The crystal structure of the TK2203 protein was determined at 1.41 Å resolution.
Keywords: aromatic compound degradation, nonhaem iron, archaea, SIRAS, homodimer, TK2203, Thermococcus kodakarensis
Abstract
The TK2203 protein from the hyperthermophilic archaeon Thermococcus kodakarensis KOD1 (262 residues, 29 kDa) is a putative extradiol dioxygenase catalyzing the cleavage of C–C bonds in catechol derivatives. It contains three metal-binding residues, but has no significant sequence similarity to proteins for which structures have been determined. Here, the first crystal structure of the TK2203 protein was determined at 1.41 Å resolution to investigate its functional role. Structure analysis reveals that this protein shares the same fold and catalytic residues as other extradiol dioxygenases, strongly suggesting the same enzymatic activity. Furthermore, the important region contributing to substrate selectivity is discussed.
1. Introduction
Dioxygenase is an enzyme that catalyzes the cleavage of C—C bonds in aromatic compounds (Dagley, 1978 ▸; Harayama et al., 1992 ▸; Vaillancourt et al., 2006 ▸; Lipscomb, 2008 ▸). Aromatic compounds are very stable and rigid, but their degradation products are useful as sources of energy and carbon. Therefore, this degrading enzyme is essential for many microorganisms. Dioxygenases catalyze the incorporation of two O atoms from O2 into catechol derivatives, resulting in ring-open, muconic acid products or 2-hydroxymuconate semialdehyde products. They are largely classified into two groups (Harayama & Rekik, 1989 ▸). Intradiol dioxygenases utilize nonhaem iron(III) to cleave aromatic rings between two hydroxylated C atoms (so-called ortho cleavage). On the other hand, extradiol dioxygenases utilize nonhaem iron(II) or other divalent metal ions to cleave aromatic rings between a hydroxylated carbon and an adjacent nonhydroxylated carbon (meta cleavage). Several crystal structures of extradiol dioxygenases have been reported (Han et al., 1995 ▸; Senda et al., 1996 ▸; Kita et al., 1999 ▸; Sugimoto et al., 1999 ▸; Titus et al., 2000 ▸) and have revealed that the 2-His-1-carboxylate (Asp or Glu residue) facial triad motif is important for metal binding in the active site.
The TK2203 protein from the hyperthermophilic archaeon Thermococcus kodakarensis KOD1 has 262 amino-acid residues and its mass is approximately 29 kDa. It contains three putative metal-binding residues, His10, His48 and Asp222, as elucidated by BLAST (Altschul et al., 1990 ▸), and is annotated as an extradiol dioxygenase. However, the TK2203 protein does not display significant sequence similarity to proteins for which three-dimensional structures are available in the Protein Data Bank, including extradiol dioxygenases, and its overall structure and function remain unclear. In this paper, we present the first crystal structure of the TK2203 protein at 1.41 Å resolution to investigate its functional role. The result clarifies that this protein is structurally similar to other extradiol dioxygenases in spite of its low sequence similarity, and the active-site residues interacting with the metal ion are also conserved. Furthermore, based on the shape of the catalytic pocket, we predicted the important region contributing to the substrate selectivity.
2. Materials and methods
2.1. DNA manipulation
The TK2203 overexpression plasmid was prepared as follows (Table 1 ▸). Using T. kodakarensis genomic DNA as a template, the two oligonucleotide primers listed in Table 1 ▸ were used to amplify the TK2203 gene. The amplified fragment was digested by NdeI and BamHI and inserted into pET-21a(+) (Novagen, Madison, Wisconsin, USA) at the respective sites.
Table 1. Macromolecule-production information.
| Source organism | T. kodakarensis KOD1 |
| DNA source | T. kodakarensis genomic DNA |
| Forward primer† | GGGGCTGCAGCATATGCTCTTTGGAATAGGCCTC |
| Reverse primer† | AAAAGGATCCTACTCCCTCACCCACAGCG |
| Cloning vector | pET-21a(+) |
| Expression vector | pET-21a(+) |
| Expression host | E. coli Rosetta2(DE3)pLysS |
| Complete amino-acid sequence of the construct produced | MLFGIGLMPHGNPALSPEDKETEKLAGVLKDIGKAFSDADSYVLISPHNVRISDHLGVIMAQHLISWLGFEGVELPGEWETDRGLAEEVYNAWKGAEIPTVDLHFASRSGRYSRWPLTWGELIPLQFLEKKPLVLLTPARRLSRETLIKAGEVLGEVLEGSEKKIALIVSADHGHAHDENGPYGYRKESEEYDRLIMELINESRLEELPEIPDELIEKALPDSYWQMLIMLGAMHRVPVKLVESAYACPTYFGMAGALWVRE |
The restriction sites used in the primer sequences are underlined.
