Abstract
Motivation: RNA thermometers (RNATs) are cis-regulatory elements that change secondary structure upon temperature shift. Often involved in the regulation of heat shock, cold shock and virulence genes, RNATs constitute an interesting potential resource in synthetic biology, where engineered RNATs could prove to be useful tools in biosensors and conditional gene regulation.
Results: Solving the 2-temperature inverse folding problem is critical for RNAT engineering. Here we introduce RNAiFold2T, the first Constraint Programming (CP) and Large Neighborhood Search (LNS) algorithms to solve this problem. Benchmarking tests of RNAiFold2T against existent programs (adaptive walk and genetic algorithm) inverse folding show that our software generates two orders of magnitude more solutions, thus allowing ample exploration of the space of solutions. Subsequently, solutions can be prioritized by computing various measures, including probability of target structure in the ensemble, melting temperature, etc. Using this strategy, we rationally designed two thermosensor internal ribosome entry site (thermo-IRES) elements, whose normalized cap-independent translation efficiency is approximately 50% greater at 42 °C than 30 °C, when tested in reticulocyte lysates. Translation efficiency is lower than that of the wild-type IRES element, which on the other hand is fully resistant to temperature shift-up. This appears to be the first purely computational design of functional RNA thermoswitches, and certainly the first purely computational design of functional thermo-IRES elements.
Availability: RNAiFold2T is publicly available as part of the new release RNAiFold3.0 at https://github.com/clotelab/RNAiFold and http://bioinformatics.bc.edu/clotelab/RNAiFold, which latter has a web server as well. The software is written in C ++ and uses OR-Tools CP search engine.
Contact: clote@bc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
RNA thermometers (RNATs), also known as thermosensors, are cis-regulatory elements that change secondary structure upon temperature shift. Examples include (i) repression of heat shock gene expression (ROSE) elements (Nocker et al., 2001), which control the expression of small heat shock genes, such as hspA in Bradyrhizobium japonicum and ibpA in Escherichia coli; (ii) FourU elements (Waldminghaus et al., 2007b), such as the virulence factor LcrF in Yersinia pestis; and (iii) Hsp17 thermosensor (Kortmann et al., 2011; Torok et al., 2001), which controls membrane integrity of the cyanobacterium Synechocystis sp. PCC6803 under stress conditions, critical for photosynthetic activity. Additional examples are described in Kortmann and Narberhaus (2012). ROSE elements and FourU elements operate as temperature-sensitive, reversible zippers, while the Listeria monocytogenes prfA thermosensor (Johansson et al., 2002), phage λ cIII thermoswitch (Altuvia et al., 1989) and E.coli CspA cold shock thermometer (Bae et al., 2000) operate in a switch-like fashion. Here, the helix of a zipper melts gradually with increasing temperature, returning to the original structure when temperature is reduced, while a switch consists of two mutually exclusive structures determined by temperature.
Several bioinformatics search methods exist to identify and predict candidate RNA thermometers. In Waldminghaus et al. (2007a) the database RNA-SURIBA (Structures of Untranslated Regions In BActeria) was created; using regular expressions, particular structural motifs were detected in the minimum free energy (MFE) structure, as determined by mfold (Zuker, 2003). In contrast, the RNAtips web server (Chursov et al., 2013) and the RNAthermsw (Churkin et al., 2014) web server both rely on base pairing probabilities computed at different temperatures using RNAfold from the Vienna RNA Package (Lorenz et al., 2011).
For some time now, RNA thermosensors have been recognized as an attractive target for rational design (Waldminghaus et al., 2008; Wieland and Hartig, 2007). Indeed, within the broader context of synthetic biology, rationally designed thermometers could be used as a thermogenetic tool to control expression by temperature regulation (i.e. on-demand protein translation), or even as a multifunctional nanoscale devices to measure temperature in the context of hyperthermic treatment of cancer cells, imaging or drug delivery (Lee and Kotov, 2007).
In Neupert et al. (2008), synthetic (zipper) thermosensors were manually designed to sequester the Shine-Dalgarno sequence AAGGAG within a single stem-loop structure containing 4–9 base pairs, several of which contained 1–2 bulges of size 1. In Waldminghaus et al. (2008), synthetic (switch) thermosensors were computationally designed to switch between a single stem-loop structure that sequesters the Shine-Dalgarno sequence GGAGG, and two shorter stem-loop structures where the Shine-Dalgarno sequence is found in the apical loop of the second stem-loop. In particular, the 2-temperature inverse folding (adaptive walk) program SwitchDesign (Flamm et al., 2001) was used to obtain 300 candidate sequences; only two candidate sequences survived after the application of several computational filters including the computation of melting curves with RNAheat (Lorenz et al., 2011). Since neither of these sequence displayed any temperature-dependent control of a reporter gene (bgaB) fusion, the top candidate sequence was used as a template in two rounds of error-prone mutagenesis followed by selection, resulting in a successful thermosensor—see Figure 5 of Waldminghaus et al. (2008). In Ogawa (2011), a non-temperature-dependent riboswitch was manually designed, which promotes cap-independent translation in wheat germ cell lysate only upon binding of the ligand theophylline. In Wachsmuth et al. (2013), a synthetic theophylline riboswitch was designed by a computational pipeline including inverse folding and experimentally shown to perform transcriptional regulation in Escherichia coli Top10 cells. In Saragliadis et al. (2013), a thermozyme was created by fusing a Salmonella RNA thermometer (RNAT) to a hammerhead ribozyme, followed by in vivo screening—thus showing that naturally occurring hammerheads and RNATs appear to be modules that can be combined. In Hoynes-O’Connor et al. (2015) small, heat-repressible RNA thermosensors (zippers) were manually designed in E.coli, which at low temperature sequester a cleavage site for RNaseE, and at high temperatures unfold to allow mRNA degradation.
Despite these impressive results, Kortmann and Narberhaus (2012) state that: ‘RNATs have little, if any sequence conservation and are difficult to predict from genome sequences. … Therefore, the bioinformatic prediction and rational design of functional RNATs has remained a major challenge.’
