Abstract
Pseudomonas sp. VLB120 uses styrene as a sole source of carbon and energy. The first step in this metabolic pathway is catalyzed by an oxygenase (StyA) and a NADH-flavin oxidoreductase (StyB). Both components have been isolated from wild-type Pseudomonas strain VLB120 as well as from recombinant Escherichia coli. StyA from both sources is a dimer, with a subunit size of 47 kDa, and catalyzes the enantioselective epoxidation of C=C double bonds. Styrene is exclusively converted to S-styrene oxide with a specific activity of 2.1 U mg−1 (kcat = 1.6 s−1) and Km values for styrene of 0.45 ± 0.05 mM (wild type) and 0.38 ± 0.09 mM (recombinant). The epoxidation reaction depends on the presence of a NADH-flavin adenine dinucleotide (NADH-FAD) oxidoreductase for the supply of reduced FAD. StyB is a dimer with a molecular mass of 18 kDa and a NADH oxidation activity of 200 U mg−1 (kcat [NADH] = 60 s−1). Steady-state kinetics determined for StyB indicate a mechanism of sequential binding of NADH and flavin to StyB. This enzyme reduces FAD as well as flavin mononucleotide and riboflavin. The NADH oxidation activity does not depend on the presence of StyA. During the epoxidation reaction, no formation of a complex of StyA and StyB has been observed, suggesting that electron transport between reductase and oxygenase occurs via a diffusing flavin.
The initial reaction of many aerobic metabolic pathways consists of an epoxidation step (1, 18, 36, 53). Generally, this reaction is monooxygenation, in which molecular oxygen is reductively activated to perform the epoxidation with concomitant formation of water. Enzyme prosthetic groups or cofactors, e.g., transition metals, pterins, and flavins, react with dioxygen, giving rise to highly reactive forms of oxygen such as oxyferryl and peroxides. Enzymes catalyzing epoxidation reactions can be found in the heme-containing cytochrome P-450 family (43), members of which are often involved in detoxification processes. Thus, especially mammalian P-450 enzymes have been thoroughly studied (15). Other examples include the nonheme iron methane monooxygenases (12, 47) and the haloperoxidases (24, 50).
Various microorganisms, including species of Xanthobacter, Rhodococcus, Nocardia, Enterobacter, and yeast (14, 16, 34), are utilizing styrene as a sole source of carbon and energy, and different styrene degradation pathways have been described (33, 48). The epoxidation of the vinyl side chain of styrene catalyzed by a monooxygenase is the initial reaction in one microbial aerobic styrene degradation pathway. Further metabolic steps include isomerization of epoxystyrene into phenylacetaldehyde and the subsequent oxidation into phenylacetic acid (16, 48). Although styrene degradation by the upper pathway has been studied extensively, especially in Pseudomonas and Xanthobacter strains such as Xanthobacter strain 124X (17), Pseudomonas fluorescens ST (3), Pseudomonas sp. strain Y2 (46), Pseudomonas sp. strain VLB120 (38), and Pseudomonas putida CA-3 (35), the current understanding of these processes is focused on organization and regulation of the genes responsible for this metabolic pathway and on exploitation of the epoxidation reaction in whole-cell systems for biotechnological applications (9, 39). The biochemical properties of the isolated enzymes have thus far not been reported.
The genes responsible for styrene degradation in the styrene-mineralizing microbe Pseudomonas sp. strain VLB120 have been cloned and characterized (38). From these studies it has been concluded that the enzyme catalyzing the initial reaction is a two-component monooxygenase encoded by styA and styB. However, studies describing the biochemistry and the catalytic properties of the isolated proteins are still missing. In particular, the molecular architecture of the monooxygenase and its function on the protein level as well as the mechanism of oxygen activation and substrate epoxidation have been only a matter of speculation.
Here, we report the detailed characterization of StyA and StyB and their physiological interaction as a two-component styrene monooxygenase system, StyAB. Biochemical evidence is presented that StyAB is a new member of the two-component flavin-dependent monooxygenase family.
MATERIALS AND METHODS
Chemicals, bacterial strains, and plasmids.
All chemicals were purchased from Fluka AG (Buchs, Switzerland). 18O2-enriched air was obtained from Eurisotop (Gif-sur-Yvette, France). Catalase from beef liver, Taq polymerase, restriction enzymes, flavin mononucleotide (FMN)-NADH oxidoreductase from Photobacterium fischeri (formerly Vibrio fischeri), and T4 DNA ligase were obtained from Roche Molecular Biochemicals (Basel, Switzerland). The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Source or reference(s) |
|---|---|---|
| Strains | ||
| Pseudomonas sp. strain VLB120 | Wild-type Pseudomonas; styrene prototroph | 38 |
| E. coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ (traD36 proAB+lacIqlacZ ΔM15) | 42 |
| E. coli JM101 | supE thi-1 Δ(lac-proAB) F′(traD36 proAB+lacqlacZΔM15 | 42 |
| Plasmids | ||
| pSPZ10 | pBRKmΔ derivative with a pBR322 ori harboring the styAB genes as an EcoRI/HindIII fragment; Apr | 37, 39 |
| pSPW1 | pZero2.1 derivative carrying a 5.7-kb genomic DNA fragment from Pseudomonas strain VLB120; lacZp styScRABCD Kmr | 38 |
| pSPZ2Not | Multiple cloning site flanked by two Not restriction sites; Apr | 37 |
| pTEZ302 | pSPZ2Not derivative harboring the styB gene as an NdeI/AscI fragment; Apr | This study |
Media and microbiological methods.
Escherichia coli strains were grown in Luria-Bertani (LB) medium (42) or M9* (38) supplemented with 150 μg of ampicillin ml−1 at 37°C for StyB production or 50 μg of kanamycin ml−1 at 30°C for StyA production, respectively.
Plasmid DNA was purified with a QIAprep spin plasmid kit (QIAGEN, Basel, Switzerland). DNA fragments and PCR products were separated by agarose gel electrophoresis and isolated using the QIAquick gel extraction kit (QIAGEN). Digestion with restriction endonucleases and ligation experiments were carried out by using standard procedures (42).
PCRs were performed in a volume of 100 μl containing 10 μl of PCR buffer for Taq polymerase, 10 μl of dimethyl sulfoxide, 200 μM (each) deoxynucleoside triphosphates, 1 μM (both) primers (see below), and 100 ng of DNA template. This mix was preincubated at 95°C in a PCR thermocycler (Perkin-Elmer, Emeryville, Calif.) for 4 min before addition of 2 U of Taq polymerase. Twenty-five cycles were performed as follows: 94°C, 1 min; 45°C, 1 min; 72°C, 1 min; and 72°C, 7 min. Negative controls were based on template DNA from pUC18. Cells competent for transformation were prepared using a CaCl2-based method (31). Transformation of the cells was done by using a standard procedure based on heat shock (42).
Cultivation of E. coli JM101(pSPZ10).
E. coli JM101(pSPZ10) was cultivated for production of recombinant StyA in a 3-liter reactor with 2 liters of M9* medium as described elsewhere (39). After overnight batch growth, a linear feed of a 45% (wt/vol) glucose solution at a rate of 10 g liter−1 h−1 was initiated. The reactor was run in fed-batch mode until the end of the cultivation. After 1 h of glucose feed, the culture was induced with 0.05% (vol/vol) dicyclopropylketone. Typically, 60 g (wet weight) of cells was harvested 7 h after induction by centrifugation (15 min; 2,700 × g; 4°C), washed, frozen as a pellet in liquid nitrogen, and stored at −80°C. Pseudomonas strain VLB120 was cultivated for the production of wild-type StyA. The organism was grown in the same 3-liter reactor containing 2 liters of M9* medium and styrene as the single carbon source. After inoculation, styrene was continuously fed to the culture by running the air stream through a bottle filled with bis(2-ethylhexyl)phthalate containing 10% (vol/vol) styrene (aeration rate, 0.4 liters min−1). After 23 h (optical density at 450 nm, 2.5), the aeration rate was increased to 1.2 liters min−1 and 400-μl portions of styrene were added directly every 30 min in addition to the constant feed via aeration. At 27.5 h (optical density at 450 nm, 8.3), the amount of styrene added directly was increased to 800 μl every 20 min. The fermentation was stopped after 32 h (optical density, 27.5) and yielded 46 g (wet weight) of cells.
Purification of the oxygenase StyA.
