Abstract
Xylella fastidiosa is a phytopathogenic bacterium that causes serious diseases in a wide range of economically important crops. Despite extensive comparative analyses of genome sequences of Xylella pathogenic strains from different plant hosts, nonpathogenic strains have not been studied. In this report, we show that X. fastidiosa strain J1a12, associated with citrus variegated chlorosis (CVC), is nonpathogenic when injected into citrus and tobacco plants. Furthermore, a DNA microarray-based comparison of J1a12 with 9a5c, a CVC strain that is highly pathogenic and had its genome completely sequenced, revealed that 14 coding sequences of strain 9a5c are absent or highly divergent in strain J1a12. Among them, we found an arginase and a fimbrial adhesin precursor of type III pilus, which were confirmed to be absent in the nonpathogenic strain by PCR and DNA sequencing. The absence of arginase can be correlated to the inability of J1a12 to multiply in host plants. This enzyme has been recently shown to act as a bacterial survival mechanism by down-regulating host nitric oxide production. The lack of the adhesin precursor gene is in accordance with the less aggregated phenotype observed for J1a12 cells growing in vitro. Thus, the absence of both genes can be associated with the failure of the J1a12 strain to establish and spread in citrus and tobacco plants. These results provide the first detailed comparison between a nonpathogenic strain and a pathogenic strain of X. fastidiosa, constituting an important step towards understanding the molecular basis of the disease.
Xylella fastidiosa is a gram-negative bacterium, limited to the plant xylem vessels, which is responsible for worldwide economic losses due to diseases caused in a variety of plants of agricultural relevance. This bacterium is transmitted to new host plants during xylem sap feeding by insect vectors and spreads from the site of infection to colonize the xylem, a water transport network of vessels composed of lignified dead cells. Bacterial cells attach to the vessel wall, forming biofilm-like colonies that, depending on the size, can occlude the xylem vessels, blocking water transport and causing water stress symptoms (22, 35).
Different strains of X. fastidiosa have been reported to infect a wide range of plants, including grapevines and citrus, almond, and pear trees, among others (26). In the United States for instance, Pierce's disease prevents profitable viticulture if leafhopper vectors are present at high densities (1, 14). In Brazil, citrus variegated chlorosis (CVC) is responsible for great financial losses to the citrus agroindustry, being detected in one-third of the citrus trees. Orange production quickly decreases in orchards affected by CVC, as fruits become hardened and of no commercial value. Interestingly, within the majority of host plants, X. fastidiosa behaves as a harmless endophyte (27).
Several X. fastidiosa strains have had their genomes completely or partially sequenced, and genome comparative analysis with different pathogenic strains of X. fastidiosa pointed to common candidate virulence determinants as well as strain-specific genomic signatures (4, 24, 32, 37). However, no information is available about the genome composition of nonpathogenic Xylella strains, which would contribute to more direct insights on pathogenicity mechanisms.
Genome-wide comparison between pathogenic and nonpathogenic strains within a species is a useful strategy for identifying candidate genes important for virulence. DNA microarray-based genome composition analysis is a good alternative to full genome sequencing and has been used in comparative studies to analyze various bacterial pathogens including Mycobacterium tuberculosis (3), Helicobacter pylori (29), Pseudomonas aeruginosa (38), Bacillus anthracis (28), Yersinia pestis, and Yersinia pseudotuberculosis (12).
In this report, we show that X. fastidiosa strain J1a12, which was isolated from citrus and is suitable for genetic transformation (6, 23), elicits few or no CVC symptoms when inoculated into citrus and tobacco plants. Furthermore, a DNA microarray-based genome composition analysis was performed by comparing J1a12 with strain 9a5c, which produces typical CVC symptoms (20) but is resistant to transformation with DNA in vitro (23), a drawback for its genetic manipulation. Our microarray data revealed that the great majority of the coding sequences (CDS) are highly conserved on both strains. However, 14 CDS were shown to be absent or highly divergent in the nonpathogenic strain. Expression profiling of both strains, PCR and reverse transcription (RT)-PCR with CDS-specific primers, and DNA sequence analysis were used to validate the genomic differences observed.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and pathogenicity tests.
Triply cloned X. fastidiosa strains 9a5c (20) and J1a12 (23) isolated from CVC symptomatic Citrus sinensis (L.) Osbeck trees (sweet orange) were grown in periwinkle wilt broth medium (7) at 28°C in the dark with 100 rpm rotatory agitation. As shown in Table 1, strain J1a12 is nonpathogenic, despite its isolation from a citrus plant with CVC symptoms. This is possibly due to a mixed population of bacteria infecting the plant (see “Final remarks” below). A culture started from a single colony was weekly passaged through serial transfers at a 1/100 dilution in periwinkle wilt medium. For citrus plant experiments, 9a5c and J1a12 strains with 9 and 24 weekly passages, respectively, were used; for tobacco plant experiments, 11 weekly passages were used for both strains. Mechanical inoculation of plants was performed essentially as described in reference 20 for C. sinensis and as described in reference 21 for Nicotiana tabacum (accession clevelandii). Further details are found in the supplemental material. The detection of X. fastidiosa in host plants followed the procedure described in reference 25, which consists of PCR experiments with a pair of primers (CVC-1 and 272-int) designed to identify Xylella strains isolated from citrus plants.
TABLE 1.
Evaluation of C. sinensis and N. tabacum plants inoculated with X. fastidiosa CVC strains 9a5c and J1a12
| Strain | Test | No. of infected plants/total no. of plants at time (mo) postinoculation
|
||||
|---|---|---|---|---|---|---|
|
C. sinensis
|
N. tabacum
|
|||||
| 5 | 8 | 15 | 1.5 | 3 | ||
| 9a5c | PCRa | 8/14 | 14/14 | 13/13b | 5/5 | 5/5 |
| Symptom evaluation | 0/14 | 3/14 | 10/13b | 5/5 | 5/5 | |
| J1a12 | PCR | 0/14 | 1/14 | 0/14 | 0/5 | 0/5 |
| Symptom evaluation | 0/14 | 0/14 | 0/14 | 0/5 | 0/5 | |
PCR experiments were performed on samples drawn from the plant xylem by using primers designed to identify Xylella citrus isolates (25).
