-1) and checked if they are in the latest DrugBank. The interactions were sorted by how many times they are predicted in the top ten out of the 10 models. Tables 5 and 6 show the results obtained by target-based and drug-based cross-validation, respectively. Surprisingly, six out of the top seven predicted interactions in Table 5 and five out of the seven predicted interactions in Table 6 can be found in the latest DrugBank (see supplementary materials for more results). This result also indicates the high predictive performance of DrugE-Rank.
Table 5.
New interactions predicted by using target-based 10 × 5-fold cross validation results (models) on Data-1
| UniProt ID (Target name) | DrugBank ID (Drug name) | #Times | Found? |
|---|---|---|---|
| Q8N1C3 (Gamma-aminobutyric acid receptor subunit gamma-1) | DB00898 (Ethanol) | 8 | Yes |
| Q99928 (Gamma-aminobutyric acid receptor subunit gamma-3) | DB00898 (Ethanol) | 8 | Yes |
| P78334 (Gamma-aminobutyric acid receptor subunit epsilon) | DB00898 (Ethanol) | 8 | Yes |
| Q13639 (5-hydroxytryptamine receptor 4) | DB00408 (Loxapine) | 7 | No |
| O94956 (Solute carrier organic anion transporter family member 2B1) | DB01045 (Rifampicin) | 7 | Yes |
| Q9UM07 (Protein-arginine deiminase type-4) | DB00759 (Tetracycline) | 7 | Yes |
| P19320 (Vascular cell adhesion protein 1) | DB00898 (Ethanol) | 7 | Yes |
Note: Predicted (top 7) interactions were sorted by how many times they are predicted out of 10 times (shown by #times), and the last column shows Yes or No, indicating if the interaction was found in the latest DrugBank database or not.
Table 6.
New interactions found by using drug-based 10 × 5-fold cross validation on Data-1
| UniProt ID (Target name) | DrugBank ID (Drug name) | #Times | Found? |
|---|---|---|---|
| Q16850 (Lanosterol 14-alpha demethylase) | DB01045 (Rifampicin) | 10 | Yes |
| O94956 (Solute carrier organic anion transporter family member 2B1) | DB01045 (Rifampicin) | 10 | Yes |
| P48051 (G protein-activated inward rectifier potassium channel 2) | DB00898 (Ethanol) | 10 | Yes |
| P20309 (Muscarinic acetylcholine receptor M3) | DB01339 (Vecuronium) | 10 | No |
| P21728 (D(1A) dopamine receptor) | DB00933 (Mesoridazine) | 10 | No |
| Q12809 (Potassium voltage-gated channel subfamily H member 2) | DB00537 (Ciprofloxacin) | 10 | Yes |
| Q02641 (Voltage-dependent L-type calcium channel subunit beta-1) | DB00898 (Ethanol) | 9 | Yes |
4.4.2. Independent test on Data-2 and Data-3
Table 7 shows the AUPR results obtained by applying models trained by Data-1 to independent datasets, Data-2 and Data-3. We can first see that the performance of all methods dropped significantly from the cross-validation results of Table 4. This implies that the data distribution drastically changed between training and testing, being different from cross-validation. However, the relative performance order of different methods were still kept. First, BLM-svr achieved the higher AUPR than all other similarity-based methods and also two feature-based methods. Second, two feature-based methods did not perform well, against not only BLM-svr but also most of other similarity-based methods. Third, DrugE-Rank with all features was the best performer among all methods, being followed by DrugE-Rank with pair features. For example, for new drug prediction, DrugE-Rank with all features achieved the highest AUPR of 0.2031 with standard deviation of 0.0127, which is amazingly 61% higher than that by the best similarity-based method, BLM-svr with 0.1263. Again this result confirmed the importance of pair features and the advantage of DrugE-Rank.
Table 7.