In this article, we introduce the software RNAiFold2T, capable of solving the inverse folding problem for two or more temperatures, i.e. generating one or more RNA sequences whose MFE secondary structures at temperatures T1 and T2 (resp. ) are user-specified target structures S1 and S2 (resp. ), or which reports that no such solution exists. RNAiFold2T is unique in that it implements two different algorithms—Constraint Programming (CP) and Large Neighborhood Search (LNS). CP is an exact, non-heuristic method that uses an exhaustive yet efficient branch-and-prune process, and is the only currently available software capable of generating all solutions or determining that no solution exists (since there are possibly exponentially many solutions, a complete solution is feasible only for structures of modest size). LNS uses a local search heuristic, complemented with local calls of CP to explore solutions of substructures of the target structures. We use RNAiFold2T to rationally design two thermo-IRES elements, whose normalized cap-independent translation efficiency is approximately 50% greater at 42°C than 30°C, when tested in reticulocyte lysates. We then benchmark RNAiFold2T with the only two other programs that solve the 2-temperature inverse folding problem: the adaptive walk SwitchDesign (SD) (Flamm et al., 2001) and the genetic algorithm Frnakenstein (FRNA) (Lyngso et al., 2012). RNAiFold2T CP generates two orders of magnitude more solutions than either, when all programs are run for 24 h, while the number of distinct problem instances that can be solved by RNAiFold2T LNS within 30 (resp. 60) min for short (resp. long) target structures is roughly comparable for each program. Finally, by analyzing existent RNATs found in the Rfam 12.0 database (Nawrocki et al., 2014), we determine that naturally occurring RNATs appear not to be optimized for the cost function [Equation (7) of Flamm et al. (2001)], and that both SD and FRNA appear to generate solutions whose cost function value is substantially lower than that of naturally occurring RNATs, in contrast to solutions returned by RNAiFold2T.
1 Methods
1.1 Biochemical methods
1.1.1. Thermo-IRES activity assay
Thermo-IRES constructs were created by replacement of wild-type nucleotides at positions 417–462 in domain 5 of the foot-and-mouth disease virus (FMDV) IRES element, whose secondary structure is depicted in Figure 1 of Fernandez et al. (2011) and Figure 2 of Lozano et al. (2014). Six computationally designed thermo-IRES elements were selected for validation, along with a negative control and the wild-type FMDV IRES element as positive control. Specifically, synthetic oligonucleotides containing the designed sequence (46 nts) in either positive or negative orientation were annealed in Tris 50 mM, pH 7.5, NaCl 100 mM, MgCl2 10 mM, 15 min at 37°C and subsequently inserted into the HindIII and XhoI restriction sites of pBIC, which harbors the wild-type IRES, linearized with the same enzymes. Colonies that carried the correct insert were then selected, and prior to expression analysis, the nucleotide sequence of the entire length of each region under study was determined (Macrogen).
In vitro transcription was performed for 1 h at 37°C using T7 RNA polymerase, as described in Lozano et al. (2015). RNA was extracted with phenol–chloroform, ethanol precipitated and then resuspended in TE. Using gel electrophoresis, the transcripts were checked for integrity. Equal amounts of the RNAs synthesized in vitro were translated in 70% rabbit reticulocyte lysate (RRL) (Promega) supplemented with 35S-methionine (10 μCi), as described in Pineiro et al. (2013). Each experiment was independently repeated in triplicate, using the wild-type RNA as a control in all assays. Luciferase (LUC) and chloramphenicol acetyl transferase (CAT) activities were measured for the bicistronic plasmid, as previously described (Fernandez-Chamorro et al., 2014). In particular, intensity of the LUC band, as well as the CAT band, produced by each transcript was determined in a densitometer, and normalized against the intensity of LUC and CAT bands produced by the wild type RNA, set at 100%. Values represent the mean ± stdev.
Luciferase activity reflects the efficiency of IRES-dependent translation, while CAT activity reflects the efficiency of -dependent translation; thus the ratio LUC/CAT was determined at 30°C and 42°C for wild-type FMDV IRES, the negative control and two thermo-IRES constructs. Selective -hydroxyl acylation analyzed by primer extension (SHAPE) was performed for wild-type IRES element, as described in Dotu et al. (2013).
1.2 Computational methods
RNAiFold2T uses CP to determine those sequences, whose MFE structure at temperature T1 (resp. T2) is identical to a user-specified target structure S1 (resp. S2). The target structures S1, S2 can also be hybridization complexes of two RNAs, rather than single secondary structures. CP performs a complete, exhaustive (branch and prune) exploration of the search space and therefore, it can return all possible solutions of the thermoswitch design problem or prove that no solution exists (given an unlimited amount of time). In addition to CP, RNAiFold2T also supports LNS, a fast local (not complete) search metaheuristic that employs CP to exhaustively explore large neighborhoods of every candidate solution at each iteration step. Moreover, since it is written in C++ using the OR-Tools engine https://github.com/google/or-tools, together with plug-ins to Vienna RNA Package (Lorenz et al., 2011) and RNAstructure (Reuter and Mathews, 2010), the user can install and run RNAiFold2T locally, thus permitting much longer execution times than supported by our web server.
The overall methodology of RNAiFold2T is similar to its precursor, RNAiFold 2.0 (Garcia-Martin et al., 2012, 2015); however, as explained below, there are a number of algorithmic details that are new and not present in RNAiFold 2.0—decomposition tree for 2 or more target structures, novel constraints that are underlined below, variable (helix) and value heuristics that are proper only to RNAiFold2T, the introduction of two types of restart heuristic in LNS to ensure a good trade-off between exploration of promising regions of the search space versus the exploration of remote portions of the search space. RNAiFold2T cleanly separates all constraints from the CP or LNS solver, thus permitting our software to be extended to support any future desirable constraints. Current constraints supported by RNAiFold2T include the following (novel constraints underlined): full and/or partial target structures or hybridization complexes at two temperatures; a plug-in to determine MFE structure using either RNAfold (Lorenz et al., 2011) or RNAstructure (Reuter and Mathews, 2010) with the Turner99, Turner2004 (Turner and Mathews, 2010) or Andronescu (Andronescu et al., 2007) energy model with dangle treatment (-d0, -d1, -d2, -d3 corresponding respectively to no dangle, max of and dangle, sum of and dangle, sum of and dangle with coaxial stacking); IUPAC nucleotide constraints, IUPAC amino acid constraints that require all returned RNA sequences to code specified peptides in one or more overlapping reading frames, structural compatibility and structural incompatibility constraints, etc. Additionally, RNAiFold2T can determine a user-specified number of solutions, all solutions (given sufficient time), or whether no solution exists. Indeed, memory requirements for RNAiFold2T are minimal, and since there are no memory leaks, the software can be run for weeks.
In developing a CP solution to a given problem the main tasks are to define the problem (specify variables, domains and constraints) and to define the search procedure. The extension of the model from RNAiFold2.0 is trivial and consists of adding new constraints for the helices corresponding to the second structure. The search procedure, however, must be adapted to the new difficulties imposed by the 2-temperature problem. New variable and value ordering heuristics are needed in order to solve 2-temperature inverse folding efficiently (Supplementary Tables S1 and S2). The algorithmic details related to the new search procedure are explained below:
1.3 Structure decomposition
As in other inverse folding methods, such as RNAinverse (Lorenz et al., 2011), NUPACK (Zadeh et al., 2011), etc., we rely on a decomposition tree of the structure into independent helices, called extended helices (EHs) and extended helices with dangles (EHwDs). See Supplementary Material for definitions of EH, EHwD and decomposition tree, and see Figure 1 for an illustration of the EHwD decomposition tree for a FourU thermometer. Decomposition trees play a special role in RNAiFold2T for the following reasons: (i) each node in the decomposition tree is a constraint; (ii) the helix and variable heuristics (described later) cause the search tree to be searched in a specific order. To improve efficiency in solving multi-temperature inverse folding, we investigated various helix and value heuristics, which steer the search within a search space defined by a composite decomposition tree, comprising subtrees for each target structure at the corresponding temperature. The helix and value heuristics are new and not part of RNAiFold 2.0.
