Abstract
Motivation: Disordered flexible linkers (DFLs) are disordered regions that serve as flexible linkers/spacers in multi-domain proteins or between structured constituents in domains. They are different from flexible linkers/residues because they are disordered and longer. Availability of experimentally annotated DFLs provides an opportunity to build high-throughput computational predictors of these regions from protein sequences. To date, there are no computational methods that directly predict DFLs and they can be found only indirectly by filtering predicted flexible residues with predictions of disorder.
Results: We conceptualized, developed and empirically assessed a first-of-its-kind sequence-based predictor of DFLs, DFLpred. This method outputs propensity to form DFLs for each residue in the input sequence. DFLpred uses a small set of empirically selected features that quantify propensities to form certain secondary structures, disordered regions and structured regions, which are processed by a fast linear model. Our high-throughput predictor can be used on the whole-proteome scale; it needs <1 h to predict entire proteome on a single CPU. When assessed on an independent test dataset with low sequence-identity proteins, it secures area under the receiver operating characteristic curve equal 0.715 and outperforms existing alternatives that include methods for the prediction of flexible linkers, flexible residues, intrinsically disordered residues and various combinations of these methods. Prediction on the complete human proteome reveals that about 10% of proteins have a large content of over 30% DFL residues. We also estimate that about 6000 DFL regions are long with ≥30 consecutive residues.
Availability and implementation: http://biomine.ece.ualberta.ca/DFLpred/.
Contact: lkurgan@vcu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Intrinsically disordered proteins (IDPs) lack stable tertiary structure under physiological conditions, either along their entire sequence or in localized regions. IDPs are abundant in eukaryotes (Peng et al., 2015; Ward et al., 2004) and are typically involved in signaling, regulation, control, storage of small molecules, sites for post-translational modifications (PTMs), transcription, translation and assembly of multi-protein complexes (Dunker et al., 2008; Dyson and Wright, 2005; Peng et al., 2014; Tompa, 2005; Wright and Dyson, 1999; Xie et al., 2007). Their functions complement functions of structured proteins, which include small molecule binding, transport and catalysis (Radivojac et al., 2007). The newest release of DisProt database (Sickmeier et al., 2007), the main source of functionally annotated IDPs, lists >30 functions (Dunker et al., 2002) that have been assigned to about 1200 disordered regions. About 74% of the functionally characterized disordered regions in DisProt concern binding to a variety of partners: proteins, DNAs, RNAs, metals and lipids. The most populated non-binding functions include flexible linkers (9%), sites for several types of PTMs (7%), transactivation (4%), nuclear localization signals (1%) and electron transfer (1%). Several methods have been developed for the prediction of binding disordered regions from protein sequences. They include methods that address protein binding (Disfani et al., 2012; Dosztanyi et al., 2009; Fang et al., 2013; Jones and Cozzetto, 2014; Khan et al., 2013; Malhis and Gsponer, 2015; Meszaros et al., 2009; Peng and Kurgan, 2015; Yan et al., 2015) and RNA and DNA binding (Peng and Kurgan, 2015). However, methods for the prediction of the other functions of the intrinsically disordered regions are lacking.
The disordered flexible linkers (DFLs) are disordered regions that serve as linkers or spacers between domains in multi-domain proteins and between structured constituents in domains (Dunker et al., 2002). Experimental annotation of DFLs primarily relies on the X-ray crystallography, NMR spectroscopy and circular dichroism. We consider these regions for several reasons. First, this is the most annotated and not related to binding function of disordered regions. Second, DFLs are important for a variety of cellular processes. A few recent examples include formation of amyloid fibrils (Shvadchak and Subramaniam, 2014), linking multiple disordered protein binding regions (Oldfield and Dunker, 2014), and movement of structured domains between catalytic sites (Anand and Mohanty, 2012). Third, there is no computational methods that predict this class of disordered regions. DFLs are cousins of linkers, which are regions that connect domains and maintain inter-domain interactions (Chen et al., 2013; George and Heringa, 2002). A sub-class of linkers are flexible linkers, defined as flexible inter-domain regions that allow two domains to move relatively to each other (Chen et al., 2013). DFLs differ from linker regions in two aspects: (1) DFLs are characterized by extreme level of flexibility and lack of defined structure (they form conformational ensembles) as compared with linkers and flexible linkers that have more defined structures; and (2) linkers are shorter (avg length of 10 residues) and localized between domains (George and Heringa, 2002), while DFLs tend to be longer (avg length of 25 residues in our dataset) and could be localized in domains, for instance to link structured elements in a domain; we show empirically that they are frequently localized in domains and give examples of intra- and inter-domain DFLs.
Although there are no computational methods that directly predict DFLs in protein sequences, Udwary–Merski algorithm (UMA) (Udwary et al., 2002) can be used to predict flexible linker regions. It assumes that flexible linkers are less likely to be conserved in the sequence and secondary structure and to be depleted in hydrophilic residues. Thus, UMA quantifies every residue as a weighted sum of hydrophobicity score and conservation scores for sequence and secondary structure. The two conservation scores are derived using ClustalX (Thompson et al., 1997) and PHDsec (Rost and Sander, 1993; Rost and Sander, 1994), and the hydrophobicity score is assigned using the Kyte and Doolittle’s hydropathy index (Kyte and Doolittle, 1982). A low UMA score indicates that a residue is more likely to be a flexible linker. Because flexible linkers are a subset of flexible residues, they could be also potentially identified with sequence-based predictors of flexible residues. These predictors include PROFbval (Schlessinger and Rost, 2005; Schlessinger et al., 2006), FlexPred (Kuznetsov and McDuffie, 2008; Kuznetsov, 2008), PredBF (Pan and Shen, 2009), PredyFlexy (de Brevern et al., 2012) and DynaMine (Cilia et al., 2013, 2014). PROFbval predicts B-factors using a neural network model, where a low/high real B-factor value indicates a low/high propensity of a residue being flexible. PredBF also predicts B-factors but using a two-layer support vector regression model. FlexPred predicts conformationally variable positions in the input protein chain using a support vector machine model. PredyFlexy classifies every input residue as rigid, intermediate or flexible and also outputs putative normalized B-factors and root mean square fluctuations, from molecular dynamic simulations. DynaMine quantifies backbone flexibility in terms of N-H S2 order parameter values using regression where smaller S2 means that a given residue is more likely to be flexible.
The UMA method and protein flexibility predictors predict flexible linkers/residues, but they do not accommodate for the disordered state of these residues. Moreover, UMA requires that the input sequence has homologous sequences to generate multiple sequence alignment profiles, which means that it may not generate predictions for some proteins, and is tedious to execute because its implementation requires manual processing. To this end, we have developed DFLpred, the first method that predicts DFLs. DFLpred does not need alignment profiles, is fast to execute and is provided to the end users as a fully automated webserver at http://biomine.ece.ualberta.ca/DFLpred/.