2.2. Gene expression and protein purification
For expression of the TK2203 protein, the plasmid was introduced into Escherichia coli Rosetta2(DE3)pLysS cells (Novagen) and overexpression of the gene was induced with isopropyl β-d-1-thiogalactopyranoside. The cells were harvested by centrifugation and resuspended in sonication buffer (50 mM sodium phosphate pH 7.0, 300 mM NaCl). The suspension with 1%(v/v) EDTA-free protease-inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) was sonicated on ice. After sonication, the soluble fraction was heat-treated for 20 min at 85°C. The solution was applied onto a Resource Q anion-exchange column (GE Healthcare, Uppsala, Sweden) and the target protein was eluted using a gradient of 0–1 M NaCl in elution buffer [50 mM Tris–HCl pH 8.0, 1 mM dithiothreitol (DTT)] at 20°C. The fraction was applied onto a Superdex 200 10/300 GL gel-filtration column (GE Healthcare) with a mobile phase of buffer consisting of 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM DTT at 20°C. The size of the molecule in solution was estimated using this column with a Gel Filtration Calibration Kit (GE Healthcare) comprising aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa) and ribonuclease A (13.7 kDa). The final purity of the fraction was tested by SDS–PAGE and native PAGE.
2.3. Crystallization, X-ray data collection and structure determination
The protein solution was concentrated to 10 mg ml−1. Crystallization attempts were performed at 20°C with the hanging-drop vapour-diffusion method or the sitting-drop vapour-diffusion method using a rotary shaker to shake the plates (50 rev min−1). Mixtures of the protein solution and reservoir solution in a 1 µl:1 µl ratio were equilibrated against 500 µl reservoir solution. Crystals were obtained using a reservoir solution consisting of 100 mM imidazole pH 8.0, 350–400 mM calcium acetate, 20–22.5%(w/v) polyethylene glycol 1000 and reached dimensions of 0.7 × 0.2 × 0.02 mm within a few days. Crystals were soaked in a cryoprotectant prepared by the addition of 15%(w/v) glycerol to the reservoir solution. A platinum derivative was also prepared by soaking in the cryoprotectant with 10 mM K2PtCl4 for 15 min. The crystals were mounted in a loop and flash-cooled in an N2 gas stream at −173°C.
Diffraction data were collected using synchrotron radiation at the Photon Factory (on BL1A with a Pilatus 2M-F detector and on AR-NW12A with an ADSC Quantum 270 detector). All data sets were processed and scaled using the programs DENZO and SCALEPACK from the HKL-2000 package (Otwinowski & Minor, 1997 ▸). The crystal structure of the TK2203 protein was solved by the single isomorphous replacement with anomalous scattering (SIRAS) method using SOLVE (Terwilliger & Berendzen, 1999 ▸) and RESOLVE (Terwilliger, 2000 ▸). The initial model was built with PHENIX (Adams et al., 2010 ▸) and manual modelling was performed using Coot (Emsley et al., 2010 ▸). The structure was refined using PHENIX and validated with MolProbity (Chen et al., 2010 ▸). Zn atoms were confirmed with data sets collected at the peak wavelengths of Zn (1.28 Å) and Cu (1.37 Å), and were likely to have been supplied by the E. coli cells. Superposition of the structures was performed using Coot on the basis of secondary-structure matching. Figures showing structures were prepared with PyMOL (DeLano, 2008 ▸).
2.4. Data deposition
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://www.pdb.org) as entry 5hee.
3. Results and discussion
3.1. Overall structure of the TK2203 protein
The crystal structure of the TK2203 protein was solved by the SIRAS method using the Pt derivative and was refined to 1.41 Å resolution. The R and R free values of the structure were 16.3 and 18.6%, respectively. 97.5% of the residues were included in the favoured region of the Ramachandran plot and 2.5% were in the allowed region. The final statistics for the data sets are shown in Table 2 ▸.