Fig. 1.
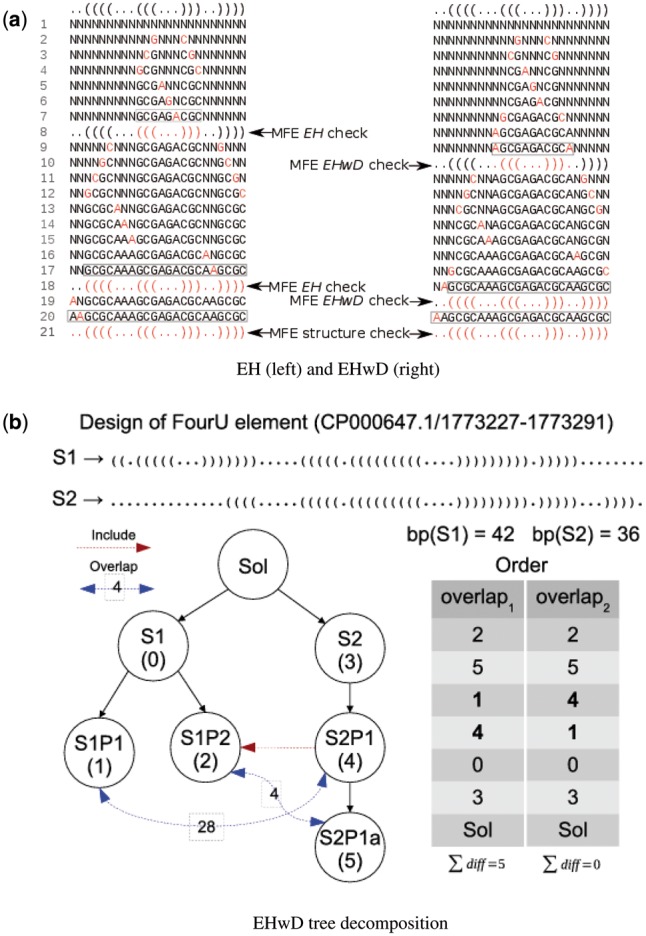
Examples of CP search for helixes versus EHwDs and EHwDs decomposition heuristic. (a) MFE structure check made for EH on the left, and EHwD on the right. In each line, either a new base pair (2 positions) or a new nucleotide (1 position) is instantiated, following the variable order heuristic. For simplification, this example assumes that correct value assignments are made in each instantiation step. (b) Tree decomposition for target structure S1 (resp. S2) at temperature T1 (resp. T2), using target structures for the FourU element CP000647.1/1773227-1773291. The table on the right presents the order of helix exploration using overlap1 and overlap2 heuristics
Consider the 65 nt FourU RNA thermometer whose MFE structures determined by RNAfold from Vienna RNA Package 2.1.9 (Lorenz et al., 2011) at 37°C and 53°C are given in dot-bracket notation by
>CP000647.1/1773227-1773291)
12345678901234567890123456789012345678901234567890123456789012345
GGACAAGCAAUGCUUGCCUUUAUGUUGAGCUUUUGAAUGAAUAUUCAGGAGGUUAAUUAUGGCAC
((.(((((…)))))))….(((((.(((((((((….))))))))).)))))…….
………….((((….(((((.(((((((((….))))))))).)))))…)))).
Let S1 (resp. S2) denote the MFE structure of this FourU thermometer at 37°C (resp. 53°C). S1 is identical to the consensus structure from Rfam 12.0 (Nawrocki et al., 2014), as well as the structure displayed in Figure 1 of Waldminghaus et al. (2007b) for the FourU sequence taken from the -UTR of the Salmonella agsA gene. Giving labels as described to nodes in the EHwD decomposition trees respectively for S1 and S2, we find the EHwD decomposition of S1 has EHwD 0 from positions 1 to 65, EHwD 1 from positions 1 to 19, and EHwD 2 from positions 23 to 58, while the EHwD decomposition of S2 has EHwD 4 from positions 1 to 65, EHwD 5 from positions 14 to 65, and EHwD 5 from positions 23 to 58. The lower portion of Figure 1 depicts the EHwD decomposition trees for temperatures T1, T2, joined together with a (dummy) root that corresponds to the solution returned by RNAiFold2T.
It can happen that the MFE structure of an EH does not agree with the target substructure, while that of the EH with dangles does. This is the reason that the EH decomposition trees of RNAiFold 1.0 (Garcia-Martin et al., 2012) were replaced by EHwD decomposition trees in RNAiFold 2.0 (Garcia-Martin et al., 2015), and why EHwD trees are used in RNAiFold2T.
In the CP and LNS search strategy, whenever a subsequence corresponding to a node of the decomposition tree has been instantiated, a check is made to determine whether the MFE structure of the subsequence is identical to the target substructure. In the case of structural disagreement, the instantiated subsequence is discarded and backtracking occurs. For any solution sequence returned by RNAiFold2T, it follows that at temperature T1 (resp. T2), each subsequence of the solution that corresponds to a node in decomposition tree (resp. ) folds into the corresponding substructure of target S1 (resp. S2). The top portion of Figure 1 illustrates the differences between EH and EHwD decomposition, and when a check is made for whether the target substructure agrees with the MFE structure of the instantiated subsequence. In the sequel, we refer to four types of elements in an EHwD:
Dangling position: Unpaired position at any side of a helix.
Unpaired position: Any other unpaired position.
Closing base pair: Outermost base pair of a helix.
Normal base pair: Any other base pair.
1.4 Heuristics for variable and value order
In a CP algorithm, one typically specifies the order in which variables are instantiated (assigned), known as the variable ordering heuristic, as well as the order in which the values belonging to the domain of each variable are to be assigned, known as the value ordering heuristic. The variable ordering heuristic is divided into two levels: first, the order in which EHwDs are to be assigned, and second, the order in which nucleotide positions within helices are to be assigned. In this section, we also discuss restarting heuristics for the Large Neighbourhood Search (LNS) variant of RNAiFold2T.
1.4.1 Variable ordering at the helix level
In the search for thermosensors, there is often an overlap between EHwDs of structure S1 and those of structure S2—this situation substantially complicates the task of finding an optimized order of exploration of the CP search space. In the leaf-to-root heuristic of RNAiFold 2.0, EHwD node H is explored before EHwD node if the height ht(H) of H is less than the height of , or if and H appears to the left of in the decomposition tree for the single target structure S. In contrast to this heuristic, RNAiFold2T implements two different approaches in order to find an adequate exploration ordering for the EHs with dangles for two target structures S1 and S2, whereby a high priority is given to solve those helices, whose sequence is determinant for other parts of the structure due to overlaps. Let N denote the number of nodes (EHwDs) in the decomposition tree for S1, plus the number of nodes (EHwDs) in the decomposition tree for S2. Suppose that are two distinct EHwDs belonging to S1 or S2, where the outermost base pair of H (resp. ) is (i, j) [resp. ]. Define the following relations:
is 1 if and , i.e. interval properly contains ; otherwise is 0.
is 1 if , or equivalently ; otherwise is 0.
is the number of positions k in H and , for which k is base-paired in both H and .