2 Materials and methods
2.1 Datasets
The functionally annotated data were collected from the newest release 6.0.2 of DisProt that includes 694 sequences. We excluded DP00688 sequence that was too long (>18 000 residues) to predict with the PSIPRED (Buchan et al., 2013) to generate secondary structure. We selected 204 sequences, which include 82 proteins that have annotations of DFLs and 122 proteins that do not have DFL annotations but for which all residues are annotated. This way we included all annotated DFLs and reduced the number of ambiguous (unannotated) residues.
We assumed that residues that are not annotated as DFLs but have other functional annotations are non-disordered flexible linker (NDFL) residues. The residues without functional annotations were excluded from the design and assessment. We divided the set of 204 proteins into five subsets and reduced sequence similarity between these subsets with BLASTClust (Altschul et al., 1990). First, we clustered the 204 sequences with sequence identity threshold at 25% and coverage of at least 10% of the sequence length. Second, the resulting 160 clusters that include similar sequences (>25% similarity) were divided at random between the five sub sets to ensure that each subset has similar number of sequences and similar ratio of DFL to NDFL residues. Four of these subsets were used in 4-fold cross-validation protocol to empirically design our predictor, i.e. to conceptualize and select inputs for the predictive model, and to select and parameterize this model. These data constitute the training dataset. The remaining fifth subset was used as an independent (never used in the design) test dataset. This way, sequences in the test dataset share low similarity with sequences in the training dataset, and also sequence in individual folds of the training dataset share low similarity with sequences in the other folds. The training and test datasets have 144 sequences and 60 sequences, respectively, and they are available at http://biomine.ece.ualberta.ca/DFLpred/.
2.2 DFLs and protein domains
We used Interpro (Mitchell et al., 2015) to annotate domains to empirically investigate whether DFLs are localized within or between domains. Interpro is a template-based method that integrates 11 databases including Pfam (Finn et al., 2014), PRINTS (Attwood et al., 2012), PROSITE (Sigrist et al., 2013) and ProDom (Servant et al., 2002) to provide prediction of proteins families and domains. The comprehensive coverage of the source databases, 15 years of history and high rate of updates make Interpro a mainstream tool for the annotation of domains (Goujon et al., 2010). The ratio of the number of intra-domain to inter-domain DFL residues in the training dataset is 905/1098 = 0.82. This indicates that a large fraction of DFLs are localized in domains. The ratio of intra-domain DFL to NDFL residues is 905/7019 = 0.13 and for inter-domain is 1098/14 132 = 0.08. To sum up, our results suggest that DFLs are often localized in protein domains.
2.3 Evaluation procedures
The prediction of DFLs results in a numeric score in the range between 0 and 1 representing propensity of each residue in the input sequence being in a DFL. We used the receiver operating characteristic (ROC) curve and the area under ROC (AUC) to examine the predictive quality. To plot ROC curves and quantify AUC values, we calculated the true-positive rates (TPRs) and false-positive rates (FPRs) by comparing predictions with native annotations at different cutoffs imposed on the predicted scores. If a score is more than or equal to a given threshold, then the corresponding residue is assumed to be in DFL, otherwise it is assumed as NDFL. TPR and FPR are defined as:
| (1) |
| (2) |
where TP is the number of true positives (correctly predicted DFL residues), FN is the number of false negatives (DFL residues that were predicted as NDFL residues), FP is the number of false positives (NDFL residues that were predicted as DFL residues), TN is the number of true negatives (correctly predicted NDFL residues). Given TPR and FPR values at different thresholds ranging between 0 and 1, we plotted the ROC curve and calculated the corresponding AUC value.
We also calculated AUClowFPR, which is the AUC for low range of FPR values, between 0 and 0.1. This part at the beginning of the ROC curve reflects ability to predict a small number of high-quality DFL residues, i.e. only a relatively small portion of these predictions are FPs. We also compute Ratio = AUClowFPR/AUCrandom where AUClowFPR is divided by the AUC of a random predictor (for which TPR always equals to FPR) computed for FPR values between 0 and 0.1. This Ratio reflects how much better a given predictor is when compared with a random prediction.
Following (Disfani et al., 2012; Peng and Kurgan, 2015; Yan et al., 2015), we tested statistical significance of differences in predictive quality offered by DFLpred and other methods, and between the selected design of DFLpred and other considered designs. This test investigates whether results on a given dataset are not biased by a subset of proteins by measuring whether the predictive performance is consistent over different subsets of the dataset. First, we randomly selected half of proteins from a given test dataset (test dataset or test folds from the cross validation on the training dataset) 10 times and we measured predictive performance of all considered methods on these 10 protein sets. We compared these 10 pairs of results between DFLpred and other methods. Given that measurements are normal, as assessed with the Anderson-Darling test (Anderson and Darling, 1952), we used paired t-test, otherwise we used the Wilcoxon signed-rank test (Wilcoxon, 1945). We considered a given difference to be significant if the p-value < 0.01.
2.4 Overall design
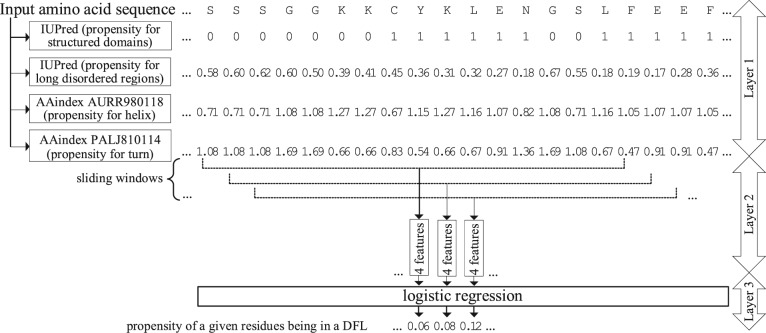
The architecture of DFLpred (Fig. 1) includes three layers:
Fig. 1.
Architecture of the DFLpred method
Represent every residue of the input sequence with its amino acid (AA) type and information predicted directly from the sequence including propensity to form structured regions, intrinsically disordered regions and helical and coil conformations.
Convert this representation into empirically selected set of numeric features that are computed using sliding windows.
Input the selected features into an empirically selected and parameterized predictive model to generate propensity scores.
2.4.1 Sequence representation
In the first layer, we represented every residue by its AA type, its physicochemical properties estimated based on the AA indices from the AAindex database (Kawashima et al., 2008), secondary structure predicted with PSIPRED (Buchan et al., 2013), intrinsically disordered and structured regions predicted with IUPred (Dosztányi et al., 2005) and sequence complexity computed with SEG (Wootton, 1994). Supplementary Figure S1 summarizes enrichment and depletion of the 20 AA types in DFL regions. We note a clear pattern where DFLs are significantly enriched in the disorder-promoting residues, such as Q, S, E and P, and depleted in the order-promoting residues, including F, Y, I, M, L and V; details are discussed in Supplementary Section 1.