AUPR for independent testing data (FDA approved, new drugs and experimental drugs)
| Methods | Data-2 | Data-3 | Data-5 |
|---|---|---|---|
| new drugs | new targets | new drugs | |
| k-NN | 0.0783 | 0.1046 | 0.0173 |
| BLM-svc | 0.1064 | 0.1955 | 0.0316 |
| BLM-svr | 0.1263 | 0.2096 | 0.0405 |
| LapRLS | 0.1155 | 0.1454 | 0.0262 |
| NetLapRLS | 0.1244 | 0.1667 | 0.0278 |
| WNN-GIP | 0.1081 | 0.1680 | 0.0338 |
| RF | 0.1192 | 0.1314 | 0.0512 |
| GBDT | 0.1168 | 0.1577 | 0.0392 |
| DrugE-Rank | 0.1329 | 0.1810 | 0.0717 |
| (DFV and TFV) | |||
| DrugE-Rank | 0.1803 | 0.2658 | 0.0732 |
| (PFV only) | (0.0123) | (0.0113) | (0.0029) |
| DrugE-Rank | |||
| (all features) | (0.0127) | (0.0078) | (0.0205) |
We further checked the performance of the competing methods in more details, using AUPR by each method for one drug (in Data-2) and that for one target (in Data-3). That is, for example, in Data-2, we checked the method which gave the highest AUPR for each drug and then, for each method, counted the number of drugs to which the highest AUPR was given. Table 8 shows the results. Both for drugs and targets, DrugE-Rank achieved the highest number. For example, DrugE-Rank performed the best in 74 out of all 124 targets, being followed by k-NN and BLM-svr, both having achieved the highest AUPR for 36 targets (please note that two or more methods may have achieved the same highest AUPR). This result also confirmed the performance advantage of DrugE-Rank.
Table 8.
The number of drugs and targets, for which each method achieved highest AUPR.
| Methods | Drugs (Data-2) | Targets (Data-3) |
|---|---|---|
| k-NN | 9 | 36 |
| BLM-svc | 14 | 35 |
| BLM-svr | 14 | 36 |
| LapRLS | 13 | 26 |
| NetLapRLS | 13 | 32 |
| WNN-GIP | 17 | 34 |
| DrugE-Rank | 24 | 74 |
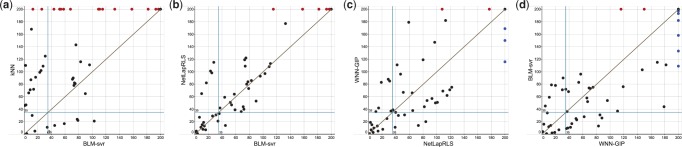
We finally explored the complementarity between different similarity-based methods, since they are used as component methods for ensemble learning of DrugE-Rank. We selected top 200 drugs predicted by each method, and plotted the rank of true interacting drugs by each method for a pair of two component methods, such as k-NN versus BLM-svr. Figure 4 shows the results by (a) k-NN versus BLM-svr, (b) NetLapRLS versus BLM-svr, (c) WNN-GIP versus NetLapRLS and (d) BLM-svr versus WNN-GIP. So each point in the figure is a drug ranked higher than 200 in at least either of the competitive two methods, where red and blue points show those ranked lower than 200 by only one method. The points close to the diagonal line are the drugs for which two competing methods perform similarly, while the points far from the diagonal line are the drugs for which one method performs much better than the other method. Figure 4 shows that no methods can beat other methods clearly. Even between the worst component method k-NN and the best component method BLM-svr, shown in (a), there are a few true interacting drugs on the side of BLM-svr, meaning that they are ranked higher by k-NN than BLM-svr. Also shown in (c), NetLapRLS and WNN-GIP, which achieved almost similar AUPR values of around 0.167 in Table 7, have very few drugs close to the diagonal line, meaning that their rankings are totally different. All these results confirm the complementarity of different component methods, which must bring diverse effects to allow DrugE-Rank to improve the predictive performance by their ensemble.
Fig. 4.
Complementarity between component methods. The 200 drugs correctly predicted for a new given target by each method is compared with that of another. The comparison is between (a) k-NN versus BLM-svr, (b) NetLapRLS versus BLM-svr, (c) WNN-GIP versus NetLapRLS and (d) BLM-svr versus WNN-GIP
4.4.3. Independent test on Data-5
Table 7 shows the AUPR results obtained by applying models trained by Data-1 to Data-4 to independent dataset Data-5. The values are further decreased from those obtained when Data-2 and Data-3 were used for independent test sets. This indicates that interactions with experimental drugs in Data-5 have totally different nature from interactions in other subsets. However, the performance order was again kept the same as that in the cross-validation experiment over Data-1. Concretely, BLM-svr achieved the best performance among six similarity-based methods, and DrugE-Rank outperformed BLM-svr. In particular DrugE-Rank with drug and target features outperformed BLM-svr rather clearly, and are rather close to the other two DrugE-Rank settings. Overall we can say that the framework of DrugE-Rank is useful for predicting unknown, independent drug–target interactions with new drugs (or targets).