All buffers contained 1 mM dithiothreitol (DTT) and 1 mM MgCl2. The entire procedure was performed at 4°C unless indicated otherwise. Fractions containing StyA were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). To prepare the cell extract, 6 g (wet weight) of cells was dissolved in 20 mM Tris-Cl buffer (pH 7.5) containing 10% (vol/vol) glycerol and cells were disrupted by two passages (1,000 lb/in2) through a French press unit (AMINCO; SLM Instruments, Inc.). Insoluble cell debris was removed by ultracentrifugation for 35 min at 150,000 × g. The supernatant was loaded onto an anion-exchange XK16/20 column (Amersham Biosciences, Dübendorf, Switzerland) filled with 12 ml of EMD trimethylaminoethyl (TMAE) 650(s) Fractogel (Merck, Darmstadt, Germany) at a flow rate of 1.5 ml min−1 in starting buffer (20 mM Tris-Cl, pH 6.5). Elution was done using a linear gradient of 0 to 360 mM NaCl in starting buffer. Fractions containing StyA were pooled, and ammonium sulfate was added to a final concentration of 1.6 M. The StyA pool was subjected to chromatography using a hydrophobic-interaction SOURCE15ETH column (volume, 35 ml; Amersham Biosciences) at a flow rate of 1.5 ml min−1 in starting buffer [20 mM Tris-Cl (pH 7.5), 1.6 M (NH4)2SO4] at 20°C. Elution of StyA was achieved by using a linear gradient of 1.6 to 0.8 M ammonium sulfate over 110 min. StyA eluted at an ammonium sulfate concentration of 1.1 M. Fractions containing StyA were identified by SDS-12% PAGE, pooled, and concentrated by ultrafiltration in a centrifugal filter device (Centricon-Biomax 30K; Millipore Corporation, Bedford, Mass.) to a volume of less than 5 ml. The pooled fractions were subjected to chromatography via size exclusion on a Superdex 200 HiLoad 16/60 column (Amersham Biosciences) at 1 ml min−1 in 20 mM KPi, pH 7.5. Fractions containing purified StyA were immediately supplemented with 10% (vol/vol) glycerol and 1 mM Pefabloc (Fluka, Buchs, Switzerland) and stored at −20°C.
Expression of styB in E. coli.
Two PCR primers (UpNdeI, 5′-CTG GGT GAT TCA TAT GAC GTT AAA AAA AG-3′, and LowAscI, 5′-GTT GTT TTG TTG GCG CGC CAT CAA TT-3′) were designed to introduce an NdeI restriction site upstream and an AscI restriction site downstream of the styB gene (bases changed in order to introduce restriction sites are in bold). The DNA fragment containing the styB gene was amplified by PCR from pSPW1 (38) by using the described primers. The PCR product was purified by agarose gel electrophoresis, digested with NdeI and AscI, and cloned into the NdeI/AscI site of plasmid pSPZ2Not (37) under the control of the alk promoter, which is induced by addition of octane or dicyclopropylketone, resulting in pTEZ302. The correct gene sequence was confirmed by DNA sequencing.
Cultivation of E. coli JM109(pTEZ302).
E. coli JM109(pTEZ302) was grown in LB medium (150 μg of ampicillin ml−1) at 37°C. For 1-liter cell culture, 100 ml of LB was inoculated with one colony of freshly transformed E. coli and incubated for 8 h. Cells were harvested by centrifugation and resuspended in 1 liter of fresh LB medium. After 40 min, 0.05% (vol/vol) dicyclopropylketone was added to induce the alk regulatory system of pTEZ302. This procedure was followed by overnight incubation of the culture at 37°C. Cells were harvested by centrifugation, and the resulting pellet was stored at −20°C.
Purification of the reductase unit StyB.
The purification of recombinant StyB (StyBrec) was based on inclusion body isolation. All steps of the procedure were performed at room temperature. Crude extract was prepared by washing 2 g (wet weight) of cells twice in 100 ml of buffer A (50 mM Tris, pH 7.5). After centrifugation, the pellet was resuspended at 100 g (wet weight) of cells liter−1 in lysis buffer (buffer A, 25% [wt/vol] sucrose, 10 mM EDTA) and incubated for 15 min on ice. Subsequently, 10 mg of lysozyme ml−1 was added and the suspension was incubated for 30 min. Afterwards, the cells were disrupted by two passages (1,000 lb/in2) through a French press (AMINCO; SLM Instruments, Inc.). The crude extract was centrifuged for 30 min at 2,000 × g. The resulting pellet was washed six times sequentially using the following buffers: W1, buffer A, 0.5% (vol/vol) Triton X-100, and 10 mM EDTA; W2, buffer A and 2 M urea; and W3, buffer A, 0.15 M NaCl, and 1 mM EDTA. Each washing step consisted of two 40-min incubations in the appropriate buffer followed by centrifugation at 2,000 × g. After washing, the inclusion bodies were solubilized in 300 ml of buffer (buffer A, 5 mM DTT, 8 M Urea) to a concentration of <0.5 mg of protein ml−1. The solution was stirred overnight at 30°C and was then submitted to ultracentrifugation (150,000 × g; 45 min; 15°C) to separate any particulate material. Refolding of StyB was achieved by diluting the supernatant dropwise 1:10 into refolding buffer (25 mM Tris, 1 mM DTT [pH 7.5]) while stirring the solution continuously. Aggregates were then removed by centrifugation (18,000 × g; 30 min). Refolded StyB was concentrated by anion-exchange chromatography using a XK16/20 column (Amersham Biosciences) filled with 12 ml of EMD TMAE 650(s) Fractogel (Merck) at 10°C. The protein was loaded onto the column at a rate of 6 ml min−1 in refolding buffer and eluted with buffer B (25 mM Tris, 0.5 mM DTT, 0.5 mM MgCl2, 1 M NaCl, 5% glycerol) at a flow rate of 2 ml min−1. Fractions showing StyB activity were pooled and kept at 4°C or were frozen at −20°C for later use.
For the enrichment of wild-type StyB, Pseudomonas strain VLB120 was grown as described above. The complete purification procedure was performed at 4°C. To prepare the cell extract, 6 g (wet weight) of cells was resuspended in 30 ml of 20 mM Tris, pH 6.5, with 10% glycerol, 1 mM DTT, and 1 mM MgCl2 and the cells were disrupted by French press treatment. Insoluble cell debris was removed by ultracentrifugation for 35 min at 150,000 × g. The supernatant was loaded onto an anion-exchange XK16/20 column (Amersham Biosciences) filled with 12 ml of EMD TMAE 650(s) Fractogel (Merck) at a flow rate of 1 ml min−1 in starting buffer (20 mM Tris-Cl, pH 6.5). Elution was done using a linear gradient of 0 to 100 mM NaCl in starting buffer for 46 ml, followed by 100 to 300 mM NaCl for an additional 216 ml. Fractions with NADH oxidation activity were pooled and resubjected to chromatography at pH 5.9 under otherwise similar conditions.
Molecular mass determination and characterization of StyAB interaction.
The molecular mass of StyA was determined by gel filtration on a calibrated Superdex 200 high-load 16/60 column (the molecular mass marker kit [range, 12 to 200 kDa] was from Sigma-Aldrich, Steinheim, Germany). The molecular mass of StyB was calculated from its nucleotide sequence and confirmed by SDS-PAGE and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis (42). Sedimentation velocity measurements were performed to determine the native molecular mass of StyB by using a Beckman Optima XL-I analytical ultracentrifuge. Sedimentation of the enzyme was measured by absorption (280 nm) and interference (42,000 rpm; 20°C; 100 scans; 6.5 h) simultaneously. Samples consisted of 1 mg of protein ml−1 in a buffer containing sodium phosphate buffer (50 mM; pH 7.3), 100 mM NaCl, 1 mM DTT, and 5% (vol/vol) glycerol. The calculation of the mass and the sedimentation coefficient was performed with the program SedFit (Peter Schuck, Division of Bioengineering and Physical Sciences, Office of Research Sciences, Office of the Director, National Institutes of Health, Bethesda, Md.). To investigate StyAB interaction, StyA and StyB were added to a concentration of 50 μM each into the above-mentioned buffer. Optionally, samples additionally contained 1 mM flavin adenine dinucleotide (FAD), 2 mM styrene, and 20 mM NADH.
Spectral measurements.
All experiments requiring photometric measurements were carried out in a temperature-controlled Cary 1E UV-visible spectrophotometer (Varian AG, Zug, Switzerland). To determine enzyme activities and substrate concentrations, the following absorption coefficients were used: NADH, 6.22 mM−1 cm−1 (340 nm); NADPH, 6.22 mM−1 cm−1 (340 nm); FAD, 11.3 mM−1 cm−1 (450 nm) and 4.78 mM−1 cm−1 (340 nm); FMN, 12.2 mM−1 cm−1 (450 nm) and 5.45 mM−1 cm−1 (340 nm); and riboflavin, 12.2 mM−1 cm−1 (450 nm) and 5.85 mM−1 cm−1 (340 nm) (40).
Samples for measuring the CO difference spectrum were prepared in 50 mM sodium phosphate buffer, pH 7.6. The StyA concentration was 1 mg ml−1. Before measurement of the spectrum between 300 and 650 nm, the protein was reduced by the addition of 15 mg of DTT and subsequently aerated with carbon monoxide for 12 min.
Determination of StyA activity.
