Crystal structures of a fragment (residues 132–251) of human Caprin-1 shed lights onto the homodimerization of Caprin-1 and its interactions with partner proteins.
Keywords: Caprin-1, Caprin-2, G3BP1, FMRP, JEV core protein, RNA stress granule
Abstract
Caprin-1 plays roles in many important biological processes, including cellular proliferation, innate immune response, stress response and synaptic plasticity. Caprin-1 has been implicated in several human diseases, including osteosarcoma, breast cancer, viral infection, hearing loss and neurodegenerative disorders. The functions of Caprin-1 depend on its molecular-interaction network. Direct interactions have been established between Caprin-1 and the fragile X mental retardation protein (FMRP), Ras GAP-activating protein-binding protein 1 (G3BP1) and the Japanese encephalitis virus (JEV) core protein. Here, crystal structures of a fragment (residues 132–251) of Caprin-1, which adopts a novel all-α-helical fold and mediates homodimerization through a substantial interface, are reported. Homodimerization creates a large and highly negatively charged concave surface suggestive of a protein-binding groove. The FMRP-interacting sequence motif forms an integral α-helix in the dimeric Caprin-1 structure in such a way that the binding of FMRP would not disrupt the homodimerization of Caprin-1. Based on insights from the structures and existing biochemical data, the existence of an evolutionarily conserved ribonucleoprotein (RNP) complex consisting of Caprin-1, FMRP and G3BP1 is proposed. The JEV core protein may bind Caprin-1 at the negatively charged putative protein-binding groove and an adjacent E-rich sequence to hijack the RNP complex.
1. Introduction
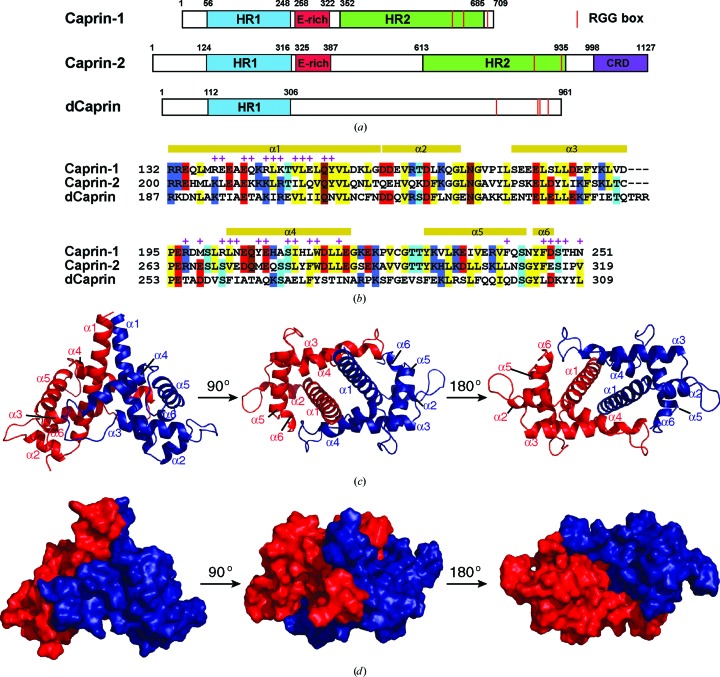
The Caprin (cytoplasmic activation/proliferation-associated protein) family has two members, Caprin-1 and Caprin-2, which are characterized by two highly conserved homologous regions (HR1 and HR2; Grill et al., 2004 ▸; Shiina et al., 2005 ▸; Fig. 1 ▸ a). A Drosophila protein, dCaprin, also contains an HR1 region. In both Caprin-1 and Caprin-2, a less conserved E-rich region is present between HR1 and HR2. Caprin-1, Caprin-2 and dCaprin all contain RGG boxes and RG-rich sequences characteristic of RNA-binding proteins (Fig. 1 ▸ a). Caprin-2 also contains an identifiable domain (CRD; C1q-related domain) at its C-terminus (Shapiro & Scherer, 1998 ▸; Gaboriaud et al., 2003 ▸; Garlatti et al., 2010 ▸; Venkatraman Girija et al., 2013 ▸; Miao et al., 2014 ▸).
Figure 1.
Caprin proteins and structure of the dimerization domain of Caprin-1. (a) Schematic representation of human Caprin-1 and Caprin-2, as well as of Drosophila Caprin (dCaprin). HR1 and HR2, homologous regions 1 and 2. CRD, C1q-related domain. The thin red lines indicate the locations of the RGG boxes. (b) Sequence of the human Caprin-1 dimerization domain aligned with the homologous sequences of Caprin-2 and dCaprin. The secondary structures are indicated above the Caprin-1 sequence. Residues involved in dimerization are indicated with a + sign. (c, d) Structure of the Caprin-1 homodimer rendered in cartoon mode and surface mode, respectively, viewed from three different angles. The two protomers are colored red and blue, respectively.
Caprin-1 is a positive regulator of cell proliferation. Its level is highest in thymus and spleen and lowest in kidney and muscle. A high level of Caprin-1 was observed in proliferating mouse T or B lymphocytes and haemopoietic progenitors (Grill et al., 2004 ▸). Caprin-1-null chicken B lymphocyte DT40 cells showed a marked reduction in the proliferation rate, which could be restored by the expression of human Caprin-1 (Wang et al., 2005 ▸). Suppression of Caprin-1 expression resulted in defective proliferation, with a prolonged G1 phase. Caprin-1 formed a stable complex with Ras GTPase-activating protein-binding protein 1 (G3BP1; Solomon et al., 2007 ▸). This complex localizes in cytoplasmic RNA granules and selectively binds mRNAs of c-Myc and cyclin D2, two proteins with essential roles in the G1/S transition (Chiles, 2004 ▸; Kaczmarek et al., 1985 ▸; Pardee, 1989 ▸).
In the brain, Caprin-1 is highly expressed in the neuronal RNA granules (Shiina et al., 2005 ▸), which contain mRNAs of key proteins for synaptic plasticity, such as Ca2+/calmodulin-dependent kinase IIα (CaMKIIα; Burgin et al., 1990 ▸), brain-derived neurotrophic factor (BDNF; Tongiorgi et al., 1997 ▸), tyrosine receptor kinase B (TrkB; Tongiorgi et al., 1997 ▸) and cAMP response element-binding protein (CREB; Crino et al., 1998 ▸). The importance of Caprin-1 in brain development is demonstrated by the observation that Caprin-1-knockout mice died soon after birth owing to respiratory failure, suggesting defects in the brain stem (Shiina et al., 2010 ▸).
Caprin-1 is also involved in the formation of cytoplasmic RNA stress granules (SGs). Caprin-1 and G3BP1 localized into SGs in response to oxidative stress. Overexpression of Caprin-1 promoted SG formation (Solomon et al., 2007 ▸). The formation of Caprin-1-containing SGs has been implicated in several diseases.