2.4.2 Considered features
In the second layer, we generated numerical features that quantify the considered structural and sequence-based properties for each residue of the input AA sequence. We represented every residue by a feature vector calculated from a sliding window centered on that residue. The sliding window aggregates structural and sequence-based information by considering characteristics of AAs adjacent in the sequence. The concept of the sliding window has been adopted in other relevant predictors such as MoRFpred, fMoRFpred, DisoRDPbind, PROFbval, FlexPred, PredBF, PredyFlexy and DynaMine. We set the length of the sliding window to 17, which is the median value of the length of longest per protein DFLs in our dataset. This way the selected window size covers the full length of at least half of DFLs without recruiting much of potential noise (NDFL residues) when used to predict shorter regions. We did not pad the window for the residues located at either terminus of the sequence, and correspondingly the length of the sliding window is reduced on one of its sides, i.e. window size is 8 for the first and last residues in the sequence. Consequently, we normalize values of features computed over the residues in the window by the size of the window. In total, we considered 2236 features including 40 features derived directly from the sequence, 2124 features derived from physicochemical properties of AAs quantified based on the AAindex database, 22 features generated from the putative secondary structure, 40 features from putative intrinsic disorder and structured regions and 10 features from the sequence complexity. These features quantify composition of AAs; composition, counts and length of putative secondary structures, intrinsically disordered regions, structured regions and high sequence complexity regions in the sliding window; average physicochemical properties of residues in the sliding windows; and AAs type, secondary structure, disorder status, sequence complexity status and physicochemical properties of the residue in the center of the window. We decided to use all physicochemical properties from AAindex (thus the large number of these features) and our empirical feature selection to remove irreverent and redundant features because there are no prior results that we could use to pre-select a subset of these properties. Detailed list and description of the considered features are included in Supplementary Section 2.
2.4.3 Selected features and predictive model
Our vector of 2236 features likely includes features that are irrelevant to the prediction of DFLs and features that have high mutual correlations. We used a two-step empirical feature selection to select a subset of features characterized by high predictive value and low mutual correlations.
In the first step we removed low-quality features that have low correlation with the annotation of the DFLs. We have two types of features: real-valued (e.g. features computed as an average over the sliding window) and binary (e.g. disordered versus ordered status of the residue in the center of the window). Inspired by (Disfani et al., 2012; Yan et al., 2015), we used point-biserial correlation coefficient (rpb) and φ coefficient (φ), respectively, for these two feature types:
| (3) |
| (4) |
In formula (3), MDFL and MNDFL (nDFL and nNDFL) are the means (numbers) of values a given real-valued feature for the residues annotated as DFLs and NDFLs, respectively; n = nDFL + nNDFL and Sn is the standard deviation of all values of that feature. In formula (4), countFiAk is the number of values i = {0, 1} of binary feature F corresponding to residues with values k = {NDFL, DFL} of the annotation A; countFi and countAk are the number of values i = {0, 1} of binary feature F and the number of residues with values k = {NDFL, DFL} of the annotation A, respectively. We calculated average rpb (for the real-valued features) and φ (for the binary features) for all considered features from four correlations computed on the training folds from the 4-fold cross-validation on the training dataset. We normalized the values of the average rpb and φ correlations to the −1 to 1 range using min–max normalization, and removed the features for which the absolute normalized rpb or φ value is less than threshold Tstep1. Next, we ranked the remaining features by their absolute normalized rpb or φ values.
In the second step, inspired by (Disfani et al., 2012; Yan et al., 2015), we eliminated mutually correlated features using the Pearson correlation coefficient (rpc). First, a set of selected features is initialized with the top-ranked in the first step feature. Next, we calculated rpc between the next-ranked feature and all selected features. If the absolute value of this rpc is less than threshold Tstep2, then we add this next-ranked features into the set of selected features, otherwise we do not add it. We apply this procedure through the entire list of ranked features passed from the first step.
We vary values of each of the two thresholds, Tstep1and Tstep2 between 0.1 and 0.9 with step of 0.05, to obtain 17 × 17 = 289 different feature sets. The corresponding feature sets vary in size between 1 and 884 features. Each feature set is used with three classifiers: logistic regression, naive Bayes and k-nearest neighbor, in the 4-folds cross-validation on the training dataset to select the design that offers the highest AUC value. We also parameterized logistic regression and k-nearest neighbor classifiers for each of these experiments by selecting their parameters that correspond to the highest AUC in the 4-folds cross-validation on the training dataset. Naive Bayes has no parameters. For the logistic regression, we considered ridge = 10x, where x ranges from −4 to 4 with step of 1. For the k-nearest neighbor, we consider the number of neighbors k ranging from 50 to 800 with the step of 50. Supplementary Table S1 summarizes results with the highest AUC value for each of the three classifiers, which are selected from across the experiments that correspond to 7514 combinations of the two thresholds and different parameters of classifiers (289 combinations for Naïve Bayes + 9*298 for logistic regression + 16*289 for k-nearest neighbor).
We selected the logistic regression classifier with four features that gives the highest values of AUC, AUClowFPR and ratio. The differences in these three measures of predictive quality between the logistic regression and the other two classifiers are statistically significant. The ratio reveals that the selected design is 3.3 times better than a random predictor when predicting with low FPR, i.e. when a high fraction of predictions of DFL residues (predicted positive residues) is correct. The architecture of this model is shown in Figure 1. Given an input AA sequence, it uses putative annotations of structured and long disordered regions generated with IUPred and two physicochemical properties of residues that quantify propensity for formation of helices and turns.
3 Results
3.1 Analysis of the DFLpred’s predictive model
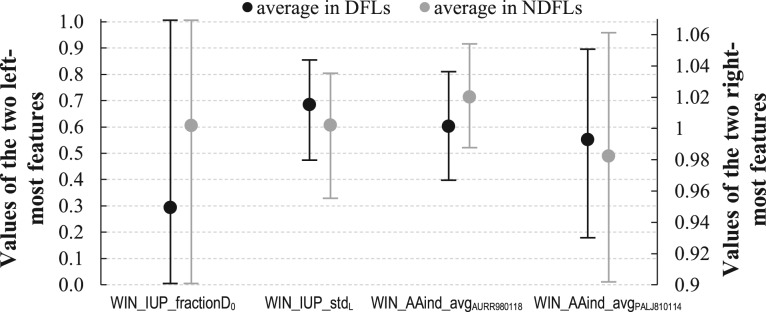
DFLpred’s model combines values of four empirically selected features using a linear function to generate the output propensities. These features were computed from the sequence using sliding windows on putative annotations generated with IUPred and two AA indices; their details are summarized in Table 1. Figure 2 compares values of the four features between the native DFL and NDFL residues in the test dataset (these results were not used to design the model, which is based on the training dataset).
Table 1.