One of the plants died 15 months after inoculation.
Microarray construction and hybridization.
A 6,152-element DNA microarray was printed containing unique internal fragments of 2,692 CDS spotted at least in duplicate, representing 94.5% of all of the 2,848 CDS annotated by Simpson et al. (32). DNA fragments ranging from 200 to 1,000 bp were PCR amplified with CDS-specific primers (18- to 23-mers) designed with PRIMER3 (http://www-genome.wi.mit.edu/genome_software/other/primer3.html) and based on the annotated genome sequence of X. fastidiosa strain 9a5c (http://aeg.lbi.ic.unicamp.br/xf). A full list of primers, PCR product sizes, and their nucleotide sequences are available at the project site (http://verjo19.iq.usp.br/xylella/microarray/). The arrays were hybridized with DNA fragments from strain 9a5c in combination with itself, J1a12, or grapevine strain Temecula (kindly provided by Marie-Anne Van Sluys, University of São Paulo) labeled separately with either Cy3- or Cy5-dCTP analogs (see the supplemental material). Expression profiling studies were carried out by labeling total RNA with the Cy-Scribe post labeling kit (Amersham Biosciences) according to the manufacturer's instructions.
Data acquisition and normalization.
Microarray slides were scanned by using a Generation III DNA scanner (Amersham Biosciences), and fluorescence intensity values (ICy3 and ICy5) from each spot were extracted by using ArrayVision, version 6.0, software (Imaging Research, Inc.). Raw fluorescence intensity and normalized data are available at the project site (http://verjo19.iq.usp.br/xylella/microarray/). Data normalization was carried out by LOWESS fitting on an M versus A plot (39), where M is the ratio of fluorescence intensities of the two measurements for each spot [defined as M = log2(ICy5/ICy3)] and A is the geometric mean of the fluorescence intensities [defined as A = 1/2 × log2(ICy5 × ICy3)]. The normalization script is available at the project site.
CDS classification process.
To determine hybridization noise and to estimate dynamic cutoff values for classifying a CDS as equally present in both strains (9a5c and J1a12) we used the hybridization data collected from three independent homotypic experiments (9a5c versus 9a5c). For this kind of experiment, also called self-self hybridization, the microarrays were cohybridized with strain 9a5c DNA separately labeled with either Cy3- or Cy5-dCTP analogs. As verified in the M versus A plot, there is a dependence of the hybridization intensity log ratio of each spot (M) with the mean log intensity of each spot (A). Thus, we have determined a cutoff value for each interval in the A axis by using kernel density estimators. We chose kernel density estimators (31) instead of the normal probability density function (16) because we experimentally derived the null distribution as the result of homotypic experiments and verified that it does not present a Gaussian behavior (further information is available at the project site [http://verjo19.iq.usp.br/xylella/microarray/]). The density distribution was integrated around the mode peak until 0.995 probability was reached, defining intensity-dependent noise threshold cutoff values (credibility intervals) based on experimental data from 9a5c versus 9a5c homotypic experiments, thus setting an interval where the hybridization ratio is considered to be 1:1 (e.g., −2.5 < M < 1.8, for the lowest accepted intensities at A = 2). These credibility intervals were subsequently used in the analysis of replicas of 9a5c versus J1a12 hybridization experiments to nonparametrically estimate the null distribution of the statistical test H0: CDS is present in both J1a12 and 9a5c strains. Spots outside the credibility intervals present strong evidence against a 1:1 ratio. Using these criteria, four categories were defined for the CDS in the J1a12 genome based on its orthologous 9a5c counterpart: (i) equally present in both strains, (ii) divergent in strain J1a12, (iii) highly divergent or absent in J1a12, and (iv) higher copy number in J1a12. Category i includes all of the CDS for which ≥60% of the replicas were inside the credibility intervals.
CDS presenting negative log ratio values outside the credibility intervals could be classified as divergent in strain J1a12 (category ii) or highly divergent or absent in J1a12 (category iii). To distinguish between these categories, we performed four control experiments where the microarrays were cohybridized with DNA from the sequenced grapevine strain Temecula (37) and strain 9a5c labeled with either Cy3- or Cy5-dCTP analogs. Next, we derived a correspondence between the hybridization intensity log ratio for each CDS amplicon and its respective nucleotide sequence identity in both strains. Amplicon sequences exhibiting nucleotide identities smaller than 20% were defined as category iii, highly divergent or absent. Their respective log ratio M was taken as the cutoff threshold (Mcutoff) of category iii. The log ratio value with the least false callings was an Mcutoff value of −1.7. At this threshold, 19 false positives and 23 false negatives were observed (0.76 and 0.92%, respectively). Category iii includes CDS with an M value of <−1.7 and a P value smaller than 0.05 in a t test against the null hypothesis H0: M ≥ −1.7. Sequencing some amplicons from J1a12 that were outside the credibility intervals and checking the divergence between 9a5c and J1a12 sequences further supported the adequacy of the threshold. The remaining CDS that have a negative log ratio value, with an M value of ≥−1.7, were considered divergent (category ii). Those CDS with positive log ratio values were considered to be present at higher copy numbers in J1a12 (category iv). CDS with low reproducibility (less than 60% of replicas in a single category) were excluded from the analysis.
Validation of microarray data.