, for .
For , is equal to , provided that ; otherwise, is 0.
For , is equal to , provided that ; otherwise is 0.
The value label(H) is defined in Supplementary Material, and corresponds to visitation order in breadth-first traversal of tree , and is a permutation that minimizes , subject to the constraint that if H properly includes , then . Note that the partition σ orders the EHwDs of S1 and S2 in order to minimize the total incremental overlap. Before the search for thermosensors begins, RNAiFold2T executes a very fast CP search to determine the optimal ordering permutation σ. Finally, by setting the index α to either 1 or 2 in the definition of , we obtain the first or second search heuristic. (Two additional helix ordering heuristics are explained in Supplementary Material.) An example of each helix order heuristic 1, 2 is shown in Figure 1—note that for even small structures, there are several differences in the helix exploration order when α is 1 or 2.
1.4.2 Variable ordering at the nucleotide level
The second level of variable ordering heuristic deals with the exploration of nucleotide positions within a given EHwD structure. This second level of variable ordering can be stated as follows:
First, non-outermost base-paired positions (x, y) of a given EHwD are instantiated from the innermost base pair to the outermost base pair.
Second, unpaired positions in a given EHwD are grouped together in consecutive runs, and these runs are ordered from largest to smallest and then instantiated from left to right.
Third, the outermost, closing base pair of a given EHwD is instantiated.
Finally, dangling positions of a given EHwD (if any) are instantiated (note that not all EHwDs contain dangling positions).
The small example shown in Figure 2 graphically depicts the differences between constraint checks and variable ordering for both RNAiFold and RNAiFold2T. Supplementary Tables S1 and S2 describe and benchmark variable and value heuristics in RNAiFold2T.
Fig. 2.
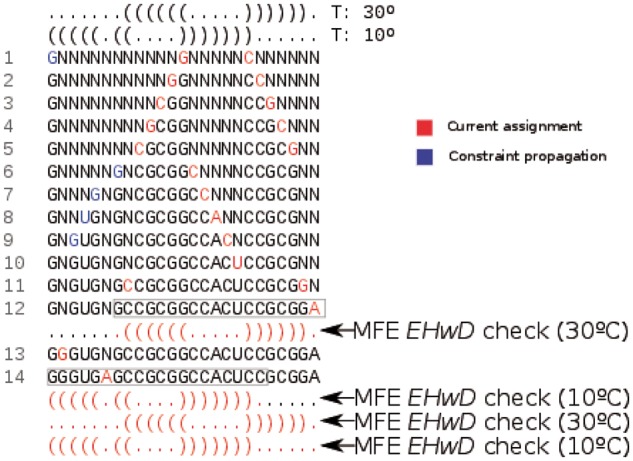
Example of CP search for target structures S1 (top) at temperature 30 °C, and S2 (bottom) at temperature 10 °C. Undetermined positions are assigned (red) in the following order: base pairs are instantiated in lines 1,2,3,4,5,13; unpaired positions in lines 6,7,8,9,10,14; a closing base pair in line 11; a dangling position in line 12. When the left nucleotide of a base pair is instantiated to C (resp. A), then propagation of the base pairing constraint reduces the domain of possible values of the right nucleotide of the base pair from to the set [resp. ]—this happens in lines 1, 6–9. For simplification, this example assumes that correct value assignments always occur at each search step
1.4.3 Value ordering
Value ordering establishes the order in which values are assigned to variables—in our case, this means values GC,CG, AU, UA, GU, UG for base-paired positions and A,C,G,U for unpaired positions. The underlying idea for ordering domain values for variables for base-paired positions and unpaired positions is to allow the creation of thermodynamically stable helices and to take into account the nature of the overlap in overlapping positions. This requires specific value orderings for base-paired positions, depending on whether the position is a dangle, mismatch, normal or closing base pair of an EHwD for both targets S1, S2, or only one of the target structures. For each of these cases, we define different ordering heuristics, described in Supplementary Table S1.
1.4.4 LNS restart heuristics
Similarly as in the LNS variant of RNAiFold, the restart condition is a given amount of time, proportional to the length of the target structures, after which search is stopped and some variables are fixed in order to explore exhaustively a large neighborhood of the current solution. After the first restart, full exploration of the remaining space with no solution found is also a restart condition.
In RNAiFold2T, when a restart is triggered, a set of positions is selected as candidates to be fixed. The MFE structure for each EHwD of the current candidate solutions is evaluated independently. If the MFE structure of an EHwD matches with the target structure at the desired temperature, and the MFE structure of all the overlapping helices in the second target structure (at the corresponding temperature) also matches, then all the EHwD positions are included in the set of candidates. When all the EHwDs have been evaluated, candidate positions are fixed with a probability of 0.9, and the set of candidate positions is stored.
Since the order of exploration is similar in each round, it could be possible that fixing similar parts of the sequence results in an exploration of almost the same region of the search space in subsequent searches, so two mechanisms are implemented to avoid this behavior: (i) in subsequent restarts, if the candidate positions to be fixed are the same as in the previous restart, then the probability of fixing positions decreases by 0.05, if not, then the initial probability of 0.9 is restored. (ii) There is a hard restart (no nucleotide position is fixed) in the case that, after 10 restarts, the set of candidate positions remains unchanged, or if all possible solutions for the current subproblem have been explored.
In local search algorithms, there is always a trade-off between exploitation and exploration. Exploitation means focusing the search on promising regions, as reflected in our choice of probability 0.9 to remain close to currently instantiated portions of the sequence. Exploration means covering different, remote regions of the search space, as reflected in our choice to decrement the probability by 0.05 and our choice to perform a hard restart after 10 restarts.