Osteosarcoma (OS) is the most common primary bone malignancy in children and adolescents (Sabile et al., 2013 ▸). In cultured OS cells, Caprin-1 interacts with the OS-promoting protein Cyr61. Overexpression of Caprin-1 led to the formation of Caprin-1–Cyr61-containing SGs and decreased the sensitivity of the OS cells to cisplatin-induced apoptosis. In SCID mice, the overexpression of Caprin-1 dramatically enhanced primary tumor growth and remarkably increased lung metastasis and mortality. In a group of OS patients, increased Caprin-1 expression was correlated with shortened survival. Moreover, patients with tumors co-expressing Caprin-1 and Cyr61 showed poorer survival than patients expressing one or neither of the proteins.
Caprin-1-containing SGs constitute a host antiviral response against Japanese encephalitis virus (JEV). JEV suppresses SG formation by using its core protein to bind Caprin-1 and recruit several SG-promoting proteins to the perinuclear region (Katoh et al., 2013 ▸). Mutations of Lys97 and Arg98 to alanines (9798A mutant) in the core protein abolish its interaction with Caprin-1. In cells infected with a mutant JEV containing the 9798A mutant, viral inhibition of SG formation was abrogated and viral propagation was impaired.
Caprin-1 may participate in the protective response to ototoxic insults. In cochlear hair cells, Caprin-1 is down-regulated by the transcription factor Pou4f3 (Xiang et al., 1997 ▸; Towers et al., 2011 ▸). Pou4f3 is essential for the survival of auditory sensory hair cells. Several mutations in the human Pou4f3 gene cause hearing loss (Collin et al., 2008 ▸; Pauw et al., 2008 ▸). Aminoglycoside-induced hair-cell damage results in a decrease in the Pou4f3 mRNA level and an increase in the Caprin-1 mRNA and protein levels, leading to the formation of Caprin-1-containing SGs.
Huntington’s disease (HD) is a neurodegenerative disorder caused by an expansion of a polyglutamine repeat within huntingtin, the HD gene product (Ross et al., 1997 ▸). It was found that expanded huntingtin interacted with Caprin-1 and was redistributed to Caprin-1–G3BP1-containing SGs (Ratovitski et al., 2012 ▸), suggesting a role of Caprin-1 in HD pathogenesis.
The functions of Caprin-1 depend on its molecular interactions. The Caprin-1–G3BP1 complex selectively interacts with c-Myc and cyclin D2 mRNAs (Solomon et al., 2007 ▸). Interestingly, although Caprin-1 and G3GP1 are both RNA-binding proteins, Caprin-1 is mainly responsible for binding the mRNAs through the C-terminal RGG and RG-rich sequences.
Caprin-1 also interacts with many protein partners. Direct interactions of Caprin-1 with G3BP1, the fragile X mental retardation protein (FMRP) and the JEV core protein were established by in vitro affinity pull-down assays (Solomon et al., 2007 ▸; Katoh et al., 2013 ▸; El Fatimy et al., 2012 ▸). The Caprin-1–G3BP1 complex has been implicated in virtually all reported functions of Caprin-1. G3BP1 contains a nuclear transport factor 2-like (NTF2) domain, an acidic region, an RNA-binding domain (RBD) and a C-terminal RGG/G-rich sequence. The crystal structure of the NTF2 domain has been reported (Vognsen et al., 2013 ▸). Residues 352–380 of Caprin-1 and the NTF2 domain of G3BP1 are required for the interaction of Caprin-1 and G3BP1 (Solomon et al., 2007 ▸). FMRP is present in all three major classes of RNA granules (SGs, neuronal granules and P-bodies) and plays important roles in mRNA metabolism (Kao et al., 2010 ▸; Darnell, 2011 ▸). The absence of FMRP causes fragile X syndrome (FXS), the most common form of inherited mental retardation (O’Donnell & Warren, 2002 ▸). FMRP contains two Agenet-like domains, two KH (hnRNP K homology) domains and an RGG region. Several NMR or crystal structures are available for various fragments of FMRP (Ramos et al., 2006 ▸; Valverde et al., 2007 ▸; Musco et al., 1996 ▸, 1997 ▸; Phan et al., 2011 ▸). Residues 231–245 of Caprin-1 and residues 427–442 of FMRP are required for the interaction between Caprin-1 and FMRP (El Fatimy et al., 2012 ▸).
While significant progress has been made in unravelling the functions and mechanisms of Caprin-1, no structural study of Caprin-1 has been reported to date. This lack of structural knowledge has hampered research efforts in related fields, especially in view of the fact that Caprin-1 does not have sequence similarities to any protein with a known structure. In this paper, we present the crystal structures of a human Caprin-1 fragment consisting of residues 132–251.
2. Materials and methods
2.1. Protein sample preparation and crystallization
The detailed procedures for molecular cloning, protein expression, purification and crystallization have been reported elsewhere (Wu et al., 2015 ▸). Briefly, a fragment of human Caprin-1 (containing residues 112–260) was expressed as a fusion protein with N-terminal Halo and His tags in Escherichia coli BL21(DE3) cells. An octapeptide that can be specifically recognized and cleaved by human rhinovirus (HRV) 3C protease was engineered between the tags and the Caprin-1 fragment. For the native protein, bacterial cells were grown in LB medium. Recombinant protein production was induced by isopropyl β-d-1-thiogalactopyranoside (IPTG; at a final concentration of 0.2 mM) when the cultures reached an OD600 of 0.6–0.8. Protein expression was carried out at 10°C for 24 h before harvesting. The fusion protein was purified using cobalt-affinity resin. The tags were cleaved by His-tagged HRV 3C protease and removed from the target protein by a reverse immobilized metal-affinity chromatography (IMAC) process. The untagged, purified protein was concentrated to a final concentration of ∼10 mg ml−1 in a buffer consisting of 25 mM Tris pH 7.0, 200 mM NaCl. For selenomethionine (SeMet)-labelled protein, the bacterial cells were grown in M9 minimal culture. When the cultures reached an OD600 of 0.6–0.8, the six amino acids leucine, isoleucine, lysine, phenylalanine, threonine and valine were added to the culture to a final concentration of 50–100 mg l−1. After half an hour of growth at 37°C, l-selenomethionine was added to the cell culture to a final concentration of 50 mg l−1. The subsequent steps were identical to those for the native protein. The crystallization trials and optimization were carried out using 96-well plates at 22°C by sitting-drop vapour diffusion. Seven sets of in-house-prepared crystallization screening solutions (each containing 96 different conditions) were used in the initial screen. To obtain native crystals in space group C121, the well solution consisted of 22% PEG 750 MME, 0.1 M Tris pH 8.0, 0.1 M potassium fluoride, 10% glycerol. To obtain SeMet-labelled crystals in space group P3121, the well solution consisted of 12% PEG 600, 0.1 M Tris pH 8.0, 0.1 M calcium acetate, 10% glycerol.