Table 2. Crystallographic statistics.
Values in parentheses are for the outermost resolution shell.
| Data set | TK2203 | K2PtCl4 | Zn_ano | Cu_ano |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 1.1000 | 1.0700 | 1.2800 | 1.3700 |
| Resolution (Å) | 50–1.41 (1.44–1.41) | 50–2.18 (2.23–2.18) | 50–2.80 (2.90–2.80) | 50–2.80 (2.90–2.80) |
| Total No. of reflections | 546546 | 224928 | 73406 | 74327 |
| No. of unique reflections | 83940 | 23799 | 11030 | 11264 |
| Mosaicity (°) | 0.25–0.61 | 0.21–0.89 | 0.81–1.01 | 0.86–1.04 |
| Multiplicity | 6.5 | 9.5 | 6.7 | 6.6 |
| Completeness (%) | 97.0 (89.7) | 100 (99.6) | 99.8 (99.4) | 99.8 (99.5) |
| 〈I/σ(I)〉 | 23.8 (2.0) | 20.8 (5.8) | 7.7 (2.4) | 6.7 (2.0) |
| R r.i.m. (%) | 8.0 (84.3) | 10.3 (35.3) | 15.2 (50.9) | 17.3 (59.8) |
| Wilson plot B factor (Å2) | 16.3 | 28.0 | 36.1 | 36.0 |
| Space group | C2 | C2 | C2 | C2 |
| Unit-cell parameters | ||||
| a (Å) | 74.4 | 74.4 | 74.3 | 74.2 |
| b (Å) | 42.5 | 42.7 | 42.4 | 42.3 |
| c (Å) | 143.5 | 143.7 | 143.8 | 143.8 |
| β (°) | 94.5 | 94.7 | 94.6 | 94.6 |
| Phasing | ||||
| Mean figure of merit (SIRAS) | 0.39 [20–3 Å] | |||
| Refinement | ||||
| Resolution limits (Å) | 47.69–1.41 | |||
| No. of reflections (F > 0) | 83930 | |||
| No. of reflections used for the R free set | 4233 | |||
| R/R free † (%) | 16.3/18.6 | |||
| R.m.s.d., bond distances (Å) | 0.007 | |||
| R.m.s.d, bond angles (°) | 1.090 | |||
| No. of molecules per asymmetric unit | 2 | |||
| Protein atoms | 4192 | |||
| Ligands | 5 Zn2+, 1 glycerol | |||
| Solvent molecules | 544 waters | |||
| Average B factor (Å2) | ||||
| Protein | 25.8 | |||
| Ligand | 31.1 | |||
| Water | 33.5 | |||
| Ramachandran plot | ||||
| Favoured (%) | 97.5 | |||
| Allowed (%) | 2.5 | |||
| Rotamer outliers (%) | 0.0 | |||
R = 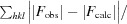
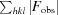 . R
free is the same as R but for a 5% subset of all reflections that were never used in crystallographic refinement.
. R
free is the same as R but for a 5% subset of all reflections that were never used in crystallographic refinement.