4.5. Two cases
We present two specific cases with sample drugs and targets, which are correct and were ranked high by DrugE-Rank, with their rankings by other competing methods. Table 9 shows the top three drugs ranked by DrugE-Rank, meaning that they were predicted to interact with a given new target, ‘inhibitor of nuclear factor kappa-B kinase subunit alpha’. Also Table 10 shows the top five targets, predicted by DrugE-Rank, and the rankings obtained by other competing methods. We can see that even if the rankings of drugs (or targets) by component methods were very low by multiple component methods, DrugE-Rank could identify these drugs successfully. For example, in Table 10, given Nintedanib, DrugE-Rank successfully identify the targets which interact with Nintedanib, such as Vascular endothelial growth factor receptor 3, which was ranked out of top 50 by four component methods, k-NN, BLM-svc, BLM-svr and WNN-GIP. Overall, this result also shows the good predictive ability of DrugE-Rank.
Table 9.
Ranks by competing methods for top three drugs predicted by DrugE-Rank for new protein target ‘inhibitor of nuclear factor kappa-B kinase subunit alpha’ (UniProt ID: O15111, in Data-3)
| DrugBank ID (Name) | DrugE-Rank | k-NN | BLM-svc | BLM-svr | LapRLS | NetLapRLS | WNN-GIP |
|---|---|---|---|---|---|---|---|
| DB00244 (Mesalazine) | 1 | 3 | 3 | 3 | 3 | 4 | 35 |
| DB00795 (Sulfasalazine) | 2 | 4 | 1177 | 4 | 4 | 5 | 3 |
| DB00233 (Aminosalicylic Acid) | 3 | 25 | 269 | 382 | 308 | 960 | 13 |
Table 10.
Ranks by competing methods for top five targets predicted by DrugE-Rank for drug ‘Nintedanib’ (DrugBank ID: DB09079, in Data-2)
| UniProt ID (Name) | DrugE-Rank | k-NN | BLM-svc | BLM-svr | LapRLS | NetLapRLS | WNN-GIP |
|---|---|---|---|---|---|---|---|
| P35916 (Vascular endothelial growth factor receptor 3) | 1 | 78 | 105 | 72 | 7 | 13 | 60 |
| P17948 (Vascular endothelial growth factor receptor 1) | 2 | 79 | 113 | 73 | 11 | 14 | 61 |
| P36888 (Receptor-type tyrosine-protein kinase FLT3) | 3 | 38 | 13 | 17 | 67 | 39 | 4 |
| P35968 (Vascular endothelial growth factor receptor 2) | 4 | 77 | 32 | 77 | 17 | 22 | 65 |
| P21802 (Fibroblast growth factor receptor 2) | 5 | 40 | 156 | 18 | 93 | 72 | 15 |
5 Discussion
Identifying drug–target interactions of new, unknown drugs or targets is a very challenging task, where interactions of new drugs or targets are unavailable. Previous methods can be divided into two groups, feature- and similarity-based methods. DrugE-Rank incorporates the outputs of similarity-based methods as features to improve the prediction performance. This point has not been considered in any existing work including those using LTR. Our experimental results clearly demonstrated that the novel combination of similarity- and feature-based methods such as DrugE-Rank is significantly better than independent approaches. Moreover, DrugE-Rank provides a robust framework to integrate other types of information to improve the accuracy of drug–target interaction prediction.