1.5 Benchmarking
1.5.1 Benchmarking data
We retrieved the sequences of seven families of non-coding RNA thermometers from Rfam: RF00038, RF00433, RF00435, RF01766, RF01795, RF01804, RF01832. These families include both cold and heat shock RNA thermometers, taken from diverse organisms including phages, prokaryotes and eukaryotes, with sequence length ranging from 60 nt to 450 nt. The benchmarking was divided into two groups: sequences shorter than and longer than 130 nt. For each sequence, we used RNAfold (Lorenz et al., 2011) with the Turner99 energy parameters (Turner and Mathews, 2010) to determine the MFE structures at temperatures T1 and T2, where temperatures were chosen (essentially) according to published experimental studies for each thermometer family (Altuvia et al., 1989; Chowdhury et al., 2003; Rinnenthal et al., 2011)—in particular, we increased the temperature difference from the published values to ensure that RNAfold produced distinct structures at T1 and T2 if possible (Supplementary Material). Turner99 rather than Turner2004 energies were used, since it required less distortion from published temperatures T1, T2. All sequences, whose MFE structures at T1 and T2 were identical, were subsequently removed. See Supplementary Tables S3–S5 and Supplementary Excel files for benchmarking results, and Supplementary Table S6 (resp. Supplementary Excel file) for a list of sequences, structures, and temperatures for sequences of length less than 130 nt (resp. greater than 130 nt). The resulting benchmarking set includes all 5 Lambda phage CIII thermoregulator elements (Lambda_thermo), all 3 FourU thermometers, 11 of 13 repression of heat shock (ROSE) elements, 8 of 14 sequences from a second family of repression of heat shock (ROSE_2) elements, 3 of 13 thermoregulators of PrfA virulence genes (PrfA), 4 of 6 HSP90 cis regulatory elements (HSP_CRE) and 14 of 15 cold shock protein regulator sequences (CspA).
1.5.2 Software used in benchmarking tests
SwitchDesign (SD) (Flamm et al., 2001), Frnakenstein (FRNA) (Lyngso et al., 2012), and RNAiFold2T were benchmarked on Rfam family RF01804 of Lambda phage CIII thermoregulator elements to determine the maximum number of distinct solutions over 24 h, restarting when no new solution is found within 1 h. Since neither FRNA nor SD output more than one solution, we made the following modifications of each program. The genetic algorithm Frnakenstein was run as many times as possible over 24 h (each time with a time limit of 1 h); we then output all sequences found in the most recent (internally stored) population which fold into the target structures S1, S2 at temperatures T1, T2. SD returns a single sequence which minimizes a cost function described in Flamm et al. (2001); thus we modified SD source code in order to test whether any sequence explored in the search was a solution. Sequences were checked at two different points in SD: when a new sequence is generated by a single mutation (SD update), and when a sequence is selected by minimization of the cost function (SD selected). In all cases, SwitchDesign was restarted if no new sequence was found in 1 h.
For each solution set obtained, additional solutions were generated by testing all single point mutations of any solution returned. Additionally, a reference solution set was produced by running RNAiFold2T for several days.
2 Results
2.1 Design of thermo-IRES switches
Domain 5 of wild-type FMDV IRES element contains the 46 nucleotides AUAGGUGACC GGAGGUCGGC ACCUUUCCUU UACAAUUAAU GACCCU at positions 417–462, as shown in Figure 1 of Fernandez et al. (2011) and Figure 2 of Lozano et al. (2014). The domain 5 stem-loop at positions 419–440 and unpaired polypyrimidine tract (PPT) region at positions 441–447, are both known to be essential for IRES activity (Kuhn et al., 1990). Using the following pipeline, two candidate thermo-IRES elements were tested, along with a negative control and a positive control (wild-type IRES).
-
As shown in Figure 3a and b, the inactive target structure S1 at was chosen to destroy domain 5 stem-loop and unpaired PPT region, while target active structure S2 at is the experimentally determined structure of wild-type domain 5 FMDV IRES element. Sequence constraints were chosen in accordance with conservation observed in a multiple alignment of 183 IRES elements (Fernandez et al., 2011), where we added AUG start codon at positions 47–49 (corresponding to IRES positions 463–465).

Using RNAiFold2T, 24,410 solutions were generated, although an additional 45 442 sequences were generates using variants of target S1.
- RNAiFold2T solutions were discarded if any of the following criteria were not met:
- Wild-type structures for domain 4 and domain 5 appear as stable substructures using Vienna Package RNALfold -L 110 -T 42.
- Domain 4 appears as a stable substructure using RNALfold -L 110 -T 30.
- Probability of active conformation at exceeds 0.2.
- Probability of inactive conformation at exceeds 0.2.
- Probability of intended target structure at intended temperature is more than double that of unintended target, i.e. .and .
Retained solutions were further filtered using various measures. For instance, candidate 1 (Seq1) had the highest value of A + B, where A is and B is d + e + – , and . This measure was designed to select sequences where the probability of the intended target at the intended temperature is high, while probability of the unintended target is low. The measure is weighted to increase the likelihood of not having the inactive conformation at . In contrast, Candidate 2 (Seq2) is one of two sequences satisfying and . See Supplementary Material for a spread sheet of measures and their values.
Seq1 and Seq2 consist of the following 46 nt: Seq1 is AUAGGUGACC GGAGGGCGGC ACCUUUUUUC CAGAAAAGUA GUCGUC (15/46 positions differ from wild-type) and Seq2 is GUAGGUGACC GGAGGACGGC ACCUUUUUUC CAGAAAAGUA GUCGUC (16/46 positions differ from wild-type). Figure 3c shows that Seq1 and Seq2 displayed an increase of approximately 50% normalized IRES-dependent translation efficiency in RRL at 42°C versus 30°C. Seq1 and Seq2 IRES elements displayed about 20% lower normalized activity than the wild-type IRES. Nonetheless, the wild-type IRES was equally active at all temperatures tested (30°C, 37°C and 42°C).
Fig. 3.
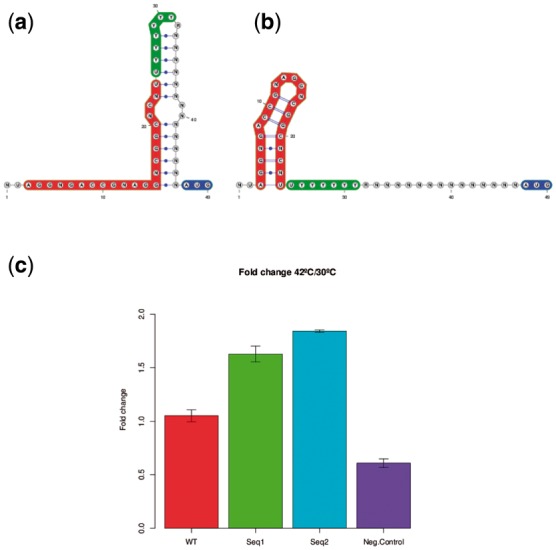
(a and b) Target structures S1 at temperature 30 °C (a) and S2 at temperature 42 °C (b) for domain 5 thermo-IRES element with added AUG codon (blue). IUPAC sequence constraints are determined from an alignment of 183 IRES sequences as shown in Figure 1 of Fernandez et al. (2011). Domain 5 stem-loop (positions 3-24 in red) and unpaired polypyrimidine tract (PPT positions 25–32 in green) are known to be essential for IRES activity (Kuhn et al., 1990). Target structure S1 was designed to sequester the PPT at low temperature, thus creating a thermo-IRES which should be functional only at high temperature. (c) Ratio of normalized IRES activity at 42 °C over that at 30 °C for wild-type FMDV IRES, a negative control, and two thermosensors designed using RNAiFold2T
2.2 Comparison of RNAiFold2T with other software
Here we compare the CP and LNS programs of RNAiFold2T with the adaptive walk program SwitchDesign (SD) (Flamm et al., 2001) and the genetic algorithm Frnakenstein (FRNA) (Lyngso et al., 2012). Below, we describe benchmarking results for datasets of thermosensor target structures S1 resp. S2 at temperatures T1 resp. T2, constructed as described in Section 1. All benchmarking was carried out on a Core2Duo PC (2.8 GHz; 2 Gbyte memory; CentOS 5.5).