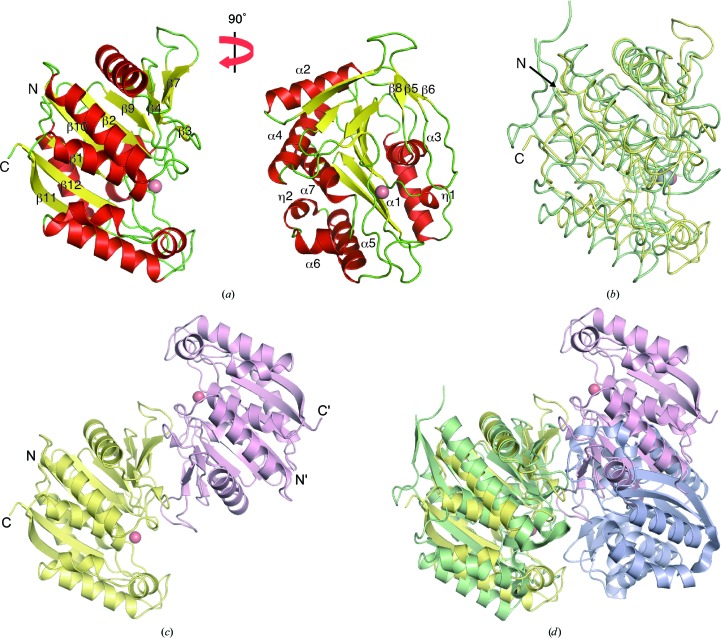
The TK2203 structure is composed of seven α-helices, 12 β-strands and two 310-helices (Fig. 1 ▸ a). The major β-sheet containing β1, β2, β4, β7, β9, β10, β11 and β12 is sandwiched between two helical clusters (cluster 1 containing α1, α3 and η1 and cluster 2 containing the other helices). This structure is common to members of the extradiol dioxygenase superfamily. Superposition of the TK2203 protein onto a β subunit of 2-aminophenol 1,6-dioxygenase (APD) from Comamonas sp. strain (PDB entry 3vsi; Li et al., 2013 ▸) is shown in Fig. 1 ▸(b). APD catalyzes the opening of 2-aminophenol analogues and generates a 2-aminomuconic 6-semialdehyde product. It consists of two α and two β subunits, which are structurally similar to each other; the β subunit has 312 residues and shares only 17% sequence identity with the TK2203 protein as calculated by BLAST. The superposition reveals that the two structures are essentially identical, resulting in a root-mean-square deviation (r.m.s.d.) of 2.5 Å for 242 Cα atoms out of a total of 302 residues (Fig. 1 ▸ b) and a Z-score of 26.4 as calculated by the DALI sever (Holm & Rosenström, 2010 ▸). In spite of the very low sequence identity, the structures are surprisingly similar.
Figure 1.
Overall structure of the TK2203 protein. (a) Monomer structure. A Zn ion is shown by a red sphere. (b) Comparison with the APD β subunit. The TK2203 protein and APD are shown in yellow and green, respectively. An Fe ion in APD is shown by a grey sphere. (c) Dimer structure. Each monomer is shown in a different colour. (d) Comparison with the APD α-subunit and β-subunit dimer. The APD α subunit is shown in blue.
Two TK2203 molecules are present in the asymmetric unit of the crystal and form a dimer via interaction mainly between the β2–β4 region of one molecule and the region around β7 of the other (Fig. 1 ▸ c). Five Zn ions are also observed (Supplementary Fig. S1). Two are in the active site (described in the next section), two are at the borders with the neighbouring molecules in the crystal and one is in the dimer interface. Calculations using the PISA server (Krissinel & Henrick, 2007 ▸) show that the interface between the two molecules buries 3280 Å2, suggesting that this assembly is stable in solution. Gel-filtration chromatographic analysis also supports this result (66 kDa; Supplementary Fig. S2). Superposition of the TK2203 homodimer onto the α-subunit and β-subunit heterodimer of APD reveals that the two dimers are quite similar and the putative active site of the TK2203 protein is more open and exposed to the solvent region (Fig. 1 ▸ d). The β-subunit homodimer of APD is completely different (Li et al., 2013 ▸). This suggests that the TK2203 protein may function by itself as a homodimer. The APD subunit genes are located adjacent to each other and contain LigB motifs (Li et al., 2013 ▸), but T. kodakarensis has no gene with a LigB motif around the TK2203 gene.