PCR and RT-PCR experiments were performed with CDS-specific primers to further investigate the status of CDS in the genome of strain J1a12 classified by using the microarray data. The reactions were carried out with genomic DNA or cDNA from strain 9a5c or J1a12 with 35 cycles of amplification. A sample of CDS presenting log ratios outside the credibility intervals, as determined by homotypic hybridization experiments (9a5c versus 9a5c), was chosen to perform the validation. Among the 64 CDS with negative log ratios (categories ii and iii), the following 33 CDS were randomly chosen for PCR validation: XF0077, XF0496, XF0497, XF0500, XF0663, XF0684, XF0696, XF0890, XF1250, XF1306, XF1581, XF1588, XF1589, XF1646, XF1663, XF1664, XF1686, XF1708, XF1709, XF1860, XF1863, XF1874, XF1877, XF1878, XF1884, XF1968, XF2193, XF2195, XF2307, XF2722, XF2768, XF2772, and XFb0001. In addition, the following CDS with positive log ratios (category iv) were also tested by PCR: XF0513, XF0514, XF0515, XF0516, XF0518, XF0519, XF0521, XF1933, XF1934, XF1935, XF1936, and XF1937. A 4-μl sample of each reaction mixture was electrophoresed in agarose gels, and DNA was stained with ethidium bromide. The amplicons were then classified by visual inspection as absent, same copy number, or more abundant in strain J1a12 in relation to strain 9a5c. In addition, DNA sequence determination was carried out for a few CDS. For that, PCR products obtained with primers based on neighboring CDS were cloned in pGEM-TEasy vector (Promega) and dideoxy sequencing reactions were performed with 100 ng of plasmid DNA in Big Dye Terminator sequencing reactions (Applied Biosystems) according to the manufacturer's instructions. Sequencing reactions were carried out with either CDS-specific or T7 promoter primers.
RESULTS AND DISCUSSION
X. fastidiosa CVC strain J1a12 is nonpathogenic.
In planta pathogenicity tests were carried out with strains 9a5c and J1a12, and results are presented in Table 1. Inoculation of citrus plants with strain 9a5c resulted in multiplication of X. fastidiosa in all plants analyzed 8 months after infection. Plants were also evaluated visually for characteristic CVC symptoms, and positive symptoms were observed in the leaves of 77% of the citrus plants 15 months after inoculation. In contrast, no symptoms were observed in the leaves of plants inoculated with strain J1a12 up to 15 months after infection.
Similar results were observed with tobacco plants as the experimental host. As shown in Table 1, none of the plants inoculated with J1a12 presented symptoms or were colonized by the bacteria. In contrast, all tobacco plants inoculated with 9a5c presented the lesions characteristic of X. fastidiosa CVC infection, as previously described (21).
These results indicate that strain J1a12 shows a nonpathogenic phenotype, in contrast to the highly pathogenic behavior of strain 9a5c. In addition, Table 1 shows that plant colonization by strain J1a12 is very inefficient, as no bacteria were detected in the plant xylem by PCR experiments with a pair of primers (CVC-1 and 272-int) which are specific for Xylella isolates from citrus plants (25). This pair of primers amplifies a genomic region of approximately 500 bp, from chromosome position 1051239 to 1051745 of the 9a5c genome, encompassing 195 bp of CDS XF1100 and an intergenic region upstream of this CDS. It is important to stress that these primers can amplify the correct DNA fragment when directly tested in J1a12 in vitro cultures (6, 23).
Genotyping by DNA microarray analysis.
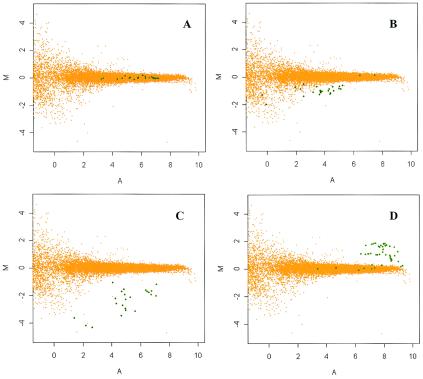
To investigate whether the differences in phenotype observed between strains 9a5c and J1a12 could be associated with differences at the DNA level, we have constructed a DNA microarray encompassing 2,692 CDS, which represents 94.5% of all CDS described in the reference strain 9a5c (32). Total DNAs isolated from strains 9a5c and J1a12 were separately labeled with either Cy3- or Cy5-dCTP fluorescent analogs, and competitive microarray hybridizations were performed. Raw and normalized hybridization data are available at the project site (http://verjo19.iq.usp.br/xylella/microarray/). An initial screening revealed that 292 CDS presented either low signal intensity or poor reproducibility and were excluded from further analysis. As detailed in Material and Methods, the remaining 2,400 CDS were classified into four categories according to the normalized hybridization fluorescence intensity ratios of J1a12 over 9a5c DNA samples determined for each CDS. Among the 2,400 CDS, approximately 96% were found to have a log ratio (M) around 0 and were classified as equally present in both strains (category i). One example is shown in Fig. 1A. This figure shows an M versus A plot, i.e., normalized intensities log ratios (M) versus the average of log intensities (A) of all the replicas for a given CDS. This kind of graph shows the dependence of the ratio on the overall intensity of each spot, indicating that, for genes with low hybridization intensity signals (A values below 2), the observed ratios have a higher intrinsic dispersion, as determined by homotypic hybridizations. As a result, different cutoff values for M were used for different ranges of intensity (A) when classifying a CDS as equally present in both strains. M versus A plots showing the reproducibility of the data for each CDS are available at the project site.
FIG. 1.