Summary of DFLpred’s predictive model
| Feature name | Description | rpb | Coeff |
|---|---|---|---|
| WIN_IUP_fractionD0 | Number of residues predicted with IUPred_struct not to be in structured regions in a sliding window divided by the window length. | −1.00 | −1.10 |
| WIN_IUP_stdL | Standard deviation of propensity scores from IUPred _long for residues in the sliding window. | 0.70 | 6.60 |
| WIN_AAind_avgAURR980118 | Average value of AURR980118 AA index for all residues in the sliding window. | −0.66 | −4.58 |
| WIN_AAind_avgPALJ810114 | Average value of AURR980118 AA index for all residues in the sliding window. | 0.57 | 3.32 |
| Intercept of the linear function | N/A | −0.92 | |
rpb: Normalized point-biserial correlation coefficient of a given feature with the annotation of DFLs in the training dataset; Coeff: coefficient of a given feature in the linear model generated with logistic regression using training dataset.
Fig. 2.
Comparison of values of features used in the DFLpred model between the native DFL residues (black lines) and native NDFL residues (gray lines) in the test dataset. The features are ranked by their absolute rpb values from the highest on the left to the lowest on the right (see Table 1). Values of the WIN_IUP_stdL are multiplied by 10 to better fit the range of values of the other features. Dots are the averages and the error bars show the first and third quantiles
The WIN_IUP_fractionD0 feature quantifies fraction of residues predicted with IUPred_struct not to be in structured regions in a window centered on the predicted residue. Its average for DFL residues is 0.29, for the NDFL residues is 0.60 and for the NFDL residues annotated as structured is 0.11. The structured residues have the lowest value because they should be primarily predicted to be in structured regions; the value > 0 because some of the residues in the surrounding window could lack structure. The high value of mean for NFDL residues is driven by the fact that these residues include disordered residues that are not DFLs which have a large number of nearby (in the sequence) unstructured residues. The average for DFLs is in between the other two averages. This reveals that propensity of these residues to be nearby putative structured regions is lower than for other disordered regions but higher than for structured regions. This makes sense because residues in DFLs link primarily structured domains, and thus their neighbors in the sequence should include a sizable fraction of structured residues, but not as large as for the structured residues.
The plot of the WIN_IUP_stdL feature in Figure 2 suggests that putative propensities for disorder of residues in DFLs have higher standard deviation in the window compared with the other residues. This means that these propensities fluctuate more in residues adjacent to DFL residues. This is reasonable given that DFLs link structured domains where propensity for disorder should be substantially lower compared with DFLs. In contrast, residues located in structured or in disordered regions would experience less variability in the propensities for disorder in these regions.
The last two features are computed by averaging values of the selected two AA indices in the sliding window. Higher values of the AURR980118 index (Aurora and Rosee, 1998) indicate higher likelihood of a given residue to be included in a helical conformation. Thus, the corresponding feature can be used as a proxy to quantify likelihood of helical conformations in the window. Figure 3 shows that residues in DFLs have lower values of this feature, which suggests that they are less likely to include helices nearby in the sequence compared with NDFLs. This again is rational because DFLs are unstructured. The second, PALJ810114 index (Palau et al., 1982), quantifies likelihood of forming turns. Here, DFLs have higher values compared with the other residues, which is sensible given that turns are relatively flexible, which is also characteristic for DFLs.
Fig. 3.
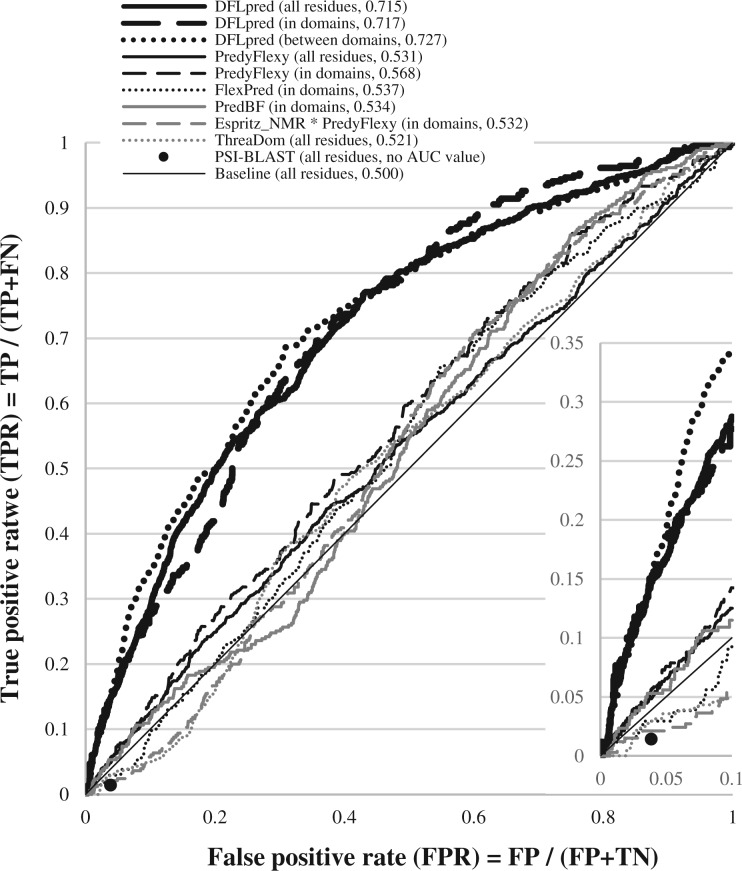
ROC curves on the test dataset for methods that achieved AUC ≥ 0.52 in Table 2 (on the whole test dataset) or in Supplementary Table S2 (for residues in or between domains). Insert in the bottom right corner focuses on the ROCs for FPR between 0 and 0.1. The scope of the prediction (all residues, in domains or between domains) and AUC values are shown inside brackets next to the names of methods in the figure legend
We also investigated predictive quality of these features individually and in different sets to find out how much they contribute to the predictive model and whether they complement each other. The AUC values based on the 4-fold cross-validations on the training dataset for the individual features sorted in the order shown in Table 1 are 0.599, 0.604, 0.584 and 0.580. The fact that AUC values are similar suggests that these features contribute equally to our model. When we use the top n = 1, 2, 3, 4 features (ranked as in Table 1, which is based on the normalized point-biserial correlation rpb), the corresponding AUC values are 0.599, 0.646, 0.672 and 0.702. This means that each feature adds to the model and demonstrates that they complement each other.
Overall, we demonstrated that the four features are meaningful markers of DFLs. They complement each other by using a different type of information to pinpoint location of DFLs. This agrees with our empirical approach to design DFLpred in which we explicitly selected highly predictive features (first step of feature selection) that are characterized by low mutual correlation (second step of feature selection).