SD, the first algorithm capable of designing thermoswitches, achieves this by optimizing the cost function given in Equation (7) of Flamm et al. (2001)—see also Equation (1) in Supplementary Material. In this context, we wanted to ascertain whether natural thermoswitches are optimized for this cost. Surprisingly, Figure 4 and Supplementary Figure S1 show that natural thermosensor sequences from Rfam appear not to be optimized for the cost function used in SD. In particular, SD and FRNA return solutions having substantially lower cost values (i.e. more optimal) than those of natural thermosensors, whose cost value appears to be the mean value returned by RNAiFold2T. Therefore, our benchmarking comparison of SD, Frnakenstein and RNAiFold2T compares the number of problems solved by each algorithm within the same amount of time.
Fig. 4.
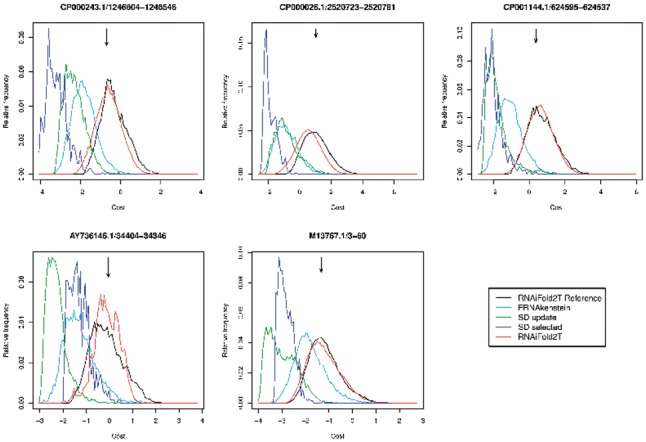
Relative frequency of the cost function optimized by SwitchDesign, for solutions returned by RNAiFold2T, SwitchDesign and Frnakenstein, given target structure S1 (resp. S2) at temperature T1 (resp. T2) for λ phage CIII thermoregulators from Rfam family RF01804. This figure is a more generous representation of the data from SwitchDesign and Frnakenstein, since all single point mutant solutions have been added to the raw output. (Supplementary Figure S1 presents histograms for the raw output of these programs.) The reference distribution for RNAiFold2T Reference (black curve), was produced by running RNAiFold2T for several days. Remaining curves are for Frnakenstein (light green), SwitchDesign (dark green and purple) and RNAiFold2T (red). Arrows indicate cost values for the real λ phage CIII thermoregulators from Rfam RF01804. Distribution for SD and FRNA without additional single point mutants shown in Supplementary Material. Supplementary Figures S1 and S3 show clearly that cost function values for Rfam sequences approximately equal the reference distribution mean
Supplementary Table S1 describes the value ordering heuristics used in RNAiFold2T; Supplementary Table S2 benchmarks RNAiFold2T with respect to different helix ordering and value heuristics, using a cutoff time of 10 min. Since the data clearly demonstrate the superiority of overlap2 helix ordering, this is taken as the default for all other benchmarks and for the web server. Supplementary Table S3 (resp. Supplementary Table S4) presents benchmarking data for LNS from RNAiFold2T and SwitchDesign and Frnakenstein, each with a cutoff time of 30 min, using Rfam thermosensor target structures of length less than 130 nt (resp. greater than 130 nt)—see Section 1 for construction of target structures and temperatures. RNAiFold2T has essentially the same performance as SD and FRNA for shorter sequences, while SD performs better than other methods for longer sequences. Target structures S1, S2 and temperatures T1, T2 for this test are given in Supplementary Table S5 displays the number of solutions for λ phage CIII thermoswitches from Rfam family RF01804. CP from RNAiFold2T, adaptive walk SwitchDesign (Flamm et al., 2001) and genetic algorithm Frnakenstein (Lyngso et al., 2012) were run on each thermosensor for 24 h, forcing a restart if no new solution was found within 1 h. Since both SwitchDesign and Frnakenstein return only a single solution, for this test we modified each as described in Section 1, resulting in two versions of SwitchDesign and one of Frnakenstein, each of which returns two orders of magnitude less solutions than RNAiFold2T. Supplementary Table S6 lists the target structures S1, S2 and temperatures T1, T2 of RNA thermometers of length less than 130 nt used in the benchmarking tests. The structures for thermometers of length 130–447 nt are too large for display, hence are available in an Excel file in Supplementary Material.
3 Discussion
In this article, we introduce the software RNAiFold2T, capable of solving the inverse folding problem for two or more temperatures, i.e. generating one or more RNA sequences whose MFE secondary structures at temperatures T1 and T2 (resp. ) are user-specified target structures S1 and S2 (resp. ), or which reports that no such solution exists. RNAiFold2T is unique in that it implements two different algorithms—CP and LNS. CP is an exact, non-heuristic method that uses an exhaustive yet efficient branch-and-prune process, and is the only currently available software capable of generating all solutions or determining that no solution exists (since there are possibly exponentially many solutions, a complete solution is feasible only for structures of modest size). CP differs from what one might call a ‘brute-force’ approach only in that it relies on a highly efficient branch-and-prune search engine, which propagates the effects of currently instantiated variables held within a constraint store. LNS uses a local search heuristic, complemented with local calls of CP to explore solutions of substructures of the target structures.
There exists an RNA sequence compatible with any two given secondary more than two structures (Reidys et al., 1997)—nevertheless, RNAiFold2T solves inverse folding problem for more than two temperatures. RNAiFold2T is modular software, with a clear separation between search procedure and constraint descriptions, thus permitting the future addition of sequence and structural constraints. In its current form, RNAiFold2T includes constraints for full and/or partial target structures or hybridization complexes at two temperatures; a plug-in to use RNAfold or RNAstructure for MFE structure computation; IUPAC nucleotide constraints, IUPAC amino acid constraints that require all returned RNA sequences to code specified peptides in one or more overlapping reading frames, structural compatibility and structural incompatibility constraints, etc. These constraints support the design of temperature-sensitive selenocysteine insertion (SECIS) elements, precursor microRNAs and mRNA domains that are targeted by microRNAs, etc. Since CP is not a heuristic, unlike other methods such as adaptive walk, genetic algorithm, etc., RNAiFold2T can in principle return all 2-temperature inverse folding solutions, or prove that none exist. The LNS algorithm of RNAiFold2T returns a single solution with approximately the same performance as state-of-the-art approaches SD and FRNA.