2.2. Data collection, data processing and structure determination
Data collection was carried out on LS-CAT beamline 21-ID-G at the Advanced Photon Source, Argonne National Laboratory. The data were processed using MOSFLM and SCALA in CCP4 (Battye et al., 2011 ▸). The structure in space group P3121 was solved by SAD using a data set collected at the peak wavelength of Se (0.97872 Å) from a single crystal of SeMet-labelled protein. Locating the Se atoms and building the initial structure were performed using the PHENIX package (Adams et al., 2011 ▸). Interactive model building was carried out with Coot (Emsley et al., 2010 ▸). The structure was refined using phenix.refine (Afonine et al., 2012 ▸). The structure in space group C121 was solved by molecular replacement using Phaser (Storoni et al., 2004 ▸) as implemented in the PHENIX package. Structure-determination statistics are shown in Table 1 ▸. The figures were prepared with PyMOL (v.1.5.0.4; Schrödinger). Atomic coordinates and diffraction data for the structures have been deposited in the Protein Data Bank with accession codes 4wbp and 4wbe.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Native | SeMet | |
|---|---|---|
| Data collection | ||
| Space group | C121 | P3121 |
| Unit-cell parameters | ||
| a (Å) | 106.2 | 66.0 |
| b (Å) | 77.31 | 66.0 |
| c (Å) | 58.01 | 117.7 |
| α = β (°) | 90.0 | 90.0 |
| γ (°) | 90.0 | 120.0 |
| Resolution (Å) | 42.6–2.05 (2.16–2.05) | 41.0–2.65 (2.79–2.65) |
| No. of unique reflections | 29397 (4258) | 9085 (1292) |
| Completeness (%) | 99.0 (98.8) | 100 (100) |
| R merge † (%) | 9.6 (54.4) | 7.6 (60.6) |
| 〈I/σ(I)〉 | 5.4 (1.6) | 11.5 (2.8) |
| CC1/2 | 0.99 (0.72) | 1.0 (0.89) |
| Multiplicity | 3.1 (3.0) | 5.4 (5.4) |
| Anomalous completeness (%) | N/A | 100 (100) |
| Anomalous multiplicity | N/A | 2.9 (2.8) |
| Refinement | ||
| Resolution (Å) | 42.6–2.05 | 41.0–2.65 |
| No. of reflections | 29397 | 9085 |
| R work/R free ‡ (%) | 19.6/23.1 | 20.9/26.5 |
| No. of atoms | ||
| Protein | 2989 | 1992 |
| Water | 269 | 38 |
| B factors (Å2) | ||
| Protein | 37.9 | 51.0 |
| Water | 38.6 | 35.7 |
| R.m.s.d., bonds (Å) | 0.005 | 0.007 |
| R.m.s.d., angles (°) | 0.940 | 1.170 |
| Ramachandran plot | ||
| Favored (%) | 97.0 | 97.0 |
| Outliers (%) | 0 | 0 |
R
merge = 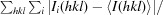
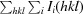 , where 〈I(hkl)〉 is the average intensity of reflection hkl.
, where 〈I(hkl)〉 is the average intensity of reflection hkl.
R
work = 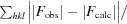
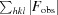 , where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is calculated as for R
work but using a randomly selected subset of data (7%) which were excluded from refinement.
, where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is calculated as for R
work but using a randomly selected subset of data (7%) which were excluded from refinement.
3. Results and discussion
3.1. The HR1 region of Caprin-1 contains a dimerization domain
The sequence of Caprin-1 has no significant homology to other protein sequences with known structure (a BLAST search against the PDB matched fragments of the Caprin-1 sequence to several sequences with E-values worse than the threshold). There are also no identifiable protein domains in Caprin-1. Identification of the HR1 and HR2 regions was solely based on sequence homology within the Caprin proteins (Grill et al., 2004 ▸; Shiina et al., 2005 ▸).
To study the structures of human Caprin-1, we used a divide-and-conquer approach. We prepared many different protein constructs corresponding to different regions of Caprin-1. As a first attempt, we tried a protein construct corresponding to the HR1 fragment, as shown in Fig. 1 ▸(a) (residues 56–248), using many different fusion tags and E. coli host strains. However, this construct did not express. We then tested a series of protein constructs starting at residues 64, 72, 80, 88, 96, 104, 112 or 120 and ending at residue 260 (at the N-terminus of the E-rich region). Most of these constructs were somehow problematic (no expression, low solubility or unstable). The protein construct corresponding to residues 112–260 was successfully expressed as a fusion protein with either a Halo or an MBP tag. The protein was stable after removal of the tag and was purified and crystallized.
The SeMet-labelled and native proteins crystallized in two different space groups. The structures were determined by the SAD (in space group P3121, with two molecules in the asymmetric unit) and molecular-replacement (in space group C121, with three molecules in the asymmetric unit) methods, respectively (Table 1 ▸). Electron densities were observed for residues 132–251 but not for residues 112–131 and 252–260, indicating that residues 132–251 of human Caprin-1 contain a stable domain. This domain mediates homodimerization (see below for details) of Caprin-1. We therefore refer to it here as the dimerization domain. As shown in Fig. 1 ▸(a), the dimerization domain comprises the C-terminal two-thirds of the HR1 region. The lack of density for some residues suggests that these residues are not well ordered. By SDS–PAGE analysis of the protein from washed crystals, we confirmed that the crystallized protein was not proteolytically cleaved.
The dimerization domain assumes a novel all-α-helical protein fold (with no similar structure in the Protein Data Bank) consisting of six α-helices (the last helix α6 has less than one helical turn). The overall structure of the domain somewhat resembles a land snail. The N-terminal half and C-terminal half of the longest helix α1 resemble the head and foot, respectively, and the other helices (α2–α6) form the shell (Figs. 1 ▸ c and 1 ▸ d). The structure forms a head-to-head dimer, which is mediated by residues on the ‘base’ side of the snail-shaped molecule (Figs. 1 ▸ c and 1 ▸ d). The same dimer is present in the two different crystal forms, although the crystal packings are different (Supplementary Fig. S1).