Xylella strain J1a12 CDS classification based on DNA microarray hybridization ratios. Examples of CDS classified in each of the four categories are shown. (A) XF1621, classified as equally present in both 9a5c and J1a12 strains, category i; (B) XF0262, classified as divergent in J1a12, category ii; (C) XF0496, classified as absent or highly divergent in strain J1a12, category iii; (D) XF1937, classified as higher copy number in J1a12, category iv. Orange dots represent the results for all CDS in the microarray from three homotypic control experiments (9a5c labeled with Cy5 versus 9a5c labeled with Cy3). Graphs show the fluorescence intensity ratio M = log2(ICy5/ICy3) versus the fluorescence intensity mean A = 1/2 × log2(ICy5 × ICy3). Green dots represent the hybridization data (9a5c labeled with Cy3 versus J1a12 labeled with Cy5 or vice versa) from multiple replicas for the indicated CDS, where the fluorescence intensity ratio M = log2(IJ1a12/I9a5c). Similar graphs for each of the CDS are available at the project site (http://verjo19.iq.usp.br/cagexylella/private/).
Fifty CDS were found to have a hybridization intensity log ratio of −1.7 < M < −0.3 and were classified as divergent in strain J1a12 (category ii, an example is shown in Fig. 1B). Table 2 lists only the CDS with hybridization intensity log ratios between −0.5 and −1.7. For the complete list of CDS in this category see Table S1 in the supplemental material. Fourteen CDS were found to have the log ratios of <−1.7 and were classified as absent or highly divergent in strain J1a12 (Table 3). A typical example is shown in Fig. 1C. Within this group, three CDS, namely XF0077, XF1250, and XF1646, were especially interesting due to their putative involvement in pathogenesis and will be discussed in more detail later. Most of the CDS in this category belong to the previously defined flexible gene pool of Xylella, which includes integrated prophages and genomic islands (4, 24). For instance, CDS XF1860, XF1874, XF1878, and XF1884 belong to genomic island 4 (24). This region has a different GC content and altered codon bias, which are common features of laterally transferred elements (15). However, CDS XF0077, XF1250, XF1646, XF1707, and XF1708 are not mapped within any genomic island and do not show altered GC content or codon bias.
TABLE 2.
CDS divergent in strain J1a12
| Genea | Product |
|---|---|
| XF0075 | Hypothetical protein |
| XF0078 | Fimbrial adhesin precursor |
| XF0157 | Hypothetical protein |
| XF0262 | Colicin V precursor |
| XF0263 | Colicin V precursor |
| XF0500 | Phage-related repressor protein |
| XF0501 | Conserved hypothetical protein |
| XF0626 | Hypothetical protein |
| XF0663 | Hypothetical protein |
| XF0665 | Hypothetical protein |
| XF0666 | Hypothetical protein |
| XF0668 | Hemolysin-type calcium binding protein |
| XF0684 | Phage-related protein |
| XF0696 | Phage-related repressor protein |
| XF1057 | Hypothetical protein |
| XF1306 | Hypothetical protein |
| XF1588 | Hypothetical protein |
| XF1589 | Plasmid stabilization protein |
| XF1590 | Plasmid stabilization protein |
| XF1609 | Glucose/galactose transporter |
| XF1664 | Hypothetical protein |
| XF1746 | Alcohol dehydrogenase |
| XF1756 | Hypothetical protein |
| XF1758 | Hypothetical protein |
| XF1851 | Serine protease |
| XF1859 | Hypothetical protein |
| XF1862 | Conserved hypothetical protein |
| XF1873 | Conserved hypothetical protein |
| XF1877 | Hypothetical protein |
| XF1883 | Hypothetical protein |
| XF1968 | Methyltransferase |
| XF2193 | Hypothetical protein |
| XF2194 | Hypothetical protein |
| XF2195 | Hypothetical protein |
| XF2217 | Imidazoleglycerolphosphate dehydratase/histidinol-phosphate phosphatase/bifunctional enzyme |
| XF2307 | Hypothetical protein |
| XF2406 | Hypothetical protein |
| XF2407 | Bacteriocin |
| XF2542 | Fimbrial protein |
| XF2722 | Type I restriction-modification system specificity determinant |
| XF2726 | Type I restriction-modification system specificity determinant |
| XF2768 | Hypothetical protein |
| XF2770 | Hypothetical protein |
| XF2772 | Hypothetical protein |
Only 44 CDS with log hybridization intensity ratios between −1.7 and −0.5 are shown in this table.
TABLE 3.
CDS absent or highly divergent in strain J1a12
| Genea | Product | Mb |
|---|---|---|
| XF0077 | Fimbrial adhesion precursor | −2.77 |
| XF0496 | Conserved hypothetical protein | −2.31 |
| XF0497 | Conserved hypothetical protein | −2.08 |
| XF0667 | Hypothetical protein | −1.99 |
| XF1250 | Arginine deaminase | −1.96 |
| XF1646 | UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase | −2.01 |
| XF1707 | Hypothetical protein | −2.30 |
| XF1708 | Conserved hypothetical protein | −2.22 |
| XF1860 | Hypothetical protein | −2.79 |
| XF1874 | Hypothetical protein | −2.13 |
| XF1878 | Hypothetical protein | −3.11 |
| XF1884 | Hypothetical protein | −2.21 |
| XFb0001 | Replication protein | −3.55 |
| XFb0002 | Hypothetical protein | −4.18 |
CDS shown here have a P value smaller than 0.05 in a t test for the null hypothesis H0: M ≥ −1.7.
DNA hybridization intensity ratios [M = log2(IJ1a12/I9a5c)] were calculated. For each gene, the values shown are the means of the results from at least eight replicates.
In addition, 40 CDS (see Table S2 in the supplemental material) were found to have a log ratio of >0 and were considered as possibly presenting a higher number of copies in J1a12 (an example is shown in Fig. 1D). The majority of the CDS in this category are of unknown function or have phage-related functions. Among them, there are 2 groups of contiguous CDS (XF0512 to XF0523 and XF1932 to XF1937). The first set is within genomic island 1 (24). The other group includes genes that belong to different functional categories such as DNA metabolism and transport, suggesting them as possible newborn paralogs in the J1a12 genome.
Despite all the information obtained with DNA microarray genotyping, it is necessary to stress that frameshifts and point mutations cannot be identified by this method. In addition, our DNA microarray experiments will not detect genes present exclusively in the unsequenced strain J1a12, impairing detection of genes that would eventually attenuate pathogenicity.