3.2 Comparison of predictive performance with closest alternative methods
We compared the predictive performance of DFLpred with closest alternative methods that could be used to find DFLs. These approaches include the UMA method that finds flexible linkers, predictors of flexible residues and disordered residues, and a domain predictor given the fact that classical linkers are localized between domains. We also combined the results of UMA with the results of the disorder predictors and the results of the flexibility predictors with the disorder predictors. This was motivated by the fact that these combinations could potentially find flexible linkers or flexible residues localized in disordered regions, which is the hallmark of the DFLs. We used two ways to combine their predictions, by multiplying the scores predicted with UMA and flexibility predictors by the binary disorder predictions and by the predicted real-valued propensity for the disorder. In the first case, the UMA and flexibility scores remain the same for the predicted disordered residues and are set to zero for the residues that are not predicted to be disordered. In the second scenario, the UMA and flexibility scores are scaled by the predicted propensity for disorder. We used a comprehensive set of predictors of flexible residues including PROFbval, FlexPred, PredBF, PredyFlexy and Dynamine. We also considered several predictors of disorder including two versions of IUPred (short and long), MFDp (Mizianty et al., 2010) and three versions of Espritz (NMR, X-Ray and DisProt; Walsh et al., 2012). We applied ThreaDom (Xue et al., 2013) to predict domains, given its strong predictive performance and availability of a webserver. Details on the computation of predictions with the other methods are given in Supplementary Section 3.
Table 2 summarizes results of DFLpred and the other methods on the test dataset. We show results for DFLpred, UMA, five methods for prediction of flexible residues, three methods for prediction of disordered residues (we show results for one version of IUPred and Espritz that secures the highest AUC) and ThreaDom for the prediction of domains. We also include result for each of the two ways to combine these methods, as described above, which secured the highest AUC value. DFLpred secures the highest AUC, AUClowFPR and Ratio values. The improvement offered by DFLpred are significant at p-value < 0.01 when compared with all considered methods. The Ratio indicates that DFLpred is 3.3 times better than a random predictor in AUC for the low values of FPR ≤ 0.1. The insert in Figure 3 visualizes this difference between this AUC of a random predictor (thin diagonal line) and DFLpred (thick black line).
Table 2.
Comparison of predictive quality on the test dataset
| Prediction target | Method | AUC | AUClowFPR | Ratio |
|---|---|---|---|---|
| DFLs | DFLpred | 0.715 | 0.016 | 3.265 |
| Flexible linkers | UMA | 0.384a | 0.003a | 0.531a |
| Flexible residues | PredyFlexy | 0.531a | 0.007a | 1.307a |
| FlexPred | 0.486a | 0.004a | 0.768a | |
| PROFbval | 0.453a | 0.007a | 0.337a | |
| PredBF | 0.445a | 0.005a | 0.988a | |
| Dynamine | 0.396a | 0.003a | 0.573a | |
| Disordered residues | Espritz_NMR | 0.399a | 0.001a | 0.218a |
| IUPred_short | 0.359a | 0.000a | 0.092a | |
| MFDp | 0.325a | 0.004a | 0.201a | |
| Domains | ThreaDom | 0.521a | 0.003a | 0.569a |
| DFLs | Espritz_NMR & PredyFlexy (the best based on binary disorder) | 0.459a | 0.006a | 1.154a |
| Espritz_NMR & PredyFlexy (the best based on disorder propensity) | 0.429a | 0.003a | 0.653a |
The methods were ranked by AUC value in each category.
aDenotes that difference in predictive quality when compared with DFLpred is significant at p < 0.01.
We use two proteins from the test dataset to visualize prediction of DFLs localized between domains (chemotaxis cheA protein; Supplementary Fig. S2A) and inside of a domain (troponin I protein; Supplementary Fig. S2B). CheA includes five domains and we focus on the C-terminus that includes Hpt, CheY binding and signal transducing H kinase domains that are connected by two inter-domain DFLs. Troponin I includes two domains: troponin I N-terminus domain, which is disordered, and troponin I domain, which is composed of two sub-domains: IT arm and regulatory head. The IT arm sub-domain has DFL, which links two of its helices that interact with troponins T and C that compose the troponin complex. The second DFL links the two sub-domains. Both of these intra-domain DFLs enable movement of several structural elements of the troponin I domain, allowing it to interact with the other members of the troponin complex (Takeda et al., 2003). The figures show predictions from DFLpred, UMA, domain predictor ThreaDom and the best-performing (based on Table 2) disorder predictor Espritz and flexibility predictor PredyFlexy. DFLpred generates higher propensities in the vicinity of the two inter-domain DFLs in CheA, with the second one predicted less accurately. It also finds the first intra-domain DFL and to some extent, given the lower values of propensity, the second intra-domain DFL in troponin I. ThreaDom accurately finds the inter-domain residues that overlap with the two DFLs in CheA, but its prediction also includes residues at the N-terminus that are not DFLs. This method finds the inter-domain region in troponin I, which is not a DFL, and has difficulty with the troponin I domain, given it is fragmented into sub-domains composition. Espritz accurately predicts the disordered residues, which coincide with the inter-domain DFLs in CheA, but it also finds disorder at the N-terminus. It correctly finds the first two disordered regions in troponin I but it misses the second intra-domain DFL and predicts the disordered region at the N-terminus the highest propensity while this region is not a linker. UMA finds three flexible linkers (residues with high scores) in CheA and only the last one coincides to some extent with the second DFLs. For the second protein, this method annotates only the N-terminus as a flexible linker and fails to identify the intra-domain DFLs. Finally, PredyFlexy does not find inter-domain residues, DFLs or disordered regions, but rather it estimates local flexibility, which fluctuates widely along the sequence of both proteins. These observations provide context to interpret results of the other methods in Table 2.
The UMA method secures low AUC and this could be explained by the fact that UMA predicts flexible linkers that likely exclude linkers located in disordered regions; the latter stems from the low value of AUClowFPR and is confirmed by our examples. The low AUC of UMA is owing to a concave shape of ROC curve that in turn results from high levels of false positives on the left side of the curve. These false positives are the residues predicted as DFLs for the purpose of our evaluation but which in fact correspond to the putative flexible linkers predicted by UMA with the highest values of propensity. The predictors of disordered residues have similar weakness. They predict all disordered residues, irrespective of their function, while most of them are not DFLs. Their low values of AUClowFPR and our troponin I example suggest that propensities generated for DFLs are lower than the propensities for other disordered regions. This results in high FPRs, concave shapes of ROC curves and consequently low AUC values. Predictors of flexible residues also secure relatively low AUC values. These methods were built using crystallographic data that exclude disordered residues and thus they cannot accurately find disordered residues, which was shown in (Peng and Kurgan, 2012). Our example reveals that they focus on local, in the sequence, flexibility that does not correlate with the localization of DFLs. ThreaDom finds the inter-domain regions but only some of them are DFLs and it in general fails to find the intra-domain DFLs. Consequently, its AUC value is relatively low. Interestingly, combining UMA/flexibility predictors with the disorder predictors also does not produce high-quality predictions. This is likely because neither UMA nor predictors of flexibility provide accurate estimates for the disordered regions. Overall, we conclude that while currently there are no approaches that can accurately predict DFLs, our DFLpred offers such accurate predictions.