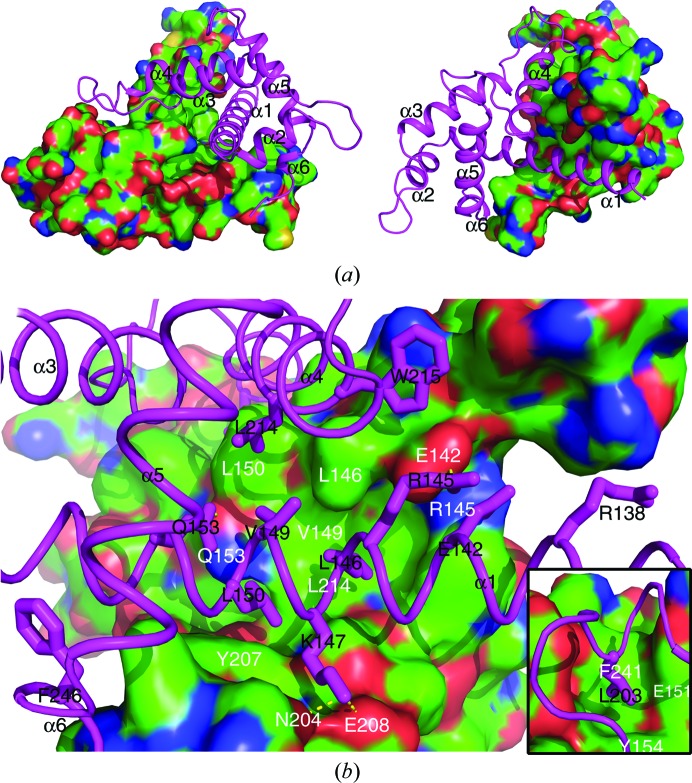
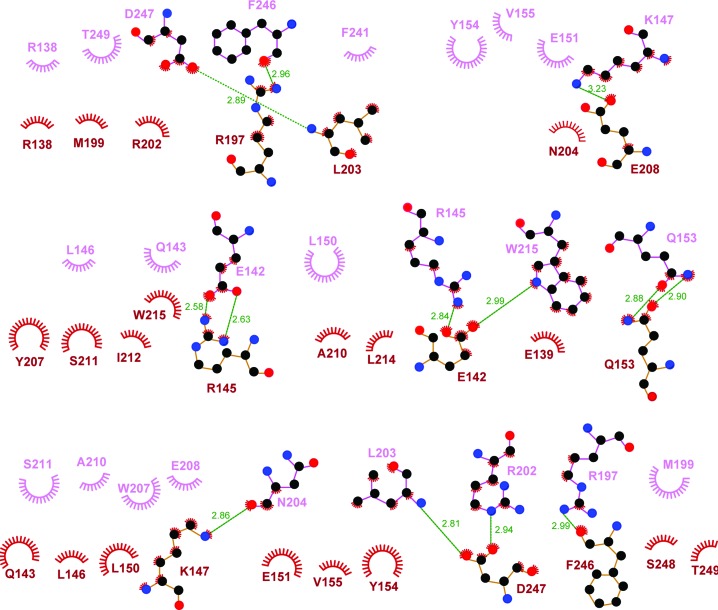
The dimerization interface is defined by about 60 residues (30 in each protomer) and buries a total of 3398 Å2 of solvent-accessible surface area from the two protomers. The extensive dimerization interface is mainly defined by residues from the α1 and α4 helices, as well as the loop regions N-terminal to the α4 helix and C-terminal to the α6 helix (Figs. 1 ▸ b, 3a and 3b). The dimerization interface exhibits a high degree of shape complementarity (Fig. 2 ▸ a). Moreover, the charged and polar residues form intermolecular salt bridges and hydrogen bonds (such as Glu142–Arg145, Lys147–Glu208 and Gln153–Gln153), and many hydrophobic residues (including Leu146, Val149, Leu150, Tyr154, Leu203, Tyr207, Leu214, Trp215, Phe241 and Phe246, as well as the aliphatic parts of the Arg145 and Lys147 side chains) mediate intermolecular hydrophobic interactions (Figs. 2 ▸ b, 3 ▸ a and 3 ▸ b). Most of the residues involved in dimerization are conserved in Caprin-2 and dCaprin (Fig. 1 ▸ b), suggesting that the corresponding regions of Caprin-2 and dCaprin may be able to form similar dimeric structures as seen in Caprin-1.
Figure 2.
Molecular interactions that mediate the homodimerization of Caprin-1. (a) Shape complementarity at the homodimerization interface. The two protomers are rendered in surface mode (colored by element, with C, N, O and S in green, blue, red and yellow, respectively) and cartoon mode (colored magenta), respectively. (b) Intermolecular hydrophobic contacts, salt bridges and hydrogen bonds at the homodimerization interface. The side chains of some of the key residues belonging to the protomer rendered in cartoon mode are shown as sticks. The residues involved in the intermolecular interactions are indicated differently for the two protomers: one-letter abbreviations and residue numbers in black for residues belonging to the protomer rendered in cartoon mode and one-letter abbreviations and residue numbers in white for residues belonging to the protomer rendered in surface mode. Hydrogen bonds are represented as yellow dashed lines. The small inserted figure at the bottom right shows another area of the homodimerization interface that is not visible in the view in the large figure.
Figure 3.
LIGPLOT presentation of molecular interactions in the dimerization interface. Hydrogen bonds are designated with green dashed lines (the distance between the two heavy atoms is indicated by a number in Å). Hydrophobic interactions are represented as starbursts.
3.2. Caprin-1 homodimerization creates a highly acidic putative protein-binding groove
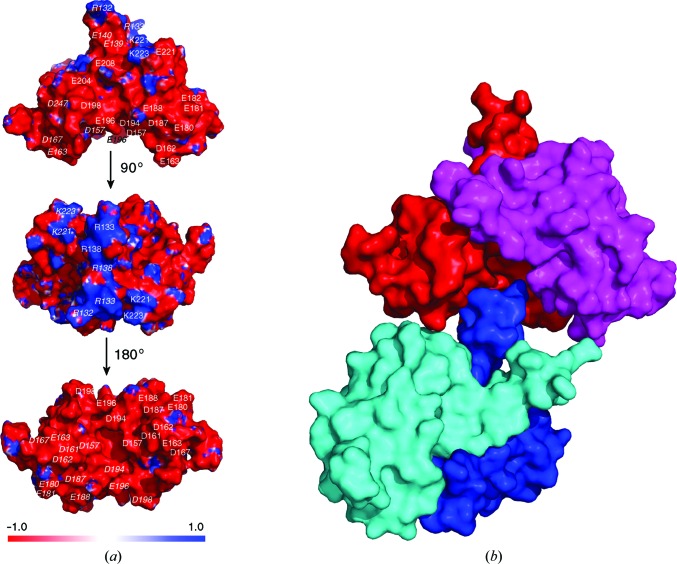
The Caprin-1 dimer has an overall shallow V-shaped structure with a short tail (Figs. 1 ▸ d and 4 ▸ a). The tail is composed of the N-terminal ends of the two α1 helices. The molecular surface of the dimeric structure is predominantly negatively charged, with a few positively charged patches at the end of the tail and in the region of Lys221 and Lys223 (Fig. 4 ▸ a). Of particular interest is the large concave surface of the V-shaped dimeric structure. This surface is dominated by 24 negatively charged residues, with 12 residues from each protomer, including Asp157, Asp161, Asp162, Glu163, Asp167, Glu180, Glu181, Asp187, Glu188, Asp194, Glu196 and Asp198 (Fig. 4 ▸ a). The presence of these residues defines a highly acidic molecular surface. This concave surface has the potential to function as a protein–protein binding groove.