Activities are generally given in units, where 1 U is defined as the activity which converts 1 μmol of substrate per minute. StyA activity was determined by measuring the formation of S-styrene oxide by reverse-phase high-pressure liquid chromatography on a LiChrospher125-4 RP-18 (5-μm) column (Machery-Nagel, Oensingen, Switzerland) at a flow rate of 0.750 ml min−1 (retention time [RT], 2.8 min). The conversion of all other substrates was determined using a CC259/4 Nucleosil 100-3 C18 HD column (Machery-Nagel) under conditions that were otherwise the same (RT: 1.2-dihydronaphtalene oxide, 5.1 min; 3-chlorostyrene oxide, 5.86 min; α-methylstyrene oxide, 5.37 min; β-methylstyrene oxide, 5.08 min; p-methylstyrene oxide, 5.38 min). The mobile phase consisted of an acetonitrile-water mixture (ratio, 60:40). A typical assay mixture contained 3 μM StyA and 3 μM StyB, 15 μM FAD, 50 mM NADH, 650 U of catalase, 150 mM sodium formate, 0.5 U of formate dehydrogenase, and 2 mM aromatic substrate in a total volume of 1,000 μl of 20 mM Tris-Cl, pH 7.5, containing 1 mM DTT and 5% glycerol. Alternatively, StyB was replaced by the FMN-NADH oxidoreductase of Photobacterium fischeri. The reaction was started by the addition of NADH. Reaction mixtures were incubated at 37°C on a shaker at 13,000 rpm. After 5 min, 0.5 ml of acetonitrile was added to stop the reaction. The samples were stored at room temperature for 20 min, followed by centrifugation (20 min; 10,000 × g; 15°C) to remove precipitated proteins. Methyl phenyl sulfoxide was extracted with diethyl-ether and analyzed by normal-phase high-pressure liquid chromatography on a CHIRALCEL OB-H column (Daicel, Deventer, Holland) at a flow rate of 0.5 ml min−1. The mobile phase consisted of an octane-isopropanol mixture (ratio, 95:5) (RTs for racemate methyl phenyl sulfoxide, 25.6 and 45.4 min).
Determination of StyB activity.
Enzyme assays were carried out at 30°C in reaction buffer consisting of sodium phosphate (50 mM NaPi, pH 7.2) supplemented with 1 mM DTT and 5% glycerol. StyB activity was determined spectrophotometrically by measuring the rate of NADH oxidation at 340 nm. A standard reaction mixture contained 0.5 μg of StyB and 200 μM FAD, FMN, or riboflavin in 1,000 μl of buffer. The assay was started by the addition of NADH to a final concentration of 200 μM. Steady-state kinetic parameters were calculated by weighted nonlinear regression analysis (Enzfitter; Elsevier-Biosoft, Cambridge, United Kingdom).
Stoichiometry of FAD binding.
The stoichiometry of FAD binding to StyB was determined by ultrafiltration. StyB at a concentration of 36 μM was incubated for 30 min at room temperature with 100 μM FAD in 2 ml of reaction buffer. Afterwards, the sample was loaded onto a CentriCon-10 centrifugal filter device (Mr cutoff, 10,000; YM-10 membrane; Millipore Corporation) and centrifuged for 10 min at 4,000 × g. The protein content and the FAD concentration in the retentate and the filtrate were determined spectrophotometrically at 450 nm by assuming that enzyme-bound FAD has the same absorption coefficient as free FAD (40). To determine the Kd of FAD binding to StyB and StyA by equilibrium dialysis, a mixture of StyB or StyA (24 μM) and FAD (620 μM) in 1.5 ml of buffer D (20 mM Tris-HCl, 20 mM imidazole, 100 mM NaCl [pH 7.3], 5% [wt/vol] glycerol) was dialyzed for 2 days against 500 ml of buffer C (20 mM Tris-HCl, 100 mM NaCl [pH 7.3], 5% [wt/vol] glycerol) by using a dialysis membrane (Mr cutoff, 10,000; Sigma-Aldrich).
Detection of hydrogen peroxide.
Hydrogen peroxide was detected using a modified assay developed by Saito et al. (41). A standard assay mixture contained 0.5 μg of StyB, 100 μM FAD, and 200 μM NADH in a volume of 1,000 μl of potassium phosphate buffer (50 mM; pH 7.0) supplemented with 1 mM 4-amino antipyrine, 12 mM phenol, and 6 U of peroxidase from Coprinus cinereus ml−1. The reaction was started by the addition of NADH. The resulting quinoid dye concentration was measured spectrophotometrically at 550 nm (ɛ550 = 22.224 mM−1 cm−1).
Metal analysis of StyAB.
Metal analysis was done using a Perkin-Elmer 1100 atomic absorption spectrometer (AAS). Solutions of StyA from recombinant E. coli JM101(pSPZ10) and wild-type Pseudomonas sp. strain VLB120 and of StyB from recombinant E. coli JM109(pTEZ302) were prepared in double-deionized water in a concentration of 2 mM protein. As a reference, the different metals were dissolved in double-deionized water at a concentration of 10 to 1 μg ml−1.
Incorporation of 18O2.
A standard StyAB activity assay was performed in 18O2-enriched air as described by Bühler et al. for xylene monooxygenase (5). The epoxidation reaction was started by addition of styrene through a rubber cap. Samples were incubated for 15 min in a shaker at 250 rpm. The reaction was stopped on ice by addition of 1 ml of ether. Products were extracted into the ether phase, which was analyzed by gas chromatography-mass spectrometry using a Fision 8000 gas chromatograph with a Supelco 24044 column. The program was as follows: 4 min at 40°C, a rate of change of 5°C min−1, and 2 min at 110°C. Controls were made with air and under anaerobic conditions. The molar ratio of 18O2 to 16O2 in the gas used for the labeling experiment was determined to be 60:40 by gas chromatography-mass spectrometry analysis.
RESULTS
Purification and characterization of the reductase subunit StyB.
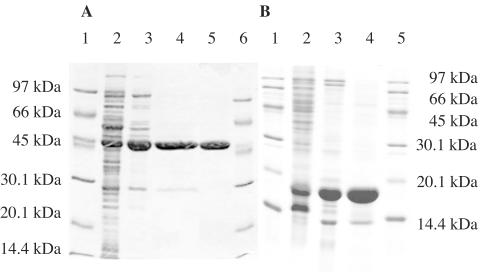
StyB was expressed under control of the alk regulatory system of Pseudomonas putida GPO1 (38) in E. coli JM109(pTEZ302) by using 0.05% dicyclopropyl ketone as an inducer. A 1-liter culture typically yielded 2 g (wet weight) of cells, with StyB accounting for approximately 45% of total cell protein. Phase-contrast microscopy showed that StyB formed inclusion bodies. We purified StyBrec from these inclusion bodies (Table 2; Fig. 1B). Purified, refolded StyB was colorless, indicating that no flavin was bound to the protein. The additional band in the purified StyB sample can be attributed to the addition of lysozyme during the purification procedure.
TABLE 2.
Data from purification of StyB from recombinant E. coli JM109 (and wild-type Pseudomonas strain VLB120)
| Purification step | Total protein (mg) | Sp acta (U mg−1) | Total activity (U) | Purification factor |
|---|---|---|---|---|
| Preparation of crude extract | 200 (780)b | NDc (0.3) | NDc (234) | NDc (1) |
| IB solubilizationd | 40 | 23 | 932 | 1 |
| Refoldingd | 20 | 197 | 3,930 | 8.5 |
| Pooling after IECe | 15 (61.2) | 200 (2) | 3,000 (104) | 8.7 (6.6) |
Activity was determined by measuring the NADH oxidation rate as described in Materials and Methods.
The purification utilized 2.3 (6) g (wet weight) of cells.
ND, Not determined because StyB was in inclusion body form and therefore not active.
Pseudomomas strain VLB120 did not form inclusion bodies (IB); therefore; no data are given for wild-type StyB.
IEC, ion-exchange chromatography.
FIG. 1.
Purification of StyA and StyB. Protein samples were analyzed at different stages of purification by SDS-12% PAGE (A) and SDS-15% PAGE (B) under reducing and denaturing conditions and stained with Coomassie brilliant blue. (A) Lanes: 1 and 6, marker; 2, crude extract from induced E. coli JM101(pSPZ10); 3, pool of fractions containing StyA after ion-exchange chromatography; 4, pool of fractions containing StyA after hydrophobic-interaction chromatography; 5, pure StyA after size exclusion chromatography. (B) Lanes: 1 and 5, marker; 2, crude extract from induced E. coli JM109(pTEZ302); 3, purified inclusion bodies; 4, StyB after ion-exchange chromatography.
Subunit size and composition of StyBrec were estimated by SDS-PAGE. StyBrec migrated as a single band of approximately 18 kDa, corresponding to the subunit size of 18.364 kDa, which was deduced from the primary sequence. MALDI-TOF analysis indicated a protein size of 18.234 kDa, compatible with the removal of a methionine residue. Edman sequencing showed that the N-terminal methionine was indeed missing. The size of the native protein was determined to be 37 kDa by analytical ultracentrifugation, suggesting a homodimeric structure for native StyBrec.