We also evaluate the considered methods separately for inter- and intra-domain residues, see Supplementary Table S2. DFLpred can accurately predict DFLs both inside and outside of domains, with AUC = 0.72 and 0.73 and Ratio = 3.3 and 3.8, respectively. These results are significantly better than the results of all other considered methods. The UMA method has substantially higher AUC for prediction of DFLs outside of domains compared with the results for residues localized inside the domains. This difference reflects the fact that flexible linkers that are targeted by UMA are localized between domains. On the other hand, flexibility predictors secure slightly better predictive quality for residues in domains compared with the residues outside the domains. Finally, disorder predictors are equally inaccurate for both inter- and intra-domain residues and the best combinations of flexibility predictors and disorder predictors work better inside the domains. The latter is perhaps because these combinations use PredyFlexy predictors that perform better inside the domains.
We plot ROC curves for all methods for which the AUC > 0.52 based on the evaluations on the entire test dataset and separately for the inter- and intra-domain residues, see Figure 3. Supplementary Figure S3 shows the ROC curves in larger format for methods with the AUC > 0.5. We also include the result when using sequence alignment. The alignment-based predictor copies DFL annotations of aligned residues from the most similar sequence in the training dataset based on its alignment to a query sequence from the test dataset. The alignment is done with PSI-BLAST using default parameters (Altschul et al., 1997). Because alignment transfers binary annotations of DFLs from the training proteins, we can show only one point for the ROC curve for this simple predictor. The FPR and TPR values of alignment-based predictors equal 0.039 and 0.014, respectively. In other words, it predicts only 1.4% of DFL residues with the cost of predicting 3.9% of NDFL residues as DFL residues. This is because our test dataset is designed to share low (25% or lower) sequence similarity with the training dataset. In contrast, DFLpred can produce high-quality results even in the absence of sequence similarity. When considering the same FPR = 0.039, DFLpred’s TRP is 10 times higher and equals 0.148. Moreover, DFLpred’s ROC curves are substantially above the curves of other methods over the entire range of FPR values. We also note a large improvement for the low FPR values (see insert in Fig. 3).
3.3 Relation between predicted propensity and predictive quality
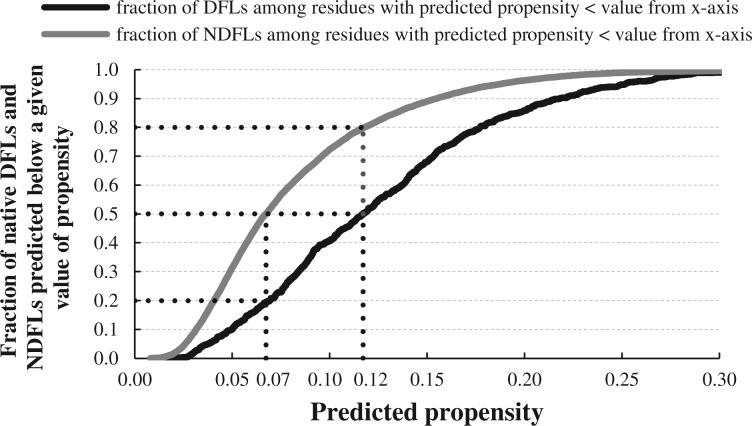
DFLpred outputs real-valued propensities for each residue in the input protein chain where higher values of propensity denote higher likelihood that a given residue is a part of a DFL. Figure 4 shows relation between the values of propensity and the predictive quality by calculating fraction of residues in native DFLs (black line) and native NDFLs (gray line), which are predicted with propensities below a value shown on the x-axis. Larger vertical separation between the two lines corresponds to a better separation between DFLs and NDFLs by outputs produced with DFLpred. The propensities generated by DFLpred are constrained to the 0–0.3 range and offer a large separation. We picked two representative values of propensities (vertical dotted lines in Fig. 3) to demonstrate how to interpret DFLpred’s predictions. Half of the NDFLs are found among the residues predicted with propensity < 0.07, while these residues include only about 20% of DFLs. Similarly, half of DFLs and only 20% of NDFLs are found among the residues for which the predicted propensity >0.12.
Fig. 4.
Relation between propensities generated by DFLpred and predictive quality on the test dataset. The predictive quality is quantified by fraction of residues in native DFLs (black line) and native NDFLs (gray line), which are predicted with propensities below a value shown on the x-axis
3.4 Runtime
DFLpred is implemented as a linear function of features computed directly from sequence and from sequence-derived predictions generated with IUPred. IUPred’s predictions are calculated from pairwise energy profile without a time-consuming alignment or predictive model. Consequently, DFLpred is fast.
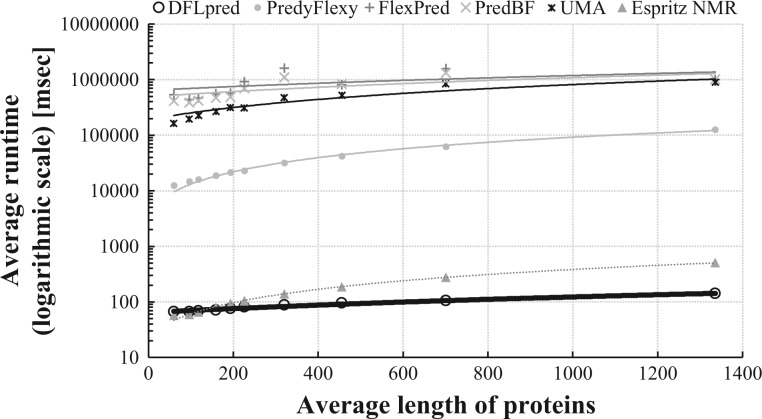
We quantify and compare DFLpred’s runtime with the runtime of UMA and all individual methods that obtained AUC > 0.5 in Table 2 or Supplementary Table S2. The predictions were run on the same 64-bit computer with 3.5 GHZ CPU and 4 GB of RAM running Ubuntu operating system. UMA is run manually and requires computation of hydrophobicity, finding homologous sequences and prediction of secondary structure. We estimate a lower bound on the UMA’s runtime by computing time to complete the most time-consuming finding of homologs. This is based on executing BLAST against the NR database using the suggested by the author e-value = 1e-20. We used stand-alone version of PredyFlexy that was provided by the authors. FlexPred and PredBF do not provide stand-alone versions but both methods use PSI-BLAST to generate position-specific scoring matrix against the NR database. We use this calculation to estimate lower bounds of their runtime. As suggested by the authors, we compute the runtime of running PSI-BLAST with five iterations and default e-value for FlexPred, and with three iterations and e-value = 0.001 for PredBF. We did not include ThreaDom because it requires running LOMETS (Local Meta-Threading-Server) framework, which takes substantially longer time than the other methods. We collected the runtime of the considered methods for 204 proteins from the training and test datasets. We sorted the proteins by their size quantified with the sequence length, divided them into 10 equally sized groups based on the size and computed average runtime for each group.