Figure 4.
Surface properties of the homodimeric Caprin-1 structure. (a) Electrostatic surface representation of the Caprin-1 dimer. Blue and red represent regions of positive and negative potential, respectively. The top and bottom figures show a large concave surface which is predominantly negatively charged. The locations of the negatively charged (D/E) or positively charged (K/R) residues are indicated by one-letter residue abbreviations and residue numbers. Residues belonging to the protomer on the left side are indicated in italics. (b) The negatively charged groove mediates a protein–protein contact between two adjacent Caprin-1 dimers (red/magenta and cyan/blue) in the crystals. The positively charged N-terminal end of the protomer in blue contacts the negatively charged groove of the red/magenta dimer.
In the crystal containing SeMet-labeled protein, the positively charged N-terminal end of one of the α1 helices (from the protomer colored in blue in Fig. 4 ▸ b) contacts a small area in the putative protein-binding groove of a neighboring dimeric structure, mainly on the surface area of the protomer colored in red (Fig. 4 ▸ b). The protein–protein interaction is not substantial (burying only 781 Å2 of solvent-accessible surface area from the two protomers) and it is not present in the crystal containing the native protein. Therefore, this protein–protein contact is probably owing to crystal packing. Although this protein–protein contact is unlikely to be biologically relevant, it shows the potential of the negative concave surface of the dimeric Caprin-1 structure to participate in protein–protein interactions. Interestingly, most of the Asp/Glu residues in this surface are not conserved in the Caprin-2 and dCaprin sequences (Fig. 1 ▸ b). Comparing the Caprin-1 and Caprin-2 sequences, nine of the 12 Asp/Glu residues in the negatively charged groove of Caprin-1 change to neutral or positively charged residues in Caprin-2 (Asp157→Gln, Asp161→Gln, Glu163→His, Glu180→Ser, Glu181→Lys, Asp187→Ile, Glu188→Lys, Asp194→Cys and Asp198→Asn). Assuming that Caprin-2 and dCaprin can form a homodimeric structure similar to the Caprin-1 homodimer, the electrostatic surface properties of the concave surfaces of the Caprin-2 and dCaprin dimers would be substantially different from the Caprin-1 dimer. Dimerization of Caprin proteins has not been reported. Our study suggests that the Caprin proteins all have the potential to homodimerize. Homodimerization may be critical for the biological functions of the Caprin proteins, such as mediating protein–protein interactions.
3.3. The FMRP-interacting motif locates on the opposite side to the homodimer interface
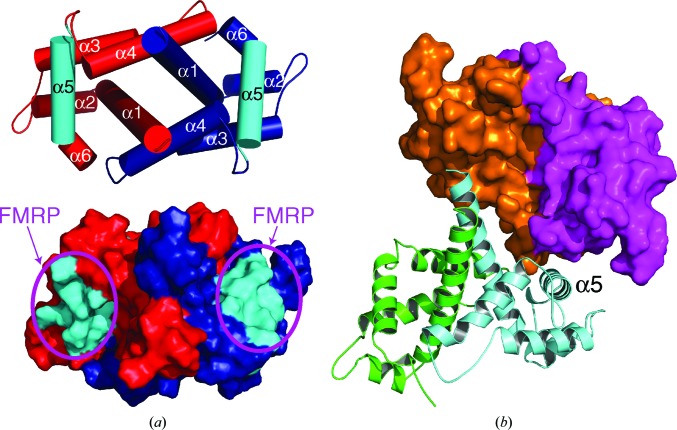
The region of Caprin-1 (residues 231–245) reported to be required for interaction with FMRP (El Fatimy et al., 2012 ▸) is located within the α5 helix (residues 230–245) (Figs. 1 ▸ b and 5 ▸ a). One face of the α5 helix is fully exposed in the dimeric structure, readily accessible for protein–protein interaction. In the crystals in both space groups P3121 and C121, the exposed surface of the α5 helix is involved in direct contact with an adjacent dimeric structure (Fig. 5 ▸ b). The connecting loop between helices α3 and α4 in the orange protomer, together with the sequence C-terminal to the α6 helix in the magenta protomer, form the binding pocket for the α5 helix in the cyan protomer. This protein–protein contact buries 791 Å2 of solvent-accessible surface area from the three protomers involved, and is present in the two different crystal forms. The exposed surface of the α5 helix is located on the opposite side to the dimerization interface of each protomer and the acidic putative protein-binding groove of the dimeric structure (Fig. 5 ▸ a). Protein–protein interaction through the α5 helix does not interfere with the homodimerization interaction of Caprin-1, and also should not interfere with protein binding in the acidic putative protein-binding groove.
Figure 5.
The α5 helix may represent a protein-binding site. (a) The known FMRP-interacting motif forms the α5 helix in the homodimeric Caprin-1 structure. The helices are rendered as cylinders. The two protomers are colored red and blue, respectively, except for the two α5 helices, which are highlighted in cyan. The exposed surface of the α5 helix may interact with an FMRP molecule. (b) The α5 helix mediates a protein–protein contact between two adjacent Caprin-1 dimers (orange/magenta and green/cyan) in the crystals.
It is not known whether the α5 helix-mediated protein–protein interaction as depicted in Fig. 5 ▸(b), which would lead to the multimerization of Caprin-1, is biologically relevant. However, this interaction observed in the crystal structures not only clearly shows the potential of the α5 helix to participate in protein–protein interaction, but also suggests a possible way that Caprin-1 might interact with FMRP. The dimeric structure of Caprin-1 can interact with two FMRP molecules on opposite molecular surfaces of the structure, with each protomer using the α5 helix as the major binding site to interact with an FMRP molecule (Fig. 5 ▸ a).
3.4. The JEV core protein may interact with Caprin-1 through the negatively charged groove of the Caprin-1 dimer and the flanking E-rich regions
The JEV core protein contains only 105 residues. It is a highly basic protein, with 25 Arg/Lys residues and only four Asp/Glu residues. While the structure of this protein has not been solved, a reasonable homologous structural model can be constructed using the NMR structure of the dimeric Dengue virus core protein (61% homologous and 31% identical) as a template (Ma et al., 2004 ▸). Guided by such a structural model, alanine-scanning mutagenesis revealed that a double replacement of Lys97 and Arg98 (9798A) completely abrogated the interaction with Caprin-1 (Katoh et al., 2013 ▸). Moreover, the 9798A mutant failed to pull down G3BP1 in immunoprecipitation assays.