To compare the catalytic properties of StyBrec and the wild-type enzyme, StyB was partially purified from Pseudomonas strain VLB120 (StyBwt+). After a first separation via anion-exchange chromatography at pH 6.5, we detected NADH-flavin oxidoreductase activities in three different fractions. Rechromatography of the pooled active fractions at pH 5.9 (near the calculated pI of StyB of 5.25) under otherwise identical conditions resulted in a single fraction with oxidoreductase activity. As the biochemical properties determined for this fraction were similar to those of StyB (see below) and because the salt concentration at which this protein eluted corresponded well to the elution profile of soluble StyBrec (data not shown), we concluded that this fraction contained StyB. A NADH oxidation activity of 2 U mg−1 was determined for the fraction containing StyB.
Kinetic data for the reductase subunit StyB.
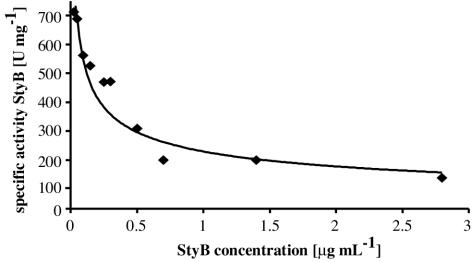
The specific activities of the oxidoreductase in the fractions from anion-exchange chromatography varied with the StyBrec concentrations. Figure 2 shows that the specific enzyme activity increased in dilute StyB preparations. At a StyB content of 1 to 2 μg of protein ml−1, the specific activity was constant at about 200 U mg−1.
FIG. 2.
StyB activity as a function of enzyme concentration. Activity was determined by measuring the NADH oxidation rate at 30°C.
The substrate spectrum and the steady-state kinetics of StyBrec and StyBwt+ were examined in detail in a series of initial rate studies (Table 3). The oxidoreductase shows Michaelis-Menten kinetics towards all its substrates when the flavin concentration is below 200 μM. Flavin concentrations above 200 μM inhibited StyBrec and StyBwt+ significantly. While StyB was exclusively dependent on NADH as a source of reducing equivalents (NADPH was not converted at all), it revealed a more comprehensive spectrum of possible electron acceptors. The results suggest identical catalytic features of StyB enzymes isolated from recombinant E. coli JM109 and wild-type Pseudomonas strain VLB120.
TABLE 3.
kcat and Km values for StyBrec (and StyBwt+) for different flavins and electron donorsa
| Substrate | Second substrate (200 μm) | kcat (s−1) | Km (μM) | kcatKm−1 (s−1 μM−1) |
|---|---|---|---|---|
| FMN | NADH | 48 | 7.1 ± 0.7 (7.0 ± 0.5) | 6.78 |
| FAD | NADH | 47 | 11.6 ± 1.4 (10.0 ± 0.9) | 4.05 |
| Riboflavin | NADH | 60 | 14.7 ± 2.8 (15.0 ± 1.4) | 4.08 |
| NADH | FMN | 60 | 21.06 ± 2.6 (25.0 ± 2.0) | 2.84 |
| NADPH | FMN | No activity |
Data were calculated by weighted nonlinear regression analysis (Enzfitterφ; Elsevier-Biosoft. Activities were determined by the standard reductase assay described in Materials and Methods, with 0.7 μg of StyB ml−1 or 80 μg of the StyB-containing fraction from Pseudomonas strain VLB120.
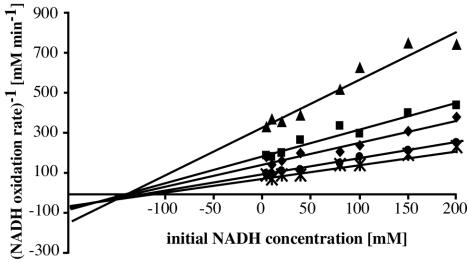
The effect of the various concentrations of FMN and NADH on the StyBrec reaction rate are shown in a Hanes plot (Fig. 3). The intersecting pattern suggests the formation of a ternary complex of StyB, NADH, and the flavin cofactor in the course of the catalytic reaction.
FIG. 3.
Hanes plot of kinetic data for StyB. Enzyme activity was measured at a fixed concentration of StyB (0.27 μmol ml−1) and various concentrations of NADH and FMN. The concentrations of FMN used were 20 (✻), 10 (•), 5 (⧫), 2.5 (▪), and 1 (▴) μM.
Purification and characterization of the monooxygenase subunit StyA.
StyA was purified from recombinant E. coli JM101(pSPZ10) (StyArec) and from the wild-type Pseudomonas strain VLB120 (StyAwt+). Details are given in Table 4 and Fig. 1A. To evaluate whether expression of styA in E. coli resulted in structural or functional modifications, both proteins were compared with respect to their basic characteristics (size, N-terminal sequence, and substrate spectrum).
TABLE 4.
Data from purification of StyArec (and StyAwt+)
| Purification stepa | Total protein (mg) | sp actb (U mg−1) | Total activity (U) | Activity yield (%) | Purification factor |
|---|---|---|---|---|---|
| Preparation of crude extract | 707 (780)c | 0.45 (0.1) | 318 (78) | 100 (100) | 1 (1) |
| Pooling after IEC | 245 (100) | 1.2 (0.31) | 294 (31) | 92 (40) | 2.7 (3.1) |
| Pooling after HIC | 180 (11) | 1.51 (1.1) | 270 (12) | 85 (15) | 3.5 (11) |
| Pooling after SEC | 95 (6) | 2.1 (1.9) | 177 (11) | 55 (14) | 4.1 (19) |
IEC, ion-exchange chromatography; HIC, hydrophobic-interaction chromatography; SEC, size exclusion chromatography.
The activity determination was based on styrene epoxide formation as described in Materials and Methods.
The purification was performed with 6 g (wet weight) of cells.
A 2-liter fermentation mixture yielded approximately 47 g (wet weight) of cells of Pseudomonas sp. strain VLB120 and 60 g (wet weight) of cells of recombinant E. coli JM101. StyA accounted for approximately 5% of the total cell protein in the wild-type strain and 25% in the recombinant strain. The cells could be stored at −80°C for over a year without significant loss of StyA activity. StyArec and StyAwt+ were purified to near homogeneity by the three-step procedure described in Materials and Methods. Purification parameters are shown in Table 4. The recombinant as well as the wild-type form of StyA was colorless upon purification, indicating that no FAD was bound to either protein.
After purification, the specific styrene epoxidation activities were 2.1 and 1.9 U mg−1 for StyArec and StyAwt+, respectively. The enzyme activity had a half-life of 2 days in buffer at 4°C. Addition of 10% (vol/vol) glycerol and 1 mM Pefabloc to the storage buffer improved enzyme stability: the activity decreased by only 20% during the first 2 weeks of storage at 4°C. Even after 3 months at 4°C, StyA still retained 60% of its initial activity.
Gel filtration chromatography on a calibrated Superdex 200 column indicated a molecular mass of 80 ± 10 kDa for the native StyArec as well as for the native StyAwt+. Both proteins migrated as a single band with a subunit size of approximately 47 kDa in an SDS-PAGE gel. These results were confirmed by MALDI-TOF analysis (molecular mass, 46.350 kDa) and suggest a homodimeric structure for both native enzyme species. The N-terminal amino acid sequences of StyAwt+ and StyArec determined via Edman analysis were identical and corresponded to that expected from the nucleic acid sequence of the styA gene (TrEMBL accession number O50214). A pI of 5.3 was calculated based on the deduced amino acid sequence of StyA.
In addition, several styrene derivates were tested as substrates for StyAwt+ (for the substrate spectrum of the recombinant enzyme, refer to Hollmann et al. [20]). The wild-type enzyme catalyzed the epoxidation of p-, α-, and β-methylstyrene, 1,2-dihydronaphthalene, methyl phenyl sulfide, and 3-chlorostyrene at rates comparable to those achieved with the recombinant form of the enzyme (data not shown).
Characteristics of the StyAB system.
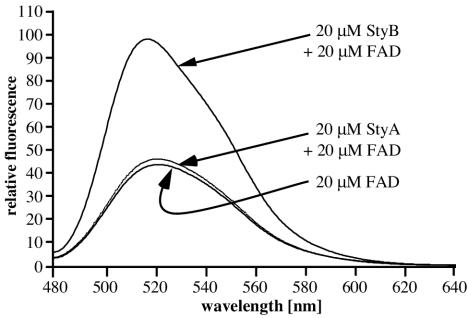
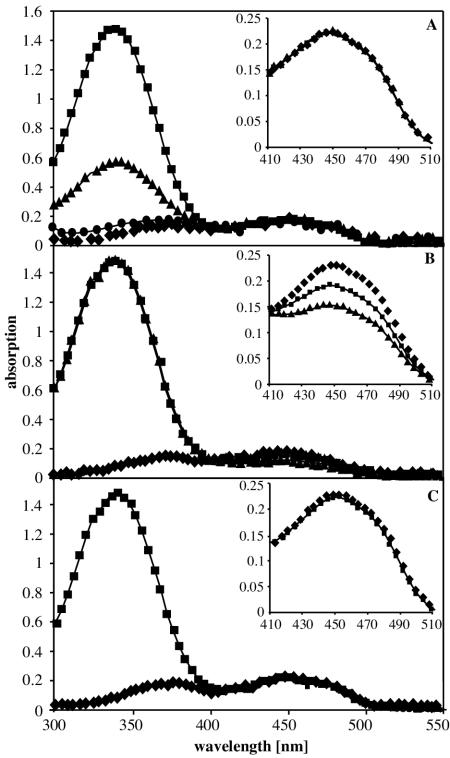
To determine whether purified recombinant StyAB (StyABrec) and wild-type StyAB (StyABwt+) contain a bound FAD or a heme as a prosthetic group, UV spectra and a CO difference spectrum of the enzyme subunits were recorded. Significant absorption maxima were not observed either for FAD or for the heme group, implying that neither subunit contains a bound cofactor after purification. To further examine the binding of FAD to StyA and StyB, we measured the fluorescence of FAD in the presence of either subunit. FAD fluoresces at 520 nm upon excitation at 450 nm. Binding of this molecule by a protein results in a change in the fluorescence emission intensity (2). Adding 20 μM StyB to 20 μM FAD led to a significant increase in the intensity of the FAD fluorescence, whereas only a minor effect was observed after the addition of 20 μM StyA to free FAD (Fig. 4). These findings indicate that FAD binds more efficiently to StyB than to StyA.