As with the synthetic hammerhead design in Dotu et al. (2015), our synthetic RNA design strategy consists of generating many solutions, which are prioritized for experimental validation by applying various computational filters. In our opinion, this strategy presents advantages over methods using SD or NUPACK, each of which returns a relatively small number of sequences that are optimized with respect to a single criterion—in the case of SD, this is the cost function (Flamm et al., 2001), and in the case of NUPACK, this is ensemble defect (Zadeh et al., 2011).
In order to ascertain the viability of our approach of not committing to a particular cost function, we used the capabilities of RNAiFold2T to find hundreds of thousands of solutions to the 2-temperature inverse folding problem for target structures from Rfam family RF01804 (λ phage CIII thermoregulators). Figure 4 and Supplementary Figure S1 show that that the cost function value of (real) Rfam sequences is not close to the minimum, but rather close to the average of the distribution. Other figures can be found in Supplementary Material, where we investigated a variant of the cost function defined using ensemble defect. So, although SD benchmarking results in Supplementary Material indicate that cost function minimization is a good strategy to find sequences whose MFE structures at temperatures T1 resp. T2 are the target structures S1 resp. S2, it appears that naturally occurring RNA thermoswitches are not optimized for the SD cost function. This observation may be important for the future design of functional synthetic thermoregulators.
In designing thermo-IRES elements, we solved the 2-temperature inverse folding problem depicted in Figure 3, where the AUG start codon was appended at the end of the 46 IUPAC constraint mask NUAGGNGACC GNAGGNCGGC NCNUUYYYYY YRNNNNNNNN NNNNNN. This was done since we assumed that the AUG start codon was located at position 463, as it occurs in the viral RNA. However, experimental validation was performed by using a construct containing the 35 nt spacer GAGCUCGAGC UUGGCAUUCC GGUACUGUUG GUAAA at the end of the designed 46 nt sequences, followed by the luciferase AUG start codon. It is possible that the secondary structure of the spacer might have rendered the polypyridine tract less accessible, thus explaining the low efficiency of thermo-IRES constructs Seq1 and Seq2. Another issue is that IRES elements are complex molecules, both in sequence and structure, which respond to more host factors than previously reported RNA thermometers (Lozano and Martinez-Salas, 2015). Secondary structure predictions could differ from the physical structure due to unmodeled protein-RNA interactions, possibly unreliable free energy parameters at temperatures different than 37°C, and due to variations in structure prediction software as shown in Figure 5. Panels (a) resp. (b) of Figure 5 show that the polypyrimidine tract (PPT) is unpaired in the secondary structure of domain 5 of wild-type IRES at 37°C (hence consistent with experimental data), when determined by RNAfold (without SHAPE) resp. RNAsc (with SHAPE) (Zarringhalam et al., 2012), while panels (c) and (d) of Figure 5 show the PPT is paired in the structure determined by RNAstructure, both with and without SHAPE (Deigan et al., 2009) (hence inconsistent with experimental data). Due to this variation in MFE structure prediction, it may be advisable in future synthetic RNA design projects to run RNAiFold2T using plug-ins for both RNAfold and RNAstructure, and to select sequences which have the same desired target structures as predicted by both algorithms. Another issue is that for many RNA thermometers, the MFE structures at low temperature T1 and high temperature T2 are almost identical, where T1, T2 are the temperatures for which a conformational change is reported in the literature. For instance, the MFE structures for the ROSE element CP000009.1/1450710-1450627 at and are nearly identical. This could be an important issue for certain software (Churkin et al., 2014; Chursov et al., 2013) that predict RNA thermometers, but is of less importance in the synthetic design of thermometers, where it is possible to exaggerate the temperature difference . Perhaps future experimental work will provide more reliable energy parameters at temperatures different than , an issue that affects both RNA and DNA melting temperature predictions (Rouzina and Bloomfield, 1999).
Fig. 5.
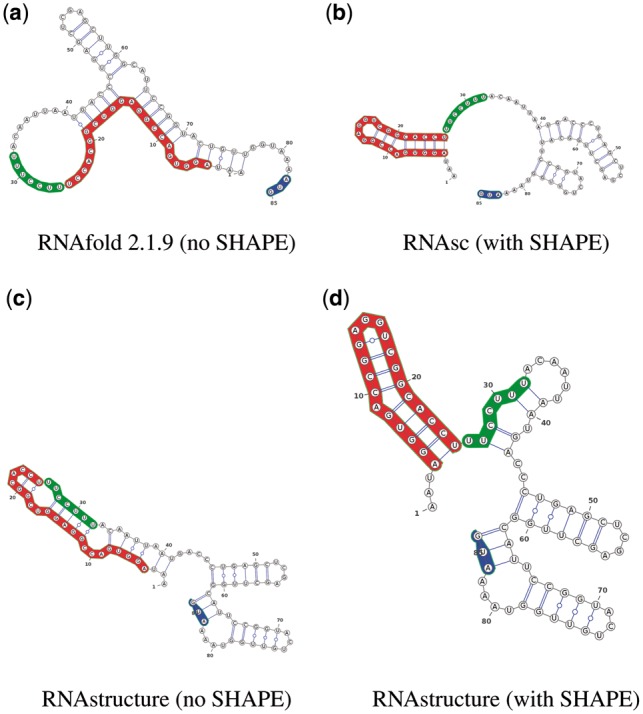
Secondary structure predictions of domain 5 of wild-type FMDV IRES, including the first functional AUG start codon, both with and without integration of probing data using selective -hydroxyl acylation analyzed by primer extension (SHAPE). (a) RNAfold 2.1.9, which does not incorporate SHAPE data. (b) RNAsc (Zarringhalam et al., 2012), which penalizes deviations from SHAPE data for both base paired and loop regions. (c) RNAstructure (Reuter and Mathews, 2010) without SHAPE data. (d) RNAstructure (Deigan et al., 2009), which penalizes deviations from SHAPE data only for base paired regions. The polypyrimidine tract (PPT) is unbound in only structures (a) from RNAfold 2.1.9 and (b) from RNAsc, compatible with the full IRES structure appearing in Figure 1 of Fernandez et al. (2011)
4 Conclusions
We present the software RNAiFold2T for the multi-temperature inverse folding problem, used to design functional thermoswitches. The CP variant of RNAiFold2T returns two orders of magnitude more solutions than other software, while the LNS variant, which returns a single solution, exhibits comparable performance with that of existent methods. The software design of RNAiFold2T currently supports a much greater variety of user-defined structural and sequence constraints than other methods, and moreover can be extended to support future constraints.