The region of Caprin-1 that is required for interaction with the JEV core protein has not been identified. Because the JEV core protein is predominantly positively charged, it may bind to the putative negatively charged protein-binding groove formed by the homodimerization of Caprin-1. Moreover, the E-rich region of Caprin-1, which is C-terminal to the dimerization domain (Fig. 1 ▸), should be positioned in close proximity to the negatively charged groove and may contribute to the interaction with the JEV core protein (Fig. 6 ▸). The sequence of the dimerization domain and E-rich region of Caprin-1 (residues 132–322) is relatively rich in negatively charged residues, with 51 Asp/Glu residues and 24 Arg/Lys residues. About 60% of the negatively charged residues of Caprin-1 reside within this sequence, which accounts for only 31% of the full-length protein.
Figure 6.
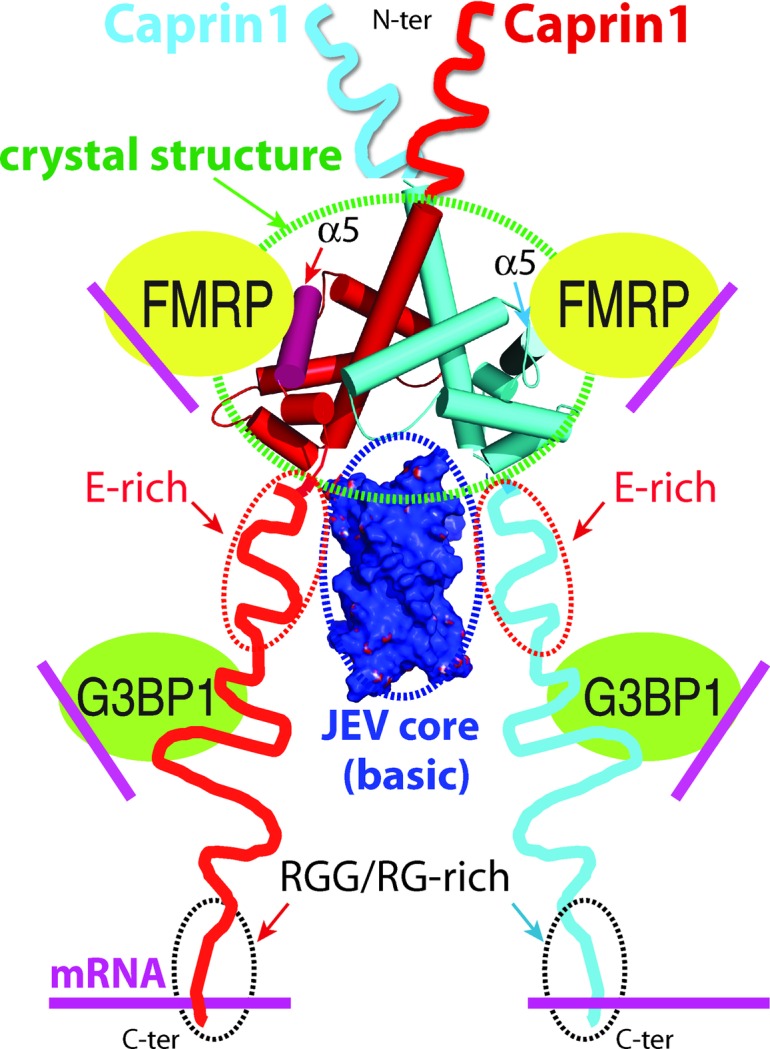
A proposed model for a Caprin-1-mediated ribonucleoprotein complex containing the three RNA-binding proteins Caprin-1, FMRP and G3BP1. A possible mechanism for the JEV core protein to hijack the complex by directly interacting with Caprin-1 is also proposed.
In the model depicted in Fig. 6 ▸, binding of the JEV core protein to the negatively charged groove and flanking E-rich regions of the Caprin-1 dimer should not disrupt the interaction of Caprin-1 with FMRP and G3BP1 in the ribonucleoprotein complex. This mode of interaction between Caprin-1 and the JEV core protein is consistent with the observation that the JEV core protein binds Caprin-1 directly, leading to the suppression of SG formation through the recruitment of several effector molecules promoting SG assembly (including FMRP and G3BP1) to the perinuclear region (Katoh et al., 2013 ▸).
3.5. The possible existence of an evolutionarily conserved ribonucleoprotein complex consisting of Caprin-1, G3BP1 and FMRP
Two cellular proteins, G3BP1 and FMRP, are known to directly interact with Caprin-1. All three of these proteins are RNA-binding proteins that are known to be components of ribonucleoprotein (RNP) complexes such as RNA stress granules. Insights from the crystal structures of the Caprin-1 dimer, in conjunction with existing protein–protein interaction data, have led us to posit the existence of a distinct RNP complex consisting of Caprin-1, G3BP1 and FMRP. As discussed in the previous section, the interaction of Caprin-1, through the α5 helix, with FMRP should not disrupt the dimeric Caprin-1 structure. The Caprin-1 sequence (residues 352–380) reported to be required for G3BP1 interaction (Solomon et al., 2007 ▸) is separated from the dimerization domain by ∼100 residues (Fig. 1 ▸ a). Therefore, the interaction of Caprin-1 with G3BP1 should not disrupt the dimeric Caprin-1 structure and the interaction between Caprin-1 and FMRP. The Caprin-1 homodimer should be able to bind two FMRP molecules and two G3BP1 molecules simultaneously (Fig. 6 ▸). Each of the Caprin-1-bound FMRP and G3BP1 molecules might also be able to further engage in other homomeric or heteromeric protein–protein interactions.