Figure 5 compares the average runtime against the average length of sequences for the five methods. The runtime of DFLpred, Espritz_NMR, PredyFlexy, UMA, FlexPred and PredBF is in the range of 102, 102 to 103, 104 to 105, 105 to 106 and 106, respectively, measured in milliseconds. DFLpred is up to 4 orders of magnitude faster than the alternatives, which is a significant advantage. To put this into perspective, if these methods would be used to predict the complete reviewed human proteome from UniProt (20 193 sequences with an average length of 561), DFLpred, ESpritz, PredyFlexy, UMA, FlexPred and PredBF would take about 40 min, 80 min, 11.5 days, 185 days, 231.5 days and 231.5 days, respectively. This estimate is based on a linear fit into the measured data that are shown in Figure 5 and assuming use of the same computer that we used to measure the runtime. The measured runtime of DFLpred on the human proteomes using this computer was 38 min, which is close to the estimated 40 min and which demonstrates that our estimates are accurate. To summarize, DFLpred is faster than the less accurate alternatives and is capable of providing predictions for the complete human proteome (and any other proteome, which by definition, would be smaller) using a modern personal computer in under an hour.
Fig. 5.
Comparison of runtime in the function of the length of the input protein sequences for UMA and all individual methods (except ThreaDom) that secured AUC > 0.5 based on the evaluations on the entire test dataset or separately for the inter- and intra-domain residues. The runtime is shown using base 10 logarithmic scale. Lines denote linear fits into the measured data that are shown using markers
3.5 Analysis of putative DFLs in human proteome
We analyzed putative annotations of DFLs generated with DFLpred in the complete reviewed human proteome collected from UniProt. We considered a given residue to form DFL is its propensity generated by DFLpred > 0.18. This cutoff corresponds to low 0.05 FPR based on the results from the cross-validation on the training dataset.
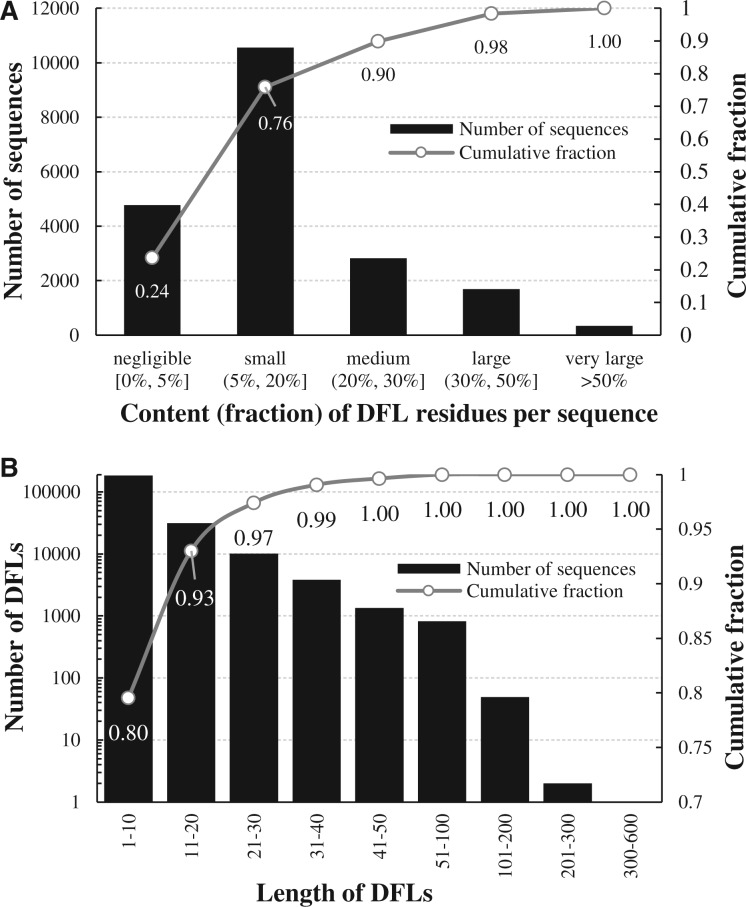
Figure 6A shows a histogram of the content of putative DFLs residues per sequence (fraction of these residues in a sequence). About 24% of proteins have no DFLs, i.e. the content is <5%, while our estimated FPR is at the same level, and another 52% have small amount of DFL residues. About 10% and 1.8% of proteins have the content >30% and >50%, respectively. We found 341 and 152 proteins that have the content of DFL residues at >50% and 60%, respectively.
Fig. 6.
Histograms with content of putative DFL residues per sequence (panel A) and length of putative DFLs (panel B) found with DFLpred in the complete reviewed human proteome. Black bars show the value of fraction (panel A) and length (panel B) and lines show the cumulative fraction for a given range of the content (panel A) and length (panel B)
Figure 6B is a histogram of the length of putative DFLs. Most of these regions are relatively short, with about 80% of them being shorter than 10 residues; some of them could be spurious predictions given the assumed 5% FPR. However, about 7% and 2.6% of these regions span at least 20 and 30 consecutive residues, respectively. We found 6029 DFL regions that that are at least 30 residues long.
3.6 Webserver
DFLpred is freely available as a webserver at http://biomine.ece.ualberta.ca/DFLpred. It requires the end user only to provide the input protein sequence(s) in FASTA format and email address. The email is used to deliver a notification of the finished prediction and URL of results that are available for download. The same information is available in the browser window given that this window will not be closed until the prediction is finished. The server automatically generates the corresponding propensities and binary predictions (DFL versus NDFL residue). The binary predictions are computed from the propensities using the cutoff = 0.18 (residues with propensity > 0.18 are assumed to form DFLs), which corresponds to the 5% FPR. The webserver allows for batch predictions of datasets with up to 5000 proteins.
4 Summary
We conceptualized, designed, tested and deployed a novel computational method, DFLpred, for the prediction of the DFLs from protein sequences. We developed four strong and complementary sequence-derived markers of DFLs and combined them using a linear function to build DFLpred. Empirical tests on independent (blind) test dataset demonstrated that our method provides relatively accurate predictions even for proteins that share low sequence identity. DFLpred outperformed the closest related methods including UMA, which predicts flexible linkers, several protein flexibility predictors and their combinations with disorder predictors. We note that the value of AUC = 0.72 secured by DFLpred is affected by the challenging nature of the dataset (proteins in this dataset share low, <25%, identity with the training sequences) and the fact that the annotations of DFLs could be incomplete. In other words, results on proteins that share larger sequences similarity with our training dataset should be better and some of the false positives in our test dataset could in fact corresponds to DFLs that are yet to be annotated. Another potential issue is the precision with which the boundaries of DFLs are annotated. This is shown in our first example (Supplementary Fig. S2A) where the boundaries of domains are slightly misaligned with the boundaries of DFLs. This will result in annotation noise that reduces the achievable limit of predictive performance. Our prediction is also characterized by a low runtime, with prediction of the entire proteome taking <1 h on a modern desktop computer. Finally, our analysis of putative DFLs in human proteome generated with DFLpred shows that DFLs likely can be found in many human proteins. About 10% of human proteins have a significant content of >30% of DFL residues and a few thousand of these regions are >30 consecutive residues.