FIG. 4.
Fluorescence emission spectra of FAD, StyA, and StyB in various combinations (excitation wavelength, 450 nm). Measurements were done in reaction buffer at room temperature.
This result was confirmed by determining the apparent Kd values for FAD binding to StyA and StyB via equilibrium dialysis. StyA at a concentration of 24 μM was incubated with 620 μM FAD as described in Materials and Methods. After 2 days of dialysis, UV spectra (280 to 600 nm) from the protein-containing compartment and the outer compartment were recorded. The difference spectrum revealed a maximum of 0.016 at 450 nm. Assuming that the absorption coefficient for enzyme-bound FAD is the same as that for free FAD (40), the concentration of enzyme-bound FAD was determined to be 1.4 μM. By considering the concentration of free FAD (1.2 μM), a Kd of 21.2 ± 2 μM was determined for FAD binding to StyA. The same experiment was done to determine the Kd of FAD binding to StyB. The corrected UV spectrum of the protein-containing fraction showed an absorption maximum of 0.161 at 450 nm. The concentration of enzyme-bound FAD was determined to be 14.2 μM, corresponding to 67.6% saturation of StyB with FAD. The Kd for FAD binding to StyB was determined to be 2.3 ± 0.3 μM.
The StyAB reaction system was incubated with different compounds known to inhibit oxidoreductases. Experiments were performed either with StyABrec and StyABwt+ to determine the styrene oxide formation rate or with StyBrec to measure the NADH oxidation rate. StyBrec activity was fully inhibited in the presence of 200 μM p-hydroxymercuribenzoate. Ninety percent of the initial StyB activity was recovered upon addition of 2 mM DTT. Chelating agents such as EDTA and phenanthroline had no effect on the epoxidation activity of the StyABrec and StyAwt+-StyBrec systems, which indicates that no divalent ions are necessary for catalysis. The same is true of metyrapone and proadifen, two effective inhibitors of cytochrome P-450 enzymes, suggesting that no heme is involved in the reaction. In the presence of 20 μM CuSO4 or 100 μM AgNO3, no StyBrec activity was detectable. The presence of StyA did not prevent the inhibitory effect of the aforementioned agents, and no formation of styrene oxide was observed. These results were confirmed by AAS analysis, in which neither StyA nor StyB was found to contain Fe2+, Cu2+, Zn2+, Mn2+, Co2+, or Mg2+.
StyAB-catalyzed epoxidation reaction.
StyA activity for the epoxidation of styrene into S-styrene oxide was restored by the addition of StyB, NADH, and FAD. The apparent Km value for styrene was determined to be 0.38 ± 0.09 mM for StyArec and 0.45 ± 0.05 mM for StyAwt+. Adding the same number of units of the flavin-NADH oxidoreductase of Photobacterium fischeri or a chemical reductant (20) instead of StyB to the reaction mixture under otherwise identical conditions resulted in 70% epoxidation activity. In contrast to the redox reaction, which is catalyzed by StyB alone, no epoxidation activity was observed for the StyAB system when FAD was replaced by FMN or riboflavin.
The potential formation of an enzyme complex between StyA and StyB was investigated using analytical ultracentrifugation. Samples were prepared as described in Materials and Methods. StyA and StyB were assayed in the presence and absence of cofactors and a substrate. In neither case was a protein population attributable to an enzyme complex detected. In fact, only two populations corresponding to native StyA and native StyB were observed.
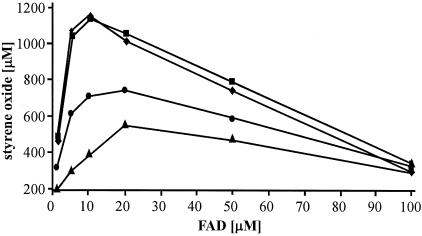
To elucidate whether StyA had any influence on the specific activity of StyB, the NADH oxidation rate was measured in the presence of different StyA concentrations, which varied from 72 nM to 23 μM at constant FAD and StyB concentrations (1 μM and 72 nM, respectively). All other parameters were as described in Materials and Methods. No change in the NADH oxidation rate was observed under these conditions, whereas the StyA activity was strongly influenced by the amount of StyB added. To avoid NADH limitation during the reaction, a NADH regeneration system based on formate dehydrogenase (26, 45) was included in the assay (see Materials and Methods). Catalase was added to prevent inactivation of the enzymes by hydrogen peroxide formation. Under these conditions, maximal epoxidation activity was measured when the molar amount of StyB present in the reaction mixture was equal to or larger than that of StyA (Fig. 5). At a FAD concentration exceeding 15 μM, the styrene oxide formation rate decreased.
FIG. 5.
Styrene oxide formation by StyAB as a function of the FAD concentration at various molar ratios of StyA to StyB. Concentrations are given in micromolar units. Ratio of StyA to StyB: ▴, 3:0.15; •, 3:0.3; ⧫, 3:3; ▪, 3:6.
The epoxidation activity of 2.1 U mg−1 persisted during the first 5 min and then rapidly decreased to 0.5 U mg−1. Assaying for product inhibition of StyAB revealed a significant decrease in the styrene epoxidation activity at styrene oxide concentrations exceeding 0.5 mM.
Changes in the UV signal of FAD at 450 nm allowed an evaluation of the binding of FADH2 to StyA (Fig. 6). When only StyB was present, the FAD concentration remained constant after addition of NADH, which was due to the spontaneous reaction of FADH2 with molecular oxygen, yielding FAD and hydrogen peroxide (550 μM) (Fig. 6A and 7, reaction 3). The presence of StyA caused a decrease in the concentration of free FAD, which was proportional to the StyA concentration (Fig. 6B). The concentration of hydrogen peroxide was determined to be 390 μM in the presence of 5 μM StyA and 220 μM in the presence of 10 μM StyA. Therefore, we conclude that StyA stabilizes a reduced or oxygenated form of FAD. Whether it is the flavoquinone or the 4α-hydroperoxoflavin remains to be clarified. Addition of the substrate styrene to a reaction mixture containing StyAB reversed the stabilizing effect of StyA, which can be ascribed to the FADH2 oxidation during styrene epoxidation (Fig. 6C).
FIG. 6.
Variation of the FAD signal upon the addition of StyA and styrene. A standard assay mixture contained 15 nM StyB, 200 μM NADH, and 20 μM FAD in 50 mM potassium phosphate buffer, pH 7.5, supplemented with 10% glycerol and 1 mM DTT. The reaction was started by adding NADH. The insets show enlargements of the FAD peaks at 450 nm. (A) Scans were made under standard conditions in the absence of StyA and styrene. ⧫, before addition of NADH; ▪, after addition of NADH; ▴, 1 min and 40 s after addition of NADH; •, 3 min and 40 s after addition of NADH. (B) Scans were made in the presence of StyA and in the absence of styrene. ⧫, before addition of NADH; ▪, after addition of NADH in the presence of 5 μM StyA; ▴, after addition of NADH in the presence of 10 μM StyA. (C) Scans were made in the presence of 10 μM StyA and 2 mM styrene. ⧫, before addition of NADH; ▪, after addition of NADH.
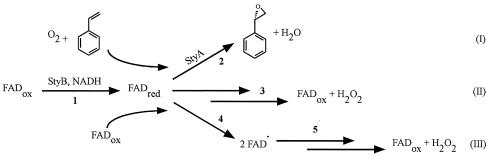
FIG. 7.
Possible reactions of reduced FAD (FADred) in an epoxidation reaction under aerobic conditions (adapted from reference 29). In reaction 1, oxidized FAD (FADox) is reduced to FADred by StyB in the presence of NADH. (I) Binding of FADred or the peroxoflavin by StyA for the epoxidation of styrene (reaction 2). (II) Auto-oxidation of FADred into hydrogen peroxide and oxidized FADox (reaction 3). (III) Radical formation by electron transfer between FADred and FADox (reaction 4) and (III) subsequent reaction with molecular oxygen, resulting in hydrogen peroxide and FADox (reaction 5).