The RNA-binding domains or sequences in Caprin-1, FMRP and G3BP1 do not participate in the protein–protein interactions as depicted in Fig. 6 ▸. In the case of Caprin-1, it was shown that the N-terminus (N-terminal to the dimerization domain) was required for binding mRNAs in vitro (Shiina et al., 2005 ▸; Shiina & Tokunaga, 2010 ▸). However, in a separate study, it was shown that the C-terminus of Caprin-1 (containing the RGG and RG-rich sequence) interacted directly and selectively with c-Myc and cyclin D2 mRNAs (Solomon et al., 2007 ▸). These RNA-binding sequences are well separated from the sequences involved in Caprin-1 dimerization or in interaction with FMRP and G3BP1. Therefore, Caprin-1 in a multi-protein assembly as depicted in Fig. 6 ▸ should be able to bind to mRNAs. In the case of G3BP1, an RNA-binding domain (RBD) and an RGG/G-rich sequence are present at the C-terminus (residues 268–466), separated from the Caprin-1 binding NTF2 domain (residues 1–141) by an acidic region (Vognsen et al., 2013 ▸; Solomon et al., 2007 ▸). The RBD and RGG/G-rich sequences of G3BP1 should be responsible for RNA binding, which should suffer no interference from protein–protein interactions mediated by the NTF2 domain. In the case of FMRP, there are two Agenet-like domains at the N-terminus, two KH domains in the central region and an RGG sequence at the C-terminus. These domains/sequences all have RNA-binding activities (Ramos et al., 2006 ▸; Valverde et al., 2007 ▸; Musco et al., 1996 ▸, 1997 ▸; Phan et al., 2011 ▸). The Caprin-1 interacting motif (residues 427–442) is located C-terminal to the KH2 domain (El Fatimy et al., 2012 ▸). Protein–protein interactions through this motif should not interfere with RNA binding in other regions of FMRP. In short, each of the three RNA-binding proteins in the complex should have the ability to bind RNA. We propose that Caprin-1 acts as a scaffolding protein to mediate the assembly of a distinct RNP complex consisting of Caprin-1, FMRP and G3BP1.
Caprin-1, FMRP and G3BP1 have all been implicated in the pathogenesis of human diseases. For examples, while Caprin-1 is required for normal brain function (Shiina et al., 2005 ▸, 2010 ▸) and the proliferation of immune B and T cells (Wang et al., 2005 ▸; Grill et al., 2004 ▸), ectopic overexpression of Caprin-1 in some other cell types has been linked to osteosarcoma (Sabile et al., 2013 ▸), breast cancer (Gong et al., 2013 ▸) and hearing loss (Towers et al., 2011 ▸). The absence of FMRP causes fragile X syndrome (FXS; O’Donnell & Warren, 2002 ▸), which is the most common form of inherited mental retardation, affecting one in 3600 males and one in 4000–6000 females (according to the World Health Organization). Similar to Caprin-1, ectopic overexpression of G3BP1 has been linked to cancer (Winslow et al., 2013 ▸; Meschenmoser et al., 2013 ▸).
Despite the physiopathological importance of these RBPs, their functional mechanisms largely remain elusive. Normally, proteins do not act alone, but interact with other proteins to form cellular networks. A Caprin-1–G3BP1 complex is present in all known Caprin-1 functions. In mouse brain neurons, Caprin-1 associates with FMRP in polyribosomes and in trafficking neuronal granules. In Drosophila embryonic cells, dCaprin and dFMRP collaborate to control the cell cycle (Papoulas et al., 2010 ▸). A functional complex containing dCaprin, dFMRP and dG3BP1 (also known as Rasputin) has been suggested based on the colocalization of the three RNA-binding proteins and their synergistic effect in cell proliferation (Baumgartner et al., 2013 ▸). It is possible that the ternary Caprin-1–FMRP–G3BP1 complex depicted in Fig. 6 ▸ is evolutionarily conserved and is involved in the functions of all three proteins.
4. Conclusion
The results of our crystallographic studies identify a stable domain within the HR1 region of Caprin-1 that is capable of mediating the tight homodimerization of Caprin-1. Based on insights from the dimeric Caprin-1 structures, in conjunction with existing biochemical data, we propose the existence of an evolutionarily conserved ribonucleoprotein complex consisting of Caprin-1, FMRP and G3BP1. Of course, the model depicted in Fig. 6 ▸ is highly speculative at this point. Further experimental work is necessary to substantiate this model.
Supplementary Material
PDB reference: HR-1 domain of human Caprin-1, space group C121, 4wbe
PDB reference: space group P3121, 4wbp
Supplementary Figure S1.. DOI: 10.1107/S2059798316004903/cb5087sup1.pdf
Acknowledgments
We thank the staff of LS-CAT at the Advanced Photon Source, Argonne National Laboratory for assistance with data collection. The work was supported by a start-up fund and a seed grant from Southern Illinois University Carbondale, as well as a grant from the National Institutes of Health (1R15GM116062-01) to ZD.
References
- Adams, P. D. et al. (2011). Methods, 55, 94–106.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Baumgartner, R., Stocker, H. & Hafen, E. (2013). PLoS Genet. 9, e1003598. [DOI] [PMC free article] [PubMed]
- Burgin, K. E., Waxham, M. N., Rickling, S., Westgate, S. A., Mobley, W. C. & Kelly, P. T. (1990). J. Neurosci. 10, 1788–1798. [DOI] [PMC free article] [PubMed]
- Chiles, T. C. (2004). J. Immunol. 173, 2901–2907. [DOI] [PubMed]
- Collin, R. W., Chellappa, R., Pauw, R. J., Vriend, G., Oostrik, J., van Drunen, W., Huygen, P. L., Admiraal, R., Hoefsloot, L. H., Cremers, F. P., Xiang, M., Cremers, C. W. & Kremer, H. (2008). Hum. Mutat. 29, 545–554. [DOI] [PMC free article] [PubMed]
- Crino, P., Khodakhah, K., Becker, K., Ginsberg, S., Hemby, S. & Eberwine, J. (1998). Proc. Natl Acad. Sci. USA, 95, 2313–2318. [DOI] [PMC free article] [PubMed]