Funding
This work was supported in part by the Discovery grant (298328) from the Natural Sciences and Engineering Research Council (NSERC) of Canada and by the Qimonda Endowed Chair to L.K. and by a scholarship from the China Scholarship Council to F.M.
Supplementary Material
References
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S., Mohanty D. (2012) Inter-domain movements in polyketide synthases: a molecular dynamics study. Mol. Biosyst., 8, 1157–1171. [DOI] [PubMed] [Google Scholar]
- Anderson T.W., Darling D.A. (1952) Asymptotic theory of certain “goodness of fit” criteria based on stochastic processes. Ann. Math. Stat., 23, 193–212. [Google Scholar]
- Attwood T.K. et al. (2012) The PRINTS database: a fine-grained protein sequence annotation and analysis resource—its status in 2012. Database, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora R., Rosee G.D. (1998) Helix capping. Protein Sci., 7, 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D.W.A. et al. (2013) Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res., 41, W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. (2013) Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev., 65, 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia E. et al. (2013) From protein sequence to dynamics and disorder with DynaMine. Nat. Commun., 4, 2741. [DOI] [PubMed] [Google Scholar]
- Cilia E. et al. (2014) The DynaMine webserver: predicting protein dynamics from sequence. Nucleic Acids Res., 42, W264–W270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brevern A.G. et al. (2012) PredyFlexy: flexibility and local structure prediction from sequence. Nucleic Acids Res., 40, W317–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disfani F.M. et al. (2012) MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics, 28, i75–i83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z. et al. (2005) The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol., 347, 827–839. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z. et al. (2009) ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics, 25, 2745–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K. et al. (2002) Intrinsic disorder and protein function†. Biochemistry, 41, 6573–6582. [DOI] [PubMed] [Google Scholar]
- Dunker A.K. et al. (2008) Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol, 18, 756–764. [DOI] [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol, 6, 197–208. [DOI] [PubMed] [Google Scholar]
- Fang C. et al. (2013) MFSPSSMpred: identifying short disorder-to-order binding regions in disordered proteins based on contextual local evolutionary conservation. BMC Bioinformatics, 14, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D. et al. (2014) Pfam: the protein families database. Nucleic Acids Res., 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R.A., Heringa J. (2002) An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng., 15, 871–879. [DOI] [PubMed] [Google Scholar]
- Goujon M. et al. (2010) A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res., 38(Suppl 2), W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov I.B., McDuffie M. (2008) FlexPred: a web-server for predicting residue positions involved in conformational switches in proteins. Bioinformation, 3, 134–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Cozzetto D. (2014) DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics, 31, 857–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S. et al. (2008) AAindex: amino acid index database, progress report 2008. Nucleic Acids Res., 36(Suppl 1), D202–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan W. et al. (2013) Predicting binding within disordered protein regions to structurally characterised peptide-binding domains. PLoS One, 8, e72838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov I.B. (2008) Ordered conformational change in the protein backbone: prediction of conformationally variable positions from sequence and low-resolution structural data. Proteins, 72, 74–87. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Malhis N., Gsponer J. (2015) Computational identification of MoRFs in protein sequences. Bioinformatics, 31, 1738–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B. et al. (2009) Prediction of protein binding regions in disordered proteins. PLoS Comput Biol, 5, e1000376.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res., 43, D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizianty M.J. et al. (2010) Improved sequence-based prediction of disordered regions with multilayer fusion of multiple information sources. Bioinformatics, 26, i489–i496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield C.J., Dunker A.K. (2014) Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem., 83, 553–584. [DOI] [PubMed] [Google Scholar]
- Palau J. et al. (1982) Protein secondary structure. Studies on the limits of prediction accuracy. Int. J. Pept. Protein Res., 19, 394–401. [PubMed] [Google Scholar]
- Pan X.Y., Shen H.B. (2009) Robust prediction of B-factor profile from sequence using two-stage SVR based on random forest feature selection. Protein and Pept. Lett., 16, 1447–1454. [DOI] [PubMed] [Google Scholar]
- Peng Z., Kurgan L. (2015) High-throughput prediction of RNA, DNA and protein binding regions mediated by intrinsic disorder. Nucleic Acids Res., 43, e121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z. et al. (2014) A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell. Mol. Life Sci., 71, 1477–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z. et al. (2015) Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell. Mol. Life Sci., 72, 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z.L., Kurgan L. (2012) Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci., 13, 6–18. [DOI] [PubMed] [Google Scholar]
- Radivojac P. et al. (2007) Intrinsic disorder and functional proteomics. Biophys. J., 92, 1439–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. (1993) Prediction of Protein Secondary Structure at Better than 70% Accuracy. J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–72. [DOI] [PubMed] [Google Scholar]
- Schlessinger A., Rost B. (2005) Protein flexibility and rigidity predicted from sequence. Proteins, 61, 115–126. [DOI] [PubMed] [Google Scholar]
- Schlessinger A. et al. (2006) PROFbval: predict flexible and rigid residues in proteins. Bioinformatics, 22, 891–893. [DOI] [PubMed] [Google Scholar]
- Servant F. et al. (2002) ProDom: Automated clustering of homologous domains. Brief. Bioinform., 3, 246–251. [DOI] [PubMed] [Google Scholar]
- Shvadchak V.V., Subramaniam V. (2014) A four-amino acid linker between repeats in the alpha-synuclein sequence is important for fibril formation. Biochemistry, 53, 279–281. [DOI] [PubMed] [Google Scholar]
- Sickmeier M. et al. (2007) DisProt: the database of disordered proteins. Nucleic Acids Res., 35(Suppl 1), D786–D793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C.J.A. et al. (2013) New and continuing developments at PROSITE. Nucleic Acids Res., 41, D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S. et al. (2003) Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature, 424, 35–41. [DOI] [PubMed] [Google Scholar]
- Thompson J.D. et al. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. (2005) The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett., 579, 3346–3354. [DOI] [PubMed] [Google Scholar]
- Udwary D.W. et al. (2002) A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J. Mol. Biol., 323, 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh I. et al. (2012) ESpritz: accurate and fast prediction of protein disorder. Bioinformatics, 28, 503–509. [DOI] [PubMed] [Google Scholar]
- Ward J.J. et al. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol., 337, 635–645. [DOI] [PubMed] [Google Scholar]
- Wilcoxon F. (1945) Individual comparisons by ranking methods. Biom. Bullet., 80–83. [Google Scholar]
- Wootton J.C. (1994) Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem., 18, 269–285. [DOI] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol., 293, 321–331. [DOI] [PubMed] [Google Scholar]
- Xie H. et al. (2007) Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res., 6, 1882–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z. et al. (2013) ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics, 29, i247–i256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. et al. (2015) Molecular Recognition Features (MoRFs) in three domains of life. Mol. Biosyst., 12, 697–710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.