Naturally occurring RNA thermometers in heat-shock and virulence genes, as well as all previously known instances of rationally designed thermometers control translation initiation by sequestering the Shine-Dalgarno ribosomal binding sequence at low temperatures, whereby this sequence becomes accessible at high temperatures due to the melting of a hairpin. In contrast, we have used RNAiFold2T to design a temperature-regulated internal ribosomal entry site (IRES) element by ensuring the presence at high temperatures of the domain 5 stem-loop and downstream single-stranded pyrimidine (Py) tract, both located upstream of the functional initiation codon and both known to be important for IRES functionality (Lozano and Martinez-Salas, 2015). At low temperatures, our thermo-IRES element is designed to adopt a conformation that down-regulates protein product by disrupting both the domain 5 stem-loop and sequestering the Py tract. We showed that our rationally designed thermo-IRES elements are functional, where the cap-independent translational efficiency is approximately 50% higher at 42°C than at 30°C; however, since the focus of this article is primarily to describe a new method, we have not taken steps to improve efficiency using error-prone mutagenesis and selection.
Supplementary Material
Funding
Research of the Clote Lab was supported by National Science Foundation grant [DBI-1262439]. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. Research of the Martinez-Salas Lab was supported by the Spanish Ministry of Economy and Competitiveness (MINECO) [CSD2009-00080, BFU2011-25437, BFU2014-54564] and by an Institutional Grant from Fundación Ramón Areces.
References
- Altuvia S. et al. (1989) Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J. Mol. Biol., 210, 265–280. [DOI] [PubMed] [Google Scholar]
- Andronescu M. et al. (2007) Efficient parameter estimation for RNA secondary structure prediction. Bioinformatics, 23, i19–i28. [DOI] [PubMed] [Google Scholar]
- Bae W. et al. (2000) Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl Acad. Sci. USA, 97, 7784–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S. et al. (2003) Temperature-controlled structural alterations of an RNA thermometer. J. Biol. Chem., 278, 47915–47921. [DOI] [PubMed] [Google Scholar]
- Churkin A. et al. (2014) RNAthermsw: direct temperature simulations for predicting the location of RNA thermometers. PLoS One, 9, e94340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chursov A. et al. (2013) RNAtips: Analysis of temperature-induced changes of RNA secondary structure. Nucleic Acids Res., 41, W486–W491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan K.E. et al. (2009) Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA, 106, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotu I. et al. (2013) Using RNA inverse folding to identify IRES-like structural subdomains. RNA Biol., 10, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotu I. et al. (2015) Complete RNA inverse folding: computational design of functional hammerhead ribozymes. Nucleic Acids Res, 42, 11752–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez N., Fernandez-Miragall O., Ramajo J., García-Sacristan A., Bellora N., Eyras E., Briones C., Martinez-Salas E. (2011) Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res., 39, 8572–8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Chamorro J. et al. (2014) Identification of novel non-canonical RNA-binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Res., 42, 5742–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm C. et al. (2001) Design of multi-stable RNA molecules. RNA, 7, 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martin J. et al. (2012) RNAiFold: A Constraint Programming algorithm for RNA inverse folding and molecular design. J. Bioinformatics Comput. Biol., 11, 1350001. [DOI] [PubMed] [Google Scholar]
- Garcia-Martin J.A. et al. (2015) RNAiFold 2.0: a web server and software to design custom and Rfam-based RNA molecules. Nucleic Acids Res., 43, W513–W521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoynes-O’Connor A. et al. (2015) De novo design of heat-repressible RNA thermosensors in E. coli. Nucleic Acids Res., 43, 6166–6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J. et al. (2002) An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell, 110, 551–561. [DOI] [PubMed] [Google Scholar]
- Kortmann J., Narberhaus F. (2012) Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol., 10, 255–265. [DOI] [PubMed] [Google Scholar]
- Kortmann J. et al. (2011) Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res., 39, 2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. et al. (1990) Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J. Virol., 64, 4625–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kotov J. (2007) Thermometer design at the nanoscale. Nano Today, 2, 48–51. [Google Scholar]
- Lorenz R. et al. (2011) Viennarna Package 2.0. Algorithms Mol. Biol., 6, 26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano G., Martinez-Salas E. (2015) Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol., 12, 113–120. [DOI] [PubMed] [Google Scholar]
- Lozano G. et al. (2014) Magnesium-dependent folding of a picornavirus IRES element modulates RNA conformation and eIF4G interaction. FEBS J., 281, 3685–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano G. et al. (2015) Local RNA flexibility perturbation of the IRES element induced by a novel ligand inhibits viral RNA translation. RNA Biol., 12, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngso R.B. et al. (2012) Frnakenstein: multiple target inverse RNA folding. BMC Bioinformatics, 13, 260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E.P. et al. (2014) Rfam 12.0: updates to the RNA families database. Nucleic Acids Res., 0, O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J. et al. (2008) Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res., 36, e124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocker A. et al. (2001) A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res., 29, 4800–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A. (2011) Rational design of artificial riboswitches based on ligand-dependent modulation of internal ribosome entry in wheat germ extract and their applications as label-free biosensors. RNA, 17, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro D. et al. (2013) Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res., 41, 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidys C. et al. (1997) Generic properties of combinatory maps: neutral networks of RNA secondary structures. Bull. Math. Biol., 59, 339–397. [DOI] [PubMed] [Google Scholar]
- Reuter J.S., Mathews D.H. (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnenthal J. et al. (2011) Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res., 39, 8258–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzina I., Bloomfield V.A. (1999) Heat capacity effects on the melting of DNA. 1. General aspects. Biophys. J., 77, 3242–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saragliadis A. et al. (2013) Thermozymes: synthetic RNA thermometers based on ribozyme activity. RNA Biol., 10, 1010–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok Z. et al. (2001) Synechocystis HSP17 is an amphitropic protein that stabilizes heat-stressed membranes and binds denatured proteins for subsequent chaperone-mediated refolding. Proc. Natl Acad. Sci. USA, 98, 3098–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.H., Mathews D.H. (2010) NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res., 38, D280–D282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth M. et al. (2013) De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res., 41, 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T. et al. (2007a) Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers. Mol. Genet. Genomics, 278, 555–564. [DOI] [PubMed] [Google Scholar]
- Waldminghaus T. et al. (2007b) Fouru: a novel type of RNA thermometer in Salmonella. Mol. Microbiol., 65, 413–424. [DOI] [PubMed] [Google Scholar]
- Waldminghaus T. et al. (2008) Generation of synthetic RNA-based thermosensors. Biol. Chem., 389, 1319–1326. [DOI] [PubMed] [Google Scholar]
- Wieland M., Hartig J.S. (2007) RNA quadruplex-based modulation of gene expression. Chem. Biol., 14, 757–763. [DOI] [PubMed] [Google Scholar]
- Zadeh J.N. et al. (2011) Nucleic acid sequence design via efficient ensemble defect optimization. J. Comput. Chem., 32, 439–452. [DOI] [PubMed] [Google Scholar]
- Zarringhalam K. et al. (2012) Integrating chemical footprinting data into RNA secondary structure prediction. PLoS One, 7, e45160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