Compared with drug and target features, pair features from the similarity based component methods are the key to improve the performance of DrugE-Rank. A common assumption behind similarity-based methods is that similar drugs (targets) are likely to interact with similar targets (drugs). We validated this assumption in DrugBank data analysis. In addition, we compared the drugs highly predicted by component methods directly, confirming that component methods in DrugE-Rank can provide diverse results and are complement to each other. This must contribute to the performance improvement of DrugE-Rank. Currently we select the component methods by considering their accuracy, diversity and efficiency. With the increase of possible component methods, an interesting problem would be how to automatically choose the most suitable component methods.
We found that identifying interactions of new drugs is more difficult than the one of new targets, which is consistent with previous study (Ding et al., 2014). This suggests that the action mechanism of drug is complicated, where one drug may have multiple types of targets and sometimes similar drugs may have different mechanisms. In this case, DrugE-Rank would be particularly useful since DrugE-Rank can integrate the diverse component methods effectively for better performance. In fact, the performance improvement over the component methods by DrugE-Rank for a new drug is much higher than the one for a new target. Another interesting discovery is that relative low prediction performance on FDA experimental drugs. One main reason is that the ratio of drug–target interaction for experimental drugs is much lower than the one for FDA approved drugs (training data). Many non-interaction pairs might be false negatives. DrugE-Rank can be used to predict top drug–target pairs as the promising candidates for further experimental verification.
6 Conclusion
We have developed DrugE-Rank to improve the performance of predicting drug–target interactions of new drugs or targets. The idea of DrugE-Rank are three folds: (i) the problem of predicting drug–target interactions can be modeled as a multilabel classification task; (ii) this problem can be suitably solved by using LTR, a powerful feature-based machine learning approach, which converts multilabel classification to label ranking; (iii) the outputs of cutting-edge similarity-based machine learning methods can be features like drug and target features, all being able to be the input of LTR. DrugE-Rank outperformed all competing methods, in both cross-validation and several independent test scenarios, which demonstrate the predictive advantage of DrugE-Rank. In addition, in contrast to previous studies with a small number of targets, all known interactions in DrugBank have been used in this work, indicating the thoroughness and validity of our results. Overall we believe that this high-performance software, DrugE-Rank will contribute to the development of pharmaceutical sciences and relevant industry.
Funding
This work was supported by the National Natural Science Foundation of China (61572139, 61379092 and 61422309), Scientific Research Starting Foundation for Returned Overseas Chinese Scholars, Ministry of Education, China. H.M. would like to thank MEXT for KAKENHI #16H02868 and Tekes for FiDiPro.
Conflict of Interest: none declared.
Supplementary Material
Footnotes
References
- Acharya C. et al. (2011) Recent advances in ligand-based drug design: relevance and utility of the conformationally sampled pharmacophore approach. Curr. Comput.-Aided Drug Des., 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S. et al. (2010) Ranking chemical structures for drug discovery: a new machine learning approach. J. Chem. Inf. Model., 50, 716–731. [DOI] [PubMed] [Google Scholar]
- Bleakley K., Yamanishi Y. (2009) Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics, 25, 2397–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E.E. et al. (2008) Pubchem: integrated platform of small molecules and biological activities. Ann. Rep. Comput. Chem., 4, 217–241. [Google Scholar]
- Boutet E. et al. (2007) Uniprotkb/swiss-prot. Methods Mol. Biol., 406, 89–112. [DOI] [PubMed] [Google Scholar]
- Burges C.J. (2010) From ranknet to lambdarank to lambdamart: an overview. Technical report MSR-TR-2010-82, Microsoft Research.
- Davis J., Goadrich M. (2006) The relationship between precision-recall and roc curves. Machine Learning, Proceedings of the Twenty-Third International Conference ICML 2006, Pittsburgh, Pennsylvania, USA, 233–240.