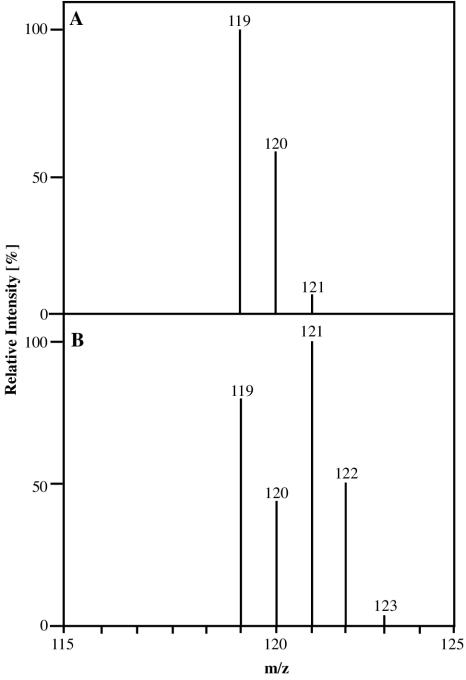
18O2 labeling experiments were performed to confirm the monooxygenating nature of the StyA-catalyzed epoxidation reaction. Figure 8 shows the mass spectra of styrene oxide formed from styrene in the presence of air (panel A) and in an atmosphere in which the molecular oxygen contained 60% 18O (panel B). The fragmentation pattern of the two spectra clearly shows that one oxygen atom from molecular oxygen was incorporated into the aromatic substrate. The ratio of labeled styrene oxide (M = 122) to nonlabeled styrene oxide (M = 120) is 52:48 and corresponds well to the ratio of 16O to 18O in the gas used. Furthermore, no product formation was detectable under otherwise identical conditions in the absence of O2 or NADH. These results clearly demonstrate that the epoxide oxygen is derived from molecular oxygen.
FIG. 8.
Mass spectra of styrene oxide synthesized by StyAB in air (A) and in 18O2-enriched atmosphere (B).
DISCUSSION
Classification of StyAB.
The epoxidation activity of StyAB was restored after addition of StyA and StyB to a reaction mixture containing NADH, FAD, O2, and styrene. This confirms earlier assumptions based on the genetic organization of the styA and styB genes (38) that StyA and StyB function together as an enzyme system. The StyAB system showed no epoxidation activity in the absence of either O2 or NADH (as a reducing agent), indicating that StyAB is a true monooxygenase. This finding was substantiated by the incorporation of 18O2 into styrene by StyAB. StyB alone exhibited NADH-flavin oxidoreductase activity and no formation of styrene oxide was detectable in the absence of StyA. StyA showed no NADH oxidation activity and epoxidized styrene in the presence of FAD and NADH and StyB or, alternatively, other reducing agents (20). Hence, we suggest that StyAB is a two-component enzyme system consisting of a monooxygenating unit (StyA) that is supplied with reducing equivalents via a reductase component (StyB). Interestingly, StyA and StyB were purified without their putative prosthetic group. Monooxygenases usually depend on a transition metal or an organic cofactor to activate dioxygen. The only known exceptions are the quinone-transforming monooxygenases, which activate oxygen without any apparent requirement for cofactors and metal ions (11). As the StyAB activity was unaffected by the addition of metal-chelating agents, we conclude that no metal ions are involved in the catalytic mechanism. Furthermore, atomic absorption spectroscopy revealed that none of the most common metal ions, such as Fe, Cu, and Co, are present in the proteins. StyAB is not a cytochrome P-450 enzyme, like the styrene monooxygenase of Exophiala jeanselmei (8), since typical P-450 CO difference spectra were not detected and the enzyme activity of StyAB was not affected by metyrapone or proadifen, which are strong inhibitors of cytochrome P-450-dependent enzymes. The epoxidation activity of isolated StyAB depended on FAD. Moreover, total inhibition was observed in the presence of CuSO4 and AgNO3, which form complexes with the flavoquinone form of free flavins (32). Based on the results discussed above, StyAB is classified as a flavin-dependent monooxygenase. It is the first soluble two-component flavin-dependent enzyme characterized in detail which is capable of epoxidizing C=C double bonds at a high rate compared to cyclohexanone monooxygenase from Acinetobacter calcoaceticus, which catalyzes the epoxidation of the highly activated C=C bond in vinylphosphonates at low rates (7, 54), or the membrane-bound squalene monooxygenase (27, 53), which is the key enzyme in the biosynthesis of cholesterol, epoxidizing squalene into 2,3-oxidosqualene.
Characterization of StyA.
StyA was purified to near homogeneity from crude extracts of recombinant E. coli JM101 and the wild-type Pseudomonas sp. strain VLB120. The specific epoxidation activities in both preparations varied only a little, with 2.1 U mg−1 for StyArec and 1.9 U mg−1 for StyAwt+. Both StyA species showed identical characteristics with respect to sizes, N-terminal sequences, substrate spectra, and Km values. Thus, expression of styA by recombinant E. coli does not result in structural or functional modifications of the enzyme compared to the wild-type protein.
The overall StyA amino acid sequence exhibits no significant homology to that of any other known enzyme. An alignment of the amino acid sequence of StyA with several sequences of two-component flavin-dependent oxygenases revealed that only the Rossmann or βαβ-fold is fully conserved in this protein. This motif (containing the GXGXXG sequence) is important for binding of the ADP moiety of FAD (49, 50).
Characterization of StyB.
StyB enzymes purified from recombinant E. coli and wild-type Pseudomonas strain VLB120 have identical substrate spectra and similar kinetic characteristics. The level of expression of styB by the wild-type strain can only be estimated. The volumetric NADH-oxidation activity was 2 U ml−1 in the enriched fraction (0.7 mg of total protein ml−1). If we assume that the specific activities of StyBwt+ and StyBrec are identical, the StyB concentration in that pool was 18 μg ml−1. Therefore, a total StyBwt+ content in the original cell extract of 0.6 mg (in 33 ml of cell extract) can be calculated. The loss of StyB during the enrichment procedure is not accounted for. This corresponds to a StyB content of approximately 0.1% of total protein. This fits well with estimations based on SDS-PAGE of the wild-type crude extract, in which we were not able to identify a protein band corresponding to StyB, meaning that the protein content had to be much lower than 1%. Thus, styB was expressed at a much lower level in the wild type than styA (5%), which is in agreement with findings reported for 4-hydroxyphenylacetate 3-monooxygenase (29).
The only modification upon expression of styB in E. coli JM109(pTEZ302) was the excision of the N-terminal methionine residue. This is a common phenomenon among recombinant proteins expressed by this host strain (4, 19) and was also reported for NAD(P)H-flavin oxidoreductase from Vibrio fischeri ATCC 7744 (21).
Incubation of StyB with NADH and FAD, riboflavin, or FMN resulted in NADH oxidation, confirming that StyB is an independent NADH-flavin oxidoreductase. It strictly depends on NADH as an electron donor, an ability shared with many other described oxidoreductases, most of which have significantly higher Km values for the other nicotinamide if they are able to convert it at all. The capability to utilize different flavins as electron acceptors is found in all NADH-flavin oxidoreductases that do not possess a protein-bound flavin. At flavin concentrations exceeding 200 μM, StyB seems to be inhibited by its substrate. Therefore, we assume an ordered sequential binding mode for NADH and FAD, with NADH binding as the first substrate. High FAD concentrations may lead to binding of FAD as the first substrate instead, and this may block the enzyme for NADH binding.
Diluted StyB exhibited specific NADH oxidation activities of up to 700 U mg−1. The rise in activity upon dilution may be due to formation of insoluble microaggregates at higher protein concentrations or various oligomer populations displaying different activities. The latter has been described, e.g., for pyruvate decarboxylase, whose specific activity depended on the oligomeric state of the enzyme (23).
The catalytic activity of StyB is up to 1 order of magnitude higher than those of most reductases in other two-enzyme monooxygenase systems, such as alkane sulfonate monooxygenase (SsuE) of E. coli (32 U mg−1) (9) and the EDTA monooxygenase (EmoB) of strain BNC1 (105 U mg−1) (43). Only for the recently described flavin-NADH oxidoreductase PheA2 of the phenol hydroxylase system was a higher activity reported (802 U mg−1) (24). The steady-state kinetics measured for StyB at various concentrations of FMN and NADH displayed an intersecting pattern when analyzed in a Hanes plot (Fig. 3). This kind of pattern suggests the formation of a ternary complex among StyB, NADH, and the flavin cofactor. A change in the concentration of the fixed substrate influenced both the slope and the intercept of the plot. In the case of a ping-pong binding mechanism, the slope would not be affected by varying of substrate concentrations. Therefore, a sequential binding mode is plausible.
Enzyme ratio.