Validation of CDS classification.
To further validate the distinction between CDS potentially absent or highly divergent and those classified as divergent, a sample of 33 CDS (listed in Materials and Methods) were analyzed by PCR with CDS-specific primers. All CDS tested were PCR negative for strain J1a12, including those with log hybridization intensity ratios between −0.5 and −1.7.
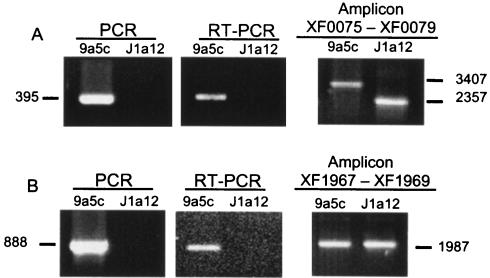
Figure 2A shows an example of a CDS (XF0077, encoding a fimbrial adhesin precursor) classified as absent or highly divergent by microarray analysis. PCR and RT-PCR experiments with CDS-specific primers corroborated its classification in this category. In addition, a PCR experiment with a pair of specific primers flanking XF0077 encompassing CDS XF0075 to XF0079 produced a smaller amplicon in strain J1a12 than in strain 9a5c (Fig. 2A). The exact position of the deleted region was determined by DNA sequence analysis, as described in the next section.
FIG. 2.
Validation of CDS classification. CDS-specific primers for XF0077 (A) and XF1968 (B) were employed to perform PCR amplifications with DNA from strains 9a5c and J1a12 (left panels) or to perform RT-PCR amplifications with total RNA (central panels). PCRs were also carried out with DNA of both strains and primers based on the sequence of the CDS flanking XF0077 (amplicon XF0075 to XF0079) or XF1968 (amplicon XF1967 to XF1969) (right panels). The sizes of the amplicons are shown in base pairs.
Data for XF1968, which encodes a putative methyltransferase classified as divergent by microarray analysis (mean log ratio of −1.4), is shown in Fig. 2B. The PCR and RT-PCR results with CDS-specific primers were negative, suggesting the absence of the CDS. However, PCR experiments with a pair of specific primers flanking XF1968 (encompassing XF1967 to XF1969) showed amplicons with the same size in both strains (Fig. 2B). Nucleotide sequencing of both amplicons confirmed that the methyltransferase encoded by XF1968 is divergent between 9a5c and J1a12 (Table 4).
TABLE 4.
Nucleotide and amino acid identity of selected divergent CDS of strain J1a12 compared to strain 9a5c
| Gene | Product | % Nucleotide identitya | % Amino acid identitya |
|---|---|---|---|
| XF0078 | Fimbrial adhesin precursor | 92.2 | 86.4 |
| XF1968 | Methyltransferase | 78.8 | 75.5 |
| XF2542 | Fimbrial protein | 73.6 | 65.8 |
| XF2726 | Type I restriction-modification system specificity determinant | 54.9 | 40.4 |
Comparison was carried out with the complete sequence of each CDS.
Three other examples of CDS classified as divergent according to microarray experiments and found to be PCR negative are also depicted in Table 4. Sequence analysis of these CDS cloned from strain J1a12 has confirmed their divergence, as they present nucleotide identities between 92 and 55% when compared to strain 9a5c (Table 4). These results show that PCR validation can be misleading, given that the primers used were based on the genome sequence of strain 9a5c. A few mismatches in the regions where the primers should anneal in the J1a12 DNA template can give PCR-negative results, leading to a wrong conclusion about the presence or absence of a gene.
RNA expression studies by microarray hybridization comparing strains J1a12 and 9a5c were performed as an additional validation of CDS classification. RNA hybridization data are available at the project site (http://verjo19.iq.usp.br/cagexylella/private/). Hybridization signals were found to be at the background level for the 14 CDS classified as absent or highly divergent in strain J1a12. On the other hand, all of these CDS showed significant expression levels in strain 9a5c.
The expression levels of CDS that were classified as equally present in both strains (category i, see Materials and Methods) were studied under standard bacterial growth conditions. This class of CDS was chosen to eliminate possible hybridization artifacts due to sequence divergence. We found that among the 2,296 CDS in this category, which presented detectable hybridization intensity values, approximately 97% exhibited comparable RNA expression levels on both strains, i.e., differences in expression were smaller than twofold, with P values smaller than 0.05 in a t test. However, about 1% of the CDS presented a higher RNA expression level (twofold or more) in strain 9a5c and about 2% had higher expression in strain J1a12 (see Tables S3 and S4 in the supplemental material). No obvious correlation could be made between the CDS presenting differential expression and the phenotypes of each strain.
Functional characteristics of genes absent or highly divergent in strain J1a12.
Among the 14 CDS classified as absent or highly divergent (Table 3), 10 have no similarity to known genes and no function could be assigned. Therefore, they will not be further discussed, although their involvement in pathogenesis cannot be excluded.