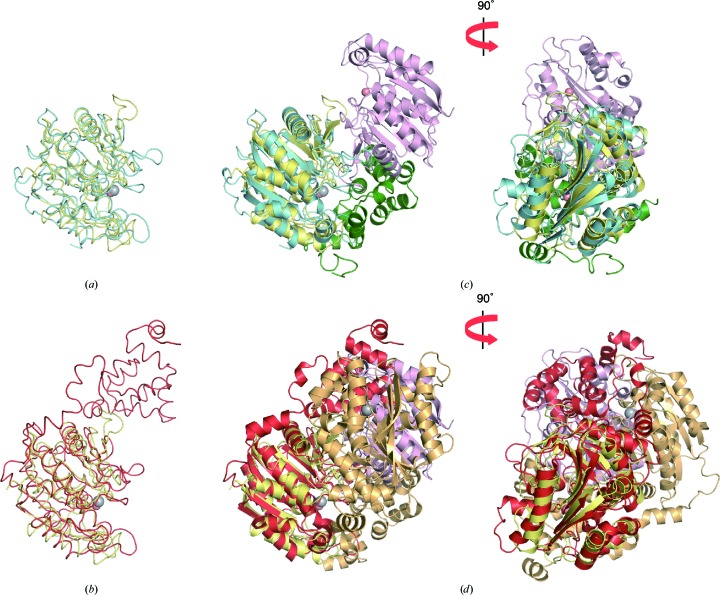
Some extradiol dioxygenases adopt different α subunits or domains containing LigA motifs (Sugimoto et al., 1999 ▸, 2014 ▸). LigAB and DesB catalyze ring-opening reactions of protocatechuate and gallate, respectively. LigAB forms a tetramer consisting of two LigA and two LigB proteins, and DesB contains both LigA and LigB domains and forms a homodimer. Although LigB from Sphingomonas paucimobilis SYK-6 (298 residues; PDB entry 1b4u; Sugimoto et al., 1999 ▸) and DesB from Sphingobium sp. SYK-6 (408 residues; PDB entry 3wr8; Sugimoto et al., 2014 ▸) share only 14 and 17% sequence identities, respectively, with the TK2203 protein as calculated by BLAST, superpositions of the TK2203 protein onto LigB and DesB reveal that their structures are identical, resulting in an r.m.s.d. of 2.5 Å for 209 Cα atoms out of a total of 262 residues for LigB (Fig. 2 ▸ a) and an r.m.s.d. of 2.3 Å for 224 Cα atoms out of a total of 262 residues for DesB (Fig. 2 ▸ b). However, the relative arrangement of the TK2203 homodimer is completely different from those of the LigAB heterodimer (Fig. 2 ▸ c) and the DesB homodimer (Fig. 2 ▸ d). Moreover, T. kodakarensis possesses no corresponding gene with the LigA motif in its genome. These results also support the homodimeric activity of the TK2203 protein.
Figure 2.
Comparison with LigAB and DesB. (a, b) Superpositions of (a) LigB (blue) and (b) DesB (red) on the TK2203 protein. (c, d) Superpositions of (c) the LigAB heterodimer (blue and green) and (d) the DesB homodimer (red and orange) on the TK2203 homodimer.
3.2. Active site
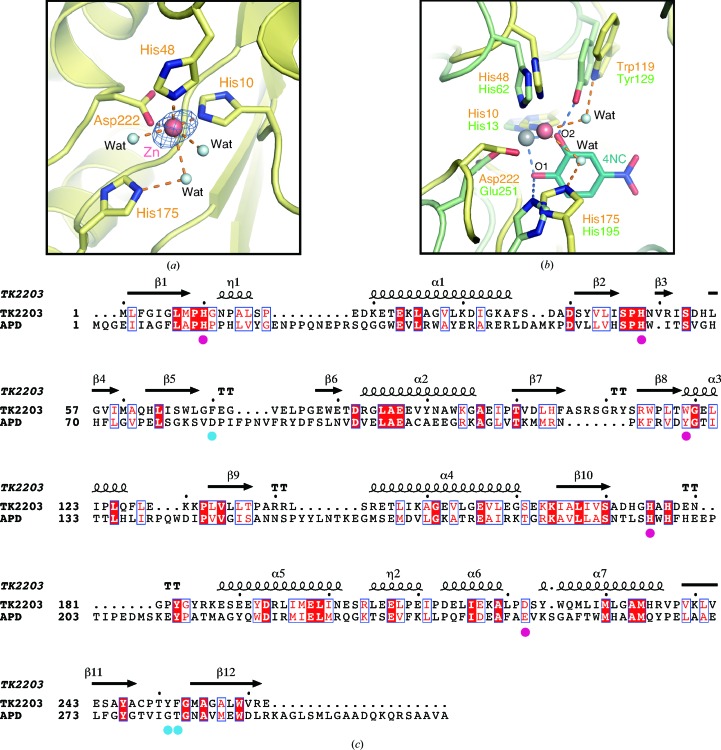
In the active site of the TK2203 protein, a Zn ion supplied by E. coli is observed (Fig. 3 ▸ a). This ion is coordinated by His10 N∊, His48 N∊, Asp222 Oδ and three water molecules. Comparison of the TK2203 protein with the β subunit of APD reveals that the coordination is almost the same as that of the Fe ion in APD, utilizing His13, His62 and Glu251 (Li et al., 2013 ▸; Figs. 3 ▸ b and 3 ▸ c). Moreover, His175 and Trp119 of the TK2203 protein, which interact with the Zn ion via water molecules, are also observed (corresponding to His195 and Tyr129 in the β subunit of APD; Fig. 3 ▸ b). In APD, His195 interacts with O1 of the inhibitor 4-nitrocatechol (4NC) and serves as a catalytic base (Li et al., 2013 ▸). On the other hand, Tyr129 is hydrogen-bonded to the equatorially coordinated O2 of 4NC and plays a possible role in the deprotonation of the hydroxyl group of the substrate and the formation of the hydrogen bond. These structural features suggest that the TK2203 protein may have the potential to function as an