The molar ratio of StyA to StyB was found to strongly influence the epoxidation rate. Highest styrene conversion rates were measured when the molar amounts of StyB present were equal to or higher than those of StyA. Considering the molar ratio of reductase to hydroxylase in other two-component systems gives a very diverse picture. For phenol hydroxylase from Bacillus thermoglucosidasius (24), the highest oxygenase activity is observed at a 100-fold excess in favor of the hydroxylase, while in other cases such as that of alkane sulfonate monooxygenase from E. coli this ratio is inverse (reductase to hydroxylase, 4.2:1) (10). Why there is such diversity in this respect is difficult to say at this stage. For the StyAB system, we assume that the amount of FADH2 is limiting for the StyA-catalyzed reaction. Thus, addition of more reductase (at sufficient FAD and NADH concentrations) increases the in situ FADH2 concentration and therefore all FADH2-dependent reaction rates are enhanced.
FAD binding.
The flavin plays a crucial role in the reaction catalyzed by StyAB. It is a substrate for the oxidoreductase StyB, which reduces it to FADH2. The reduced FADH2 is a substrate for the oxygenase StyA, where it activates oxygen for the epoxidation of the aromatic substrate. Thus, FADH2 is the functional link between the two enzyme components. The mechanism of transfer of the reduced flavin from the oxidoreductase to the oxygenase is still a matter of discussion. We have observed no complex formation between the two components so far. Considering the StyB activity in the presence of StyA, we noticed a change in the NADH oxidation rate upon the addition of the hydroxylase only when working with low FAD concentrations and high StyA concentrations (data not shown). Under nonlimiting FAD concentrations, no change in the oxidoreductase activity was detected upon the addition of StyA. We conclude that due to the fact that FADH2 is bound by the hydroxylase it cannot be reoxidized by molecular oxygen to hydrogen peroxide and FAD as fast as it can without StyA and thus the reductase runs into a FAD limitation, which can be easily overcome by the addition of more flavin. A similar observation was reported for 4-hydroxyphenylacetate monooxygenase from E. coli (29). Hence, it seems reasonable to conclude that StyA and StyB do not interact and FADH2 diffuses from one active site to the other. This kind of FADH2 transfer can be controversially discussed because of the rapid auto-oxidation of unbound FADH2 in the presence of molecular oxygen. Galan et al. (13) proposed the two-component flavin-diffusible monooxygenase enzyme family based on their findings for 4-hydroxyphenylacetate 3-monooxygenase of E. coli W and showed that an electron transfer via a freely diffusing FADH2 is possible. They suggested StyAB (among others) to be a member of this family. Unfortunately, the biochemical processes behind this electron transport mechanism have been poorly investigated so far. Only for the bacterial luciferase system has an in vivo flavin transfer been further elucidated. Tu and coworkers (22, 28) recently showed that the oxidases (luciferases) of Vibrio fischeri and Vibrio harveyi interact with the corresponding NADH-flavin oxidoreductases, leading to direct transfer of flavin between the two components. This system differs significantly from StyAB. Most importantly, the binding mechanism and the enzyme kinetics of the NADH-flavin oxidoreductase change upon addition of the luciferase (22, 28), whereas we did not observe such influences in the case of StyAB.
The FAD concentration in the reaction mixture had a significant influence on the rate of the epoxidation reaction catalyzed by StyAB: maximal epoxidation rates were observed at FAD concentrations between 10 and 20 μM (Fig. 5). Higher FAD concentrations resulted in a decrease of the styrene oxide formation rate as a consequence of reduced StyA activity. The same findings were reported for the p-hydroxyphenylacetate hydroxylase from Acinetobacter baumannii (6) and 4-hydroxyphenylacetate-3-hydroxylase from E. coli (52). Others have discussed this inhibiting effect as a competitive inhibition of FAD at the hydroxylase component. In our case, we suggest a different possibility, since almost no interaction between FAD and StyA was found. Reduced flavins can auto-oxidize in two different ways, as outlined in Fig. 7 (30). Either FADH2 reacts with molecular oxygen to yield hydrogen peroxide and FAD or FADH2 reacts with FAD to form a highly reactive flavin radical. The overall formation rate of the FAD radical depends directly on the concentration of FAD. Therefore, increasing concentrations of FAD shift the reaction equilibrium towards the unproductive FADH2 oxidation, thereby explaining the lower epoxidation activity in the presence of higher FAD concentrations. This mechanism would involve free FADH2. We therefore propose a reaction sequence wherein StyB catalyzes the NADH-promoted reduction of FAD into FADH2, which diffuses to StyA. FAD binds much tighter to StyB than to StyA. Due to the high instability of FADH2 under reaction conditions, a more detailed characterization of FADH2 binding to StyA is not possible using UV-visible spectroscopy. However, stabilization of FADH2 in O2-containing media was observed only in the presence of StyA, suggesting that StyA tightly binds reduced FAD and thus slows down its oxidative degradation (Fig. 6).
In summary, the results presented here demonstrate that StyA and StyB form a functional enzyme system in which the StyB-catalyzed reaction is not influenced by StyA. We conclude that StyAB is a two-component flavin-dependent monooxygenase capable of epoxidizing C=C double bonds. We now expect fast kinetic measurements to add to a more detailed understanding of the epoxidation mechanism, especially regarding reaction rates of individual redox steps.
Acknowledgments
This work was funded by BBW 00.0309 and the European Commission (QLRT2000-00725).
We are indebted to Frank Hollmann for most helpful discussions and to Willem van Berkel and Bruno Bühler for critical reading of the manuscript. We thank Yvonne Bruderer and Baron Lumani for excellent assistance in purification of StyA and StyB, Hans Peter Kohler for the gift of 18O2, Thomas Schalch and Bernadetta Rodriges for their help during the analytical ultracentrifugation experiments, Ildiko Fonyo for the AAS analysis, and Ulrich Bauer for the help during the fermentation of the organisms.
REFERENCES
- 1.Archelas, A., and R. Furstoss. 1999. Biocatalytic approaches for the synthesis of enantiopure epoxides, p. 159-191. In W.-D. Fessner (ed.), Topics in current chemistry, vol. 200. Biocatalysis: from discovery to application. Springer, Berlin, Germany.
- 2.Barrio, J. R., G. L. Tolman, N. J. Leonard, R. D. Spencer, and G. Weber. 1973. Flavin 1, N6-ethenoadenin dinucleotide: dynamic and static quenching of fluorescence. Proc. Natl. Acad. Sci. USA 70:941-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrametti, F., A. M. Marconi, G. Bestetti, C. Colombo, E. Galli, M. Ruzzi, and E. Zennaro. 1997. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl. Environ. Microbiol. 63:2223-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Bassat, A., and K. Bauer. 1987. Amino-terminal processing of proteins. Nature 326:315.
- 5.Bühler, B., A. Schmid, B. Hauer, and B. Witholt. 2000. Xylene monooxygenase catalyzes the multistep oxygenation of toluene and pseudocumene to corresponding alcohols, aldehydes, and acids in Escherichia coli JM101. J. Biol. Chem. 275:10085-10092. [DOI] [PubMed] [Google Scholar]
- 6.Chaiyen, P., C. Suadee, and P. Wilairat. 2001. A novel two-protein component flavoprotein hydroxylase: p-hydroxyphenylacetate hydroxylase from Acinetobacter baumannii. Eur. J. Biochem. 268:5550-5561. [DOI] [PubMed] [Google Scholar]
- 7.Colonna, S., N. Gaggero, G. Carrea, G. Ottolina, P. Pasta, and F. Zambianchi. 2002. First asymmnetric epoxidation catalysed by cyclohexanone monooxygenase. Tetrahedron Lett. 43:1797-1799. [Google Scholar]
- 8.Cox, H. H. J., B. W. Faber, W. N. M. Van Heiningen, H. Radhoe, H. J. Doddema, and W. Harder. 1996. Styrene metabolism in Exophilia jeanselmei and involvement of a cytochrome P-450-dependent styrene monooxygenase. Appl. Environ. Microbiol. 62:1471-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Gennaro, P., A. Colmegna, E. Galli, G. Sello, F. Pelizzoni, and G. Bestetti. 1999. A new biocatalyst for production of optically pure aryl epoxides by styrene monooxygenase from Pseudomonas fluorescens ST. Appl. Environ. Microbiol. 65:2794-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkansulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274:26639-26646. [DOI] [PubMed] [Google Scholar]
- 11.Fetzner, S. 2002. Oxygenases without requirement for cofactors or metal ions. Appl. Microbiol. Biotechnol. 60:243-257. [DOI] [PubMed] [Google Scholar]
- 12.Fox, B. G., J. G. Borneman, L. P. Wackett, and J. D. Lipscomb. 1990. Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry 29:6419-6427. [DOI] [PubMed] [Google Scholar]
- 13.Galan, B., E. Diaz, M. A. Prieto, and J. L. Garcia. 2000. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new flavin:NAD(P)H reductase subfamily. J. Bacteriol. 182:627-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grbic-Galic, G., N. Churchman-Eisel, and I. Mrakovic. 1990. Microbial transformation of styrene by anaerobic consortia. J. Appl. Bacteriol. 69:247-260. [DOI] [PubMed] [Google Scholar]
- 15.Guengerich, F. 1991. Reactions and significance of cytochrome P-450 enzymes. J. Biol. Chem. 266:10019-10022. [PubMed] [Google Scholar]
- 16.Hartmans, S. 1995. Microbial degradation of styrene, p. 227-239. In V. P. Singh (ed.), Biotransformations: microbial degradation of health risk compounds. Elsevier Science, Amsterdam, The Netherlands.