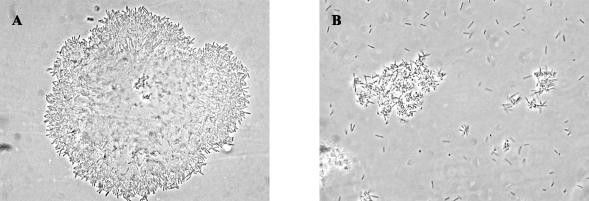
The X. fastidiosa strain 9a5c genome encodes three fimbrial adhesin subunits from type III pilus (XF0077, XF0078, and XF0080). Our microarray data classified XF0077 as absent or highly divergent, and we have confirmed its deletion in strain J1a12 by DNA sequence analysis. The deleted region, encompassing 1,050 nucleotides (Fig. 2A), extends from position 76845 to 77895 (numbers from the strain 9a5c main chromosome). XF0078 was classified as divergent in J1a12, and DNA sequence analysis has shown a 92.2% nucleotide identity with its ortholog in 9a5c (Table 4). The third fimbrial adhesin paralog (XF0080) was classified as equally present in both strains (category i). The three paralogs from 9a5c are similar to mrkD from Klebsiella pneumoniae and share high similarity with each other (XF0077 and XF0078 share 70% amino acid sequence identity and both display around 60% amino acid identity to XF0080). As reported for the adhesins of K. pneumoniae (30), the N-terminal region exhibits a greater degree of variability, probably conferring on strain 9a5c the ability to adhere to different bacterial and/or host cell components or even producing an extracellular matrix with greater bonding capacity. In K. pneumoniae, mrkD null mutants are fimbriate but nonadhesive (34) and the mrkD gene product is not required for biofilm formation (18). Proteomic and mass spectrometric analyses of whole-cell lysates and extracellular components have demonstrated that structural and adhesive subunits of fimbriae are ubiquitous in cultures of Xylella strain 9a5c (33). Despite the presence of two CDS encoding the adhesion subunit precursors XF0078 and XF0080 in J1a12, we have observed that this strain displays a much less aggregated phenotype in vitro than 9a5c cells, as shown in Fig. 3. One possible explanation for this phenotype is that the presence of the adhesin encoded by XF0077 is important for the adhesion of X. fastidiosa cells.
FIG. 3.
X. fastidiosa strain J1a12 has lost the ability to aggregate. Optical microscopy of X. fastidiosa strains 9a5c (A) and J1a12 (B). Magnification, ×1,000.
Interestingly, another CDS confirmed to be deleted from strain J1a12 is XF1250, which encodes a putative arginase. The deleted region of 963 nucleotides extends from position 1203200 to 1204163 (numbers from the strain 9a5c main chromosome). This enzyme catalyzes the first step of arginine degradation in the urea cycle, which may thus be incomplete in strain J1a12. Besides being a substrate for arginases, arginine is also a substrate for nitric oxide (NO) synthase, which converts l-arginine to l-citrulline, releasing NO. In H. pylori, it has recently been shown that the arginase (rocF) encoded by this bacterium inhibits NO production by macrophages at physiologic concentrations of l-arginine. H. pylori rocF mutants cocultured with macrophages were killed due to the restoration of normal levels of NO. These results indicate that bacterial arginase down-regulates NO production, acting as a survival mechanism that contributes to the successful infection of the host (10). Thus, it is tempting to speculate that the absence of arginase in X. fastidiosa strain J1a12 is linked to its reduced growth in planta and its incapacity to colonize the xylem vessels; J1a12 cells would not be able to inhibit NO production by the plant, analogous to the rocF mutant of H. pylori (10). In fact, it is known that in plants, NO has an immune protective role, mediating the plant defense against pathogens and serving as a signal in hormonal responses. Indeed, two NO-synthesizing enzymes have recently been found in plants, one of which is pathogen inducible (5, 11).
XF1646, classified as absent or highly divergent in J1a12, was annotated as a putative UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase, which is similar to the lpxD gene from Rickettsia rickettsii. This enzyme catalyzes the third step in lipid A biosynthesis, a constituent of lipopolysaccharides from the outer membrane. In Escherichia coli, an lpxD mutant had its susceptibility to various antibiotics increased as high as 512-fold, indicating alterations in the outer membrane permeability barrier (36). An altered outer membrane structure due to the incomplete lipid A biosynthesis may render J1a12 more sensitive to antimicrobial agents eventually produced by the plant host. The increased permeability of its outer membrane may also explain why J1a12 is amenable to DNA transformation, whereas 9a5c is not (6, 17, 23).
The two highest DNA hybridization intensity ratios obtained when comparing 9a5c versus J1a12 were derived from the two CDS encoded by the mini plasmid pXF1.3 (XFb0001 and XFb0002). This result reflects the presence of multiple copies of this plasmid in strain 9a5c (32) and its absence in strain J1a12, as previously reported (6).
Final remarks.
Among the 14 genes classified as absent or highly divergent, three CDS encoding a fimbrial adhesin precursor, an arginase, and a UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase are conspicuously absent in the nonpathogenic strain J1a12. Due to their putative role in bacterial survival in infected hosts, they emerge here as important players in Xylella pathogenicity. The observation that several other genes are missing in J1a12 gives support to the hypothesis that bacterial pathogenesis is a multifactorial process and that each of these factors may contribute somewhat quantitatively to the development of disease. In fact, inactivation of a single fimbrial adhesin gene (PD0058) or a single fimbriae protein gene (PD0062) in X. fastidiosa grapevine strain Temecula was not sufficient to decrease bacterial pathogenicity, causing only a slight reduction in the bacterial population (9).
Recent microarray expression studies comparing X. fastidiosa 9a5c cells freshly isolated from citrus with bacteria attenuated after several passages in axenic culture have shown that most genes found to be induced in the freshly isolated condition were associated with adhesion and with possible adaptation to the host environment (8). However, the set of genes observed in that study is different from the genes found to be absent in the nonpathogenic strain analyzed in the present report, reinforcing the multifactorial hypothesis of bacterial pathogenesis. Furthermore, our results obtained with J1a12 are independent of the number of passages in culture, since the microarray experiments were performed with DNA from bacterial cells obtained after either 14 or 24 passages and no differences were observed (data not shown).
Our plant colonization assays showing that strain J1a12 is unable to induce CVC symptoms or even sustain itself in host plants raise the question of how strain J1a12 was originally isolated from a citrus tree. A recent report about the diversity of the endophytic bacterial community in citrus trees (2) can shed light upon this intriguing question. Possibly, the presence of strain J1a12 in symptomatic plants is dependent on other microorganisms and/or other X. fastidiosa pathogenic strains eventually present in the biofilms formed by the aggregated cells and clogging the xylem vessels of infected plant hosts (13, 19).