extradiol dioxygenase like APD. Two hydroxyl groups (or one hydroxyl group and one amino group) of the substrate interact with the Fe ion at the positions of the two water molecules. Next, an O2 molecule is also bonded to the Fe ion. Trp119 is hydrogen-bonded to the deprotonated hydroxyl group of the substrate and positions the substrate in the proper position. His175 then interacts with the O2 molecule and functions as a catalytic base. This structure contains the Zn ion from E. coli, but it is suggested that the reaction of the TK2203 protein may require an Fe ion.
Figure 3.
Active site. (a) Metal-binding site. Water molecules are shown by light-blue spheres. An Zn anomalous difference Fourier map at 4.5σ is shown in blue. (b) Comparison with the APD β subunit. The inhibitor 4-nitrocatechol in the APD stucture is shown in dark blue. (c) Sequence alignment between the TK2203 protein and the β subunit of APD. The sequences were aligned by ClustalW (Larkin et al., 2007 ▸) with slight modifications and the figure was prepared by ESPript (Gouet et al., 1999 ▸). The secondary structure of the TK2203 protein is shown above the sequences. Helices and β-strands are indicated by coils and arrows, respectively. Identical and similar residues are indicated by white letters on a red background and red letters, respectively. Metal-binding residues and aromatic residues in the active site are indicated by magenta and blue circles, respectively.
The active-site pocket of the TK2203 protein is composed of some large side-chain (aromatic) residues: Phe70, Trp119, Tyr251 and Phe252 (Fig. 4 ▸). These residues may contribute to substrate selectivity. In APD, Tyr129 (corresponding to Trp119 in the TK2203 protein; Fig. 3 ▸ b) is predicted to have an influence on the substrate specificity towards noncatecholic compounds because other extradiol dioxygenases preferring catechol analogues as substrates instead possess histidine residues (Li et al., 2013 ▸). The TK2203 protein may have the same feature as APD. Superposition of the TK2203 protein with the β subunit of APD clarifies two largely different loop regions (orange loops in Fig. 4 ▸). As the loop containing Tyr251 and Phe252 and that containing Phe70 are near the nitro group of 4NC in the APD structure (distances of 0.9 and 3.8 Å, respectively), it is reasonable to assume that these loops affect substrate binding. The loop regions are not conserved in APD (Fig. 3 ▸ c) and the Cα traces are also completely different from those in APD. These results suggest that the three aromatic residues and the shapes of the loops may play an important role in selecting or accepting substituents on the substrate.
Figure 4.