- 17.Hartmans, S., M. J. van der Werft, and J. A. M. de Bont. 1990. Bacterial degradation of styrene involving a novel flavin adenine dinucleotide-dependent styrene monooxygenase. Appl. Environ. Microbiol. 56:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins, D. R. (ed.). 1994. A survey of the biotransformations of drugs and chemicals in animals, vol. 1-5. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 19.Hirel, P.-H., J.-M. Schmitter, P. Dessen, G. Fayat, and S. Blanquet. 1989. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. USA 86:8247-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollmann, F., P.-C. Lin, B. Witholt, and A. Schmid. 2003. Stereospecific biocatalytic epoxidation: the first example of direct regeneration of a FAD-dependent monooxygenase for catalysis. J. Am. Chem. Soc. 125:8209-8217. [DOI] [PubMed] [Google Scholar]
- 21.Inouye, S. 1994. NAD(P)H-flavin oxidoreductase from the bioluminescent bacterium, Vibrio fischeri ATCC 7744, is a flavoprotein. FEBS Lett. 347:163-168. [DOI] [PubMed] [Google Scholar]
- 22.Jeffers, C. E., and S.-C. Tu. 2001. Differential transfers of reduced flavin cofactor and product by bacterial flavin reductase to luciferase. Biochemistry 40:1749-1754. [DOI] [PubMed] [Google Scholar]
- 23.Killenberg-Jabs, M., A. Jabs, H. Lilie, R. Golbik, and G. Hubner. 2001. Active oligomeric states of pyruvate decarboxylase and their functional characterization. Eur. J. Biochem. 268:1698-1704. [PubMed] [Google Scholar]
- 24.Kirchner, U., A. H. Westphal, R. Müller, and W. J. H. van Berkel. 2003. Phenol hydroxylase from Bacillus thermoglucosidans A7: a two-protein component monooxygenase with a dual role for FAD. J. Biol. Chem. 278:47545-47553. [DOI] [PubMed] [Google Scholar]
- 25.Krenn, B. E., H. Plat, and R. Wever. 1988. Purification and some characteristics of a non-haem bromoperoxidase from Streptomyces aureofaciens. Biochim. Biophys. Acta 952:255-260. [DOI] [PubMed] [Google Scholar]
- 26.Kula, M.-R., and U. Kragl. 2000. Dehydrogenases in the synthesis of chiral compounds, p. 839-863. In R. N. Patel (ed.), Stereoselective biocatalysis. Marcel Dekker, New York, N.Y.
- 27.Laden, B. P., Y. Tang, and T. D. Porter. 2000. Cloning, heterologous expression, and enzymological characterization of human squalene monooxygenase. Arch. Biochem. Biophys. 374:381-388. [DOI] [PubMed] [Google Scholar]
- 28.Lei, B., and S.-C. Tu. 1998. Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry 37:14623-14629. [DOI] [PubMed] [Google Scholar]
- 29.Louie, T. M., X. S. Xie, and L. Xun. 2003. Coordinated production and utilization of FADH2 by NAD*P(H)-flavin oxidoreductase and 4-hydroxyphenylacetate 3-monooygenase. Biochemistry 42:7509-7517. [DOI] [PubMed] [Google Scholar]
- 30.Massey, V. 1994. Activation of molecular oxygen by flavins and flavoproteins. J. Biol. Chem. 269:22459-22462. [PubMed] [Google Scholar]
- 31.Meyer, A., A. Schmid, M. Held, A. H. Westphal, M. Roethlisberger, H.-P. E. Kohler, W. J. H. vanBerkel, and B. Witholt. 2002. Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. J. Biol. Chem. 277:5575-5585. [DOI] [PubMed] [Google Scholar]
- 32.Müller, F. 1991. Chemistry and biochemistry of flavoenzymes, vol. 1, p. 1-71. CRC Press, Inc., Boca Raton, Fla.
- 33.O'Connor, K. E., and A. D. W. Dobson. 1996. Microbial degradation of alkenylbenzenes. World J. Microbiol. Biotechnol. 12:207-212. [DOI] [PubMed] [Google Scholar]
- 34.O'Leary, N. D., K. E. O'Connor, and A. D. W. Dobson. 2002. Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol. Rev. 26:403-417. [DOI] [PubMed] [Google Scholar]
- 35.O'Leary, N. D., K. E. O'Connor, W. Duetz, and A. D. W. Dobson. 2001. Transcriptional regulation of styrene degradation in Pseudomonas putida CA-3. Microbiology 147:973-979. [DOI] [PubMed] [Google Scholar]
- 36.Onumonu, A. N., A. Colocoussi, C. Matthews, M. P. Woodland, and D. J. Leak. 1994. Microbial alkene epoxidation—merits and limitation. Biocatalysis 10:211-218. [Google Scholar]
- 37.Panke, S., V. de Lorenzo, A. Kaiser, B. Witholt, and M. G. Wubbolts. 1999. Engineering of a stable whole-cell biocatalyst capable of (s)-styrene oxide formation for continuous two-liquid-phase applications. Appl. Environ. Microbiol. 65:5619-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panke, S., B. Witholt, A. Schmid, and M. G. Wubbolts. 1998. Towards a biocatalyst for (s)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VBL120. Appl. Environ. Microbiol. 64:2032-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panke, S., M. G. Wubbolts, A. Schmid, and B. Witholt. 2000. Production of enantiopure styrene oxide by recombinant Escherichia coli synthesizing a two-component styrene monooxygenase. Biotechnol. Bioeng. 69:91-100. [DOI] [PubMed] [Google Scholar]
- 40.Parry, R. J., and W. Li. 1997. An NADH:FAD oxidoreductase from the valanimycin producer, Streptomyces viridifaciens. J. Biol. Chem. 272:23303-23311. [DOI] [PubMed] [Google Scholar]
- 41.Saito, Y., M. Mifune, S. Nakashima, J. Odo, and Y. Tanaka. 1987. Determination of hydrogen peroxide with N,N-diethylaniline and 4-aminoantipyrine by use of an anion-exchange resin modified with manganese-tetrakis(sulphophenyl)porphyrine, as a substitute for peroxidase. Talanta 34:667-669. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sono, M., M. P. Roach, E. D. Coulter, and J. H. Dawson. 1996. Heme-containing oxygenases. Chem. Rev. 96:2841-2887. [DOI] [PubMed] [Google Scholar]
- 44.Thibaut, D., N. Ratet, D. Bisch, D. Faucher, L. Debussche, and F. Blanche. 1995. Purification of the two-component system catalyzing the oxidation of the d-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J. Bacteriol. 177:5199-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tishkov, V. I., A. G. Galkin, V. V. Feorchuk, P. A. Savitsky, A. M. Rojkova, H. Gieren, and M.-R. Kula. 1999. Pilot scale production and isolation of recombinant NAD+- and NADP+-specific formate dehydrogenases. Biotechnol. Bioeng. 64:187-193. [PubMed] [Google Scholar]
- 46.Velasco, A., S. Alonso, J. L. Garcia, J. Perera, and E. Diaz. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 180:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallar, B. J., and J. D. Libscomb. 1996. Dioxygen activation by enzymes containing binuclear non-heme iron clusters. Chem. Rev. 96:2625-2657. [DOI] [PubMed] [Google Scholar]
- 48.Warhurst, A. M., K. F. Clarke, R. A. Hill, R. A. Holt, and C. A. Fewson. 1994. Metabolism of styrene by Rhodococcus rhodochrous NCIMB13259. Appl. Environ. Microbiol. 60:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wierenga, R. K., M. C. H. De Maeyer, and W. G. J. Hol. 1985. Interaction of pyrophosphate moieties with α-helixes in dinucleotide-binding proteins. Biochemistry 24:1346-1357. [Google Scholar]
- 50.Wierenga, R. K., P. Terpstra, and W. G. J. Hol. 1985. Prediction of the occurrence of the ADP-binding αβα-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]
- 51.Wiesner, W., K. van Pee, and F. Lingens. 1988. Purification and characterization of a novel bacterial non-heme chloroperoxidase from Pseudomonas pyrrocinia. J. Biol. Chem. 263:13725-13732. [PubMed] [Google Scholar]
- 52.Xun, L., and E. R. Sandvik. 2000. Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl. Environ. Microbiol. 66:481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto, S., and K. Bloch. 1970. Studies on squalene epoxidase of rat liver. J. Biol. Chem. 245:1670-1674. [PubMed] [Google Scholar]
- 54.Zambianchi, F., P. Pasta, G. Carrea, S. Colonna, N. Gaggero, and J. M. Woodley. 2002. Use of isolated cyclohexanone monooxygenase from recombinant Escherichia coli as a biocatalyst for baeyer-villiger and sulfide oxidations. Biotechnol. Bioeng. 78:489-496. [DOI] [PubMed] [Google Scholar]