Among the CDS found to be absent in J1a12 and proposed here to play a role in disease development, four CDS do have orthologs in the three other pathogenic Xylella strains that have been sequenced (see Table S5 in the supplemental material). Thus, despite the diversity of hosts, geographical location, and disease symptoms, different Xylella strains may present similar mechanisms of pathogenesis. Our results suggest that common strategies could be undertaken to control the diseases that are caused by different X. fastidiosa strains infecting various host plants. Given the importance of the set of genes found here, further functional characterization is warranted. Towards this end, we are currently trying to complement strain J1a12 with the CDS shown to be absent from its genome. Different from 9a5c, the nonpathogenic J1a12 strain is amenable to DNA transformation with plasmid vectors and the transposome system (6, 17, 23). Thus, we believe that with complementation studies it will be possible to evaluate the role of these CDS in bacterial virulence and the ability to colonize xylem vessels.
Supplementary Material
Acknowledgments
This work was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
We are greatly indebted to Hugo A. Armelin for coordinating the Cooperation for Analysis of Gene Expression (CAGE) Project and for strongly supporting this work. We thank João Carlos Setubal and João Kitajima for providing information about reannotation of the Xylella genomic sequence and Apuã C. M. Paquola, Milton Y. Nishiyama, Jr., and Abimael A. Machado for providing bioinformatic tools. We thank Jesus Ferro for coordinating our PCR amplification data bank and for the Xylella cosmid library. We also thank Sanvai R. P. Rocha, Mateus de Almeida Santos, and Anelise G. Mariano for help in pathogenicity tests and Helder Nakaya and Ari J. S. Ferreira for valuable help in the beginning of this work.
A.M.d.S., H.E.-D., S.L.G., and S.V.-A. were partially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico, (CNPq). T.K., L.M.M., R.Z.N.V., and P.A.Z. are FAPESP doctoral fellows.
REFERENCES
- 1.Almeida, R. P. P., and A. H. Purcell. 2003. Transmission of Xylella fastidiosa to grapevines by Homalodisca coagulata (Hemiptera: Cicadellidae). J. Econ. Entomol. 96:264-271. [DOI] [PubMed] [Google Scholar]
- 2.Araujo, W. L., J. Marcon, W. Maccheroni, Jr., J. D. Van Elsas, J. W. Van Vuurde, and J. L. Azevedo. 2002. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl. Environ. Microbiol. 68:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya, A., S. Stilwagen, G. Reznik, H. Feil, W. S. Feil, I. Anderson, A. Bernal, M. D'Souza, N. Ivanova, V. Kapatral, N. Larsen, T. Los, A. Lykidis, E. Selkov, Jr., T. L. Walunas, A. Purcell, R. A. Edwards, T. Hawkins, R. Haselkorn, R. Overbeek, N. C. Kyrpides, and P. F. Predki. 2002. Draft sequencing and comparative genomics of Xylella fastidiosa strains reveal novel biological insights. Genome Res. 12:1556-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandok, M. R., A. J. Ytterberg, K. J. van Wijk, and D. F. Klessig. 2003. The pathogen-inducible nitric oxide synthase (iNOS) in plants is a variant of the P protein of the glycine decarboxylase complex. Cell 113:469-482. [DOI] [PubMed] [Google Scholar]
- 6.da Silva Neto, J. F., T. Koide, S. L. Gomes, and M. V. Marques. 2002. Site-directed gene disruption in Xylella fastidiosa. FEMS Microbiol. Lett. 210:105-110. [DOI] [PubMed] [Google Scholar]
- 7.Davis, M. J., W. J. French, and N. W. Schaad. 1981. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr. Microbiol. 6:309-314. [Google Scholar]
- 8.de Souza, A. A., M. A. Takita, H. D. Coletta, C. Caldana, G. H. Goldman, G. M. Yanai, N. H. Muto, R. C. de Oliveira, L. R. Nunes, and M. A. Machado. 2003. Analysis of gene expression in two growth states of Xylella fastidiosa and its relationship with pathogenicity. Mol. Plant-Microbe Interact. 16:867-875. [DOI] [PubMed] [Google Scholar]
- 9.Feil, H., W. S. Feil, J. C. Detter, A. H. Purcell, and S. E. Lindow. 2003. Site-directed disruption of the fimA and fimF fimbrial genes of Xylella fastidiosa. Phytopathology 93:675-682. [DOI] [PubMed] [Google Scholar]
- 10.Gobert, A. P., D. J. McGee, M. Akhtar, G. L. Mendz, J. C. Newton, Y. Cheng, H. L. Mobley, and K. T. Wilson. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98:13844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, F. Q., M. Okamoto, and N. M. Crawford. 2003. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302:100-103. [DOI] [PubMed] [Google Scholar]
- 12.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. F. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins, D. L. 1989. Xylella fastidiosa-xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 27:271-290. [Google Scholar]
- 14.Hopkins, D. L., and A. H. Purcell. 2002. Xylella fastidiosa: cause of Pierce's disease of grapevine and other emergent diseases. Plant Dis. 86:1056-1066. [DOI] [PubMed] [Google Scholar]
- 15.Karlin, S. 2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9:335-343. [DOI] [PubMed] [Google Scholar]
- 16.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:RESEARCH0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koide, T., J. F. da Silva Neto, S. L. Gomes, and M. V. Marques. 2004. Insertional transposon mutagenesis in the Xylella fastidiosa citrus variegated chlorosis strain with transposome. Curr. Microbiol. 48:247-250. [DOI] [PubMed] [Google Scholar]