- Ding H. et al. (2014) Similarity-based machine learning methods for predicting drug–target interactions: a brief review. Brief. Bioinfo., 15, 734–747. [DOI] [PubMed] [Google Scholar]
- Gönen M. (2012) Predicting drug–target interactions from chemical and genomic kernels using bayesian matrix factorization. Bioinformatics, 28, 2304–2310. [DOI] [PubMed] [Google Scholar]
- Jacob L., Vert J.P. (2008) Protein–ligand interaction prediction: an improved chemogenomics approach. Bioinformatics, 24, 2149–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser M. et al. (2009) Predicting new molecular targets for known drugs. Nature, 462, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V. et al. (2014) Drugbank 4.0: shedding new light on drug metabolism. Nucleic Acids Res., 42,-D1, D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011) A short introduction to learning to rank. IEICE Trans., 94-D, 1854–1862. [Google Scholar]
- Liu B. et al. (2015a) Application of learning to rank to protein remote homology detection. Bioinformatics, 31, 3492–3498. [DOI] [PubMed] [Google Scholar]
- Liu K. et al. (2015b) Meshlabeler: improving the accuracy of large-scale mesh indexing by integrating diverse evidence. Bioinformatics, 31, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Y. (2009) Learning to rank for information retrieval. Found. Trends Inf. Retriev., 3, 225–331. [Google Scholar]
- Lounkine E. et al. (2012) Large-scale prediction and testing of drug activity on side-effect targets. Nature, 486, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne P.D. (2002) Structure-based virtual screening: an overview. Drug Discov. Today, 7, 1047–1055. [DOI] [PubMed] [Google Scholar]
- Mei J.P. et al. (2013) Drug–target interaction prediction by learning from local information and neighbors. Bioinformatics, 29, 238–245. [DOI] [PubMed] [Google Scholar]
- Mousavian Z., Masoudi-Nejad A. (2014) Drug-target interaction prediction via chemogenomic space: learning based methods. Expert Opin. Drug Metabol. Toxicol., 10, 1273–1287. [DOI] [PubMed] [Google Scholar]
- Nagamine N., Sakakibara Y. (2007) Statistical prediction of protein–chemical interactions based on chemical structure and mass spectrometry data. Bioinformatics, 23, 2004–2012. [DOI] [PubMed] [Google Scholar]
- Nunez S. et al. (2012) Target-drug interations: first principles and their application to drug discovery. Drug Discov. Today, 17, 10–22. [DOI] [PubMed] [Google Scholar]
- Overington J. et al. (2006) How many drug targets are there. Nat. Rev. Drug Discov., 5, 993–996. [DOI] [PubMed] [Google Scholar]
- Qeli E. et al. (2014) Improved prediction of peptide detectability for targeted proteomics using a rank-based algorithm and organism-specific data. J. Proteomics., 108, 269–283. [DOI] [PubMed] [Google Scholar]
- Rao H. et al. (2011) Update of profeat: a web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res., 39(suppl 2), W385–W390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke F. et al. (2010) Structrank: a new approach for ligand-based virtual screening. J. Chem. Inf. Model., 51, 83–92. [DOI] [PubMed] [Google Scholar]
- Scannell J. et al. (2012) Diagnosing the decline in pharmaceutical r&d efficiency. Nat. Rev. Drug Discov., 11, 191–200. [DOI] [PubMed] [Google Scholar]
- Shi J. et al. (2015) Predicting drug–target interaction for new drugs using enhanced similarity measures and super-target clustering. Methods, 83, 98–104. [DOI] [PubMed] [Google Scholar]
- Tabei Y. et al. (2012) Identification of chemogenomic features from drug–target interaction networks using interpretable classifiers. Bioinformatics, 28, i487–i494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Laarhoven T. et al. (2011) Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics, 27, 3036–3043. [DOI] [PubMed] [Google Scholar]
- van Laarhoven T., Marchiori E. (2013) Predicting drug-target interactions for new drug compounds using a weighted nearest neighbor profile. PloS One, 8, e66952.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z. et al. (2010) Semi-supervised drug-protein interaction prediction from heterogeneous biological spaces. BMC Syst. Biol., 4, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi H. et al. (2011) Analysis of multiple compound–protein interactions reveals novel bioactive molecules. Mol. Syst. Biol., 7, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi Y. (2013) Chemogenomic approaches to infer drug-target interaction networks. Methods Mol. Biol., 939, 97–113. [DOI] [PubMed] [Google Scholar]
- Yamanishi Y. et al. (2008) Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics, 24, i232–i240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. (2015) When drug discovery meets web search: learning to rank for ligand-based virtual screening. J. Cheminf., 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. et al. (2013) Collaborative matrix factorization with multiple similarities for predicting drug–target interactions In ACM KDD, 1025–1033. ACM. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.