- Darnell, J. C. (2011). Curr. Opin. Genet. Dev. 21, 465–473. [DOI] [PMC free article] [PubMed]
- El Fatimy, R., Tremblay, S., Dury, A. Y., Solomon, S., De Koninck, P., Schrader, J. W. & Khandjian, E. W. (2012). PLoS One, 7, e39338. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gaboriaud, C., Juanhuix, J., Gruez, A., Lacroix, M., Darnault, C., Pignol, D., Verger, D., Fontecilla-Camps, J. C. & Arlaud, G. J. (2003). J. Biol. Chem. 278, 46974–46982. [DOI] [PubMed]
- Garlatti, V., Chouquet, A., Lunardi, T., Vivès, R., Païdassi, H., Lortat-Jacob, H., Thielens, N. M., Arlaud, G. J. & Gaboriaud, C. (2010). J. Immunol. 185, 808–812. [DOI] [PubMed]
- Gong, B., Hu, H., Chen, J., Cao, S., Yu, J., Xue, J., Chen, F., Cai, Y., He, H. & Zhang, L. (2013). Biomed. Pharmacother. 67, 629–636. [DOI] [PubMed]
- Grill, B., Wilson, G. M., Zhang, K.-X., Wang, B., Doyonnas, R., Quadroni, M. & Schrader, J. W. (2004). J. Immunol. 172, 2389–2400. [DOI] [PubMed]
- Kaczmarek, L., Hyland, J. K., Watt, R., Rosenberg, M. & Baserga, R. (1985). Science, 228, 1313–1315. [DOI] [PubMed]
- Kao, D.-I., Aldridge, G. M., Weiler, I. J. & Greenough, W. T. (2010). Proc. Natl Acad. Sci. USA, 107, 15601–15606. [DOI] [PMC free article] [PubMed]
- Katoh, H., Okamoto, T., Fukuhara, T., Kambara, H., Morita, E., Mori, Y., Kamitani, W. & Matsuura, Y. (2013). J. Virol. 87, 489–502. [DOI] [PMC free article] [PubMed]
- Ma, L., Jones, C. T., Groesch, T. D., Kuhn, R. J. & Post, C. B. (2004). Proc. Natl Acad. Sci. USA, 101, 3414–3419. [DOI] [PMC free article] [PubMed]
- Meschenmoser, K., Kim, Y., Franken, S., Nowak, M., Feldmann, G., Bendas, G., Wolfgarten, M., Messmer, D. & Schmidt-Wolf, I. G. (2013). In Vivo, 27, 431–442. [PubMed]
- Miao, H., Jia, Y., Xie, S., Wang, X., Zhao, J., Chu, Y., Zhou, Z., Shi, Z., Song, X. & Li, L. (2014). J. Biol. Chem. 289, 34104–34113. [DOI] [PMC free article] [PubMed]
- Musco, G., Kharrat, A., Stier, G., Fraternali, F., Gibson, T. J., Nilges, M. & Pastore, A. (1997). Nature Struct. Biol. 4, 712–716. [DOI] [PubMed]
- Musco, G., Stier, G., Joseph, C., Castiglione Morelli, M. A., Nilges, M., Gibson, T. J. & Pastore, A. (1996). Cell, 85, 237–245. [DOI] [PubMed]
- O’Donnell, W. T. & Warren, S. T. (2002). Annu. Rev. Neurosci. 25, 315–338. [DOI] [PubMed]
- Papoulas, O., Monzo, K. F., Cantin, G. T., Ruse, C., Yates, J. R. III, Ryu, Y. H. & Sisson, J. C. (2010). Development, 137, 4201–4209. [DOI] [PMC free article] [PubMed]
- Pardee, A. B. (1989). Science, 246, 603–608. [DOI] [PubMed]
- Pauw, R. J., van Drunen, F. J., Collin, R. W., Huygen, P. L., Kremer, H. & Cremers, C. W. (2008). Arch. Otolaryngol. Head Neck Surg. 134, 294–300. [DOI] [PubMed]
- Phan, A. T., Kuryavyi, V., Darnell, J. C., Serganov, A., Majumdar, A., Ilin, S., Raslin, T., Polonskaia, A., Chen, C., Clain, D., Darnell, R. B. & Patel, D. J. (2011). Nature Struct. Mol. Biol. 18, 796–804. [DOI] [PMC free article] [PubMed]
- Ramos, A., Hollingworth, D., Adinolfi, S., Castets, M., Kelly, G., Frenkiel, T. A., Bardoni, B. & Pastore, A. (2006). Structure, 14, 21–31. [DOI] [PubMed]
- Ratovitski, T., Chighladze, E., Arbez, N., Boronina, T., Herbrich, S., Cole, R. N. & Ross, C. A. (2012). Cell Cycle, 11, 2006–2021. [DOI] [PMC free article] [PubMed]
- Ross, C. A., Margolis, R. L., Rosenblatt, A., Ranen, N. G., Bêcher, M. W. & Aylward, E. (1997). Medicine, 76, 305–338. [DOI] [PubMed]
- Sabile, A. A., Arlt, M. J., Muff, R., Husmann, K., Hess, D., Bertz, J., Langsam, B., Aemisegger, C., Ziegler, U., Born, W. & Fuchs, B. (2013). Biochim. Biophys. Acta, 1832, 1173–1182. [DOI] [PubMed]
- Shapiro, L. & Scherer, P. E. (1998). Curr. Biol. 8, 335–338. [DOI] [PubMed]
- Shiina, N., Shinkura, K. & Tokunaga, M. (2005). J. Neurosci. 25, 4420–4434. [DOI] [PMC free article] [PubMed]
- Shiina, N. & Tokunaga, M. (2010). J. Biol. Chem. 285, 24260–24269. [DOI] [PMC free article] [PubMed]
- Shiina, N., Yamaguchi, K. & Tokunaga, M. (2010). J. Neurosci. 30, 12816–12830. [DOI] [PMC free article] [PubMed]
- Solomon, S., Xu, Y., Wang, B., David, M. D., Schubert, P., Kennedy, D. & Schrader, J. W. (2007). Mol. Cell. Biol. 27, 2324–2342. [DOI] [PMC free article] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Tongiorgi, E., Righi, M. & Cattaneo, A. (1997). J. Neurosci. 17, 9492–9505. [DOI] [PMC free article] [PubMed]
- Towers, E. R., Kelly, J. J., Sud, R., Gale, J. E. & Dawson, S. J. (2011). J. Cell Sci. 124, 1145–1155. [DOI] [PMC free article] [PubMed]
- Valverde, R., Pozdnyakova, I., Kajander, T., Venkatraman, J. & Regan, L. (2007). Structure, 15, 1090–1098. [DOI] [PubMed]
- Venkatraman Girija, U., Gingras, A. R., Marshall, J. E., Panchal, R., Sheikh, M. A., Gál, P., Schwaeble, W. J., Mitchell, D. A., Moody, P. C. E. & Wallis, R. (2013). Proc. Natl Acad. Sci. USA, 110, 13916–13920. [DOI] [PMC free article] [PubMed]
- Vognsen, T., Møller, I. R. & Kristensen, O. (2013). PLoS One, 8, e80947. [DOI] [PMC free article] [PubMed]
- Wang, B., David, M. D. & Schrader, J. W. (2005). J. Immunol. 175, 4274–4282. [DOI] [PubMed]
- Winslow, S., Leandersson, K. & Larsson, C. (2013). Mol. Cancer, 12, 156. [DOI] [PMC free article] [PubMed]
- Wu, Y., Zhu, J., Huang, X. & Du, Z. (2015). Acta Cryst. F71, 324–329. [DOI] [PMC free article] [PubMed]
- Xiang, M., Gan, L., Li, D., Chen, Z.-Y., Zhou, L., O’Malley, B. W. Jr, Klein, W. & Nathans, J. (1997). Proc. Natl Acad. Sci. USA, 94, 9445–9450. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HR-1 domain of human Caprin-1, space group C121, 4wbe
PDB reference: space group P3121, 4wbp
Supplementary Figure S1.. DOI: 10.1107/S2059798316004903/cb5087sup1.pdf