- 18.Langstraat, J., M. Bohse, and S. Clegg. 2001. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect. Immun. 69:5805-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leite, B., M. L. Ishida, E. Alves, H. Carrer, S. F. Pascholati, and E. W. Kitajima. 2002. Genomics and X-ray microanalysis indicate that Ca2+ and thiols mediate the aggregation and adhesion of Xylella fastidiosa. Braz. J. Med. Biol. Res. 35:645-650. [DOI] [PubMed] [Google Scholar]
- 20.Li, W. B., L. Zreik, N. G. Fernandes, V. S. Miranda, D. C. Teixeira, A. J. Ayres, M. Garnier, and J. M. Bove. 1999. A triply cloned strain of Xylella fastidiosa multiplies and induces symptoms of citrus variegated chlorosis in sweet orange. Curr. Microbiol. 39:106-108. [DOI] [PubMed] [Google Scholar]
- 21.Lopes, S. A., D. M. Ribeiro, P. G. Roberto, S. C. Franca, and J. M. Santos. 2000. Nicotiana tabacum as an experimental host for the study of plant-Xylella fastidiosa interactions. Plant Dis. 84:827-830. [DOI] [PubMed] [Google Scholar]
- 22.Machado, M. A., A. A. Souza, H. D. Coletta-Filho, E. E. Kuramae, and M. A. Takita. 2001. Genome and pathogenicity of Xylella fastidiosa. Mol. Biol. Today 2:33-43. [Google Scholar]
- 23.Monteiro, P. B., D. C. Teixeira, R. R. Palma, M. Garnier, J. M. Bove, and J. Renaudin. 2001. Stable transformation of the Xylella fastidiosa citrus variegated chlorosis strain with oriC plasmids. Appl. Environ. Microbiol. 67:2263-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunes, L. R., Y. B. Rosato, N. H. Muto, G. M. Yanai, V. S. da Silva, D. B. Leite, E. R. Goncalves, A. A. de Souza, H. D. Coletta-Filho, M. A. Machado, S. A. Lopes, and R. C. de Oliveira. 2003. Microarray analyses of Xylella fastidiosa provide evidence of coordinated transcription control of laterally transferred elements. Genome Res. 13:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pooler, M. R., and J. S. Hartung. 1995. Specific PCR detection and identification of Xylella fastidiosa strains causing citrus variegated chlorosis. Curr. Microbiol. 31:377-381. [DOI] [PubMed] [Google Scholar]
- 26.Purcell, A. H., and D. L. Hopkins. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34:131-151. [DOI] [PubMed] [Google Scholar]
- 27.Purcell, A. H., and S. R. Saunders. 1999. Fate of Pierce's disease strains of Xylella fastidiosa in common riparian plants in California. Plant Dis. 83:825-830. [DOI] [PubMed] [Google Scholar]
- 28.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. X. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 29.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebghati, T. A. S., T. K. Korhonen, D. B. Hornick, and S. Clegg. 1998. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect. Immun. 66:2887-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silverman, B. W. 1986. Density estimation. Chapman and Hall, London, England.
- 32.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. Leite, E. G. Lemos, M. V. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. M. Madeira, H. M. Madeira, C. L. Marino, M. V. Marques, E. A. Martins, E. M. Martins, A. Y. Matsukuma, C. F. Menck, E. C. Miracca, C. Y. Miyaki, C. B. Monteriro-Vitorello, D. H. Moon, M. A. Nagai, A. L. Nascimento, L. E. Netto, A. Nhani, Jr., F. G. Nobrega, L. R. Nunes, M. A. Oliveira, M. C. de Oliveira, R. C. de Oliveira, D. A. Palmieri, A. Paris, B. R. Peixoto, G. A. Pereira, H. A. Pereira, Jr., J. B. Pesquero, R. B. Quaggio, P. G. Roberto, V. Rodrigues, A. J. de M. Rosa, V. E. de Rosa, Jr., R. G. de Sa, R. V. Santelli, H. E. Sawasaki, A. C. da Silva, A. M. da Silva, F. R. da Silva, W. A. da Silva, Jr., J. F. da Silveira, M. L. Silvestri, W. J. Siqueira, A. A. de Souza, A. P. de Souza, M. F. Terenzi, D. Truffi, S. M. Tsai, M. H. Tsuhako, H. Vallada, M. A. Van Sluys, S. Verjovski-Almeida, A. L. Vettore, M. A. Zago, M. Zatz, J. Meidanis, and J. C. Setubal. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 33.Smolka, M. B., D. Martins, F. V. Winck, C. E. Santoro, R. R. Castellari, F. Ferrari, I. J. Brum, E. Galembeck, H. Della Coletta Filho, M. A. Machado, S. Marangoni, and J. C. Novello. 2003. Proteome analysis of the plant pathogen Xylella fastidiosa reveals major cellular and extracellular proteins and a peculiar codon bias distribution. Proteomics 3:224-237. [DOI] [PubMed] [Google Scholar]
- 34.Tarkkanen, A. M., R. Virkola, S. Clegg, and T. K. Korhonen. 1997. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect. Immun. 65:1546-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyson, G. E., B. J. Stojanovic, R. F. Kuklinski, T. J. Divittorio, and M. L. Sullivan. 1985. Scanning electron-microscopy of Pierce's disease bacterium in petiolar xylem of grape leaves. Phytopathology 75:264-269. [Google Scholar]
- 36.Vaara, M., and M. Nurminen. 1999. Outer membrane permeability barrier in Escherichia coli mutants that are defective in the late acyltransferases of lipid A biosynthesis. Antimicrob. Agents Chemother. 43:1459-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. Camargo, A. C. da Silva, D. H. Moon, M. A. Takita, E. G. Lemos, M. A. Machado, M. I. Ferro, F. R. da Silva, M. H. Goldman, G. H. Goldman, M. V. Lemos, H. El-Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, V. E. Rosa, Jr., F. T. Sassaki, J. A. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, G. M. Yanai, L. G. Zaros, E. L. Civerolo, A. J. Simpson, N. F. Almeida, Jr., J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfgang, M. C., B. R. Kulasekara, X. Y. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.