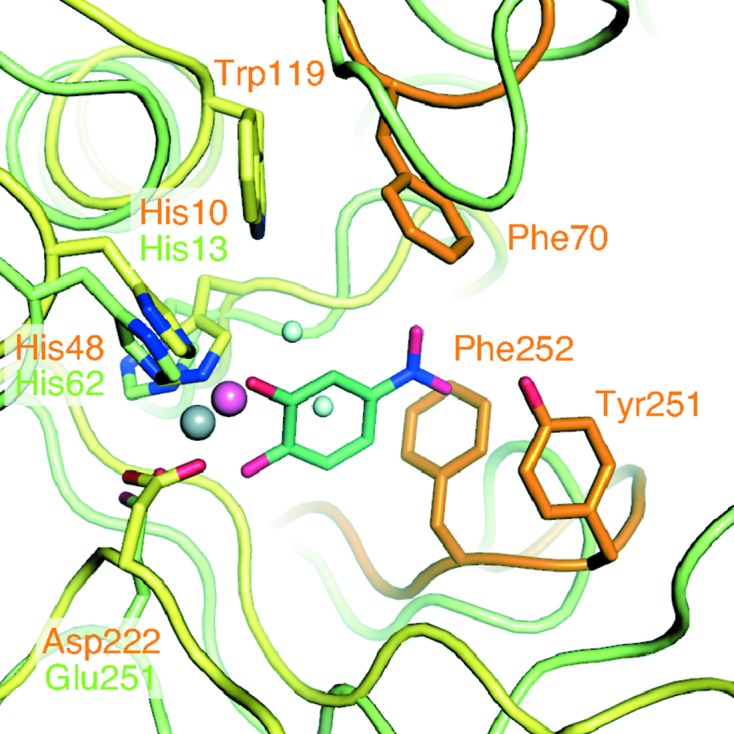
Aromatic residues around the active site. Two loops of the TK2203 protein related to substrate selectivity are shown in orange.
In summary, we determined the crystal structure of the TK2203 protein at 1.41 Å resolution. This revealed that the TK2203 protein has the same structural fold and the same catalytic residues as APD, strongly suggesting that it may have extradiol dioxygenase activity. Moreover, we discussed the important regions related to substrate selectivity. The substrate of the TK2203 protein remains unclear, but further studies on the reaction mechanism, including the catalytic metal, will be elucidated by future biochemical analyses based on this structural study.
Supplementary Material
PDB reference: TK2203 protein, 5hee
Supporting Information: Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X16006920/nw5037sup1.pdf
Acknowledgments
We thank the beamline scientists at the Photon Factory for their help. This work was partly supported by Grants-in-Aid for Scientific Research (26291012 to KM) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The synchrotron-radiation experiments were performed at the Photon Factory with the approval of the Photon Factory Advisory Committee.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol. 215, 403–410. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Dagley, S. (1978). Q. Rev. Biophys. 11, 577–602. [DOI] [PubMed]
- DeLano, W. L. (2008). PyMOL. http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Han, S., Eltis, L. D., Timmis, K. N., Muchmore, S. W. & Bolin, J. T. (1995). Science, 270, 976–980. [DOI] [PubMed]
- Harayama, S., Kok, M. & Neidle, E. L. (1992). Annu. Rev. Microbiol. 46, 565–601. [DOI] [PubMed]
- Harayama, S. & Rekik, M. (1989). J. Biol. Chem. 264, 15328–15333. [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Kita, A., Kita, S., Fujisawa, I., Inaka, K., Ishida, T., Horiike, K., Nozaki, M. & Miki, K. (1999). Structure, 7, 25–34. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Li, D.-F., Zhang, J.-Y., Hou, Y.-J., Liu, L., Hu, Y., Liu, S.-J., Wang, D.-C. & Liu, W. (2013). Acta Cryst. D69, 32–43. [DOI] [PubMed]
- Lipscomb, J. D. (2008). Curr. Opin. Struct. Biol. 18, 644–649. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Senda, T., Sugiyama, K., Narita, H., Yamamoto, T., Kimbara, K., Fukuda, M., Sato, M., Yano, K. & Mitsui, Y. (1996). J. Mol. Biol. 255, 735–752. [DOI] [PubMed]
- Sugimoto, K., Senda, T., Aoshima, H., Masai, E., Fukuda, M. & Mitsui, Y. (1999). Structure, 7, 953–965. [DOI] [PubMed]
- Sugimoto, K., Senda, M., Kasai, D., Fukuda, M., Masai, E. & Senda, T. (2014). PLoS One, 9, e92249. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Titus, G. P., Mueller, H. A., Burgner, J., Rodríguez De Córdoba, S., Peñalva, M. A. & Timm, D. E. (2000). Nature Struct. Biol. 7, 542–546. [DOI] [PubMed]
- Vaillancourt, F. H., Bolin, J. T. & Eltis, L. D. (2006). Crit. Rev. Biochem. Mol. Biol. 41, 241–267. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: TK2203 protein, 5hee
Supporting Information: Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X16006920/nw5037sup1.pdf